Abstract
Conventional diagnostic tests for legionellosis were negative for a 61-year-old immunocompromised man with pneumonia. However, coculture of a sputum sample with Acanthamoeba polyphaga amoebae led to the recovery of Legionella anisa. This procedure may be a sensitive and convenient diagnostic method, especially for non-Legionella pneumophila species infections that can be diagnosed only by culture.
Legionella pneumophila, the agent of Legionnaires' disease, was first recognized during an outbreak of pneumonia in 1976 (9). This agent has been found to be a common cause of community-acquired and nonsocomial pneumonia (13). Improved culture techniques have also led to infections due to other Legionella species being increasingly reported, and 20 species have now been shown to be pathogenic for humans (14). The definitive method for the diagnosis of legionellosis is culture of the organism, with a sensitivity varying from 32 to 80% (3, 13). This approach remains the sole available procedure for uncommonly encountered species. Isolation of Legionella spp. is based on inoculation of clinical samples onto a buffered charcoal yeast extract agar base enriched with α-ketoglutarate and l-cysteine (BCYE) (14, 15). Coculture with cells and amoebae has also been demonstrated to be convenient (8, 11). Moreover, some Legionella species can be recovered only by using this approach (1). We herein describe the application of the amoebic coculture method to the clinical isolation of a Legionella anisa strain that could not be recovered by agar plating.
A 61-year-old nonsmoking man with a chronic myeloblastic leukemia diagnosed 2 years previously was admitted to the hospital for an acute febrile syndrome. This patient was being treated with melphalan (12 mg/day for 4 days every month). The last treatment had been administered 3 weeks before admission. On admission, the patient was confused and had a fever of 39°C. Chest radiography revealed widespread pulmonary infiltrates. Laboratory data revealed a leukocyte count of 19,160/mm3 with 69% granulocytes, 2% lymphocytes, 1% monocytes, 8% myeloneutrophils, and 20% myeloblastic cells. A type 7 acute myeloblastic leukemia was diagnosed. An intravenous antibiotic therapy of ticarcillin-clavulanic acid, ofloxacin, vancomycin, and amphotericin D (12 and 0.8 g, 400 mg, 2 g, and 50 mg daily, respectively) was administered. Three days later, intravenous erythromycin (3 g daily) was added. The patient's condition progressively worsened, and he died 10 days later. Standard axenic cultures of blood and urine specimens remained sterile. Culture on a sputum sample taken before any antibiotic therapy yielded only bacteria from the oral flora. L. pneumophila antigen detection in urine was negative, as was L. pneumophila serology. A sputum sample was inoculated onto BCYE and BMPA (BCYE with cefamandole, polymyxin B, and anisomycine) agar plates (Oxoid, Dardilly, France) and into amoebic microplates. After 6 days of culture, intra-amoebic bacilli were detected and were later identified as L. anisa. Twenty days after inoculation, BCYE agar and BMPA cultures did not yield Legionella.
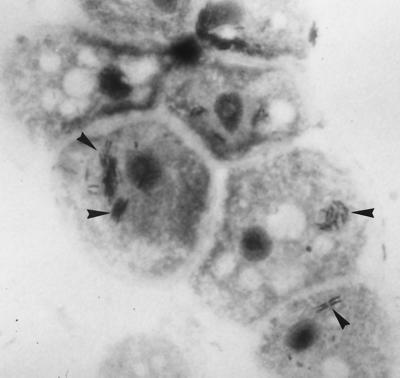
Amoebic coculture was performed as follows. An Acanthamoeba polyphaga strain, Linc AP-1 (provided by T. J. Rowbotham, Leeds Public Health Laboratory, Leeds, United Kingdom), was grown in a 150-cm2 cell culture flask with 30 ml of peptone-yeast extract-glucose broth at 30°C (11). These amoebae are routinely maintained in our laboratory and grow well under the conditions described above (doubling time, approximately 24 h). When their concentration, determined by count in a Nageotte cell with trypan blue, reached 105/ml, the amoebae were harvested and pelleted by centrifugation. The supernatant was removed, and the amoebae were resuspended in 50 ml of Page's amoebic saline (PAS) (11). Centrifugation and resuspension in PAS were repeated twice. After the last centrifugation, the amoebae were resuspended in 30 ml of PAS. Next, 1.5 ml of this suspension was distributed into each well of a 12-well Costar (Corning, N.Y.) microplate. The patient's fresh sputum sample was divided in two aliquots. One aliquot was inoculated directly onto BCYE and BMPA agar plates (Oxoid) and incubated at 35°C in a 2.5 to 5% CO2 atmosphere (Gengag CO2; Biomérieux, Marcy l'Étoile, France) for 20 days. The other was mixed with an equivalent volume of a sputum lytic solution of 2,3-hydroxy-1,4-dithiolbutane (digest-EUR; Eurobio, les Ullis, France) and kept at room temperature for 10 min; then it was mixed with 10 ml of sterile distilled water in order to disrupt cells and centrifuged at 200 × g for 10 min. The supernatant was removed and centrifuged at 8,000 × g for 10 min. The supernatant was removed, and the pellet was resuspended in 200 μl of PAS. Then, 100 μl of suspension was inoculated into one well of an amoebic microplate, and 100 μl was inoculated onto BCYE and BMPA agar plates that were incubated as described above. After inoculation, the amoebic microplate was centrifuged at 1,000 × g for 30 min and incubated at 32°C. At 3 and 6 days after inoculation, the microplate was gently shaken in order to suspend amoebae, and 100 μl of the suspension was used for cytocentrifugation. Slides were Giemsa stained. At day 6, numerous intra-amoebic bacteria were detected (Fig. 1). These were subcultured (100 μl of infected amoebic suspension) on BCYE medium, where they were detectable, among normal bacteria of the oral flora, after 3 days of incubation at 35°C in a 2.5 to 5% CO2 atmosphere. Another subculture using 100 μl of infected amoebic suspension was obtained on a fresh amoebic microplate inoculated as for the sputum sample. These bacteria were oxidase, catalase, and gelatinase positive and grew on BMPA but not on Columbia sheep blood agar. Illumination with long-wavelength UV light (Wood's lamp) showed blue-white fluorescence of colonies. DNA extracts suitable for use as templates in PCR assays were prepared from 10 bacterial colonies suspended in 100 μl of sterile water by using the QIAmp tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. These DNA extracts were amplified and sequenced by using mip gene primers Legmip-f and Legmip-r as previously described (10). The 590-bp sequence obtained was compared with DNA sequence databases using the program BLAST 2.0 (National Center for Biotechnology Information) and showed 100% similarity with the mip gene sequence of L. anisa (GenBank accession number U91607). Immunofluorescence techniques detected no antibodies against the isolate in the patient's serum.
FIG. 1.
Giemsa-stained aggregates (arrowheads) of L. anisa within A. polyphaga amoebae (magnification, ×1,000).
L. anisa is commonly encountered in the environment (6), but to our knowledge, this is only its fourth implication in legionellosis (2, 4, 7). Two previous cases occurred also in immunocompromised patients. It has also been implicated in cases of Pontiac fever (5). Nevertheless, the rarity of cases diagnosed as caused by this species could be due to the failure to isolate it. Diagnostic approaches available for Legionella infections are isolation of the bacterium in culture, direct detection of bacterial antigens or nucleic acids in clinical specimens, and detection of a serological response to the bacterium. Culture remains the method of choice (15) and, when samples are processed correctly, has a sensitivity comparable to or higher than those of other methods (about 80%). However, all diagnostic procedures have been developed and evaluated only for L. pneumophila. For rarely encountered Legionella species, the sole available procedure is culture.
The use of coculture of specimens with amoebae has led to the isolation of L. pneumophila in some instances where inoculated BCYE agar plates remained sterile, and it has allowed isolation of several fastidious Legionella species from clinical and environmental samples (1). The higher sensitivity of this procedure is probably due to a culture amplification phenomenon. When a clinical sample is inoculated on an agar plate, the number of detectable colonies is at best equal to the number of bacteria originally present in the sample. In one of the three cases of L. anisa infection reported, a single colony was detected on agar plate (4). The use of amoebae allows intense multiplication within amoebae of Legionella sp. organisms that become detectable by means of microscopic examination of amoebae or by subculturing on agar plates as previously described (11). Legionella bacteria become detectable even among contaminants, as in the case reported herein. The high sensitivity of this procedure has also allowed the isolation of L. pneumophila from a stool specimen (12).
Clearly, we are unable as yet to draw any meaningful conclusions regarding the comparative performances of coculture and BCYE agar plates. A major drawback could be the delay involved in culture. Nevertheless, we are currently evaluating the prospective merits of both methods for the diagnosis of pneumonia.
REFERENCES
- 1.Birtles R J, Rowbotham T J, Raoult D, Harrison T G. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology. 1996;142:3525–3530. doi: 10.1099/13500872-142-12-3525. [DOI] [PubMed] [Google Scholar]
- 2.Bornstein N, Mercatello A, Marmet D, Surgot M, Deveaux Y, Fleurette J. Pleural infection caused by Legionella anisa. J Clin Microbiol. 1989;27:2100–2101. doi: 10.1128/jcm.27.9.2100-2101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edelstein P H. Legionnaires' disease. Clin Infect Dis. 1993;16:741–747. doi: 10.1093/clind/16.6.741. [DOI] [PubMed] [Google Scholar]
- 4.Fallon R J, Stack B H R. Legionnaires' disease due to Legionella anisa. J Infect. 1990;20:227–229. doi: 10.1016/0163-4453(90)91144-3. [DOI] [PubMed] [Google Scholar]
- 5.Fenstersheib M D, Miller M, Diggins C, Liska S, Detwiler L, Benson Werner S, Lindquist D, Lanier Thacker W, Benson R. Outbreak of Pontiac fever due to Legionella anisa. Lancet. 1990;336:35–37. doi: 10.1016/0140-6736(90)91532-f. [DOI] [PubMed] [Google Scholar]
- 6.Gorman G W, Feeley J C, Steigerwalt A, Edelstein P H, Moss C, Brenner D J. Legionella anisa: a new species of Legionella isolated from potable waters and a cooling tower. Appl Environ Microbiol. 1985;49:305–309. doi: 10.1128/aem.49.2.305-309.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanier Thacker W, Benson R F, Hawes L, Mayberry W R, Brenner D J. Characterization of a Legionella anisa strain isolated from a patient with pneumonia. J Clin Microbiol. 1990;28:122–123. doi: 10.1128/jcm.28.1.122-123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Scola B, Michel G, Raoult D. Isolation of Legionella pneumophila by centrifugation of shell vial cell cultures from multiple liver and lung abscesses. J Clin Microbiol. 1999;37:785–787. doi: 10.1128/jcm.37.3.785-787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDade J E, Shepard C C, Fraser D W, Tsai T R, Redus M A, Dowle W R Laboratory Investigation Team. Legionnaires' disease. Isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 10.Ratcliff R M, Lanser J A, Manning P A, Heuzenroeder M W. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J Clin Microbiol. 1998;36:1560–1567. doi: 10.1128/jcm.36.6.1560-1567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowbotham T J. Isolation of Legionella pneumophila from clinical specimens via amoebae and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36:978–986. doi: 10.1136/jcp.36.9.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowbotham T J. Isolation of Legionella pneumophila serogroup 1 from human feces with use of amoebic cocultures. Clin Infect Dis. 1998;26:502–503. doi: 10.1086/517095. [DOI] [PubMed] [Google Scholar]
- 13.Stout J E, Yu V L. Legionellosis. N Engl J Med. 1997;337:682–687. doi: 10.1056/NEJM199709043371006. [DOI] [PubMed] [Google Scholar]
- 14.Winn W C. Legionella. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 533–544. [Google Scholar]
- 15.Zuravleff J J, Yu V L, Shonnard J W, Davis B K, Rihs J D. Diagnosis of Legionnaires' disease. An update of laboratory methods with new emphasis on isolation by culture. JAMA. 1983;250:1981–1985. doi: 10.1001/jama.250.15.1981. [DOI] [PubMed] [Google Scholar]