Abstract
Bryophytes are nonvascular spore-forming plants. Unlike in flowering plants, the gametophyte (haploid) generation of bryophytes dominates the sporophyte (diploid) generation. A comparison of bryophytes with flowering plants allows us to answer some fundamental questions raised in evolutionary cell and developmental biology. The moss Physcomitrium patens was the first bryophyte with a sequenced genome. Many cell and developmental studies have been conducted in this species using gene targeting by homologous recombination. The liverwort Marchantia polymorpha has recently emerged as an excellent model system with low genomic redundancy in most of its regulatory pathways. With the development of molecular genetic tools such as efficient genome editing, both P. patens and M. polymorpha have provided many valuable insights. Here, we review these advances with a special focus on polarity formation at the cell and tissue levels. We examine current knowledge regarding the cellular mechanisms of polarized cell elongation and cell division, including symmetric and asymmetric cell division. We also examine the role of polar auxin transport in mosses and liverworts. Finally, we discuss the future of evolutionary cell and developmental biological studies in plants.
A review of the cell biological and developmental mechanisms of bryophytes, including Physcomitrium patens and Marchantia polymorpha.
Introduction
During 470 million years of evolution, the body plans of different groups of land plants diversified independently among the gametophyte and sporophyte generations (Nishiyama et al., 2003; Puttick et al., 2018). In extant bryophytes, the sister group of the tracheophytes, the gametophyte generation is dominant over the sporophyte generation. The gametophyte comprises an apical–basal axis with an apical stem cell producing 3D shoot tissues in which the gametes (antheridia and archegonia) develop (Bennici, 2008). In contrast, the sporophyte in bryophytes consists of a single terminal sporangium (monosporangiate), which is remarkably different from the polysporangiophyte in, for example, flowering plants (Kenrick and Crane, 1991; Ligrone et al., 2012). Three taxonomic bryophyte groups have been recognized and are represented by the three main divisions: Marchantiophyta (liverworts), Bryophyta (mosses), and Anthocerotophyta (hornworts) (Wickett et al., 2014). It has been proposed that extant bryophytes are paraphyletic and liverworts may retain many more ancestral characteristics than other lineages (Bowman et al., 2017). Recent phylogenetic studies, however, overturned this view (Puttick et al., 2018). These studies suggested that bryophytes are monophyletic and that the ancestral embryophytes possessed more complex structures than had been envisaged. Therefore, the current simple body plan of liverworts is considered to be the result of the loss of many characteristics and transformations (Wickett et al., 2014; Puttick et al., 2018; de Sousa et al., 2019; Harris et al., 2020). Bryophyte monophyly suggests that all extant bryophytes and tracheophytes share a last common ancestor, from which they diverged and evolved the characteristics specific to their lineage. Therefore, comparing several bryophyte lineages is crucial to obtain insights into plant evolutionary developmental biology. Among bryophytes, the liverwort Marchantia polymorpha and the moss Physcomitrium patens (previously known as Physcomitrella patens) are established as model species (Prigge and Bezanilla, 2010; Rensing et al., 2020; Kohchi et al., 2021). In contrast, the development of tools and resources for studying hornworts is still in progress, despite their importance for understanding the evolution of key land plant traits (Frangedakis et al., 2021a, 2021b).
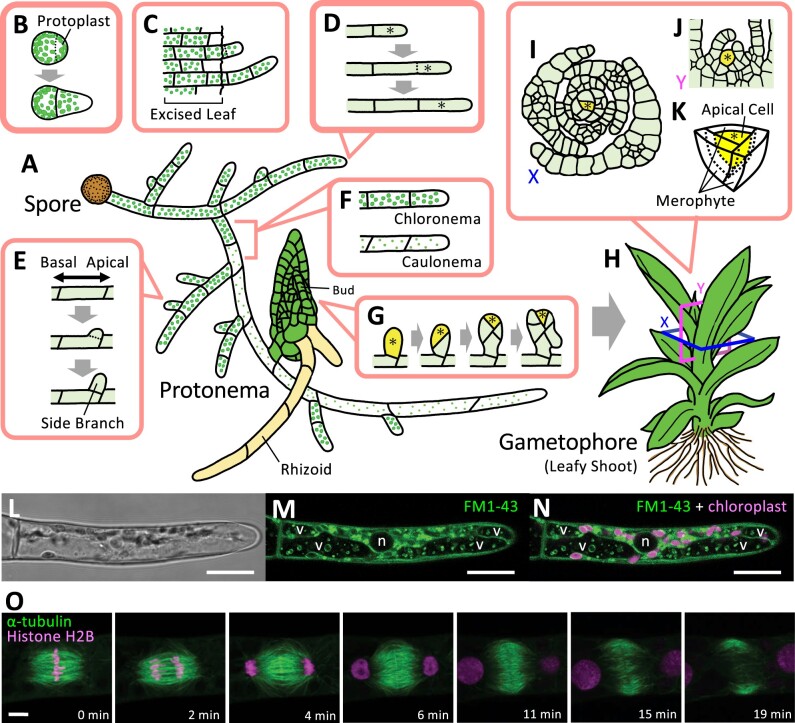
Physcomitrium patens generates filamentous protonemata, the juvenile phase of the gametophyte, which emerge on spore germination (Figure 1A). The protonema possesses a stem cell at the tip (apical stem cell) and undergoes tip growth by performing polarized cell expansion and asymmetric cell division (Figure 1, B–D). Protonemata also branch by generating new apical stem cells (Figure 1E; Menand et al., 2007; Kofuji and Hasebe, 2014). Throughout these processes, the apical stem cell undergoes asymmetric cell division in a single plane, resulting in 1-D filamentous growth (Figure 1, A–F). In contrast, protonemata subsequently develop a leafy gametophore that exhibits 3D growth by generating a different type of apical stem cell (Figure 1G). This apical cell divides asymmetrically with three cleavage planes, which generates leaf initials in a spiral pattern (Figure 1, H–K; Harrison et al., 2009).
Figure 1.
Illustration of gametophyte development in P. patens. Protonemata germinate from spores (A) or emerge from protoplasts (B) and exhibit filamentous tip growth, which is dependent on polarized cell elongation and subsequent asymmetric cell divisions of apical stem cells located at the tip of each filament. Protonemata can also regenerate from excised cells and tissues upon physical damage (C). Concomitant with tip growth (D), protonemata undergo side branching by performing additional asymmetric cell divisions at the apical end of each cell (except for the apical cell) (E). At a still poorly defined time frame during filamentous growth, chloronemata, a type of juvenile protonemata, transition to caulonemata, which serve as precursor cells that have the potential to generate buds (F). While branching produces new apical stem cells that exhibit 1D growth, bud formation accompanies the formation of a single tetrahedral apical stem cell that undergoes 3D growth, a critical step for gametophore formation (G–K). This single apical stem cell in the shoot displays a spiral pattern of asymmetric cell division and produces merophytes (K), whose subsequent cell division activity results in the formation of leaves (I and J). Most of the tissues generated under these developmental processes have simple tissue structures and are a single cell layer thick (Menand et al., 2007; Kofuji and Hasebe, 2014; Falz and Müller-Schussele, 2019). These characteristics present significant advantages for studies in cell and developmental biology and prompted researchers to study the mechanisms of tip growth, asymmetric cell divisions, cell differentiation, and other cell biological phenomena. For example confocal images, a protonemata tip cell stained with FM1–43 (L–N) and time-lapse images of a dividing protonemata tip cell co-expressing α-tubulin:GFP and histoneH2B:RFP (O) are shown (Hiwatashi et al., personal communication). Bright-field images (L) are shown in gray, fluorescence images of FM1-43 (M and N) and α-tubulin:GFP (O) are shown in green, and autofluorescence of chloroplasts (N), as well as fluorescence images of histone H2B:RFP (O), are shown in magenta. “v” and “n” indicates vacuoles and nucleus, respectively. Yellow-colored cells with an asterisk indicate apical stem cells. The square labeled X or Y in (H) indicates a horizontal or vertical section of the shoot apical meristem. The schematic illustration of the horizontal section (I) or vertical section (J) was drawn based on Harrison et al., 2009 and it is labeled as X or Y, respectively. Scale bars 20 µm (L–N) and 5 µm (O).
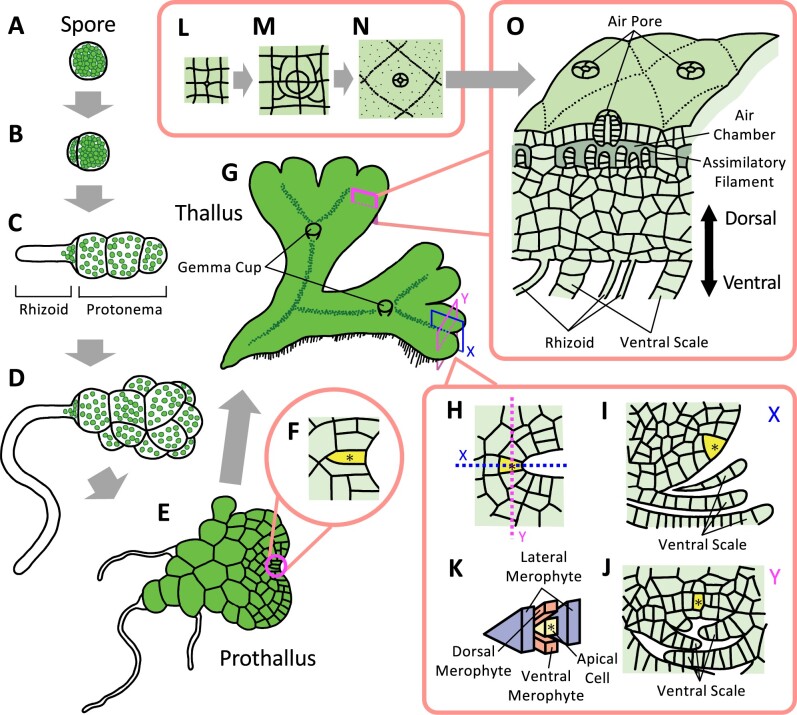
Marchantia polymorpha also develops protonemata upon spore germination (Figure 2, A–C). These protonemata, however, exhibit 1D growth only for a short period before differentiating into a flat mat-like tissue called a prothallus, which possesses an apical stem cell at the tip (Figure 2, C–F). While the prothallus apical stem cell performs asymmetric cell division in two planes, which leads to growth in two dimensions, this apical cell later begins to undergo asymmetric cell division with four planes and generates 3D tissues called thalli (Figure 2, G–O; Bowman et al., 2016; Shimamura, 2016; Kohchi et al., 2021). Thalli differentiate gemma cups on their dorsal tissues, where gemmae can be clonally propagated. Due to the ease of propagation of gemmae through vegetative growth, gemmalings (immature thalli that develop from gemmae) are one of the main targets of cell and developmental biological studies (Kato et al., 2020; Kohchi et al., 2021).
Figure 2.
Illustration of gametophyte development in M. polymorpha. Marchantia polymorpha produces protonemata following spore germination (A–C). Spores exhibit asymmetric cell division, producing one large and one small cell (A and B). While the small cell directly differentiates into a rhizoid, the large cell undergoes several rounds of transverse cell division, leading to the generation of small protonemata (C). In contrast to the filamentous protonemata seen in P. patens, protonemata in M. polymorpha subsequently divide irregularly and generate cell clusters with spherical structures (D). They further generate an apical stem cell that undergoes oblique cell division with two cutting faces, leading to the formation of a 2D heart-shaped body called the prothallus (E and F). Alongside the growth of the prothallus, the apical stem cell eventually begins to exhibit cell division with four cutting faces (G–K) and produces 3D vegetative thalli containing dorsal and ventral tissues (L–O). Rhizoids differentiate in ventral tissues and undergo tip growth (O), while air chambers and gemma cups differentiate in the dorsal tissues (G, L–N). Thalli also undergo periodic dichotomous branching via doubling apical meristems (G). Marchantia polymorpha has recently established as a model organism. Its simple genome structure with low gene redundancy as well as the availability of Agrobacterium-mediated T-DNA transformation provide it with significant advantages for performing molecular biology, including the isolation of large populations of mutants and the comprehensive establishment of fluorescent protein-tagged organelle marker lines (Kanazawa et al., 2016; Minamino et al., 2018). Yellow-colored cells with an asterisk indicate an apical stem cell. The square labeled X or Y in (G) indicates a vertical longitudinal or vertical transverse section of the apical meristem. K, Shows the apical cell and the merophytes that are cut off from the apical cell. X and Y in (H–J) indicate a vertical longitudinal (I) and vertical transverse (J) section of the apical stem cell. H, Shows a horizontal section of the apical stem cell. L–N, Show the developmental process of air pores, which undergo several rounds of regular cell division. The schematic illustrations of the prothallus are drawn based on Kny 1890.
With the numerous approaches available in P. patens and M. polymorpha, the study of these model plants can yield fundamental advances in our understanding of cellular and developmental mechanisms in an evolutionarily informative plant lineage (Rensing et al., 2020; Kohchi et al., 2021). Here, we review current advances obtained by cell biological studies in P. patens and M. polymorpha with an emphasis on polarity formation during cell differentiation and tissue morphogenesis. We first examine mechanisms of polarized elongation in protonemata and rhizoids because cell polarity is best understood in these cell types. We also describe the mechanisms and evolution of symmetric and asymmetric cell divisions, the latter accomplished based on polar signals within a cell, which play crucial roles in the specification of differential cell fate during morphogenesis. Finally, we examine how bryophytes utilize the molecular machinery of polar auxin transport, a mechanism that coordinates cell and tissue polarity in flowering plants.
Part I: polarized cell elongation in bryophytes
Cell shape changes and cell proliferation are two fundamental strategies for morphogenesis in both unicellular and multicellular organisms. Polarized cell elongation contributes to changing cell shapes by orienting cellular processes. Tip growth, an extreme form of polarized cell growth, is a common mode of cell expansion and morphogenesis seen in various organisms. Examples of tip growth include hyphal growth in fungi, the directional cell extension of root hairs and pollen tubes in flowering plants, and the growth of neural axons in animals (Manck et al., 2015; Yogev and Shen, 2017; Orr et al., 2020). The central questions regarding the mechanisms of tip growth are how cells specify the growth domain and determine the direction of cell elongation, and how this directional growth is maintained.
Polarized cell elongation in P. patens
Physcomitrium patens protonemata are an excellent system to address these questions due to their simple filamentous structures in which the apical stem cell undergoes tip growth (Menand et al., 2007; Vidali and Bezanilla, 2012). Studies in P. patens have revealed that the actin cytoskeleton plays an essential role in tip growth (Figure 3A). Pharmacological interference of actin polymerization severely inhibits tip growth (Harries et al., 2005). Loss-of-function of actin-depolymerizing factor (cofilin) and profilin, which are actin-binding proteins involved in the turnover and reconstruction of the actin cytoskeleton, induces the formation of stunted, small, depolarized cells (Vidali et al., 2007; Augustine et al., 2008). Formins, which nucleate and elongate actin filaments, are also involved in tip growth (van Gisbergen et al., 2012). Formins are grouped into two classes in flowering plants and are conserved in bryophytes. RNAi-based approaches in P. patens suggested that class II formins with a phosphatase and tensin homologous (PTEN) domain are involved in tip growth, while class I formins are not (Vidali et al., 2009). PTEN domains in class II formin specifically bind to phosphatidylinositol-3,5-bisphosphate, and this binding appears to be indispensable for controlling tip growth (Figure 3A). Essential roles of actin in tip growth have also been reported in flowering plants (Rounds and Bezanilla, 2013). These findings highlight the evolutionarily conserved function of the actin cytoskeleton in regulating tip growth (Table 1).
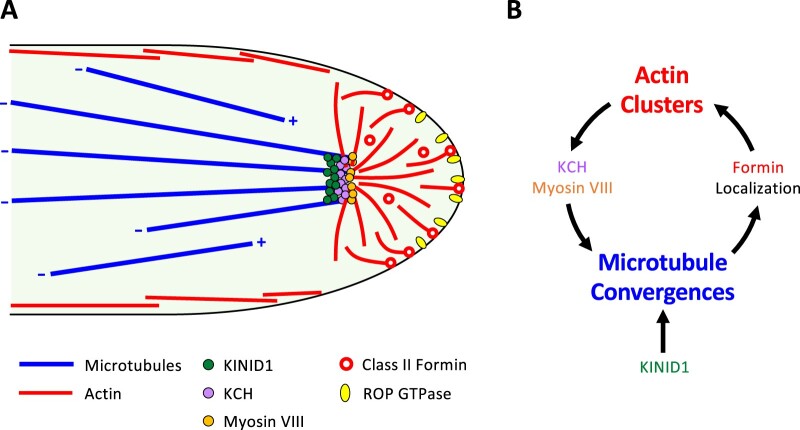
Figure 3.
Current model for tip growth in P. patens. A, Cytoskeletal structures and their regulatory proteins in a protonemata apical stem cell. Cytoskeletons play crucial roles in tip growth of protonemata. While actin filaments mediate polarized elongation, microtubules regulate the directionality of tip growth. Actin filaments are enriched at apical regions and form cluster-like structures. In contrast, microtubules align along the longitudinal axis of protonemata with their plus end directed to the apical region. The plus ends of microtubules are converged by kinesin motor protein KINID1 and KCH just beneath apical actin clusters. Actin clusters and microtubule convergence are thought to be interconnected, where the microtubule spatially restricts actin polymerization and vesicle clustering. The interconnection between actin clusters and microtubule convergence is mediated by cytoskeletal motor proteins, including myosin VIII and KCH. B, Proposed model for the positive feedback loop between actin filaments and microtubules. The convergence of forward-oriented microtubules confines the distribution of class II formins that generate actin filaments. Actin filaments thereafter induce microtubule convergence through the function of myosin VIII. This positive feedback loop ensures persistent polarized growth.
Table 1.
Key findings of Part I: polarized cell elongation in bryophytes
|
The actin cytoskeleton could function in tip growth by providing the tracks for the movement of exocytic vesicles toward the apical tip (Vidali and Bezanilla, 2012). Actin filaments accumulate strongly near the cell apex, where myosin XI and secretory vesicles spatially colocalize. Loss of myosin XI does not affect actin dynamics but causes defective tip growth; similar phenotypes have been observed in mutants of the actin-related genes mentioned above (Vidali et al., 2010). Although the cargos of vesicles transported by myosin XI remain to be elucidated, myosin XI presumably mediates the polarized secretion of new extensible cell wall compounds and membranes to the apex of the cell.
Microtubules are also involved in controlling tip growth in P. patens. Microtubules align along the longitudinal axis of protonemata (Figure 3A). The depolymerization of microtubules by drug treatments produces bent, swollen, or even multiple tips (Doonan et al., 1988). The plus end (the end that grows more rapidly) of each microtubule grows toward the apical expansion zone and converges to a single point just behind the cell apex (Hiwatashi et al., 2014). Importantly, plant-specific kinesins (KINID1) accumulate in this region where they control the organization of microtubules by bundling their plus ends during tip growth (Figure 3A). A recent study also identified the involvement of kinesin-8, kinesin-13, and kinesin-14 family kinesins with a calponin homology domain (KCH) in controlling the directionality of tip growth (Yamada and Goshima, 2018; Leong et al., 2020; Li et al., 2021). While KINID1 and KCH play essential roles in the formation and maintenance of microtubule convergence, kinesin-8 and kinesin-13 appear to be involved in stabilizing the position of microtubule convergence. These findings suggest that microtubules play a critical role in determining the directionality of cell expansion (Table 1).
The interactions between actin filaments and microtubules play crucial roles in diverse cellular functions (Takeuchi et al., 2017). During tip growth in protonemata, microtubule convergence at the cell apex intersects the apical actin clusters (Figure 3A). Myosin VIII proteins are localized to the regions between converging microtubules and actin clusters. Loss-of-function of myosin VIII genes induces the collapse of microtubule convergence, as well as the instability of both the sizes and shapes of actin clusters (Wu and Bezanilla, 2018). Furthermore, microtubule convergence requires actin and KCH motor proteins, which can also bind to actin filaments (Yamada and Goshima, 2018). These findings suggest that myosin VIII and KCH crosslink the actin filaments and microtubules (Figure 3A). Importantly, microtubules also direct the localization of formins that mediate actin nucleation and elongation, which in turn specifies the tip localization of microtubules (Wu and Bezanilla, 2018). This positive feedback loop between actin polymerization and microtubule localization ensures persistent cell elongation in defined locations during the growth of protonemata (Figure 3B).
Rho/Rac of plants (ROP) small GTPase has been implicated in polarized tip growth of root hairs and pollen tubes in flowering plants (Feiguelman et al., 2018). Similarly, in P. patens, ROP GTPase plays an essential role in tip growth of protonemata (Burkart et al., 2015; Cheng et al., 2020). ROP GTPase localizes to the apical plasma membranes (PMs) of tip-growing protonemata (Figure 3A), and its loss-of-function mutant fails to initiate tip growth. The dynamics of actin filaments, as well as cell wall depositions, are also affected in this loss-of-function mutant (Burkart et al., 2015). Notably, systematic loss-of-function analysis of ROP regulators and effectors, such as the GTPase-activating proteins and guanine nucleotide exchange factors for ROP GTPase, further confirmed the crucial role of ROP GTPase in protonemal tip growth (Bascom et al., 2019). Several studies in animals and yeasts have uncovered the roles of the Rho/RAC/CDC42 family of small GTPases in regulating tip growth (Etienne-Manneville and Hall, 2002). These findings suggest that the mechanisms underlying tip growth are evolutionarily conserved across different eukaryotic organisms (Table 1).
Polarized cell elongation in M. polymorpha
Rhizoid growth in M. polymorpha is also a good experimental system for studying tip growth. These rhizoids are unicellular, with a filamentous shape and few chloroplasts, and undergo persistent polarized cell elongation (Shimamura, 2016; Kohchi et al., 2021). As in other systems, microtubules in M. polymorpha rhizoids play a crucial role in specifying the directionality of tip growth. Microtubules align along the longitudinal axis and converge at the apical tip. NIMA (never in mitosis gene a)-related protein kinase, which is known to be involved in regulating microtubule organization in Arabidopsis, specifically localizes at microtubule convergence and controls the direction of cell elongation (Takatani et al., 2017; Otani et al., 2018; Takatani et al., 2020). A T-DNA mutant screening of M. polymorpha sporelings (immature thalli that develop from spores) with defective rhizoid growth and subsequent sequence polymorphism-based genetic mapping identified 33 genes involved in rhizoid growth (Honkanen et al., 2016). The responsible genes encode proteins involved in cell wall biosynthesis (i.e. cellulose synthase-like class D and rhamnose biosynthesis) and cell wall integrity sensing (i.e. CrRLK1L family receptor kinase and PTI-like serine/threonine kinase). They also encode proteins involved in vesicle transport and the cytoskeleton (i.e. Rab GEF, Rho GAP, and class XI myosin) along with proteins that have not yet been functionally characterized in plants. In addition to being a tour de force of genetic analysis, these findings further confirmed the notion that the mechanisms of tip growth are conserved among land plants and were active in the earliest land plants (Table 1).
Part II: symmetric and asymmetric cell division in bryophytes
Polarized cell elongation contributes to changing cell shapes, representing fundamental strategies for morphogenesis in both unicellular and multicellular organisms. Besides polarized cell elongation, cell division (both symmetric and asymmetric) is another fundamental strategy for morphogenesis in living organisms (Tajbakhsh et al., 2009). Symmetric cell division expands the mass of similar cells within tissues, while asymmetric cell divisions presage cellular differentiation, generating new cell types: daughter cells with distinct sizes, shapes, and developmental fates based on positional and polarity information within the mother cells (Sunchu and Cabernard, 2020). Asymmetric cell division plays pivotal role in controlling pattern formation and maintenance of stem cells (Munoz-Nortes et al., 2014; Santoro et al., 2016). Therefore, understanding the mechanism of symmetric and asymmetric cell division is crucial for gaining insights into plant development (Pillitteri et al., 2016; Shao and Dong, 2016). In this section, we first explain the basic mechanisms of plant cell division and its evolution. Next, we introduce current concepts regarding the mechanisms of asymmetric cell division in bryophytes. Finally, we describe how the changes in division planes are associated with the elaboration of the body plan in bryophytes. We also discuss the underlying mechanism that mediates the change in bryophyte body plans.
Cell division machinery in flowering plants
Cell division in plants requires the building of a new cell wall (cell plate) between two daughter nuclei. In flowering plants, the cell plate is formed at the midline of the cell. The cytoskeletal cellular structure known as the phragmoplast mediates the trafficking of vesicles carrying cell wall components toward the cell plate (Jürgens, 2005; Smertenko et al., 2017) (Figure 4A). Cell wall deposition is initiated by the homotypic fusion of vesicles delivered in the region within the bipolar phragmoplast where phragmoplast microtubules overlap. Microtubule overlap zones are thought to be established by the microtubule cross-linking protein MAP65 (Figure 4B; Müller et al., 2004; Ho et al., 2011a). The phragmoplast that formed at the center of the cells expands toward the edges of the cell as cytokinesis progresses, finally reaching the mother cell wall and completing cell division (Rasmussen et al., 2013; Livanos and Müller, 2019). The following three steps, which are repeated cyclically, are proposed as the mechanism for this centrifugal expansion of the phragmoplast microtubule array: (1) attachment of γ-tubulin complexes onto the microtubules at the leading edge; (2) the promotion of microtubule nucleation by γ-tubulin complexes, followed by microtubule elongation; and (3) bundling of nascent microtubules (Murata et al., 2013).
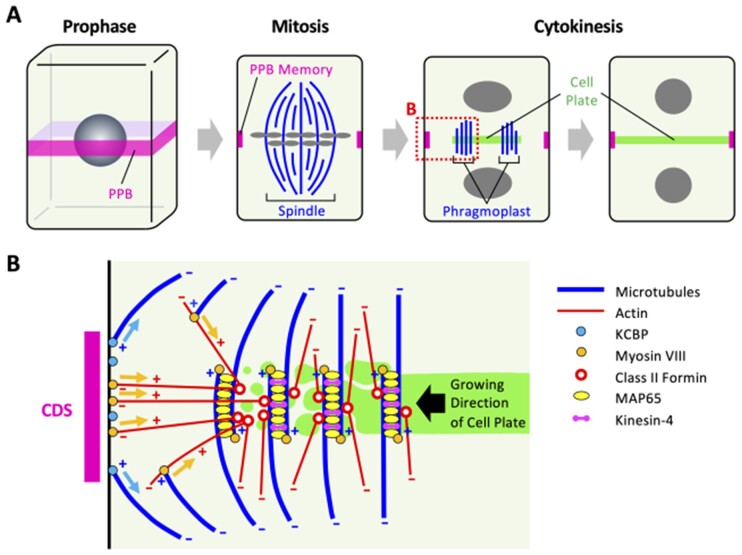
Figure 4.
Illustration of the cytoskeletal organization in a dividing plant cell. A, Stages of plant cytokinesis in flowering plants. The PPB forms during the G2 phase at the future division site and persists throughout prophase. Although the PPB disassembles as the mitotic spindle forms during the transition to metaphase, the PPB leaves behind information that determines the future division site. This information, the PPB memory, is composed of several proteins, such as the motor proteins POK1, KCBP, and myosin VIII. During cytokinesis, two antiparallel cytoskeletal structures form a phragmoplast with their plus ends facing the nascent cell plate. The cell plate and phragmoplast expand gradually until they reach the PM. When the phragmoplast reaches the PM, the cell plate and parental membrane fuse, completing cytokinesis. γ-tubulin complexes and Augmin are thought to be involved in organizing the spindle and phragmoplast microtubule arrays, where Augmin activates the γ-tubulin ring complex to generate spindle and phragmoplast microtubules (Ho et al., 2011b; Hofmann, 2012). B, Proposed mechanism of phragmoplast guidance. MAP65 resides between interdigitating (antiparallel overlapping) microtubules in the phragmoplast midplane, where it confines deposition of the cell plate membrane to a well-defined plane. Class II formin, localized at the phragmoplast midzone, generates actin filaments, which interact with myosin VIII localized at the CDS and peripheral microtubules. While peripheral microtubules with plus ends associated with myosin VIII translocate on actin filaments and are incorporated into the expanding phragmoplast, cortical myosin VIII reels the growing phragmoplast toward the CDS based on its motor activity. In addition to myosin VIII, the myosin/kinesin-chimera protein KCBP is also involved in phragmoplast guidance. KCBP localizes at the CDS and interacts with microtubules at the phragmoplast periphery, whereby KCBP reels in the growing phragmoplast to ensure the correct landing of the phragmoplast at the CDS. In P. patens, microtubule overlap zones are thought to be established by the microtubule cross-linking protein Map65 and their regions confined by kinesin-4, which in turn recruits the exocyst complex to define the region where vesicles are delivered to build the new cell plate (Kosetsu et al., 2013; de Keijzer et al., 2017; Tang et al., 2019). Although PPBs do not exist in some cell types in P. patens, the cellular functions and structures of phragmoplasts are conserved between P. patens and flowering plants. This suggests that homologs of the kinesin-4 and exocyst complex in flowering plants are also involved in defining the regions of cell plate formation.
The mechanism that attracts the phragmoplast to the PM at the cortical division site (CDS), a specialized layer of cytoplasmic proteins on the inner face of the PM where the cell plate fuses with the parental cell walls, is called phragmoplast guidance. This process plays an important role in determining the position and orientation of division planes (Livanos and Müller, 2019). The guidance of the phragmoplast is mediated by ring-like structures called preprophase bands (PPBs) that encircle the premitotic cell in flowering plants (Figure 4A). PPBs contain microtubules and actin filaments and are enriched in endoplasmic reticulum (ER) (Zachariadis et al., 2001). PPBs form during the G2 phase at the future division plane and are known to be an active site of endocytosis (Karahara et al., 2009). PPBs persist throughout prophase but disassemble as the mitotic spindle forms at the transition to metaphase, suggesting that PPBs leave behind a “memory” that attracts the phragmoplast during cytokinesis (Figure 4A). In fact, motor proteins such as POK1 kinesin, the myosin/kinesin-chimera protein KCBP, and myosin VIII remain at the CDS from prophase to the end of cytokinesis in flowering plants and function as the PPB memory (Xu et al., 2008; Lipka et al., 2014; Wu and Bezanilla, 2014; Buschmann et al., 2015). Moreover, the RAN GTPase-activating protein and microtubule-associated TANGLED also function as the PPB memory in flowering plants (Walker et al., 2007; Xu et al., 2008). The exact mechanisms of phragmoplast guidance remain to be elucidated, but the emerging picture is that microtubules and actin filaments work together to achieve phragmoplast guidance (Figure 4B). Phragmoplast microtubules are oriented with their plus ends toward the cell plate. The plus ends of actin filaments are anchored to the phragmoplast mid zone, while the minus ends (the slow-growing ends) are located in the cytosol in flowering plants (Figure 4B; Wu and Bezanilla, 2014). One possible mechanism is that the motor activity of KCBP (kinesin-like calmodulin) toward the minus end of the microtubule and that of myosin VIII toward the plus end of actin reel the growing phragmoplast toward the CDS collaboratively (Figure 4B). Altogether, the cooperative action of the PPB and the phragmoplast plays an important role in determining the cell division planes and cell plate formation.
In addition to the PPB, another type of microtubule called the polar cap plays a critical role in determining the cell division planes (Lloyd and Chan, 2006, Kosetsu et al., 2017). The polar cap that encases the nuclear surface is formed at opposite sides of the nuclear envelope during prophase (Lloyd and Chan, 2006). A lack of polar caps induces misorientation of spindles and division planes, indicating that the polar caps play important roles in determining spindle and cell division axes (Kosetsu et al., 2017). Until recently, the PPB has thought to play dominant role in determining division plane orientation, but reassessment of the function of PPB on division plane orientation has recently advocated (Schaefer et al., 2017). A study of trm678 mutants (lacking PPBs) suggested that the PPB is not required for determining division orientation but is instead required for stabilizing or fine-tuning division orientation (Schaefer et al., 2017). Taking this finding into account, it can be concluded that polar caps play a major role in determining division orientation.
Cell division machineries in M. polymorpha and P. patens and their evolutionary implications
The mechanisms of cell division in plants have undergone significant divergence during evolution. Centrosomes are typical animal cell structures that are present in some green algae, but not in flowering plants. Notably, liverworts and mosses display intermediate evolutionary forms between those of algae and flowering plants (Buschmann and Zachgo, 2016). Marchantia polymorpha exhibits a phragmoplast and PPBs but also employs a so-called polar organizer (PO), a centrosome-like microtubule-organizing center lacking centrioles, in gemmalings (Buschmann et al., 2016). POs are formed by late interphase and localize to the opposite ends of the preprophase nucleus, where they nucleate microtubules and produce astral microtubules. Interestingly, astral microtubules are connected to the PPB, implying a functional link between them. In fact, interfering with POs by treatment isopropyl N-(3-chlorophenyl)-carbamate, which affects the splitting or replication of the microtubule-organizing centers in various eukaryotes, disturbs the formation of PPBs (Buschmann et al., 2016). This strongly suggests that POs are required for PPB formation and, in turn, in division plane alignment.
Physcomitrium patens also exhibits centrosome-like structures called gametosomes in leafy gametophore cells, while such structures are not observed in protonemata (Kosetsu et al., 2017). Unlike centrosomes and POs, microtubules are loosely focused in the gametosome, and their center cannot be unambiguously assigned (Kosetsu et al., 2017). This microtubule cloud transiently appears in the cytoplasm during prophase and while it is dispensable for spindle formation, it is necessary for metaphase spindle orientation, which contributes to division plane determination. Interestingly, both division plane determination and the biogenesis of gametosomes appear to be independent of PPBs, as PPBs are not present in early gametophore cells. The role of gametosomes in controlling division plane determination both with and without PPB formation in P. patens, as well as the presence of centrosome-like structures and PPBs in the same cells in M. polymorpha, suggest a stepwise transition between ancient and modern cell division machineries during plant evolution (Table 2).
Table 2.
Key findings of Part II: symmetric and asymmetric cell division in bryophytes
|
Asymmetric cell division in spores and protoplasts and its similarity to zygotic division in fucoid algae
Asymmetric cell division plays a crucial role in maintaining stem cells and generating a population of differentiated cells (Pillitteri et al., 2016; Shao and Dong, 2016). During their first division, P. patens protoplasts undergo asymmetric cell division, in which they produce one small apical stem cell and one large, differentiated cell (Figure 1A; Kofuji and Hasebe, 2014). Similarly, M. polymorpha spores undergo asymmetric cell division and produce one small rhizoid and one large nonrhizoid cell (Figure 2, A and B; Shimamura, 2016; Kohchi et al., 2021). Despite the obvious asymmetric cell divisions and the simple cellular structures, the mechanisms underlying these processes have not yet been studied. Interestingly, these asymmetric cell divisions are reminiscent of the first asymmetric cell division in zygotes of fucoid brown algae, although they have different evolutionary origins (Bisgrove et al., 2003). In the fucoid zygote, the initial polarity cue is provided by sperm entry, but this cue is overridden by directional environmental cues, such as light and gravity. Cortical actin patches form based on these polarity cues, whereby they initiate rhizoid growth. Notably, auxin and polar auxin transport are implicated in this polarization process, which suggests that directional environmental cues may be translated into an intracellular auxin gradient or a nonuniform pattern of auxin efflux that orients zygotic polarity (Sun et al., 2004). It is interesting to speculate that similar mechanisms controlling asymmetric cell division in spores and protoplasts may exist in P. patens and M. polymorpha.
Asymmetric cell division in the filamentous tissue and leafy gametophore of P. patens
Physcomitrium patens protonemata exhibit asymmetric cell division in tip-growing apical cells (Menand et al., 2007; Kofuji and Hasebe, 2014; Figure 1D). During this process, the place where the cell plate forms is controlled by the position of the nucleus. The migration of the nucleus during tip growth and asymmetric division depends on the cooperative activity of kinesin motor proteins, including KCBP, ARMADILLO REPEAT KINESIN1 (ARK), and KCH. KCH and ARK show minus end-directed and plus end-directed motor activity, respectively, and their mutations cause a change in nuclear positions (Jonsson et al., 2015; Miki et al., 2015; Yamada et al., 2017). Moreover, knockout mutants of KCH display an apical shift of the cell division planes according to the position of the nucleus, pointing to the role of this protein in determining the division plane position (Miki et al., 2015; Yamada and Goshima, 2018). This nuclear migration subsequently collaborates with the mechanisms that guide the phragmoplast to the CDS, whereby myosin VIII pulls the phragmoplast microtubule to the CDS along actin filaments (Figure 4B). This mechanism helps ensure that cell plate expansion occurs along a plane defined by the CDS (Table 2; Wu and Bezanilla, 2014).
Branching processes in P. patens protonemata also accompany asymmetric cell division, generating new apical stem cells at the apical ends of cells (Kofuji and Hasebe, 2014; Figure 1E). As is the case with tip-growing cells, polarized cell expansion, as well as nuclear migration, has been reported during side branch formation. A recent study revealed that ROP small GTPase localizes at the future bulging site several hours before the polarized cell expansion (bulging) occurs (Cheng et al., 2020). Notably, this process occurs earlier than the nuclear migration to the apical end of the cell, indicating that ROP GTPase acts as an upstream factor involved in controlling nuclear migration and therefore the position of asymmetric cell division. ROP GTPase regulates actin filament dynamics as well as cell wall remodeling, which induces polarized cell expansion (Cheng et al., 2020; Yi and Goshima, 2020). Although the mechanism is unknown, this bulging in turn exerts a guidance cue for controlling microtubule-dependent nuclear migration, which determines the asymmetric positioning of the division planes. Importantly, mutants with loss-of-function of ROP GTPase display defective bulge formation and abnormal cell division planes aligning in the transverse direction (Cheng et al., 2020; Yi and Goshima, 2020). These findings indicate that ROP GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during the branching of P. patens filamentous tissues (Table 2).
In addition to filamentous tissues, the leafy gametophore in P. patens also displays asymmetric cell division. During bud formation, shoot initials undergo asymmetric cell division 4 times to establish the tetrahedral shape of gametophore apical stem cells (Figure 1G). The established tetrahedral apical stem cell continuously divides asymmetrically into three planes, which generates leaf initials, a critical step in acquiring leafy structures (Figure 1, I–K; Harrison et al., 2009). The orientation of the cell division planes during tetrahedral apical cell formation is thought to be controlled by gametosomes, the acentrosomal structures that regulate spindle orientation (Table 2; Kosetsu et al., 2017). Moreover, a recent study also uncovered critical functions of the CLAVATA (CLV) peptide and receptor-like kinase pathway in controlling the division planes during this process (Whitewoods et al., 2018). Defects in gametosome formation induce the deviation of division plane angles, and loss-of-function of the CLV pathway induces cell division that is parallel rather than perpendicular to the first division, resulting in the finger-like projection of gametophores. Notably, gametosomes have not been observed in protonemata, and loss-of-function of the CLV pathway does not affect protonemal growth, suggesting that gametosomes and the CLV pathway specifically regulate asymmetric cell division during leafy gametophore development (Table 2).
The asymmetric cell division machinery in P. patens appears to vary depending on the types of cells. A single gametosome is detected during the first three asymmetric cell divisions of the gametophore initials, and the number of gametosomes eventually increases in the leaf initial cells. PPB has not been detected in protonemal cells or in early leafy gametophore cells, while PPB is present in leaf initial cells (Doonan et al., 1987; Spinner et al., 2010; Wu and Bezanilla, 2014). Moreover, loss-of-function of TONNEAU1, a cellular component implicated in PPB formation, leads to the misorientation of cell division in leaf-like gametophore cells but not in protonemal cells (Spinner et al., 2010). These findings suggest the existence of a PPB-independent cell division plane orientation mechanism in protonemata and gametophore initial cells. They also suggest that PPBs and gametosomes control cell division orientation in leaf-like gametophore cells, although their interplay is unclear (Kosetsu et al., 2017).
Asymmetric cell division and body plan transition in mosses and liverworts
The gametophyte body plans of mosses and liverworts are highly dependent on the type of division planes in the apical stem cells. A single plane cleavage of the apical stem cells generates 1D filamentous structures such as protonemata (Figures 1, A–F and 2, A–C), while a two-plane cleavage generates 2D mat-like structures such as leaves or simple thalloids (Figures 1, H–J and 2, E and F). Cleavage in three or four planes enables the onset of 3D growth, generating bushy leafy shoots or complex thalloids, respectively (Figures 1, H–K and 2, G–O; Niklas et al., 2019). The acquisition of apical stem cells with three or four cleavage planes during evolution played an important role in enabling the colonization and radiation of plants on land (Moody, 2020).
In the lifecycles of P. patens and M. polymorpha, a transition between filamentous, planar, and bushy growth is observed (Harrison et al., 2009; Shimamura, 2016; Kohchi et al., 2021). Germinated spores undergo single plane cleavage, producing protonemata with transitory filamentous structures (Figures 1, A–F and 2, A–C). In P. patens, protonemata subsequently form apical stem cells that undergo asymmetric division with three cleavage planes, spirally generating leaf initials that divide into two planes to facilitate planar growth (Harrison et al., 2009; Figure 1, H–K). In contrast, M. polymorpha protonemata develop apical stem cells with two cleavage planes, which produce a prothallus with a planar shape (Figure 2, E and F). The apical stem cell in the prothallus subsequently transforms into an apical stem cell with four cleavage planes, generating a thallus with dorsoventrality (Figure 2, G–O; Shimamura, 2016; Naramoto et al., 2019; Kohchi et al., 2021).
A study in P. patens proposed a model for the mechanism of the body plan transition. APETALA2-type transcription factors orthologous to Arabidopsis AINTEGUMENTA, PLETHORA, and BABY BOOM (APB) were revealed to function as a molecular switch to induce 3D growth (the development of a shoot-like gametophyte apical stem cell) by activating cytokinin biosynthetic genes (Aoyama et al., 2012). Ubiquitin-associated protein NO GAMETOPHORES1 (NOG1) acts antagonistically to DEFECTIVE KERNEL1, a membrane-anchored protein with a C-terminus similar to animal calpain domains II and III, to induce the degradation of a repressor of APB activity (Perroud et al., 2014; Johansen et al., 2016; Moody et al., 2018; Perroud et al., 2020). Moreover, the activation of cytokinin biosynthesis by APB induces the expression of NOG2 (encoding a shikimate o-hydroxycinnamoyl-transferase), leading to the activation of a local auxin response that is dependent on the CLV pathway (Moody et al., 2021). This auxin–cytokinin crosstalk is hypothesized to modulate the local concentrations of auxin and cytokinin to ensure the proper orientation of cell division within buds to initiate 3D growth and to prevent the formation of unnecessary buds (Moody et al., 2021).
A study in M. polymorpha provided important insights into the function of the CLV pathway in bryophytes. CLV3/EMBRYO SURROUNDING REGION-RELATED (CLE) family genes are divided into two subgroups based on the initial amino acid in the mature form of the peptide hormone, where the H-type includes Arabidopsis TDIF and the R-type includes Arabidopsis CLV3. The M. polymorpha genome contains two CLE genes, MpCLE1 and MpCLE2, belonging to the H- and R-type, respectively (Hirakawa et al., 2019). The R-type CLE peptide MpCLE2/CLV3, an ortholog of PpCLV, acts as a positive regulator of stem cell maintenance in the M. polymorpha gametophyte body (Hirakawa et al., 2020). This function is different from that of PpCLV, which controls the division plane for the initiation of 3D growth. Furthermore, MpCLE1, a H-type CLE peptide, negatively regulates stem cell activity and controls cell division orientation, which is reminiscent of the function of the P. patens R-type CLE peptide (Hirakawa et al., 2019). It appears that the M. polymorpha H-type CLE peptide possesses similar functions to the P. patens R-type CLE peptide in terms of stem cell regulation, while the biological functions of the R-type CLEs in P. patens and M. polymorpha are different. The finding that 3D growth in M. polymorpha is normal in the absence of MpCLE2 also suggests that the transition mechanism from 2D to 3D growth is at least partially different between mosses and liverworts, even though the 3D growth system is thought to have been established in their common ancestors (Harrison, 2017).
Part III: polar auxin transport in bryophytes
Morphogenesis of multicellular organisms requires coordinated cell elongation and cell division relative to the body axis (Yoshida et al., 2019). In plants, auxin regulates multiple aspects of plant development, including cell division and cell elongation. In particular, the vectorial transport of auxin within tissues, called polar auxin transport, coordinates cell and tissue polarity (the body axis), forming the basis for shoot branching, stem cell maintenance, vascular differentiation, and tropic responses in angiosperms (Peer et al., 2011). The direction of polar auxin transport is determined by the PIN-formed (PIN) proteins, which localize to the PMs in a polar manner (Krecek et al., 2009). Several genes that regulate the polar localization of PINs have been identified from studies in Arabidopsis (Adamowski and Friml, 2015; Naramoto, 2017). A recent comparative genomic analysis shows that many of these genes are conserved in liverworts, mosses, ferns, and seed plants (Suetsugu et al., 2016; Bowman et al., 2017). Moreover, polar auxin transport has been observed in various gametophytic and sporophytic tissues in mosses and liverworts (Cooke et al., 2002; Poli et al., 2003; Fujita et al., 2008). These findings lead us to ask questions about the origin and evolution of polar auxin transport and how PIN-mediated auxin transport controls development in bryophytes. Here we provide a summary of current knowledge regarding PIN-mediated auxin efflux and its role in the development of mosses and liverworts. We also examine the evolutionary process of polar auxin transport systems involving PIN-dependent and -independent auxin transport.
PIN-mediated auxin efflux during P. patens gametophyte development
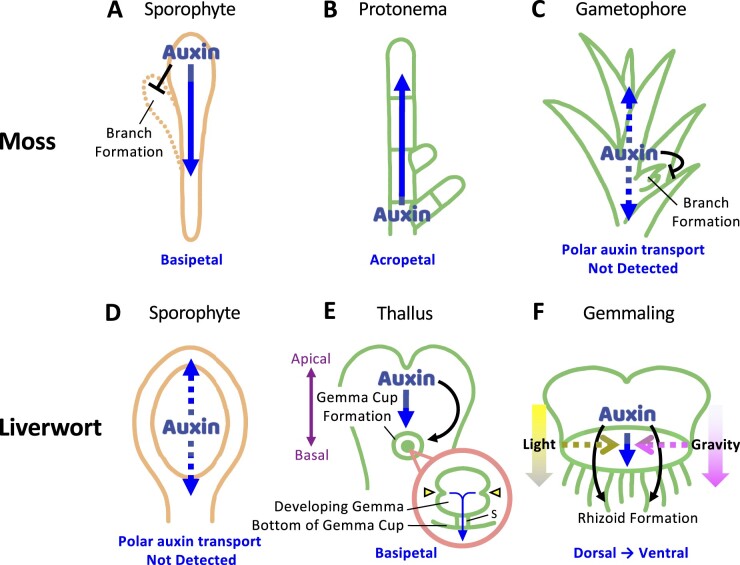
Studies in P. patens have provided some important findings concerning polar auxin transport (Figure 5, A–C). Like the angiosperm Arabidopsis, the P. patens genome also encodes two types of PIN proteins, long- and short-type PINs, which differ in the length of the hydrophilic loop between the transmembrane regions. Long-type PINs are canonical PIN proteins that efflux auxin from a cell, and short-type PINs are noncanonical PIN proteins with postulated roles in auxin metabolism and homeostasis within a cell (Viaene et al., 2013; Bennett, 2015). Auxin efflux mediated by long PINs plays crucial role in the growth and development of protonemata and leafy gametophores (Viaene et al., 2013; Bennett, 2015). Loss-of-function of the long-type PIN genes PpPINA and PpPINB leads to an earlier cell fate transition from chloronemata to caulonemata (Figure 1F; Viaene et al., 2014). The change in PIN activity affects cell division and cell elongation in the leaves of leafy gametophores. It also affects shoot tropism upon light or gravity stimuli (Table 3; Bennett et al., 2014b). Furthermore, treatment with N-1-naphthylphthalamic acid (NPA), an inhibitor of polar auxin transport, disrupts the extension of the proximodistal and mediolateral axes of the developing leaf as well as the function of the apical meristem of the gametophore. Importantly, PpPINA and PpPINB localize to the apical side of the PM (toward the growing tip of filamentous tissue) in protonemata (Table 3; Figure 5B). They also exhibit polar localization at the young leaf stage while possessing the capacity to change their localization within the PM from bipolar to symmetrical during the developmental transition to cell elongation (Viaene et al., 2014; Bennett et al., 2014b). These findings suggest that PIN-mediated auxin transport regulates multiple aspects of moss development throughout the gametophytic generations (Table 3). The recruitment of PIN-mediated auxin transport to regulate shoot tropism and meristem function has clear parallels among land plants.
Figure 5.
Auxin transport in gametophytic and sporophytic tissue of mosses and liverworts. A–C, Transport patterns of auxin and its role in moss development. Polar auxin transport has been experimentally measured in sporophytic tissue in P. patens (A) but not in gametophytic tissue in protonemata (B) or the leafy gametophore (C). Polar auxin transport inhibits branch formation in sporophytic tissues, which is reminiscent of the apical dominance mediated by polar auxin transport in flowing plants. In protonemata, acropetal auxin transport is expected due to the apical localization of PIN proteins (B). It is also thought that auxin is diffusively transported through plasmodesmata, which may inhibit shoot branching in P. patens (C). Note that bidirectional polar auxin transport has also been reported in Polytrichum (another moss genus) sporophytes. The different sensitivities of polar auxin transport inhibitors between acropetal and basipetal transport suggest that these processes are regulated by different cellular machineries (Poli et al., 2003). D–F, Transport patterns of auxin and its role in liverwort development. Polar auxin transport is not detected in sporophytic tissue (D) but is detected in gametophytic tissue in liverworts (E and F). In M. polymorpha, auxin is basipetally transported along the midvein, which is thought to regulate gemma cup formation (E). Within gemma cups, the growth and development of gemma, including the number and position of apical notches (E, inset in pink circle), are regulated by polar auxin transport. The expression pattern of ProGH3:GUS, an auxin response marker, suggests that auxin accumulates at the bottom of gemma cups (Ishizaki et al., 2012). Therefore, auxin may be transported from apical notches to the stalk attached to the bottom of the gemma cup (E, inset in the pink circle). In addition to basipetal auxin transport, auxin may be transported from dorsal tissues to ventral tissues during the germination process of gemmalings (F). This auxin transport toward ventral tissues determines dorsoventral tissue patterning, which in turn promotes rhizoid formation at ventral tissues (F). Interfering with either polar auxin transport or light and gravity signals induces rhizoid formation from both dorsal and ventral tissues (Halbsguth and Kohlenbach, 1953; Allsopp et al., 1968). This suggests that light and gravity signals are translated into polar auxin transport to specify dorsoventral patterning of gemmalings. Blue arrow indicates the direction of auxin flow. Arrowhead in the inset of (E) indicates the apical notch. Stalk is abbreviated as “S.”
Table 3.
Key findings of Part III: polar auxin transport in bryophytes
|
The conserved roles of PIN-mediated polar auxin transport in moss sporophyte development
Bryophyte sporophytes undergo polarized growth to form linear tripartite axes. Moss and liverwort sporophytes develop an apical capsule, a basal foot, and an intermediate seta (stalk), whereas hornwort sporophytes consist of an elongated apical sporangium and a basal foot with an intervening intercalary meristem (Ligrone et al., 2012). Among these, young setae from moss sporophytic tissues are the only structures that can transport auxin in a polar manner at measurable levels (Figure 5A; Rose and Bopp, 1983; Cooke et al., 2002; Poli et al., 2003; Fujita et al., 2008). In P. patens, inhibition of polar auxin transport by NPA induces the formation of multiple sporangia (Fujita et al., 2008). Knockout mutations of PpPINA and PpPINB induce a similar phenotype (Bennett et al., 2014b). These findings suggest that sporophyte development in moss is regulated by polar auxin transport, although the subcellular localization of PINs has not yet been determined (Table 3; Fujita et al., 2008; Bennett et al., 2014b). This multiple sporangium phenotype could be derived from the early embryonic duplication of the apical cell or bifurcation. Interestingly, this phenotype resembles the branching morphologies of the early prevascular fossils. Polar auxin transport is known to play a crucial role in determining the architecture of angiosperms, and the changes in apical meristem function underpin architectural divergence among plant groups (Philipson, 1990; Harrison, 2017; Harrison and Morris, 2018), suggesting that PIN-mediated auxin transport has contributed to the morphological diversification during land plant evolution.
Polar auxin transport during liverwort gametophyte development
Even though sporophytic tissues in liverworts do not exhibit clear polar auxin transport, gametophytic tissues can transport auxin in a polar manner (Figure 5, D–F; Maravolo, 1976; Gaal et al., 1982). In M. polymorpha, auxin is basipetally transported along the thallus midvein (Figure 5E), and this transport is inhibited by 2,3,5-triiodobenzoic acid (TIBA), cinnamic acid, and ethylene, known inhibitors of polar auxin transport in tracheophytes (Maravolo, 1976). Moreover, interference with energy-yielding metabolism (due to anaerobic conditions) reduces basipetal transport. These findings indicate that liverwort gametophytes carry out typical polar auxin transport, as do flowering plants (Table 3).
Polar auxin transport is thought to play crucial roles in the growth and development of gametophytes throughout the vegetative lifecycle of bryophytes. In M. polymorpha, TIBA treatment induces the production of numerous rhizoids from both the dorsal and ventral surfaces of gemma upon germination (Allsopp et al., 1968). Abnormally shaped gemmae also form within gemma cups in the presence of TIBA in M. nepalensis (Kaul et al., 1962). Whereas the gemma structures in wild-type plants are symmetrical, with a single notch on either side (Figure 5E), TIBA treatment induces abnormal numbers and positions of apical notches, such as gemmae without an apical notch or with one or three apical notches, and gemmae with two asymmetrical apical notches (Kaul et al., 1962). Moreover, fewer gemmae and gemma cups form in the presence of TIBA. Similar phenotypes are also observed in auxin signaling mutants and transgenic lines of M. polymorpha in which auxin biosynthesis has been modified. Auxin response factor1 (Mparf1) mutants show defective positions and numbers of apical notches in M. polymorpha (Kato et al., 2017). A dominant mutation of Indole-3-Acetic Acid (MpIAA) repressors, overexpression of the co-repressor MpTPL (TOPLESS), or reduced MpYUC2 (YUCCA2) auxin biosynthesis activity inhibit gemma cup formation (Eklund et al., 2015; Kato et al., 2015; Flores-Sandoval et al., 2015). The subcellular localization of MpPIN1, as well as the developmental phenotypes of Mppin1 mutants, have not yet been determined, and thus the role of MpPIN is unclear. Nevertheless, these findings suggest that polar auxin transport plays important role in gametophyte development in liverworts (Table 3).
Plasmodesmata as symplastic auxin transfer routes between tissues
Shoot branching plays a major role in determining plant architecture, which has evolved independently in sporophytic and gametophytic tissues in flowering plants and in bryophytes, respectively (Kenrick and Crane, 1997; Edwards et al., 2014; Harrison, 2017). PIN-mediated auxin transport is known to be involved in inhibiting shoot branching in vascular plant sporophytes (Prusinkiewicz et al., 2009; Shinohara et al., 2013). Similarly, auxin is thought to inhibit the branching of leafy gametophores in P. patens (Figure 5C). A mathematical modeling approach combined with physiological experiments suggested a requirement for bi-directional (or diffusion-like) auxin transport away from the apex to generate realistic branching patterns in P. patens (Figure 5C; Coudert et al., 2015). In contrast to the findings in flowering plants, however, pina pinb mutants, as well as NPA-treated plants in P. patens, exhibit mildly disrupted branching patterns. Moreover, treatment with 2-[4-(dimethylamino)-2-hydroxybenzoyl]benzoic acid, an inhibitor of ATP binding cassette class B/P-glycoproteins (ABCB/PGPs), which function as auxin transporters, does not result in any branching phenotypes. This indicates that the PIN and ABCB/PGP family transporters are not significant contributors to the branching pattern. Interestingly, the branching pattern is sensitive to the application of the callose synthesis inhibitor 2-deoxy-D-glucose (Coudert et al., 2015). Considering that auxin can move between cells through plasmodesmata in Arabidopsis (Han et al., 2014; Gao et al., 2020), these data suggest that auxin can move with diffusive properties via callose-gated plasmodesmatal connections between cells, which contributes to shoot branching by maintaining auxin concentration gradients between cells.
Evolution of the auxin transport system
It has been proposed that short-type PIN proteins that localize to the ER were the ancestral form of PIN proteins (Viaene et al., 2013). However, a subsequent phylogenetic and structural analysis, with a massive increase in taxon sampling compared to previous studies, overturned this view (Bennett et al., 2014a; Bennett, 2015). Notably, long-type PIN is present in all land plants, but short-type PIN is not present in some, such as lycophytes and monilophytes, demonstrating that long-type PIN is the ancestral form (Bennett et al., 2014a; Bennett, 2015). Moreover, contrary to previous reports (De Smet et al., 2011; Viaene et al., 2013), the primary structures of PIN proteins in charophyte algae (Klebsormidium nitens and Spirogyra pratensis) are not similar to the structures of short-type PIN proteins (Hori et al., 2014; Bennett et al., 2014a). These algal proteins have long loops with no obvious homology to the canonical long loop. This suggests that short-type PIN proteins evolved repeatedly and independently from within the canonical long-type PIN lineage and underwent neofunctionalization events (Bennett et al., 2014a; Bennett, 2015).
In addition to PIN proteins, auxin transport is also regulated by ABCB/PGPs transporters and plasmodesmata. Orthologs of ABCB/PGP family transporters have been identified in the Chlorophyta and Streptophyta (charophyte algae and all land plants) (De Smet et al., 2011). In contrast, plasmodesmata are thought to have emerged after the divergence of the charophyte alga K. nitens (Nishiyama et al., 2018). Importantly, recent analysis suggested the existence of PIN-mediated auxin transport in K. nitens, although the exact function is not clear (Skokan et al., 2019). These observations, together with the finding that PINs are not present in the Chlorophyta, suggest that ABC/PGPs may function as the most ancestral auxin transport system, although it is nearly impossible to perform sequence-based assignment of functional orthologs due to the significant functional diversification within ABCB/PGPs. A plasmodesmata-mediated transport system may have been acquired after the emergence of PIN-mediated polar auxin transport systems.
Conclusions and future perspectives
Polarized cell elongation, cell division, and polar auxin transport are critical biological processes for controlling plant morphogenesis. Studies of P. patens and M. polymorpha and their comparison with flowering plants provide important insights into the cell and developmental biological mechanisms underlying these processes. It is clear that highly conserved cytoskeletal components involving directional vesicle transport and cell wall remodeling are utilized for controlling polarized cell elongation among land plants. In addition, overall, the same molecular components involved in polarized cell elongation, such as the cytoskeletal components of myosin VIII, formin II, and ROP GTPase, are exploited to control cell division. Importantly, among land plants, the cell division plane is regulated by the analogous microtubule structures (polar caps, gametosome, and POs) that resemble centrosomes, suggesting that these centrosome-like structures act as conserved mechanisms for division plane orientation. Furthermore, studies of P. patens have suggested that polar auxin transport can act as an evolutionarily conserved biological system for regulating light and gravity responses among land plants. Studies in M. polymorpha, which identified the auxin-dependent pattern formation of gemmae, also suggested that polar auxin transport can act as an evolutionarily conserved biological mechanism to coordinate cell and tissue polarity. Altogether, studies of P. patens and M. polymorpha have contributed to the discovery of important cell and developmental biological mechanisms governing plant morphogenesis, besides contributing to bryophyte biology.
Bryophytes are attractive plants for studying cell and developmental biology in the context of evolutionary biology. Although the gametophytic tissues of bryophytes and the sporophytic tissues of flowering plants are not evolutionarily homologous, recent studies have indicated that common gene regulatory networks are utilized to regulate the growth and development of analogous structures (Coudert et al., 2015; Naramoto et al., 2019; Hata and Kyozuka, 2021; Veron et al., 2021). The tip growth of the P. patens protonemal apical cell and the M. polymorpha rhizoid cell are both mediated by similar mechanisms to those of root hair and pollen tube development in flowering plants (Table 1; Jones and Dolan, 2012; Bascom et al., 2018). Moreover, the differentiation of water-conducting cells in P. patens and that of xylem cells in flowering plants is mediated by a similar mechanism (Xu et al., 2014). Importantly, it has been suggested that the stomata mother cell in monocots, the branching filamentous cell in P. patens, and the gemma initial cell in M. polymorpha all utilize ROP GTPase machineries to execute asymmetric cell division (Humphries et al., 2011; Facette et al., 2015; Hiwatashi et al., 2019; Yi and Goshima, 2020). Besides these similarities among land plants, studies in yeast and mammalian cells, and their comparison with plant cells (i.e. the tip growth mechanism) suggested that essentially, there is a widely conserved set of cytoskeletal gene products that coordinate cell growth, shape, and division within all classes of eukaryotic organisms. Among all eukaryotic cells, P. patens has a suite of advantages for cell biological analysis. The cells are large and exposed and thus easier to access, and they are easier to culture than mammalian cells. Resources for forward genetics in P. patens have caught up with the already unsurpassable resources for reverse genetics. Based on these advantages, we conclude that P. patens could serve as an outstanding model organism for studying these essential features of eukaryotic cell structural integrity.
There also appear to be some similarities in the functions of auxin among land plants. As is the case with flowering plants (Adamowski and Friml, 2015; Naramoto, 2017), PIN regulates the responses of the leafy gametophores of P. patens to light and gravity signals (Table 3; Viaene et al., 2014; Bennett et al., 2014b). Interestingly, it is suggested that in M. polymorpha, light and gravity signals mediate the dorsoventral tissue patterning of the gemmaling, and the involvement of auxin in this process has been proposed (Figure 5F; Fitting, 1936a, 1936b; Halbsguth and Kohlenbach, 1953; Bowman, 2016). These findings imply that the elaborate regulation of PIN polar localization in the complex tissues of angiosperms can be traced back to simpler polarity changes in their ancestors. It is tempting to speculate that light and gravity signals control polar auxin transport, which in turn mediates the dorsoventral patterning of M. polymorpha gemmalings (Figure 5F). Future studies of light and gravity responses and their involvement in tissue patterning in M. polymorpha could shed light on the ancient mechanisms of polar auxin transport and its evolutionary process.
In contrast to these similarities, bryophytes also possess many unique characteristics that are not found in flowering plants. The reproduction of bryophytes relies on motile, bi-flagellated sperm cells that require water in order to fertilize the egg cell (Furuichi and Matsuura, 2016; Horst and Reski, 2017; Meyberg et al., 2020). In this process, many interesting biological phenomena exist. The recognition system between the egg and sperm as well as the biogenesis of unique intracellular structures during spermatogenesis, including the centriole and cilia, are hot research topics. In fact, the chemotaxis mechanisms of sperm involving calcium ion fluxes and glutamate receptor-like channels in P. patens have been elucidated (Ortiz-Ramirez et al., 2017). The processes of centriole biogenesis and maturation in P. patens were analyzed and novel structures, such as “naked cartwheels,” were identified (Pereira et al., 2020). Further studies into these processes in P. patens and M. polymorpha should provide important insights into the evolution of the fertilization system of land plants.
Physcomitrium patens and M. polymorpha are thought to represent the monophyletic Setaphyta (Renzaglia et al., 2018), in which M. polymorpha lost genes and regulatory complexity, while P. patens underwent gene duplication and increased its gene regulation complexity (Puttick et al., 2018). Their morphologies are somewhat different from typical mosses and liverworts: short seta in P. patens and the complex thalloid body in M. polymorpha are not observed in most mosses and liverworts (Shaw et al., 2011; Kirbis et al., 2020). As it remains unclear whether they represent typical mosses and liverworts, it is important to study both organisms in addition to other plant species. Moreover, the third major group of bryophytes, the hornwort, has emerged as a new frontier in evolutionary cell and developmental biology. The genomes of hornworts (Anthoceros angustus and Anthoceros punctatus), a sister group to liverworts and mosses, have recently sequenced (Li et al., 2020; Zhang et al., 2020). Hornworts possess some important traits for studying evolutionary developmental cell biology (Frangedakis et al., 2021a). The first cell division of the zygote in hornworts is parallel to the longitudinal direction of the archegonium, which differs from that of the other bryophytes. The development of a basal meristem with perpetual division activity within sporophyte tissue is also a unique character of hornworts. Hornworts also display monoplastidy (one or just a few chloroplasts per cell), while all other land plants possess several chloroplasts within a cell. Moreover, hornworts undergo symbiosis with cyanobacteria and with arbuscular mycorrhizal fungi, whereas M. polymorpha and P. patens do not. A technique for Agrobacterium-mediated T-DNA transformation of A. agrestis has recently established (Frangedakis et al., 2021b). Future cell and developmental studies of hornworts will help elucidate the evolutionary process of cell and developmental mechanisms of these key land plant traits.
In addition to bryophytes, studying charophyte algae is also important for obtaining evolutionary insights into gene function. The genomes of some charophytes, the sister lineage of land plants, including K. nitens, Chara braunii, Spirogloea musicola, and Mesotaenium endlicherianum, have already been sequenced (Hori et al., 2014; Nishiyama et al., 2018; Cheng et al., 2019). Importantly, a comparative genomic study suggested that many characteristics, such as tip growth, branching, plasmodesmata, and phragmoplasts, emerged in the common ancestor of Phragmoplastophyta. This division of plants comprises three lineages of charophyte algae (Charophyceae, Coleochaetophyceae, and Zygnematophyceae) and the land plants (Nishiyama et al., 2018). Future cell and developmental biological studies of A. agrestis and one of the charophyte algae, in addition to P. patens and M. polymorpha, will help elucidate the origin and evolution of the key cell biological phenomena and their relationships with plant body plans.
Acknowledgments
We apologize to the authors whose works are not cited because of space constraints. We thank Prerna Singh and Ooi-kock Teh for editing the manuscript.
Funding
This work was supported by a Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and technology, Japan (KAKENHI grant numbers 20H03286 and 20H05403 for S.N., 20H04878 to T.F., and 20H05684 for J.K.) and the Takeda Science Foundation (for S.N.).
Conflict of inte rest statement. The authors declare no conflict of interest.
Contributor Information
Satoshi Naramoto, Department of Biological Sciences, Faculty of Science, Hokkaido University, Sapporo, Hokkaido 060-0810, Japan.
Yuki Hata, Graduate School of Life Sciences, Tohoku University, Aoba-ku, Sendai 980-8577, Japan.
Tomomichi Fujita, Department of Biological Sciences, Faculty of Science, Hokkaido University, Sapporo, Hokkaido 060-0810, Japan.
Junko Kyozuka, Graduate School of Life Sciences, Tohoku University, Aoba-ku, Sendai 980-8577, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Satoshi Naramoto (satoshi.naramoto@sci.hokudai.ac.jp).
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp A, Pearman C, Rao AN (1968) The effects of some growth substances and inhibitors on the development of marchantia gemmae. Phytomorphology 18: 84–94 [Google Scholar]
- Aoyama T, Hiwatashi Y, Shigyo M, Kofuji R, Kubo M, Ito M, Hasebe M (2012) AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development 139: 3120–3129 [DOI] [PubMed] [Google Scholar]
- Augustine RC, Vidali L, Kleinman KP, Bezanilla M (2008) Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J 54: 863–875 [DOI] [PubMed] [Google Scholar]
- Bascom C Jr, Burkart GM, Mallett DR, O’Sullivan JE, Tomaszewski AJ, Walsh K, Bezanilla M (2019) Systematic survey of the function of ROP regulators and effectors during tip growth in the moss Physcomitrella patens. J Exp Bot 70: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom CS Jr, Hepler PK, Bezanilla M (2018) Interplay between ions, the cytoskeleton, and cell wall properties during tip growth. Plant Physiol 176: 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T (2015) PIN proteins and the evolution of plant development. Trends Plant Sci 20: 498–507 [DOI] [PubMed] [Google Scholar]
- Bennett T, Brockington SF, Rothfels C, Graham SW, Stevenson D, Kutchan T, Rolf M, Thomas P, Wong GK, Leyser O, et al. (2014a) Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol Biol Evol 31: 2042–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett TA, Liu MM, Aoyama T, Bierfreund NM, Braun M, Coudert Y, Dennis RJ, O’Connor D, Wang XY, White CD, et al. (2014b) Plasma membrane-targeted PIN proteins drive shoot development in a moss. Curr Biol 24: 2776–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennici A (2008) Origin and early evolution of land plants: problems and considerations. Commun Integr Biol 1: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Henderson DC, Kropf DL (2003) Asymmetric division in fucoid zygotes is positioned by telophase nuclei. Plant Cell 15: 854–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL (2016) A brief history of marchantia from dreece to genomics. Plant Cell Physiol 57: 210–229 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Araki T, Kohchi T (2016) Marchantia: past, present and future. Plant Cell Physiol 57: 205–209 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304 e215 [DOI] [PubMed] [Google Scholar]
- Burkart GM, Baskin TI, Bezanilla M (2015) A family of ROP proteins that suppresses actin dynamics, and is essential for polarized growth and cell adhesion. J Cell Sci 128: 2553–2564 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Zachgo S (2016) The evolution of cell division: from streptophyte algae to land plants. Trends Plant Sci 21: 872–883 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Holtmannspotter M, Borchers A, O’Donoghue MT, Zachgo S (2016) Microtubule dynamics of the centrosome-like polar organizers from the basal land plant Marchantia polymorpha. New Phytol 209: 999–1013 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Dols J, Kopischke S, Pena EJ, Andrade-Navarro MA, Heinlein M, Szymanski DB, Zachgo S, Doonan JH, Lloyd CW (2015) Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain. J Cell Sci 128: 2033–2046 [DOI] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y, et al. (2019) Genomes of subaerial zygnematophyceae provide insights into land plant evolution. Cell 179: 1057–1067 e1014 [DOI] [PubMed] [Google Scholar]
- Cheng X, Mwaura BW, Chang Stauffer SR, Bezanilla M (2020) A fully functional ROP fluorescent fusion protein reveals roles for this GTPase in subcellular and tissue-level patterning. Plant Cell 32: 3436–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke TJ, Poli D, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49: 319–338 [PubMed] [Google Scholar]
- Coudert Y, Palubicki W, Ljung K, Novak O, Leyser O, Harrison CJ (2015) Three ancient hormonal cues co-ordinate shoot branching in a moss. Elife 4: e06808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keijzer J, Kieft H, Ketelaar T, Goshima G, Janson ME (2017) Shortening of microtubule overlap regions defines membrane delivery sites during plant cytokinesis. Curr Biol 27: 514–520 [DOI] [PubMed] [Google Scholar]
- De Smet I, Voss U, Lau S, Wilson M, Shao N, Timme RE, Swarup R, Kerr I, Hodgman C, Bock R, et al. (2011) Unraveling the evolution of auxin signaling. Plant Physiol 155: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa F, Foster PG, Donoghue PCJ, Schneider H, Cox CJ (2019) Nuclear protein phylogenies support the monophyly of the three bryophyte groups (Bryophyta Schimp). New Phytol 222: 565–575 [DOI] [PubMed] [Google Scholar]
- Doonan J, Cove D, Lloyd C (1988) Microtubules and microfilaments in tip growth:evidence that microtubules impose polarity on protonemal growth in Physcomitrella patens. J Cell Sci 89: 533–540 [Google Scholar]
- Doonan J, Cove D, Corke F, Lloyd C (1987) Pre-prohase band of microtubules, absent from tip-growing moss filaments, arise in leafy shoots during transition to intercalary growth. Cell Motil Cytoskeleton 7: 138–153 [Google Scholar]
- Edwards D, Morris JL, Richardson JB, Kenrick P (2014) Cryptospores and cryptophytes reveal hidden diversity in early land floras. New Phytol 202: 50–78 [DOI] [PubMed] [Google Scholar]
- Eklund DM, Ishizaki K, Flores-Sandoval E, Kikuchi S, Takebayashi Y, Tsukamoto S, Hirakawa Y, Nonomura M, Kato H, Kouno M, et al. (2015) Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27: 1650–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPase in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Facette MR, Park Y, Sutimantanapi D, Luo A, Cartwright HN, Yang B, Bennett EJ, Sylvester AW, Smith LG (2015) The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat Plants 1:14024 [DOI] [PubMed] [Google Scholar]
- Falz AL, Müller-Schussele SJ (2019) Physcomitrella as a model system for plant cell biology and organelle-organelle communication. Curr Opin Plant Biol 52: 7–13 [DOI] [PubMed] [Google Scholar]
- Feiguelman G, Fu Y, Yalovsky S (2018) ROP GTPases structure-function and signaling pathways. Plant Physiol 176: 57–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting (1936a) Untersuchungen über die Induktion der Dorsiventralität bei den keimenden Brutkörpern von Marchantia und Lunularia. I. Die Induktoren und ihre Wirkungen. Jahrbüch Wiss Bot 82: 333–376 [Google Scholar]
- Fitting (1936b) Untersuchungen über die Induktion der Dorsiventralität bei den Marehantieen brutkörpern. II. Die Schwerkraft als Induktor der Dorsiventralität. Jahrbüh Wiss Bot 82: 696–740 [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL (2015) A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLoS Genet 11: e1005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangedakis E, Shimamura M, Villarreal JC, Li FW, Tomaselli M, Waller M, Sakakibara K, Renzaglia KS, Szovenyi P (2021a) The hornworts: morphology, evolution and development. New Phytol 229: 735–754 doi:org/10.1111/nph.16784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangedakis E, Waller M, Nishiyama T, Tsukaya H, Xu X, Yue Y, Tjahjadi M, Gunadi A, Van Eck J, Li FW, et al. (2021b) An Agrobacterium-mediated stable transformation technique for the hornwort model Anthoceros agrestis. New Phytol doi:org/10.1111/nph.17524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Sakaguchi H, Hiwatashi YSJW, Ito M, Deguchi H, Sato T, Hasebe M (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10: 176–186 [DOI] [PubMed] [Google Scholar]
- Furuichi T, Matsuura K (2016) Kinetic analysis on the motility of liverwort sperms using a microscopic computer-assisted sperm analyzing system. Environ Control Biol 54: 45–49 [Google Scholar]
- Gaal DJ, Dufresne SJ, Maravolo NC (1982) Transport of C-14-labeled indoleacetic-acid in the hepatic Marchantia poly-morpha. Bryogist 85: 410–418 [Google Scholar]
- Gao C, Liu X, De Storme N, Jensen KH, Xu Q, Yang J, Liu X, Chen S, Martens HJ, Schulz A, et al. (2020) Directionality of plasmodesmata-mediated transport in Arabidopsis leaves supports auxin channeling. Curr Biol 30: 1970–1977 e1974 [DOI] [PubMed] [Google Scholar]
- Halbsguth VM, Kohlenbach HW (1953) Einige versuche uber die wirkung von heteroauxin auf die symmetrieentwicklung der brutkörperkeimilinge von marachantia Polymorpha L. Planta 42: 349–366 [Google Scholar]
- Han X, Hyun TK, Zhang M, Kumar R, Koh EJ, Kang BH, Lucas WJ, Kim JY (2014) Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev Cell 28: 132–146 [DOI] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BJ, Harrison CJ, Hetherington AM, Williams TA (2020) Phylogenomic evidence for the monophyly of bryophytes and the reductive evolution of stomata. Curr Biol 30: 2001–2012 e2002 [DOI] [PubMed] [Google Scholar]
- Harrison CJ (2017) Auxin transport in the evolution of branching forms. New Phytol 215: 545–551 [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Morris JL (2018) The origin and early evolution of vascular plant shoots and leaves. Philos Trans R Soc Lond B Biol Sci 373: 20160496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Roeder AH, Meyerowitz EM, Langdale JA (2009) Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr Biol 19: 461–471 [DOI] [PubMed] [Google Scholar]
- Hata Y, Kyozuka J (2021) Fundamental mechanisms of the stem cell regulation in land plants: lesson from shoot apical cells in bryophytes. Plant Mol Biol doi: 10.1007/s11103-021-01126-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Uchida N, Yamaguchi YL, Tabata R, Ishida S, Ishizaki K, Nishihama R, Kohchi T, Sawa S, Bowman JL (2019) Control of proliferation in the haploid meristem by CLE peptide signaling in Marchantia polymorpha. PLoS Genet 15: e1007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Fujimoto T, Ishida S, Uchida N, Sawa S, Kiyosue T, Ishizaki K, Nishihama R, Kohchi T, Bowman JL (2020) Induction of Multichotomous Branching by CLAVATA Peptide in Marchantia polymorpha. Curr Biol 30: 3833–3840 e3834 [DOI] [PubMed] [Google Scholar]
- Hiwatashi T, Goh H, Yasui Y, Koh LQ, Takami H, Kajikawa M, Kirita H, Kanazawa T, Minamino N, Togawa T, et al. (2019) The RopGEF KARAPPO is essential for the initiation of vegetative reproduction in Marchantia polymorpha. Curr Biol 29: 3525–3531 e3527 [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y, Sato Y, Doonan JH (2014) Kinesins have a dual function in organizing microtubules during both tip growth and cytokinesis in Physcomitrella patens. Plant Cell 26: 1256–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CM, Hotta T, Guo F, Roberson RW, Lee YR, Liu B (2011a) Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell 23: 2909–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CM, Hotta T, Kong Z, Zeng CJ, Sun J, Lee YR, Liu B (2011b) Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR (2012) Augmin’s role in microtubule generation in plants. Plant Cell 24: 1304–1304 [Google Scholar]
- Honkanen S, Jones VAS, Morieri G, Champion C, Hetherington AJ, Kelly S, Proust H, Saint-Marcoux D, Prescott H, Dolan L (2016) The mechanism forming the cell surface of tip-growing rooting cells is conserved among land plants. Curr Biol 26: 3238–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Reski R (2017) Microscopy of Physcomitrella patens sperm cells. Plant Methods 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG (2011) ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 23: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T (2012) Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J Plant Res 125: 643–651 [DOI] [PubMed] [Google Scholar]
- Johansen W, Ako AE, Demko V, Perroud PF, Rensing SA, Mekhlif AK, Olsen OA (2016) The DEK1 calpain linker functions in three-dimensional body patterning in Physcomitrella patens. Plant Physiol 172: 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones VAS, Dolan L (2012) The evolution of root hairs and rhizoids. Ann Bot 110: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson E, Yamada M, Vale RD, Goshima G (2015) Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat Plants 1: 15087 [PMC][26322239] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G (2005) Cytokinesis in higher plants. Annu Rev Plant Biol 56: 281–299 [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Era A, Minamino N, Shikano Y, Fujimoto M, Uemura T, Nishihama R, Yamato KT, Ishizaki K, Nishiyama T, et al. (2016) SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol 57: 307–324 [DOI] [PubMed] [Google Scholar]
- Karahara I, Suda J, Tahara H, Yokota E, Shimmen T, Misaki K, Yonemura S, Staehelin LA, Mineyuki Y (2009) The preprophase band is a localized center of clathrin-mediated endocytosis in late prophase cells of the onion cotyledon epidermis. Plant J 57: 819–831 [DOI] [PubMed] [Google Scholar]
- Kato H, Yasui Y, Ishizaki K (2020) Gemma cup and gemma development in Marchantia polymorpha. New Phytol 228: 459–465 [DOI] [PubMed] [Google Scholar]
- Kato H, Ishizaki K, Kouno M, Shirakawa M, Bowman JL, Nishihama R, Kohchi T (2015) Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet 11: e1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Kouno M, Takeda M, Suzuki H, Ishizaki K, Nishihama R, Kohchi T (2017) The roles of the sole activator-type auxin response factor in pattern formation of Marchantia polymorpha. Plant Cell Physiol 58: 1642–1651 [DOI] [PubMed] [Google Scholar]
- Kaul KN, Mitra GC, Tripathi BK (1962) Responses of marchantia in aseptic culture to well-known auxins and antiauxins. Ann Bot 26: 447–466 [Google Scholar]
- Kenrick P, Crane PR (1991) Water-conducting cells in early fossil land plants: implications for the early evolution of tracheophytes. Bot Gazette 152: 335–356 [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kirbis A, Waller M, Ricca M, Bont Z, Neubauer A, Goffinet B, Szovenyi P (2020) Transcriptional landscapes of divergent sporophyte development in two mosses, physcomitrium (physcomitrella) patens and Funaria hygrometrica. Front Plant Sci 11: 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Hasebe M (2014) Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol 17: 13–21 [DOI] [PubMed] [Google Scholar]
- Kohchi T, Yamato KT, Ishizaki K, Yamaoka S, Nishihama R (2021) Development and molecular genetics of Marchantia polymorpha. Ann Rev Plant Biol 72: 677–702 [DOI] [PubMed] [Google Scholar]
- Kosetsu K, de Keijzer J, Janson ME, Goshima G (2013) MICROTUBULE-ASSOCIATED PROTEIN65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in Physcomitrella patens. Plant Cell 25: 4479–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosetsu K, Murata T, Yamada M, Nishina M, Boruc J, Hasebe M, Van Damme D, Goshima G (2017) Cytoplasmic MTOCs control spindle orientation for asymmetric cell division in plants. Proc Natl Acad Sci USA 114: E8847–E8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecek P, Skupa P, Libus J, Naramoto S, Tejos R, Friml J, Zazimalova E (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SY, Edzuka T, Goshima G, Yamada M (2020) Kinesin-13 and kinesin-8 function during cell growth and division in the moss physcomitrella patens. Plant Cell 32: 683–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Nishiyama T, Waller M, Frangedakis E, Keller J, Li Z, Fernandez-Pozo N, Barker MS, Bennett T, Blazquez MA, et al. (2020) Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat Plants 6: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Deng Z, Kamisugi Y, Chen Z, Wang J, Han X, Wei Y, He H, Terzaghi W, Cove DJ, et al. (2021) A minus-end directed kinesin motor directs gravitropism in Physcomitrella patens. Nat Commun 12: 4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS (2012) The origin of the sporophyte shoot in land plants: a bryological perspective. Ann Bot 110: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stockle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Müller S (2014) The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos P, Müller S (2019) Division plane establishment and cytokinesis. Ann Rev Plant Biol 70: 239–267 [DOI] [PubMed] [Google Scholar]
- Lloyd C, Chan J (2006) Not so divided: the common basis of plant and animal cell division. Nat Rev Mol Cell Biol 7: 147–152 [DOI] [PubMed] [Google Scholar]
- Manck R, Ishitsuka Y, Herrero S, Takeshita N, Nienhaus GU, Fischer R (2015) Genetic evidence for a microtubule-capture mechanism during polarised growth of Aspergillus nidulans. J Cell Sci 128: 3569–3582 [DOI] [PubMed] [Google Scholar]
- Maravolo NC (1976) Polarity and Localization of Auxin movement in the Hepatic, Marchantia polymorpha. Am J Bot 63: 526–531 [Google Scholar]
- Menand B, Calder G, Dolan L (2007) Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J Exp Bot 58: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Meyberg R, Perroud PF, Haas FB, Schneider L, Heimerl T, Renzaglia KS, Rensing SA (2020) Characterisation of evolutionarily conserved key players affecting eukaryotic flagellar motility and fertility using a moss model. New Phytol 227: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Nishina M, Goshima G (2015) RNAi screening identifies the armadillo repeat-containing kinesins responsible for microtubule-dependent nuclear positioning in Physcomitrella patens. Plant Cell Physiol 56: 737–749 [DOI] [PubMed] [Google Scholar]
- Minamino N, Kanazawa T, Era A, Ebine K, Nakano A, Ueda T (2018) RAB GTPases in the basal land plant Marchantia polymorpha. Plant Cell Physiol 59: 845–856 [DOI] [PubMed] [Google Scholar]
- Moody LA (2020) Three-dimensional growth: a developmental innovation that facilitated plant terrestrialization. J Plant Res 133: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody LA, Kelly S, Rabbinowitsch E, Langdale JA (2018) Genetic regulation of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr Biol 28: 473–478 e475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody LA, Kelly S, Clayton R, Weeks Z, Emms DM, Langdale JA (2021) NO GAMETOPHORES 2 is a novel regulator of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr Biol 31: 555–563 e554 [DOI] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Nortes T, Wilson-Sanchez D, Candela H, Micol JL (2014) Symmetry, asymmetry, and the cell cycle in plants: known knowns and some known unknowns. J Exp Bot 65: 2645–2655 [DOI] [PubMed] [Google Scholar]
- Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S (2017) Polar transport in plants mediated by membrane transporters: focus on mechanisms of polar auxin transport. Curr Opin Plant Biol 40: 8–14 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Jones VAS, Trozzi N, Sato M, Toyooka K, Shimamura M, Ishida S, Nishitani K, Ishizaki K, Nishihama R, et al. (2019) A conserved mechanism mediates the convergent evolution of plant shoot lateral organs. PLoS Biol 17: e3000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ, Wayne R, Benitez M, Newman SA (2019) Polarity, planes of cell division, and the evolution of plant multicellularity. Protoplasma 256: 585–599 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin-I T, Seki M, NIshide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al. (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100: 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D, et al. (2018) The chara genome: secondary complexity and implications for plant terrestrialization. Cell 174: 448–464 e424 [DOI] [PubMed] [Google Scholar]
- Orr RG, Cheng X, Vidali L, Bezanilla M (2020) Orchestrating cell morphology from the inside out - using polarized cell expansion in plants as a model. Curr Opin Cell Biol 62: 46–53 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Michard E, Simon AA, Damineli DSC, Hernandez-Coronado M, Becker JD, Feijo JA (2017) GLUTAMATE RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature 549: 91–95 [DOI] [PubMed] [Google Scholar]
- Otani K, Ishizaki K, Nishihama R, Takatani S, Kohchi T, Takahashi T, Motose H (2018) An evolutionarily conserved NIMA-related kinase directs rhizoid tip growth in the basal land plant Marchantia polymorpha. Development 145: dev154617. [DOI] [PubMed] [Google Scholar]
- Peer WA, Blakeslee JJ, Yang H, Murphy AS (2011) Seven things we think we know about auxin transport. Mol Plant 4: 487–504 [DOI] [PubMed] [Google Scholar]
- Pereira SG, Sousa AL, Nabais C, Paixão T, Holmes AJ, Schorb M, Goshima G, Tranfield EM, Becker JD, Bettencourt-Dias M (2020) The 3D architecture and molecular fundations of de novo centriole assembly via bicentrioles. bioRxiv 10.1101/2020.12.21.423647 [DOI] [PubMed]
- Perroud PF, Demko V, Johansen W, Wilson RC, Olsen OA, Quatrano RS (2014) Defective Kernel 1 (DEK1) is required for three-dimensional growth in Physcomitrella patens. New Phytol 203: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud PF, Meyberg R, Demko V, Quatrano RS, Olsen OA, Rensing SA (2020) DEK1 displays a strong subcellular polarity during Physcomitrella patens 3D growth. New Phytol 226: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Philipson WR (1990) The significance of apical meristems in the phylogeny of land plants. Plant Syst Evol 173: 17–38 [Google Scholar]
- Pillitteri LJ, Guo X, Dong J (2016) Asymmetric cell division in plants: mechanisms of symmetry breaking and cell fate determination. Cell Mol Life Sci 73: 4213–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli D, Jacobs M, Cooke TJ (2003) Auxin regulation of axial growth in bryophyte sporophytes: its potential significance for the evolution of early land plants. Am J Bot 90: 1405–1415 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Bezanilla M (2010) Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137: 3535–3543 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA 106: 17431–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. (2018) The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol 28: 733–745 e732 [DOI] [PubMed] [Google Scholar]
- Rasmussen CG, Wright AJ, Müller S (2013) The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J 75: 258–269 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M (2020) The moss physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell 32: 1361–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia KS, Villarreal Aguilar JC, Garbary DJ (2018) Morphology supports -the setaphyte hypothesis: mosses plus liverworts form a natural group. Bryophyte Divers Evol 40: 11–17 [Google Scholar]
- Rose S, Bopp M (1983) Uptake and polar transport of indoleacetic acid in moss rhizoids. Physiol Plant 58: 57–61 [Google Scholar]
- Rounds CM, Bezanilla M (2013) Growth mechanisms in tip-growing plant cells. Annu Rev Plant Biol 64: 243–265 [DOI] [PubMed] [Google Scholar]
- Santoro A, Vlachou T, Carminati M, Pelicci PG, Mapelli M (2016) Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep 17: 1700–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E, Belcram K, Uyttewaal M, Duroc Y, Goussot M, Legland D, Laruelle E, de Tauzia-Moreau ML, Pastugia M, Bouchez D (2017) The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356: 186–189 [DOI] [PubMed] [Google Scholar]
- Shao W, Dong J (2016) Polarity in plant asymmetric cell division: division orientation and cell fate differentiation. Dev Biol 419: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AJ, Szovenyi P, Shaw B (2011) Bryophyte diversity and evolution: windows into the early evolution of land plants. Am J Bot 98: 352–369 [DOI] [PubMed] [Google Scholar]
- Shimamura M (2016) Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol 57: 230–256 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokan R, Medvecka E, Viaene T, Vosolsobe S, Zwiewka M, Müller K, Skupa P, Karady M, Zhang Y, Janacek DP, et al. (2019) PIN-driven auxin transport emerged early in streptophyte evolution. Nat Plants 5: 1114–1119 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Assaad F, Baluska F, Bezanilla M, Buschmann H, Drakakaki G, Hauser MT, Janson M, Mineyuki Y, Moore I, et al. (2017) Plant cytokinesis: terminology for structures and processes. Trends Cell Biol 27: 885–894 [DOI] [PubMed] [Google Scholar]
- Spinner L, Pastuglia M, Belcram K, Pegoraro M, Goussot M, Bouchez D, Schaefer DG (2010) The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Takemiya A, Kong SG, Higa T, Komatsu A, Shimazaki K, Kohchi T, Wada M (2016) RPT2/NCH1 subfamily of NPH3-like proteins is essential for the chloroplast accumulation response in land plants. Proc Natl Acad Sci USA 113: 10424–10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Basu S, Brady SR, Luciano RL, Muday GK (2004) Interactions between auxin transport and the actin cytoskeleton in developmental polarity of Fucus distichus embryos in response to light and gravity. Plant Physiol 135: 266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunchu B, Cabernard C (2020) Principles and mechanisms of asymmetric cell division. Development 147: dev167650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocheteau P, Le Roux I (2009) Asymmetric cell divisions and asymmetric cell fates. Annu Rev Cell Dev Biol 25: 671–699 [DOI] [PubMed] [Google Scholar]
- Takatani S, Verger S, Okamoto T, Takahashi T, Hamant O, Motose H (2020) Microtubule response to tensile stress is curbed by NEK6 to buffer growth variation in the Arabidopsis hypocotyl. Curr Biol 30: 1491–1503 e1492 [DOI] [PubMed] [Google Scholar]
- Takatani S, Ozawa S, Yagi N, Hotta T, Hashimoto T, Takahashi Y, Takahashi T, Motose H (2017) Directional cell expansion requires NIMA-related kinase 6 (NEK6)-mediated cortical microtubule destabilization. Sci Rep 7: 7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Staehelin LA, Mineyuki Y (2017) Actin-microtubule interaction in plants. Cytoskeleton - Structure, Dynamics, Function and Disease, IntechOpen, London. [Google Scholar]
- Tang H, de Keijzer J, Overdijk EJR, Sweep E, Steentjes M, Vermeer JEM, Janson ME, Ketelaar T (2019) Exocyst subunit Sec6 is positioned by microtubule overlaps in the moss phragmoplast prior to cell plate membrane arrival. J Cell Sci 132: jcs222430. [DOI] [PubMed] [Google Scholar]
- van Gisbergen PA, Li M, Wu SZ, Bezanilla M (2012) Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol 198: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron E, Vernoux T, Coudert Y (2021) Phyllotaxis from a single apical cell. Trends Plant Sci 26: 124–131 [DOI] [PubMed] [Google Scholar]
- Viaene T, Delwiche CF, Rensing SA, Friml J (2013) Origin and evolution of PIN auxin transporters in the green lineage. Trends Plant Sci 18: 5–10 [DOI] [PubMed] [Google Scholar]
- Viaene T, Landberg K, Thelander M, Medvecka E, Pederson E, Feraru E, Cooper ED, Karimi M, Delwiche CF, Ljung K, et al. (2014) Directional auxin transport mechanisms in early diverging land plants. Curr Biol 24: 2786–2791 [DOI] [PubMed] [Google Scholar]
- Vidali L, Bezanilla M (2012) Physcomitrella patens: a model for tip cell growth and differentiation. Curr Opin Plant Biol 15: 625–631 [DOI] [PubMed] [Google Scholar]
- Vidali L, Augustine RC, Kleinman KP, Bezanilla M (2007) Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tuzel E, Bezanilla M (2010) Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PA, Guérin C, Franco P, Li M, Burkart GM, Augustin RC, Blanchoin L, Bezanilla M (2009) Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KL, Müller S, Moss D, Ehrhardt DW, Smith LG (2007) Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr Biol 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitewoods CD, Cammarata J, Nemec Venza Z, Sang S, Crook AD, Aoyama T, Wang XY, Waller M, Kamisugi Y, Cuming AC, et al. (2018) CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr Biol 28: 2365–2376 e2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SZ, Bezanilla M (2014) Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3:e03498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SZ, Bezanilla M (2018) Actin and microtubule cross talk mediates persistent polarized growth. J Cell Biol 217: 3531–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, et al. (2014) Contribution of NAC transcription factors to plant adaptation to land. Science 343: 1505–1508 [DOI] [PubMed] [Google Scholar]
- Xu X, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CSM, Meier I (2008) RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci USA 105: 18637–18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Tanaka-Takiguchi Y, Hayashi M, Nishina M, Goshima G (2017) Multiple kinesin-14 family members drive microtubule minus end-directed transport in plant cells. J Cell Biol 216: 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Goshima G (2018) The KCH kinesin drives nuclear transport and cytoskeletal coalescence to promote tip cell growth in Physcomitrella patens. Plant Cell 30: 1496–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Goshima G (2020) Rho of plants GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during Physcomitrella patens branching. Curr Biol 30: 2860–2868 e2863 [DOI] [PubMed] [Google Scholar]
- Yogev S, Shen K (2017) Establishing neuronal polarity with environmental and intrinsic mechanisms. Neuron 96: 638–650 [DOI] [PubMed] [Google Scholar]
- Yoshida S, van der Schuren A, van Dop M, van Galen L, Saiga S, Adibi M, Moller B, Ten Hove CA, Marhavy P, Smith R, et al. (2019) A SOSEKI-based coordinate system interprets global polarity cues in Arabidopsis. Nat Plants 5: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariadis M, Quader H, Galatis B, Apostolakos P (2001) Endoplasmic reticulum preprophase band in dividing root-tip cells of Pinus brutia. Planta 213: 824–827 [DOI] [PubMed] [Google Scholar]
- Zhang J, Fu XX, Li RQ, Zhao X, Liu Y, Li MH, Zwaenepoel A, Ma H, Goffinet B, Guan YL, et al. (2020) The hornwort genome and early land plant evolution. Nat Plants 6: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]