Abstract
Stomatal pores and the leaf cuticle regulate evaporation from the plant body and balance the tradeoff between photosynthesis and water loss. MYB16, encoding a transcription factor involved in cutin biosynthesis, is expressed in stomatal lineage ground cells, suggesting a link between cutin biosynthesis and stomatal development. Here, we show that the downregulation of MYB16 in meristemoids is directly mediated by the stomatal master transcription factor SPEECHLESS (SPCH) in Arabidopsis thaliana. The suppression of MYB16 before an asymmetric division is crucial for stomatal patterning, as its overexpression or ectopic expression in meristemoids increased stomatal density and resulted in the formation of stomatal clusters, as well as affecting the outer cell wall structure. Expressing a cutinase gene in plants ectopically expressing MYB16 reduced stomatal clustering, suggesting that cutin affects stomatal signaling or the polarity setup in asymmetrically dividing cells. The clustered stomatal phenotype was rescued by overexpressing EPIDERMAL PATTERNING FACTOR2, suggesting that stomatal signaling was still functional in these plants. Growing seedlings ectopically expressing MYB16 on high-percentage agar plates to modulate tensile strength rescued the polarity and stomatal cluster defects of these seedlings. Therefore, the inhibition of MYB16 expression by SPCH in the early stomatal lineage is required to correctly place the polarity protein needed for stomatal patterning during leaf morphogenesis.
The inhibition of MYB16 expression by SPEECHLESS in the early stomatal lineage is required to correctly place the polarity complex for proper stomatal patterning during leaf morphogenesis.
Introduction
Terrestrialization was a critical event in which organisms moved from the ocean to the land. Two pivotal features—stomata and the cuticle—evolved to enable plants to adapt to the major changes between the aqueous and aerial environments. Stomata are valves on the plant surface that control gas exchange and water loss. The cuticle is a lipidic, hydrophobic layer between the external environment and the air-exposed plant surface. The coordination of stomatal formation and cuticle deposition on the epidermis balances the tradeoff between allowing photosynthesis and preventing excessive water evaporation.
In Arabidopsis thaliana, stomata are created through a series of asymmetric and oriented cell divisions to ensure the proper stomatal density and distribution on the epidermis. Stomatal lineage cells sequentially express three basic helix–loop–helix master transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA, to drive the maturation of stomata. SPCH mediates the asymmetric cell divisions of epidermal cells to produce meristemoids and stomatal lineage ground cells (SLGCs; MacAlister et al., 2007), which may become pavement cells or reinitiate asymmetric division to produce more stomata. MUTE induces meristemoids to differentiate into guard mother cells (GMCs) (Pillitteri et al., 2007). After MUTE upregulates the expression of FAMA and other genes involved in the cell cycle (Han et al., 2018), MUTE degrades and FAMA accumulates to cause the symmetric division of the GMC into two guard cells (GCs; Ohashi-Ito and Bergmann, 2006).
Stomatal patterning follows the “one-cell-spacing” rule, which means that two stomata should never be in direct contact with each other (Geisler et al., 2000). Two pathways regulate stomatal patterning. One relies on cell–cell communication, and the other is the establishment of polarity during asymmetric cell division. The EPIDERMAL PATTERNING FACTOR (EPF) family peptide ligands, including EPF1 and EPF2, are secreted from stomatal lineage cells to restrict stomatal development in neighboring cells (Hara et al., 2007; Hunt and Gray, 2009). Upon directly binding to members of the receptor-like kinase ERECTA family (ERf; Lee et al., 2012), the EPF signal activates the mitogen-activated protein kinase (MAPK) cascade (Wang et al., 2007) to diminish SPCH stability (Lampard et al., 2008), which further prevents cells from entering the stomatal lineage. During asymmetric division, the polarity complex is formed in the SLGCs and is integrated with MAPK signaling to decrease SPCH levels, ultimately breaking the stomatal fate in the SLGCs (Zhang et al., 2015). The polarity complex includes BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION (POLAR), and members of the BREVIS RADIX (BRX) family (Dong et al., 2009; Pillitteri et al., 2011; Zhang et al., 2015; Houbaert et al., 2018; Rowe et al., 2019). These factors interact with each other and form a scaffold to recruit MAPK components in the SLGCs. Thus, cell–cell communication, MAPK signaling, and the polarity complex work in combination to regulate stomatal patterning in the epidermis.
Plant cuticles form a physical barrier between the epidermis and the environment to protect tissues against dehydration. The cuticle is biosynthesized by the epidermal cells and mainly consists of cutin, a cross-linked biopolymer, and wax, a mixture of various lipids (Yeats and Rose, 2013; Bhanot et al., 2021). Cutin monomers vary between species and among the organs within a species, but they are typically composed of oxygenated C16 or C18 fatty acids. Cutin monomers are biosynthesized at the endoplasmic reticulum (ER) membrane, transported to the outside of the cells, and polymerized into a thin layer on the epidermal cell wall. In the ER, three biosynthetic steps occur, including ω-hydroxylation, midchain hydroxylation, and the biosynthesis of acyl-CoA intermediates. ω-hydroxylation is mediated by the CYP86 subfamily of cytochrome P450s, such as CYP86A4 in Arabidopsis flowers (Li-Beisson et al., 2009), while midchain hydroxylation is mediated by the CYP77 subfamily of cytochrome P450s, such as CYP77A6 in Arabidopsis flowers (Li-Beisson et al., 2009). Among the nine Arabidopsis long-chain acyl-CoA synthetases (LACSs), both LACS1 and LACS2 are essential for transforming C16/C18 fatty acids into acyl-CoA precursors during cutin biosynthesis and for modifying very-long-chain fatty acids for wax biosynthesis (Lü et al., 2009).
The acyl-CoA precursors are then converted to cutin monomers, such as 2-monoacylglycerol, via a reaction catalyzed by glycerol-3-phosphate sn-2-acyltransferase (GPAT) family members (Yang et al., 2010, 2012). In the case of Arabidopsis floral cutin, this activity is mediated by GPAT6 (Li-Beisson et al., 2009), whereas GPAT4 and GPAT8 function in Arabidopsis leaves and stems (Li et al., 2007). After leaving the ER membrane, the cutin monomers are transported into the outer epidermal cell wall by the ATP-binding cassette (ABC) transporters ABCG11 and ABCG32 (McFarlane et al., 2010; Bessire et al., 2011). CUTIN DEFICIENT1 (CD1) in tomato (Solanum lycopersicum) is an extracellular enzyme that catalyzes the biosynthesis of cutin polymers using 2-mono(10,16-dihydroxyhexadecanoyl)glycerol (2-MHG) in vitro (Yeats et al., 2012). The putative orthologs of CD1 in Arabidopsis are cutin synthase-like (CUS) proteins (Yeats et al., 2014). CUS1 catalyzes the formation of cutin polyesters in vitro (Yeats et al., 2014; Philippe et al., 2016), while CUS2 is required for the maintenance of cuticular ridges as the Arabidopsis sepals grow (Hong et al., 2017).
Most of the known cuticle biosynthesis-related genes are known or predicted to encode biosynthetic enzymes, and few transcription factors have been found to activate the pathway. WAX INDUCER1/SHINE1 (WIN1/SHN1), an AP2 domain-containing transcription factor, was first identified from a transcription factor overexpression screen. This transcription factor controls wax and cutin formation, as well as regulating cell wall composition (Stracke et al., 2001; Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). MYB16, an R2R3 MYB transcription factor, together with MYB106 from the same MYB subgroup (Stracke et al., 2001), regulates epidermal cell shape in petals and directly upregulates the expression of WIN1/SHN1 and genes involved in cutin biosynthesis (Baumann et al., 2007; Oshima et al., 2013), which suggests a link between cuticle development and epidermal cell differentiation. In barley (Hordeum vulgare), the wax-deficient eceriferum-g mutant (cer-g) produces clustered stomata (Zeiger and Stebbins, 1972). The mutation of HIGH CARBON DIOXIDE (HIC), an Arabidopsis enzyme involved in the biosynthesis of very-long-chain fatty acids, resulted in a thinner cuticle and higher stomatal density (Gray et al., 2000). Also, the overexpression of WIN1/SHN1 decreased stomatal density (Yang et al., 2011). These findings suggest a connection between stomatal patterning and cuticle defects.
The cuticle has been proposed to carry inhibitors or directly implement an inhibitory signal for stomatal development (Bird and Gray, 2003). In the leaves, the mechanical properties of the cuticle are independent of its thickness (Wiedemann and Neinhuis, 1998), but they rely on lipidic components such as cutin and wax as well as polysaccharides, suggesting that both the amount and composition of the cuticular material influence its biomechanics (Takahashi et al., 2012; Khanal et al., 2013). In the mangrove tree Sonneratia alba, the decrease in leaf flexibility during growth correlates with changes in cuticle composition: the deposition of wax and cutin in the cuticle reaches a plateau at an early stage of leaf growth, but the amounts of cutan and polysaccharides increase during the later stages (Takahashi et al., 2012). Since most mechanical properties of the cuticle were measured from isolated plant cuticles, the correlation between the biomechanical properties and physiological behavior of the epidermis in an intact plant is yet to be fully addressed (Dominguez et al., 2011; España et al., 2014).
Cell division and cell expansion during growth deform the cell membrane and extracellular layer to generate surface mechanical force. The disruption of the extracellular matrix reduces cell adhesion and results in a disorganized, tumor-like growth on the shoot (Krupková et al., 2007); therefore, the tissue-wide coordination of cell–cell adhesion is important for maintaining its proper patterning. To study the tensile strength of plant tissues, Verger et al. (2018) used an atomic force microscope (AFM) to detect the mechanical forces in the outer cell wall of the Arabidopsis cell adhesion mutant quasimodo1 (qua1). Growing qua1 mutants in high-percentage agar plates reduced the water potential in plants and decreased pavement cell stiffness and cell wall tension (Verger et al., 2018), suggesting that the outer cell wall contributes to the mechanical properties of the epidermis.
The localization of polarized proteins could also respond to the action force generated from the growth direction in both the leaf and shoot (Heisler et al., 2010; Bringmann and Bergmann, 2017). Altering the mechanical force in the leaf epidermis using laser ablation led to the redistribution of polarity proteins (Bringmann and Bergmann, 2017). In animals, elevated mechanical strength in neutrophil cells prevented the recruitment of polar components and inhibited cell migration (Houk et al., 2012); therefore, the changes in the outer cell wall continuum could influence the mechanical properties of the epidermis and affect polarity setup, leading to the alteration of tissue patterning.
Transcriptome profiling of stomatal lineage cells and expression analysis revealed the enrichment and preferential expression of MYB16 in SLGCs, which raises the question of why MYB16 is not required in meristemoids (Ho et al., 2021). In the current study, to explore the role of MYB16 in stomatal formation, we investigated the dynamics of SPCH and MYB16 expression during asymmetric cell division. Ectopically expressing MYB16 altered stomatal patterning and the cuticle–cell wall continuum. The appropriate regulation of MYB16 activity in asymmetrically dividing cells was found to be required to establish the proper polarity necessary for stomatal patterning.
Results
MYB16 expression occurs in SLGCs but is restricted in meristemoids due to its negative regulation by SPCH
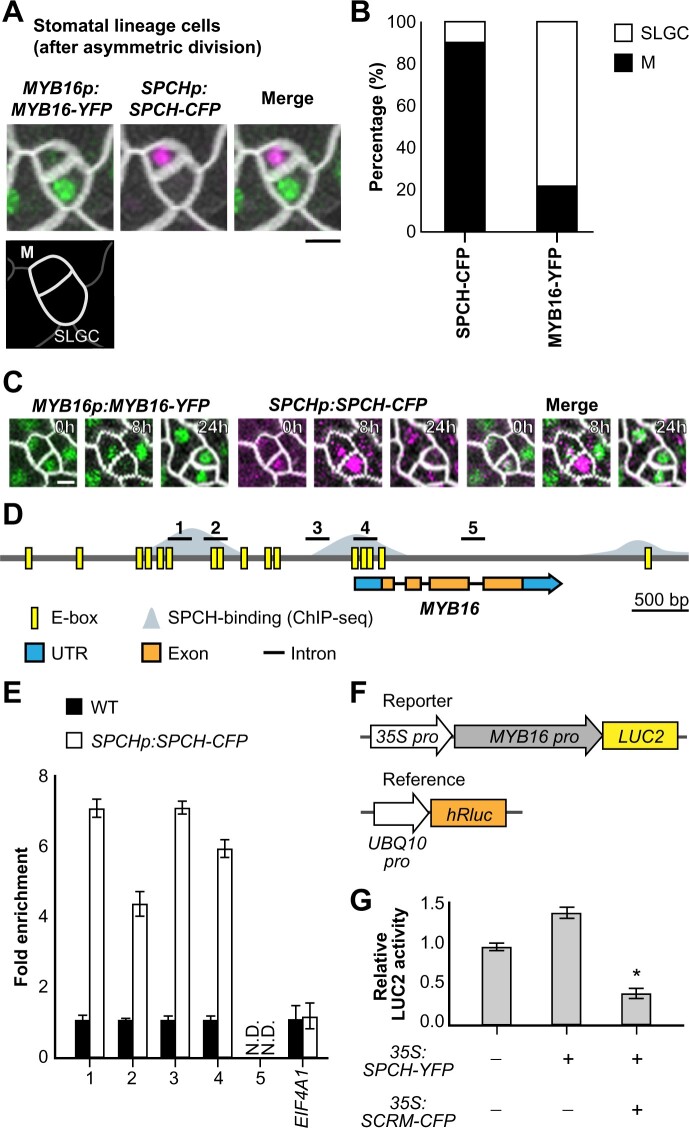
A recent study showed that MYB16 transcripts are enriched in SLGCs and that different MYB16 protein levels are present in the two daughter cells following their asymmetric division (Ho et al., 2021). Since SPCH is the master regulator of stomatal fate, we observed the expression patterns of MYB16-YFP and SPCH-CFP in wild-type (WT) young true leaves (7 days post-germination [dpg]) to further investigate MYB16 activity in stomatal lineage cells. We examined asymmetrically divided sister cells, meristemoids, and SLGCs and observed that SPCH was often present in meristemoids and that MYB16 was localized to SLGCs (Figure 1A; Supplemental Figure S1). Quantification of signals in an entire leaf showed that, in the 583 pairs of meristemoids and SLGCs analyzed, SPCH was more frequently found in meristemoids (89.6%) than in SLGCs (10.4%), whereas MYB16 was more often localized to SLGCs (78.4%) than meristemoids (21.6%; Figure 1B); however, we did observe MYB16 in a few meristemoids and pavement cells (Supplemental Figure S1). Using time-lapse imaging, we observed that MYB16 was expressed in meristemoids, but its expression was quickly replaced by SPCH before their asymmetric division (Figure 1C). These observations indicate that MYB16 is preferentially localized to SLGCs, a young cell state in the epidermis, with a possible negative relationship between SPCH and MYB16.
Figure 1.
SPCH binds to the MYB16 promoter and downregulates its expression in meristemoids. A, Still images of MYB16-YFP (green) and SPCH-CFP (magenta) in a meristemoid (M)–SLGC pair in a true leaf from a WT plant at 7 dpg. B, Frequency of SPCH or MYB16 in either cell of meristemoid–SLGC pairs. SPCH was often found in meristemoids, as predicted (89.6%). In contrast, MYB16 preferentially localized to SLGCs (78.4%). A whole leaf image was used to obtain 583 pairs. M, meristemoid. C, Time-lapse imaging of SPCH and MYB16 in a meristemoid–SLGC pair. Both SPCH and MYB16 were found in the meristemoid at 0 h, but only the SPCH signal remained at 8 h before asymmetric cell division (24 h). D, Diagram of the MYB16 genome region: E-boxes (CANNTG) predicted by PlantPAN 3.0 are shown in yellow, SPCH-binding sites obtained from SPCH ChIP-seq data (Lau et al., 2014) are shown in gray. Five regions (black bars) designed for the ChIP-qPCR assay were used in (E). E, SPCH binds to the MYB16 promoter, as revealed by ChIP-qPCR of 4 dpg SPCHp:SPCH-CFP seedlings with GFP-trap beads. Three biological replicates (independent experiments with the same experimental procedure) showed similar results. EIF4A1 is a negative control. N.D., not detected. Data are means (sd). F, The experimental design for the MYB16 luciferase assay. The MYB16 promoter was fused with a mini-35S promoter to enhance gene expression. Ratiometric luminescent reporters were used to normalize the expression difference in a given construct. G, SPCH functions with SCRM/ICE1 to downregulate MYB16 expression. The luciferase assay was performed with protoplasts from 3-week-old WT plants. Four biological replicates showed similar results. *P < 0.001, by Student’s t test. Data are means (sd). For (A) and (C), cell outlines are marked by RC12A-mCherry (gray). Scale bars, 5 µm. See also Supplemental Figures S1 and S2.
In line with our hypothesis, SPCH chromatin immunoprecipitation sequencing (ChIP-seq) and induced-expression assays previously showed that the promoter of MYB16 is bound by SPCH and that MYB16 expression is downregulated after SPCH induction (Lau et al., 2014). To confirm the direct binding of SPCH, we searched for the potential SPCH binding motif, an E-box, in the MYB16 promoter using the PlantPAN 3.0 website-based predictor (Chow et al., 2019; Figure 1D, yellow boxes). Based on the overlap between the E-box predictions and the SPCH binding sites identified by Lau et al. (2014) (Figure 1D, gray area), we designed five sets of primers for use in ChIP coupled with quantitative polymerase chain reaction (ChIP-qPCR) to test the binding of SPCH to the MYB16 promoter. Regions 1–4, but not the gene body (region 5), showed an increased fold change in binding in plants expressing SPCHp:SPCH-CFP (Figure 1E). To determine whether the binding caused the positive or negative regulation of MYB16 expression, we performed a luciferase assay with ratiometric luminescent reporters to control the expression and copy number of a given construct. A ∼3-kb MYB16 promoter was fused to the reporter gene LUCIFERASE2 (LUC2), as well as a mini-35S promoter to enhance its expression (Figure 1F). SPCH is known to form heterodimers with SCREAM(SCRM)/ICE1 (Kanaoka et al., 2008), so we included SCRM/ICE1 in the assay as well. SPCH expressed alone in protoplasts conferred no change in luciferase activity compared with the control, whereas co-expressing SPCH and SCRM/ICE1 reduced luciferase expression (Figure 1G). In summary, these findings indicate that SPCH directly binds to the MYB16 promoter and functions with SCRM/ICE1 to suppress MYB16 transcription.
Stomatal formation is regulated by a transcription factor cascade starting with SPCH and then MUTE to drive stomatal fate. The inhibition of the MYB16 promoter by SPCH led us to investigate whether MUTE also represses MYB16 expression. A previous study using a MUTE inducible line (iMUTE) revealed that MYB16 was downregulated after MUTE induction (Han et al., 2018). MUTE and SPCH have similar binding regions, as reported by Han et al. (2018); therefore, we used the same primer sets as those used in Figure 1D to test MUTE binding to the MYB16 promoter. We performed ChIP-qPCR using MUTEp:MUTE-CFP seedlings, revealing that MUTE binds to the same regions as SPCH (Supplemental Figure S2, A and B). To confirm the regulatory effect of MUTE on MYB16, we performed a ratiometric luciferase assay. Surprisingly, the co-expression of MUTE and SCRM increased LUC2 expression (Supplemental Figure S2C). This finding conflicts with what was found in the MUTE induction data set (Han et al., 2018), suggesting that additional factors function together with MUTE to repress MYB16 expression in vivo.
MYB16 overexpression increases stomatal number and cluster formation
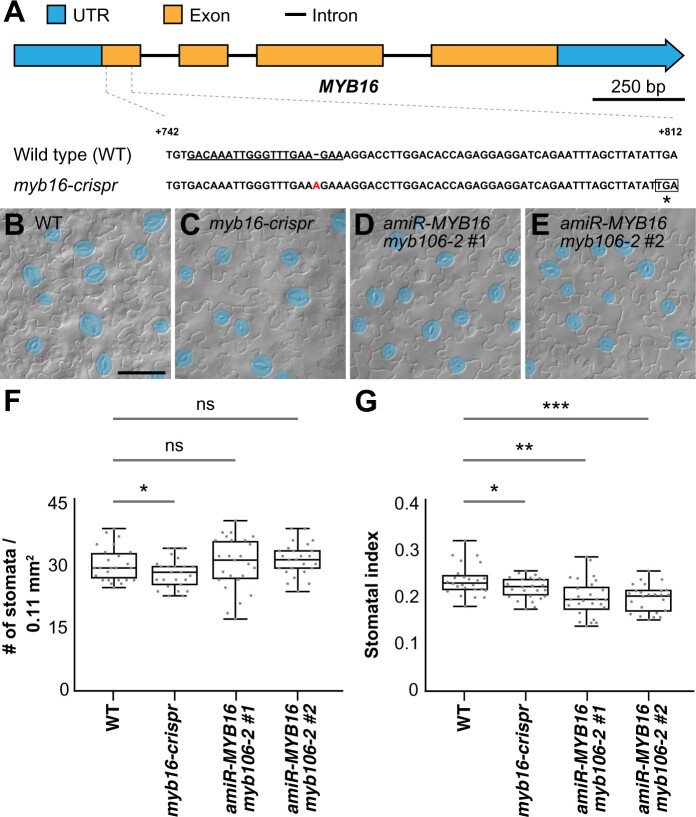
The MIXTA-like MYB transcription factors MYB16 and MYB106 function redundantly (Stracke et al., 2001; Oshima et al., 2013), which led Ho et al. (2021) to use a dominant-negative form of MYB16, MYB16-SRDX, to explore the resulting phenotypes. In contrast to its striking floral-organ-fusion phenotype, plants harboring MYB16-SRDX showed only a slightly decreased stomatal density (stomata/area). In line with this previous observation, we generated the myb16 mutant line myb16-crispr, with a premature stop codon in the first exon, which also showed a slightly reduced stomatal density and stomatal index (stomatal number divided by the total cell number; Figure 2 and Figure 3, A and B). Reduced stomatal indices were also observed in the two amiR-MYB16 myb106-2 double-mutant lines, in which the target genes were knocked down and knocked out, respectively (Figure 2). The weak stomatal phenotype in the myb16-cripsr and amiR-MYB16 myb106-2 lines suggests that, unlike the function of SPCH in promoting cell division, the MIXTA-like MYB transcription factors influence stomatal production through a different mechanism.
Figure 2.
The stomatal index is reduced in MIXTA-like loss-of-function mutants. A, Scheme of the MYB16 gene with the targeted sequence of CRISPR guide RNA (underlined). An adenine (A) insertion results in a premature stop codon (asterisk). B–E, DIC images of lower epidermis from 10-dpg WT true leaves (B), myb16-crispr (C) and two independent lines in which MYB16 artificial micro-RNA (amiR-MYB16) was introduced into the myb106-2 background (D) and (E). Mature GCs are pseudo-colored in blue. Scale bars, 50 µm. F, Quantification of stomatal density in (B) to (E). Stomatal density was reduced in the clustered regularly interspaced short palindromic repeats (CRISPR)-edited MYB16 line (myb16-crispr) but not in amiR-MYB16/myb106-2. n = 25 seedlings. *P < 0.05, by Wilcoxon signed-rank test. Data are medians (interquartile range); ns, not significant. G, Quantification of stomatal index in (B–E). Stomatal index was reduced in both myb16-crispr and amiR-MYB16/myb106-2. n = 25 seedlings. *P < 0.05; **P < 0.01; ***P < 0.001, by Wilcoxon signed-rank test. Data are medians (interquartile range).
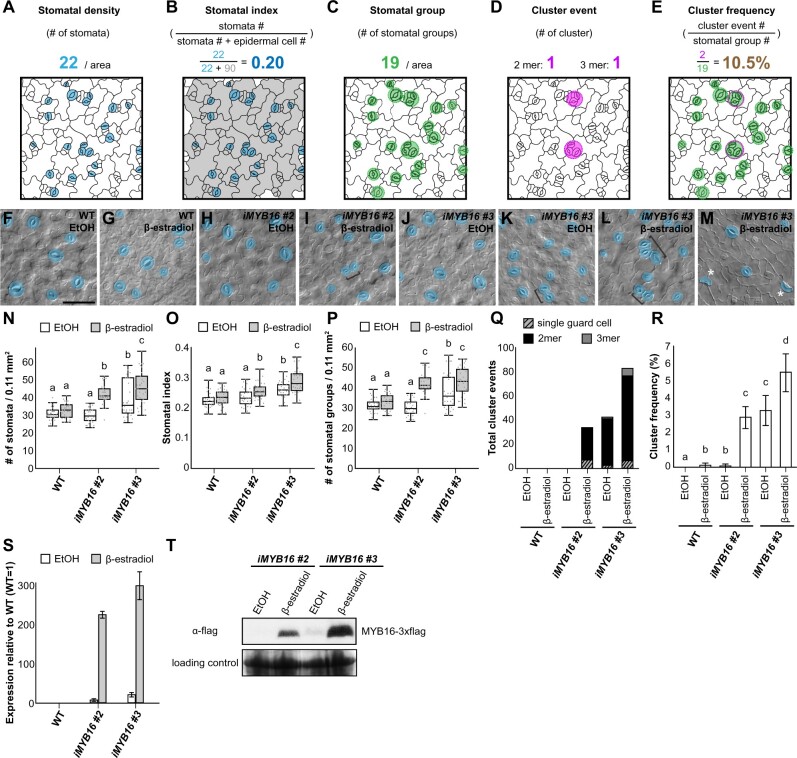
Figure 3.
Overexpression of MYB16 induces the formation of stomatal clusters. A, Stomatal density is defined by the total stomatal number in a given area. Mature stomata with pores are colored in blue. B, Stomatal index calculated by dividing the stomatal number by the total epidermal cell number (the sum of stomata, meristemoids, SLGCs, and pavement cells) in a given area, i.e. the number of blue cells divided by the number of blue and gray cells. C, Stomatal group is defined as a single stomatal island. Adjacent stomata are counted as one group. Groups are indicated by green circles. D, Cluster event represents the event number of stomatal pairs, triplets, or more than three adjacent stomata. Clusters are indicated by magenta circles. E, Cluster frequency calculated by dividing the number of cluster events by the number of stomatal groups, i.e. the number of magenta circles divided by the number of green circles. F–M, DIC images of the lower epidermis in 10-dpg WT true leaves and two MYB16 inducible lines (iMYB16#2 and #3) treated with EtOH (mock) or 50-µM β-estradiol. Stomatal pairs (brackets) were often found after β-estradiol treatment in the MYB16 induced lines. Occasionally, single GCs (asterisks) were found (M). Mature GCs are pseudo-colored in blue. Scale bar, 50 µm. N, Quantification of stomatal density (number of pores per 0.11 mm2) from (F) to (M). Stomatal density increased after β-estradiol treatment in the iMYB16 lines. Compared to iMYB16#2, iMYB16#3 had a higher stomatal number under mock treatment, which suggests leaky expression of MYB16. O, Quantification of stomatal index (the number of stomata divided by the total number of epidermal cells) from (F) to (M). Stomatal index increased after β-estradiol treatment in iMYB16 lines. P, Quantification of the number of stomatal groups (cluster as a single complex) per 0.11 mm2 from (F) to (M). The number of stomatal groups increased after β-estradiol treatment in iMYB16 lines. Q, Quantification of abnormal stomatal phenotypes shows that iMYB16 lines had more single GC and stomatal clusters (2–3 mer) than WT plants after β-estradiol treatment. Data are the sum of event numbers of 30 lower epidermis samples. R, Quantification of cluster frequency shows that stomatal clusters formed more frequently in iMYB16 than WT plants after β-estradiol treatment. Data are the means (se) of 30 lower epidermis samples. S, The expression of MYB16 was induced more than 200-fold after β-estradiol treatment, as revealed by qRT-PCR. iMYB16#3 (23.5×) was expressed at a higher level than iMYB16#2 (7.3×) under mock conditions. Data are means (sd). T, Immunoblot analysis indicating that MYB16 protein levels are tightly controlled in iMYB16#2. iMYB16#3 had leaky MYB16 expression under mock conditions, which could explain the phenotypes observed in (K). Coomassie blue staining of total protein represents a loading control. Thirty lower epidermis samples were used in (N)–(R). For (N)–(R), three biological replicates showed similar results. P < 0.05, Kruskal–Wallis test with Dunn’s test (N)–(P), P < 0.05. One-way ANOVA with Tukey post hoc test (R). Medians and interquartile ranges are shown in (N)–(P).
Stomatal phenotypes are often measured as stomatal densities and indices. To systematically describe the phenotypes of various plant lines, we used the following measurements: (1) stomatal density; (2) stomatal index; (3) stomatal group; (4) cluster event; and (5) cluster frequency. (1) Stomatal density describes the number of stomata in a given area, which is influenced by the initiation of a stomatal lineage and the expansion of the pavement cells from both stomatal and nonstomatal lineages (Figure 3A). (2) To remove the effect of pavement cell expansion, stomatal index is used to represent the number of stomata as a proportion of the total number of epidermal cells (Figure 3B). (3) Increased stomatal density or index could result from increased numbers of stomatal initial cells or stomatal clusters; therefore, we measured stomatal groups as the number of stomatal islands, which ignores the effect of clusters and considers adjacent stomata to be derived from the same origin of asymmetrically divided sister cells (a single stomatal group; Figure 3C). (4) A stomatal cluster event describes the frequency of stomatal clusters with more than two adjacent stomata (Figure 3D). (5) The cluster frequency represents the ratio between the number of stomatal cluster events and stomatal groups. This reflects the error rate of cluster formation (Figure 3E).
To examine whether MYB16 participates in stomatal development, we created several MYB16 inducible lines. Two lines, iMYB16#2 and iMYB16#3, showed abnormal stomatal patterns such as clusters and single GCs following a 4-day treatment with 50-µM β-estradiol (inducer; Figure 3, F–M). In contrast to the myb16-crispr and amiR-MYB16 myb106-2 mutants (Figure 2), the inducible lines showed a higher stomatal density and stomatal index, with more stomatal groups and cluster events, and a higher cluster frequency following their induction (Figure 3, N–R). The elevated stomatal index (Figure 3O) indicates that the increased number of stomata is due to both the initiation of stomatal production, as revealed by the stomatal group measurement (Figure 3P), and the formation of adjacent stomata, as revealed by the number of total cluster events (Figure 3Q). Note that one inducible line, iMYB16#3, produced more stomata and stomatal clusters under mock conditions (Figure 3, K, N–R). Further analyses showed higher MYB16 transcript and protein abundance in iMYB16#3 under mock treatment (Figure 3, S and T), suggesting that this line was a leaky mutant. Taken together, the overexpression of MYB16 increased the number of stomata produced and the formation of stomatal clusters.
Downregulation of MYB16 in meristemoids is required for proper stomatal patterning
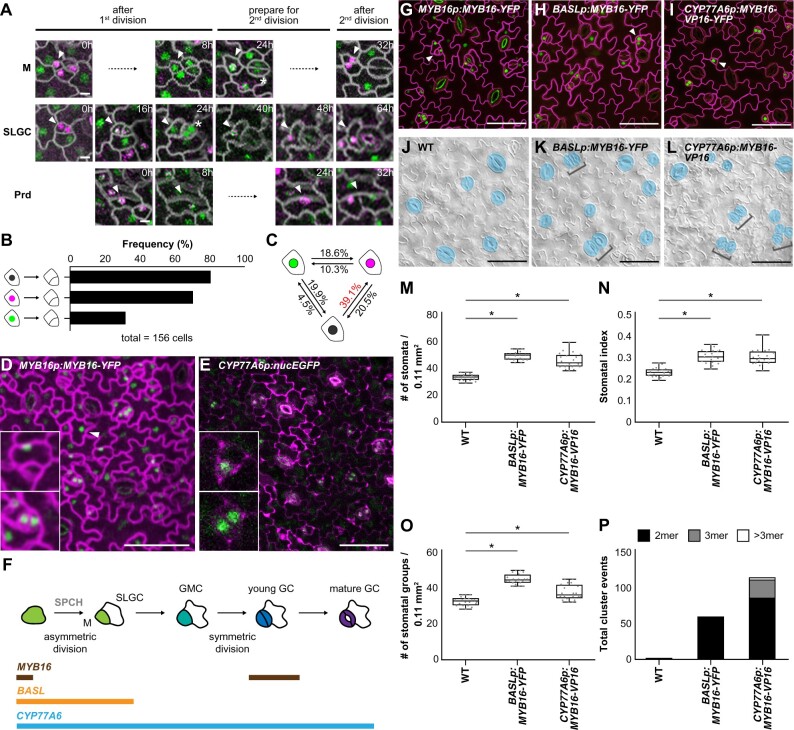
To examine the timing of MYB16 expression during stomatal development, we performed time-lapse imaging of young true leaves expressing MYB16 and SPCH under the control of endogenous promoters. The plants were imaged every 8 or 16 h for 4 days for a total of seven timepoints. We checked the cell division events and protein localizations in three types of cells: meristemoid, SLGC, and protoderm (Figure 4A). The three types of cells showed a similar pattern: the co-expression of MYB16 and SPCH or SPCH alone was often observed before an asymmetric cell division, with SPCH expression remaining after the division (Figure 4A). To quantitatively analyze the patterns in the time-lapse images, we used the PrefixSpan algorithm (Pei et al., 2001) to explore the possible sequential events of MYB16 expression, SPCH expression, and cell division. A total of 156 cells with strong MYB16-YFP and/or SPCH-CFP fluorescence were used. Four categories, MYB16 only, SPCH only, co-localization, and division event, were noted across the seven timepoints (Supplemental Figure S3A). In total, 57 patterns were generated (Supplemental Figure S3B). The pattern frequency was calculated as the number of observed cells displaying a particular pattern divided by the total number of cells analyzed (frequency/156 cells).
Figure 4.
Ectopic expression of MYB16 in early stomatal lineage cells causes stomatal cluster formation. A, Time-lapse confocal images of MYB16-YFP (green) and SPCH-CFP (magenta) in 7-dpg true leaves show that SPCH and MYB16 were expressed individually or together, depending on the sequence of cell division. Arrowheads indicate cells of interest. An asterisk shows MYB16-YFP in the pair of GCs. Timestamps indicate the time since the start of the first cell division. M, meristemoid; Prd, protoderm. B, Results from sequential pattern analysis of protein expression using the PrefixSpan algorithm. The colocalization (black) and SPCH alone (magenta) were more frequently seen than MYB16 alone (green) before cell division. A total of 156 serial events were collected from time-lapse confocal images of 7-dpg true leaves for sequential pattern analysis. C, Frequency of the state transition between SPCH alone, MYB16 alone, and colocalization using the PrefixSpan algorithm. The state transition from colocalization (black) to SPCH alone (magenta) had the highest frequency (39.1%). D, MYB16-YFP driven by the MYB16 promoter in an 8-dpg true leaf. Confocal image shows that MYB16 expression is limited to SLGCs (upper inset), young GCs (lower inset), and pavement cells (arrowhead). E, Confocal image of the CYP77A6 transcriptional reporter in an 8-dpg true leaf shows that CYP77A6 expression is stomatal lineage-specific, as seen in meristemoid (upper inset) and pairs of GCs (lower inset). Seven individual T1 lines were characterized and showed similar results. F, Summarized expression window of the MYB16, BASL, and CYP77A6 promoters. M, meristemoid. G–I, Confocal images of 10-dpg true leaves. MYB16-YFP was driven by the MYB16 (G), BASL (H), or CYP77A6 (I) promoter. Rather than being preferentially localized to SLGCs in the MYB16 native promoter lines (G), BASL (H), and CYP77A6 (I) promoter-driven MYB16 signals were seen in meristemoid cells (arrowheads). J–L, DIC images of the lower epidermis from 10-dpg true leaves of the WT (J), BASLp:MYB16-YFP (K), and CYP77A6p:MYB16-VP16 (L) lines. Stomatal clusters (brackets) were found in BASLp and CYP77A6p lines. Mature GCs are pseudo-colored in blue. M–O, Quantification of stomatal density (M), stomatal index (N), and the number of stomatal groups (O). All parameters increased when MYB16 was ectopically expressed in early stomatal lineage cells. n = 20 seedlings. *P < 0.05, by Wilcoxon signed-rank test. Data are medians (interquartile range). P, Total cluster events show that ectopically expressing MYB16 in stomatal lineage causes stomatal cluster formation. The values were obtained from the sum of the events from 20 lower-epidermis samples. Cell outlines are marked either by ML1p:RC12A-mCherry in (A), (D), and (G) to (I) (gray in (A), magenta in (D) and (G)–(I)) or stained by PI in (E). Scale bars, 5 µm in (A) and 50 µm in (D), (E), and (G)–(L). For (M)–(P), three biological replicates showed similar results. See also Supplemental Figures S3–S7.
First, we focused on the sequential pattern of MYB16 or SPCH expression followed by cell division. The most common pattern was the colocalized expression of MYB16 and SPCH before cell division (80.8%), followed by SPCH alone (70.5%), then MYB16 alone (31.4%; Figure 4B), which suggests that SPCH but not MYB16 expression drives cell division. Second, to examine the relationship between SPCH and MYB16 expression, we checked all the possible state transitions between SPCH alone, MYB16 alone, and their colocalized expression. The PrefixSpan analysis revealed that the most common phenomenon was the colocalized expression of MYB16 and SPCH, followed SPCH expression alone (39.1%; Figure 4C). Together with our findings regarding the inhibition of MYB16 expression by SPCH (Figure 1G), and the time-lapse imaging of MYB16 and SPCH expression (Figure 1C), we conclude that SPCH suppresses MYB16 expression in meristemoids before asymmetric division. Moreover, although MYB16 was expressed in the early meristemoids and disappeared before their asymmetric division, it was again expressed in young GCs (Figure 4, A [SLGC, 24 h, asterisk], D, and F). The tightly regulated expression pattern of MYB16 indicates that the timing of its downregulation is critical during stomatal development.
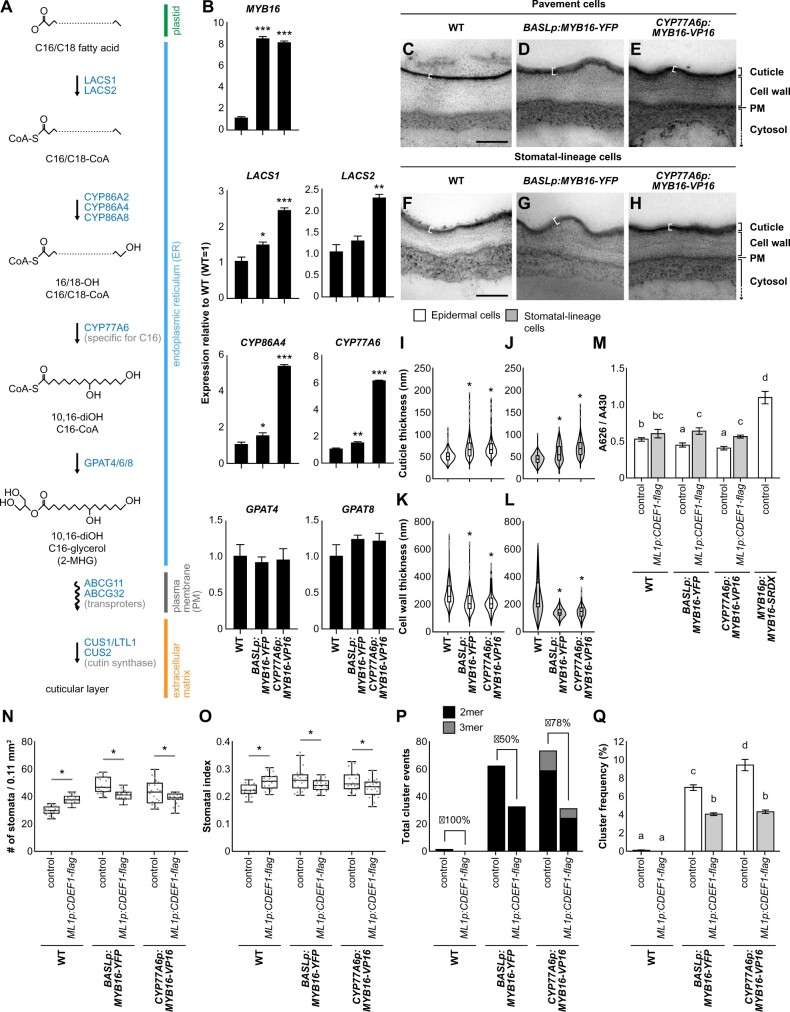
The cuticle can improve stress tolerance in plants; therefore, enhancing cuticle production by overexpressing MYB106 and SHN1/WIN1 provides an opportunity to help plants adapt to adverse environmental stresses (Oshima et al., 2013). Despite this, the overexpression of MYB106 driven by the 35S promoter resulted in severe dwarfism, which is not beneficial for commercial applications (Oshima et al., 2013). To avoid pleiotropic effects, Oshima and Mitsuda (2016) used the promoters of genes that function downstream of MYB16 and MYB106, CYP77A6 and CYP86A4, to spatiotemporally enhance cuticle production. CYP77A6- and CYP86A4-promoter-driven MYB16 expression effectively induced the accumulation of cuticular material in the leaves and petals, respectively, without inhibiting plant growth (Oshima and Mitsuda, 2016). The authors used scanning electron microscopy to demonstrate that stomatal clusters formed in plants expressing CYP77A6p:MYB16-VP16, a dominant-positive form of MYB16 fused with a transcription activation domain (VP16), but not in plants expressing CYP86A4p:MYB16-VP16 (Oshima and Mitsuda, 2016).
Since the expression of endogenous MYB16 is tightly controlled in the leaf epidermis (Figures 1, 4, D and F), we wondered whether the cluster formation in CYP77A6p:MYB16-VP16 plants but not in CYP86A4p:MYB16-VP16 plants resulted from this transcriptional control. We used the same promoter regions as Oshima and Mitsuda (2016) and fused them with a nucleus-localized fluorescent protein (nucEGFP) to generate the CYP77A6p:nucEGFP and CYP86A4p:nucEGFP transcriptional reporters. Because CYP77A6 and CYP86A4 encode well-known enzymes in cutin biosynthesis, we expected that they would be expressed in every epidermal cell. Interestingly, all seven independent CYP77A6 reporter lines showed a stomatal lineage-specific expression pattern of nucEGFP signals (Supplemental Figure S4, A–D). We quantified the fluorescent signals in one line as follows: meristemoids (25.6%), GMCs (22%), young GCs (27.4%), and mature GCs (25%; Figure 4, E and F; Supplemental Figure S4, A–D, M). For the CYP86A4 reporters, we observed two types of expression patterns: class I, in which signals were found in the stomatal lineage cells only, with preferential expression in young and mature GCs (52.9% and 47.1%, respectively; four independent lines; Supplemental Figure S4, E–H, M); and class II, in which signals were found in every epidermal cell (four independent lines; Supplemental Figure S4, I–M). The discrepancy in the expression patterns of the CYP86A4 reporters made it difficult for us to draw conclusions about the transcription pattern of CYP86A4. By analyzing stomatal single-cell RNAseq data (Lopez-Anido et al., 2021), we found that among the three MYB16 downstream targets (Oshima et al., 2013; Oshima and Mitsuda, 2016), ABCG12 transcripts were abundantly found in most stomatal lineage cells (Supplemental Figure S5), whereas CYP77A6 and CYP86A4 transcripts were much less abundant and were more commonly found in cells with a stomatal fate (Supplemental Figure S5). Further analyses using different versions of the CYP86A4 promoter are required to confirm its expression pattern in the leaf epidermis. Based on the CYP77A6 expression pattern (Figure 4E; Supplemental Figure S4, A–D) and the stomatal clusters produced by CYP77A6p:MYB16-VP16 plants (Oshima and Mitsuda, 2016), we hypothesized that the suppression of MYB16 expression during the early stage of stomatal development prevents ectopic cuticle deposition and ensures proper stomatal patterning.
BASL is expressed during asymmetric cell divisions in stomatal lineage cells (Dong et al., 2009); therefore, to test our hypothesis, we used the BASL promoter in addition to the CYP77A6 promoter to ectopically express MYB16 in meristemoids (Figure 4F). Similar to previous results (Figure 1), MYB16 expression driven by its native promoter (MYB16p:MYB16-YFP) was preferentially localized to the SLGCs (Figure 4G, arrowhead). To analyze the stomatal phenotypes, we used BASLp:MYB16-YFP plants generated in this study and CYP77A6p:MYB16-VP16 plants obtained from Oshima and Mitsuda (2016). Ectopically expressing MYB16 fused with the BASL or CYP77A6 promoter (BASLp:MYB16-YFP or CYP77A6p:MYB16-VP16-YFP) restricted the nuclear signals to stomatal-lineage cells (Figure 4, H and I, arrowheads). Three BASLp:MYB16-YFP and CYP77A6p:MYB16-VP16-YFP lines were obtained, all of which showed similar stomatal phenotypes, with pairs of clustered stomata (Figure 4K; Supplemental Figure S6). Since the CYP77A6p:MYB16-VP16 plants were found to have strong stomatal phenotypes in the previous study (Oshima and Mitsuda, 2016), we used this line for the following studies. Compared to WT seedlings, the lines with MYB16 ectopic expression (referred to as ectopic MYB16 lines hereafter) had higher stomatal densities and stomatal indices, as well as more stomatal groups and total cluster events (Figure 4, J–P), suggesting that the increased numbers of stomata in the ectopic MYB16 lines arose from an elevated number of stomatal initiation events and stomatal clustering. The high variation in the stomatal densities and numbers of stomatal groups observed in CYP77A6p:MYB16-VP16 could be attributed to two types of stomatal phenotypes in seedlings (Supplemental Figure S7): some of the seedlings had typical clustered stomata, while others showed tumor-like colonies (small cell clusters) similar to the basl-1 mutant (Dong et al., 2009). These findings, together with the localization and timing of MYB16 expression, suggest that the reduction in MYB16 expression in meristemoids is required for proper stomatal formation and the prevention of stomatal clustering.
Changes to the cuticle–cell wall continuum influence stomatal development in the ectopic MYB16 lines
MYB16 is a key regulator of cuticle development (Oshima et al., 2013). SHN1/WIN1, one of its downstream targets, modifies the outer cell wall via the production of cutin and cuticular wax (Broun et al., 2004; Kannangara et al., 2007). The SHN family also acts directly on enzymes that modify the epidermal cell wall by altering the formation of cell wall polysaccharides and structural proteins (Shi et al., 2011). The ectopic expression of MYB16 may therefore affect genes involved not only in cutin biosynthesis but also in cell wall modification.
The biosynthesis of cutin involves ω-hydroxylation, midchain hydroxylation, and the biosynthesis of acyl-CoA intermediates and glycerolipids. Figure 5A illustrates the general cutin monomer biosynthesis pathway, but the corresponding biosynthetic enzymes may vary depending on the type of monomer. To explore the expression of the corresponding genes in the ectopic MYB16 lines, we measured LACS1/2 (Lü et al., 2009) and GPAT4/8 (Li et al., 2007) expression, as well as the expression of the MYB16 downstream targets CYP77A6 and CYP86A4, using qRT-PCR. In accordance with the broader expression range of the CYP77A6 promoter (Figure 4F), all of these genes except for GPAT4 and GPAT8 were upregulated in the ectopic MYB16 lines, with higher expression levels in CYP77A6p:MYB16-VP16 than in BASLp:MYB16-YFP plants (Figure 5B). The expression levels of these genes also reflect the degree of stomatal clustering, as CYP77A6p:MYB16-VP16 plants produced more stomatal clusters than BASLp:MYB16-YFP plants (Figure 4P). The cluster phenotypes (Figures 3 and 4) and the expression patterns of the genes in the ectopic MYB16 lines were consistent with the findings for the MYB16 inducible lines (iMYB16), in which the cuticular biosynthesis-related genes (LACS1, LACS2, CYP77A6, CYP86A4, GPAT4, and GPAT8) were all upregulated by the induction of MYB16 expression (Supplemental Figure S8).
Figure 5.
Expressing the cutinase gene CDEF1 decreases stomatal cluster formation in ectopic MYB16 lines. A, A diagram showing the biosynthesis of one type of cutin monomer. The C16/C18 fatty acid from plastids is transformed into the hydroxylated acyl-CoA intermediate and then becomes cutin monomers in the ER. The transport of the monomers supplies the material required for the polymerization of the cuticle layer outside the cell walls. The polymerization of the cuticle layer is mediated by cutin synthase. Genes involved in the reaction are labeled in blue. B, Relative mRNA expression of cuticle biosynthesis genes in ectopic MYB16 lines. A 7-dpg WT is used as a reference (equal to 1). *P < 0.05; **P < 0.01; ***P < 0.001, by Student’s t test. Data are mean (sd). Three biological replicates showed similar results. C–H, TEM images of the lower epidermis from 7-dpg true leaves of the WT (C) and (F), BASLp:MYB16-YFP (D) and (G), and CYP77A6p:MYB16-VP16 (E) and (H). Both pavement cells (C)–(E) and stomatal-lineage cells (F)–(H) showed thicker cuticle deposition (white brackets) on the cell surface in BASLp and CYP77A6p lines than the WT. I–L, Quantification of cuticle (I) and (J) and cell wall (K) and (L) thickness from (C) to (H). The cuticle thickness of epidermal (I) and (K) and stomatal lineage cells (J) and (L) increased in ectopic MYB16 lines compared to the WT, but cell wall thickness decreased. Data were obtained from 200 to 300 regions of lower epidermal cells. *P < 0.05, by Student’s t test. Data are medians (interquartile range). Gray dots represent Tukey outliers. M, Quantification of penetrated TB showing that MYB16-SRDX plants are more permeable than BASLp or CYP77A6p plants. After ectopically introducing the cutinase gene CDEF1 into BASLp or CYP77A6p lines, seedlings showed more penetrated TB compared with the control in each background. The TB absorbance (A626) is normalized by chlorophyll absorbance (A430). P < 0.05. Kruskal–Wallis with Dunn’s test. Data are means (se). N, O, Quantification of stomatal density (N) and stomatal index (O) shows the partial rescue of MYB16 ectopic expression plants by ectopically expressing the cutinase gene CDEF1. n = 20 10-dpg seedlings. *P < 0.05, by Wilcoxon signed-rank test. Data are medians (interquartile range). P, Q, The number of stomatal clusters (P) and cluster frequency (Q) decreased after ectopically expressing the cutinase gene CDEF1. For (P), the values were obtained from the sum of the events in a total of 20 lower-epidermis samples. Rescue percentage is the difference in the number of cluster events between the control and ML1p:CDEF1-flag divided by the event number in the control. For (Q), cluster frequency is the cluster event number divided by the number of stomatal groups. P < 0.05. One-way ANOVA with Tukey post hoc test. Data are means (se). For (C)–(H), shared scale bars, 200 nm, in (C) and (F). For (M)–(Q), two biological replicates and two individual lines in each background showed similar results. See also Supplemental Figures S8–S10.
Next, we used TEM to directly quantify the changes to the cuticle. We measured cuticle thickness in pavement cells and stomatal-lineage cells in the abaxial side of the leaf (Figure 5, C–H; the electron-dense cuticle is indicated by a white bracket; Supplemental Figure S9, A–I). The cuticles of the ectopic MYB16 epidermal cells, including both pavement cells and stomatal-lineage cells, were thicker than those of WT plants (Figure 5, I and J). This observation was validated by the measurement of stomatal-lineage cells only (Figure 5J). We also noticed that the cell wall thickness of the stomatal-lineage cells was reduced in the BASLp:MYB16-YFP and CYP77A6p:MYB16-VP16 lines relative to the WT (Figure 5, K and L). These observations suggest that MYB16 plays a role in both cutin biosynthesis and cell wall formation.
In addition to the transmission electron microscope (TEM) observations, the toluidine blue (TB) penetration test is often used to measure the permeability of seedlings to this aqueous dye (Tanaka et al., 2004), with a greater uptake of TB in seedlings with defective cuticles. The dominant-negative MYB16p:MYB16-SRDX plants were more strongly stained with TB than the WT plants; in contrast, the BASLp:MYB16-YFP and CYP77A6p:MYB16-VP16 lines showed less TB penetration (Figure 5M; Supplemental Figure S9, J–M). Since TB also has a high affinity for acidic components such as acid groups in the cell wall, the decreased staining of TB in the ectopic MYB16 lines may also suggest the altered composition of the cell wall. In summary, the ectopic expression of MYB16 enhanced the cuticle thickness and the sealing property of the cuticle, as revealed by both TEM imaging and TB staining assays.
To confirm that the formation of stomatal clusters in the ectopic MYB16 lines was due to the accumulation of cutin, we ectopically expressed CDEF1, a cutinase gene (Takahashi et al., 2010), in epidermal cells under the control of the epidermis-specific AtML1 promoter in a BASLp:MYB16-YFP or CYP77A6p:MYB16-VP16 background. For each double transgenic combination, two lines were analyzed. First, we examined the expression of MYB16 and CDEF1 in the double transgenic lines using qRT-PCR (Supplemental Figure S10, A and B), revealing that MYB16 and CDEF1 were not silent after introducing the two transgenes. Although MYB16 expression was slightly reduced in CYP77A6p:MYB16-VP16/ML1p:CDEF1-FLAG #2, it was still higher than it was in the WT background (Supplemental Figure S10A). Second, we confirmed the presence of the MYB16-YFP protein in the BASLp:MYB16-YFP/ML1p:CDEF1-FLAG plants using confocal microscopy (Supplemental Figure S10, C and D). Finally, we used TB staining to estimate the effect of cutinase, revealing that, compared with the ectopic MYB16 lines alone, TB more strongly penetrated the ectopic MYB16 seedlings expressing CDEF1 (Figure 5M; Supplemental Figure S9, N–P).
The specific epidermal expression of CDEF1 led to a slight increase in the stomatal density and stomatal index in WT plants, while both measurements were reduced in the ectopic MYB16 lines with CDEF1 expression (Figure 5, N and O; Supplemental Figure S10, E and F). Because the changes in stomatal index could result from the alteration of the numbers of single stomate or clustered stomata, we measured the cluster events and cluster frequency to specifically probe the influence of the cuticle on stomatal cluster formation (Figure 5, P and Q; Supplemental Figure S10, G and H). The stomatal cluster events were reduced in WT plants and in the ectopic MYB16 lines expressing CDEF1 (Figure 5P; Supplemental Figure S10G). Since stomatal clusters rarely form in the WT, there was no significant difference in the cluster frequency between the WT with or without leaf epidermal CDEF1 expression (Figure 5Q; Supplemental Figure S10H); however, the cluster frequency was strongly reduced in the ectopic MYB16 lines expressing CDEF1 (Figure 5Q; Supplemental Figure S10H). This rescue of the cluster frequency phenotype suggested a link between stomatal development and cutin deposition.
EPF-mediated signaling is not affected in the ectopic MYB16 lines
The ectopic expression of MYB16 altered the cuticle–cell wall continuum (Figure 5) and increased the number of stomata and stomatal clusters (Figures 3 and 4). To elucidate the possible mechanisms affecting stomatal patterning, we first examined the expression of stomatal genes after the short-term induction (0, 6, and 8 h) of MYB16 expression using the iMYB16 line. MYB16 expression was highly upregulated, as expected (Supplemental Figure S11), while SPCH and MUTE expression remained similar at both 0 and 6 h of induction but decreased after 8 h of induction. FAMA was slightly upregulated after both 6 h and 8 h of induction. The co-factor of SPCH, MUTE, and FAMA, SCRM/ICE1, was downregulated after 8 h of induction, as were the polarity complex genes BASL and POLAR. Regarding the signaling genes establishing the one-cell-spacing rule, EPF1 was strongly downregulated and EPF2 was slightly downregulated after 8 h of induction (Supplemental Figure S11). The number of stomata in iMYB16 plants was elevated after the induction of MYB16 expression (Figure 3); however, the asymmetric division driver, SPCH, the stomatal fate determinator, MUTE, and their cofactor SCRM/ICE1 were not upregulated, indicating that the elevated number of stomata was not caused by the regulation of stomatal transcription factors. The observed elevated FAMA expression could be due to the overproduction of stomatal clusters. The reduced transcript levels of BASL, POLAR, EPF1, and EPF2 after 8 h of induction resulted from the maturation of precursor cells. Therefore, the excessive stomata produced by iMYB16 seedlings and the unchanged transcript levels of SPCH and MUTE (Supplemental Figure S11 at 6 h) suggest that, at 8 h of MYB16 induction, these stomata had reached the FAMA stage or beyond, and the altered transcript levels were not the major reason for the enhanced stomatal production.
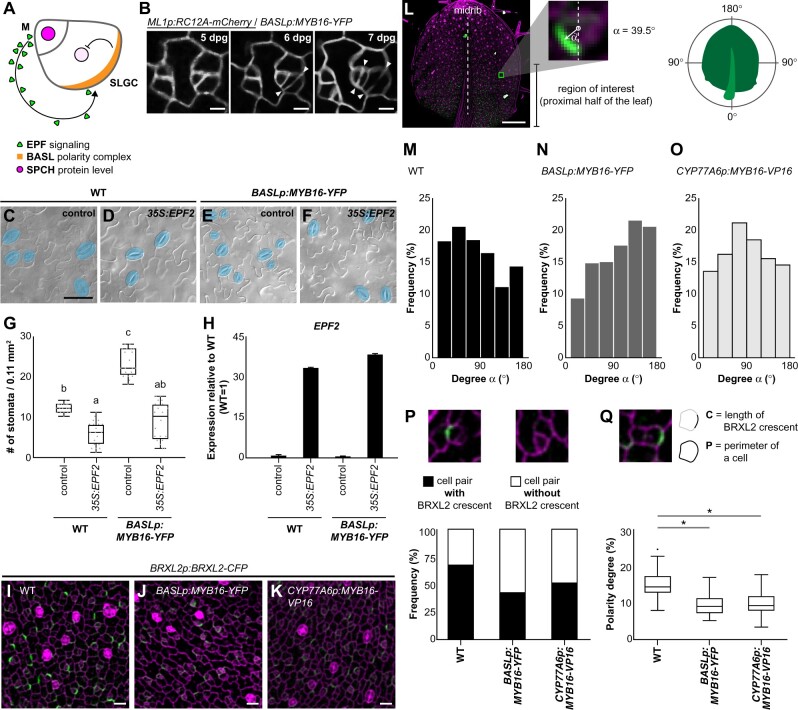
During stomatal development, to ensure proper patterning, the positional control of the polarity complex is required to faithfully place EPF-mediated MAPK signaling components in the SLGCs to inhibit stomatal fate (Dong et al., 2009; Pillitteri et al., 2011; Zhang et al., 2015; Houbaert et al., 2018; Rowe et al., 2019; Figure 6A). To explore how stomatal clusters are formed, we examined their formation in plants expressing BASL promoter-driven MYB16-YFP and RC12A-mCherry, a cell membrane marker. Under normal conditions, an SLGC undergoes an asymmetric division to form nonadjacent stomatal precursors (Figure 6B, left part); however, both daughter cells produced by divisions in the ectopic MYB16 line were frequently observed to form two adjacent stomata (Figure 6B; arrowheads indicate divisions).
Figure 6.
Stomatal cluster formation is caused by the mis-localization and reduced amounts of polarity protein in the stomatal lineage. A, Diagram shows the EPF-mediated inhibitory pathway incorporating the spatially labeled polarity complex to prevent stomatal cluster formation in Arabidopsis. EPFs are secreted from meristemoid (M) cells and activate inhibitory signaling in SLGCs, where the polarity complex recruits inhibitory components, leading to decreased SPCH levels. B, Time-lapse confocal images show stomatal cluster formation in the BASL promoter-driven ectopic MYB16 line. Left parts of the images show normal stomatal formation. Right parts show that the adjacent stomate is derived from an SLGC, resulting in stomatal clustering. Arrowheads indicate divisions. C–F, DIC images of the lower epidermis from 14-dpg cotyledons of the WT or BASLp-driven MYB16 lines with or without the overexpression of EPF2. Mature stomata are pseudo-colored in blue. Scale bar, 50 µm. G, Quantification of stomatal density showing that the overexpression of EPF2 reduces the number of stomata in both WT and BASLp:MYB16-YFP. n = 20 14-dpg plants. P < 0.001, by Kruskal–Wallis test with Dunn’s method. Data are medians (interquartile range). H, qRT-PCR analysis of relative mRNA level of EPF2 in 14-dpg cotyledons. EPF2 was highly expressed in plants with the 35S:EPF2 construct. Data are means (sd). I–K, Confocal images of the polarity marker BRXL2 in 7-dpg true leaves. Compared to the WT (I), BRXL2-CFP signal (green) is dimmer in BASLp:MYB16-YFP (J) and CYP77A6p:MYB16-VP16 (K). Four individual lines of each background showed similar results. L, Angle α indicating the angle between the vectors of the midrib and BRXL2. The vector toward the proximal part of a 7-dpg true leaves is set to 0°. α angles were measured in the bases of leaves. M–O, The orientation of polarity in the WT (M), BASLp:MYB16-YFP (N), and CYP77A6p:MYB16-VP16 (O). To avoid the PI effect, α angles were quantified from confocal images of 7-dpg true leaves expressing BRXL2 in the indicated lines without PI staining. n = 769, 324, and 615 cells in (M–O), respectively. P, The proportion of cells with BRXL2 crescents is reduced in ectopic MYB16 lines compared to the WT. n = 227, 150, and 217 cell pairs in the WT, BASLp:MYB16-YFP, and CYP77A6p:MYB16-VP16, respectively. Q, Analysis of the polarity degree of BRXL2 crescents shows that ectopic MYB16 lines have a lower polarity degree compared to the WT. Polarity degree is calculated from crescent length divided by cell perimeter. The dataset is derived from 67 cells with peripheral BRXL2 for each line. The dot shows the Tukey outlier. *P < 0.001, by Student’s t test. Data are medians (interquartile range). Cell outline marked by ML1p:RC12A-mCherry in (B, gray) and labeled by PI in (I)–(K), (P), and (Q) (magenta). Scale bars, 5 µm in (B), 20 µm in (I)–(K), and 200 µm in (L). For (M)–(Q), data combined from four individual lines of each background. See also Supplemental Figures S11 and S12.
The misregulation of EPF-mediated stomatal signaling was previously shown to result in cluster formation (Hara et al., 2007; Hunt and Gray, 2009). The cuticle has been proposed to act as an inhibitory signal or a regulator of diffusion across the outer epidermal cell wall to control stomatal numbers (Gray et al., 2000; Bird and Gray, 2003). To test whether the alteration of the cuticle in the ectopic MYB16 lines influences EPF-mediated signaling, we overexpressed EPF2, encoding a major EPF ligand involved in early stomatal development, in BASLp:MYB16-YFP plants. Similar to the WT response, the stomatal density was reduced in BASLp:MYB16-YFP (Figure 6, C–H), suggesting that EPF-mediated signaling was still functional in the ectopic MYB16 lines.
Stomatal clusters in the ectopic MYB16 lines result from the reduced and disrupted localization of polarity protein in asymmetrically dividing cells
In addition to EPF signaling, the polarity setup is also important for stomatal patterning. The establishment of stomatal polarity was previously shown to be affected by the manipulation of mechanical force using laser ablation (Bringmann and Bergmann, 2017). The tensile strength, measured using AFM, is different in the WT and the cell adhesion mutant qua1 (Verger et al., 2018), indicating that the alteration of the cell wall composition modulates the mechanical properties of the epidermis. The thicker cuticle in epidermal cells and the changes in the cell walls of the stomatal-lineage cells (Figure 5) suggested that the tensile strength in the ectopic MYB16 lines is different from that of the WT. We therefore hypothesized that the changes in the leaf cuticle influence the physical properties of the epidermis, consequently modulating the establishment of polarity.
To observe the polarity setup in stomatal-lineage cells, we used BRXL2-CFP as an indicator of the polarity of the polar complex in these cells. The intensity of the fluorescence of BRXL2-CFP was markedly lower in the ectopic MYB16 lines than in the plants with a WT background (Figure 6, I–K; Supplemental Figure S12, A–C). To further characterize these defects, we used three parameters to quantify cell polarity. First, we measured the tissue-wide orientation of BRXL2 based on the degree α of the angle between the midrib (distal–proximal axis) and the direction of BRXL2 polarity (Figure 6L). BRXL2 polarity varies depending upon the position of the leaf (Bringmann and Bergmann, 2017). The BRXL2 crescents in the leaf tip do not exhibit a specific pattern, but at the base of the leaf, they tend to face toward the midrib in both the left and right halves of the leaf, matching the symmetry of the leaf (Bringmann and Bergmann, 2017). We therefore analyzed BRXL2-CFP signals in cells located in the basal halves of the leaves (Figure 6L, region of interest). The leaves were not stained with propidium iodide (PI) because we were concerned that the PI solution could damage the cells and cause polarity changes. Most of the BRXL2 orientations were between 30° and 60° in 7-dpg WT seedlings (Figure 6M), similar to the previous observations (Bringmann and Bergmann, 2017); however, in the ectopic MYB16 lines, BRXL2 orientation peaked between 120° and 150° in BASLp:MYB16-YFP and between 60° and 90° in CYP77A6p:MYB16-VP16 (Figure 6, N and O). Second, we quantified the proportion of cells with or without a BRXL2 crescent in the meristemoid–SLGC pairs. The proportion of cells with BRXL2 crescents was reduced in BASLp:MYB16-YFP (43%) and CYP77A6p:MYB16-VP16 (52%) compared to the WT (69%) (Figure 6P). Third, the size of the faint BRXL2 signal was smaller in the ectopic MYB16 lines than in the WT (Figure 6, I–K). To quantify the crescent size, we examined the polarity degree (Zhang et al., 2015; Gong et al., 2021), i.e. crescent length normalized by the cell perimeter, and found that the BRXL2 crescents were significantly shorter in both ectopic MYB16 lines than in the WT (Figure 6Q). In summary, these findings indicate that the adjacent clustered stomata in the ectopic MYB16 lines were derived from an SLGC with reduced expression of the polarity components and aberrantly oriented BRXL2.
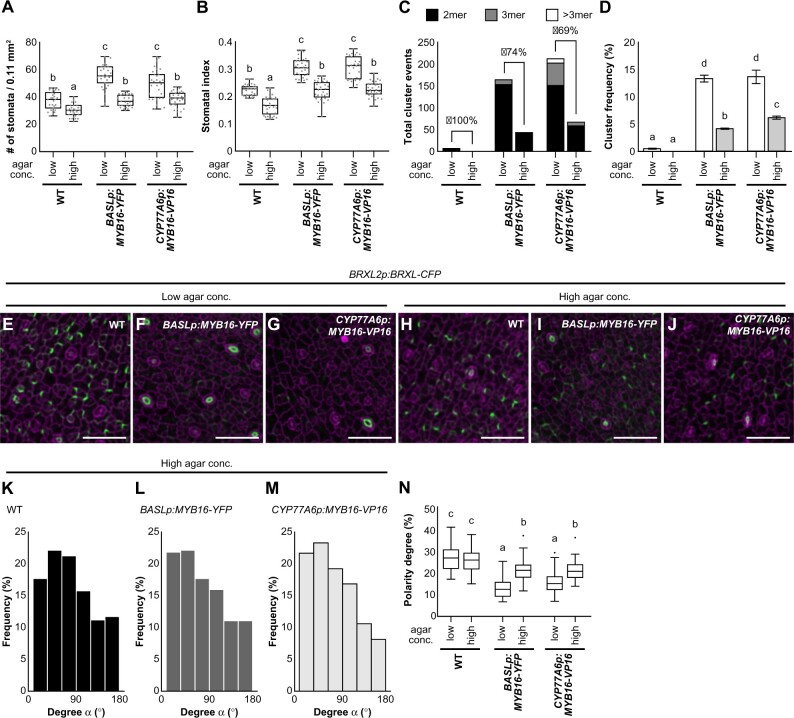
To further confirm that the disrupted establishment of polarity caused stomatal cluster formation in the ectopic MYB16 lines, we attempted to rescue the polarity defects. Reducing the water potential by growing seedlings in high-percentage agar was previously shown to decrease pavement cell stiffness and cell wall tension (Verger et al., 2018). We therefore applied this strategy to manipulate mechanical force and modulate polarity localization during stomatal development. Since the BRXL2 polarity setup was influenced by the changes in the mechanical properties of the leaf epidermis (Bringmann and Bergmann, 2017), we compared plants growing on low- (0.8%) and high-percentage (2.4%) agar plates. In the ectopic MYB16 lines, the stomatal density and stomatal index were rescued by treatment with high-percentage agar (Figure 7, A and B); cluster events and the cluster frequency were also reduced (Figure 7, C and D). Moreover, polarity in the ectopic MYB16 lines was partially rescued by this treatment (Figure 7, E–J; Supplemental Figure S12). Using the same quantifying methods used in Figure 6, we revealed that the orientation as well as the degree of polarity was rescued in the ectopic MYB16 plants grown on high-percentage agar (Figure 7, K–N). Taken together, modulating the tensile strength achieved by growing plants in high-percentage agar partially rescued the polarity and stomatal phenotypes of the ectopic MYB16 lines. These results indicate that ectopic MYB16 expression alters the composition of the cell wall and the cuticle and influences the mechanical properties of the epidermis, resulting in an aberrant polarity setup in stomatal lineage cells.
Figure 7.
The stomatal phenotype in ectopic MYB16 lines is rescued by high-percentage agar treatment. A, B, Quantification of stomatal density (A) and stomatal index (B) show the rescue of the stomatal phenotype in ectopic MYB16 lines under high-percentage agar treatment. Low, 0.8% agar (normal conditions), and high, 2.4% agar. n = 30 10-dpg seedlings. P < 0.01, Kruskal–Wallis test with Dunn’s test. Data are medians (interquartile range). Three biological replicates showed similar results. C, D, Stomatal cluster number (C) and cluster frequency (D) decrease after high-percentage agar treatment. For (C), the rescue percentage was calculated using the difference in cluster events between two types of agar treatments divided by the number of cluster events in low-percentage agar treatment. For (D), cluster frequency is the cluster event number divided by the number of stomatal groups; P < 0.05. One-way ANOVA with Tukey post hoc test. Data are means (se). Three biological replicates showed similar results. E–J, Confocal images of the polarity marker BRXL2 in 7-dpg true leaves from plants grown in two different concentrations of agar. BRXL2-CFP signal (green) is similar in the WT (E) and (H) but stronger in BASLp:MYB16-YFP (F) and (I) and CYP77A6p:MYB16-VP16 (G) and (J) after high-percentage agar treatment. K–M, The orientation of polarity is rescued in BASLp:MYB16-YFP and CYP77A6p:MYB16-VP16 after high-percentage agar treatment. The data were quantified from confocal images of 7-dpg true leaves expressing BRXL2 without PI staining. n = 587, 369, 321 cell pairs in (K)–(M), respectively. N, The rescue of the BRXL2 crescent size in ectopic MYB16 lines by high-percentage agar treatment. The calculation method is the same as in Figure 6Q. Sixty cells with peripheral BRXL2 were collected from each line under each treatment. The dot shows the Tukey outlier. P < 0.001, by one-way ANOVA with Tukey post hoc test. Data are medians (interquartile range). For (E)–(J), cell outline is labeled by PI (magenta). Scale bars, 50 µm in (E)–(J). For (K)–(N), data are combined from four individual lines of each background. See also Supplemental Figure S12.
Discussion
MYB16 expression in stomatal lineage cells
During organ morphogenesis, cell-to-cell signaling, oriented cell division, and the extracellular matrix are all required for maintaining stem cell multipotency or transforming stem cells into differentiated cells, thereby generating a specific pattern. The mechanisms by which biochemical signaling and mechanical properties are integrated to form a functional tissue are yet to be elucidated. Here, we used Arabidopsis stomatal lineage cells to investigate the coordination of the cuticle and fate determiners in leaf epidermal development. Using time-lapse imaging and quantitative analysis of sequential events in stomatal lineage progression, we revealed that MYB16, which plays a role in cutin biosynthesis and cell wall modification (Broun et al., 2004; Kannangara et al., 2007; Shi et al., 2011; Oshima et al., 2013), is transcriptionally suppressed by SPCH in meristemoids (Figure 1). Following the activity of SPCH, MUTE is expressed in meristemoids to drive stomatal development. Despite the previously finding that MYB16 is downregulated in inducible SPCH and MUTE lines (Lau et al., 2014; Han et al., 2018), we showed that MUTE and its co-factor SCRM induced MYB16 expression in vitro (Supplemental Figure S2), suggesting that other players may also function with MUTE to inhibit MYB16 expression in vivo.
One of the functions of an epidermal cell is to produce an outer epidermal cell wall/cuticle to form a physical barrier. Therefore, every protodermal cell in the epidermis might be able to express MYB16 to turn on downstream genes for cuticle production; however, the timing of expression is important. Given that stomatal development is tightly controlled, the downregulation of MYB16 expression in meristemoids and its reappearance after the symmetric division of young GCs (whose fate is specified; Ho et al., 2021; Figure 4) might be controlled by both stomatal transcription factors and the cell cycle itself. Eventually, a mature stomate with a thick cuticle is formed. These findings indicate that the formation of the cuticle is tightly regulated and integrated with stomatal lineage progression during leaf growth to ensure the proper development of stomata and the leaf epidermis.
Overexpression of MYB16 results in increased stomatal density and stomatal cluster formation
Various stomatal quantification methods are used to distinguish stomatal numbers and clusters. Stomatal phenotypes are often quantified in terms of stomatal density and stomatal index (Figure 3); however, because these measures count every stomate as a single event, it is difficult to determine whether the increased number of stomata observed in plants overexpressing or ectopically expressing MYB16 comes from multiple stomatal singlets or clusters. Thus, the stomatal group measurement is used to represent stomatal origins. When comparing two samples, if the numbers of stomatal groups are similar among samples, any increase in stomatal number will be due to cluster events. This method applies to clusters from the same origin (dividing sister cells), as shown in Figure 6B or the basl mutant (Dong et al., 2009). In mutants with severe signaling defects such as the tmm or erecta erecta-like 1 (erl1) erl2 triple mutants, whose stomatal clusters are usually large, it can be difficult to distinguish the origins of the stomatal lineages. The stomatal clusters shown in the MYB16-overexpression lines (Figures 3 and 4) were pairs and triplets, meaning that the stomatal group measurement could be used to estimate the number of stomatal origins.
The increased stomatal index, stomatal group, and cluster events in lines overexpressing or ectopically expressing MYB16 (Figures 3 and 4) suggest that overexpressing MYB16 promotes both stomatal production and cluster formation. Increasing wax accumulation by overexpressing WIN1/SHN1 reduced stomatal formation, but this effect was not mediated by the transcriptional control of stomata-related genes (Yang et al., 2011). These findings, together with the observation that WIN1/SHN1 regulate the expression of genes involved in cutin and cell wall biosynthesis (Broun et al., 2004; Kannangara et al., 2007; Oshima et al., 2013), suggest that the cell wall and cuticle influence stomatal development. Here, we made a similar observation, i.e. inducing MYB16 expression resulted in an elevated number of stomata (Figure 3). However, SPCH and MUTE were not upregulated in these lines (Supplemental Figure S11), indicating that stomatal transcription factors do not play a role in enhancing stomatal production in the ectopic MYB16 lines. In addition, the mutation of HIC, a gene involved in the biosynthesis of very-long-chain fatty acid, results in a thinner cuticle and higher stomatal density (Gray et al., 2000). These observations point to a negative relationship between the cuticle and stomatal production. This finding was also supported by expressing CDEF1 in WT plants, which led to a thinner cuticle and an increased stomatal number. However, we observed a decreased stomatal number in ectopic MYB16 plants expressing CDEF1 (Figure 5, K and L). As revealed by TEM (Figure 5; Supplemental Figure S9), the cuticle and cell wall thickness were altered in the ectopic MYB16 lines. Since CDEF1 only degrades cutin, the compositions of the cuticle and cell wall may be different in WT and ectopic MYB16 lines. These differences in cuticle and cell wall structure would lead to the difference in stomatal initiation between these lines.
Although the formation of the cuticle has been linked to stomatal density, as suggested above (Figure 5K; Gray et al., 2000; Yang et al., 2011), our study showed that, besides the increased stomatal origins revealed by measuring stomatal groups, the number of cluster events was also significantly increased in the ectopic MYB16 lines (Figures 3 and 4). After inducing MYB16 expression for 8 h, the transcript levels of BASL, POLAR, and EPF1 were significantly reduced and EPF2 was moderately downregulated (Supplemental Figure S11), suggesting that ectopic MYB16 expression, together with the changes in the expression or activity of SPCH and MUTE, transcriptionally regulates the genes involved in cell-to-cell signaling to influence stomatal patterning. It is also possible, however, that the decreased levels of BASL, POLAR, EPF1, and EPF2 expression at 8 h after induction result from the maturation of precursor cells, since SPCH and MUTE were also downregulated. In addition, the weak stomatal phenotype of amiR-MYB16 myb106-2 (Figure 2) and the low correlation between MYB16 expression and cell division (Figure 3B) imply that MYB16 is not directly involved in promoting cell division. We therefore investigated mechanisms other than transcriptional control, as shown in Figure 6 and discussed below.
The effects of MYB16 in modulating the cuticle–cell wall continuum
A transcript-level analysis showed that the cutin-related genes were upregulated in both ectopic MYB16 lines (Figure 5B), but LACS1/2, CYP77A6, and CYP86A4 were more highly expressed in plants expressing MYB16 under the control of the CYP77A6 promoter (Figure 5B). This phenomenon could be due to the difference in cell-type specificity, as the BASL promoter only functions in meristemoids (Figure 4F), while the CYP77A6 promoter (Figure 4F; Supplemental Figure S4) is active throughout stomatal development. Since GCs are covered with a thick cuticle, the CYP77A6 promoter may also be much more active than the BASL promoter in the stomatal lineage (Figure 4F). Although the transcript levels of cutin-related genes were different in the ectopic MYB16 lines, the epidermal cells had thicker cuticles in both the BASL promoter- and CYP77A6 promoter-driven ectopic MYB16 lines. It is possible that cells maintain homeostasis for cuticle production, so even the higher enzyme production in the CYP77A6 plants versus the BASL plants did not result in different cuticular thicknesses. Another possibility is that we only showed the biosynthesis pathway for a certain type of cutin monomers in Figure 5a, but perhaps MYB16 is also involved in the biosynthesis of other types of cutin monomers. In addition, although we used the promoters of genes expressed in the stomatal lineage to overexpress MYB16, enhanced cuticle formation was detected in all epidermal cells. It is possible that cutin monomers are limiting for cutin biosynthesis and are synthesized at high levels due to the increased amounts of stomatal linage cells and then diffuse to the outer cell wall in young leaves and are catalyzed by the extracellular enzymes CUS1 and CUS2 (Yeats et al., 2014; Philippe et al., 2016; Hong et al., 2017) into cutin in the epidermis. Thus, thicker cuticles were observed in all types of epidermal cells. Interestingly, changes in cell wall thickness were also observed in stomatal-lineage cells (Figure 5, K and L), providing further evidence that MYB16 expressed under the control of the BASL or CYP77A6 promoter functions in the stomatal lineage. MYB16 may modify the composition of the cell wall through its downstream genes or SHN family members, which regulate cutin and wax biosynthesis genes and other genes for cell wall modification (Shi et al., 2011). The identification of the downstream targets of MYB16 using ChIP-seq will improve our understanding of its role in establishing the cuticle–cell wall continuum.
Cuticular thickness and wax coverage are not correlated with cuticular permeability (Riederer and Schreiber, 2001; Voisin et al., 2009; Xie et al., 2020). For example, the pectin mutant qua2 has an increased amount of cutin and a thick cuticle but highly increased cuticle permeability (Lorrai et al., 2021), implying that a thicker cuticle is not always associated with the better sealing property of the cuticle-cell wall continuum. Instead, the complex relationship between the cell wall and cuticle structure is highly important for the sealing property of the epidermis. One important component of the cuticle is wax, which typically constitutes 20%–60% of the cuticle mass and is responsible for resistance to water loss (Samuels et al., 2008). Since wax cannot be observed in TEM images and wax and cutin biosynthesis could be regulated by the same transcription factor (such as WIN1/SHN1), we cannot exclude the potential contribution of wax to water permeability in the ectopic MYB16 lines. CDEF1 only degrades cutin and permeability significantly decreased after expressing CDEF1 in the ectopic MYB16 lines. These findings suggest that cutin strongly contributes to the sealing property of the epidermis and that the altered composition of cutin and wax in the cuticle might influence water permeability in the ectopic MYB16 lines.
Because the ectopic expression of MYB16 influenced the structure of the outer cell wall continuum (Figure 5), changes in cuticle thickness and cuticle properties could affect stomatal formation by affecting either biochemical signaling or mechanical properties. Since the long-distance mobility of salicylic acid is modulated by the cuticle (Lim et al., 2020), it is possible that stomatal signal transduction was affected in the ectopic MYB16 lines. However, overexpressing EPF2 still suppressed the stomatal phenotypes (Figure 6, C–H), indicating that EPF-mediated signaling is not affected by the altered cuticle structure and that the change in the mechanical properties of the epidermis is the reason for the clustering phenotype in the ectopic MYB16 lines, which is discussed below.
The outer cell wall continuum influences the establishment of stomatal polarity
The modulation of the stiffness of a tissue is able to specify the microenvironment and direct stem cell specification (Engler et al., 2006). In animals, mechanobiology studies have focused on cancer metastasis (Makale, 2007) and tissue regeneration, such as hair regeneration in the epidermis (Chen et al., 2015). In plants, the disruption of pectin metabolism reduces cell adhesion and generates disorganized tumor-like growths on the shoots (Krupková et al., 2007; San-Bento et al., 2014). Unlike direct cell adhesion, which is mediated by protein linkage and the extracellular matrix in animal systems, cell adhesion in plants is mostly mediated by the deposition of a pectin-rich middle lamella between adjacent cell walls to promote the mechanical force on the epidermis and coordinate tissue-wide growth. In contrast to the modification of pectin between cell walls, cuticle accumulation mainly occurs on top of the leaf epidermis, with a potential covalent linkage between cutin and polysaccharides (Fang et al., 2001).
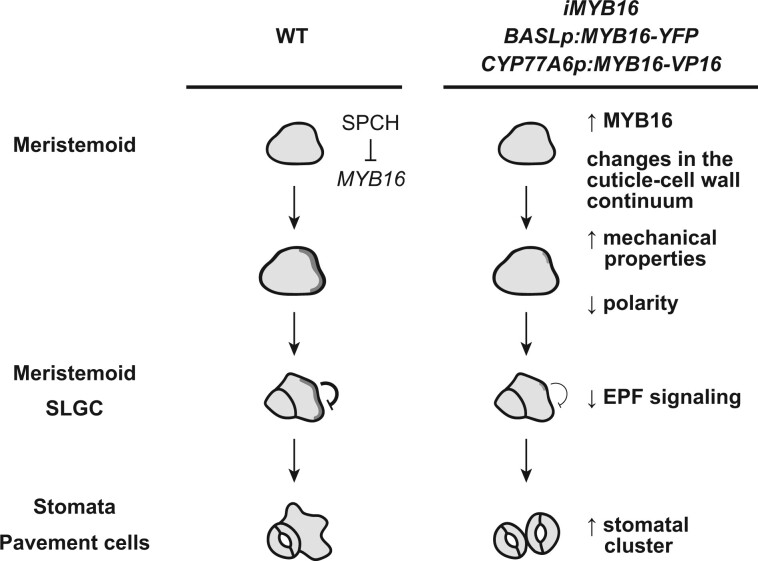
Both the amount of cuticular materials and their composition influence cuticle biomechanics (Dominguez et al., 2011; Takahashi et al., 2012; España et al., 2014). Using AFM measurements, Verger et al. (2018) probed the outer cell wall–cell wall zone to detect changes in tension in Arabidopsis and the pectin mutant qua1. In addition, qua2, a mutant similar to qua1, has an altered cuticle structure in the epidermis (Lorrai et al., 2021). These findings suggest that the cuticle contributes to the mechanical properties of the epidermis. Here, overexpressing MYB16 modified the cuticle and cell wall thickness, as shown in the TEM images (Figure 5; Supplemental Figure S9). These changes were possibly due to the transcriptional control of the cutin biosynthesis pathway by MYB16 and its downstream target SHN1 and may lead to changes in the cell wall components as well. To reveal whether the morphological change is linked to this alteration of the mechanical properties of the epidermis, we used BRXL2 as an indicator because its localization in a leaf reflects the direction of growth and tension (Bringmann and Bergmann, 2017). In ectopic MYB16 plants, the polarity of BRXL2 and the cell fate were disturbed, resulting in clustered stomata (Figures 3, 4, and 6). The rescue of polarity and stomatal clustering by growth on high-percentage agar to reduce the water potential and modulate the mechanical properties of the epidermis suggests that the tensile strength is disrupted in the ectopic MYB16 lines (Figure 7). The expression of the cutinase gene CDEF1 to degrade cutin polymers in the epidermis also rescued the cluster phenotypes in the ectopic MYB16 lines, pointing to the significant contribution of cutin to the physical properties of the epidermis (Figure 5). All lines of evidence suggest that the changes in the cuticle-cell wall continuum influence the mechanical properties of the epidermis, which further influences the establishment of polarity during stomatal development, leading to aberrant stomatal patterning. Further chemical analyses of cuticular components, transcriptome analysis, and AFM measurements could help us better understand the biomechanics of the leaf epidermis during development in the ectopic MYB16 lines. Taken together, these findings indicate that the downregulation of MYB16 in meristemoids is required for establishing proper polarity during asymmetric cell division in stomates during development (Figure 8).
Figure 8.
Ectopic MYB16 expression in meristemoids leads to stomatal cluster formation by modulating polarity protein behavior during asymmetric cell division. In WT epidermis, SPCH represses MYB16 expression in meristemoids to ensure polarity establishment for proper stomatal patterning. However, in the MYB16 overexpression and ectopic expression lines, high MYB16 expression in meristemoids enhances the thickness in the cuticle. The change in the cuticle–cell wall continuum leads to the reduction and mis-polarization of polarity protein during asymmetric cell division, which further impairs the EPF-mediated inhibitory signaling and results in the formation of stomatal clusters. The dark gray shading represents the polarity complex.
Stomata and the cuticle on the leaf epidermis
The evolutionary origins of genes that specify stomatal development and function have been resolved phylogenetically in the bryophytes (Harris et al., 2020; Philippe et al., 2020). Genes related to lipid biosynthesis can be traced back to algae; however, the basic cuticle biosynthetic machinery, such as CYP77A, GPAT, and MYB, started to evolve in the bryophytes (Kong et al., 2020). These lines of evidence, together with our findings of the CYP77A6 and MYB16 expression patterns, suggest that the stomata and cutin machinery co-evolved to specify the formation of a stomate whose ledges (lips around each stomatal pore) are coated with a thick waterproof cuticle. Besides the genes involved in stomatal function and structure, the emergence of the MYB transcription factors provided a strategy for the spatiotemporal control of gene expression in a developmental context. In Arabidopsis, MYB16 and MYB106 regulate cuticle formation in reproductive organs (Oshima et al., 2013); however, MYB16 is a major regulator of cuticle production in vegetative tissues (Oshima and Mitsuda, 2013). The specific functions of the different members of a gene family lead to a diverse transcriptional network, which is required for building a multicellular organism. Using the stomatal lineage-specific transcriptomes and single-epidermal-cell transcriptomes (Adrian et al., 2015; Ho et al., 2021; Lopez-Anido et al., 2021), we could start to explore the developmental regulation and cell type-specific expression of a particular set of genes involved in cuticle biosynthesis. Biochemical and functional analyses will further pinpoint the lipid metabolic steps in the cuticle biosynthesis pathway. The manipulation of cuticle production and stomatal numbers on the leaf epidermis could be utilized to improve plant growth and productivity under adverse conditions.
Materials and methods
Plant materials, growth conditions, and chemical treatments
Arabidopsis thaliana Col-0 was the WT used in all experiments, and all transgenic lines were created in this background. The plant reporters used in this study were AtML1p:RC12A-mCherry, SPCHp:SPCH-CFP (Davies and Bergmann, 2014), CYP77A6p:MYB16-VP16, and MYB16p:MYB16-SRDX (Oshima et al., 2013). The primers used to generate the DNA constructs are listed in Supplemental Table S1. Details about the new constructs, MYB16p:MYB16-YFP, BASLp:MYB16-YFP, the MYB16-inducible system (for iMYB16), CYP77A6p:MYB16-VP16-YFP, mini35S-MYB16p:LUC2, 35S:SPCH-YFP, 35S:SCRM-CFP, ML1p:CDEF1-FLAG, and the myb16-CRISPR allele, are provided in Supplemental Table S2. Details about the plant materials are provided in Supplemental Table S3.
Ethanol (EtOH)-sterilized seeds were sown on 1/2 Murashige and Skoog (MS) medium plates and stored at 4°C for stratification. After 24 h, the plates were transferred to a plant tissue culture room at 22°C under a 16-h light/8-h dark cycle (intensity: 80–100 µmol m−2 s−1, wavelength: 400–720 nm, bulb type: fluorescent tube). For the β-estradiol treatment, β-estradiol in absolute EtOH was added to 1/2 MS medium to a final concentration of 50 µM. An equal volume of absolute EtOH was added to 1/2 MS medium as the mock control. Plant materials used for phenotyping and the extraction of mRNAs and proteins were grown on 1/2 MS plates for six days, transferred to EtOH or β-estradiol-containing plates, and grown for four more days.
Microscopy
To quantify the stomatal phenotypes, 10-dpg seedlings were fixed overnight in a 7:1 solution of ethanol:acetic acid. Before being observed under a Leica DM2500 LED microscope with a DIC prism (Leica Microsystems), the seedlings were washed with Milli-Q water (Sigma-Aldrich) and softened with 1-M KOH until they were completely transparent. The samples were then mounted in Milli-Q water for observation. The stomata were quantified from the central regions of the cotyledons or true leaves.
To observe the fluorescent signals, live seedlings with or without PI staining by applying a vacuum for 30 min were mounted in sterile water and observed under a Leica STELLARIS 8 (for time-lapse and whole-leaf analyses; Leica Microsystems) or Zeiss LSM880 (for the expression pattern analysis; Carl Zeiss) confocal microscope. CFP, GFP, and YFP were excited with 458-nm, 488-nm, 514-nm lasers, respectively. PI and mCherry were excited with a 561-nm laser.
TEM
Whole seedlings at 7 dpg were pre-fixed with 2.5% glutaraldehyde in 0.1-M phosphate-buffered saline (PBS; pH = 7.0) at room temperature for 4 h and then post-fixed with 1% osmium tetroxide in 0.1 M PBS at room temperature for 4 h. After fixation, the samples were rinsed in three 20-min changes of buffer before being dehydrated through an acetone series and embedded in Spurr’s resin. The embedded samples were sectioned using a Leica Reichert Ultracut S or Leica EM UC6 ultramicrotome (Leica Microsystems). The ultra-thin sections (∼90 nm) were stained with 5% uranyl acetate in 50% methanol and 0.4% lead citrate in 0.1-M NaOH. For observation and imaging, an FEI Tecnai G2 Spirit Twin TEM (Thermo Fisher Scientific) at 80 kV was used with an Orius CCD camera (Gatan). For the cuticle and cell wall measurement, 5 regions of ∼60 images (total of approximately 300 regions) of each line from three leaves were acquired.
mRNA analysis and protein detection
For the detection of mRNA expression, total RNA was extracted from the plant tissue using an RNA Plus Mini Kit (LabPrep). Complementary DNA (cDNA) was synthesized from 1-µg total RNA using SuperScript III transcriptase (Thermo Fisher Scientific), after which the products were diluted with diethyl pyrocarbonate–water. The reaction solution contained the cDNA template, specific primers, and Power SYBR Green Master Mix (Applied Biosystems) for the qRT-PCR, which was performed using the QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). Relative gene expression was analyzed using QuantStudio 12K Flex software (Applied Biosystems). The primers used for qRT-PCR analysis are listed in Supplemental Table S4.
For protein detection, total protein was extracted from the plant tissues by boiling them in protein sample buffer (62.5-mM Tris–HCl [pH 6.8], 2.5% sodium dodecyl sulphate (SDS), 0.002% Bromophenol Blue, 0.7135-M β-mercaptoethanol, and 10% glycerol) at 100°C for 10 min. The extracted proteins were directly used for immunoblot analyses following 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). Following electrophoresis and protein transfer, the samples were hybridized with specific antibodies using a SNAP ID 2.0 Protein Detection System (Merck). Monoclonal ANTI-FLAG M2 antibody (Sigma, Lot# SI.CC.4005) was used in this study. The chemiluminescence signal was detected in a darkroom using film. The loading control was an SDS–PAGE gel stained with Coomassie Blue.
ChIP qRT-PCR assay
To isolate the SPCH–chromatin complex, 1.5 g of tissue from 4-dpg WT (Col-0), SPCHp:SPCH-CFP, or MUTEp:MUTE-CFP seedlings was harvested and vacuum-infiltrated with 1% formaldehyde for 10 min. The crosslinking reaction was stopped by adding 2-M glycine solution. The samples were homogenized, their nuclei were isolated, cell lysis was performed, and the DNA was fragmented, as described previously (Haring et al., 2007). The resulting protein–chromatin solution was incubated overnight with green fluorescent protein (GFP) antibody-conjugated magnetic beads (ChromoTek) at 4°C. After eluting and reverse-crosslinking the samples at 65°C, the final products were purified using a PCR cleanup column (Geneaid) and eluted in 20-µL 10-mM Tris–HCl (pH 8.0). For qRT-PCR, the reaction solution contained the purified products, specific primers, and Power SYBR Green Master Mix (Applied Biosystems). The reactions were performed on a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems) with the following cycling conditions: hold at 50°C for 2 min and 95°C for 10 min; 40 cycles of PCR at 95°C for 15 s and 60°C for 1 min; melt curve at 95°C for 15 s, 60°C for 1 min, and ramping (0.05°C·s−1) to 95°C, using EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (EIF4A1) as the negative control. The fold enrichment was calculated as follows: the Ct value from the IP products was divided by the Ct value of the input, and the ratio was normalized by setting the WT (Col-0) value to 1 for each individual detected region. The primers used for qRT PCR analysis are listed in Supplemental Table S4.
Luciferase reporter assay
Using PEG-mediated transformation (Yoo et al., 2007), the reporter (mini35S-MYB16p:LUC2), the effectors (35S:SPCH-YFP and 35S:SCRM-CFP), and naked DNA were cotransformed into mesophyll protoplasts isolated from 3-week-old WT (Col-0) leaves. After transformation, the protoplasts were incubated in W5 solution at 22°C for one 16-h light/8-h dark cycle. On the second day, the protoplasts were collected and their luciferase activities were assayed using a Dual-Luciferase System (Promega). The relative LUC2 activity was calculated as (luminescence intensity generated by LUC2)/(luminescence intensity generated by Rluc). The values were normalized to the empty vector control, which was set to 1.
PrefixSpan algorithm
Time-lapse images of 7-dpg seedlings were obtained using whole-leaf tile scanning with a Leica STELLARIS 8 confocal microscope. The interval time was 8 or 16 h. The serial changes in the fluorescence signals of cells and the cell division events were recorded (n = 156). The matrix was analyzed using the Python package PrefixSpan in generator mode (Pei et al., 2001). The output matrix is presented as event counts (Supplemental Figure S3).
Image processing
To analyze the PI-stained confocal images, several stacks were processed using SurfCut in ImageJ (Erguvan et al., 2019).
TB penetration test
The plants used for the TB penetration test were grown on 1/2 MS medium containing 0.8% Phyto agar (Duchefa Biochemie). A 0.05% (w/v) TB solution in Milli-Q water was prepared and filtered through a 0.22-µm polyvinylidene difluoride filter. The seedlings were immersed in the filtered TB solution for 2 min, after which the excess dye was washed out with Milli-Q water. The aerial tissues were transferred into tubes containing 1 mL 80% ethanol and incubated for 2 h in the dark. The ethanol solution was examined by spectrophotometry to detect TB (absorbance of 430 nm) and chlorophyll (absorbance of 626 nm).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MYB16 (AT5G15310), MYB106 (AT3G01140), SPCH (AT5G53210), BASL (AT5G60880), BRXL2 (AT3G14000), CYP77A6 (AT3G10570), CYP86A4 (AT1G01600), LACS1 (AT2G47240), LACS2 (AT1G49430), GPAT4 (AT1G01610), GPAT8 (AT4G00400), ABCG12 (AT1G51500), AtML1 (AT4G21750), and CDEF1 (AT4G30140).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. MYB16 is preferentially localized to SLGCs.
Supplemental Figure S2. MUTE binds to the MYB16 promoter and upregulates its expression in vitro.
Supplemental Figure S3. The workflow of the analysis of MYB16 and SPCH dynamics using PrefixSpan.
Supplemental Figure S4. Quantification of the CYP77A6 and CYP86A4 transcription patterns.
Supplemental Figure S5. Expression patterns of the MYB16-targeted genes from a single-cell perspective.
Supplemental Figure S6. The ectopic expression of MYB16 results in the formation of stomatal clusters.
Supplemental Figure S7. The two types of stomatal phenotypes observed in CYP77A6p:MYB16-VP16.
Supplemental Figure S8. Cutin-related genes are upregulated after induction in the iMYB16 lines.
Supplemental Figure S9. Phenotypes of ectopic MYB16 lines revealed by TEM and a TB penetration assay.
Supplemental Figure S10. Introducing the cutinase gene CDEF1 into the leaf epidermis rescues the stomatal phenotype in the ectopic MYB16 lines.
Supplemental Figure S11. Expression of genes related to stomatal development in the iMYB16 lines after a short-term induction of MYB16 expression, as revealed by qRT-PCR.
Supplemental Figure S12. Polarity in the ectopic MYB16 lines is rescued by growth on high-percentage agar growth medium.
Supplemental Table S1. Primers for DNA manipulation.
Supplemental Table S2. Details of DNA manipulation.
Supplemental Table S3. Details of plant materials.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr Yoshimi Oshima and Dr Nobutaka Mitsuda (National Institute of Advanced Industrial Science and Technology, Japan) for providing the amiR-MYB16 myb106-2, CYP77A6p\:MYB16-VP16, and MYB16p:MYB16-SRDX lines. We thank Dr Dominique Bergmann at Stanford University and the Howard Hughes Medical Institute for providing stomata-related constructs, Dr Shu-Hsing Wu at the Institute of Plant and Microbial Biology (IPMB) for providing a 3-FLAG vector, and Dr Shi-Long Tu and Ping Cheng at IPMB, Academia Sinica, for providing the Gateway-compatible destination vector (pCAMBIA1390(GW) containing 35S:XVE-LexA) for the induction system. We thank Dr Keng-Hui Lin and Dr Chih-Wen Yang at the Institute of Physics, Academia Sinica, and Dr He-Chun Chou at the Research Center for Applied Science, Academia Sinica, for consulting with us on surface tension. We thank Mei-Jane Fang and Ming-Ling Cheng at the Genomic Technology Core Lab (IPMB, Academia Sinica) for the DNA sequencing service. We thank Mei-Jane Fang and Ji-Ying Huang at the Live-Cell Imaging Division, Cell Biology Core Lab (IPMB, Academia Sinica) for advice on using the Leica STELLARIS 8 and Zeiss LSM880 confocal microscopes. We thank Dr Wann-Neng Jane, Chia-Me Hsu, and Yi-Chia Chou at the Electron Microscope Division, Cell Biology Core Lab (IPMB, Academia Sinica) for preparing samples and providing advice on using the TEM. We thank Dr Paul Verslues (IPMB, Academia Sinica) and Dr Hsou-Min Li (Institute of Molecular Biology (IMB), Academia Sinica) for their suggestions regarding this manuscript.
Funding
This work was supported by the Ministry of Science and Technology in Taiwan (MOST 108-2311-B-001-003-MY3).
Conflict of interest statement. None declared.
Contributor Information
Shao-Li Yang, Institute of Plant and Microbial Biology, Academia Sinica, Nangang, Taipei, Taiwan.
Ngan Tran, Institute of Plant and Microbial Biology, Academia Sinica, Nangang, Taipei, Taiwan.
Meng-Ying Tsai, Institute of Plant and Microbial Biology, Academia Sinica, Nangang, Taipei, Taiwan.
Chin-Min Kimmy Ho, Institute of Plant and Microbial Biology, Academia Sinica, Nangang, Taipei, Taiwan.
S.L.Y. and C.M.K.H. designed the experiments. S.L.Y., N.T., and M.Y.T. performed the experiments. S.L.Y. and C.M.K.H. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Chin-Min Kimmy Ho (chmho@gate.sinica.edu.tw).
References
- Adrian J, Chang J, Ballenger CE, Bargmann BO, Alassimone J, Davies KA, Lau OS, Matos JL, Hachez C, Lanctot A, et al. (2015) Transcriptome dynamics of the stomatal lineage: Birth, amplification, and termination of a self-renewing population. Dev Cell 33: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, Van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Bessire M, Borel S, Fabre G, Carrac L, Efremova N, Yephremov A, Cao Y, Jetter R, Jacquat AC, Métraux JP, et al. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot V, Fadanavis SV, Panwar J (2021) Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: there is more scope. Environ Exp Bot 183: 104364 [Google Scholar]
- Bird SM, Gray JE (2003) Signals from the cuticle affect epidermal cell differentiation. New Phytol 157: 9–23 [DOI] [PubMed] [Google Scholar]
- Bringmann M, Bergmann DC (2017) Tissue-wide mechanical forces influence the polarity of stomatal stem cells in Arabidopsis. Curr Biol 27: 877–883 [DOI] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wang L, Plikus MV, Jiang TX, Murray PJ, Ramos R, Guerrero-Juarez CF, Hughes MW, Lee OK, Shi S, et al. (2015) Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 161: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CN, Lee TY, Hung YC, Li GZ, Tseng KC, Liu YH, Kuo PL, Zheng HQ, Chang WC (2019) PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res 47: D1155–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KA, Bergmann DC (2014) Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc Natl Acad Sci USA 111: 15585–15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E, Heredia-Guerrero JA, Heredia A (2011) The biophysical design of plant cuticles: an overview. New Phytol 189: 938–949 [DOI] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC (2009) BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689 [DOI] [PubMed] [Google Scholar]
- Erguvan Ö, Louveaux M, Hamant O, Verger S (2019) ImageJ SurfCut: a user-friendly pipeline for high-throughput extraction of cell contours from 3D image stacks. BMC Biol 17: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- España L, Heredia-Guerrero JA, Segado P, Benítez JJ, Heredia A, Domínguez E (2014) Biomechanical properties of the tomato (Solanum lycopersicum) fruit cuticle during development are modulated by changes in the relative amounts of its components. New Phytol 202: 790–802 [DOI] [PubMed] [Google Scholar]
- Fang X, Qiu F, Yan B, Wang H, Mort AJ, Stark RE (2001) NMR studies of molecular structure in fruit cuticle polyesters. Phytochemistry 57: 1035–1042 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Varnau R, Wallner ES, Acharya R, Bergmann DC, Cheung LS (2021) Quantitative and dynamic cell polarity tracking in plant cells. New Phytol 230: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, Van Der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM (2000) The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Han S, Qi X, Sugihara K, Dang JH, Endo TA, Miller KL, Kim E-D, Miura T, Torii KU (2018) MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev Cell 45: 303–315 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BJ, Harrison CJ, Hetherington AM, Williams TA (2020) Phylogenomic evidence for the monophyly of Bryophytes and the reductive evolution of stomata. Curr Biol 30: 2001–2012 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Bringmann M, Oshima Y, Mitsuda N, Bergmann DC (2021) Transcriptional profiling reveals signatures of latent developmental potential in Arabidopsis stomatal lineage ground cells. Proc Natl Acad Sci USA 118: e2021682118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Brown J, Segerson NA, Rose JKC, Roeder AHK (2017) CUTIN SYNTHASE 2 maintains progressively developing cuticular ridges in Arabidopsis sepals. Mol Plant 10: 560–574 [DOI] [PubMed] [Google Scholar]
- Houbaert A, Zhang C, Tiwari M, Wang K, de Marcos Serrano A, Savatin D-V, Urs MJ, Zhiponova MK, Gudesblat GE, Vanhoutte I, et al. (2018) POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563: 574–578 [DOI] [PubMed] [Google Scholar]
- Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD (2012) Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148: 175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal BP, Grimm E, Finger S, Blume A, Knoche M (2013) Intracuticular wax fixes and restricts strain in leaf and fruit cuticles. New Phytol 200: 134–143 [DOI] [PubMed] [Google Scholar]
- Kong L, Liu Y, Zhi P, Wang X, Xu B, Gong Z, Chang C (2020) Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol 184: 1998–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupková E, Immerzeel P, Pauly M, Schmülling T (2007) The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J 50: 735–750 [DOI] [PubMed] [Google Scholar]
- Lampard GR, MacAlister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC (2014) Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, Mcabee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Liu H, Yu K, Liu R, Shine MB, Fernandez J, Burch-Smith T, Mobley JK, McLetchie N, Kachroo A, et al. (2020). The plant cuticle regulates apoplastic transport of salicylic acid during systemic acquired resistance. Sci Adv 6: eaaz0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Anido CB, Vatén A, Smoot NK, Sharma N, Guo V, Gong Y, Anleu Gil MX, Weimer AK, Bergmann DC (2021) Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev Cell 56: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrai R, Francocci F, Gully K, Martens HJ, De Lorenzo G, Nawrath C, Ferrari S (2021) Impaired cuticle functionality and robust resistance to Botrytis cinerea in Arabidopsis thaliana plants with altered homogalacturonan integrity are dependent on the class III peroxidase AtPRX71. Front Plant Sci 12: 696955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59: 553–564 [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Makale M (2007) Cellular mechanobiology and cancer metastasis. Birth Defects Res 81: 329–343 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Shin JJH, Bird DA, Samuelsa AL (2010) Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N (2016) Enhanced cuticle accumulation by employing MIXTA-like transcription factors. Plant Biotechnol 33: 161–168 [Google Scholar]
- Oshima Y, Mitsuda N (2013) The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signal Behav 8: e26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Han J, Mortazavi-Asl B, Pinto H, Chen Q, Dayal U, Hsu MC (2001) PrefixSpan: mining sequential patterns efficiently by prefix-projected pattern growth. In Proceedings of the 17th International Conference on Data Engineering, Heidelberg, Germany
- Philippe G, Gaillard C, Petit J, Geneix N, Dalgalarrondo M, Bres C, Mauxion JP, Franke R, Rothan C, Schreiber L, et al. (2016). Ester cross-link profiling of the cutin polymer of wild-type and cutin synthase tomato mutants highlights different mechanisms of polymerization. Plant Physiol 170: 807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe G, Sørensen I, Jiao C, Sun X, Fei Z, Domozych DS, Rose JK (2020) Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr Opin Plant Biol 55: 11–20 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Peterson KM, Horst RJ, Torii KU (2011) Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23: 3260–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Rowe MH, Dong J, Weimer AK, Bergmann DC (2019) A plant-specific polarity module establishes cell fate asymmetry in the Arabidopsis stomatal lineage. bioRxiv: DOI: 10.1101/614636 [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- San-Bento R, Farcot E, Galletti R, Creff A, Ingram G (2014) Epidermal identity is maintained by cell-cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J 77: 46–58 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, de Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Shimada T, Kondo M, Tamai A, Mori M, Nishimura M, Hara-Nishimura I (2010) Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol 51: 123–131 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Tsubaki S, Sakamoto M, Watanabe S, Azuma JI (2012) Growth-dependent chemical and mechanical properties of cuticular membranes from leaves of Sonneratia alba. Plant Cell Environ 35: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Verger S, Long Y, Boudaoud A, Hamant O (2018) A tension-adhesion feedback loop in plant epidermis. eLife 7: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin D, Nawrath C, Kurdyukov S, Franke RB, Reina-Pinto JJ, Efremova N, Will I, Schreiber L, Yephremov A (2009) Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann P, Neinhuis C (1998) Biomechanics of isolated plant cuticles. Bot Acta 111: 28–34 [Google Scholar]
- Xie L-J, Tan W-J, Yang Y-C, Tan Y-F, Zhou Y, Zhou D-M, Xiao S, Chen Q-F (2020) Long-chain acyl-CoA synthetase LACS2 contributes to submergence tolerance by modulating cuticle permeability in Arabidopsis. Plants (Basel) 9: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Isabel Ordiz M, Jaworski JG, Beachy RN (2011) Induced accumulation of cuticular waxes enhances drought tolerance in Arabidopsis by changes in development of stomata. Plant Physiol Biochem 49: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol 160: 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Huang W, Chatterjee S, Viart HMF, Clausen MH, Stark RE, Rose JKC (2014) Tomato Cutin Deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. Plant J 77: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LBB, Viart HMF, Isaacson T, He Y, Zhao L, Matas AJ, Buda GJ, Domozych DS, Clausen MH, et al. (2012). The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol 8: 609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeiger E, Stebbins GL (1972) Developmental genetics in barley: a mutant for stomatal development. Am J Bot 59: 143–148 [Google Scholar]
- Zhang Y, Wang P, Shao W, Zhu JK, Dong J (2015) The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev Cell 33: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.