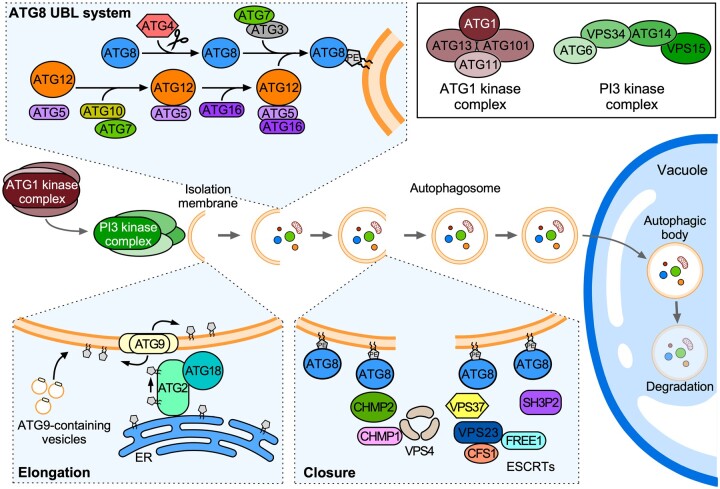
Abstract
Endomembrane trafficking is essential for all eukaryotic cells. The best-characterized membrane trafficking organelles include the endoplasmic reticulum (ER), Golgi apparatus, early and recycling endosomes, multivesicular body, or late endosome, lysosome/vacuole, and plasma membrane. Although historically plants have given rise to cell biology, our understanding of membrane trafficking has mainly been shaped by the much more studied mammalian and yeast models. Whereas organelles and major protein families that regulate endomembrane trafficking are largely conserved across all eukaryotes, exciting variations are emerging from advances in plant cell biology research. In this review, we summarize the current state of knowledge on plant endomembrane trafficking, with a focus on four distinct trafficking pathways: ER-to-Golgi transport, endocytosis, trans-Golgi network-to-vacuole transport, and autophagy. We acknowledge the conservation and commonalities in the trafficking machinery across species, with emphasis on diversity and plant-specific features. Understanding the function of organelles and the trafficking machinery currently nonexistent in well-known model organisms will provide great opportunities to acquire new insights into the fundamental cellular process of membrane trafficking.
We summarize the current knowledge on plant endomembrane trafficking with a focus on ER-to-Golgi transport, endocytosis, trans-Golgi network-to-vacuole transport and autophagy.
Introduction
Eukaryotic cells contain a multitude of highly dynamic and interconnected membrane compartments comprising the membrane trafficking system. Despite their astonishing complexity, the distribution and dynamics of these membrane compartments are highly organized and regulated. The membrane trafficking system underpins the export and uptake of extracellular material, remodeling and signaling at the cell interface, intracellular targeting, and maintenance of internal compartmentalization. Although the organization of the membrane trafficking system, including the endoplasmic reticulum (ER), the Golgi apparatus, endosomes, and lytic compartments, is conserved among eukaryotes, vascular plants display distinct organizational characteristics that may necessitate adaptive specializations. The plant endomembrane system is considerably more complex than that of unicellular yeasts and is characterized by expanded protein families that increase the specificity of diverse vesicular trafficking processes as well as by plant-specific features that have evolved as adaptation strategies to support at best the plant lifestyle.
Excellent reviews recently covering the plant ER structure (Kriechbaumer and Brandizzi, 2020), the Golgi apparatus (Robinson, 2020), and the trans-Golgi network (TGN) (Renna and Brandizzi, 2020) document the important differences between plant and mammalian organelles. Briefly, the plant Golgi apparatus is polydisperse, with individual motile Golgi stacks remaining throughout mitosis, while it is immobile in mammalian cells, disassembled during mitosis, and often appears as a perinuclear ribbon of interconnected Golgi stacks (Saraste and Prydz, 2019). Plant cells also lack an intermediate compartment between the ER and the Golgi apparatus. Nevertheless, entry into the Golgi complex in plants has been proposed to occur through the so-called Golgi Entry Core Compartment, which may be the structure that initiates the first cis-Golgi cisterna, although further characterization is still needed (Ito et al., 2018; Ito and Boutté, 2020). Another compartment with unique features in plant cells is the TGN, a multitasking and versatile organelle that is very distinct from its mammalian counterpart (Rosquete et al., 2018; Renna and Brandizzi, 2020). The TGN is a sorting hub for post-Golgi trafficking pathways that deliver cargo to the plasma membrane (PM) and the vacuole or to the cell plate during cytokinesis. In contrast to its mammalian counterpart, the plant TGN is also involved in the trafficking/recycling of endosomal material and functions as an early endosome (EE), being at the interface of the secretory and endocytic pathways (Viotti et al., 2010). Morphologically, the mammalian TGN is defined as a specific compartment of the trans-most cisterna of the Golgi apparatus, whereas the plant TGN can be described as a cluster of tubules and vesicles (mainly clathrin-coated vesicles [CCVs]) and occurs as the Golgi-associated (GA) TGN, located on the trans-side of the Golgi apparatus, and the Golgi-independent (GI) TGN, which are physically detached from the Golgi stacks and, thus, are independent organelles (Kang et al., 2011; Uemura et al., 2014; Rosquete et al., 2018; Renna and Brandizzi, 2020). In plants, the Golgi apparatus and the TGN play a specialized role in the biosynthesis and sorting of cell wall components, including biosynthetic enzymes, structural proteins, and the matrix polysaccharides hemicellulose and pectin (for a review, see Sinclair et al., 2018).
Membrane trafficking is an active area of plant research and remarkable progress has been achieved mainly by means of the multicellular plant Arabidopsis (Arabidopsis thaliana) as a model for cell biological, biochemical, and functional studies. In plant cells, the membrane trafficking system comprises several major trafficking pathways, including the biosynthetic secretory pathway, the most extensively studied clathrin-mediated endocytosis (CME) pathway, and the vacuolar transport pathway. The biosynthetic secretory pathway transports newly biosynthesized proteins from the ER through the Golgi to the PM and/or the extracellular space. Conversely, receptors and transporters are endocytosed from the PM to the TGN/EEs and are either recycled or targeted to the vacuole for degradation through prevacuolar compartments (PVCs), also named multivesicular bodies (MVBs) or late endosomes (LEs). The vacuolar transport pathway drives the newly synthesized protein to the vacuole (Figure 1).
Figure 1.
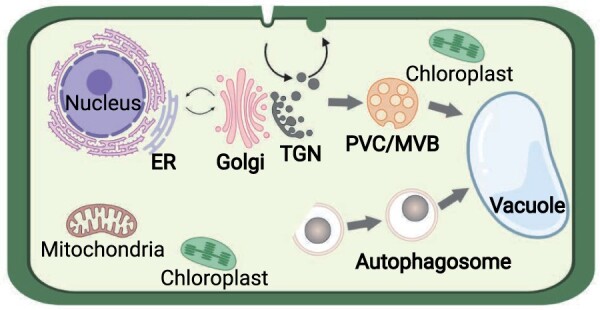
Endomembrane trafficking in plants. Major protein trafficking pathways in the endomembrane system include the biosynthetic routes for secreted proteins and those destined to the vacuole, involving vesicle trafficking and compartment maturation between the ER, Golgi apparatus, TGN, and the prevacuolar compartment (PVC)/MVB. The endocytic pathway removes PM proteins and delivers them to the TGN, where they are sorted to be either recycled back to the PM or intended for degradation in the vacuole. The endocytic and biosynthetic routes converge at the TGN, a main hub for protein sorting. Autophagosomes are formed de novo in the cytoplasm and enclose cellular components, including organelles. Fusion with the vacuole results in autophagosome cargo degradation.
This review summarizes the current knowledge on plant membrane trafficking with emphasis on four distinct membrane trafficking pathways (Figure 1). We first describe the ER–Golgi pathway and then focus on the endocytic and the TGN-to-vacuole transport pathways. We will not cover exocytosis or recycling because these topics were recently reviewed (Rodriguez-Furlan et al., 2019a). Although autophagy is an intracellular catabolic pathway important for maintaining cellular homeostasis, membrane trafficking events are involved in the biogenesis and further maturation of the autophagosomes. Therefore, we also discuss autophagy as a separate trafficking pathway in plants.
ER–Golgi trafficking
Trafficking between the ER and the Golgi apparatus involves Coat Protein I (COPI)- and COPII-coated vesicles, although the nature of COPII carriers (vesicles or tubules) is still controversial. Indeed, the existence of standard COPII vesicles was recently challenged in mammalian cells (Phuyal and Farhan, 2021; Shomron et al., 2021; Weigel et al., 2021) and is also a controversial issue in plant cells (for a detailed discussion, see Robinson et al., 2015). Export from the ER occurs at specialized regions called ER exit/export sites (ERES), which are the sites where COPII proteins are recruited to mediate transport from the ER to the Golgi apparatus (Brandizzi, 2018 and references therein). Using a dynamic three-dimensional (3D) visualization of the compartments of the early secretory pathway, the ERES of mammalian cells have been shown to consist of an interwoven, tubular network extending from the ER, with the boundary between ER and ERES defined by a COPII collar (Shomron et al., 2021; Weigel et al., 2021). COPII and COPI have been suggested to act sequentially at ERES for ER-to-Golgi transport, that is, COPII concentrates at the ERES neck and regulates cargo sorting into ERES (rather than coating transport carriers), whereas COPI acts more distally and appears to be involved in the last step of anterograde ER-to-Golgi transport (Shomron et al., 2021; Weigel et al., 2021). In plants, COPII-containing ERES are continuously associated with motile Golgi stacks, suggesting that they form a secretory unit (daSilva et al., 2004; Brandizzi, 2018). ERES are specifically labeled by SECRETORY 16A (SEC16A), which functions as a scaffold to facilitate COPII assembly and largely overlaps with COPII subunits (Takagi et al., 2013). Many ERES are tightly linked with the cis-face of Golgi stacks, as revealed by colocalization with fluorescent Golgi markers, but non-Golgi-associated ERES have been found as well (Hanton et al., 2009; Brandizzi, 2018). However, COPII budding profiles observed in plants at the ultrastructural level are solitary events and not strictly situated underneath a Golgi stack (Robinson, 2020). Recently, free ERES with forming COPII vesicles have been proposed to move around the ER membrane for cargo loading, whereafter COPII-bound ERES are captured by a Golgi stack in an ER cavity, from which COPII vesicles bud from the ERES and fuse with the cis-Golgi cisterna, with the final release of ERES from the Golgi stacks for recycling. This dynamic ERES cycling may contribute to an efficient ER–Golgi trafficking in plants (Takagi et al., 2020).
The Golgi-to-ER transport is mediated by COPI vesicles and the ER domain in which these retrograde carriers fuse has been determined by the localization of fluorescently tagged homologs of mammalian COPI-tethering factors (Lerich et al., 2012; see Robinson, 2020, for a detailed discussion). The ER import/arrival sites (ERIS/ERAS) appear to associate with the ERES at the ER membrane immediately underneath the cis-Golgi cisterna, thus generating bidirectional transport portals at the ER–Golgi interface (Roy Chowdhury et al., 2020), which is in agreement with the secretory unit concept (Brandizzi, 2018).
COPII proteins and ER export
COPII coat proteins that assemble on the ER have been shown to mediate export from the ER or the ERES. The COPII coat consists of five proteins, SECRETION-ASSOCIATED, RAS-RELATED1 (SAR1), SEC23, SEC24, SEC13, and SEC31 (for reviews, see Béthune and Wieland, 2018; Brandizzi, 2018). COPII coat assembly is initiated by the activation of the small GTPase SAR1 through the ER-localized transmembrane protein SEC12, a SAR1-specific guanine nucleotide exchange factor that can recruit SAR1. Upon GDP/GTP exchange, SAR1 integrates into the ER membrane and initiates membrane deformation, followed by the sequential recruitment of two COPII coat heterocomplexes (SEC23/SEC24 and SEC13/SEC31). Activated SAR1 first interacts with SEC23 to recruit the SEC23/SEC24 complex from the cytosol. This recruiting step then leads to cargo selection within COPII vesicles by a direct interaction of SEC24 with signals exposed on the cytosolic side of cargo proteins. Therefore, the SEC23/SEC24 complex acts as a COPII adaptor complex. Subsequently, through an interaction between SEC23 and a proline-rich domain in SEC31, a SEC13/SEC31 hetero-tetramer is also recruited from the cytosol. The SEC13/31 complex creates a cage-like lattice with a cuboctahedral shape forming COPII-coated vesicles, but also assembling on tubular structures that can accommodate giant cargoes, such as procollagen in mammalian cells (Raote and Malhotra, 2021). Recent yeast data suggest that the inner SEC23/SEC24 coat alone can already provide a membrane-remodeling function with organizational input from the outer coat (Hutchings et al., 2021). COPII uncoating is driven by GTP hydrolysis onto SAR1 with the aid of the GTPase-activating activity of SEC23 (Gomez-Navarro and Miller, 2016; Béthune and Wieland, 2018).
ER exit can be highly selective and is limited to soluble and membrane-associated secretory proteins that have reached properly folded and assembled conformations (Barlowe and Helenius, 2016). Despite their high concentrations, ER-resident proteins (including chaperones) are marginally secreted, as a consequence of either selective protein retention in the ER to prevent uptake into COPII vesicles, or efficient signal-mediated retrieval of escaped ER residents via COPI vesicles (Barlowe and Helenius, 2016). Cargo enrichment into COPII vesicles is mediated by a direct interaction between sorting signals (ER export signals) in cargo proteins and the COPII subunit SEC24, which contains multiple independent cargo-binding sites (see below). Protein sorting is also facilitated by cargo receptors that interact with both cargo (on their luminal side) and coat proteins (on their cytosolic side; Gomez-Navarro and Miller, 2016; Béthune and Wieland, 2018). In addition to selective signal-mediated ER export, proteins can also enter COPII vesicles by default, as part of fluids and membranes (bulk flow; Barlowe and Helenius, 2016). Recently, the inclusion of p24 proteins and other abundant cargoes in COPII vesicles has been proposed to generate macromolecular crowding that prevents the capture of ER-resident proteins (Gomez-Navarro et al., 2020). Cargo abundance directly influences the number and size of the ERES, which correlate with the amount of COPII cargo proteins both in mammals (Venditti et al., 2014) and plants (Hanton et al., 2009; Brandizzi, 2018).
One-third of all eukaryotic proteins enter the ER either to be exported via COPII carriers to the different compartments of the secretory pathway or else to be secreted. How can the COPII system accommodate a variety of cargoes that differ in size, structure, and topology (Phuyal and Farhan, 2021)? Many cargoes that follow the COPII pathway are not just passive passengers, but play an active role in controlling the stability, dynamics, and geometry of the COPII coat. Indeed, larger cargoes bound to SEC24 may increase the diameter of the COPII cage, whereas abundant and asymmetric cargoes containing a bulk luminal portion, such as GPI-anchored proteins and p24 family proteins, may influence the physical properties of ER membranes, thus affecting vesicle budding (Venditti et al., 2014). In this respect, the biogenesis of specialized COPII vesicles that contain GPI-anchored proteins (GPI-APs) in yeast (Saccharomyces cerevisiae) and mammals involves the recruitment of a specialized COPII machinery, including specific SEC24 isoforms (Lopez et al., 2019; Kinoshita, 2020). Another key question is how the COPII system is regulated to adapt either to the different needs for cargo secretion taking place in many specialized secretory cells, or to the various stress conditions (Lopez et al., 2019; Wang et al., 2020a). The five core COPII components are largely conserved at the protein sequence level across eukaryotes (Chung et al., 2016). The yeast genome encodes a single paralog for each COPII subunit, with the exception of SEC24, and most are essential for ER export. In contrast, mammals and plants contain multiple COPII subunit paralogs (due to gene duplication). In mammals, COPII paralogs are not completely functionally redundant and this may also be the case in plants (Chung et al., 2016). In the model species Arabidopsis, five putative SAR1, two SEC13, two SEC31, seven SEC23, and three SEC24 paralogs have been identified through protein homology searches, largely outnumbering the ones detected in other eukaryotes. As a consequence, plants have been hypothesized to use different COPII subunit paralogs to fine-tune ER export in response to various stress and developmental conditions, perhaps through different populations of COPII carriers (Chung et al., 2016; Brandizzi, 2018). Interestingly, a unique pair, consisting of the COPII Arabidopsis paralogs SAR1A and SEC23A, was found to be essential for ER export of the ER-resident stress sensor/transducer basic leucine zipper 28 (bZIP28), suggesting a specific role for these paralogs in the formation of COPII vesicles containing specific cargoes under stress conditions (Zeng et al., 2015). SEC24 is a particularly interesting COPII subunit because of its role in cargo selection. In this respect, mammalian COPII vesicles generated in vitro from semi-intact cells by the addition of different recombinant SEC24 isoformshad different protein compositions (Adolf et al., 2019). Yeast SEC24 has at least four different cargo-binding sites that can recognize a wide spectrum of cargoes. Two of these sites (the B and C sites) are functionally conserved in the mammalian paralogs (Gomez-Navarro and Miller, 2016 and references therein). Interestingly, the B site is involved in p24 protein binding via a C-terminal diaromatic (ΦC) motif, but also in the binding of LxxLE and DxE motifs that function as ER export signals (Figure 2). No comparable information is yet available in plants. In Arabidopsis, three SEC24 isoforms are present, SEC24A, SEC24B, and SEC24C. Interestingly, all residues of the B site in human SEC24A are conserved in all Arabidopsis SEC24 isoforms. Although a sec24a knockout leads to lethality during the early stages of plant growth, a missense point mutation affecting the B site in SEC24A resulted in ER morphology defects and disruption of the ER–Golgi integrity. Notably, these phenotypes were not rescued by overexpression of either SEC24B or SEC24C, suggesting that they are functionally distinct from SEC24A (Faso et al., 2009; Nakano et al., 2009). Nevertheless, it is still unknown whether different SEC24 paralogs may recruit different cargoes in plants. Therefore, additional studies are needed to test for functional redundancy or diversity between plant COPII paralogs.had
Figure 2.
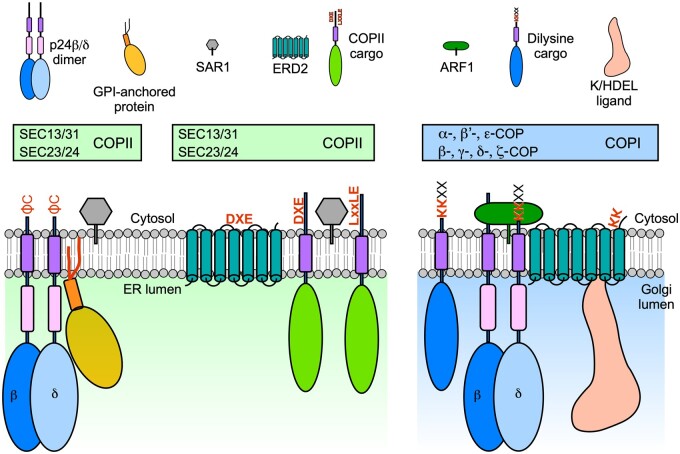
Protein sorting at the ER-Golgi interface is mediated by Coat Protein I (COPI) and COPII proteins. ER export is mediated by COPII proteins that recognize sorting signals in cargo proteins via the SEC24 subunit. ER export signals include DXE and LxxLE motifs or a C-terminal diaromatic (ΦC) motif in p24 proteins. ER export of proteins with a bulky luminal portion [p24 proteins and glycosylphosphatidylinositol (GPI)-anchored proteins] may involve specific COPII vesicles (probably including specific COPII isoforms), but this remains to be demonstrated in plants. p24 proteins are required for ER export of GPI-anchored proteins in mammals, yeast, and plants. Sorting of the K/HDEL receptor ERD2 within COPII vesicles may involve a DXE motif exposed by a conformational change occurring at the neutral pH of the ER. Retrograde Golgi-to-ER transport is mediated by COPI proteins that recognize sorting signals in cargo proteins (including a dilysine KK motif in p24 proteins and other cargoes). Sorting of ERD2 within COPI vesicles may implicate a KK motif exposed by a conformational change occurring at the acidic pH of the Golgi apparatus.
COPI vesicles for Golgi–ER trafficking and intra-Golgi transport
COPI vesicles mediate intra-Golgi trafficking and retrograde transport from the cis-Golgi back to the ER. Very recent data also suggest that COPI proteins play an active role in the last step of anterograde ER–Golgi transport in mammalian cells (Shomron et al., 2021; Weigel et al., 2021). Directionality of intra-Golgi vesicles is still a matter of debate and will not be discussed here (for a discussion on models of intra-Golgi trafficking, please see Ito et al., 2014; Mironov and Beznoussenko, 2019). The COPI coat is composed of the GTPase ADP-ribosylation factor 1 (ARF1) and a cytosolic complex (coatomer) made of seven subunits (α-, β-, β′-, γ-, δ-, ε-, and ζ-COP), which is recruited en bloc from the cytosol onto Golgi membranes (Arakel and Schwappach, 2018; Béthune and Wieland, 2018). From biochemical studies, two coatomer subcomplexes can be distinguished: the α-/β′-/ε-COP subcomplex, possibly working as a “cage-like” complex (with α- and β′-COP trimers, showing a strong structural similarity to clathrin triskelions), and the β-/γ-/δ-/ζ-COP subcomplex, possibly functioning as an “adaptor-like” complex (similar to clathrin adaptors). However, the situation may be a bit more complex, because COPI-sorting signals can be recognized by subunits from both subcomplexes: whereas classical dilysine (KKxx or KxKxx) motifs appear to be recognized by α- and β′-COP subunits, FFxxBB(x)n motifs in the p24 protein family are recognized by the γ-COP subunit, and (ϕ/ψ/R)RxR motifs by the δ-COP subunit (Arakel and Schwappach, 2018; Béthune and Wieland, 2018). COPI-coated vesicles are first formed via the recruitment of cytosolic (GDP-bound) ARF1 to the Golgi membranes by interacting with dimers of p24 family proteins. The GTP/GDP exchange allows insertion of ARF1 into the membrane and its dissociation from p24 proteins. Coatomer can then interact with ARF1-GTP (two ARF1 dimers per coatomer) and with the cytosolic tail of p24 proteins and other cargo proteins (containing COPI-sorting signals; Figure 2). Coatomer polymerization that, in contrast to clathrin and COPII, does not form an outer polyhedral layer, leads to the formation of COPI vesicles, whereas GTP hydrolysis in ARF1 results in COPI uncoating (Arakel and Schwappach, 2018; Béthune and Wieland, 2018).
Different populations of COPI vesicles may exist to mediate either intra-Golgi trafficking or Golgi-to-ER transport. In mammalian cells, these putative vesicle populations were proposed to be formed by the different isoforms of the γ-COP and ζ-COP subunits (all other subunits have a single isoform; Popoff et al., 2011). However, a proteomic analysis of COPI vesicles formed in vitro with different recombinant COPI subunit isoforms added to the reactions did not reveal significant differences in their protein composition (Adolf et al., 2019). Arabidopsis (and most plant species) possess additional COPI subunit isoforms. Except for the δ-COP and γ-COP subunits, all other COPI subunits, including the α- and β′-COP subunits, with a potential role in cargo binding, have two to three isoforms, a striking difference from yeast, in which all COPI subunits are encoded by single genes (Gao et al., 2014). Interestingly, morphologically different COPI vesicles were detected in plants (Donohoe et al., 2007). COPIa vesicles, with a lightly stained lumen and a thicker coat, are mainly found at the ER–cis Golgi interface and are probably involved in retrograde trafficking from the cis-Golgi to the ER. In contrast, COPIb vesicles, with a dense lumen and a thinner coat, bud from medial and trans- cisternae, suggesting that they are involved in intra-Golgi trafficking. Whether in plants different COPI subunit isoforms are functionally redundant or are part of different populations of COPI vesicles containing different cargo proteins is still unknown (Gao et al., 2014; Gimeno-Ferrer et al., 2017; Sánchez-Simarro et al., 2020). Mutants in plant COPI subunits exhibit an altered Golgi morphology. A knockout in the α2-COP gene is associated with fewer cisternae per Golgi stack and many abnormal vesicle clusters around the Golgi remnants (Gimeno-Ferrer et al., 2017), as also observed upon silencing of ε-COP in Arabidopsis (Woo et al., 2015) and β′-COP in tobacco (Nicotiana benthamiana) (Ahn et al., 2015). However, depletion of β-COP increased the length of the Golgi stack, in some cases seemingly the consequence of lateral fusion between adjacent Golgi stacks (Sánchez-Simarro et al., 2020). Although the plant Golgi apparatus does not form ribbon-like structures, it is possible that Golgi stacks might perhaps undergo homotypic fusion events upon COPI depletion, as observed in tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells treated with brefeldin A (Ritzenthaler et al., 2002). Loss of COPI function has also been shown to affect plant growth and tolerance to salt stress, in particular to chloride ions, possibly due to mislocalization or reduced activity of chloride channels/transporters (Sánchez-Simarro et al., 2020, 2021). Interestingly, interference with the COPI function produces a strong upregulation of SEC31A, but not of genes encoding other COPII subunit isoforms, possibly as part of the unfolded protein response activated under these conditions (Gimeno-Ferrer et al., 2017; Pastor-Cantizano et al., 2018; Sánchez-Simarro et al., 2020).
Cargo receptors for ER–Golgi trafficking
In this section, we will focus on the well-characterized K/HDEL receptor ER RETENTION DEFECTIVE2 (ERD2) and on p24 family proteins. Very little is known about other putative cargo receptors for ER-to-Golgi trafficking in plants, except for the possible role of CORNICHON in the ER export of transmembrane proteins (including PM cargoes and the Golgi-localized rice [Oryza sativa] sodium transporter OsHKT1-3; Rosas-Santiago et al., 2015, 2017) and the potential role of RETENTION IN THE ER1 (RER1) protein family in ER localization of transmembrane proteins, including SEC12p (Sato et al., 1999).
p24 family proteins
p24 proteins are part of a family of approximately 24-kDa, type-I transmembrane proteins, which are major components of COPI- and COPII-coated vesicles and cycle between the ER and the Golgi apparatus, possibly acting as cargo receptors (Langhans et al., 2008; Montesinos et al., 2012; Pastor-Cantizano et al., 2016). Their luminal part contains the Golgi Dynamics (GOLD) domain, proposed to be involved in cargo recognition, and a coiled-coil domain required for oligomerization of p24 proteins. They have a single transmembrane domain and a short cytoplasmic domain that contains COPI- and/or COPII-binding signals and, hence, is important for their trafficking and localization (Montesinos et al., 2014; Pastor-Cantizano et al., 2016, 2017). Based on sequence similarity, p24 proteins can be grouped into four subfamilies: α, β, γ, and δ. However, in contrast to yeast and mammals, plants contain only p24 proteins from the β and δ subfamilies (Pastor-Cantizano et al., 2016). As described above, p24 proteins play an established role in COPI vesicle formation. In addition, p24 proteins might function as cargo receptors during ER–Golgi transport and putative p24 cargoes have recently been identified in plants. In particular, Arabidopsis p24 proteins of the δ subfamily (containing dilysine motifs) are required for COPI-dependent transport of the K/HDEL receptor ERD2 and bound K/HDEL ligands (luminal ER-resident proteins) from the Golgi apparatus back to the ER (see below; Montesinos et al., 2014; Pastor-Cantizano et al., 2017, 2018). p24 proteins have been recently shown to be involved in the transport of GPI-APs to the cell surface (Bernat-Silvestre et al., 2020). In yeast, sorting of GPI-APs into specific ERES is lipid-based and independent of p24 proteins, although the latter are required to sort GPI-APs within COPII vesicles (Lopez et al., 2019). For the interaction between GPI-APs and p24 proteins, the glycan core of the GPI anchor needs to be completely remodeled (Lopez et al., 2019). In mammals, glycan core remodeling is also necessary for the interaction with p24 proteins, but, in contrast to yeast, it is required to concentrate GPI-APs at specific ERES (Kinoshita, 2020). In plants, p24 proteins interact with GPI-APs via their coiled-coil domain to facilitate their ER export and transport to the cell surface, a prerequisite that does not apply to other PM protein types, including transmembrane proteins (Bernat-Silvestre et al., 2020). However, whether the ER export of GPI-APs in plants necessitates specific COPII subunit paralogs, as in mammals and yeast, is still unknown. Very recently, in yeast, sorting of GPI-APs into specific ERES has been found to depend on the ceramide chain length of the GPI anchor (Rodriguez-Gallardo et al., 2020). A possible role for lipids in sorting of GPI-APs at specific ERES has not yet been established in plants.
The K/HDEL receptor ERD2
ER-resident luminal proteins are characterized by a K/HDEL motif at their C terminus. These proteins are recognized by the multispanning transmembrane receptor ERD2 in yeast and plants or the KDEL receptor (KDELR1–KDELR3) in mammals (Newstead and Barr, 2020). Binding of KDEL ligands in vitro to KDELR has been shown to depend on pH, with maximal and very low binding efficiency at acidic and neutral pH values, respectively (Wilson et al., 1993; Scheel and Pelham, 1996). Based on these properties, KDEL ligands are generally accepted to bind to KDELR/ERD2 in the slightly acidic pH of the Golgi apparatus and receptor–ligand complexes to be recycled back to the ER in COPI vesicles. Once at the ER, at a neutral pH, KDEL ligands dissociate from KDELR/ERD2 and the receptors are presumably recycled to the Golgi in COPII vesicles (Newstead and Barr, 2020).
Whether ERD2 also cycles between the ER and Golgi apparatus in plants is still a matter of controversy (Montesinos et al., 2014; Pastor-Cantizano et al., 2017, 2018; Silva-Alvim et al., 2018; for a detailed discussion, see Robinson and Aniento, 2020). Recently, the crystal structures of chicken (Gallus gallus domesticus) KDELR in its ligand-free ER state and ligand-bound Golgi state have been resolved. This elegant study provided important clues to understand the molecular basis of the pH-dependent Golgi-to-ER retrieval of KDEL ligands (ER luminal proteins) by KDELR in mammalian cells (Bräuer et al., 2019). The KDEL signal is recognized by KDELR through electrostatic interactions, with the positive side chain of the lysine residue accommodated in a negative KDELR pocket and the three carboxyl groups of aspartate, glutamate, and the C-terminus of leucine accommodated in positively charged pockets (Bräuer et al., 2019; Newstead and Barr, 2020). KDEL binding causes a conformational change in KDELR, involving movement of transmembrane domain 1 (TM1) and TM6, exposing a cluster of lysine residues in the cytosolic C terminus of KDELR, reminiscent of the KKXX or KXKXX dilysine motifs; as a result, a COPI-binding site is formed to facilitate sorting of the receptor within COPI vesicles for Golgi-to-ER retrograde transport. At the neutral pH inside the ER, deprotonation of histidine 12 triggers TM1 and TM6 to move back to their original position and to release the KDEL ligand. This conformational change now buries the lysine residues and reveals several acidic residues that may partly form a diacidic (DXE) COPII-binding ER export motif, hence, facilitating an efficient ER export of the ligand-free receptor back to the Golgi apparatus (Bräuer et al., 2019; Newstead and Barr, 2020). This mutually exclusive mechanism may be the reason for the selective recruitment of different coat complexes by KDELR (COPI and COPII by the ligand-bound and ligand-free KDELR, respectively) and the bidirectional transport of KDELR at the ER–Golgi interface (Bräuer et al., 2019; Figure 2). Moreover, the KDELR-induced conformational change on ligand binding that allows an efficient packaging of receptor–ligand complexes within COPI vesicles is consistent with a proteomics study of mammalian COPI vesicles that clearly revealed that KDELRs and p24 proteins are among the top hits of their core proteome (Adolf et al., 2019). In contrast, efficient packaging of free KDELR in COPII vesicles may account for its predominant Golgi localization at steady state. Strikingly, most of the residues involved in binding of KDEL and COPI or COPII are highly conserved in Arabidopsis ERD2 proteins, hinting at a similar mechanism for the plant proteins, although this suggestion needs to be demonstrated. In any case, data from both mammalian and plant cells imply the following model for the function of p24 proteins and ERD2 in the Golgi-to-ER transport of ERD2-bound K/HDEL ligands. The p24 family of proteins interacts with both ARF1 and ERD2 in the acidic pH of the Golgi apparatus (Majoul et al., 1998, 2001; Montesinos et al., 2014; Pastor-Cantizano et al., 2017), whereas ERD2 can also oligomerize and interact with ARF1 (Majoul et al., 1998, 2001; Xu and Liu, 2012). Whether the ERD2-induced conformational change upon ligand binding leads to ERD2 oligomerization or favors its interaction with p24 proteins still needs to be tested. These interactions may facilitate the recruitment of the COPI coatomer complex to Golgi membranes, through its interaction with ARF1 and with dilysine motifs in the cytosolic tail of p24 proteins or other membrane cargoes, including ERD2 bound to K/HDEL ligands. Once in the ER, the conformational change of ERD2 allows the release of KDEL ligands and perhaps the dissociation of ERD2 from p24 proteins (Robinson and Aniento, 2020).
Clathrin-mediated endocytosis in plants: same, but different
Endocytosis is defined as the uptake of extracellular molecules together with membrane-bound proteins into the cell, through the formation of a vesicle from the PM. Although endocytosis had initially been described as a simple transport process, this notion quickly changed after the discovery that endocytosis is linked to almost all cellular signaling aspects. In all eukaryotes, endocytosis provides nutrients and/or other types of molecules to the cell and, simultaneously, regulates the PM composition to modulate signaling. Depending on the machinery involved, endocytosis is classified as clathrin-mediated and clathrin-independent. In mammals, CME is initiated from the PM and recruits the cargo and coat machinery into foci, designated clathrin-coated pits (CCPs) that eventually mature and scission off to form CCVs. The CCVs are uncoated in seconds to form vesicles that fuse with the EE, where the cargo is further sorted for either recycling back to the PM, or degradation to the lysosome (Kaksonen and Roux, 2018; Mettlen et al., 2018). In plants, CME is the best studied and the major mode of endocytosis (Paez Valencia et al., 2016; Reynolds et al., 2018), but studies have lagged behind other models due to the erroneous perception that turgor pressure would prevent PM invagination (Cram, 1980) and because of lack of cargoes. However, CCVs have been identified at the PM, first in protoplasts (Hillmer et al., 1986) and later in walled plant cells using electron microscopy (EM) (Hawes and Martin, 1986; Coleman et al., 1987). These discoveries and the saturable uptake of a labeled elicitor fraction into soybean (Glycine max) cells (Horn et al., 1989) further strengthened the view that endocytosis also takes place in plants, whereafter CME in plants was expected to operate in an evolutionarily conserved manner as in yeast and mammals. Furthermore, advanced imaging methods suitable for plant cells had to be developed and adapted, because visualization of endocytosis in plant cells was challenging due to the rigid cell wall, the high turgor pressure, and the exclusive study in the context of multicellular organisms (Dragwidge and Van Damme, 2020; Johnson et al., 2020). Now, nearly 35 years after the first report of CCVs in plants, our understanding of some of the basic CME mechanisms has rapidly progressed (Figure 3; Paez Valencia et al., 2016; Qi et al., 2018; Reynolds et al., 2018; Schwihla and Korbei, 2020). Whereas some evolutionary conservation of endocytic molecular components between plants and other model systems exists, plant CME possesses many unique features that are absent in the currently well-characterized CME models (More et al., 2020). Below, we summarize recent knowledge of CME in plants (Figure 3) with emphasis on the similarities with yeast and mammals and the uniqueness of plant CME.
Figure 3.
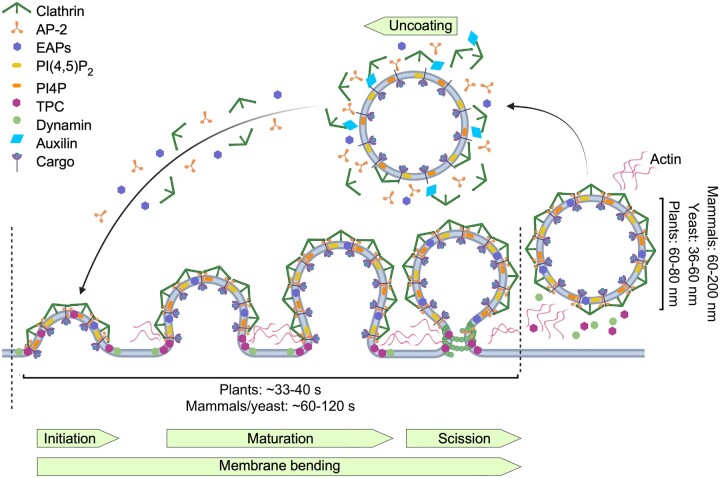
Clathrin-mediated endocytosis (CME) in plants. CME is a multistep process involving initiation and stabilization of clathrin-coated pits (CCPs), maturation and membrane bending, followed by dynamin-catalyzed scission and uncoating. Clathrin-coated vesicles (CCVs) are wider in mammalian cells than in yeast and plants, but plant CCVs form faster than in yeast, where CCVs are pinched off from a narrow tubular invagination. In plants, internalized CCVs display a delayed uncoating, whereas actin is not essential for CME. Adaptor and accessory proteins involved in plant CME are shown. AP-2, AP-2; TPC, TPLATE complex; EAPs, endocytic accessory proteins; PIP, phosphatidylinositol phosphate.
Clathrin and CCVs in plants
Clathrin subunits are recruited from the cytoplasm to the nascent budding sites in the form of hexameric, three-legged triskelions composed of three self-polymerizing clathrin heavy chains (CHCs) and three light chains (CLCs). Clathrin homologs exist in plants, with two CHCs (CHC1 and CHC2) and three CLCs (CLC1, CLC2, and CLC3) in Arabidopsis (Kitakura et al., 2011; Wang et al., 2013). Unlike yeast and similar to mammals, clathrin is essential for plants because chc1 chc2 double knockout mutants in Arabidopsis are not viable (Kitakura et al., 2011). Although the presence of clathrin-type lattices and CCPs at the PM of plant cells was visualized (Dragwidge and Van Damme, 2020), the successful capture of ongoing CME events by EM was rare, and live-cell fluorescence imaging methods have only been adopted of late to look directly at the dynamics of CME events in plants (Johnson et al., 2020). Recently, high-resolution images of CCPs have been obtained by means of scanning electron microscopy (SEM) on metal replicas of unroofed Arabidopsis root protoplast cells, allowing the direct examination of clathrin-coated structures budding from the PM (Narasimhan et al., 2020). Such an SEM analysis revealed that, as in mammals, plant CME follows the constant curvature model of CCV formation, in which the curvature is generated by continuous clathrin polymerization (Bucher et al., 2018; Scott et al., 2018) and produces vesicles predominantly of a hexagonal basket type. However, plant CCVs are formed much faster, namely in 33–42 s compared to 60–120 s in yeast and mammals; plant CCVs are also larger, with an average diameter of 60–80 nm compared to 60–200 nm (Narasimhan et al., 2020) and 36–60 nm in mammals and yeast, respectively (Kaksonen and Roux, 2018; Figure 3). Therefore, plant-specific mechanisms of membrane bending must exist to allow overcoming a turgor pressure higher than that found in yeast, while producing larger CCVs.
Endocytic adaptor protein complexes
In addition to clathrin, CCVs contain nonclathrin components collectively named endocytic accessory proteins (EAPs). These EAPs ensure efficient coated-pit assembly, cargo recruitment, and prompt coat disassembly after budding. The EAPs are divided into heterotetrameric and monomeric adaptors (Kirchhausen et al., 2014).
Adaptor protein complex-2
The most abundant and major EAP in mammals is the heterotetrameric Assembly Polypeptide/Adaptor Protein (AP) complex-2 (AP-2), which binds phosphatidylinositol-4,5-bis-phosphate [PI(4,5)P2] at the PM, cargo, as well as clathrin, and other EAPs. AP-2 consists of two large (α, [AP2A] and β2, [AP2B]), one medium (µ2, [AP2M]) and one small (σ2, [AP2S]) subunits. AP2M contains an N-terminal longin-related domain followed by a C-terminal μ homology domain (µHD; Traub and Bonifacino, 2013). Crystallography revealed that mammalian AP-2 adopts closed (inactive) and open (active) conformations (Jackson et al., 2010). The active state is promoted by PI(4,5)P2 binding at the PM and by phosphorylation (Traub and Bonifacino, 2013; Kelly et al., 2014). Recent cryo-EM studies confirmed that AP-2 initially contacts the membrane in a closed conformation via PI(4,5)P2-binding sites on its α and β2 subunits. Binding of PI(4,5)P2 to the C-terminal domain of the µ2 subunit (µ2-C) shifts the equilibrium to an open state, in which the cargo-binding sites become exposed (Kovtun et al., 2020). The Fer/CIP4 Homology domain only (FCHo) proteins (Syp1p in yeast) can also promote the open conformation of AP-2 (Hollopeter et al., 2014; Umasankar et al, 2014). Open AP-2 can then bind the cargo, further stabilizing the adaptor complex at the membrane. The β2 appendage of AP-2 interacts with CHC and provides a platform for binding other EAPs to regulate membrane curvature, assembly, and/or disassembly (Kovtun et al., 2020). In mammals, AP-2 is essential for CME, because AP-2 deficiency in mice leads to embryonic lethality (Zizioli et al., 1999; Mitsunari et al., 2005) and AP-2 knockdowns result in an approximately 12-fold reduction in CCP formation in HeLaM cells (Motley et al., 2003). In contrast, AP-2 is partially required, but not essential, for CME in yeast (Weinberg and Drubin, 2012), whereas in the nematode Caenorhabditis elegans AP-2 does not need all four of its subunits and can function as a hemicomplex (Gu et al., 2013). AP-2 is conserved in plants (Bashline et al., 2013; Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013) and Arabidopsis plants with loss of function in a single AP-2 subunit are viable, but display defects in vegetative and floral organ development (Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013), effector-triggered immunity (Hatsugai et al., 2016), growth under nutrient-limiting conditions (Yoshinari et al., 2019), and phytohormone responses (Di Rubbo et al., 2013; Kim et al., 2013). Altogether, similarly to nematodes (Gu et al., 2013), AP-2 hemicomplexes might be functional in plants as well.
In Arabidopsis, AP-2 is implicated in the CME of several PM-localized proteins, including the PIN-FORMED (PIN) auxin transporters (Fan et al., 2013; Kim et al., 2013), the brassinosteroid (BR) receptor BR INSENSITIVE1 (BRI1; Di Rubbo et al., 2013; Liu et al., 2020a), the boron transporter BOR1 (Yoshinari et al., 2019), the boric acid channel NODULIN26-LIKE INTRINSIC PROTEIN5;1 (NIP5;1; Wang et al., 2017), and cellulose synthase (Bashline et al., 2015). However, the mechanisms that govern the recruitment of these proteins into CCVs are unclear. In mammalian cells, AP-2 recognizes endocytic cargoes by binding to two highly conserved motifs (Traub and Bonifacino, 2013). The first motif is the tyrosine motif, YXXΦ, where Y is tyrosine, X is any amino acid, and Φ is a bulky hydrophobic residue such as leucine (L), isoleucine (I), methionine (M), valine (V), or phenylalanine (F), which is recognized and directly bound by the µHD of AP2M (Ohno et al., 1995). The second motif type is the acidic dileucine motif [DE]XXXL[LI], (aspartic [D] and glutamic [E] acids), which is recognized by the small σ2 subunit (Kelly et al., 2008). YXXΦ motifs are found in the cytoplasmic parts of several plant PM proteins (Geldner and Robatzek, 2008; Arora and Van Damme, 2021). Various observations pointed out that the functional tyrosine motifs in plants that might also be recognized by other adaptor complexes (Zuo et al., 2000; Ron and Avni 2004; Takano et al., 2010; Kleine-Vehn et al., 2011; Yoshinari et al., 2012, 2019; Sancho-Andrés et al., 2016). Importantly, the direct binding of cargo to Arabidopsis AP2M in an YXXΦ motif-dependent manner needed for CME has only been demonstrated for the Agrobacterium (Agrobacterium tumefaciens)-derived virulence protein VirE2 (Li and Pan, 2017) and for the BRI1 receptor (Liu et al., 2020a). The internalization of VirE2 and Agrobacterium infection are facilitated by this interaction (Li and Pan, 2017) and mutations in the Y898XXΦ motif of BRI1 significantly reduce, but do not abolish, its internalization (Liu et al., 2020a). Although these examples hint at the possibility that AP-2 binds cargoes through their YXXΦ motifs, the underlying mechanisms of cargo recognition by AP-2 and cargo recruitment into CCVs in plants remain largely elusive.
The TPLATE complex
In addition to canonical AP-2, plants possess a second endocytic eight-core-subunit adaptor complex, designated the TPLATE complex (TPC; Van Damme et al., 2006, 2011; Gadeyne et al., 2014). TPC comprises the TPLATE protein, originally discovered as a cell plate-localized protein-containing adaptin-like features, and seven previously unknown proteins named TPLATE-ASSOCIATED SH3 DOMAIN CONTAINING PROTEIN (TASH3), LOLITA, TWD40-1, TWD40-2, TPLATE COMPLEX MUNISCIN-LIKE (TML), A. thaliana EH DOMAIN CONTAINING PROTEIN1 (AtEH1)/Pan1, and AtEH2/Pan1, of which the first six proteins can be traced back to the last eukaryotic common ancestor, whereas AtEH1/Pan1 and AtEH2/Pan1 are plant-specific constituents with similarity to the yeast Pan1p and each containing two EPIDERMAL GROWTH FACTOR RECEPTOR PATHWAY SUBSTRATE15 (EPS15) HOMOLOGY (EH) domains (Gadeyne et al., 2014; Hirst et al., 2014). TPC is an evolutionarily ancient protein complex that has only been experimentally characterized in plants and in the slime mold Dictyostelium discoideum (Gadeyne et al., 2014; Hirst et al., 2014). Notably, the TPC of Dictyostelium (designated TSET) was identified as a hexameric complex lacking the AtEH/Pan1 homologs (Hirst et al., 2014). Although Amoebozoa contains proteins with similarity to AtEH/Pan1, they resemble more closely the yeast Eps15-like protein Ede1p (EH Domains and Endocytosis 1), which is not found in plants (Gadeyne et al., 2014; Wang et al., 2019). A unique feature of the TPC is the presence of a µHD in the TML subunit that is absent from the TSET complex, but is present in the muniscin (FCHo/Syp1) family from mammals and yeast, but not in plants (Reider et al., 2009; Henne et al., 2010). TSET also lacks the TPLATE subunit extension with an additional anchor domain. In contrast to the TASH3 TPC subunit, the corresponding TSET subunit, designated TSAUCER, does not possess the linker domain ending with a Src homology3 (SH3) domain present in TASH3. Although the two AtEH/Pan1 subunits have been hypothesized to be peripherally associated with a hexameric TPC subcomplex, possibly resembling the TSET of Dictyostelium, a colocalization analysis revealed that the TPC is recruited to the PM as an octameric complex (Wang et al., 2020b). Recently, an integrative structural approach identified the TPC architecture (Yperman et al., 2021a). The TPLATE subunit acts as a hub for TPC assembly and the AtEH/Pan1 proteins are linked to the complex via µHD of the TML subunit, similarly to the interactions between yeast and mammalian muniscins that possess an µHD, and the EH domain-containing proteins (Yperman et al., 2021a). Thus, the ancestral TPC might be speculated to have been an octamer and that the AtEH/Pan1 subunits had been lost in the Amoebozoa lineage, concomitantly with the loss of µHD. In the TPC model, the µHD of TML is exposed and can recruit membranes in planta, as do the EH domains of AtEH1/Pan1. Therefore, these domains contribute largely to the PM recruitment of the TPC (Yperman et al., 2021a, 2021b). The generated model for the TPC architecture implies many overall structural similarities between the TPC and other coatomer complexes, such as COPI and AP-2, but with striking differences in the domain positioning regarding complex orientations toward the membrane (Yperman et al., 2021a). In contrast to AP-2, TPC is essential in plants, because knockout mutations in all tested TPC subunits led to pollen lethality and the downregulation of two individual TPC subunit genes caused seedling lethality. Induced knockdowns of TPC subunit genes or conditional destabilization of TPC impairs CME (Gadeyne et al., 2014; Wang et al., 2021). TPC components interact with clathrin, AP-2, AP180-amino-terminal-homology (ANTH) domain-containing proteins, and dynamin-related proteins (DRPs). TPC has been proposed to function as a classical CME adaptor that either binds cargo directly and drives CCV formation or recruits the AP-2 in plant cells (Zhang et al., 2015a). In agreement, the TPC is implicated in the trafficking of cellulose synthase (Bashline et al., 2015; Sánchez-Rodríguez et al., 2018) and SECRETORY CARRIER MEMBRANE PROTEIN5 (Yperman et al., 2021b). However, a more recent study argued that the TPC acts in membrane bending instead (Johnson et al., 2021). A total internal reflection fluorescence microscopy study of CME in plant cells revealed that the TPC dissociates prior to clathrin after the departure of CCVs from the PM (Narasimhan et al., 2020). A more detailed follow-up analysis using structured illumination microscopy (SIM) indicated that the TPC and clathrin overlap at the beginning of the CME, but that, as the CCV increases in diameter, the TPC is excluded from the CCV coat and localizes at the rim around the CCV (Johnson et al., 2021). Moreover, a conditional TPC disruption resulted in the formation of flat clathrin lattices. In agreement, in vitro experiments showed that AtEH1/Pan1 could bend membranes (Johnson et al., 2021). Although a plausible hypothesis, it remains to be determined whether the entire TPC complex can deform membranes.
Other endocytic adaptors and scaffolds in plants
Another endocytic adaptor in the mammalian system is the monomeric assembly protein 180 (AP180), present in neurons, and its paralog found in other tissues, the clathrin assembly lymphoid myeloid (CALM) leukemia protein (also named phosphatidylinositol-binding clathrin assembly protein or PICALM; Maritzen et al., 2012). AP180 and CALM both contain an ANTH domain and can bind PI(4,5)P2, clathrin, and AP-2. Deletion of the two yeast homologs, Yap1801 and Yap1802, does not affect CME, but rather the sorting of the yeast homolog of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein, the vesicle-associated membrane protein (VAMP)/Snc1 (Burston et al., 2009). Similarly, a knockout in the single AP180/CALM gene in the fruitfly (Drosophila melanogaster) and the nematode C. elegans and a knockdown by RNA interference in HeLa cells lead to the formation of aberrant synaptic vesicles and coated structures, respectively, but without any effect on general CME (Nonet et al., 1999, Bao et al., 2005; Meyerholz et al., 2005). In mammals, AP180/CALM has been proposed to be necessary for the internalization of the SNARE protein subset consisting of the brevin-type secretory R-SNAREs, VAMP2, VAMP3, and VAMP8 into CCVs (Miller, et al., 2011; Maritzen et al., 2012).
In plants, the ANTH domain protein family is extended with 18 PICALM members in Arabidopsis (Zouhar and Sauer, 2014), but the function of only a very few is known. The first ANTH domain protein identified in Arabidopsis, AP180/PICALM6 (Barth and Holstein, 2004) binds PI(4,5)P2, AP-2, and clathrin and regulates CCV size. Another four ANTH proteins, EPSIN-LIKE CLATHRIN ADAPTOR1 (ECA1)/PICALM1a, ECA2/PICALM5a, ECA4/PICALM4a, and CLATHRIN-ASSOCIATED PROTEIN1 (CAP1)/PICALM4b are linked to CME in plants (Song et al., 2012; Gadeyne et al., 2014; Adamowski et al., 2018; Muro et al., 2018; Nguyen et al., 2018; Kaneda et al., 2019). Only recently, a function for plant PICALMs in retrieving R-SNAREs was demonstrated, as in other model systems (Fujimoto et al., 2020). Indeed, ECA1/PICALM1a and PICALM1b act as APs for CME of the longin-type R-SNARE VAMP72, interacting with the SNARE domain of VAMP72 and clathrin at the PM. The loss of PICALM1 function resulted in faulty retrieval of VAMP72, but general CME remained unaffected. Interestingly, during CME in the mammalian system, VAMP7, the homolog to plant VAMP72, is recovered from the PM, not via its interaction with PICALM, but with another clathrin adaptor, the HIV Rev-binding protein (Pryor et al., 2008), which is absent in plants. These results indicate that ANTH domain-containing proteins recycle different types of R-SNAREs in plants and mammals, hinting at a divergence in the endocytosis mechanism between these two kingdoms.
Several other proteins, including amphiphysins and endophilins, which contain N-Bin, amphiphysin and Rvs (N-BAR) domains, and SH3 domains, are implicated in mammalian CME. The N-BAR domains bind membranes upon dimerization and can induce membrane bending (Kirchhausen et al., 2014). In Arabidopsis, SH3 domain-containing proteins were identified (Mosesso et al., 2019), of which the TPC subunit TASH3 (Gadeyne et al., 2014) and the SH3 DOMAIN-CONTAINING PROTEIN1 (SH3P1), SH3P2, and SH3P3 proteins (Lam et al., 2001; Nagel et al., 2017) have been implicated in CME. All SH3P proteins have a shortened plant-specific version of the BAR domain at their N terminus and a SH3 domain at their C terminus. The SH3 domain of SH3P proteins has been reported to also bind ubiquitin and to interact with components of the endosomal sorting complex required for the transport (ESCRT) complex and autophagosome (Zhuang et al., 2013; Nagel et al., 2017). The question remains to be addressed whether plant SH3P proteins function as CME adaptors to recruit DRPs (Rosendale et al., 2019) or ubiquitinated cargoes, or whether they act in the ESCRT pathway (Mosesso et al., 2019).
Role of membrane composition in CCV formation
Many EAPs bear interaction sites for polyphosphatidyl inositol polyphosphate (PIP) head groups. The lipid membrane signature provides spatial landmarks for cargo selection and recruitment of the endocytic machinery to the correct membranes. Mammals and yeast utilize, in particular, PI(4,5)P2, the principal species found at the PM, to recruit clathrin through interactions with the AP-2 lipid-binding domains and accessory APs (Traub and Bonifacino, 2013; Kelly et al., 2014). Dynamin binds PI(4,5)P2 through its PH domain, while AP180, epsin, endophilin, and amphiphysin do so through their ANTH, ENTH, and BAR domains, respectively (Kirchhausen et al., 2014). Acute elimination of PI(4,5)P2 from the PM induces a nearly complete loss of clathrin-coated structures (Boucrot et al., 2006; Zoncu et al., 2007). Although information about EAP-to-PM recruitment via their interaction with PIPs in plants is scarce, mapping of the phosphoinositide locations in plants revealed notable differences between plants and mammals (Simon et al., 2016; Noack and Jaillais, 2017). A prime example is the massive accumulation of phosphatidylinositol 4-phosphate (PI4P) at the plant PM, instead of the prominent presence of PI4P in the mammalian Golgi/TGN compartments and, to a lesser extent, at the mammalian PM. In addition, phosphatidylinositol 3-phosphate (PI3P) labels LEs in plants and EEs in mammals. In contrast, the localization of PI(4,5)P2 at the PM and of PI(3,5)P2 in LEs/MVBs is conserved in eukaryotes (Simon et al., 2016). Several examples in plants support the view that, as in mammals, PI(4,5)P2 serves as a recruiting platform for the CME machinery. For example, PI(4,5)P2 levels directly influence the mobilization of clathrin to the PM (Zhao et al., 2010), the presence of the lipid-binding PH domains in a number of endocytic regulatory proteins, such as DRPs, and the binding of the accessory APs SH3Ps to PI(4,5)P2 (Lam et al., 2001; Zhuang et al., 2013). Inducible and tunable depletion of PI(4,5)P2 in plants using a synthetic Inducible Depletion of PI(4,5)P2 in Plants (iDePP) system confirmed that PI(4,5)P2 is needed to recruit various endocytic proteins, including AP-2 to the PM, and, hence, to regulate plant CME (Doumane et al., 2021). These results are fully in line with the previous findings that CME is partially impaired upon loss- or gain-of-function of the PI monophosphate 5-kinase PIP5K (Mei et al., 2012; Ischebeck et al., 2013), but, unlike mammalian systems, in which PI(4,5)P2 is absolutely required for CME (He et al., 2017), it can still occur upon PI(4,5)P2 depletion, albeit at a reduced rate (Doumane et al., 2021). Given that PI4P is more abundant in the plant PM (Simon et al., 2016), it is possible that it plays a more prominent role in the recruitment of EAPs during CME. Accordingly, TPC recruitment to the PM depends more on the interaction of the TML µHD with PI4P and, to a lesser degree, with PI(4,5)P2 (Yperman et al., 2021a). Interestingly, the EH domains of AtEH1/Pan1 bind strongly to phosphatidic acid (PA; Yperman et al., 2021b), which is required to maintain the electrostatic PM field (Platre et al., 2018), suggesting a role for PA in plant CME.
Role of actin in CCV formation
Actin assembly is a prerequisite for CME in yeast, whereas actin is needed only under conditions of high membrane tension or when large cargoes are taken up in mammalian cells (Galletta et al., 2010). Plant cells, like yeast, are enclosed within a cell wall and possess an even higher turgor pressure; therefore, actin has been assumed to play a key role in membrane invagination during plant endocytosis (Šamaj et al., 2004; Robatzek et al., 2006; Chen et al., 2011), but its inhibition has no visible influence on CME kinetics or efficiency (Narasimhan et al., 2020). Whereas yeast and mammals have an actomyosin network that strongly accumulates at the endocytic spot (Collins et al., 2011; Picco et al., 2015), actin and its associated proteins are not stored at plant CME loci. Moreover, plants do not possess the myosin class involved in yeast CME (Narasimhan et al., 2020). Altogether, these observations challenge the existing models for membrane invagination under turgor and simply that plants have evolved a unique mechanism of membrane bending against turgor. Clathrin polymerization energy, in addition to other membrane deforming proteins, such as the TPC, might provide enough force to overcome turgor pressure (Narasimhan et al., 2020; Johnson et al., 2021). Actin, nonetheless, is important for the “collection” of post-endocytic vesicles, for the movement of EEs, and for cargo sorting (Narasimhan et al., 2020). Interestingly, in addition to CME, the TPC subunit AtEH/Pan1 was demonstrated to function in actin-regulated autophagy and in recruiting several components of the endocytic machinery to the AtEH/Pan1-positive autophagosomes (Wang et al., 2019), suggesting that autophagy from the ER–PM contact sites depends on actin.
Scission for CCV departure and uncoating
The pinching off CCPs from the PM to form CCVs is controlled by DRPs (large GTPases) through an oligomerization cycle and GTP hydrolysis (Kaksonen and Roux, 2018; Mettlen et al., 2018). Two DRP families in plants are involved in endocytosis, of which the DRP2 family is the most similar to metazoan DRPs and is involved in CME, whereas the DRP1 family is plant-specific (Bednarek and Backues, 2010; Fujimoto and Tsutsumi, 2014). Live-cell imaging revealed that DRP2 and DRP1 arrive and depart to and from CCPs in tandem (Fujimoto et al., 2010), suggesting that these two distinct DRP families function coordinately in CME through their ability to self-associate (Fujimoto et al., 2008) and through their association with SH3 domain-containing proteins and PM lipids. In mammals, the SH3 domain-containing proteins bind dynamin and recruit it to CCPs (Rosendale et al., 2019), whereas in plants, the TASH3 subunit of the TPC interacts with members of the DRP2 and DRP1 families (Gadeyne et al., 2014). Furthermore, SH3P2 forms a complex with DRP1, affecting its accumulation at the cell plate (Ahn et al., 2017). Whether such an interaction between SH3P2 and DRP1 might be important for CME remains to be resolved. However, despite their coordinated function in CME, the DRP1 and DRP2 proteins also play clathrin-independent roles and might be regulated in distinct manners (Reynolds et al., 2018).
In both yeast and mammals, following scission, CCVs rapidly shed their clathrin coat through the coordinated action of the ATPase/chaperone heat shock cognate 70 (Hsc70) and its cofactor proteins, such as auxilin (Merrifield and Kaksonen, 2014). Two auxilin-related proteins have been identified in Arabidopsis (Lam et al., 2001; Adamowski et al., 2018), with AUXILIN-LIKE1 and AUXILIN-LIKE2 being part of a complex with clathrin and the ANTH domain protein CAP1/PICALM4b. AUXILIN-LIKE1 interacts with SH3P1 and stimulates vesicle uncoating in the presence of Hsc70 (Lam et al., 2001; Adamowski et al., 2018). Overexpression of either the AUXILIN-LIKE1 or AUXILIN-LIKE2 gene inhibits CME most probably by destabilizing clathrin recruitment to CCPs. In contrast, loss-of-function of AUXILIN-LIKE1 and AUXILIN-LIKE2 did not visibly interfere with plant development or CME, implying that uncoating may function differently in plants (Adamowski et al., 2018). Live imaging showed that the removal of the clathrin coat of plant CCVs is delayed and that coat components are only gradually discarded on their path to EEs (Narasimhan et al., 2020). This observation is in contrast to the existing models of CME in mammals and yeast and remains another unique feature of plant CME.
Post-Golgi traffic via maturation of endomembrane organelles
Post-Golgi trafficking to the vacuole involves organelle maturation and membrane fusion, implicating the TGN, plant PVCs, and the vacuole. At least four pathways have been identified for vacuolar cargo targeting in plants, some of which appear to be specific to plants, but only a few targeting domains and vacuolar sorting receptors are known (reviewed in Di Sansebastiano et al., 2017; Minamino and Ueda, 2019). The best-characterized vacuolar targeting pathway includes a conserved recruitment of cargo via the TGN, PVCs/MVBs, and the sequential action of the GTPases RAB5 and RAB7 (Figure 4; Xiang et al., 2013; Uemura and Ueda, 2014; Uemura, 2016). Rab GTPases function as molecular switches between an inactive GDP-bound and an active GTP-bound state, interacting with downstream effectors. Guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) promote the GDP-to-GTP exchange of Rab GTPases and GTP hydrolysis, respectively. The expansion of the Rab GTPase family in plants is thought to contribute to specificity in endomembrane fusion reactions (Minamino and Ueda, 2019). A second pathway that also involves the TGN and PVCs and requires RAB5, but not RAB7, is necessary for the targeting of SYNTAXIN OF PLANTS22 (SYP22; Ebine et al., 2014) and the vacuolar H+ ATPase subunit VHA-a3 (Feng et al., 2017a). A third pathway that is independent of the Golgi entails the delivery of vacuolar cargoes directly from the ER to the vacuole. Only a few proteins have been described to be targeted in this manner based on the lack of Golgi-specific glycosylation or insensitivity to brefeldin A (Park et al, 2004; Isayenkov et al., 2011). A fourth pathway depends on the AP-3, a multimeric coat adaptor involved in cargo selection between the TGN and LEs or vacuoles (Feraru et al., 2010; Zwiewka et al., 2011). The AP-3 pathway is independent of RAB5 and RAB7 and, thus far, shown to target the SNARE protein VAMP713 (Ebine et al., 2014; Uemura and Ueda, 2014), the sucrose transporter SUC4, and PROTEIN S-ACYL TRANSFERASE10 (PAT10; Wolfenstetter et al., 2012; Feng et al., 2017b). How different proteins are sorted into these various pathways and which other regulatory elements are involved are still unknown.
Figure 4.
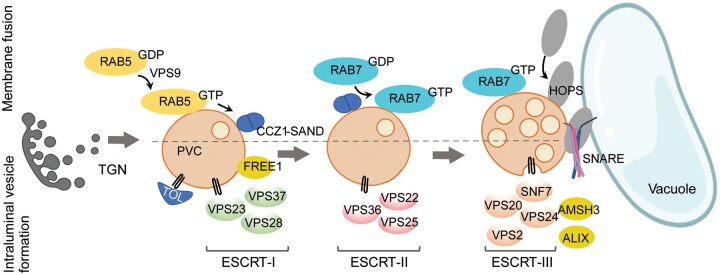
Post-Golgi trafficking to the vacuole. Trafficking to the vacuole involves the maturation of prevacuolar compartments (PVCs) in preparation for fusion. The recruitment of fusiogenic proteins starts with the activation of the GTPase RAB5 and is followed by RAB7 activation and HOPS recruitment. The SNARE complex mediates the final membrane fusion step. PVC maturation also involves the formation of intraluminal vesicles carrying ubiquitinated cargo proteins for vacuolar degradation. This process initiates with the recruitment of ubiquitinated cargo by TOL proteins and the activity of the plant-specific component FREE1, followed by the sequential action of the ESCRT-I to III complexes, resulting in the inward bending of the membrane to form internal vesicles.
The RAB5- and RAB7-dependent pathways for vacuolar cargo are conserved in all eukaryotes and involve protein transport via the TGN and PVCs/MVBs. Vacuolar cargo trafficking from the TGN to the PVC was thought to occur by vesicle budding and fusion, but recent studies have provided support for the maturation of specific TGN domains and bulk flow of cargo proteins (Scheuring et al., 2011; Cui et al., 2016). Due to the complexity of vacuolar trafficking pathways, more research is needed to determine in which organelles cargoes are recruited and sorted (Di Sansebastiano et al., 2017). PVC maturation entails the budding of intraluminal vesicles (ILVs) that carry integral membrane proteins destined for degradation in the vacuole. Targeting of these proteins to the ILVs is regulated by ubiquitination. The ESCRT machinery is responsible for the recruitment of the ubiquitinated cargo and membrane budding for ILV formation (Zhuang et al., 2015; Cui et al., 2016). Plant genomes appear to encode most subunits of the ESCRT-I, ESCRT-II, and ESCRT-III subcomplexes and the AAA ATPase VACUOLAR PROTEIN SORTING4 (VPS4), but not ESCRT-0 (Gao et al., 2017), whose role is seemingly fulfilled by TOM-Like (TOL) proteins. TOL2 and TOL6 bind polyubiquitin chains as well as VPS23, a subunit of ESCRT-I, and degrade ubiquitinated proteins (Moulinier-Anzola et al., 2020). In addition, plant-specific components have been identified, such as FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING1 (FREE1) needed for vacuole traffic and biogenesis, degradation of membrane-associated ubiquitinated proteins, and seedling viability (Gao et al., 2015; Kolb et al., 2015; Zhuang et al., 2015). FREE1, also named FYVE1, binds to the ESCRT-I and ESCRT-III subunits VPS23A and SNF7A, respectively, and has been suggested to function in ESCRT-I–III bridging (Belda-Palazon et al., 2016). Unlike other eukaryotes, where ILVs form one by one, plant cells develop a network of concatenated vesicles, in which the ESCRT-III component SNF7 remains attached to the ILV membrane (Buono et al., 2017). SNF7 typically oligomerizes into spirals at the ILV budding site and its retention at the concatenated vesicles in plants might possibly form a diffusion barrier to prevent cargo protein escape (Buono et al., 2017). Furthermore, SNF7 is stabilized at the endosomal membrane by FYVE4, a novel FYVE-domain-containing plant-specific protein (Liu et al., 2021). Significant crosstalk between the ESCRT function, phytohormonal, and stress response pathways are starting to emerge (Ge et al., 2019; Li et al., 2019; Xiao et al., 2020).
The RAB5- and RAB7-dependent vacuolar pathways, which are important for the trafficking of the storage proteins aleurain, phaseolin, and 12S globulin, involve the sequential activity of RAB5/RabF2a and RAB7/RabG3c GTPases (Figure 4). RAB5 is activated by its GEF, VPS9a, resulting in the recruitment of the GTPase to the PVC membrane (Goh et al., 2007). ENDOSOMAL RAB EFFECTOR WITH PX-DOMAIN (EREX), a protein-containing the Phox homology (PX) domain, is recruited to the PVC by its interaction with the GTP-bound form of the RAB5 protein ARA7 and with PI3P. However, the role of EREX, EREX-LIKE1 (EREL1), or EREL2 in PVC maturation is still unknown (Sakurai et al., 2016). A homolog of EREX, GLUTELIN PRECURSOR ACCUMULATION5 (GPA5), is required for storage protein trafficking to vacuoles in rice endosperm via dense vesicles. GPA5 was proposed as a RAB5 effector in rice that mediates the fusion of dense vesicles with protein storage vacuoles (PSVs) (Ren et al., 2020). A RAB5 for RAB7 exchange is promoted by a conserved complex formed by CALCIUM CAFFEINE ZINC SENSITIVITY1 (CCZ1) and MONENSIN SENSITIVITY1 (MON1, also named SAND) (Uemura and Ueda, 2014) that acts as an effector of RAB5-GTP and as the GEF of RAB7. Activation of RAB7 by CCZ1/MON1 is the last step prior to PVC fusion to the vacuole, which is mediated by the HOMOTYPIC FUSION AND VACUOLE PROTEIN SORTING (HOPS)-tethering complex and SNAREs.
Tethering factors are large multi-subunit complexes that mediate the initial contact between two organellar membranes prior to fusion (Ravikumar et al., 2017). The plant vacuolar traffic is regulated by two tethering complexes, CLASS C CORE VACUOLE/ENDOSOME TETHERING (CORVET) and HOPS (Figure 4; Rojo et al., 2003; Niihama et al., 2009; Hao et al., 2016; Vukašinović and Žárský, 2016; Brillada et al., 2018; Takemoto et al., 2018). Both complexes are conserved in eukaryotes and consist of six subunits, of which four, VPS16, VPS18, VPS11, and VPS33, form a shared core (Nickerson et al., 2009; Wickner, 2010; Solinger and Spang, 2013; van der Kant et al., 2015) with the addition of the two specific subunits VPS39 and VPS41 (HOPS) and VPS3 and VPS8 (CORVET), as detected in Arabidopsis by co-immunoprecipitation experiments (Takemoto et al., 2018). The specific regulatory functions of each complex are expected to be encoded in the complex-specific subunits. HOPS plays an important role in the promotion of a trans-SNARE complex assembly between two fusogenic membranes and this function is mainly mediated by the Sec1/Munc18 (SM) protein VPS33 (Baker et al., 2015; Baker and Hughson, 2016). HOPS may further facilitate membrane fusion by lowering the energy barrier for membrane pore formation (D'Agostino et al., 2017). Interactions between different subunits of HOPS and SNAREs support a conserved role for HOPS in the plant SNARE complex association and have been detected in planta between VPS16 (also named VACUOLELESS [VCL1]) and the Qa SNARE protein SYP22 (Rojo et al., 2001, 2003; Hicks et al., 2004), between VPS33 and SYP22 (Brillada et al., 2018) or between all HOPS subunits and the R SNARE VAMP713 (Takemoto et al., 2018). Furthermore, interactions between VPS39 and RAB7, co-localization studies, and genetic analyses have led to the suggestion that HOPS might be involved in the RAB5- and RAB7-dependent pathways for vacuolar traffic (Takemoto et al., 2018). All plant HOPS subunits are essential in Arabidopsis (Rojo et al., 2003; Niihama et al., 2009; Hao et al., 2016; Vukašinović and Žárský, 2016; Brillada et al., 2018; Takemoto et al., 2018) and loss-of-function alleles for VPS33 and VPS41 are male-sterile (Hao et al., 2016; Brillada et al., 2018; Takemoto et al., 2018). The mechanisms that regulate HOPS function in plants is not well characterized, but its interaction with RAB7 suggests that it functions as a RAB7 effector (Takemoto et al., 2018). How this interaction regulates the recruitment to the membrane and the link with SNAREs is still unknown. Recently, the interaction between RAB7/RABG3f-GTP and the retromer subunit VPS35 at the PVC has been shown to be necessary for HOPS complex assembly, highlighting a putative checkpoint between anterograde and retrograde pathways (Rodriguez-Furlan et al., 2019b). Silencing of most HOPS subunit genes by dexamethasone-inducible artificial microRNAs results in vacuolar fragmentation phenotypes that substantiate a role for HOPS in homotypic vacuole fusion in plants (Brillada et al., 2018; Takemoto et al., 2018).
Whereas HOPS and CORVET are conserved in eukaryotes and tether similar organellar membranes, CORVET-mediated fusion events have shifted in plants. In yeast and metazoans, CORVET tethers endosomal membranes by binding to RAB5-GTP and, therefore, appears to mediate homotypic endosomal fusion before endosomes mature into MVBs (Balderhaar and Ungermann, 2013). In contrast, plant CORVET functions in membrane tethering between LEs/PVC and the vacuole (Takemoto et al., 2018) downstream of RAB5 activation, but independently from the CCZ1/SAND complex. Therefore, CORVET mediates tethering for the RAB5-dependent and RAB7-independent pathways. Mistargeting of SYP22 in vps3 mutants is consistent with this model.
PVC and vacuolar homotypic fusions
Fusion between the PVC and the vacuole is the last step in vacuolar transport (Figure 4; Uemura and Ueda, 2014; Zhuang et al., 2015; Peixoto et al., 2016). SNARE complex assembly follows the membrane tethering by HOPS or CORVET. SNAREs belong to a highly conserved protein family that mediates membrane fusion between two organelles. According to the SNARE hypothesis, one SNARE from the vesicle membrane (R SNARE) and two or three SNAREs from the target membrane (Qa, Qb, and Qc SNAREs) form a four-helical bundle designated a trans-SNARE complex. By zippering of the trans-SNARE complex, the two opposing membranes are brought together to promote fusion and the transition to a cis-SNARE complex at the newly formed membrane (Chen and Scheller, 2001; Wickner, 2010). SEC17 and the SEC18 ATPase unfold the cis-SNARE complex into individual SNAREs, allowing SNARE recycling for another round of fusion. The identity of the SNARE protein complement contributes to the specificity of the organellar fusion events. In plants, a vacuolar SNARE complex has been identified that contained SYP22 (Qa), VTI11 (Qb), SYP51 (Qc), and either VAMP727 (R) (Ebine et al., 2008) or VAMP713/VAMP711 (Ebine et al., 2011; Fujiwara et al., 2014; Uemura and Ueda, 2014). Mutant phenotypes indicate functional redundancy between SYP21 and SYP22 (Uemura et al., 2010), but either protein is necessary, because the double mutant could not be isolated. A large paralog expansion of the SNARE complement in seed plants hints at SNARE function diversification (Kanazawa et al., 2016). Loss-of-function mutants in VTI11 have fragmented vacuolar phenotypes due to the abnormal activity of the vacuolar SNARE complex and results in VPS41-green fluorescent protein (GFP) accumulation at small aggregated vacuoles (Zheng et al., 2014b; Brillada et al., 2018; Cui et al., 2019). As HOPS has been shown to proofread the fidelity of the SNARE complex in yeast (Starai et al., 2008), these phenotypes may be the consequence of VPS41 within the HOPS complexes binding to mismatched SNARE complexes at vacuolar or prevacuolar membranes in these mutants (Brillada et al., 2018).
Membrane lipids are key regulators of vacuole traffic, biogenesis, and dynamics (Heilmann and Heilmann, 2015). Phosphoinositides play important roles in pollen development (Heilmann and Ischebeck, 2016; Ugalde et al., 2016), homotypic vacuole fusion (Zheng et al., 2014a), endosome maturation (Hirano et al., 2015), and vacuole morphology (Hirano et al., 2017). Sterols and sphingolipids are also essential for vacuolar transport (Li et al., 2015). Biosynthesis of these lipids is induced by auxin and is regulated by the large cytosolic ribosomal protein subunit RPL4. As information about the abundance of sterols and sphingolipids along with the endomembrane system is lacking, it is still not clear how lipid composition regulates protein trafficking to the vacuole (Zhang et al., 2015b), implying that additional studies are required to understand how phosphoinositides and other lipids control vacuolar biogenesis and dynamics. Phosphoinositides are critical for HOPS recruitment in both yeast (Stroupe et al., 2006) and plants (Brillada et al., 2018), but membrane curvature may couple with phosphoinositides to act as proxies to control VPS41 recruitment. In a protein overlay assay, recombinant glutathione-S-transferase (GST)-VPS41 binds to phosphoinositides, including PI3P and PI(3,5)P2 (Brillada et al., 2018), suggesting that VPS41 is recruited to the vacuole by phosphoinositides, but in a liposome assay, both VPS41 and VPS33 bound only in their absence. Live-cell imaging demonstrated that VPS41-GFP normally accumulates at the vacuolar membrane and revealed that VPS41 dissociates from the vacuole after wortmannin treatment, which inhibits phosphatidylinositol 3-kinase, an enzyme that synthesizes PI3P (Stack and Emr, 1994) found in LEs and vacuoles. Remarkably, VPS41-GFP did not dissociate from LEs after treatment with wortmannin. These observations suggest that the role of PI3P role recruiting VPS41 depends on membrane curvature, with phosphoinositide promoting recruitment to the flat vacuolar membranes and abolishing recruitment to the highly curved endosome membrane. In plants, wortmannin induces the fusion of vacuoles (Zheng et al., 2014a) and LEs (Wang et al., 2009), but this effect was not observed in yeast (Wang et al., 2009). Regulation of vacuole fusion by phosphoinositides may be important for stomatal movements because wortmannin triggers rapid vacuole fusion in guard cells (Zheng et al., 2014a).
Trafficking to PSVs or lytic vacuoles
Plant cell vacuoles are unique because they are dynamic in structure and composition depending on the plant developmental stage and cell type. The vacuolar pH in developing embryos is neutral, consistent with their function for the storage of protein reserves. These PSVs are the main storage compartments of proteins from seeds and are, therefore, very important for human nutrition. Seed germination triggers the transition from PSVs to lytic vacuoles (LVs), involving changes in protein composition and acidification, resulting in the breakdown of storage proteins. The mechanisms that mediate this transition are not well understood, but changes in gene expression induce the accumulation of hydrolytic enzymes and transporters typical of the LVs. An important question is how plants manage to sort events to vacuoles that are undergoing this transition, because a shift in cargo starts to occur. Previous work in Arabidopsis embryos showed that vacuolar markers carrying PSV or LV targeting signals as well as PSV- or LV-resident Tonoplast Intrinsic Proteins (TIPs) are transported to either type of vacuole and that their accumulation into a specific vacuole type is driven by the timing of expression of their encoding genes during development (Hunter et al., 2007). Several recent reviews describe ongoing research to characterize vacuolar sorting signals and cognate receptors for PSV and LV cargoes (Zhang et al., 2021; Ashnest and Gendall, 2018). The Arabidopsis protein CALCIUM-DEPENDENT PROTEIN KINASE1 ADAPTOR PROTEIN2 (CAP2), important for LV traffic during the LV-to-PSV transition in germinating seeds has been identified (Kwon et al., 2018). The CAP2 proteins of Arabidopsis and crystalline ice-plant (Mesembryanthemum crystallinum) are predicted to form a coil–coil structure and were proposed as members of the syntaxin family (Chehab et al., 2007), but this activity has yet to be tested, because no conserved domains are apparent. cap2 mutant phenotypes in Arabidopsis included delayed vacuole maturation and acidification, delayed degradation of the 12S-globulin α and β subunits, and reduced trafficking of LV cargoes, including the two soluble proteins sporamin and aleurein, and the two membrane proteins β fructosidase 4 and the aquaporin γTIP. Furthermore, CAP2 also recruits GAPC2 to the PVC. GAPC2 is a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) isoform that converts glyceraldehyde-3-phosphate to 1,3-bisphospho-d-glycerate in the presence of NAD and inorganic phosphate (Pi) and is involved in the glycolytic pathway. Glycolytic enzymes have been implicated in protein trafficking in plant and mammalian cells (Jaquinod et al., 2007; Parsons et al., 2012; Heard et al., 2015; Hinckelmann et al., 2016), but none of the plant proteins have been shown to actively regulate trafficking. As gapc2 mutants delay vacuolar protein trafficking, the phenotypes of cap2 mutants might in part result from abnormal GAPC2 accumulation at the PVC.
Is the function of GAPC2 at the PVC related to its metabolic activity as part of glycolysis? A complete glycolytic pathway, including GAPDH, was detected in neuronal secretory vesicles, in which the ATP generated by these enzymes is used by motors to propel these vesicles along microtubules (Hinckelmann et al., 2016). In the case of plant LV biogenesis, a large demand of energy would arise from the membrane fusion events between the PVC and the vacuole, especially in the PSV-to-LV transition during seed germination. Interestingly, other glycolytic enzymes in the same pathway have been discovered in organellar proteomes, including phosphoglycerate kinases in the Golgi, LEs, and vacuoles (Jaquinod et al., 2007; Parsons et al., 2012; Heard et al., 2015), enolase in LEs and vacuoles (Shimaoka et al., 2004; Yoshida et al., 2013; Heard et al., 2015), and pyruvate kinases in the TGN and vacuoles (Shimaoka et al., 2004; Drakakaki et al., 2012). The identification of GAPC2 points to the possibility that plants may also have co-opted energy-generating pathways to energize the demanding mechanisms of vacuolar biogenesis.
Autophagy is a membrane trafficking pathway
Macroautophagy (hereafter autophagy) is a potent quality control mechanism by which damaged or unwanted cellular components are rapidly degraded to maintain cellular homeostasis (Marshall and Vierstra, 2018). Autophagy is induced by various biotic and abiotic stress responses and keeps the cell in tune with a changing environment (Rodriguez-Gallardo et al., 2020). Studies over the last decade have shown that autophagy is highly selective. Modular proteins, termed selective autophagy receptors or cargo receptors, recognize specific cargoes and mediate their degradation (Stolz et al., 2014). Although a plethora of cargo receptors have been identified in mammals, cargo receptors that mediate selective organelle recycling in plants have remained elusive thus far (Stephani and Dagdas, 2020). As selective autophagy receptors have been reviewed in detail (Stephani and Dagdas, 2020), we will focus on the membrane trafficking aspects of autophagy, namely the biogenesis and maturation of the autophagic organelle. We will also highlight the crosstalk mechanism between autophagy and other endomembrane trafficking pathways.
Two multiprotein kinase complexes initiate autophagosome biogenesis
The hallmark of autophagy is autophagosomes, the double-membrane vesicles formed de novo that engulf the autophagic cargoes and deliver them to the vacuoles for recycling (Nakatogawa, 2020; Chang et al., 2021). Autophagosomes start with a cup-shaped isolation membrane or phagophore that expands and surrounds the cargo. Upon closure, autophagosomes are then delivered to the vacuole, where the outer membrane fuses with the vacuolar membrane to supply a single membrane autophagic body to the vacuolar lumen. Resident hydrolytic enzymes degrade the autophagic body and the building blocks are released back to the cytosol to feed various anabolic reactions. Here we summarize recent advances in autophagosome biogenesis, highlighting the crosstalk between autophagy and the endomembrane trafficking pathways.
Pioneering work has shown that the core autophagy machinery is also conserved in plants (Marshall and Vierstra, 2018). Autophagosome biogenesis is coordinated by a set of approximately 20 autophagy-related (ATG) proteins that had previously been discovered in yeast (Matoba and Noda, 2021) and is initiated by a kinase complex, including the serine–threonine kinase ATG1 and the APs ATG11 and ATG13 (Figure 5; Li et al., 2014; Huang et al., 2019). The ATG1 kinase complex is enriched in intrinsically disordered proteins that form liquid-like phase-separated compartments to orchestrate the autophagy machinery (Fujioka et al., 2020). Upstream regulators, such as the Target of Rapamycin kinase or AMP-activated protein kinase, control these phase separation events via phosphorylation, thereby modulating autophagy (Fujioka et al., 2020).
Figure 5.
Overview of autophagosome biogenesis. Autophagy-inducing conditions activate signaling pathways with the ATG1 kinase (ATG1K) complex (consisting of ATG1, ATG11, ATG13, and ATG101) as a common target to initiate autophagosome biogenesis. The ATG1 complex phosphorylates the PI3 kinase (PI3K) complex (consisting of ATG6, ATG14, VPS15, and VPS34) that drives the nucleation of the isolation membrane, also termed the phagophore. Phosphatidylinositol 3-phosphate (PI3P) production leads to the recruitment of the PI3P-binding proteins ATG18 and ATG2. ATG9-containing vesicles serve as membrane seeds for the phagophore, whose development is maintained by the lipid transfer activity of ATG2, while the lipid scramblase activity of ATG9 ensures the even distribution of transferred phospholipids between the inner and outer phagophore membranes. During phagophore elongation, ATG8 is covalently bound to phosphatidylethanolamine (PE) via the ubiquitin-like (UBL) protein conjugation system, composed of an E1 (ATG7), E2 (ATG3), and E3 (ATG5-ATG12-ATG16 complex), allowing its insertion into the growing phagophore. Phagophore closure is mediated by ESCRT complexes: the ESCRT-I subunit VPS37 can recruit the ESCRT-III subunits CHMP1/CHMP2 and the ATPase VPS4, while VPS23, another ESCRT-I subunit, interacts with the FYVE domain-containing proteins CFS1 and FREE1, enabling the recruitment of SH3P2 proteins. Upon closure, autophagosomes are delivered to the vacuole. The inner membrane-bound autophagosome structure, known as an autophagic body, is released into the vacuolar lumen and is degraded by resident hydrolases, allowing recycling of its constituents.
An emerging theme in the mammalian autophagy field is that autophagosome biogenesis is induced by the cargo macromolecules (Zaffagnini and Martens, 2016). Cargo receptors that recognize the bulky autophagic cargo interact with the mammalian ATG11 counterpart, the focal adhesion kinase family interacting protein of 200 kDa (FIP200; Hara et al., 2008), to drive the cargo in close proximity to the core autophagy machinery (Turco et al., 2020). Thus far, aggrephagy, mitophagy, and ER-phagy receptors have been shown to interact with FIP200 (Smith et al., 2018; Ravenhill et al., 2019; Turco et al., 2019; Vargas et al., 2019; Stephani et al., 2020). A cargo-induced autophagosome biogenesis model elucidates how the cell selects and recycles only the damaged or unwanted cellular components within a crowded cytoplasm.
Until very recently, the source and the growth of the phagophore membrane had remained controversial. Previous studies in yeast, human cells, and plants, have revealed that another multiprotein kinase complex, designated phosphoinositide 3-kinase (PI3K) complex, generates PI3P on autophagosome biogenesis sites (Nakatogawa, 2020; Liu et al., 2020b). The PI3K complex comprises several proteins, including VPS34, ATG6, and ATG14. VPS34 can also associate with another protein, VPS38 (also named UV radiation resistance-associated gene protein or UVRAG), to form the PI3K complex II that regulates the endosomal function, illustrating the close connection between autophagy and endosomal trafficking (Chang et al., 2021). PI3P production recruits several PI3P-binding proteins, such as the β-PROPELLERS THAT BIND POLYPHOSPHOINOSITIDES (PROPPIN) proteins ATG18 and ATG2 (Fracchiolla et al., 2020; Sawa-Makarska et al., 2020). Recent studies in yeast indicated that ATG2 bridges the ER with the developing phagophore and transfers lipid from the ER to the phagophore for sustained growth around the cargo (Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019). In vitro reconstitution studies with purified yeast ATG proteins and liposomes revealed that the phagophore is seeded by ATG9 vesicles originating from the TGN (Figure 5; Sawa-Makarska et al., 2020). ATG9 is the only transmembrane domain protein among ATG proteins; cryo-EM analysis of yeast, plant, and human ATG9 proteins showed that they all form a homotrimeric complex with two distinct pores on each side of the membrane (Guardia et al., 2020; Lai et al., 2020; Maeda et al., 2020). Further reconstitution assays showed that ATG9 functions as a lipid scramblase equilibrating the transferred phospholipids on each membrane of the autophagosome (Maeda et al., 2020; Matoba et al., 2020). Altogether, these dynamic lipid fluxes and distribution events underlie the rapid growth of the phagophore around the cargo.
Phagophores originate from ER sub-compartments that are rich in phospholipids. Recent studies in mammalians and plants have revealed that ER-derived COPII vesicles also contribute to autophagosome biogenesis. In mammals, a specific SEC23 isoform, SEC23B, is redirected to phagophores under autophagy-inducing conditions (Jeong et al., 2018). Similarly, in plants, a unique SAR1 isoform, SAR1D, participates in autophagosome biogenesis in concert with the Rab GTPase RABD2A (Zeng et al., 2021). Although the molecular details remain scarce, the exocyst complex also plays a role in autophagosome biogenesis in both mammals and plants. In mammals, the GTPase RalB interacts with EXOCYST84 (EXO84) and promotes autophagosome biogenesis by recruiting its machinery (Bodemann et al., 2011), whereas in Arabidopsis, another exocyst component, SEC5, and another GTPase, RHO-RELATED PROTEIN FROM PLANTS8 (ROP8), localize to phagophores under autophagy-inducing conditions (Lin et al., 2021). These examples clearly highlight the intricate balance between the autophagic and exocytic trafficking pathways.
Two conjugation systems ensure rapid growth of the phagophore
One of the key steps in autophagosome biogenesis is the lipidation of the ubiquitin-like protein ATG8 with phosphatidylethanolamine (PE). ATG8 lipidation is mediated by two ubiquitination-like conjugation reactions: the E1-like and E2-like enzymes ATG7 and ATG10 catalyze isopeptide bond formation between ATG5 and the ubiquitin-like protein ATG12 (Thompson et al., 2005; Phillips et al., 2008). The ATG5–ATG12 conjugate then interacts with ATG16 to form a complex that is carried to the phagophore by PI3P-binding proteins and functions as the E3 enzyme for the next ubiquitination reaction, culminating in the lipidation of ATG8 family proteins (Nakatogawa, 2020; Chang et al., 2021). ATG7, ATG3, and the ATG5–ATG12–ATG16 complex act as E1, E2, and E3 enzymes, respectively, and mediate the conjugation of ATG8 to a lipid moiety, PE, allowing the insertion of ATG8 proteins into the developing phagophore (Phillips et al., 2008; Chung et al., 2010). Before conjugation, ATG8 is activated by ATG4 cysteine proteases that cleave the ATG8 C terminus and exposes glycine residues for lipidation (Figure 5; Woo et al., 2014).
ATG8 acts as an organizer of the autophagy pathway. Through a short linear motif, designated ATG8-interacting motif (AIM), ATG8 interrelates with various proteins, including (1) core autophagy proteins, such as ATG1 or ATG4; (2) SNARE proteins and adaptors that mediate vesicle fusion and trafficking; and (3) selective autophagy receptors that are responsible for selective cargo recruitment (Kellner et al., 2017). In yeast and mammals, atg8 mutants still form autophagosomes, but they are reduced and less efficient in terms of cargo degradation (Martens, 2016). In conjunction with the cargo-induced autophagosome biogenesis model, the ATG8 family of proteins probably facilitates autophagosome expansion by concentrating critical proteins at the autophagosome biogenesis sites. In addition, this protein family plays key roles in compartmentalization of autophagic degradation. Unlike yeast, in which autophagosome biogenesis takes place at the vacuolar membrane, mammals and plants have multiple ATG8 isoforms that interact with different sets of receptor and APs (Wirth et al., 2019; Zess et al., 2019). Functional specialization of these ATG8 isoforms allows compartmentalization of different selective autophagy mechanisms that occur simultaneously in a cell while sharing the core autophagy machinery.
Recent research has challenged the long-standing view that ATG8 is conjugated only to double-membrane phagophores. Although thus far not studied in plants, in mammals, ATG8 conjugation complexes can attach ATG8 proteins to single-membrane compartments, such as phagosomes or endosomes (Codogno et al., 2012; Jacquin et al., 2017). The ATG1 kinase complex is dispensable for these noncanonical autophagy processes. Accumulating evidence suggests that the WD40 domain of ATG16 recruits the ATG8 conjugation machinery to different endolysosomal compartments to facilitate ATG8 conjugation to these membranes (Fletcher et al., 2018; Durgan et al., 2021). During noncanonical autophagy, ATG8 can be modified by phosphatidylserine instead of PE (Durgan et al., 2021). In light of the crucial roles of phosphatidylserine in plant PM organization (Platre et al., 2019), it will be interesting to investigate whether noncanonical autophagy plays a role in plant signaling processes.
ESCRTing autophagosome closure
For the phagophore to completely encapsulate the cargo, the membrane between the two leaflets needs to be severed, so that the opposing membranes can fuse together. This membrane remodeling process is topologically identical to the membrane scission events that occur during MVB biogenesis (Isono, 2021). Recently, in mammals, similarly to MVB biogenesis, membrane scission during phagophore closure has shown to be mediated by ESCRT complexes. The ESCRT-I subunit VPS37A localizes at the phagophore edges and recruits other ESCRT proteins, such as the ESCRT-III subunit Charged Multivesicular Body Protein 2A (CHMP2A) or the ATPase VPS4 (Takahashi et al., 2018, 2019; Zhen et al., 2020).
Although the molecular details are still limited, ESCRT-mediated phagophore closure appears to be conserved in plants as well (Isono, 2021). Two FYVE domain-containing proteins, FREE1 and CELL DEATH-RELATED ENDOSOMAL FYVE/SYLF PROTEIN1 (CFS1), interact with the ESCRT-I subunit VPS23 (Gao et al., 2015; Sutipatanasomboon et al., 2017). Autophagic flux assays revealed that both APs are critical for autophagic degradation. Another FREE1-interacting protein, SH3P2, also plays a crucial role in autophagy (Zhuang and Jiang, 2014). As in mammalian cells, the ESCRT-III component CHMP1 or the ATPase VPS4 is also associated with selective autophagy processes (Figure 5; Spitzer et al., 2015; Zess et al., 2019). Altogether, these studies indicate that plant ESCRT complexes mediate phagophore closure as well. Whether there are autophagy-specific ESCRT complexes or how ESCRT recruitment to the phagophore edges is coordinated with the phagophore expansion machinery is still unknown.
Autophagosome–vacuole fusion is a black box in plants
Once autophagosomes are formed, they need to be delivered to the vacuolar membrane and fuse with the tonoplast. These processes have been studied in depth in yeast and mammals. Autophagy adaptors, which directly interact with ATG8 proteins, facilitate the trafficking and fusion of autophagosomes. Autophagic SNARE proteins that directly interact with ATG8 proteins and the HOPS complex mediate the fusion of autophagosomes with lytic compartments (Zhao and Zhang, 2019).
However, how autophagosomes are transported to and fuse with the tonoplast remains mostly unknown. Systematic analyses of the autophagy adaptors or autophagy-specific SNARE proteins are missing in plants. Considering the large volume occupied by vacuoles in plants cells, it will be interesting to know whether the tonoplast is compartmentalized in terms of autophagic and endocytic cargo deliveries as well as to understand how vacuolar trafficking regulates the autophagosome biogenesis.
Autophagy as a quality control mechanism for endomembrane trafficking
In addition to the crosstalk mechanisms that coordinate autophagosome biogenesis and trafficking, autophagy also oversees membrane trafficking pathways to remove malfunctioning units. In mammals, several studies revealed that autophagy mediates the degradation of damaged or superfluous ER, Golgi, lysosomes, or endocytic vesicles. Recently, autophagy has been found to play a role in endomembrane quality control in plants and AtEH1/Pan1, a component of the TPC essential for CME in Arabidopsis, has been shown to regulate autophagosome biogenesis. Under autophagy-inducing conditions, AtEH1/Pan1 recruits other TPC members and endocytic protein trafficking regulators to autophagosomes (Wang et al., 2019). Interestingly, AtEH1/Pan1 autophagosomes are localized in close proximity to VESICLE-ASSOCIATED PROTEIN27 (VAP27)-labeled ER-PM contact sites. Genetic perturbation of both the VAP27 and AtEH1/Pan1 genes seem to affect autophagy dynamics (Wang et al., 2019). How and when AtEH1/Pan1 shifts from an endocytic regulator to an autophagy inducer remains unknown. Similarly, EXO70, a part of the exocytosis-regulating octameric exocyst complex, can also be deployed as an autophagy receptor. EXO70B and EXO70D interact with ATG8 via AIMs to control the immune and cytokinin signaling pathways, respectively (Acheampong et al., 2020; Brillada et al., 2021). EXO70B isoforms colocalize with ATG8 upon immune signaling activation and probably dampen secretory activity to monitor pathogen infections (Brillada et al., 2021). In contrast, EXO70D mediates the degradation of type-A response regulators, which negatively regulate cytokinin signaling (Acheampong et al., 2020). These examples illustrate how plant cells utilize known membrane trafficking players as autophagy receptors to control adaptive signaling responses.
Conclusions and perspectives
Although our understanding of plant endomembrane trafficking pathways has increased significantly over the last decade, large gaps are still waiting to be filled. Recent advances in imaging, the adoption of new genetics and biochemical techniques, and the establishment of new model systems, offer unprecedented opportunities to address these challenges in plant cell biology. In Table 1, we highlight some of the key questions and suggest applications by which their answers might be facilitated.
Table 1.
Key questions in plant cell biology and approaches or model systems that may facilitate their investigations
| Question | Approach/Model System | References |
|---|---|---|
| What is the nature of COPII carriers and ER–Golgi transport in plants? | Whole-cell focused ion beam (FIB)-SEM, combined with cryogenic-structured illumination microscopy used recently in mammalian cells to re-examine the ultrastructural features of ERES and transport intermediates at the ER–Golgi interface at isotropic nanoscale resolution through an entire cell. These techniques have challenged the current view of ER–Golgi transport and the function of COPII proteins and might be applied to further dissect the peculiarities of ER–Golgi transport in plants | Hoffman et al. (2020); Xu et al. (2017); Weigel et al. (2021) |
| Are COPI and COPII isoforms functionally specialized? | The diversity of COPI and COPII isoforms in widely used model plants, such as Arabidopsis, adds complexity to functional studies. The use of alternative model systems, with less complex genetics, such as the liverwort Marchantia polymorpha, may help elucidate whether gene expansion has functional implications | Kanazawa et al. (2016) |
| Can cargo trafficking be visualized in live cells? | Retention Using Selective Hooks can be used to synchronize cargo trafficking under physiological conditions without stressing cells and may be a promising tool in plants | Pacheco-Fernandez et al. (2021) |
| How can essential processes be studied in a spatiotemporally controlled manner? | In mammalian cells, inducible degron systems based on auxin degrons or plant photoreceptors are available, although their equivalent does not exist for plant cells. Inducible systems in plants include inducible clustered regularly interspaced short palindromic repeats (CRISPRs) or gene silencing modules to probe the function of essential proteins in a time-dependent manner | Nishimura et al. (2009); Tischer and Weiner (2014) |
| Can endomembrane trafficking be analyzed in crop model systems? | Studies of endomembrane trafficking in crop plants are challenging, partly due to the difficulty of plant transformation. The maize (Zea mays) fluorescence tagging collection from the Cold Spring Harbor Laboratory (Cold Spring Harbor, NY, USA) includes 117 translational fusions, many of which represent cargo targeted to different organelles that may be used as a starting point for protein trafficking studies in this important crop | Krishnakumar et al. (2015) |
| Are there organ- or cell-type-specific endocytosis and autophagy trafficking pathways? | There is a wealth of knowledge in Arabidopsis regarding cell type-specific promoters. With the invention of CRISPR-Tissue-specific genome editing tools, we can now address cell type-specific endocytic and autophagic pathways | Decaestecker et al. (2019) |
| How do autophagosomes fuse with the tonoplast? | A reductionist approach, such as in vitro reconstitution using purified proteins and compartments, may help address this highly challenging but fundamental question | Bas et al. (2018) |
Accession numbers
For reference, all proteins mentioned in this review are listed in Supplemental Data Set S1 with their full names, abbreviations and accession numbers.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Data Set S1. List of all proteins mentioned in this review.
Supplementary Material
Acknowledgments
We thank Daniël Van Damme and David Robinson for critical reading of the CME section and the ER–Golgi section, respectively, Karel Van Lierde for preparation of Figure 3, and Martine De Cock for help in preparing the manuscript. Figures 1 and 3–5 were prepared with BioRender (BioRender.com).
Funding
This work was supported by grants from Ministerio de Economía y Competitividad (BFU-2016-76607-P) to F.A., the Austrian Science Fund Special Research Program (SFB F79 Targeted Protein Degradation to Y.D. and V.S de M. H., National Science Foundation (MCB-1918746) to M.R.-P. and the Research Foundation-Flanders (G.0E57.18N) to E.R.
Conflict of interest statement. None declared.
Contributor Information
Fernando Aniento, Departamento de Bioquímica y Biología Molecular, Instituto Universitario de Biotecnología y Biomedicina (BIOTECMED), Universitat de València, E-46100 Burjassot, Spain.
Víctor Sánchez de Medina Hernández, Gregor Mendel Institute, Austrian Academy of Sciences, Vienna BioCenter, 1030 Vienna, Austria; Vienna BioCenter PhD Program, Doctoral School of the University of Vienna and Medical University of Vienna, A-1030, Vienna, Austria.
Yasin Dagdas, Gregor Mendel Institute, Austrian Academy of Sciences, Vienna BioCenter, 1030 Vienna, Austria.
Marcela Rojas-Pierce, Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC 27514, USA.
Eugenia Russinova, Department of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium; Center for Plant Systems Biology, VIB, 9052 Ghent, Belgium.
F. A., Y. D., M. R.-P., V.S.de M. H., and E. R. wrote the manuscript and prepared figures.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plcell) is: Eugenia Russinova (eugenia.russinova@psb.vib-ugent.be).
References
- Acheampong AK, Shanks C, Cheng C-Y, Schaller GE, Dagdas Y, Kieber JJ (2020) EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc Natl Acad Sci USA 117: 27034–27043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Narasimhan M, Kania U, Glanc M, De Jaeger G, Friml J (2018) A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30: 700–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf F, Rhiel M, Hessling B, Gao Q, Hellwig A, Béthune J, Wieland FT (2019) Proteomic profiling of mammalian COPII and COPI vesicles. Cell Rep 26: 250–265 [DOI] [PubMed] [Google Scholar]
- Ahn H-K, Kang YW, Lim HM, Hwang I, Pai H-S (2015) Physiological functions of the COPI complex in higher plants. Mol Cells 38: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn G, Kim H, Kim DH, Hanh H, Yoon Y, Singaram I, Wijesinghe KJ, Johnson KA, Zhuang X, Liang Z, et al. (2017) SH3 Domain-containing protein 2 plays a crucial role at the step of membrane tubulation during cell plate formation. Plant Cell 29: 1388–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakel EC, Schwappach B (2018) Formation of COPI-coated vesicles at a glance. J Cell Sci 131: jcs209890 [Erratum J Cell Sci 131: jcs.218347]. [DOI] [PubMed] [Google Scholar]
- Arora D, Van Damme D (2021) Motif-based endomembrane trafficking. Plant Physiol 186: 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashnest JR, Gendall AR (2018) Trafficking to the seed protein storage vacuole. Funct Plant Biol 45: 895–910 [DOI] [PubMed] [Google Scholar]
- Baker RW, Hughson FM (2016) Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol 17: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM (2015) A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349: 1111–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJk, Ungermann C (2013) CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci 126: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Bao H, Daniels RW, MacLeod GT, Charlton MP, Atwood HL, Zhang B (2005) AP180 maintains the distribution of synaptic and vesicle proteins in the nerve terminal and indirectly regulates the efficacy of Ca2+-triggered exocytosis. J Neurophysiol 94: 1888–1903 [DOI] [PubMed] [Google Scholar]
- Barlowe C, Helenius A (2016) Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol 32: 197–222 [DOI] [PubMed] [Google Scholar]
- Barth M, Holstein SE (2004) Identification and functional characterization of Arabidopsis AP180, a binding partner of plant αC-adaptin. J Cell Sci 117: 2051–2062 [DOI] [PubMed] [Google Scholar]
- Bas L, Papinski D, Licheva M, Torggler R, Rohringer S, Schuschnig M, Kraft C (2018) Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol 217: 3656–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Li S, Anderson CT, Lei L, Gu Y (2013) The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol 163: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Li S, Zhu X, Gu Y (2015) The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc Natl Acad Sci USA 112: 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek SY, Backues SK (2010) Plant dynamin-related protein families DRP1 and DRP2 in plant development. Biochem Soc Trans 38: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B, Rodriguez L, Fernandez MA, Castillo M-C, Anderson EM, Gao C, Gonzalez-Guzman M, Peirats-Llobet M, Zhao Q, De Winne N, et al. (2016) FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat-Silvestre C, De Sousa Vieira V, Sanchez-Simarro J, Pastor-Cantizano N, Hawes C, Marcote MJ, Aniento F (2020) p24 family proteins are involved in transport to the plasma membrane of GPI-anchored proteins in plants. Plant Physiol 184: 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béthune J, Wieland FT (2018) Assembly of COPI and COPII vesicular coat proteins on membranes. Annu Rev Biophys 47: 63–83 [DOI] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou Y-H, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144: 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M (2006) Role of lipids and actin in the formation of clathrin-coated pits. Exp Cell Res 312: 4036–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F (2018) Transport from the endoplasmic reticulum to the Golgi in plants: where are we now? Semin Cell Dev Biol 80: 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer P, Parker JL, Gerondopoulos A, Zimmermann I, Seeger MA, Barr FA, Newstead S (2019) Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science 363: 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillada C, Teh O-K, Ditengou FA, Lee C-W, Klecker T, Saeed B, Furlan G, Zietz M, Hause G, Eschen-Lippold L, et al. (2021) Exocyst subunit Exo70B2 is linked to immune signaling and autophagy. Plant Cell 33: 404–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillada C, Zheng J, Krüger F, Rovira-Diaz E, Askani JC, Schumacher K, Rojas-Pierce M (2018) Phosphoinositides control the localization of HOPS subunit VPS41, which together with VPS33 mediates vacuole fusion in plants. Proc Natl Acad Sci USA 115: E8305–E8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Frey F, Sochacki KA, Kummer S, Bergeest J-P, Godinez WJ, Kräusslich H-G, Rohr K, Taraska JW, Schwarz US, et al. (2018) Clathrin-adaptor ratio and membrane tension regulate the flat-to-curved transition of the clathrin coat during endocytosis. Nat Commun 9: 1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buono RA, Leier A, Paez-Valencia J, Pennington J, Goodman K, Miller N, Ahlquist P, Marquez-Lago TT, Otegui MS (2017) ESCRT-mediated vesicle concatenation in plant endosomes. J Cell Biol 216: 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Báez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E (2009) Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol 185: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Jensen LE, Hurley JH (2021) Autophagosome biogenesis comes out of the black box. Nat Cell Biol 23: 450–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Patharkar OR, Cushman JC (2007) Isolation and characterization of a novel v-SNARE family protein that interacts with a calcium-dependent protein kinase from the common ice plant, Mesembryanthemum crystallinum. Planta 225: 783–799 [DOI] [PubMed] [Google Scholar]
- Chen X, Irani NG, Friml J (2011) Clathrin-mediated endocytosis: the gateway into plant cells. Curr Opin Plant Biol 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2: 98–106 [DOI] [PubMed] [Google Scholar]
- Roy Chowdhury S, Bhattacharjee C, Casler JC, Jain BK, Glick BS, Bhattacharyya D (2020) ER arrival sites associate with ER exit sites to create bidirectional transport portals. J Cell Biol 219: e201902114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung KP, Zeng Y, Jiang L (2016) COPII paralogs in plants: functional redundancy or diversity? Trends Plant Sci 21: 758–769 [DOI] [PubMed] [Google Scholar]
- Codogno P, Mehrpour M, Proikas-Cezanne T (2012) Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 13: 7–12 [DOI] [PubMed] [Google Scholar]
- Coleman J, Evans D, Hawes C, Horsley D, Cole L (1987) Structure and molecular organization of higher plant coated vesicles. J Cell Sci 88: 35–45 [Google Scholar]
- Collins A, Warrington A, Taylor KA, Svitkina T (2011) Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol 21: 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram WJ (1980) Pinocytosis in plants. New Phytol 84: 1–17 [Google Scholar]
- Cui Y, Cao W, He Y, Zhao Q, Wakazaki M, Zhuang X, Gao J, Zeng Y, Gao C, Ding Y, et al. (2019). A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat Plants, 5: 95–105 [DOI] [PubMed] [Google Scholar]
- Cui Y, Shen J, Gao C, Zhuang X, Wang J, Jiang L (2016) Biogenesis of plant prevacuolar multivesicular bodies. Mol Plant 9: 774–786 [DOI] [PubMed] [Google Scholar]
- D'Agostino M, Risselada HJ, Lürick A, Ungermann C, Mayer A (2017) A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature 551: 634–638 [DOI] [PubMed] [Google Scholar]
- daSilva LLP, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F (2004) Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, Van Isterdael G, Beeckman T, Nowack MK, Jacobs TB (2019) CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31: 2868–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu Z-Y, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, et al. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of BRASSINOSTEROID INSENSITIVE1 in Arabidopsis. Plant Cell 25: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano GP, Barozzi F, Piro G, Denecke J, de Marcos Lousa C (2017) Trafficking routes to the plant vacuole: connecting alternative and classical pathways. J Exp Bot 69: 79–90 [DOI] [PubMed] [Google Scholar]
- Donohoe BS, Kang B-H, Staehelin LA (2007) Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc Natl Acad Sci USA 104: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumane M, Lebecq A, Colin L, Fangain A, Stevens FD, Bareille J, Hamant O, Belkhadir Y, Munnik T, Jaillais Y, et al. (2021) Inducible depletion of PI(4,5)P2 by the synthetic iDePP system in Arabidopsis. Nat Plants 7: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragwidge JM, Van Damme D (2020) Visualising endocytosis in plants: past, present, and future. J Microsc 280: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al. (2012) Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan J, Lystad AH, Sloan K, Carlsson SR, Wilson MI, Marcassa E, Ulferts R, Webster J, Lopez-Clavijo AF, Wakelam MJ, et al. (2021) Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Mol Cell 81: 2031–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Ebine K, Inoue T, Ito J, Ito E, Uemura T, Goh T, Abe H, Sato K, Nakano A, Ueda T (2014) Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr Biol 24: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J (2013) Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837 [DOI] [PubMed] [Google Scholar]
- Faso C, Chen Y-N, Tamura K, Held M, Zemelis S, Marti L, Saravanan R, Hummel E, Kung L, Miller E, et al. (2009) A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q-N, Song S-J, Yu S-X, Wang J-G, Li S, Zhang Y (2017b) Adaptor protein-3-dependent vacuolar trafficking involves a subpopulation of COPII and HOPS tethering proteins. Plant Physiol 174: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q-N, Zhang Y, Li S (2017a) Tonoplast targeting of VHA-a3 relies on a Rab5-mediated but Rab7-independent vacuolar trafficking route. J Integr Plant Biol 59: 230–233 [DOI] [PubMed] [Google Scholar]
- Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, Kleine-Vehn J, Friml J (2010) The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, et al. (2018) The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J 37: e97840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracchiolla D, Chang C, Hurley JH, Martens S (2020) A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol 219: e201912098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Nakazono M, Tsutsumi N (2008) Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep 27: 1581–1586 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S-i, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N (2010) Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci USA 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Ebine K, Nishimura K, Tsutsumi N, Ueda T (2020) Longin R-SNARE is retrieved from the plasma membrane by ANTH domain-containing proteins in Arabidopsis. Proc Natl Acad Sci USA 117: 25150–25158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Tsutsumi N (2014) Dynamin-related proteins in plant post-Golgi traffic. Front Plant Sci 5: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, May AI, Knorr RL, Suzuki K, Ohsumi Y, et al. (2020) Phase separation organizes the site of autophagosome formation. Nature 578: 301–305 [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Uemura T, Ebine K, Nishimori Y, Ueda T, Nakano A, Sato MH, Fukao Y (2014) Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol 55: 781–789 [DOI] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G, et al. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704 [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Mooren OL, Cooper JA (2010) Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol 21: 604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Cai Y, Wang Y, Kang B-H, Aniento F, Robinson DG, Jiang L (2014) Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci 19: 508–515 [DOI] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Cui Y, Fu X, He Y, Zhao Q, Zeng Y, Shen J, Luo M, Jiang L (2015) Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci USA 112: 1886–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L (2017) Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci 22: 986–998 [DOI] [PubMed] [Google Scholar]
- Ge C, Gao C, Chen Q, Jiang L, Zhao Y (2019) ESCRT-dependent vacuolar sorting and degradation of the auxin biosynthetic enzyme YUC1 flavin monooxygenase. J Integr Plant Biol 61: 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Robatzek S (2008) Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno-Ferrer F, Pastor-Cantizano N, Bernat-Silvestre C, Selvi-Martínez P, Vera-Sirera F, Gao C, Perez-Amador MA, Jiang L, Aniento F, Marcote MJ (2017) α2-COP is involved in early secretory traffic in Arabidopsis and is required for plant growth. J Exp Bot 68: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, Melero A, Li X-H, Boulanger J, Kukulski W, Miller EA (2020) Cargo crowding contributes to sorting stringency in COPII vesicles. J Cell Biol 219: e201806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, Miller E (2016) Protein sorting at the ER-Golgi interface. J Cell Biol 215: 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Liu Q, Watanabe S, Sun L, Hollopeter G, Grant BD, Jorgensen EM (2013) AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. eLife 2: e00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia CM, Tan X-F, Lian T, Rana MS, Zhou W, Christenson ET, Lowry AJ, Faraldo-Gómez JD, Bonifacino JS, Jiang J, et al. (2020) Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep 31: 107837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Chatre L, Brandizzi F (2009) Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J 57: 963–974 [DOI] [PubMed] [Google Scholar]
- Hao L, Liu J, Zhong S, Gu H, Qu L-J (2016) AtVPS41-mediated endocytic pathway is essential for pollen tube-stigma interaction in Arabidopsis. Proc Natl Acad Sci USA 113: 6307–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S-i, Natsume T, Guan J-L, Mizushima N (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Hillmer R, Yamaoka S, Hara-Nishimura I, Katagiri F (2016) The μ subunit of Arabidopsis adaptor protein-2 is involved in effector-triggered immunity mediated by membrane-localized resistance proteins. Mol Plant-Microbe Interact 29: 345–351 [DOI] [PubMed] [Google Scholar]
- Hawes C, Martin B (1986) Deep etching of plant cells: cytoskeleton and coated pits. Cell Biol Int Rep 10: 985–992 [Google Scholar]
- He K, Marsland R III, Upadhyayula S, Song E, Dang S, Capraro BR, Wang W, Skillern W, Gaudin R, Ma M, et al. (2017) Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 552: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard W, Sklenář J, Tomé DFA, Robatzek S, Jones AME (2015) Identification of regulatory and cargo proteins of endosomal and secretory pathways in Arabidopsis thaliana by proteomic dissection. Mol Cell Proteomics 14: 1796–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann M, Heilmann I (2015) Plant phosphoinositides-complex networks controlling growth and adaptation. Biochim Biophys Acta 1851: 759–769 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Ischebeck T (2016) Male functions and malfunctions: the impact of phosphoinositides on pollen development and pollen tube growth. Plant Reprod 29: 3–20 [DOI] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT (2010) FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 328: 1281–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV (2004) Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol 134: 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S, Depta H, Robinson DG (1986) Confirmation of endocytosis in higher plant protoplasts using lectin-gold conjugates. Eur J Cell Biol 41: 142–149 [Google Scholar]
- Hinckelmann M-V, Virlogeux A, Niehage C, Poujol C, Choquet D, Hoflack B, Zala D, Saudou F (2016) Self-propelling vesicles define glycolysis as the minimal energy machinery for neuronal transport. Nat Commun 7: 13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Munnik T, Sato MH (2015) Phosphatidylinositol 3-phosphate 5-kinase, FAB1/PIKfyve kinase mediates endosome maturation to establish endosome-cortical microtubule interaction in Arabidopsis. Plant Physiol 169: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Munnik T, Sato MH (2017) Inhibition of phosphatidylinositol 3,5-bisphosphate production has pleiotropic effects on various membrane trafficking routes in Arabidopsis. Plant Cell Physiol 58: 120–129 [DOI] [PubMed] [Google Scholar]
- Hirst J, Schlacht A, Norcott JP, Traynor D, Bloomfield G, Antrobus R, Kay RR, Dacks JB, Robinson MS (2014) Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife 3: e02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020) Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367: eaaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollopeter G, Lange JJ, Zhang Y, Vu TN, Gu M, Ailion M, Lambie EJ, Slaughter BD, Unruh JR, Florens L, et al. (2014) The membrane-associated proteins FCHo and SGIP are allosteric activators of the AP2 clathrin adaptor complex. eLife 3: e03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn MA, Heinstein PF, Low PS (1989) Receptor-mediated endocytosis in plant cells. Plant Cell 1: 1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zheng C, Liu F, Yang C, Zheng P, Lu X, Tian J, Chung T, Otegui MS, Xiao S, et al. (2019) Genetic analyses of the Arabidopsis ATG1 Kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31: 2973–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings J, Stancheva VG, Brown NR, Cheung ACM, Miller EA, Zanetti G (2021) Structure of the complete, membrane-assembled COPII coat reveals a complex interaction network. Nat Commun 12: 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isayenkov S, Isner J-C, Maathuis FJM (2011) Rice two-pore K+ channels are expressed in different types of vacuoles. Plant Cell 23: 756–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Werner S, Krishnamoorthy P, Lerche J, Meijón M, Stenzel I, Löfke C, Wiessner T, Im YJ, Perera IY, et al. (2013) Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25: 4894–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E (2021) ESCRT is a great sealer: non-endosomal function of the ESCRT machinery in membrane repair and autophagy. Plant Cell Physiol 62: 766–774 [DOI] [PubMed] [Google Scholar]
- Ito Y, Boutté Y (2020) Differentiation of trafficking pathways at Golgi entry core compartments and post-Golgi subdomains. Front Plant Sci 11: 609516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Uemura T, Nakano A (2014) Formation and maintenance of the Golgi apparatus in plant cells. Int Rev Cell Mol Biol 310: 221–287 [DOI] [PubMed] [Google Scholar]
- Ito Y, Uemura T, Nakano A (2018) The Golgi entry core compartment functions as a COPII-independent scaffold for ER-to-Golgi transport in plant cells. J Cell Sci 131: jcs203893. [DOI] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Höning S, Evans PR, Owen DJ (2010) A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141: 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin E, Leclerc-Mercier S, Judon C, Blanchard E, Fraitag S, Florey O (2017) Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy 13: 854–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y-T, Simoneschi D, Keegan S, Melville D, Adler NS, Saraf A, Florens L, Washburn MP, Cavasotto CN, Fenyö D, et al. (2018) The ULK1-FBXW5-SEC23B nexus controls autophagy. eLife 7: e42253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Dahhan DA, Gnyliukh N, Kaufmann WA, Zheden V, Costanzo T, Mahou P, Hrtyan M, Wang J, Aguilera-Servin J, et al. (2021) The TPLATE complex mediates membrane bending during plant clathrin-mediated endocytosis. bioRxiv 2021.04.26.441441 [Advance Access publication on 26 March 2021] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Gnyliukh N, Kaufmann WA, Narasimhan M, Vert G, Bednarek SY, Friml J (2020). Experimental toolbox for quantitative evaluation of clathrin-mediated endocytosis in the plant model Arabidopsis. J Cell Sci 133: jcs248062. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Roux A (2018) Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19: 313–326 [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Era A, Minamino N, Shikano Y, Fujimoto M, Uemura T, Nishihama R, Yamato KT, Ishizaki K, Nishiyama T, et al. (2016) SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol 57: 307–324 [DOI] [PubMed] [Google Scholar]
- Kaneda M, van Oostende-Triplet C, Chebli Y, Testerink C, Bednarek SY, Geitmann A (2019) Plant AP180 N-terminal homolog proteins are involved in clathrin-dependent endocytosis during pollen tube growth in Arabidopsis thaliana. Plant Cell Physiol 60: 1316–1330 [DOI] [PubMed] [Google Scholar]
- Kang B-H, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kellner R, De la Concepcion JC, Maqbool A, Kamoun S, Dagdas YF (2017) ATG8 Expansion: a driver of selective autophagy diversification? Trends Plant Sci 22: 204–214 [DOI] [PubMed] [Google Scholar]
- Kelly BT, Graham SC, Liska N, Dannhauser PN, Höning S, Ungewickell EJ, Owen DJ (2014) AP2 controls clathrin polymerization with a membrane-activated switch. Science 345: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Späte K, Miller SE, Evans PR, Höning S, Owen DJ (2008) A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456: 976–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Xu Z-Y, Song K, Kim DH, Kang H, Reichardt I, Sohn EJ, Friml J, Juergens G, Hwang I (2013) Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T (2020) Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol 10: 190290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Owen D, Harrison SC (2014) Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol 6: a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J (2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. (2011) Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb C, Nagel M-K, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E (2015) FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol 167: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Dickson VK, Kelly BT, Owen DJ, Briggs JAG (2020) Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles. Sci Adv 6: eaba8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Brandizzi F (2020) The plant endoplasmic reticulum: an organized chaos of tubules and sheets with multiple functions. J Microsc 280: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V, Choi Y, Beck E, Wu Q, Luo A, Sylvester A, Jackson D, Chan AP (2015) A maize database resource that captures tissue-specific and subcellular-localized gene expression, via fluorescent tags and confocal imaging (Maize Cell Genomics Database). Plant Cell Physiol 56: e12. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Shen J, Lee MH, Geem KR, Jiang L, Hwang I (2018) AtCAP2 is crucial for lytic vacuole biogenesis during germination by positively regulating vacuolar protein trafficking. Proc Natl Acad Sci USA 115: E1675–E1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LTF, Yu C, Wong JSK, Lo HS, Benlekbir S, Jiang L, Lau WCY (2020) Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy 16: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam BC-H, Sage TL, Bianchi F, Blumwald E (2001) Role of SH3 domain-containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell 13: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-López G, Robinson DG, Aniento F (2008) In vivo trafficking and localization of p24 proteins in plant cells. Traffic 9: 770–785 [DOI] [PubMed] [Google Scholar]
- Lerich A, Hillmer S, Langhans M, Scheuring D, van Bentum P, Robinson DG (2012) ER import sites and their relationship to ER exit sites: a new model for bidirectional ER-Golgi transport in higher plants. Front Plant Sci 3: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) Autophagy-related11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li Y, Zhao Q, Li T, Wei J, Li B, Shen W, Yang C, Zeng Y, Rodriguez PL, et al. (2019) The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat Plants 5: 512–524 [DOI] [PubMed] [Google Scholar]
- Li X, Pan SQ (2017) Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci Adv 3: e1601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Sun R, Hicks GR, Raikhel NV (2015) Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc Natl Acad Sci USA 112: E89–E98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zeng Y, Zhu Y, Shen J, Ye H, Jiang L (2021) Plant Rho GTPase signaling promotes autophagy. Mol Plant 14: 905–920 [DOI] [PubMed] [Google Scholar]
- Liu F, Hu W, Li F, Marshall RS, Zarza X, Munnik T, Vierstra RD (2020b) AUTOPHAGY-RELATED14 and Its associated phosphatidylinositol 3-kinase complex promote autophagy in Arabidopsis. Plant Cell 32: 3939–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Kumar R, Claus LAN, Johnson AJ, Siao W, Vanhoutte I, Wang P, Bender KW, Yperman K, Martins S, et al. (2020a) Endocytosis of BRASSINOSTEROID INSENSITIVE1 is partly driven by a canonical Tyr-based motif. Plant Cell 32: 3598–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zeng Y, Li H, Yang C, Shen W, Xu M, Xiao Z, Chen T, Li B, Cao W, et al. (2021) A plant-unique ESCRT component, FYVE4, regulates multivesicular endosome biogenesis and plant growth. New Phytol 231: 193–209 [DOI] [PubMed] [Google Scholar]
- Lopez S, Rodriguez-Gallardo S, Sabido-Bozo S, Muñiz M (2019) Endoplasmic reticulum export of GPI-anchored proteins. Int J Mol Sci 20: 3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Otomo C, Otomo T (2019) The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8: e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T (2020) Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 27: 1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Sohn K, Wieland FT, Pepperkok R, Pizza M, Hillemann J, Söling H-D (1998) KDEL receptor (Erd2p)-mediated retrograde transport of the cholera toxin A subunit from the Golgi involves COPI, p23, and the COOH terminus of Erd2p. J Cell Biol 143: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Straub M, Hell SW, Duden R, Söling H-D (2001) KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev Cell 1: 139–153 [DOI] [PubMed] [Google Scholar]
- Maritzen T, Koo SJ, Haucke V (2012) Turning CALM into excitement: AP180 and CALM in endocytosis and disease. Biol Cell 104: 588–602 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69: 173–208 [DOI] [PubMed] [Google Scholar]
- Martens S (2016) No ATG8s, no problem? How LC3/GABARAP proteins contribute to autophagy. J Cell Biol 215: 761–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al. (2020) Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Matoba K, Noda NN (2021) Structural catalog of core Atg proteins opens new era of autophagy research. J Biochem 169: 517–525 [DOI] [PubMed] [Google Scholar]
- Mei Y, Jia W-J, Chu Y-J, Xue H-W (2012) Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Kaksonen M (2014) Endocytic accessory factors and regulation of clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol 6: a016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Chen P-H, Srinivasan S, Danuser G, Schmid SL (2018) Regulation of clathrin-mediated endocytosis. Annu Rev Biochem 87: 871–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz A, Hinrichsen L, Groos S, Esk P-C, Brandes G, Ungewickell EJ (2005) Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Höning S, Robinson MS, Peden AA, Owen DJ (2011) The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell 147: 1118–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino N, Ueda T (2019) RAB GTPases and their effectors in plant endosomal transport. Curr Opin Plant Biol 52: 61–68 [DOI] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV (2019) Models of intracellular transport: pros and cons. Front Cell Dev Biol 7: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunari T, Nakatsu F, Shioda N, Love PE, Grinberg A, Bonifacino JS, Ohno H (2005) Clathrin adaptor AP-2 is essential for early embryonal development. Mol Cell Biol 25: 9318–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Pastor-Cantizano N, Robinson DG, Marcote MJ, Aniento F (2014) Arabidopsis p24δ5 and p24δ9 facilitate Coat Protein I-dependent transport of the K/HDEL receptor ERD2 from the Golgi to the endoplasmic reticulum. Plant J 80: 1014–1030 [DOI] [PubMed] [Google Scholar]
- Montesinos JC, Sturm S, Langhans M, Hillmer S, Marcote MJ, Robinson DG, Aniento F (2012) Coupled transport of Arabidopsis p24 proteins at the ER-Golgi interface. J Exp Bot 63: 4243–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- More K, Klinger CM, Barlow LD, Dacks JB (2020) Evolution and natural history of membrane trafficking in eukaryotes. Curr Biol 30: R553–R564 [DOI] [PubMed] [Google Scholar]
- Mosesso N, Nagel M-K, Isono E (2019) Ubiquitin recognition in endocytic trafficking—with or without ESCRT-0. J Cell Sci 132: jcs232868. [DOI] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MNJ, Robinson MS (2003) Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulinier-Anzola J, Schwihla M, De-Araújo L, Artner C, Jörg L, Konstantinova N, Luschnig C, Korbei B (2020) TOLs function as ubiquitin receptors in the early steps of the ESCRT pathway in higher plants. Mol Plant 13: 717–731 [DOI] [PubMed] [Google Scholar]
- Muro K, Matsuura-Tokita K, Tsukamoto R, Kanaoka MM, Ebine K, Higashiyama T, Nakano A, Ueda T (2018) ANTH domain-containing proteins are required for the pollen tube plasma membrane integrity via recycling ANXUR kinases. Commun Biol 1: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel M-K, Kalinowska K, Vogel K, Reynolds GD, Wu Z, Anzenberger F, Ichikawa M, Tsutsumi C, Sato MH, Kuster B, et al. (2017) Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci USA 114: E7197–E7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano RT, Matsushima R, Ueda H, Tamura K, Shimada T, Li L, Hayashi Y, Kondo M, Nishimura M, Hara-Nishimura I (2009) GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21: 3672–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H (2020) Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458 [DOI] [PubMed] [Google Scholar]
- Narasimhan M, Johnson A, Prizak R, Kaufmann WA, Tan S, Casillas-Pérez B, Friml J (2020) Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 9: e52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead S, Barr F (2020) Molecular basis for KDEL-mediated retrieval of escaped ER-resident proteins—SWEET talking the COPs. J Cell Sci 133: jcs.250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, Lee MH, Song K, Ahn G, Lee J, Hwang I (2018) The A/ENTH domain-containing protein AtECA4 is an adaptor protein involved in cargo recycling from the trans-Golgi network/early endosome to the plasma membrane. Mol Plant 11: 568–583 [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ (2009) Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol 21: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M, Takemoto N, Hashiguchi Y, Tasaka M, Morita MT (2009) ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2017) Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr Opin Plant Biol 40: 22–33 [DOI] [PubMed] [Google Scholar]
- Nonet ML, Holgado AM, Brewer F, Serpe CJ, Norbeck BA, Holleran J, Wei L, Hartwieg E, Jorgensen EM, Alfonso A (1999) UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol Biol Cell 10: 2343–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier M-C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS (1995) Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269: 1872–1875 [DOI] [PubMed] [Google Scholar]
- Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, Noda NN (2019) Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26: 281–288 [DOI] [PubMed] [Google Scholar]
- Pacheco-Fernandez N, Pakdel M, Von Blume J (2021) Retention Using Selective Hooks (RUSH) cargo sorting assay for protein vesicle tracking in HeLa cells. Bio Protoc 11: e3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS (2016) Endocytosis and endosomal trafficking in plants. Annu Rev Plant Biol 67: 309–335 [DOI] [PubMed] [Google Scholar]
- Park M, Kim SJ, Vitale A, Hwang I (2004) Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol 134: 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons HT, Christiansen K, Knierim B, Carroll A, Ito J, Batth TS, Smith-Moritz AM, Morrison S, McInerney P, Hadi MZ, et al. (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Bernat-Silvestre C, Marcote MJ, Aniento F (2018) Loss of Arabidopsis p24 function affects ERD2 trafficking and Golgi structure, and activates the unfolded protein response. J Cell Sci 131: jcs203802. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, García-Murria MJ, Bernat-Silvestre C, Marcote MJ, Mingarro I, Aniento F (2017) N-linked glycosylation of the p24 family protein p24δ5 modulates retrograde Golgi-to-ER transport of K/HDEL ligands in Arabidopsis. Mol Plant 10: 1095–1106 [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Montesinos JC, Bernat-Silvestre C, Marcote MJ, Aniento F (2016) p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253: 967–985 [DOI] [PubMed] [Google Scholar]
- Peixoto B, Pereira S, Pissarra J (2016) Plant vacuolar sorting: an overview. In Cánovas F, Lüttge U, Matyssek R, eds, Progress in Botany, Vol 78. Springer, Cham, Switzerland, pp 67–94 [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuyal S, Farhan H (2021) Want to leave the ER? We offer vesicles, tubules, and tunnels. J Cell Biol 220: e202104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco A, Mund M, Ries J, Nédélec F, Kaksonen M (2015) Visualizing the functional architecture of the endocytic machinery. eLife 4: e04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, del Mar Marquès-Bueno M, Creff A, Maneta-Peyret L, Fiche J-B, Nollmann M, Miège C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62 [DOI] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018) A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell 45: 465–480 [DOI] [PubMed] [Google Scholar]
- Popoff V, Adolf F, Brügger B, Wieland F (2011) COPI budding within the Golgi stack. Cold Spring Harb Perspect Biol 3: a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Pleskot R, Irani NG, Van Damme D (2018) Meeting report—cellular gateways: expanding the role of endocytosis in plant development. J Cell Sci 131: jcs.222604. [DOI] [PubMed] [Google Scholar]
- Raote I, Malhotra V (2021) Tunnels for protein export from the endoplasmic reticulum. Annu Rev Biochem 90: 605–663 [DOI] [PubMed] [Google Scholar]
- Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, Randow F (2019) The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell 74: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar R, Steiner A, Assaad FF (2017) Multisubunit tethering complexes in higher plants. Curr Opin Plant Biol 40: 97–105 [DOI] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B (2009) Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J 28: 3103–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Pan T, Wang Y, Wang Y, Gan L, Wei Z, Wang F, Wu M, Jing R, et al. (2020) GPA5 encodes a Rab5a effector required for post-Golgi trafficking of rice storage proteins. Plant Cell 32: 758–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna L, Brandizzi F (2020) The mysterious life of the plant trans-Golgi network: advances and tools to understand it better. J Microsc 278: 154–163 [DOI] [PubMed] [Google Scholar]
- Reynolds GD, Wang C, Pan J, Bednarek SY (2018) Inroads into internalization: five years of endocytic exploration. Plant Physiol. 176: 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenführ A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG (2002) Reevaluation of the effects of Brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG (2020) Plant Golgi ultrastructure. J Microsc 280: 111–121 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Aniento F (2020) A model for ERD2 function in higher plants. Front Plant Sci 11: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Brandizzi F, Hawes C, Nakano A (2015) Vesicles versus tubes: is endoplasmic reticulum-Golgi transport in plants fundamentally different from other eukaryotes? Plant Physiol 168: 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Furlan C, Domozych D, Qian W, Enquist P-A, Li X, Zhang C, Schenk R, Winbigler HS, Jackson W, Raikhel NV, et al. (2019b) Interaction between VPS35 and RABG3f is necessary as a checkpoint to control fusion of late compartments with the vacuole. Proc Natl Acad Sci USA 116: 21291–21301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Furlan C, Minina EA, Hicks GR (2019a) Remove, recycle, degrade: regulating plasma membrane protein accumulation. Plant Cell 31: 2833–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gallardo S, Kurokawa K, Sabido-Bozo S, Cortes-Gomez A, Ikeda A, Zoni V, Aguilera-Romero A, Perez-Linero AM, Lopez S, Waga M, et al. (2020) Ceramide chain length-dependent protein sorting into selective endoplasmic reticulum exit sites. Sci Adv 6: eaba8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV (2003) The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol Biol Cell 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Santiago P, Lagunas-Gómez D, Barkla BJ, Vera-Estrella R, Lalonde S, Jones A, Frommer WB, Zimmermannova O, Sychrová H, Pantoja O (2015) Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT13. J Exp Bot 66: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Santiago P, Lagunas-Gomez D, Yáñez-Domínguez C, Vera-Estrella R, Zimmermannová O, Sychrová H, Pantoja O (2017) Plant and yeast cornichon possess a conserved acidic motif required for correct targeting of plasma membrane cargos. Biochim Biophys Acta 1864: 1809–1818 [DOI] [PubMed] [Google Scholar]
- Rosendale M, Van TNN, Grillo-Bosch D, Sposini S, Claverie L, Gauthereau I, Claverol S, Choquet D, Sainlos M, Perrais D (2019) Functional recruitment of dynamin requires multimeric interactions for efficient endocytosis. Nat Commun 10: 4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, Davis DJ, Drakakaki G (2018) The plant trans-Golgi network: not just a matter of distinction. Plant Physiol 176: 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai HT, Inoue T, Nakano A, Ueda T (2016) Endosomal RAB effector with PX-domain, an interacting partner of RAB5 GTPases, regulates membrane trafficking to protein storage vacuoles in Arabidopsis. Plant Cell 28: 1490–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šamaj J, Baluška F, Voigt B, Schlicht M, Volkmann D, Menzel D (2004) Endocytosis, actin cytoskeleton, and signaling. Plant Physiol 135: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Shi Y, Kesten C, Zhang D, Sancho-Andrés G, Ivakov A, Lampugnani ER, Sklodowski K, Fujimoto M, Nakano A, et al. (2018) The cellulose synthases are cargo of the TPLATE adaptor complex. Mol Plant 11: 346–349 [DOI] [PubMed] [Google Scholar]
- Sánchez-Simarro J, Bernat-Silvestre C, Aniento F, Marcote MJ (2021) ß-COP mutants show specific high sensitivity to chloride ions. Plant Signal Behav 16: 1858629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Simarro J, Bernat-Silvestre C, Gimeno-Ferrer F, Selvi-Martínez P, Montero-Pau J, Aniento F, Marcote MJ (2020) Loss of Arabidopsis β-COP function affects Golgi structure, plant growth and tolerance to salt stress. Front Plant Sci 11:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Andrés G, Soriano-Ortega E, Gao C, Bernabé-Orts JM, Narasimhan M, Müller AO, Tejos R, Jiang L, Friml J, Aniento F, et al. (2016) Sorting motifs involved in the trafficking and localization of the PIN1 auxin efflux carrier. Plant Physiol 171: 1965–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Prydz K (2019) A new look at the functional organization of the Golgi ribbon. Front Cell Dev Biol 7: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ueda T, Nakano A (1999) The Arabidopsis thaliana RER1 gene family: its potential role in the endoplasmic reticulum localization of membrane proteins. Plant Mol Biol 41: 815–824 [DOI] [PubMed] [Google Scholar]
- Sawa-Makarska J, Baumann V, Coudevylle N, von Bülow S, Nogellova V, Abert C, Schuschnig M, Graef M, Hummer G, Martens S (2020) Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 369: eaaz7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel AA, Pelham HR (1996) Purification and characterization of the human KDEL receptor. Biochemistry 35: 10203–10209 [DOI] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, et al. (2011) Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwihla M, Korbei B (2020) The beginning of the end: initial steps in the degradation of plasma membrane proteins. Front Plant Sci 11: 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BL, Sochacki KA, Low-Nam ST, Bailey EM, Luu Q, Hor A, Dickey AM, Smith S, Kerkvliet JG, Taraska JW, et al. (2018) Membrane bending occurs at all stages of clathrin-coat assembly and defines endocytic dynamics. Nat Commun 9: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K-I, Maeshima M, Yokota A, Tomizawa K-I, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45: 672–683 [DOI] [PubMed] [Google Scholar]
- Shomron O, Nevo-Yassaf I, Aviad T, Yaffe Y, Zahavi EE, Dukhovny A, Perlson E, Brodsky I, Yeheskel A, Pasmanik-Chor M, et al. (2021) COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J Cell Biol 220: e201907224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Alvim FAL, An J, Alvim JC, Foresti O, Grippa A, Pelgrom A, Adams TL, Hawes C, Denecke J (2018) Predominant Golgi residency of the plant K/HDEL receptor is essential for its function in mediating ER retention. Plant Cell 30: 2174–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MLA, Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud M-C, Jaillais Y (2016) A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R, Rosquete MR, Drakakaki G (2018) Post-Golgi trafficking and transport of cell wall components. Front Plant Sci 9: 1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S (2018) CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell 44: 217–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Spang A (2013) Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J 280: 2743–2757 [DOI] [PubMed] [Google Scholar]
- Song K, Jang M, Kim SY, Lee G, Lee G-J, Kim DH, Lee Y, Cho W, Hwang I (2012) An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol 159: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Li F, Buono R, Roschzttardtz H, Chung T, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS (2015) The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Emr SD (1994) Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem 269: 31552–31562 [PubMed] [Google Scholar]
- Starai VJ, Hickey CM, Wickner W (. 2008) HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell 19: 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephani M, Dagdas Y (2020) Plant selective autophagy—still an uncharted territory with a lot of hidden gems. J Mol Biol 432: 63–79 [DOI] [PubMed] [Google Scholar]
- Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Sanchez de Medina Hernandez V, Mohseni A, Clavel M, Zeng Y, Naumann C, et al. (2020) A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 9: e58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16: 495–501 [DOI] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W (2006) Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J 25: 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutipatanasomboon A, Herberth S, Alwood EG, Häweker H, Müller B, Shahriari M, Zienert AY, Marin B, Robatzek S, Praefcke GJK, et al. (2017) Disruption of the plant-specific CFS1 gene impairs autophagosome turnover and triggers EDS1-dependent cell death. Sci Rep 7: 8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Kimori Y, Shimada T, Hara-Nishimura I (2020) Dynamic capture and release of endoplasmic reticulum exit sites by Golgi stacks in Arabidopsis. iScience 23: 101265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Renna L, Takahashi H, Koumoto Y, Tamura K, Stefano G, Fukao Y, Kondo M, Nishimura M, Shimada T, et al. (2013) MAIGO5 functions in protein export from Golgi-associated endoplasmic reticulum exit sites in Arabidopsis. Plant Cell 25: 4658–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, He H, Tang Z, Hattori T, Liu Y, Young MM, Serfass JM, Chen L, Gebru M, Chen C, et al. (2018) An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9: 2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Liang X, Hattori T, Tang Z, He H, Chen H, Liu X, Abraham T, Imamura-Kawasawa Y, et al. (2019) VPS37A directs ESCRT recruitment for phagophore closure. J Cell Biol 218: 3336–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Ebine K, Askani JC, Krüger F, Gonzalez ZA, Ito E, Goh T, Schumacher K, Nakano A, Ueda T (2018) Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc Natl Acad Sci USA 115: E2457–E2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer D, Weiner OD (2014) Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol 15: 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Bonifacino JS (2013) Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol 5: a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco E, Fracchiolla D, Martens S (2020) Recruitment and activation of the ULK1/Atg1 kinase complex in selective autophagy. J Mol Biol 432: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco E, Witt M, Abert C, Bock-Bierbaum T, Su M-Y, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, et al. (2019) FIP200 Claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol Cell 74: 330–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T (2016) Physiological Roles of plant post-Golgi transport pathways in membrane trafficking. Plant Cell Physiol 57: 2013–2019 [DOI] [PubMed] [Google Scholar]
- Uemura T, Morita MT, Ebine K, Okatani Y, Yano D, Saito C, Ueda T, Nakano A (2010) Vacuolar/pre-vacuolar compartment Qa-SNAREs VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis. Plant J 64: 864–873 [DOI] [PubMed] [Google Scholar]
- Uemura T, Suda Y, Ueda T, Nakano A (2014) Dynamic behavior of the trans-Golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol 4: 694–703 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T (2014) Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr Opin Plant Biol 22: 116–121 [DOI] [PubMed] [Google Scholar]
- Ugalde J-M, Rodriguez-Furlán C, De Rycke R, Norambuena L, Friml J, León G, Tejos R (2016) Phosphatidylinositol 4-phosphate 5-kinases 1 and 2 are involved in the regulation of vacuole morphology during Arabidopsis thaliana pollen development. Plant Sci 250: 10–19 [DOI] [PubMed] [Google Scholar]
- Umasankar PK, Ma L, Thieman JR, Jha A, Doray B, Watkins SC, Traub LM (2014) A clathrin coat assembly role for the muniscin protein central linker revealed by TALEN-mediated gene editing. eLife 3: e04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ (2019) ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218: 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Coutuer S, De Rycke R, Bouget F-Y, Inzé D, Geelen D (2006) Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18: 3502–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MCE, De Jaeger G, Russinova E, Geelen D (2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci USA 108: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R, Jonker CTH, Wijdeven RH, Bakker J, Janssen L, Klumperman J, Neefjes J (2015) Characterization of the mammalian CORVET and HOPS complexes and their modular restructuring for endosome specificity. J Biol Chem 290: 30280–30290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, Youle RJ (2019) Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell 74: 347–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti R, Wilson C, De Matteis MA (2014) Exiting the ER: what we know and what we don't. Trends Cell Biol 24: 9–18 [DOI] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof Y-D, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukašinović N, Žárský V (2016) Tethering complexes in the Arabidopsis endomembrane system. Front Cell Dev Biol 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Miao Y, Lam SK, Jiang L (2009) Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mylle E, Johnson A, Besbrugge N, De Jaeger G, Friml J, Pleskot R, Van Damme D (2020b) High temporal resolution reveals simultaneous plasma membrane recruitment of TPLATE complex subunits. Plant Physiol 183: 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, et al. (2019) Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun 10: 5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu M, Gao C, Zeng Y, Cui Y, Shen W, Jiang L (2020a) The roles of endomembrane trafficking in plant abiotic stress responses. J Integr Plant Biol 62: 55–69 [DOI] [PubMed] [Google Scholar]
- Wang C, Yan X, Chen Q, Jiang N, Fu W, Ma B, Liu J, Li C, Bednarek SY, Pan J (2013) Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25: 499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yoshinari A, Shimada T, Hara-Nishimura I, Mitani-Ueno N, Feng Ma J, Naito S, Takano J (2017) Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. Plant Cell 29: 824–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yperman K, Grones P, Jiang Q, Dragwidge J, Mylle E, Mor E, Nolf J, Eeckhout D, De Jaeger G, et al. (2021) Conditional destabilization of the TPLATE complex impairs endocytic internalization. Proc Natl Acad Sci USA 118: e2023456118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel AV, Chang C-L, Shtengel G, Xu CS, Hoffman DP, Freeman M, Iyer N, Aaron J, Khuon S, Bogovic J, et al. (2021) ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184: 2412–2429 [DOI] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG (2012) Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol 22: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 26: 115–136 [DOI] [PubMed] [Google Scholar]
- Wilson DW, Lewis MJ, Pelham HR (1993) pH-dependent binding of KDEL to its receptor in vitro. J Biol Chem 268: 7465–7468 [PubMed] [Google Scholar]
- Wirth M, Zhang W, Razi M, Nyoni L, Joshi D, O'Reilly N, Johansen T, Tooze SA, Mouilleron S (2019) Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat Commun 10: 2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenstetter S, Wirsching P, Dotzauer D, Schneider S, Sauer N (2012) Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell 24: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CH, Gao C, Yu P, Tu L, Meng Z, Banfield DK, Yao X, Jiang L (2015) Conserved function of the lysine-based KXD/E motif in Golgi retention for endomembrane proteins among different organisms. Mol Biol Cell 26: 4280–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Park E, Dinesh-Kumar SP (2014) Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA 111: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280: 979–993 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Yang C, Liu C, Yang L, Yang S, Zhou J, Li F, Jiang L, Xiao S, Gao C, et al. (2020) SINAT E3 ligases regulate the stability of the ESCRT component FREE1 in response to iron deficiency in plants. J Integr Plant Biol 62: 1399–1417 [DOI] [PubMed] [Google Scholar]
- Xu CS, Hayworth KJ, Lu Z, Grob P, Hassan AM, García-Cerdán JG, Niyogi KK, Nogales E, Weinberg RJ, Hess HF (2017) Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6: e25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Liu Y (2012) Plant ERD2s self-interact and interact with GTPase-activating proteins and ADP-ribosylation factor 1. Plant Signal Behav 7: 1092–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Shimono Y, Shirakawa M, Fukao Y, Kawase T, Hatsugai N, Tamura K, Shimada T, Hara-Nishimura I (2013) Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25: 2958–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ohnishi M, Fukao Y, Okazaki Y, Fujiwara M, Song C, Nakanishi Y, Saito K, Shimmen T, Suzaki T, et al. (2013) Studies on vacuolar membrane microdomains isolated from Arabidopsis suspension-cultured cells: local distribution of vacuolar membrane proteins. Plant Cell Physiol 54: 1571–1584 [DOI] [PubMed] [Google Scholar]
- Yoshinari A, Hosokawa T, Amano T, Beier MP, Kunieda T, Shimada T, Hara-Nishimura I, Naito S, Takano J (2019) Polar localization of the borate exporter BOR1 requires AP2-dependent endocytosis. Plant Physiol 179: 1569–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari A, Kasai K, Fujiwara T, Naito S, Takano J (2012) Polar localization and endocytic degradation of a boron transporter, BOR1, is dependent on specific tyrosine residues. Plant Signal Behav 7: 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yperman K, Papageorgiou AC, Merceron R, De Munck S, Bloch Y, Eeckhout D, Jiang Q, Tack P, Grigoryan R, Evangelidis T, et al. (2021b) Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding. Nat Commun 12: 3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yperman K, Wang J, Eeckhout D, Winkler J, Vu LD, Vandorpe M, Grones P, Mylle E, Kraus M, Merceron R, et al. (2021a) Molecular architecture of the endocytic TPLATE complex. Sci Adv 7: eabe7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S (2016) Mechanisms of selective autophagy. J Mol Biol 428: 1714–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong K-B, Schekman R, Jiang L (2015) Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 14360–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Li B, Ji C, Feng L, Niu F, Deng C, Chen S, Lin Y, Cheung KCP, Shen J, et al. (2021) A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 118: e2021293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zess EK, Jensen C, Cruz-Mireles N, De la Concepcion JC, Sklenar J, Stephani M, Imre R, Roitinger E, Hughes R, Belhaj K, et al. (2019) N-terminal β-strand underpins biochemical specialization of an ATG8 isoform. PLoS Biol 17: e3000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hicks GR, Raikhel NV (2015b) Molecular composition of plant vacuoles: important but less understood regulations and roles of tonoplast lipids. Plants 4: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li H, Lu H, Hwang I (2021) The trafficking machinery of lytic and protein storage vacuoles: how much is shared and how much is distinct? J Exp Bot 72:3504–3512 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Persson S, Hirst J, Robinson MS, Van Damme D, Sánchez-Rodríguez C (2015a) Change your TPLATE, change your fate: plant CME and beyond. Trends Plant Sci 20: 41–48 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yan A, Feijó JA, Furutani M, Takenawa T, Hwang I, Fu Y, Yang Z (2010) Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 22: 4031–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Zhang H (2019) Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol 218: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Spangenberg H, Munson MJ, Brech A, Schink KO, Tan K-W, Sørensen V, Wenzel EM, Radulovic M, Engedal N, et al. (2020) ESCRT-mediated phagophore sealing during mitophagy. Autophagy 16: 826–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Han SW, Munnik T, Rojas-Pierce M (2014a) Multiple vacuoles in impaired tonoplast trafficking3 mutants are independent organelles. Plant Signal Behav 9: e972113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Han SW, Rodriguez-Welsh MF, Rojas-Pierce M (2014b) Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol Plant 7: 1026–1040 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Cui Y, Gao C, Jiang L (2015) Endocytic and autophagic pathways crosstalk in plants. Curr Opin Plant Biol 28: 39–47 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Jiang L (2014) Autophagosome biogenesis in plants: roles of SH3P2. Autophagy 10: 704–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizioli D, Meyer C, Guhde G, Saftig P, von Figura K, Schu P (1999) Early embryonic death of mice deficient in γ-adaptin. J Biol Chem 274: 5385–5390 [DOI] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV (2007) Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci USA 104: 3793–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Sauer M (2014) Helping hands for budding prospects: ENTH/ANTH/VHS accessory proteins in endocytosis, vacuolar transport, and secretion. Plant Cell 26: 4232–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Nishizawa N, Wu Y, Kost B, Chua N-H (2000) KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M, Feraru E, Möller B, Hwang I, Feraru MI, Kleine-Vehn J, Weijers D, Friml J (2011) The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.