Abstract
Epigenomics is the study of molecular signatures associated with discrete regions within genomes, many of which are important for a wide range of nuclear processes. The ability to profile the epigenomic landscape associated with genes, repetitive regions, transposons, transcription, differential expression, cis-regulatory elements, and 3D chromatin interactions has vastly improved our understanding of plant genomes. However, many epigenomic and single-cell genomic assays are challenging to perform in plants, leading to a wide range of data quality issues; thus, the data require rigorous evaluation prior to downstream analyses and interpretation. In this commentary, we provide considerations for the evaluation of plant epigenomics and single-cell genomics data quality with the aim of improving the quality and utility of studies using those data across diverse plant species.
Useful consideration for quality control and evaluation of plant epigenomics data are provided.
Introduction
High-throughput sequencing has revolutionized the study of plant epigenomes. The cost of sequencing has never been cheaper, which has shifted the bottleneck to sample collection, epigenomic assays, sequencing library preparation, and data analysis. Plant samples are readily evaluated for transcript and small RNA (smRNA) abundance, as well as the patterns and distribution of transcription factor (TF) binding sites, DNA methylation, histone modifications, and chromatin accessibility. Some of these assays have even been adopted for use in single cells. However, with the democratization of sequencing, challenges arise with regard to the proper evaluation of experimental and data quality. Data quality can have numerous definitions, but here we use this term to refer to the quality and outcome of the overall experiment and its usefulness for uncovering biological phenomena and not to the “sequencing quality” scores generated by sequencing instruments. The aim of this commentary is to raise awareness of data quality standards, define common pitfalls and issues associated with large-scale experiments, and provide considerations that we suggest colleagues embrace when generating and analyzing plant epigenomics data.
What distinguishes transcriptome and epigenome assays from genome sequencing is biological (as opposed to genotypic) and experimental variability (dependent on endogenous and exogenous stimuli as well as the complexity of the assay itself). As a result, whole-genome sequencing (WGS) typically does not require biological replicates, whereas the quantitative nature of transcriptomics and epigenomics typically does. For example, for genome sequencing, the identification of single-nucleotide (nt) polymorphisms can be viewed as a qualitative difference at a single position between two genotypes. Given sufficient sequencing depth and uniform genome coverage, these binary differences between genotypes are readily identified in the absence of replication. However, comparisons of transcript or smRNA abundance between samples are often used in a quantitative manner and, as a result, must be accompanied by biological replication to support the major claims.
Replicates from distinct biological samples are required to exclude technical and biological variance not associated with the variable of interest, especially in the context of studies that draw conclusions based on quantitative changes in gene expression (Schurch et al., 2016). Technical replicates (within an experiment) provide insight into variation associated with an assay, but they do not describe the range of results expected within a population (Vaux et al., 2012). While unreplicated studies have been published for many types of “-seq” data, such as single-cell transcriptome sequencing and early studies of chromatin immunoprecipitation (ChIP) coupled with sequencing, most fields have matured to the point in which the need for proper biological replication outweighs the “wow factor” of the new technology; similar shifts have occurred going back to microarray studies, RNA-seq, and a wide variety of “-seq” studies. That is, the first few papers describing a new technology were often accepted with minimal biological replication, because the method is novel, likely to be of high value to the scientific community, and therefore deemed worthy of rapid publication. Scientific advances often require a delicate balance between the development and application of new methods. In the early days of most technologies and data-rich approaches, costs are high. However, at a certain point, as the approach becomes more routine for biological exploration and the associated costs (typically) decline, the essential components of good experimental design, including biological replication, must come to the fore.
Evaluation of enrichment and additional quality controls
A distinguishing feature of transcriptome/epigenome sequencing approaches compared to genome sequencing is the reliance on the enrichment of molecules from a larger pool, often using biochemical approaches. For example, most RNA-sequencing (RNA-seq) experiments rely on the enrichment of mRNA from total RNA by the capture of mRNA via poly(A) tails. For smRNAs, size exclusion is used as a form of enrichment. To detect TF binding or histone modification abundance using ChIP-sequencing (ChIP-seq), antibodies are required to capture specific protein : DNA interactions. Similarly, the detection of accessible chromatin requires the isolation of chromatin, followed by enzymatic treatment to enrich nucleosome-depleted regions. The need to enrich for RNA, smRNA, and/or chromatin increases the complexity of these assays, resulting in an increase in both technical and biological variability in the data linked to the assay materials (i.e. kit components, chemicals, or plant tissue quality), the individual experimenter(s), or the location where the experiment was performed.
In this commentary, we provide some useful considerations for replication and quality control metrics that can be used to evaluate enrichment/data quality for common transcriptome and epigenome assays (Table 1). The evaluation of these metrics will help researchers determine if experiments should be repeated or if caution should be taken with interpretation of the data. Adding quality control metrics to supplemental material is commonly accepted by journals for some assays, yet lacking for others. We strongly believe that including some of these quality control metrics for high-throughput sequencing-based experiments will improve the ability of readers to accurately evaluate the results both pre and postpublication. We describe several issues that are common among multiple types of assay, followed by a description of data type-specific considerations.
Table 1.
Recommendations for important checks and controls for plant epigenome assays
| Important Checks and Controls | RNA-seq | smRNA-seq | WGBS | ChIP-seq | Chromatin Accessibility | Single-cell RNA-seq | Single-cell ATAC-seq |
|---|---|---|---|---|---|---|---|
| Low level of duplicate reads | x | x | x | x | x | x | x |
| Appropriate normalization | x | x | x | x | x | x | x |
| Report number of sequenced and aligned reads | x | x | x | x | x | x | x |
| Show representative genome browser screen shots (including replicates) | x | x | x | x | x | x | x |
| Evaluate RNA/DNA quality | x | x | x | ||||
| High and/or consistent alignment rates | x | x | x | x | x | x | x |
| Show Venn diagram or Upset plots of identified clusters/regions/peaks between replicates | x | x | x | x | |||
| Aligned smRNA sequences are enriched for 21–24 nt sizes | x | ||||||
| Report bisulfite conversion rates | x | ||||||
| Report read coverage of the genome | x | ||||||
| Report SPOT or FRiP scores | x | x | x | ||||
| Implement IDR with replicates | x | ||||||
| Include input or IP background control | x | ||||||
| Plot read coverage around genomic features (genes/TEs/TSSs, etc.) | x | x | x | ||||
| Consider a spike-in control | x | x | |||||
| Evaluate enzymatic bias using genomic DNA control | x | ||||||
| Report number of cells targeted | x | x | |||||
| Report number of unique transcripts/Tn5 integrations per cell | x | x | |||||
| Evaluate marker genes | x | x | |||||
| Filter cells with high proportion of organellar reads | x |
PCR bias
When possible, the amount of input material used for each epigenomic library should be standardized to reduce variation that can be attributed to polymerase chain reaction (PCR) bias. The number of PCR cycles should be minimized to reduce PCR bias and duplicate reads. Duplicate reads are commonly used as a measure of library quality and complexity. A duplicate read is one that has the same start and end point and is often the result of PCR duplication as opposed to an independent biological event. In-depth analyses of sequenced libraries require careful inspection of the level of enrichment achieved. Unfortunately, although there are methods to evaluate enrichment and data quality, they are rarely included in the presentation of data in manuscripts, which shifts the burden of evaluating data quality to the reader or user, postpublication.
Sequence alignments
For all transcriptome and epigenome data types generated from sequencing, it is important to have an understanding of the limitations of sequence alignments. There are numerous strategies for the alignment of sequenced reads, depending on the scientific question being pursued as well as downstream analyses and data interpretation. Arguably, just as important is an understanding of the limitations of current genome assemblies and annotations as well as computational packages designed to work with well-assembled genomes. We are fortunate to be able to conduct these experiments in an era when high-quality reference genomes exist for many plants, and new assemblies and versions are continually being released. However, even for these high-quality genomes, assembly and annotation errors still exist that can lead to erroneous results (Mendieta et al., 2021).
Challenges with “peak” identification
Analysis of transcriptomic and epigenomic data requires dedicated pipelines for which the subject of the analysis dictates how the data should be processed. For example, in RNA-seq, comparisons are typically made at the level of transcripts or gene loci, which provides a fixed set of regions that are generally uniquely mapped and supported by prior analysis and data, such as genome assembly, annotation, cDNA support from Expressed Sequence Tags, and previous RNA-seq data. Analysis of epigenomic data is typically more challenging, given that regions of sequencing coverage enrichment (i.e. regions that resolve into peaks) have rarely been preidentified using a gold standard approach and can be associated with repetitive DNA regions that are difficult to assemble. That is, an epigenomic experiment may identify novel peaks that were not previously observed and reported, thus making it challenging to cross-check or validate those results using existing data.
Another major consideration is the evaluation of peak quality and quantity between samples. The main goal of smRNA-seq, ChIP-seq, and chromatin accessibility mapping experiments is to identify regions in the genome that are enriched for biological signals (i.e. have a significant density of stacked sequenced reads; Figure 1). Upon sequencing, these regions are often referred to as peaks; an array of software packages have been designed to identify these regions in a genome wide-fashion (Ji et al., 2008; Zhang et al., 2008; Rozowsky et al., 2009; Heinz et al., 2010; Guo et al., 2012; Xu et al., 2014; Ibrahim et al., 2015; Meers et al., 2019). These software packages perform signal processing steps in an attempt to approximate human judgment and generally involve smoothing and cutoff parameters. Given the wide variety of signal shapes that can occur within and between data types, there is no gold standard for these parameter selections. However, when biological replicates are available, there are recommended procedures to perform after a peak caller is applied. When biological replicates are sequenced, correlations between datasets can be used to evaluate data quality and to estimate high-confidence target peaks. For example, it is common practice to use correlation analyses between biological replicates to evaluate RNA-seq samples. Comparisons of genome-wide transcript abundance from entire organs for each gene often reveal correlations ˃0.95. However, for epigenomic assays, such as ChIP-seq and chromatin accessibility mapping, correlations of reads in peaks (as opposed to genes) should be used to evaluate consistency between biological replicates, because read coverage of large predefined genomic bins (e.g. 10 or 100 kilobase [kb]) obfuscates technical and experimental variation.
Figure 1.

Accounting for mappability and genome assembly artifacts. A schematic diagram of typical aligned data obtained by ATAC-seq. Regions of chromatin accessibility are indicated by peaks. A region that appears enriched for sequencing coverage (labeled “false peak”) is actually due to a collapsed repeat in the genome assembly. Tn5-treated genomic DNA helps to identify these problematic regions. A k-mer-based approach is used to reveal regions of the genome that are uniquely mappable for a given sequence fragment length.
Another method used to evaluate peak quality and sample-to-sample variation involves the irreproducible discovery rate (IDR; Li et al., 2011). This method uses a statistical framework to compare the signal ranks of peaks, providing IDR values for all peaks across biological replicates. A comparison of the number of peaks passing the determined IDR threshold between true and re-sampled pseudo-replicates allows the numeric quality of replicated enrichment experiments to be assessed. According to the standards set by the ENCODE project, the ratio between peaks enriched in replicates and pseudo-replicates should be between 0.5 and 2.0 (Landt et al., 2012). A large proportion of peaks above the IDR threshold indicate substantial discrepancies between signals among replicates, which is indicative of poor reproducibility. We note that large variation in the number of sequencing reads between biological replicates can result in variable “peak” detection due to the sensitivity of identification of rare or cell-type-specific peaks (Jung et al., 2014).
Controls that account for assembly quality and the sequence alignment strategy
Some of the most challenging-to-study regions of any genome are the repeats (e.g. transposons [TEs] or centromeric regions), as they confound most assembly algorithms and are often collapsed into single units in assemblies, even though they exist as complex arrays in the genome. Due to their repetitive nature, these regions often appear as “peaks” in any coverage-based assay such as ChIP-seq and chromatin accessibility mapping, even if the actual result is not an enrichment or peak. In other words, the collapse of the repeats in the genome assembly can yield misleading artifacts. Peaks occurring as a result of a genome assembly or sequence alignment strategy obscure true peaks that result from biological signals. One strategy to evaluate the sequence alignment approach and to manage these problematic regions of the genome is to determine sequence mappability using a k-mer-based approach that matches the read length from a library (Derrien et al., 2012). An alternative approach is to compare these problematic regions that result from mis-assembly or repeat regions to a genomic DNA sequencing library, such as WGS or ChIP-seq/chromatin accessibility mapping input (Figure 1). Next, for coverage-based enrichment assays, determine how many “peaks” are identified from the genomic DNA library (Figure 1). Any peaks found in this control library within mappable regions of the genome are likely artifacts of the alignment strategy and/or the genome assembly (Figure 1; Pickrell et al., 2011). An alternative approach is to use the input or WGS alignments as a baseline to measure enrichment (Figure 1). By knowing which regions of the genome behave in this manner, those regions can be excluded from downstream analyses, reducing associated erroneous results.
Data visualization
A valuable tool that should be used to quickly assess the quality of transcriptomic and epigenomics data is a genome browser, such as the Integrative Genomics Viewer (Thorvaldsdottir et al., 2013) or JBrowse (Buels et al., 2016; Hofmeister and Schmitz, 2018). These genome browsers allow aligned data and/or processed files to be visualized. The visualized data can be used to roughly evaluate the uniformity of coverage or the enrichment of transcripts, smRNAs, DNA methylation, or peaks around expected genomic regions. The data can also be used to evaluate the accuracy of software used to identify differentially methylated regions and/or differential peaks. It should be noted that genome browsers and associated screenshots do not supplant quantitative measurements of the data, but they are important tools for data presentation and are recommended for the additional evaluation of data quality.
Data type-specific considerations
RNA-seq
There are numerous applications of RNA-seq technologies, but in this section, we will focus specifically on the sequencing of mRNAs. Quantitative and qualitative measurement of RNA is a cornerstone of genome biology. Measuring mRNAs is the most common: the data are used to annotate genomes, discover splice isoforms, and most often to estimate steady-state transcript levels. The estimates of transcript levels are typically compared across treatments, genotypes, or tissue types to infer or determine which genes contribute to key differences in the compared materials.
Important checks and controls
First and foremost, RNA integrity should be evaluated prior to library preparation (e.g. with a fragment bioanalyzer), as poor-quality RNA can lead to erroneous artifacts. In most cases, biological triplicates are sufficient for the identification of differentially expressed genes, although the number can vary depending on the study system, the question being posed, and the sensitivity needed from the analysis. Key indicators of high-quality RNA-seq libraries include high and consistent sequence alignment percentages, low levels of duplicate reads, and uniform distribution of reads across transcript annotations. Low or inconsistent alignment percentages between samples likely indicate a problem with RNA quality. These types of libraries often possess many more nonpoly(A)-tailed RNAs compared to high-quality libraries. One emerging method that is useful for evaluating duplicate reads is to use unique molecular identifiers, which are commonly used in single-cell genomics. In parallel, incorporating spike-in RNAs is useful for estimating transcript abundance. A variety of methods are used to normalize RNA-seq data, but the two main approaches are normalizing by library size and transcript length or implementing a quantile normalization approach. The latter is typically used when changes in transcript abundance are not normally distributed.
smRNA-seq
Plant smRNAs (here defined as RNAs produced by Dicer-like [DCL] proteins and bound to Argonaute [AGO] effector proteins) are categorized into discrete classes based on their size (generally 21- 24-nt long) and mode of action. The two major classes are small interfering RNAs (siRNAs) and microRNAs (miRNAs). The functions of siRNAs in RNA-directed DNA methylation (to silence repeats and TEs) and miRNAs or secondary siRNAs in the posttranscriptional regulation of mRNAs have been extensively reviewed (Axtell, 2013a; Holoch and Moazed, 2015).
Important checks and controls
Upon sequence alignment, the majority of DCL/AGO-related smRNAs from most plants range between 21- and 24-nt long. The absence of discernible peaks of 21- and/or 24-nt RNAs in the total size distribution of a smRNA-seq alignment could indicate issues with the quality of the input RNA or library (Mathioni et al., 2017). smRNA library preparation is often accompanied by a size exclusion step to separate these RNAs from mRNAs, tRNAs, and rRNAs. However, poor RNA quality can lead to the degradation of these longer RNAs, which leads to their abundance within fractionations of smRNA populations. These RNAs can easily be filtered, especially for tRNAs and rRNAs; however, degraded mRNAs are more challenging to filter, as genes are often potential targets of the smRNAs under investigation. Therefore, to evaluate if an mRNA is a true source of siRNAs, the strandedness and size of the aligned RNAs can be examined. For example, a sign of mRNA degradation versus true smRNAs is the presence of numerous sequenced reads that are outside the expected size ranges (21–24 nt) that are derived primarily from the sense strand of the transcript. Computational analyses can be performed to identify individual loci where aligned smRNAs are not predominantly in the 21–24 nt size range (Axtell, 2013b; Johnson et al., 2016). A handful of such loci does not necessarily indicate a fatal flaw in the library; some mRNAs, especially highly abundant ones, likely have a true in vivo population of semi-degraded fragments.
Other possible issues include amplification bias, which is observed as a paucity of distinct or unique reads; this is most commonly observed when the input quantities of RNA are well below the levels recommended for the protocol (Wright et al., 2019). Differential expression of smRNAs using sequencing should require a minimum of three biological replicates and the use of proper statistical procedures that robustly model dispersion and control the false discovery rate (Schurch et al., 2016). Additionally, the choice of an appropriate normalization method is critical when quantifying smRNAs and identifying differentially accumulated smRNAs between samples. While normalization against total smRNA reads or rRNA/tRNA-filtered total smRNA reads is most common, this can generate biased results when changes in a class of smRNAs are found in a particular genotype or sample. In fact, rRNA/tRNA fragments constitute a fair proportion of total smRNA reads and can serve as an internal control for normalization (McCormick et al., 2011), provided that all samples were prepared at the same time with the same methodology and sequenced in the same sequencing run. When sRNA-seq data are to be used quantitatively, we recommend that several normalization methods be applied and the outcomes validated by qRT-PCR or northern blotting against select miRNAs or siRNAs known to be stable across all samples being analyzed.
Cytosine DNA methylation
DNA methylation can be examined at single nt resolution using whole-genome bisulfite sequencing (WGBS) or enzymatic methyl-seq (Cokus et al., 2008; Lister et al., 2008; Feng et al., 2020). These single-base resolution technologies are especially useful in plants, as DNA methylation occurs in three distinct contexts (CG, CHG, and CHH, where H = A, C, or T) that reflect the activities of distinct DNA methylation pathways (Law and Jacobsen, 2010). DNA methylation is highly enriched at transcriptionally silent regions of plant genomes such as TEs and repeats (Cokus et al., 2008; Lister et al., 2008; Feng et al., 2010; Zemach et al., 2010; Niederhuth et al., 2016). It is also found within gene bodies of a subset of actively transcribed genes in angiosperms as well as some ferns and gymnosperm species (Tran et al., 2005; Takuno and Gaut, 2012; Takuno et al., 2016; Bewick et al., 2017). Methylome data are useful for a variety of reasons; their intended use for describing qualitative versus quantitative aspects of the samples influences the need for biological replication. For example, methylome data that accompanies a genome assembly and is used to broadly characterize and annotate the methylation patterns around genes, transposon, repeats and other genomic features doesn’t typically require replication. However, the identification of differentially methylated regions requires biological replication. Here, the analysis is often focused on comparing the same genomic region across a variety of samples derived from a different genotype (mutant or natural isolate), a different tissue, or subjected to different environmental conditions. Similar to other quantitative methods like RNA-seq, a variety of software packages are publicly available that are specifically tailored to process, align, and identify differentially methylated positions and/or regions between samples from a treatment versus control group and/or a population (Xi and Li, 2009; Krueger and Andrews, 2011; Schultz et al., 2015).
Important checks and controls
Input genomic DNA used for methylome sequencing should be of high quality, although it does not need to be as pure as samples that are prepared for genome assemblies. Poor input quality of genomic DNA can lead to uneven uniformity of coverage across the genome, making comparisons between samples and biological replicates challenging. WGBS depends on either the chemical or enzymatic conversion of unmethylated cytosine to uracil (Frommer et al., 1992; Feng et al., 2020). Upon PCR, uracil is converted to thymine. An important consideration for methylome sequencing is the conversion rate of the treatment. This can be measured by evaluating the percentage of cytosines detected as “methylated” in the chloroplast genome or in an unmethylated spike-in DNA control, such as lambda phage DNA. To detect small changes in methylation, conversion rates should be ˃99%. If using unmethylated plastid genomes as controls, one should be certain to ensure that sequences are not duplicated in the nuclear genome, as this could interfere with measuring the conversion efficiency. This conversion rate, along with sequence alignment rates (most often for uniquely aligned reads), should always be reported, typically in a supplemental table. Lastly, the expected genome coverage per sample will depend on the experiment. Typically, >15× coverage per biological replicate is sufficient for most analyses, as suggested by the Human Epigenome Roadmap Consortium (http://www.roadmapepigenomics.org/protocols). As a reminder, because methylome sequencing results in strand-specific data, 15× coverage corresponds to 7.5× per strand. Measuring the percentage of duplicate reads is also useful in evaluating library complexity. For low input or low-quality genomic DNA, a higher rate of duplicate reads is often observed. In most cases, duplicate reads should be reduced to a single read for downstream analyses, and coverage estimates should be calculated after they are removed. Otherwise, the genome coverage will be artificially inflated due to technical issues.
ChIP-seq
ChIP-seq is a useful method for evaluating the enrichment of protein–DNA interactions (Johnson et al., 2007). In this method, plant tissue is treated with formaldehyde to crosslink and preserve protein–DNA interactions. After crosslinking, antibodies or epitope tags specific to the protein or hybrid protein of interest are used to detect and isolate the protein along with the associated DNA, which is then sequenced. This assay is commonly used to detect TF DNA binding sites or locations of posttranslationally modified or variant histones. The most important consideration for ChIP-seq is recognizing that it is an enrichment assay and that the quality of the enrichment is dependent on numerous experimental factors, such as the quality of the antibody and/or the precipitation protocol, in addition to biological factors such as the tissue specificity of the signal or its response to biological variation. A key goal of this assay is to maximize the antibody-tagged DNA signal over background. In the ideal experiment, there would be no background, meaning that no sequenced reads were detected from regions of the genome that were not associated with the protein being assayed. Unfortunately, background is essentially impossible to eliminate from this procedure, and therefore it must be accounted for using an input or other control library.
A high-quality ChIP-seq experiment typically results in a high coverage of sequenced reads at discrete regions throughout the genome, again referred to as “peaks,” although in low-quality ChIP-seq datasets, peaks are hard to distinguish from background signal (Figure 2). The shape, location, magnitude, and sensitivity to identify peaks differ depending on the protein–DNA interaction being measured and the complexity and sequencing coverage of the sequencing library (Hower et al., 2011; Jung et al., 2014). For example, TF peaks are typically sharp and enriched near the transcriptional start site (TSS), upstream and downstream of genes, whereas histone modifications and histone variant peaks are generally broader. The patterns and distribution of histone modifications have been well characterized in certain plant genomes (Zhang et al., 2007; Bernatavichute et al., 2008; Zhang et al., 2009; Li et al., 2019; Lu et al., 2019; Ricci et al., 2019; Montgomery et al., 2020; Zhao et al., 2020). There is a high similarity in the patterns and distributions of histone modifications among plant genomes. For example, trimethylation of lysine 4 of histone 3 (H3K4me3) is highly enriched around the TSS in plant genomes, whereas H3K4me2 and H3K4me1 are found within gene bodies (Zhang et al., 2009). These expected patterns and distributions at genes or other genomic features can be used to evaluate the quality of the enrichment. A novel method referred to as Cleavage Under Targets and Release Using Nuclease or Cleavage Under Targets and Tagmentation was recently developed to measure protein–DNA interactions (Skene and Henikoff, 2017; Kaya-Okur et al., 2019). Although the method is unique, most of the same methods used for the evaluation of data quality are the same as ChIP-seq. Additionally, Tn5-based chromatin profiling methods are biased toward detecting accessible chromatin, which requires extra scrutiny in downstream analyses (Wang and Zhang, 2021).
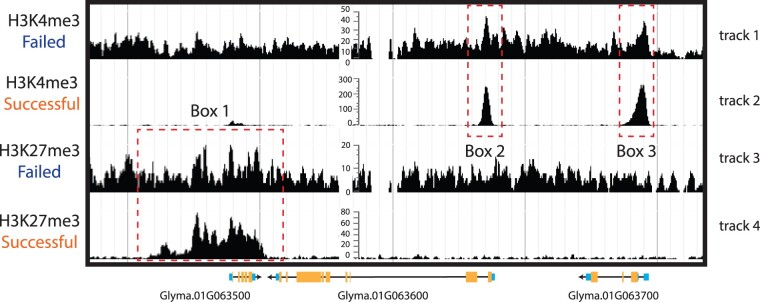
Figure 2.
Visualization of ChIP-seq enrichment of histone modifications. Low-quality/failed versus high-quality ChIP-seq data are shown for H3K4me3 and H3K27me3 from soybean (Glycine max) leaves. The first and third tracks show low-quality and/or failed ChIP-seq data, whereas tracks 2 and 4 show high-quality data. Box 1 shows a region of H3K27me3 enrichment in track 4, whereas the same region shows almost no enrichment in track 3. As is typical for H3K27me3, enrichment is present throughout the gene body into the upstream region. Boxes 2 and 3 show enrichment for H3K4me3 at TSSs in track 2, whereas weak enrichment is detected in track 1.
Important checks and controls
Sequence alignment percentages should be evaluated and reported for each sample. The percentage of duplicate reads can be used to determine if the sample is of low complexity. This is a common issue with ChIP-seq, as sub-nanogram amounts of DNA are typically immunoprecipitated from each ChIP, especially in the case of TF mapping. The preparation of a sequencing library does not necessarily indicate that the assay worked as intended; it only indicates that input DNA was used in the library preparation. To evaluate the quality of the ChIP experiment, the enrichment of sequenced reads relative to known genomic regions such as genes, repeats, promoters, and so on should be used. Additionally, genome-wide metrics for measuring signal-to-noise, such as Signal Portion of Tags (SPOT) and Fraction of Reads in Peaks (FRiPs), are used to establish the extent of enrichment, where greater values indicate lower background (Ji et al., 2008). However, SPOT/FRiP scores are largely qualitative and dependent on the heterogeneity of the sample, varying widely depending on the species, tissue, or cell type. In cases where biological replicates are performed, IDR can be implemented to establish the reproducibility of the experiments by comparing the ratio of identified peaks between and within biological replicates (see Landt et al. (2012) for more information on IDR and recommended thresholds). It is useful to present a meta-analysis of the coverage distribution of reads across these features for all biological replicates, as well as heat maps, which reveal locus-specific patterns of enrichment. These two data types are complementary in that one provides an average score across all genomic features, whereas the other shows individual data points for the studied features. It is also recommended that authors present representative genome browser screenshots of the biologically replicated data, typically as a supplemental figure. Together, this information can be used by readers to rapidly evaluate the quality of the experiment, as strong signals are expected compared to background regions of the genome. Lastly, the goal of most ChIP-seq experiments is peak identification, which often requires an input control for comparative purposes. It is typically recommended that a portion of the isolated chromatin be used as an input control for mapping TFs. To identify peaks from histone modifications, the use of an antibody to unmodified Histone H3 is recommended, as this reflects the distribution of nucleosomes across the genome. In both cases, the input DNA can be used to control for “mappability” of the genome as described above. In certain cases, adding spike-in chromatin from another species (e.g. fruit fly [Drosophila melanogaster] or mouse [Mus musculus]) to each sample prior to performing the ChIP experiment can be useful for absolute measurements of enrichment, particularly in genomes of lesser quality. A spike-in ChIP approach is invaluable for quantifying global changes in features across biological samples, for example, when comparing a reduction in a given histone modification between strongly affected mutants and wild-type plants.
Chromatin accessibility
Regions of the genome that are depleted of nucleosomes often reflect accessible chromatin; these regions are enriched for cis-regulatory elements (CREs) and TF binding (Stalder et al., 1980). A variety of methods are used to identify chromatin accessibility, such as micrococcal nuclease (MNase) sequencing (Liu et al., 2015; Zhang et al., 2015), DNase I sequencing (Zhang et al., 2012b; Cumbie et al., 2015), and Assay for Transposase Accessible Chromatin sequencing (ATAC-seq; Buenrostro et al., 2013; Lu et al., 2017; Maher et al., 2018). Each of these methods utilizes an enzyme that releases accessible chromatin fragments, either via enzymatic digestion (MNase or DNase) or sequencing adapter integration (ATAC-seq; Zhang et al., 2012b; Zhang et al., 2012a; Sullivan et al., 2014). These assays have been invaluable during the last decade at improving the ability to investigate the noncoding regions of the genome for candidate CREs (Rodgers-Melnick et al., 2016; Reynoso et al., 2019; Ricci et al., 2019). They are especially useful in plants with large genomes in which CREs can be located ˃100 kb away from their target gene(s) (Oka et al., 2017; Lu et al., 2019). These assays are similar to ChIP-seq in that they are evaluated based on sequencing coverage at distinct regions throughout the genome, again referred to as “peaks.” These peaks represent DNA fragments released from accessible chromatin and are highly enriched at TSSs of genes in plant genomes. However, as mentioned above, the identified regions can be proximal or distal to their target gene(s). They can even be located in introns, exons, 5′- or 3′-untranslated regions, or downstream of their target genes and interact with the target promoter through 3D chromatin interactions.
Important checks and controls
Unlike other epigenomic assays, accessible chromatin profiling techniques capture the signal from the ends of sequencing fragments rather than the center (e.g. ChIP-seq). As such, aligned reads from accessible chromatin profiling must be reformatted prior to the identification of peaks. For example, analysis of ATAC-seq data often includes post alignment steps that identify Tn5 integration sites at base resolution by initially collecting 5′-coordinates. These coordinates are then shifted by +5/−4 for forward and reverse alignments, respectively, to account for the 9-bp binding footprint of Tn5. As most peak callers assume that the biological signal originates from the centers of paired reads, naïve application on unprocessed chromatin accessibility sequencing data will lead to false-positive peaks, especially for sequencing libraries with generally larger insert sizes.
The percentage of aligned reads and the duplication read rate are again useful metrics. Similar to ChIP-seq, high-quality chromatin accessibility assays have a high signal-to-background ratio, often measured using FRiPs. For a detailed explanation of how to evaluate data quality from chromatin accessibility mapping assays, this article provides useful guidelines (Bubb and Deal, 2020). A key consideration is that genomic DNA should be treated with the appropriate enzyme (MNase, DNase I, or Tn5) for each new species under study to evaluate enzymatic bias and the overrepresentation of specific fragments. Another key point is that high-quality ATAC-seq data from Arabidopsis thaliana typically has a FRiP score >35%, whereas in maize (Zea mays), a sufficient score is typically >20%. The evaluation of FRiP scores is dependent on the species being studied, but in general, a higher percentage of reads in peaks reflects a higher quality experiment. FRiP scores or other metrics that evaluate signal-to-noise ratios are often influenced by the genome size and the number of sequenced reads. With a greater genome size, there is a higher probability of background reads, given that the number of chromatin-accessible regions does not scale with genome size. A sufficient number and complexity of sequenced reads should be generated such that the number of identified peaks becomes saturated. Another way to evaluate chromatin accessibility data is to plot the enrichment of read coverage around the TSSs of genes, as most expressed genes possess chromatin. If a TSS-sequencing method has not been applied to precisely identify TSS locations in a sample, they can be roughly approximated using annotated TSSs. A strong enrichment should be observed around the TSSs of genes compared to input controls. This should be performed for all biological replicates of all samples. Displaying data in a genome browser for all biological replicates are also a highly recommended approach to quickly evaluate the quality of the data and to present it to a broader audience.
Single-cell genomics
The advent of single-cell genomics is one of the most exciting recent technological developments in genomics that will significantly influence plant biology research. The ability to survey molecular profiles of populations of individual cells from any plant species without the need to generate transgenic reporter lines will undoubtedly lead to breakthrough discoveries. Single-cell RNA-seq (scRNA-seq; Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019; Satterlee et al., 2020; Xu et al., 2021; Zhang et al., 2021b, 2021a) and single-cell ATAC-seq (scATAC-seq; Dorrity et al., 2021; Farmer et al., 2021; Marand et al., 2021) have been applied to a few plant species and tissue types. This work has thus far demonstrated the ability to resolve cell type information from bulk populations by aggregating cells of the same type, typically leveraging dimensionality reduction (principal component analysis, non-negative matrix factorization, t-distributed stochastic neighbor embedding, uniform manifold approximation and projection, etc.) and graph-based clustering techniques (Leiden and Louvain) (Blondel et al., 2008; van der Maaten and Hinton, 2008; McInnes et al., 2018; Traag et al., 2019). Single-cell methods are particularly useful for pinpointing genes expressed in a cell-type-specific manner and identifying cell-type-specific CREs. Together, scRNA-seq and scATAC-seq hold great promise for studying gene regulatory networks at cellular resolution.
There are numerous methods for preparing single-cell genomic libraries, but the most commonly used thus far in plants relies on instrumentation and reagents provided by 10× Genomics. Unlike other sequencing libraries described above, the cost of preparing single-cell libraries easily varies by 10- to-20-fold depending on the type of library. As a result, preparing the sequencing library can be more expensive than the sequencing itself, which has major implications for experimental design. Unfortunately, single-cell genomics is susceptible to both biological and technical variation, even more so than bulk cell-based assays, considering that cellular heterogeneity is masked in bulk assays by averaging signals across profiled cells. To resolve this variation, biological replicates are required, especially for studies that rely almost solely on single-cell data as the core of the story. The current standard of reproducibility for single-cell approaches relies on qualitative evaluation of replicate mixing in reduced dimensional representations (e.g. within PCA/tSNE/UMAP embeddings), or quantitatively through comparisons of cell-type proportions per biological replicate. Additional quantitative metrics of reproducibility for single-cell techniques are likely to emerge as costs continue to fall and single-cell methods become more widely adopted by the plant science community.
Important checks and controls
Single-cell genomic analysis results in sparse data across thousands of individual cells. As a result, single-cell genomic libraries are often sequenced to saturation, for example, more than 500 million reads per 10,000 cells/nuclei, resulting in a high rate of read duplication. The percentage of aligned sequences should be reported for all samples. Additionally, due to the single-cell resolved nature of the data, the number of target cells profiled and the number of recovered cell profiles as well as the number of processed reads per cell must be reported. For example, a typical high-quality scRNA-seq or scATAC-seq library from plants using the 10× Genomics platform will capture >40% of input cells/nuclei with at least 1,000 unique transcripts or Tn5 integrations per cell. However, lower numbers of events per cell are acceptable for species with fewer genes (e.g. the liverwort Marchantia polymorpha). We also recommend the quantification of ambient RNA and chromatin from lysed cells and nuclei to distinguish true cells from background noise (Young and Behjati, 2020). Additionally, scRNA-seq experiments in plants have generally relied on generating protoplasts to create single-cell suspensions. For such approaches, it is imperative to generate bulk-scale RNA-seq control datasets to account for enzyme treatment-induced changes to cell states. Caution should be taken to minimize the number of droplets and/or barcodes that possess multiple nuclei or cells, which can be evaluated empirically by genotype/species mixing (Marand et al., 2021) or through the use of cell hashing (Stoeckius et al., 2018) prior to scaling up library preparations. Additionally, for scATAC-seq, the FRiP score and enrichment of reads around the TSSs of genes can be used to evaluate the quality of the data (Marand and Schmitz, 2021). Data quality from scATAC-seq is often much better than that of bulk ATAC-seq experiments, and FRiP scores >75% and >45% have been achieved for scATAC-seq in Arabidopsis and maize, respectively (Dorrity et al., 2021; Marand et al., 2021). The exact reason for the superior data quality is not known, but the ability to exclude cells that do not have enrichment of reads around the TSS or within peaks improves downstream analyses. Lastly, cells with high amounts of reads that align to the chloroplast and/or mitochondria from scATAC-seq data can be removed, as they likely represent broken nuclei or supernatant generated from the isolation of cells. The ability to remove data from suspect cells is likely one reason for the superior data quality and improved downstream analyses using single-cell compared to bulk ATAC-seq approaches.
Conclusions
Transcriptomics and epigenomics are revealing features of plant genomes at an unprecedented pace and providing a major source of as-yet untested hypotheses. The ability to produce high-quality sequencing libraries from these assays ranges in difficulty from relatively straightforward (RNA-seq) to challenging (ChIP-seq). However, the production of a sequencing library from any of these assays does not ensure the assay worked as intended; it only indicates that a library has been prepared and is ready for sequencing. Appropriate computational analysis of the sequenced library is required to evaluate the quality of the experiment used to produce the input RNA and/or DNA for library preparation. The major goal of this commentary is to raise awareness of the complexity of data quality issues associated with plant epigenomics research. We recognize that many of the techniques and/or considerations will need to be modified or adapted over time as novel technological and/or computational/statistical methods are developed. The considerations presented above are common methods that are currently used by many in the field to assess experiment and data quality. Presenting these key metrics as supplemental data during manuscript submission and publication will assist in the reader’s ability to appropriately evaluate interpretations pre and postpublication. Finally, just as the range of “-seq” experiments has exploded over the last decade, along with the ways to isolate tissues or cells for these methods, there are surely many new data types and acquisition approaches yet to emerge. These are likely to come with their own pitfalls, analysis quirks, and statistical challenges. We hope that authors will rise to the occasion, deeply probing and questioning the quality of their data and including their tests, concerns, and conclusions within their manuscript files to assist readers in understanding the limits of their observations and claims.
Acknowledgments
We would like to thank G. Alex Mason for valuable input and discussions on this commentary.
Funding
This commentary was funded with support from the National Science Foundation (NSF IOS-1856627 and MCB-1856143) and the National Institutes of Health (NIH R01GM134682) to R.J.S., an NSF Postdoctoral Fellowship in Biology (DBI-1905869) to A.P.M., and NSF (MCB-1552455 and MCB-2043544) and NIH (R35GM124806) grants to X.Z.
Conflict of interest statement. None declared.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Robert J. Schmitz (schmitz@uga.edu).
References
- Axtell MJ (2013a) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159 [DOI] [PubMed] [Google Scholar]
- Axtell MJ (2013b) ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19:740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute Y, Zhang X, Cokus S, Pellegrini M, Jacobsen S, Dilkes B (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3:e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Niederhuth CE, Ji L, Rohr NA, Griffin PT, Leebens-Mack J, Schmitz RJ (2017) The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol 18:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guilamume J, Lambiotte R, Lefebvre E (2008) Fast unfolding of communities in large networks. J Stat Mech 2008 DOI:10.1088/1742-5468/2008/10/P10008 [Google Scholar]
- Bubb KL, Deal RB (2020) Considerations in the analysis of plant chromatin accessibility data. Curr Opin Plant Biol 54:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al. (2016) JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumbie JS, Filichkin SA, Megraw M (2015) Improved DNase-seq protocol facilitates high resolution mapping of DNase I hypersensitive sites in roots in Arabidopsis thaliana. Plant Methods 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, Timmermans MCP (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev Cell 48:840–852 e845 [DOI] [PubMed] [Google Scholar]
- Derrien T, Estelle J, Marco Sola S, Knowles DG, Raineri E, Guigo R, Ribeca P (2012) Fast computation and applications of genome mappability. PLoS One 7:e30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrity MW, Alexandre CM, Hamm MO, Vigil AL, Fields S, Queitsch C, Cuperus JT (2021) The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nat Commun 12:3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A, Thibivilliers S, Ryu KH, Schiefelbein J, Libault M (2021) Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant 14:372–383 [DOI] [PubMed] [Google Scholar]
- Feng S, Zhong Z, Wang M, Jacobsen SE (2020) Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing. Epigenet Chromatin 13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107:8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89:1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, Gifford DK (2012) High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol 8:e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister BT, Schmitz RJ (2018) Enhanced JBrowse plugins for epigenomics data visualization. BMC Bioinformatics 19:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16:71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hower V, Evans SN, Pachter L (2011) Shape-based peak identification for ChIP-Seq. BMC Bioinformatics 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim MM, Lacadie SA, Ohler U (2015) JAMM: a peak finder for joint analysis of NGS replicates. Bioinformatics 31:48–55 [DOI] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, Cuperus JT (2019) Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell 31:993–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26:1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502 [DOI] [PubMed] [Google Scholar]
- Johnson NR, Yeoh JM, Coruh C, Axtell MJ (2016) Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6:2103–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YL, Luquette LJ, Ho JW, Ferrari F, Tolstorukov M, Minoda A, Issner R, Epstein CB, Karpen GH, Kuroda MI, et al. (2014) Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res 42:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S (2019) CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22:1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ (2011) Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5:1752–1779 [Google Scholar]
- Li Z, Wang M, Lin K, Xie Y, Guo J, Ye L, Zhuang Y, Teng W, Ran X, Tong Y, et al. (2019) The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Seddon AE, Tsai ZT, Major IT, Floer M, Howe GA, Shiu SH (2015) Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res 25:1182–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ (2017) Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Marand AP, Ricci WA, Ethridge CL, Zhang X, Schmitz RJ (2019) The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat Plants 5:1250–1259 [DOI] [PubMed] [Google Scholar]
- Maher KA, Bajic M, Kajala K, Reynoso M, Pauluzzi G, West DA, Zumstein K, Woodhouse M, Bubb K, Dorrity MW, et al. (2018) Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30:15–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Schmitz RJ (2021) Single-cell analysis of cis-regulatory elements. Curr Opin Plant Biol 65:102094. [DOI] [PubMed] [Google Scholar]
- Marand AP, Chen Z, Gallavotti A, Schmitz RJ (2021) A cis-regulatory atlas in maize at single-cell resolution. Cell 184:3041–3055 e3021 [DOI] [PubMed] [Google Scholar]
- Mathioni SM, Kakrana A, Meyers BC (2017) Characterization of plant small RNAs by next generation sequencing. Curr Protocol Plant Biol 2:39–63 [DOI] [PubMed] [Google Scholar]
- McCormick KP, Willmann MR, Meyers BC (2011) Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J (2018) UMAP: uniform manifold approximation and projection for dimension reduction. arXiv [Google Scholar]
- Meers MP, Tenenbaum D, Henikoff S (2019) Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenet Chromatin 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendieta JP, Marand AP, Ricci WA, Zhang X, Schmitz RJ (2021) Leveraging histone modifications to improve genome annotations. G3 11: jkab263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SA, Tanizawa Y, Galik B, Wang N, Ito T, Mochizuki T, Akimcheva S, Bowman JL, Cognat V, Marechal-Drouard L, et al. (2020) Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Curr Biol 30:573–588 e577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, et al. (2016) Widespread natural variation of DNA methylation within angiosperms. Genome Biol 17:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka R, Zicola J, Weber B, Anderson SN, Hodgman C, Gent JI, Wesselink JJ, Springer NM, Hoefsloot HCJ, Turck F, et al. (2017) Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biol 18:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gaffney DJ, Gilad Y, Pritchard JK (2011) False positive peaks in ChIP-seq and other sequencing-based functional assays caused by unannotated high copy number regions. Bioinformatics 27:2144–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso MA, Kajala K, Bajic M, West DA, Pauluzzi G, Yao AI, Hatch K, Zumstein K, Woodhouse M, Rodriguez-Medina J, et al. (2019) Evolutionary flexibility in flooding response circuitry in angiosperms. Science 365:1291–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejia-Guerra MK, Colome-Tatche M, et al. (2019) Widespread long-range cis-regulatory elements in the maize genome. Nat Plants 5:1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Vera DL, Bass HW, Buckler ES (2016) Open chromatin reveals the functional maize genome. Proc Natl Acad Sci USA 113:E3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB (2009) PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27:66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Huang L, Kang HM, Schiefelbein J (2019) Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol 179:1444–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee JW, Strable J, Scanlon MJ (2020) Plant stem-cell organization and differentiation at single-cell resolution. Proc Natl Acad Sci USA 117:33689–33699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. (2015) Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523:212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch NJ, Schofield P, Gierlinski M, Cole C, Sherstnev A, Singh V, Wrobel N, Gharbi K, Simpson GG, Owen-Hughes T, et al. (2016) How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22:839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M, Turco GM, Zhu Y, O’Malley RC, Brady SM, et al. (2019) High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep 27:2241–2247 e2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H (1980) Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20:451–460 [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, Satija R (2018) Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep 8:2015–2030 [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2012) Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol 29:219–227 [DOI] [PubMed] [Google Scholar]
- Takuno S, Ran JH, Gaut BS (2016) Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants 2:15222. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ (2019) From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S (2005) DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol 15:154–159 [DOI] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605 [Google Scholar]
- Vaux DL, Fidler F, Cumming G (2012) Replicates and repeatswhat is the difference and is it significant? A brief discussion of statistics and experimental design. EMBO Rep 13:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang Y (2021) Tn5 transposase-based epigenomic profling methods are prone to open chromatin bias. bioRxiv doi:10.1101/2021.07.09.451758 [Google Scholar]
- Wright C, Rajpurohit A, Burke EE, Williams C, Collado-Torres L, Kimos M, Brandon NJ, Cross AJ, Jaffe AE, Weinberger DR, et al. (2019) Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genom 20:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W (2009) BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, Peng W (2014) Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150:97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu Z, Wang L, Fox N, et al. (2021) Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56:557–568 e556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Behjati S (2020) SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9: giaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919 [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhang W, Jiang J (2015) Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiol 168:1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Chen Y, Wang JW (2021a) A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev Cell 56:1056–1074 e1058 [DOI] [PubMed] [Google Scholar]
- Zhang TQ, Xu ZG, Shang GD, Wang JW (2019) A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol Plant 12:648–660 [DOI] [PubMed] [Google Scholar]
- Zhang TQ, Chen Y, Liu Y, Lin WH, Wang JW (2021b) Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat Commun 12:2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang T, Wu Y, Jiang J (2012a) Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24:2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu Y, Schnable JC, Zeng Z, Freeling M, Crawford GE, Jiang J (2012b) High-resolution mapping of open chromatin in the rice genome. Genome Res 22:151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xie L, Zhang Q, Ouyang W, Deng L, Guan P, Ma M, Li Y, Zhang Y, Xiao Q, et al. (2020) Integrative analysis of reference epigenomes in 20 rice varieties. Nat Commun 11:2658. [DOI] [PMC free article] [PubMed] [Google Scholar]