Abstract
A simple and precise genotyping system based on PCR using type-specific primers was developed for the determination of genotypes A through F of hepatitis B virus (HBV). This assay system is considered to be a useful tool for the molecular diagnosis of HBV infection and for large-scale surveys.
Hepatitis B virus (HBV) is a well-known agent of acute and chronic hepatitis, with an estimated 350 million chronic carriers around the world. HBV has a circular and partially double-stranded DNA genome of 3.2 kb containing four overlapping open reading frames. HBV strains isolated worldwide have been classified into six genomic groups deduced from genome comparisons and designated genotypes A to F (3, 6, 7). The HBV genotypes have a characteristic geographic distribution. HBV genotyping by phylogenetic analysis based on nucleotide sequences produces the most reliable and certain genotyping results. However, this is not an appropriate method for large-scale genotyping. On the other hand, several groups have reported the genotyping of HBV by the restriction fragment length polymorphism method (1, 2, 4, 8). However, their methods were not so sensitive and specific compared with our PCR genotyping method. In this paper, we report a simpler, more rapid, and more specific genotyping system for HBV involving PCR using type-specific primers.
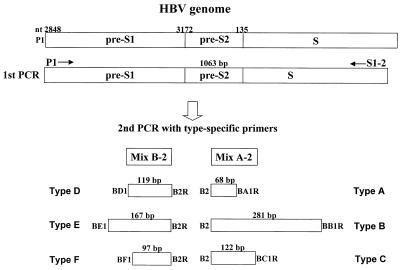
A total of 55 HBV DNA-positive serum samples obtained from individuals in six different countries, including Japan, Vietnam, the United States, Egypt, Ghana, and Bolivia, were used for the evaluation of our genotyping system. We selected the HBV DNA-positive samples by nested PCR. The sequences of PCR primers used in this study are shown in Table 1. The first-round PCR primers (outer primer pairs) and second-round PCR primers (inner primer pairs) were designed on the basis of the conserved nature of nucleotide sequences in regions of the pre-S1 through S genes, irrespective of the six HBV genotypes. P1 (sense) and S1-2 (antisense) were universal outer primers (1,063 bases). B2 was used as the inner primer (sense) with a combination called mix A for genotypes A, B, and C. Mix A consisted of antisense primers BA1R (type A specific), BB1R (type B specific), and BC1R (type C specific). B2R was used as the inner primer (antisense) with a combination called mix B for genotypes D, E, and F. Mix B consisted of sense primers BD1 (type D specific), BE1 (type E specific), and BF1 (type F specific). These primer combinations for second-round PCR were designed on the basis of the differences in the sizes of the genotype-specific bands. The type-specific primers were designed on the basis of the conserved nature of those sequences within a genotype and on the basis of their poor homology with the sequences derived from other HBV genotypes. The strategy for HBV genotyping is illustrated in Fig. 1.
TABLE 1.
Primer sequences used for HBV genotyping by nested PCR
| Primer | Sequencea (position, specificity, and polarity) |
|---|---|
| First PCR | |
| P1b | 5′-TCA CCA TAT TCT TGG GAA CAA GA-3′ (nt 2823–2845, universal, sense) |
| S1-2 | 5′-CGA ACC ACT GAA CAA ATG GC-3′ (nt 685–704, universal, antisense) |
| Second PCR | |
| Mix A | |
| B2 | 5′-GGC TCM AGT TCM GGA ACA GT-3′ (nt 67–86, types A to E specific, sense) |
| BA1R | 5′-CTC GCG GAG ATT GAC GAG ATG T-3′ (nt 113–134, type A specific, antisense) |
| BB1R | 5′-CAG GTT GGT GAG TGA CTG GAG A-3′ (nt 324–345, type B specific, antisense) |
| BC1R | 5′-GGT CCT AGG AAT CCT GAT GTT G-3′ (nt 165–186, type C specific, antisense) |
| Mix B | |
| BD1 | 5′-GCC AAC AAG GTA GGA GCT-3′ (nt 2979–2996, type D specific, sense) |
| BE1 | 5′-CAC CAG AAA TCC AGA TTG GGA CCA-3′ (nt 2955–2978, type E specific, sense) |
| BF1 | 5′-GYT ACG GTC CAG GGT TAC CA-3′ (nt 3032–3051, type F specific, sense) |
| B2R | 5′-GGA GGC GGA TYT GCT GGC AA-3′ (nt 3078–3097, types D to F specific, antisense) |
An “M” represents a nucleotide that could be either an A or a C; a “Y” represents a nucleotide that could be either a C or a T. nt, nucleotide.
The sequence for primer P1 was determined by Lindh et al. (2).
FIG. 1.
Strategy for genotyping of HBV by PCR using type-specific primers. nt, nucleotide.
The nucleic acid was extracted from 100-μl serum samples using a nucleic acid extraction kit (SepaGene RV-R; Sanko Junyaku Co., Ltd., Tokyo, Japan). The resulting pellet was resuspended in RNase-free water and then subjected to nested PCR. We amplified the HBV genome by nested PCR using the universal primers (P1 and S1-2) for the outer primers, followed by two different mixtures containing type-specific inner primers as described above. The first PCR was carried out in a tube containing 40 μl of a reaction buffer made up of the following components: 50 ng of each outer primer, a 200 μM concentration of each of the four deoxynucleotides, 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Norwalk, Conn.), and 1× PCR buffer containing 1.5 mM MgCl2. We used AmpliTaq Gold DNA polymerase to obtain an automatic hot-start reaction. The thermocycler (GeneAmp PCR system 2400, 9600, and 9700; Perkin-Elmer) was programmed to first incubate the samples for 10 min at 95°C, followed by 40 cycles consisting of 94°C for 20 s, 55°C for 20 s, and 72°C for 1 min. As illustrated in Fig. 1, two second-round PCRs were performed for each sample, with the common universal sense primer (B2) and mix A for types A through C and the common universal antisense primer (B2R) and mix B for types D through F. A 1-μl aliquot of the first PCR product was added to two tubes containing the second sets of each of the inner primer pairs, each of the deoxynucleotides, AmpliTaq Gold DNA polymerase, and PCR buffer, as in the first reaction. These were amplified for 40 cycles with the following parameters: preheating at 95°C for 10 min, 20 cycles of amplification at 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s, and an additional 20 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 30 s. Genotypes of HBV for each sample were determined by identifying the genotype-specific DNA bands. The two different second-round PCR products from one sample were separately electrophoresed on a 3% agarose gel, stained with ethidium bromide, and evaluated under UV light. The sizes of PCR products were estimated according to the migration pattern of a 50-bp DNA ladder (Pharmacia Biotech, Uppsala, Sweden).
To test the validity of our PCR genotyping system, genotypes of HBV were also determined by phylogenetic analysis of pre-S1 through S genes in 40 samples. Amplified PCR products were subjected to direct sequencing, and then phylogenetic analysis was performed as reported previously (5).
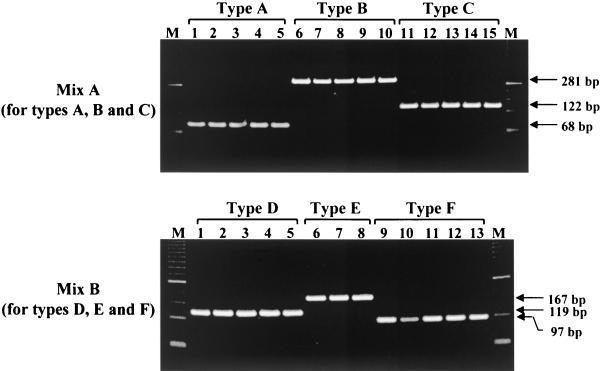
Mix A allows for the specific detection of PCR products for types A, B, and C, and mix B allows for detection of types D, E, and F. As shown in Fig. 2, type-specific PCR products were recognized clearly by their distinct sizes in gel electrophoresis. When 28 isolates in the panel (5 samples from each genotype except for type E), for which serum samples were available, were typed by PCR, the results were in complete accord with the sequences corresponding to their type-specific primers. In the second stage of PCR, type A HBV DNA was amplified with the type A-specific primer, but not with other type-specific primers. Furthermore, to confirm the specificity of our PCR assay, we compared the genotyping results between typing by PCR and by phylogenetic analysis for the 40 samples examined. The results showed 100% concordance between the two assays. In addition, the alignment of the representative pre-S1 through S genes of HBV isolates in the present study revealed that there was a consensus sequence at the same nucleotide positions among different isolates from each genotype. Using this new assay system, we investigated the geographic distribution of HBV genotypes in various countries. The data showed that the distribution of the HBV genotypes in this study population was in accord with the known geographic distribution of HBV genotypes (Table 2).
FIG. 2.
The typical electrophoresis patterns of PCR products from different HBV genotypes as determined by our PCR genotyping system. In each genotype group except for type E, five serum samples that had genotypes already determined by phylogenetic analysis were used. M, molecular size standards.
TABLE 2.
Genotypic distribution of HBV among different countries, determined by PCR using type-specific primers
| Country | n | No. of samples of genotypea:
|
||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | UCb | ||
| Japan | 10 | 0 | 0 | 10 (100) | 0 | 0 | 0 | 0 |
| Vietnam | 30 | 0 | 10 (33) | 19 (63) | 0 | 0 | 0 | 1 (3) |
| United States | 5 | 3 (60) | 0 | 2 (40) | 0 | 0 | 0 | 0 |
| Egypt | 2 | 0 | 0 | 0 | 2 (100) | 0 | 0 | 0 |
| Ghana | 4 | 1 (25) | 0 | 0 | 0 | 3 (75) | 0 | 0 |
| Bolivia | 4 | 0 | 0 | 0 | 0 | 0 | 4 (100) | 0 |
| Total | 55 | 4 (7) | 10 (18) | 31 (56) | 2 (4) | 3 (6) | 4 (7) | 1 (2) |
Numbers in parentheses are percentages.
UC, unclassified.
The genotyping of HBV is important to clarify the route and pathogenesis of the virus. In particular, the examination of sequence diversity among different isolates of the virus is important, because variants may differ in their patterns of serologic reactivity, pathogenicity, virulence, and response to therapy. On the other hand, HBV has genetic variations which correspond to the geographic distribution, and it has been proposed that HBV can be classified into six major genotypes (3, 6, 7). In designing the genotype-specific PCR primers, it is well established that not only higher matching in the entire sequences but also the matching of the two to three nucleotides at the 3′ ends is one of the important parameters for specific priming. Based on this fact, we designed type-specific PCR primers. Sequences within the same genotype were different by ≤2 nucleotides among the entire sequences of the genotype-specific primer, while the sequence within the different genotypes had a difference of ≥3 nucleotides. In the present study, a new genotyping method, based on type-specific primers for PCR, by which HBV isolates can be classified into genotypes A through F is described. To confirm the specificity of the results of PCR typing, phylogenetic analysis in the pre-S1 through S genes of HBV was also performed, and we confirmed the specificity of the results obtained with our PCR genotyping system. This method is very convenient and will assist research workers in conducting large-scale epidemiological studies. Additional investigations, using the serum samples from other geographic regions, are required for the further classification and characterization of HBV. In fact, Stuyver et al. (9) reported recently the identification of a novel genotype of HBV (designated genotype G). For this purpose, our genotyping system using the PCR method introduced here will be useful.
In conclusion, we reported on a newly developed precise PCR genotyping system using type-specific primers, allowing the identification of types A through F. This assay system may be useful for rapid and sensitive genotyping of the HBV genome when either epidemiological, pathological, or transmission studies of this agent are carried out in large scale.
Acknowledgments
We thank Tetsutaro Sata (National Institute of Infectious Diseases) for his continuous encouragement during this study. We also thank Chiaki Miyoshi (International Medical Center of Japan), Ko-ichi Ishikawa and Yutaka Takebe (National Institute of Infectious Diseases), Vo Xuan Ouang and Banh Vu Dien (Cho Ray Hospital, Ho Chi Minh City, Vietnam), Abdel Rahman El-Zayadi (Cairo Liver Center, Cairo, Egypt), and Alfred M. Prince (New York Blood Center, New York, N.Y.) for providing valuable serum samples.
This study was supported in part by a Grant-in-Aid for Science Research of the Ministry of Education, Science and Culture of Japan, by the Ministry of Health and Welfare of Japan, and by an International Medical Cooperation Research Grant of Japan.
REFERENCES
- 1.Lindh M, Anderson A S, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J Infect Dis. 1997;175:1285–1293. doi: 10.1086/516458. [DOI] [PubMed] [Google Scholar]
- 2.Lindh M, Gonzalez J E, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163–174. doi: 10.1016/s0166-0934(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 3.Magnius L O, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 4.Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, Robertson B H. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66–71. doi: 10.1016/s0014-5793(99)00471-8. [DOI] [PubMed] [Google Scholar]
- 5.Naito H, Hayashi S, Abe K. The entire nucleotide sequence of two hepatitis G virus isolates belonging to a novel genotype: isolation in Myanmar and Vietnam. J Gen Virol. 2000;81:189–194. doi: 10.1099/0022-1317-81-1-189. [DOI] [PubMed] [Google Scholar]
- 6.Nordor H, Couroucee A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R I, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 8.Shih J W-K, Cheung L C, Alter H J, Lee L M, Gu J R. Strain analysis of hepatitis B virus on the basis of restriction endonuclease analysis of polymerase chain reaction products. J Clin Microbiol. 1991;29:1640–1644. doi: 10.1128/jcm.29.8.1640-1644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuyver L, Gendt S D, Geyt C V, Zoulim F, Fried M, Schinazi R F, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]