Abstract
Ralstonia paucula (formerly CDC group IV c-2) can cause serious human infections. Confronted in 1995 with five cases of nosocomial bacteremia, we found that pulsed-field gel electrophoresis could not distinguish between the isolates and that randomly amplified polymorphic DNA analysis was poorly discriminatory. In this study, we used PCR-ribotyping and PCR-restriction fragment length polymorphism analysis of the spacer 16S-23S ribosomal DNA (rDNA); both methods were unable to differentiate R. paucula isolates. Eighteen strains belonging to other Ralstonia species (one R. eutropha strain, six R. pickettii strains, three R. solanacearum strains, and eight R. gilardii strains) were also tested by PCR-ribotyping, which failed to distinguish between the four species. The 16S-23S rDNA intergenic spacer of R. paucula contains the tRNAIle and tRNAAla genes, which are identical to genes described for R. pickettii and R. solanacearum.
Many changes have been made to the taxonomy of gram-negative environmental bacilli since 1995. Yabuuchi et al. created the novel genus Ralstonia and included three species: R. pickettii, R. solanacearum, and R. eutropha (27). In 1999, another species, Ralstonia gilardii, was described by Coenye et al. (4). Among these species, R. pickettii and R. gilardii have been isolated in human infections, while R. solanacearum remains a phytopathogenic organism (19) and R. eutropha is apparently nonpathogenic (4). CDC group IV c-2 was assigned in 1999 to the genus Ralstonia (14, 18, 25) and is now named Ralstonia paucula sp. nov. (25). R. paucula has been isolated from pool water, groundwater, and bottled mineral water (2, 9, 17) and from clinical specimens. Despite its low pathogenicity, it is now recognized as an opportunistic pathogen which can generate serious infections such as septicemia, peritonitis, abscess, and tenosynovitis, particularly in immunocompromised patients (14). Treatment is mainly based on beta-lactams such as cefotaxime and imipenem, which showed the best in vitro activity (14).
In 1996, we reported five cases of CDC group IV c-2 nosocomial bacteremia at our children's hospital. Comparison between these isolates and eight other blood isolates obtained from five Paris hospitals by means of randomly amplified polymorphic DNA (RAPD) analysis showed a single pattern (13, 14). Genotyping with pulsed-field gel electrophoresis was unsuccessful (14). In contrast, parallel processing of four reference strains obtained from the Centers for Disease Control and Prevention showed four distinct patterns with both techniques.
Recently, several authors have reported the use of PCR-ribotyping (3, 5, 7, 8, 12, 22, 23) and PCR-restriction fragment length polymorphism (PCR-RFLP) analysis (20, 21) for strain differentiation within various bacterial species. In this work, we evaluated the ability of these two techniques to distinguish between R. paucula strains and between strains belonging to other Ralstonia species (R. eutropha, R. pickettii, R. gilardii, and R. solanacearum). As the ribosomal DNA (rDNA) intergenic spacer can contain genes coding for tRNA (15, 24), we also explored this region in R. paucula and compared it to those of other Ralstonia species.
Nine R. paucula strains isolated by blood culture were studied, two from Armand-Trousseau Hospital and the other seven from five Paris hospitals. We also studied four R. paucula reference strains (CDC 104521, CDC 104522, CDC 104523, and CDC 104524), one R. eutropha strain (ATCC 17697), and 17 strains belonging to three other species of Ralstonia, comprising 6 R. pickettii strains (ATCC 27511T, LMG 7012, LMG 7008, LMG 7005, LMG 7011, CIP 104062), 8 R. gilardii strains (LMG 3400, LMG 3399, LMG 5886T, LMG 5887, LMG 5888, LMG 5910, LMG 5913, and CIP 105966T), and 3 R. solanacearum strains (ATCC 11696T, CIP 104762T, and CIP 74.18).
PCR-ribotyping was performed as described by Kostman et al. (7). The sequences of the 16S and 23S primers were 5′-TTGTACACACCGCCCGTCA-3′ and 5′-GGTACCTTAGATGTTTCAGTTC-3′ (7), respectively (Unité de Chimie Organique, Institut Pasteur, Paris, France).
DNA sequencing of the amplification product of one isolate from Trousseau Hospital was performed by Euro Sequence Genes Service (Evry, France).
Sequences of the 16S-23S rDNA spacer regions of other bacteria similar to R. paucula were extracted from the GenBank database by using the BLAST algorithm (1). Sequences were aligned with CLUSTALW 1.61 (6). Secondary structures of potential tRNA sequences were studied using Mfold software version 3.0 (http://www.ibc.wustl.edu/∼zuker).
For RFLP analysis of the 16S-23S rDNA intergenic spacer, two enzymes, AciI and AlwI, with seven and three restriction sites, respectively, were selected. 16S-23S rDNA spacer amplicons were digested with 4 U of each endonuclease (New England BioLabs, Saint-Quentin-en-Yvelines, France) and fractionated by electrophoresis in 2.5% high-resolution agarose gels (MetaPhor; TEBU, Le Perray en Yvelines, France) at 100 V for 3 h.
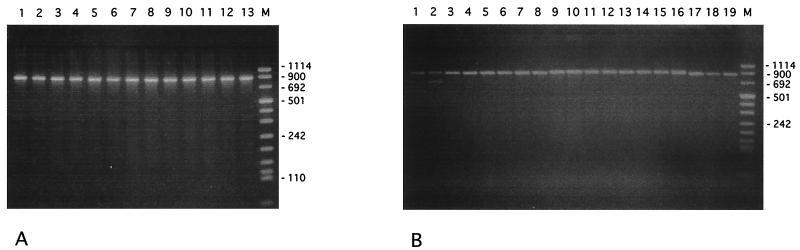
With PCR-ribotyping, all of the strains of R. paucula exhibited a single fragment of between 831 and 947 bp in size, based on the use of DNA Molecular Weight Marker III. A similar pattern was observed with the six R. pickettii strains and the three R. solanacearum strains. The seven R. gilardii strains also showed a similar pattern, with a single fragment of slightly larger size. In contrast, the R. eutropha strain showed a distinct pattern with two fragments between 900 and 692 bp (Fig. 1).
FIG. 1.
PCR-ribotyping patterns of Ralstonia species. (A) R. paucula strains. Lanes 1 to 4, type strains; lanes 5 and 6, Trousseau Hospital strains; lanes 7 to 13, other French hospital strains. (B) Ralstonia strains. Lane 1, R. paucula strain; lane 2, R. eutropha strain; lanes 3 to 8, R. pickettii strains; lanes 9 to 16, R. gilardii strains; lanes 17 to 19, R. solanacearum strains. Lanes M, molecular size markers; sizes are in base pairs.
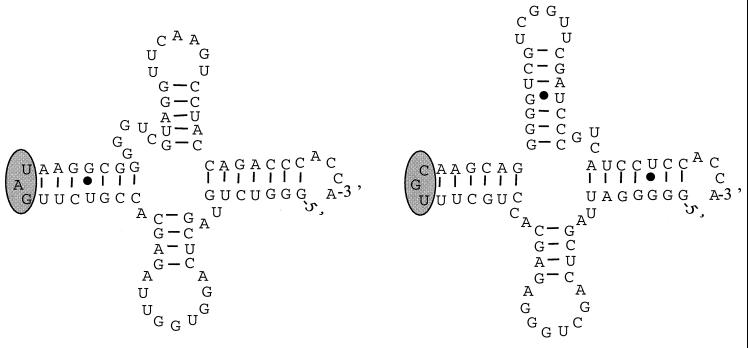
Sequencing of the amplified fragment of one R. paucula isolate from Trousseau Hospital produced an 850-bp sequence, with 153 bp belonging to the 16S rDNA region, 521 bp corresponding to the intergenic region, and the last 176 bp belonging to the 23S rDNA region (Fig. 2). A search for sequence similarities in the GenBank database identified sequences homologous to tRNA genes for alanine and isoleucine on the intergenic spacer sequence of R. paucula. Folding of these sequences produced typical three-stem-loop structures of tRNA with anticodons on loop 2 corresponding to alanine and isoleucine (10). The sizes of the tRNAIle and tRNAAla genes were, respectively, 76 and 77 bp. Possible secondary structures of tRNAIle and tRNAAla of R. paucula are shown in Fig. 3. Alignment of the tRNAIle and tRNAAla gene sequences with those of R. pickettii, R. solanacaerum, and Burkholderia cepacia showed identical sequences for R. paucula, R. solanacaerum, and R. pickettii (except for one deletion in the tRNAAla of R. pickettii) but four or five substitutions for B. cepacia (four substitutions in tRNAIle and five in tRNAAla) (data not shown).
FIG. 2.
Intergenic spacer region sequence of R. paucula (850 bp) (accession no. AF237657), with 153 bp belonging to the 16S portion (first set of boldface type), 521 bp corresponding to the intergenic spacer region, and the last 176 bp belonging to the 23S portion (last set of boldface type). The tRNAIle and tRNAAla genes (77 and 76 bp, respectively) are underlined in boldface.
FIG. 3.
Possible secondary structures of tRNAIle (left) and tRNAAla (right) of R. paucula with the three stem-loops and the anticodon region (shaded).
The amplicons of R. paucula isolates were digested with AciI and AlwI. All of the strains shared a single pattern with each of the two endonucleases. The AciI pattern was composed of eight fragments (340, ∼200, 122, 119, 92, 75, 49, and 44 bp), and the AlwI pattern was composed of four fragments (418, 218, 130, and 75 bp) (data not shown).
We had previously found that RAPD analysis had poor discriminatory power to type Paris isolates of R. paucula and that pulsed-field gel electrophoresis was unsuccessful, probably owing to DNA degradation caused by strong DNase activity, which was not blocked by formaldehyde fixation or boiling (14).
In 1992, Kostman et al. developed a PCR-ribotyping method for B. cepacia which could detect significant polymorphisms in the intergenic 16S-23S spacer of rRNA genes (7). In 1997, Shreve et al. used PCR-RFLP analysis for the 16S and 23S regions of the rRNA genes in an epidemiological study of B. cepacia infection (21). Segonds et al. used PCR-RFLP analysis of the 16S rRNA gene to differentiate Burkholderia species (20).
PCR-ribotyping analysis was not discriminatory for all of the R. paucula clinical isolates, or even for the Centers for Disease Control and Prevention reference strains. Moreover, R. pickettii and R. solanacearum strains could not be differentiated, whereas R. eutropha and R. gilardii strains were clearly distinguished. In our hands, this technique seems to be inappropriate for typing R. paucula isolates and for distinguishing between strains belonging to three other Ralstonia species (R. paucula, R. pickettii, and R. solanacearum).
R. paucula possesses tRNAAla and tRNAIle genes in the 16S-23S spacer of the rRNA operon. The locations of these genes have been established for several species (11, 15, 24, 26). In 1995, Tyler et al. showed that several Pseudomonas species, and particularly Pseudomonas pickettii (now Ralstonia pickettii [27]), had 16S-23S spacer regions which contained potential tRNA sequences for both isoleucine and alanine (24). The tRNAAla and tRNAIle genes were similar in R. paucula, R. solanacaerum, and R. pickettii. These sequences, which are highly conserved in the three Ralstonia species, are probably also well conserved in the genus Ralstonia. In contrast, some differences were observed in the gene sequences of other species, such as B. cepacia.
We did not use RFLP analysis to test 16S rDNA in R. paucula isolates, as all of the 16S rDNA sequences of the reference strains and Paris isolates were identical (14). The identical RFLP profiles of R. paucula isolates obtained with PCR-RFLP analysis of the 16S-to-23S regions of the rRNA genes suggest that the 16S-23S rRNA internal transcribed spacer is well conserved in this species.
Our R. paucula blood isolates remain indistinguishable, despite the use of three genotyping tools (RAPD analysis, PCR-ribotyping, and PCR-restriction of the intergenic spacer region). This lack of diversity of French strains remains unexplained; in particular, the geographic origins and dates of clinical isolation are quite distinct. Shortly, the amplified fragment length polymorphism technique will be applied to the French R. paucula strains.
Nucleotide sequence accession number.
The 16S-23S rDNA spacer sequence described here has been registered with GenBank under accession number AF237657.
Acknowledgments
We thank Chantal Bizet (Collection de l'Institut Pasteur, Paris, France) for providing us with one R. pickettii strain (CIP 104062), two R. solanacearum strains (CIP 104762T and CIP 74.18), and one R. gilardii strain (CIP 105966T) and J. M. Sénèque (bioMérieux Marcy-l'Etoile, France) for providing us with five R. pickettii strains (ATCC 27511T, LMG 7012, LMG 7008, LMG 7005, and LMG 7011), seven R. gilardii strains (LMG 3400, LMG 3399, LMG 5886T, LMG 5887, LMG 5888, LMG 5910, and LMG 5913), and one R. solanacearum strain (ATCC 11696T).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspinall S T, Graham R. Two sources of contamination of a hydrotherapy pool by environmental organisms. J Hosp Infect. 1989;14:285–292. doi: 10.1016/0195-6701(89)90068-6. [DOI] [PubMed] [Google Scholar]
- 3.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 4.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan J R W, Kersters K, Vandamme P. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–413. doi: 10.1099/00207713-49-2-405. [DOI] [PubMed] [Google Scholar]
- 5.Forman W, Axelrod P, St John K, Kostman J, Khater C, Woodwell J, Vitagliano R, Truant A, Satishchandran A, Fekete T. Investigation of a pseudo-outbreak of orthopedic infections caused by Pseudomonas aeruginosa. Infect Control Hosp Epidemiol. 1994;15:652–657. doi: 10.1086/646828. [DOI] [PubMed] [Google Scholar]
- 6.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Kostman J R, Edlind T D, Lipuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagatolla C, Dolzani L, Tonin E, Lavenia A, Di Michele M, Tommasini T, Monti-Bragadin C. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J Clin Microbiol. 1996;34:2440–2443. doi: 10.1128/jcm.34.10.2440-2443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manaia C M, Nunes O C, Morais P V, da Costa M S. Heterotrophic plate counts and the isolation of bacteria from mineral waters on selective and enrichment media. J Appl Bacteriol. 1990;69:871–876. doi: 10.1111/j.1365-2672.1990.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 10.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 11.McClelland M, Petersen C, Welsh J. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J Clin Microbiol. 1992;30:1499–1504. doi: 10.1128/jcm.30.6.1499-1504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin G L, Howe D K, Biggs D R, Smith A R, Ludwinsski P, Fox B C, Tripathy D N, Frasch C E, Wenger J D, Carey R B, Hassan-King M, Vodkin M H. Amplification of rDNA loci to detect and type Neisseria meningitidis and other eubacteria. Mol Cell Probes. 1993;7:7–17. doi: 10.1006/mcpr.1993.1002. [DOI] [PubMed] [Google Scholar]
- 13.Moissenet D, Tabone M D, Girardet J P, Leverger G, Garbarg-Chenon A, Vu-Thien H. Nosocomial CDC group IV c-2 bacteremia: epidemiological investigation by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1996;34:1264–1266. doi: 10.1128/jcm.34.5.1264-1266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moissenet D, Goujon C P, Garbarg-Chenon A, Vu-Thien H. CDC group IV c-2: a new Ralstonia species close to Ralstonia eutropha. J Clin Microbiol. 1999;37:1777–1781. doi: 10.1128/jcm.37.6.1777-1781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan E A, Ikemura T, Post L E, Nomura M. tRNA genes in rRNA operons of Escherichia coli. In: Soll D, Abelson J, Schimmel P, editors. Transfer RNA: biological aspects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1979. pp. 259–266. [Google Scholar]
- 16.Neefs J M, Van de Peer Y, Hendriks L, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990;18:2237–2242. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oie S, Oomaki M, Yorioka K, Tatsumi T, Amasaki M, Fukuda T, Hakuno H, Nagano K, Matsuda M, Hirata N, Miyano N, Kamiya A. Microbial contamination of ‘sterile water’ used in Japanese hospitals. J Hosp Infect. 1998;38:61–65. doi: 10.1016/s0195-6701(98)90175-x. [DOI] [PubMed] [Google Scholar]
- 18.Osterhout G J, Valentine J L, Dick J D. Phenotypic and genotypic characterization of clinical strains of CDC group IVc-2. J Clin Microbiol. 1998;36:2618–2622. doi: 10.1128/jcm.36.9.2618-2622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palleroni N J, Doudoroff M. Phenotypic characterization and deoxyribonucleic acid homologies of Pseudomonas solanacearum. J Bacteriol. 1971;107:690–696. doi: 10.1128/jb.107.3.690-696.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shreve R M, Johnson S J, Milla C E, Wielinski C L, Regelmann W E. PCR ribotyping and endonuclease subtyping in the epidemiology of Burkholderia cepacia infection. Am J Respir Crit Care Med. 1997;155:984–989. doi: 10.1164/ajrccm.155.3.9117036. [DOI] [PubMed] [Google Scholar]
- 22.Smith-Vaughan H C, Sriprakash K S, Mathews J D, Kemp D J. Long PCR-ribotyping of nontypeable Haemophilus influenzae. J Clin Microbiol. 1995;33:1192–1195. doi: 10.1128/jcm.33.5.1192-1195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sontakke S, Farber J M. The use of PCR ribotyping for typing strains of Listeria spp. Eur J Epidemiol. 1995;11:665–673. doi: 10.1007/BF01720301. [DOI] [PubMed] [Google Scholar]
- 24.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonas on the basis of the sequencing of genes coding for 16S–23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, De Vos P, Falsen E. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol. 1999;49:663–669. doi: 10.1099/00207713-49-2-663. [DOI] [PubMed] [Google Scholar]
- 26.Welsh J, McClelland M. PCR-amplified length polymorphisms in tRNA intergenic spacers for categorizing staphylococci. Mol Microbiol. 1992;6:1673–1680. doi: 10.1111/j.1365-2958.1992.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 27.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]