F-box protein MdAMR1L1 promotes GDP-mannose pyrophosphorylase degradation via the ubiquitination pathway to inhibit Asc synthesis in apple.
Abstract
Ascorbate (Asc) is an important antioxidant in plants and humans that plays key roles in various physiological processes. Understanding the regulation of Asc content in fruit plants is important for improving plant resiliency and optimizing Asc in food. Here, we found that both the transcript level and protein abundance of Asc Mannose pathway Regulator 1 Like 1 (MdAMR1L1) was negatively associated with Asc levels during the development of apple (Malus × domestica) fruit. The overexpression or silencing of MdAMR1L1 in apple indicated that MdAMR1L1 negatively regulated Asc levels. However, in the leaves of MdAMR1L1-overexpressing apple lines, the transcript levels of the Asc synthesis gene Guanosine diphosphate-mannose pyrophosphorylase MdGMP1 were increased, while its protein levels and enzyme activity were reduced. This occurred because the MdAMR1L1 protein interacted with MdGMP1 and promoted its degradation via the ubiquitination pathway to inhibit Asc synthesis at the post-translational level. MdERF98, an apple ethylene response factor, whose transcription was modulated by Asc level, is directly bound to the promoter of MdGMP1 to promote the transcription of MdGMP1. These findings provide insights into the regulatory mechanism of Asc biosynthesis in apples and revealed potential opportunities to improve fruit Asc levels.
Introduction
L-Ascorbate (Asc; also known as vitamin C) is an important antioxidant and acts as a redox buffer in almost all organisms. Asc plays important roles in aspects of plant growth and development, signal transduction, and stress defense networks (Broad et al., 2020). Moreover, humans cannot synthesize Asc and must obtain it through dietary uptake, primarily from fruits and vegetables (Li and Schellhorn, 2007). The concentration of Asc not only varies between different species or cultivars of plants but also varies greatly between different tissues of the same plant. For example, apple (Malus × domestica) leaves can exhibit Asc concentrations over 500 mg 100 g−1 fresh weight (FW), but its concentration in the flesh of a ‘Gala’ apple fruit is below 5 mg 100 g−1 FW (Li et al., 2008).
Although several biosynthetic routes for Asc have been proposed in plants, the D-mannose/L-galactose (D-Man/L-Gal) pathway appears to be predominant (Wheeler et al., 1998; Lorence et al., 2004; Ishikawa and Shigeoka, 2008). In the D-Man/L-Gal pathway (Figure 1A), Asc is synthesized from D-Man-1-P via GDP-D-Man, GDP-L-Gal, L-Gal, and L-Gal-1,4-lactone as intermediates (Wheeler et al. 1998; Ishikawa and Shigeoka, 2008). All of the genes in this biosynthetic pathway have been identified (Broad et al., 2020). GDP-mannose pyrophosphorylase (GMP, also known as VTC1) is involved in the conversion of GDP-Man-1-phosphate to GDP-D-Man, and several studies involving mutants or transgenic plants have demonstrated that this enzyme is involved in Asc synthesis (Conklin et al., 1999; Badejo et al., 2008).
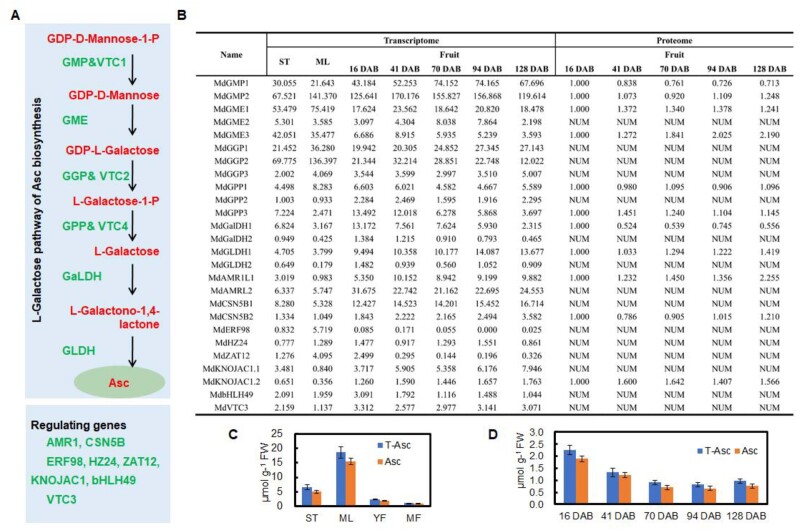
Figure 1.
Expression patterns of key genes involved in Asc biosynthesis in different tissues and during apple fruit development. A, The important synthetic and regulatory genes in the D-Man/L-Gal pathway of Asc biosynthesis. GME, GDP-mannose-3',5'-epimerase; GPP/VTC4, L-galactose-1-P phosphatase; GalDH, L-galactose dehydrogenase; GLDH, L-galactose-1,4-lactone dehydrogenase. B, mRNA and protein expression levels of genes in the D-Man/L-Gal pathway of Asc biosynthesis in different tissues and developing apple fruit of “Greensleeves” apple based on RNA-seq (Zhu et al., 2021) and proteomic data (Li et al., 2016a). NUM represents undetected proteins. Levels of Asc in (C) different tissues and (D) developing apple fruit of “Greensleeves”. Bars represent the mean value ±se (n = 3); ST, shoot tips; ML, mature leaves; YF, young fruit (16 d after bloom, DAB); MF, mature fruit (122 DAB); T-Asc, total Asc; Asc, reduced Asc.
Asc concentrations in plant tissues are tightly regulated through a balance between synthesis and catabolism and can be affected by numerous factors that are internal or external to the plants (Bulley and Laing, 2016), such as developmental processes (Bulley et al., 2009; Li et al., 2011), light conditions (Yabuta et al., 2007; Li et al., 2008), and oxidative stress (Yoshimura et al., 2014). Changes in Asc concentrations in response to environmental stimuli and internal development are related to the expression of certain transcripts critical to Asc metabolic pathways (Mellidou and Kanellis, 2017). Asc-mediated feedback inhibition of GDP-L-galactose phosphorylase (GGP) translation via a highly conserved upstream open reading frame may represent a prominent mechanism for rapidly controlling Asc accumulation (Laing et al., 2015; Fenech et al., 2021). In response to environmental stimuli, Asc biosynthesis can be regulated by AtERF98, an ethylene response factor, in Arabidopsis (Zhang et al., 2012), bHLH59, a natural variation in basic helix–loop–helix transcription factor 59, in tomato (Ye et al., 2019) and HZ24, an HD-Zip I transcription factor 24, in tomato (Hu et al., 2016), which positively regulates the transcription of genes in the L-galactose pathway to increased Asc concentrations and were associated with enhanced tolerance to environmental stress. Additionally, CSN5B, as a component of the photomorphogenic COP9 signalosome, promotes the ubiquitination and degradation of GMP through the 26S proteasome pathway to reduce the Asc concentration and the salt tolerance in Arabidopsis (Wang et al., 2013). And Asc Mannose pathway Regulator 1 (AtAMR1), an F-box protein that negatively regulates the transcription of genes in the L-galactose pathway, deletion mutant in Arabidopsis increased Asc concentrations and antioxidant stress (Zhang et al., 2009). Although Asc biosynthesis and its regulation have been well studied in plants and the variation in Asc concentrations in different cultivars shows relatively high heritability (Mellidou et al., 2012; Bulley and Laing, 2016; Ye et al., 2019), very little is known regarding the molecular mechanisms involved in the perception and/or transduction of internal signals for the regulation of Asc concentrations in different tissues and developmental stages.
Previously, we carried out proteomic (Li et al., 2016a) and transcriptomic analyses (Zhu et al., 2021) in developing apple fruits. In these previously published datasets, we found that the mRNA and protein levels of Asc Mannose pathway Regulator 1 Like 1 (MdAMR1L1), a homolog of AtAMR1 in Arabidopsis (Zhang et al., 2009) were high, and these high levels were negatively associated with Asc levels in the developing fruits. In this work, the overexpression of MdAMR1L1 caused a decrease in the Asc level, while silencing the gene increased the Asc level, which indicated that MdAMR1L1 negatively impacted Asc levels in apple. Furthermore, the MdAMR1L1 protein interacts with MdGMP1 to promote its degradation via the ubiquitination pathway. Additionally, the expression of the transcription factor MdERF98 is induced by reduced Asc level and increases MdGMP1 transcription. The findings provide insights into the regulatory mechanism of mannose metabolism in apple and reveal potential opportunities for improving Asc content in fruit.
Results
AMR1-Like gene expression and its relationship with Asc levels in apple
To identify major genes involved in controlling Asc biosynthesis in apple, homologs of known genes involved in the synthesis and regulation of genes of the D-Man/L-Gal pathway (Bulley and Laing, 2016; Figure 1A) were sought in the Malus × domestica genome. The transcriptional patterns of these genes were analyzed in developing fruit and other tissues, including shoot tips and mature leaves, based on our RNA-seq data (Figure 1B;Supplementary Data Excel File S1). Additionally, protein abundance profiling in developing fruits was analyzed based on our previous proteomics data (Figure 1B). Among the genes in the D-Man/L-Gal pathway, the mRNA expression patterns of MdGME1 and MdGGP2 were similar to the Asc levels in different tissues (Figure 1, B and C). On the other hand, MdGMP1, MdGME2, and MdGLDH1 exhibited high transcript levels in mature fruit, which showed the lowest Asc level (Figure 1, B and C). However, the changes in the GMP1 mRNA level in developing fruits were opposite to the changes in GMP1 protein abundance, and the changes in its protein abundance were consistent with the changes in Asc levels (Figure 1, B and D). Among the regulatory genes, the transcript level and protein abundance of MdAMR1L1 were inversely related to the Asc levels in different tissues and developmental stages of fruits (Figure 1).
In the Malus genome, MdAMR1L1 and MdAMR1L2 share 92.8% similarity at the amino acid level and were homologous to the AtAMR1 gene of Arabidopsis according to phylogenetic analysis (Supplemental Figure 1A), but MdAMR1L1 only had 31% similarity with AtAMR1 at the amino acid level. Both sequences included the conserved F-box domain (Supplemental Figure 1B) and showed percentage sequence similarity to several known F-box proteins (Supplemental Figure S1C). The Real Time Quantitative PCR (RT-qPCR) results further indicated that the transcript abundance of MdAMR1L1 was inversely proportional to the Asc levels in different tissues and developing fruit and leaves under different light conditions. Similar results were not obtained for MdAMR1L2 (Supplemental Figures S2 and S3).
Virus-induced MdAMR1L1 silencing increased Asc level in apple fruit
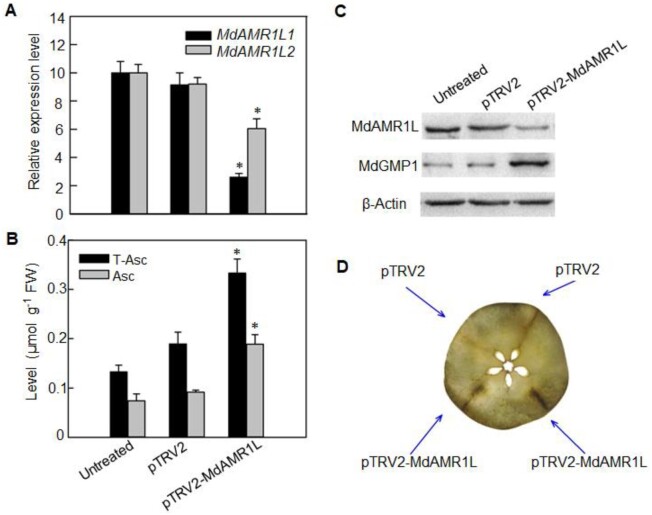
To explore the potential role of MdAMR1L1 in regulating the Asc level in apple fruit, an MdAMR1L1 fragment (sharing 98% nucleic acid sequence similarity with MdAMR1L2) was selected for virus-induced gene silencing (VIGS) experiments in “Starking Delicious” apple fruit. The transcription levels of MdAMR1L1/2 were significantly decreased 6 d after the injection of the VIGS vector pTRV2-MdAMR1L1 compared with the levels in the control (Figure 2A), and western blot analysis showed that the protein abundance of MdAMR1L1/2 was also substantially decreased (Figure 2C). As expected, the Asc level was increased around the site of pTRV2-MdAMR1L1 injection (Figure 2B), which was also confirmed by the histochemical localization of Asc (Figure 2D).
Figure 2.
MdAMR1L silencing increased Asc levels and MdGMP1 protein level in apple flesh. A, Relative expression levels of MdAMR1L1 (black) and MdAMR1L2 (gray) in “Starking Delicious” flesh inside the injection area 6 d after VIGS treatments based on qRT-PCR. For each sample, transcript levels were normalized with those of MdActin. Relative expression levels for each gene were obtained via the ΔΔCT method. Fruit inoculated with Agrobacterium tumefaciens containing empty vectors (pTRV2) and without injection (untreated) were taken as the controls. B, Levels of total Asc (T-Asc, black) and reduced Asc (Asc, grey) in ‘Starking Delicious’ flesh 6 d after VIGS treatments. C, Expression abundances of MdAMR1L1/2 and MdGMP1 proteins in ‘Starking Delicious’ flesh inside the injection areas 6 d after VIGS treatments. β-Actin was used as the reference protein. Untreated was not injected with any treatment. D, Asc localization using AgNO3 in transverse sections of ‘Starking Delicious’ flesh after silencing MdAMR1L1/2 (pTRV2-MdAMR1L1/2) for 6 d. The image was digitally extracted. Injection areas are marked. Bars represent the mean value ±se (n = 3). Asterisks indicate significantly different values (P < 0.05, independent t tests).
The overexpression of MdAMR1L1 decreased the Asc level in apple leaves
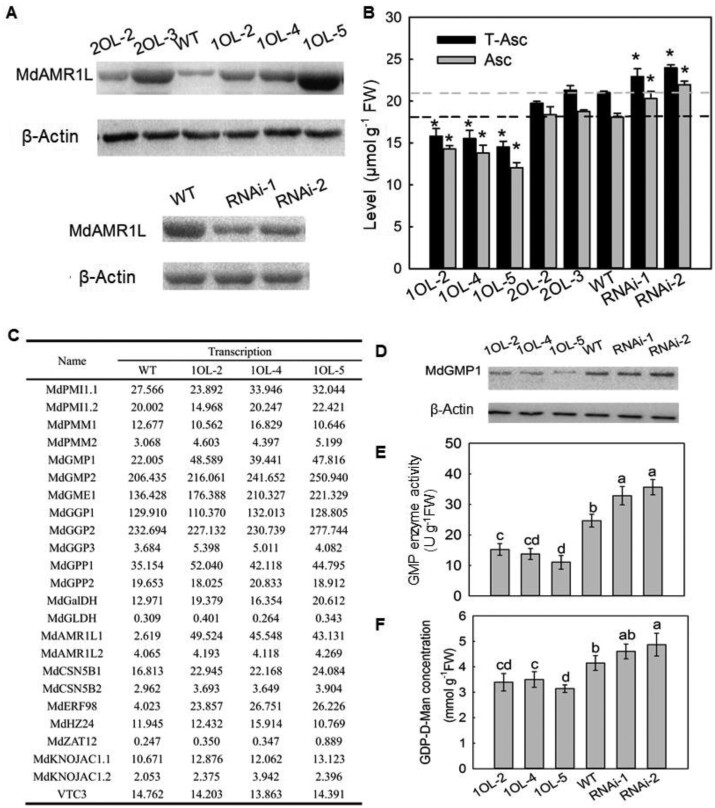
To identify the function of MdAMR1L1/2 in Asc biosynthesis, we constructed MdAMR1L1 and MdAMR1L2 overexpression and co-suppression vectors. These vectors were transformed into the leaves of wild-type (WT) “GL3” apple. After RT-PCR, RT-qPCR and western blot analysis (Supplemental Figure S4; Figure 3A), we obtained three independent MdAMR1L1 overexpression (MdAMR1L1-OE) lines (1OL-2, 1OL-4, and 1OL-5), two independent MdAMR1L2-OE lines (2OL-2 and 2OL-3), and two independent MdAMR1L1/2 co-suppression lines RNA interference (RNAi-1 and RNAi-2). In the transgenic apple lines overexpressing MdAMR1L1 or MdAMR1L2, the corresponding mRNA level was increased 6–15 times in the leaves, without significantly affecting the expression level of the other gene (Supplemental Figure S4B). In contrast, the MdAMR1L1/2 co-suppression lines showed <50% of the expression of MdAMR1L1 and MdAMR1L2 found in the WT apple (Supplemental Figure S4B). Although the photosynthesis rates of the transgenic apple lines under controlled environmental conditions were unchanged (Supplemental Figure S5B), plant growth was inhibited in the MdAMR1L1-OE lines (Supplemental Figure S5A). Since it will take 4–6 years for the transgenic apple to reach the reproductive phase for fruiting, Asc levels were not yet measured in the transgenic fruits. The Asc levels in the leaves of transgenic apple plants were decreased by 21%, 28%, and 34% in 1OL-2, 1OL-4, and 1OL-5, respectively, compared with the concentrations in WT plants, whereas the levels of Asc in 2OL-2 and 2OL-3 leaves remained unchanged (Figure 3B). Accordingly, the Asc levels were higher in the RNAi lines than in WT (Figure 3B). These results indicated that overexpression of MdAMR1L1 decreased the Asc level in apple leaves, whereas overexpression of MdAMR1L2 did not.
Figure 3.
MdAMR1L1 expression is negatively associated with MdGMP1 protein levels in leaves of MdAMR1L transgenic apple lines. A, Expression levels of MdAMR1L1/2 proteins in leaves of WT and MdAMR1L transgenic lines. β-Actin was used as the reference protein. B, Levels of Asc in leaves of WT and MdAMR1L overexpression (OE) and MdAMR1L/2 RNAi transgenic lines. C, mRNA expression levels of genes involved in Asc biosynthesis in ‘GL3’ apple leaves of WT and MdAMR1L1 overexpression transgenic lines based on RNA-seq. Fold-difference is designated as a log2 value. D, Expression levels of MdGMP1 protein in leaves of WT, MdAMR1L1 overexpression, and RNAi transgenic lines. β-Actin was used as the reference protein. E, The enzyme activity of GMP in leaves of WT, MdAMR1L1 overexpression, and MdAMR1L1/2 RNAi transgenic lines. F, Concentration of GDP-D-Mannose in leaves of WT, MdAMR1L1 overexpression, and RNAi transgenic lines. GDP-D-Mannose is the catalytic product of the GMP enzyme. The transgenic lines including overexpressing MdAMR1L1 (1OL-2, 1OL-4, and 1OL-5), MdAMR1L2 (2OL-2 and 2OL-3), and both MdAMR1L1 and MdAMR1L2 silent lines (RNAi-1 and RNAi-2). Bars represent the mean value ±se (n = 3). Asterisks/lowercase letters indicate significantly different values (P < 0.05, independent t tests or one-way ANOVA test).
The expression patterns of genes related to Asc biosynthesis in MdAMR1L1 transgenic apple plants
To explore the reason for decreased Asc level in the MdAMR1L1-OE transgenic lines, mRNA-seq was used to analyze the changes in gene expression in the leaves of MdAMR1L1-OE apple lines (1OL-2, 1OL-4, and 1OL-5). Among the differentially expressed genes (DEGs) compared with WT (log2-fold changes greater than 1, P < 0.01), 239 genes were common to 1OL-2, 1OL-4, and 1OL-5 (Supplementary Data Excel File S2). Of these DEGs, 154 were upregulated and 85 were downregulated (Supplemental Figure S6). RT-qPCR analysis showed 7.85-, 11.43-, and 13.76-fold higher MdAMR1L1 expression levels in 1OL-2, 1OL-4, and 1OL-5 leaves than WT (Supplemental Figure S4B), which were consistent with the data of mRNA-seq (Figure 3C).
According to the annotation of the mRNA-seq data, among the 47 genes putatively related to Asc (Figure 3C;Supplemental Figure S7, B), only 5 genes showed differential expression in the MdAMR1L1-OE apple lines compared with the WT. And four of these genes were upregulated, namely MdAMR1L1, MdGMP1, MdERF98, and MdMIOX4, the remaining gene MdNAT12.2 was downregulated. Although the expectation was decreased expression, many genes in the D-Man/L-Gal pathway (such as MdGMP1, MdGME1, MdGGP1, MdGPP1, MdGalDH, and MdGLDH) showed increased expression in the MdAMR1L1-OE apple lines, especially MdGMP1 (Figure 3C). These increased transcript levels were confirmed by RT-qPCR (Supplemental Figure S7A). These results suggested that MdAMR1L1 regulates Asc biosynthesis is not by negatively affecting gene expression related to Asc biosynthesis in the D-Man/L-Gal pathway in apple, which is different from the report in Arabidopsis that AtAMR1 negatively affected the expression of genes in L-Gal pathway (Zhang et al., 2009).
To further confirm a role for MdAMR1L1/2 in determining Asc levels, we expressed MdAMR1L1/2 in WT Arabidopsis. The expression of MdAMR1L1 in Arabidopsis did not substantially change the phenotype, while MdAMR1L2 expression led to small plant size (Supplemental Figure S8A). The overexpression of either gene in WT Arabidopsis resulted in a reduced Asc level (Supplemental Figure S8B). However, the expression levels of genes in the D-Man/L-Gal pathway were not significantly changed (Supplemental Figure S8C), which further indicated that the regulatory mechanism by MdAMR1L1 in Asc biosynthesis in apple is different from that of AtAMR1 in Arabidopsis.
MdGMP1 protein abundance is related to the Asc level in MdAMR1L1 transgenic apple
To determine whether MdGMP1 protein abundance was altered when MdGMP1 mRNA expression levels were upregulated in transgenic apple overexpressing MdAMR1L1, MdGMP1 protein level was evaluated by western blotting with a specific MdGMP1 antibody. Interestingly, the protein abundance (Figure 3D) and enzyme activity of MdGMP (Figure 3E) were significantly reduced, and the level of GDP-D-Man was also decreased (Figure 3F). Correspondingly, MdAMR1L1/2 silencing in WT apple leaves and fruits increased the abundance of the MdGMP1 protein (Figures 2, C and 3, D). MdGMP1 protein abundance and enzyme activity were much higher in mature leaves, which also exhibited much higher Asc levels (Figure 1, E;Supplemental Figure S9, C) and lower MdAMR1L1 expression (Figure 1, B;Supplemental Figure S2, D), than apple fruits and was lowest in mature fruit (Supplemental Figure S9). Overexpression in apple calli further showed that increased MdGMP1 expression could increase the Asc level (Supplemental Figure S10).
MdAMR1L1 interacts with MdGMP1 to negatively regulate its protein level
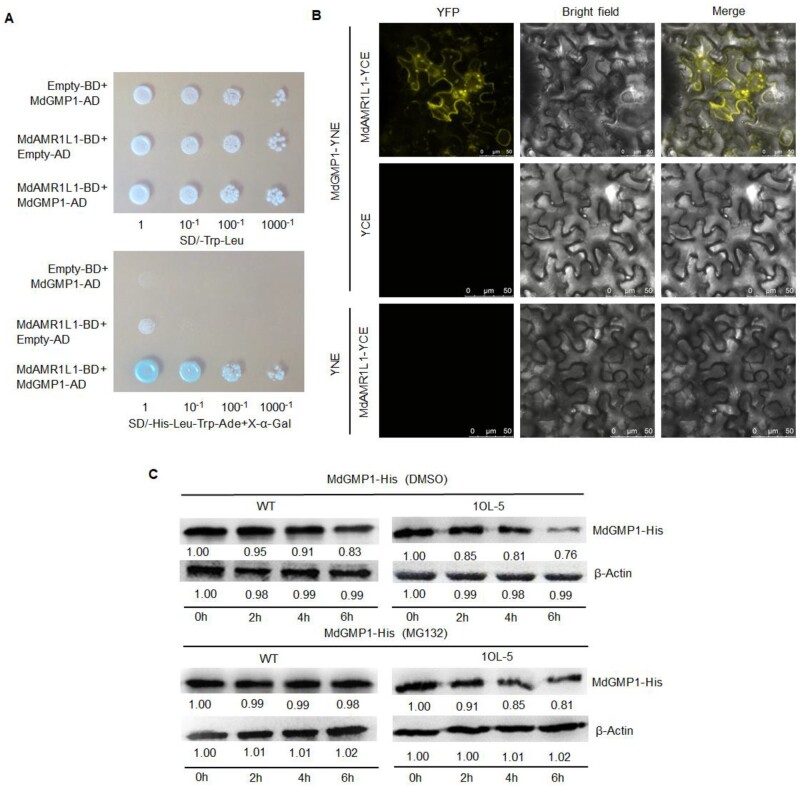
To determine the roles of MdAMR1L1 in regulating Asc levels, we screened for its interactors in apple by combining immunoprecipitation with mass spectrometry (IP-MS). Total proteins were extracted from MdAMR1L1-OE lines (1OL-2 and 1OL-5) and subjected to immunoprecipitation with anti-MdAMR1L1 agarose beads. After mass spectrometry analysis, MdGMP1 was detected in 235 candidate interacting proteins. This was confirmed by in vitro and in vivo experiments, including yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (Bi-FC) assays (Figure 4, A and B). In the Y2H system, the interaction of MdGMP1 and MdAMR1L1 was indicated by the induction of β-galactosidase activity and growth on media lacking Leucine, Tryptophan, Histidine, and Adenine (-Leu/-Trp/-His/-Ade) (Figure 4A). In the Bi-FC assay, the reconstituted fluorescence of yellow fluorescent protein (YFP) generated by MdAMR1L1-YC and MdGMP1-YN in Nicotiana benthamiana cells also indicated the interaction between these proteins (Figure 4, B).
Figure 4.
MdAMR1L1 interacts with MdGMP1 and promotes the degradation of MdGMP1 through the ubiquitination pathway. A, MdAMR1L1 interacts with MdGMP1 in a Yeast Two-Hybrid assay. pGBKT7-MdAMR1L1 (MdAMR1L1-BD) and pGADT7-MdGMP1 (MdGMP1-AD), MdAMR1L1-BD, and the empty pGADT7 vector (Empty-AD, Control), MdGMP1-AD and the empty pGBKT7 vector (Empty-BD, Control) were co-transformed into Y2H Gold yeast cells. The upper panel shows the growth of the transformed yeast on SD/–Leu–Trp medium. The lower panel shows an assay for X-α-Gal activity of the transformants grown on SD/-Leu–Trp–His–Ade. B, MdAMR1L1 interacts with MdGMP1 in a Bi-FC assay. The N-terminus of YFP was fused to MdAMR1L1 and the C-terminus of YFP was fused to MdGMP1. Combinations of the plasmids and controls (pEarleyGate202-YC Empty Vector (YCE) and pEarleyGate201-YN Empty Vector (YNE)) were transformed into N. benthamiana. The topmost panels indicated the presence of YFP signal due to the reconstitution of YFP through protein–protein interactions of the tested pairs. Bar = 10 mm. C, MdAMR1L1 promotes the degradation of MdGMP1-His protein (upper) and the addition of MG132 (a proteasome inhibitor, in DMSO) enhanced the stability of MdGMP1-His protein (lower) in vitro. The total protein extracted from the leaves of WT and MdAMR1L1-OE (1OL-5) transgenic apple were mixed with purified MdGMP1-His protein in equal content and then treated with either DMSO or 50 µM MG132 for 0 h, 2 h, 4 h, and 6 h. MdGMP1-His protein level was detected by Western blot using anti-His antibodies. β-Actin was used as the reference protein.
To investigate whether MdAMR1L1 promotes the degradation of MdGMP1 protein via the ubiquitination pathway, total protein extracted from WT and MdAMR1L1-OE (1OL-5) apple leaves were incubated with purified MdGMP1-His protein. The abundance of the MdGMP1-His protein gradually decreased over time, but the degradation was accelerated in the extract from 1OL-5 compared with that from WT (Figure 4C). The application of MG132, a proteasome inhibitor, inhibited MdGMP1-His protein degradation (Figure 4C, bottom panels). To further confirm if MdGMP1 degradation was mediated by proteasomal degradation, we used Murashige and Skoog (MS) culture medium supplemented with 5 μM MG132/DMSO for the culture of WT and MdAMR1L1-OE apple seedlings (1OL-5). After 1 month, MdGMP1 protein levels in the leaves of WT and 1OL-5 plants treated with MG132 were increased compared with those in the Dimethyl sulfoxide (DMSO)-treated plants (control) (Supplemental Figure S11A). Additionally, the MG132-treated plants showed higher Asc levels than the DMSO-treated plants, and the Asc level in 1OL-5 leaves recovered to the WT level (Supplemental Figure S11B). These results indicated that MdAMR1L1 negatively regulates the MdGMP1 protein via the ubiquitination pathway and affects the biosynthesis of Asc in apple plants.
MdERF98 directly regulates MdGMP1 transcription and Asc biosynthesis
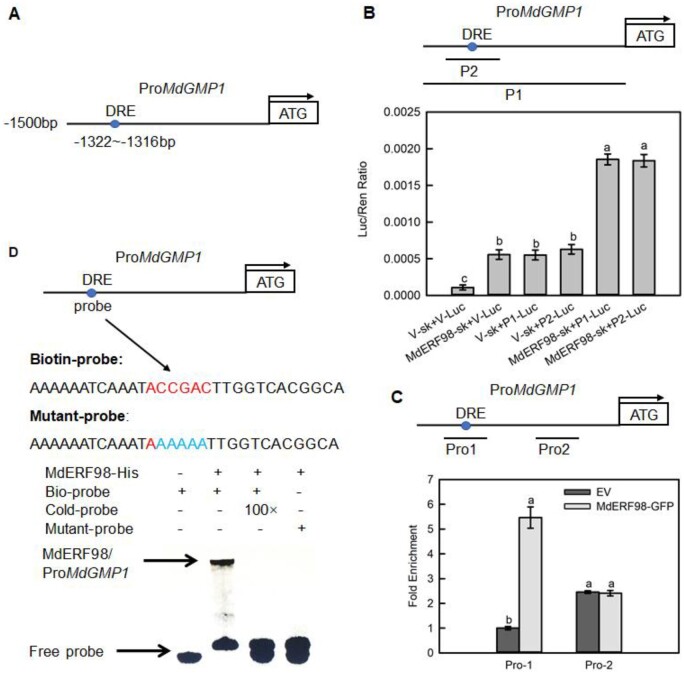
Although the MdGMP1 protein level was reduced in the apple overexpressing MdAMR1L1, the mRNA level of MdGMP1 was significantly upregulated. MdERF98, which encodes a transcription factor that exhibits transactivation activity in yeast cells and localizes to the nucleus (Supplemental Figure S12), is a homologous gene of AtERF98, whose product activates the expression of GMP and GME (Guanosine diphosphate-mannose-3',5'-epimerase) in Arabidopsis (Zhang et al., 2012). Therefore, we assumed that the upregulation of MdGMP1 expression might be due to the increased MdERF98 transcript levels in apple leaves overexpressing MdAMR1L1 (Figure 3, C and E; Supplemental Figures 7, A and 13). The MdGMP1 promoter contains one Dehydration-responsive element (DRE) core sequence (ACCGAC) for ERF transcription factor binding, located between −1316 and −1322-bp upstream of the transcription start site (Figure 5A). Separate dual-luciferase assays, chromatin immunoprecipitation-qPCR assays, and electrophoretic mobility shift assays (EMSA) were used to verify that MdERF98 binds to the MdGMP1 promoter. The presence of both MdERF98 and an MdGMP1 promoter carrying the DRE motif could induce high luciferase activity in N. benthamiana leaves (Figure 5B). Overexpression of MdERF98 substantially increased the PCR-based detection of the MdGMP1 promoter containing the DRE element but not one lacking it (Figure 5C). MdERF98 could only bind to the MdGMP1 promoter when a labeled probe containing the DRE element was added in the EMSA experiment (Figure 5D).
Figure 5.
MdERF98 directly binds to the promoter of MdGMP1. A, Diagram of MdGMP1 promoter region containing a MdERF98 potential DRE binding site, which is located at 1,322- to 1,316-bp upstream of the MdGMP1 initiation codon (ATG). B, Dual luciferase assays in tobacco leaves showed that MdERF98 binds to the promoter of MdGMP1. P1 represents the 1,500-bp promoter of MdGMP1, P2 represents the fragment of the MdGMP1 promoter containing the DRE motif, V-sk represents empty pGreenII 62-SK vector, V-Luc represents empty pGreenII 0800-LUC vector. C, ChIP-qPCR assay in transgenic apple calli. Cross-linked chromatin samples were extracted from 35S:MdERF98-GFP or EV-GFP (Empty pMDC83-GFP vector) transgenic apple calli and precipitated with an anti-GFP antibody. Eluted DNA was subjected to PCR for amplification of sequences neighboring the DRE by qPCR. Pro1 and Pro2, two regions of MdGMP1 promoter, were investigated. The value of Pro-1 EV-GFP was set to 1. The ChIP assay was repeated three times, and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. D, EMSA showing that MdERF98 binds to the DRE motif of the MdGMP1 promoter. The biotin-labeled probe was a fragment of the MdGMP1 promoter containing the DRE motif, and the cold-probe was the nonlabeled probe sequence (at 100-fold that of the bio-probe). The mutant-probe was the biotin-labeled sequence with five nucleotides mutated. MdERF98-His was a purified fusion protein. Bars represent the mean value ±se (n = 3). Lowercase letters indicate significantly different values (P < 0.05, one-way ANOVA test).
In apple callus overexpressing MdERF98, the expression of MdGMP1 was significantly upregulated, and Asc level was increased compared with callus carrying the empty vector (Supplemental Figure S14). We also generated two transgenic apple lines with suppressed expression of MdERF98 (RNAi-MdERF98-1 and RNAi-MdERF98-2) (Supplemental Figure S15A). In the leaves of RNAi-MdERF98 transgenic lines, the expression levels of MdGMP1 and MdGME1 were significantly lower than those in the leaves carrying the empty vector (Supplemental Figure S15, B and C). As a result, the Asc levels were significantly reduced (Supplemental Figure S15D). Together, these results indicated that MdERF98 binds to the DRE motif of the MdGMP1 promoter and increases both MdGMP1 transcription and Asc biosynthesis.
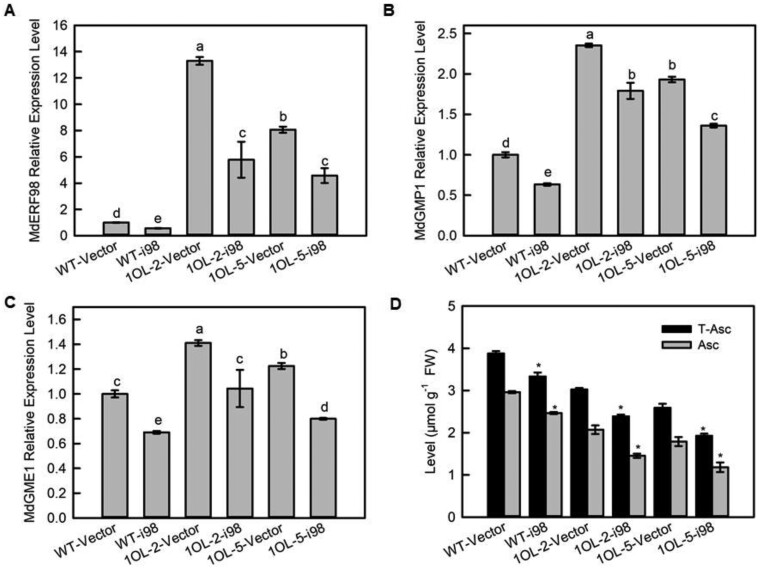
To confirm if the increase in MdERF98 transcript levels was one of the reasons for the increase in MdGMP1 transcripts, we suppressed the expression of MdERF98 in WT and MdAMR1L1-OE transgenic apple leaves (1OL-2 and 1OL-5) (Figure 6A). The silencing of MdERF98 in WT apple reduced the mRNA levels of MdGMP1 and MdGME1 and the Asc levels, confirming the role of MdERF98 in regulating MdGMP1 expression (Figure 6, B–D). In the MdAMR1L1-OE background (1OL-2-i98 and 1OL-5-i98), the reduction in MdERF98 expression led to a similar reduction in MdGMP1 and a greater decrease in Asc levels in the silenced lines compared with the MdAMR1L1-OE lines (1OL-2 and 1OL-5) (Figure 6). These observations indicated that MdGMP1 upregulation in the MdAMR1L1-OE lines was due to increased MdERF98 expression, which mitigated the decrease in Asc level caused by the overexpression of MdAMR1L1.
Figure 6.
The MdGMP1 mRNA level is related to the MdERF98 transcript level in MdAMR1L1 overexpression transgenic lines. A, Relative expression levels of MdERF98 mRNA in leaves of WT, WT-i98, 1OL-2 and 5, and 1OL-2/5-i98 lines based on qRT-PCR. For each sample, transcript levels were normalized with those of MdActin. Relative expression levels for each gene were obtained via the ΔΔCT method. The value of WT-Vector was set to “1”. MdERF98 expression was silenced in “GL3” leaves of WT and MdAMR1L1-OE (1OL-2 and 1OL-5) transgenic lines. WT-i98, MdERF98 RNAi silencing in WT background; 1OL-2/5-i98, MdERF98 RNAi silencing in MdAMR1L1 overexpression backgrounds. B and C, Relative expression levels of MdGMP1 and MdGME1 transcripts in leaves of WT, WT-i98, 1OL-2/5, and 1OL-2/5-i98 lines based on qRT-PCR. For each sample, transcript levels were normalized with those of MdActin. Relative expression levels for each gene were obtained via the ΔΔCT method. The value of WT-Vector was set to “1”. D, Levels of Asc in leaves of WT, WT-i98, 1OL-2/5, and 1OL-2/5-i98 lines. Bars represent the mean value ±se (n = 3). Lowercase letters/asterisks indicate significantly different values (P < 0.05, one-way ANOVA test or independent t tests).
MdERF98 expression responds to changes in Asc level in apple resulting in feedback regulation of Asc biosynthesis
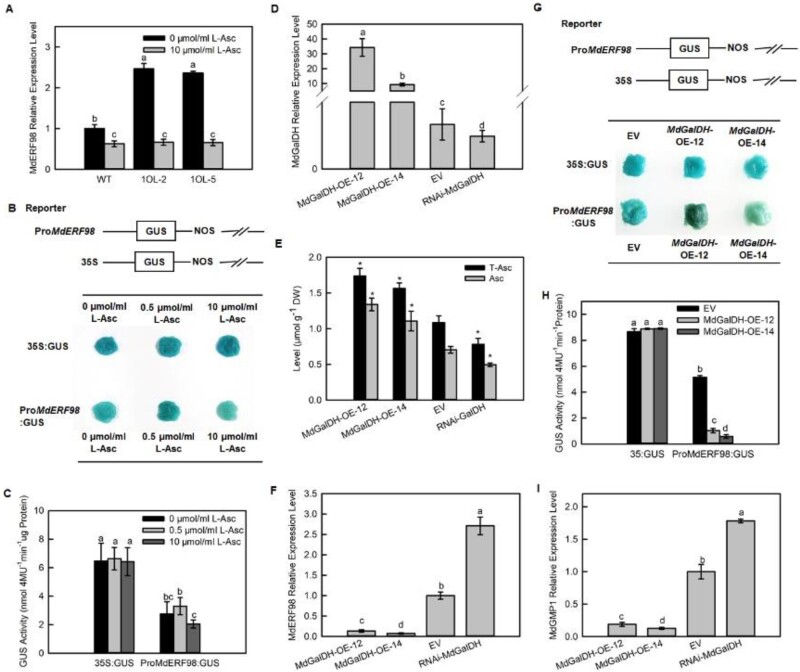
We hypothesized that the increased MdERF98 and MdGMP1 transcript levels in transgenic apple overexpressing MdAMR1L1 might be in response to decreased Asc levels. To confirm this hypothesis, we first showed that the transcriptional level of MdERF98 was significantly downregulated after the supply of exogenous L-Asc to WT and transgenic apples. With Asc treatment, MdERF98 transcripts in the MdAMR1L1-OE apple lines (1OL-2 and 1OL-5) were reduced to levels below those in WT apples (Figure 7A). A similar response was detected for MdGMP1 transcript levels (Supplemental Figure S16). The activity of the MdERF98 promoter in the apple calli system under different exogenous L-Asc levels was measured via a GUS activation assay (Figure 7, B and C). The addition of high levels of exogenous L-Asc decreased the staining and activity of GUS compared with the apple calli without L-Asc treatment after 6 h of incubation (Figure 7, B and C).
Figure 7.
MdERF98 transcript levels are regulated by feedback from Asc levels. A–C, MdERF98 transcript levels change with exogenous Asc concentration. A, Relative expression levels of MdERF98 transcripts in leaves of WT and MdAMR1L1 overexpression lines treated with exogenous L-Asc. One-month-old WT and MdAMR1L1 overexpression (1OL-2 and 1OL-5) apple seedlings grown in tissue culture were treated with exogenous 10 μmol/mL L-Asc or 0 μmol/mL L-Asc, as control, for 12 h. The value of WT-0 μmol/ml L-Asc was set to “1”. B and C, GUS staining and activity to analyze MdERF98 transcript levels in response to changes in exogenous Asc concentration. The ProMdERF98:GUS fusion protein and 35S:GUS (control) were transiently expressed in apple fruit calli of WT “WangLin”, then the transgenic calli were treated with 0, 0.5, or 10 μmol/mL L-Asc for 6 h. D–H, MdERF98 expression responded to increasing endogenous Asc levels. D, Relative expression levels of MdGalDH in apple fruit calli of EV and MdGalDH overexpression (MdGalDH-OE-12/14) and MdGalDH RNAi transgenic lines (RNAi-MdGalDH). The expression of MdGalDH in apple fruit calli of WT “WangLin” was increased or decreased by Agrobacterium infection as described in “Materials and Methods”. E, Levels of Asc in apple fruit calli of EV and MdGalDH overexpression and MdGalDH RNAi transgenic lines. F, Relative expression levels of MdERF98 in apple fruit calli of EV and MdGalDH overexpression and MdGalDH RNAi transgenic lines. G and H, GUS staining and activity to analyze the transcription of MdERF98 in response to MdGalDH overexpression. The ProMdERF98:GUS fusion protein and 35S:GUS (control) were transiently expressed in EV and MdGalDH overexpression transgenic lines. I, Expression levels of MdGMP1 in apple fruit calli of EV and MdGalDH overexpression and MdGalDH RNAi transgenic lines. The expression levels of MdERF98, MdGalDH, and MdGMP1 were investigated by qRT-PCR. For each sample, transcript levels were normalized with those of MdActin. Relative expression levels for each gene were obtained via the ΔΔCT method. The value of EV was set to “1”. Bars represent the mean value ±se (n = 3). Lowercase letters/asterisks indicate significantly different values (P < 0.05, one-way ANOVA test or independent t tests). EV, empty vector; DW, dry weight.
To further confirm our hypothesis, we generated transgenic apple calli overexpressing MdGalDH and interfering MdGalDH (Figure 7D), which is a gene in the L-Gal pathway of Asc biosynthesis. The MdGalDH-OE-12 and MdGalDH-OE-14 transgenic lines showed significantly increased Asc levels, while the RNAi-MdGalDH transgenic lines showed significantly decreased Asc levels compared with the empty vector control (Figure 7E). However, the changes of MdERF98 transcription level were opposite to the changes of Asc levels in the MdGalDH transgenic lines (MdGalDH-OE-12, MdGalDH-OE-14, and RNAi-MdGalDH) (Figure 7F). GUS staining and activity measurement also indicated that the overexpression of MdGalDH decreased the activity of the MdERF98 promoter compared with that in calli expressing the EV (Figure 7, G and H). In addition, the MdGMP1 transcript levels were similar to MdERF98 in MdGalDH transgenic lines (Figure 7I). Our results indicated that MdERF98 could respond to changes in Asc levels to regulate its own expression and thereby affect the transcription of the downstream gene MdGMP1. This is the reason that the transcript levels of MdERF98 and MdGMP1 were significantly higher when Asc levels were decreased in transgenic apple leaves overexpressing MdAMR1L1.
Discussion
Asc biosynthesis in apple is subject to double regulation: at both the mRNA and protein levels
Asc is an important water-soluble antioxidant in plants and is regulated by both biosynthesis and recycling from its oxidized form (Broad et al., 2020). In proteomic (Li et al., 2016a) and transcriptomic datasets (Zhu et al., 2021) of developing apple fruit, the mRNA expression patterns of only MdGME1 and MdGGP2 (among all genes of the D-Man/L-Gal pathway) were similar to the Asc levels in different tissues (Figure 1). At the genetic level, the GGP/VTC2 putative paralogs were consistently linked with the total Asc concentrations in apple flesh (Mellidou et al., 2012), suggesting that Asc contents are primarily determined by the extent of expression of GGP in different genotypes of apple. Although the GGP and VTC2 paralog proteins could not be detected, our proteomic data revealed that the MdGMP1 protein abundance was consistent with the concentration of Asc during fruit development, showing gradually decreases(Figure 1, B and D), but that the mRNA expression pattern of MdGMP1 was upregulation (Figure 1B). These results indicated that the GMP protein, an enzyme in the D-Man/L-Gal pathway that is involved in Asc biosynthesis (Conklin et al., 1997; Badejo et al., 2008; Sawake et al., 2015; Li et al., 2019), is subject to post-transcriptional regulation or post-translational modification when it participates in apple Asc biosynthesis. The photomorphogenic factor CSN5B links light modulation with Asc synthesis at the post-translational level via the degradation of polyubiquitinated VTC1 (Wang et al., 2013).
MdAMR1L1 decreases Asc biosynthesis by controlling MdGMP1 protein level
The expression of MdAMR1L1, at both the mRNA and protein levels, was also inversely related to the Asc level in different apple tissues and in developing fruits (Figure 1; Supplemental Figure S2). Silencing of MdAMR1L1 resulted in an increased Asc level in apple fruit and leaves (Figures 2, C and 3, B), while the overexpression of MdAMR1L1 decreased the Asc level in apple leaves (Figure 3B). These results indicated that MdAMR1L1 negatively regulates the Asc level in apple, as observed in Arabidopsis (Zhang et al., 2009). In the MdAMR1L1-OE apple lines, many genes in the D-Man/L-Gal pathway (such as MdGMP1, MdGME1, and MdGalDH1) showed increased expression (Figure 3, C;Supplemental Figure S7, A). We also heterologously expressed MdAMR1L1/2 in Arabidopsis and found that the expression of MdAMR1L1 or MdAMR1L2 decreased the Asc level in Arabidopsis (Supplemental Figure S8, B), but that the mRNA levels of genes in the D-Man/L-Gal pathway were not significantly changed (Supplemental Figure S8, C). These results indicated that MdAMR1L1 negatively regulates Asc biosynthesis not by inhibiting the transcription level of Asc synthesis genes in the D-Man/L-Gal metabolic network.
The protein domains encoded by MdAMR1L1 include the conserved F-box domain and show sequence homology to several known F-box proteins (Supplemental Figure S1). F-box proteins function as a component of an SCF ubiquitin ligase complex and interact specifically with other proteins to control their quantity and stability through ubiquitination (Kim et al., 2019). Our identification of MdAMR1L1 interacting proteins by IP-MS and other methods strongly indicated that MdAMR1L1 could interact with MdGMP1 and promote its degradation via the ubiquitination system (Figure 4; Supplemental Figure S11). The protein abundance of MdGMP1, the enzyme activity of GMP, and the concentration of GDP-Man were decreased in the MdAMR1L1-OE lines (Figure 3). Although GMP is required for the biogenesis of cell walls and protein glycosylation (Hancock and Viola, 2005), several studies involving mutants or transgenic plants have demonstrated that the GMP gene participates in the synthesis of Asc through the D-Man/L-Gal pathway (Conklin et al., 1997; Badejo et al., 2008; Sawake et al., 2015; Li et al., 2019), including our results showing that MdGMP1 overexpression in apple calli can increase Asc level (Supplemental Figure S10). To date, almost all identified regulatory genes associated with Asc synthesis have been related to GMP [e.g., the transcription factor AtERF98 (Zhang et al., 2012) in Arabidopsis, SlbHLH59 (Ye et al., 2019) and HD-ZIP (Hu et al., 2016) in tomato, and the post-translational factors KONJAC1/2 (Sawake et al., 2015) and CSN5B (Wang et al., 2013) in Arabidopsis]. These advances imply that GMP activity must be finely regulated at the transcriptional and post-transcriptional/translational levels in the synthesis of Asc.
MdERF98 regulates MdGMP1 transcription and is involved in the feedback regulation of Asc biosynthesis
Our study showed that the expression of the ethylene response factor MdERF98 increased significantly with the overexpression of MdAMR1L1, which was consistent with the change in MdGMP1 mRNA levels (Figure 3; Supplemental Figure S13). In Arabidopsis, the homologous gene AtERF98 can directly interact with the DRE-containing region of the VTC1 (GMP) promoter and promote its transcription (Zhang et al., 2012). Our results showed that MdERF98 could bind to the DRE-containing region of the MdGMP1 promoter and positively regulate both its expression and Asc synthesis (Figures 5 and 6; Supplemental Figure S14 and S15). In addition to GMP, AtERF98 also regulates the expression of GME and MIOX (Zhang et al., 2012). In transgenic apple lines in which either MdAMR1L1 was overexpressed or MdERF98 was silenced, MdGME1 and MdMIOX4 both showed similar expression patterns to MdERF98 and MdGMP1 (Figure 3, C; Supplemental Figures S7, B, S15, C, and S17), which implied that in apple MdERF98 might also regulate the expression of MdGME1 and MdMIOX4.
Interestingly, the transcript levels of MdERF98 and MdGMP1 were upregulated in MdAMR1L1-OE apple leaves with decreased Asc levels. We hypothesized that this might be related to feedback regulation by Asc. Feedback regulation of Asc biosynthesis by Asc has been indicated to involve GME (Wolucka and Van, 2003), GalDH (Mieda et al., 2004), and GGP (Laing et al., 2015). This was confirmed by our results (Figure 7). First, supplying exogenous L-Asc inhibited the increased expression of MdERF98 and MdGMP1 in MdAMR1L1-OE apple leaves (Figure 7A;Supplemental Figure S15). Second, MdERF98 expression and promoter activity were inhibited by increased exogenous L-Asc, which significantly inhibited the activity of proMdERF98:GUS (Figure 7, B and C). Additionally, MdERF98 expression was negatively associated with the concentration of Asc in MdGalDH transgenic apple fruit calli (Figure 7, E–H). In addition, the expression pattern of MdGMP1 was the same as MdERF98 in MdGalDH transgenic apple fruit calli (Figure 7I). Asc oxidation products are toxic and can cause metabolic damage (Wang et al., 2020), so Asc feeding can cause stress and may also have a certain impact on our results. However, we obtained transgenic fruit calli with altered endogenous Asc levels through genetic transformation to further confirm our conclusion. These observations indicate that MdERF98 expression is subject to feedback regulation by Asc levels in apple and affects the transcription of the downstream gene MdGMP1 to regulate the homeostasis of Asc. Given that Asc is an essential antioxidant that affects ROS metabolism, hormone signaling, redox status, and other biological processes in plant cells (Broad et al., 2020), the feedback regulation of Asc biosynthesis might be an adaptive mechanism for meeting the demand for Asc under altered growth, development and environmental conditions. The mechanism of the feedback regulation mediated by MdERF98 might be related to hormone or ROS (Reactive oxygen species) signaling, and many DEGs in both types of signaling pathways were detected in transgenic apple overexpressing MdAMR1L1. More work on this topic needs to be carried out in future research.
Taken together, our data demonstrate a mechanism of Asc biosynthesis regulation through the transcriptional and post-translational regulation of MdGMP1 and Asc feedback regulation, according to the following findings. Overexpression of MdAMR1L1 in apple leads to increased transcript and protein abundance of MdAMR1L1. Because MdAMR1L1 can interact with MdGMP1 and target this protein to degradation by the ubiquitin-proteasome pathway, the protein abundance of MdGMP1 in MdAMR1L1-OE apple lines was decreased, consequently decreasing the concentration of Asc. Asc biosynthesis is thought to be regulated by compensation machinery, and the feedback regulation of Asc plays an important role in Asc biosynthesis. When plants perceive a decrease in their Asc levels, a signal of this decrease will be transmitted to MdERF98 to promote its expression, perhaps to cope with environmental stress. Then, MdERF98 will bind directly to the promoter of MdGMP1 and activates its expression to maintain Asc homeostasis (Figure 8). This could be interpreted as a compensatory machinery for Asc synthesis and accumulation to maintain Asc homeostasis in plants.
Figure 8.
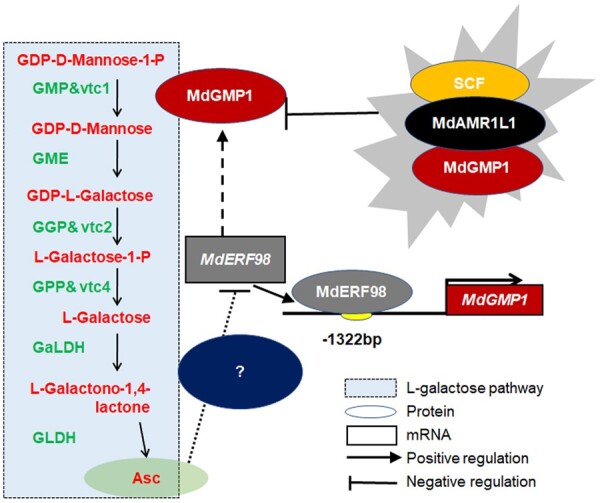
Model of MdAMR1L1 and MdERF98 regulation of ascorbate biosynthesis in apple through participating in the transcription and posttranslational modification of MdGMP1. MdAMR1L1 could interact with and target the MdGMP1 protein to the ubiquitination pathway for degradation. MdAMR1L1 overexpression in apple decreased both MdGMP1 protein abundance and Asc level. MdERF98, a transcription factor, can positively regulate MdGMP1 expression and is involved in feedback regulation of Asc biosynthesis in apple. Decreased Asc caused by overexpressing MdAMR1L1 would cause increased expression of MdERF98 that leads to increased transcript levels of MdGMP1. On the other hand, an increasing Asc level would inhibit the expression of MdERF98 and MdGMP1. This feedback regulation, modulated by MdERF98, is useful to avoid a huge variation of Asc level in plant cells.
Materials and methods
Plant materials and growth conditions
As in our previous report (Li et al., 2016a and Zhu et al., 2021), proteomic data and RNA-seq data were generated from “Greensleeves” apple (Malus × domestica) fruit collected at five developmental stages (16, 41, 70, 94, and 128 d after bloom). Fully expanded leaf and shoot tip samples were also subjected to RNA-seq.
Tissue-cultured WT and MdAMR1L1-, MdAMR1L2-, and MdERF98-transformed “GL3” apple plantlets were initially grown on MS medium supplemented with 25 mg L−1 kanamycin, 0.2 mg L−1 IAA, and 0.3 mg L−1 6-BA for four weeks. They were then transferred to rooting MS medium supplemented with 0.5 mg L−1 IAA and 0.5 mg L−1 IBA. After rooting, plants of these genotypes were transferred to a culture room maintained at 23°C under a 14-h photoperiod, supplemented with fluorescent light (60 μmol m−2 s−1). After these plants had grown for more than two months, their fully mature leaves were sampled. The samples were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
Vector construction and plant transformation
For the co-silencing of MdAMR1L1/2, a 418-bp common fragment of MdAMR1L1 and MdAMR1L2 (which share 98.5% nucleic acid sequence similarity) was amplified and cloned into pTRV2 to generate pTRV2-MdAMR1L. The constructs were transformed into Agrobacterium strain GV3101 (pMP90) for the VIGS experiments, as described previously (Lin et al., 2008). The histochemical localization of Asc was carried out according to a previously reported method (Li et al., 2008).
To construct the vector for the MdAMR1L1/2-overexpression lines, we introduced the coding region of MdAMR1L1/2 into the pGWB411 binary vector by Gateway cloning technology. To silence the MdAMR1L/MdERF98 genes in the leaves of WT “GL3” apple, the specific fragments of them were cloned into the pK7GWIWG2D(II) vector by Gateway cloning technology, respectively. The recombinant plasmid was transformed into Agrobacterium strain LBA4404, which was subsequently introduced into the leaves of WT “GL3” via Agrobacterium-mediated transformation using a method described in Dai et al. (2013).
To determine the effect of MdGMP1 and MdGalDH, the full-length coding sequence of MdGMP1 or MdGalDH was inserted into the PacI (TTAATTAA) and AscI (GGCGCGCC) sites the pMDC83 binary vector under the control of the CaMV 35S promoter. A 300-bp specific sequence of MdGalDH was cloned into the pK7GWIWG2D(II) vector by Gateway cloning technology. The recombinant plasmid was transformed into Agrobacterium strain EHA105, which was subsequently introduced into WT “WangLin” apple fruit calli via Agrobacterium-mediated transformation using a method described in Li et al. (2016b).
All primers used are shown in Supplemental Table S1.
RNA extraction and RT-qPCR
Total RNA was extracted from samples via a modified CTAB method (Gasic et al., 2004) and treated by DNase before reverse transcription. Gene-specific primers were designed from the CDS (Coding sequence) of apple genes. The amplified PCR products were quantified with an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Their expression levels were normalized to the expression level of the internal control actin (CN938023) in the quantitative PCR experiment. Data were analyzed via the ΔΔCT method using the iQ5 2.0 standard optical system analysis software.
Western blot analysis
The procedures for total protein sample extraction and Western blotting followed Sun et al. (2018), and total protein concentrations were determined with protein assay kits (Bio-Rad) using bovine serum albumin as a standard. Specific monoclonal antibodies against a peptide (CMETQWSDLPK) from a highly conserved region of MdAMR1L1 and another peptide (CLDKIELRPTSIEKE) from a highly conserved region of MdGMP1 (Genscript, Nanjing, China) were generated in rabbits. β-Actin was monitored with a monoclonal antibody (CWBIO). The antigen–antibody complexes were detected using the ClarityTM Western ECL Substrate (Bio-Rad) according to the manufacturer’s instructions.
Determination of reduced and total Asc
The reduced Asc and total Asc (T-Asc) levels were determined according to Cheng and Ma (2004) with slight modification. This assay is based on the oxidation of Asc by Asc oxidase in an acidic solution. The Asc level was calculated as the difference in the absorption at 265 nm before and after the addition of Asc oxidase. The Asc level was quantified by comparing the difference in absorption relative to a standard curve.
RNA sequencing and data analysis
RNA-seq data were generated from fully expanded leaves of MdAMR1L1 transgenic apple plants, with three biological replicates. Sequencing was performed using an Illumina HiSeq 2500 sequencer with 250-bp single-end sequencing (Illumina, San Diego, CA, USA) at Biomarker Technologies, China.
The RNA-seq reads were processed by first removing barcode and adaptor sequences, followed by alignment to the SILVA rRNA database using Bowtie, allowing up to three mismatches to remove potential contaminating reads. The clean reads were then aligned to the Malus GDDH13 version 1.1 genome using TopHat, allowing one mismatch. After alignment, the raw counts of the mapped reads for each Malus gene model from GDDH13 version 1.1 were derived and then normalized to reads per kilobase of exon model per million mapped reads (RPKM) values.
Determination of GMP enzyme activity and GDP-Man concentration
The GMP enzyme activity was determined according to the previously described method (Sawake et al., 2015) with slight modification. To extract GMP, 0.2 g sample was homogenized in 1.5 mL lysis buffer (50 mmol/L; 20 v/v glycerin; 1 mmol/L EDTA; 1 w/v PVP). The extract was centrifuged at 9,000 rpm for 10 min at 2°C. The activity of GMP was determined by monitoring the formation of GDP-Man. A 3 mL reaction mixture contained 50 mM Tris (pH 7.5) buffer, 2 mM MgCl2, 10 mM GTP, 1 mM Man 1-P, and 100 μL extracted enzyme solution. After being reacted for 30 min at 28°C, the reaction product GDP-Man were detected and quantified with an Agilent 1260 Infinity HPLC with a CarboPac PA1 column (4 × 250 mm; Dionex) according to the method described by Pauly et al. (2000).
GDP-Man was assayed using liquid chromatography–tandem mass spectrometry (LC–MS/MS) methods as previously described method (Sawake et al., 2015). The 100-mg powder samples were used to extract GDP-Man in the pre-cold solution (stored at 4°C) containing 300 mL of chloroform, 600 mL of methanol, and 1 nmol of TDP-Glc (Sigma-Aldrich, as an internal) at 4°C for 2 h. Then 1 mL of water was added to each sample, which was mixed by vortexing and centrifuged at 14,000 g at 4°C for 5 min. The upper aqueous phase was collected as soluble GDP-Man extraction, and dried with a centrifugal vacuum concentrator, and resuspended in 1 mL of water. GDP-Man was separated with a porous graphitic carbon LC column (5 μm particle size, 1 × 150 mm; Thermo Fisher Scientific) using an Agilent 1260 Infinity HPLC. GDP-Man was detected using a QTRAP 5500 (AB Sciex) with a Turbo V Ion Source (AB Sciex). Mass spectrometry parameters for TDP-Glc (Q1 mass, 563 D; Q3 mass, 321 D; collision energy, 30 eV) were determined using direct infusion of 1 mM of TDP-Glc in 50% (v/v) acetonitrile into the electrospray ion source using a syringe pump at a flow rate of 20 mL/min. GDP-Man abundance was determined by signal peak area integration using MultiQuant 2.1 (AB Sciex).
Protein–protein interaction validation
For the IP-MS assay, total leaf protein was extracted using lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 4 M urea, and 1 mM PMSF (Phenylmethanesulfonyl fluoride)). The IP procedure was performed according to the instructions of the Pierce Classic Magnetic IP/Co-IP Kit (Thermo Scientific). The monoclonal antibodies against MdAMR1L1 were described above. The eluate was subjected to SDS–PAGE (Sodium dodecyl sulfate- Polyacrylamide gel electrophoresis) followed by Coomassie Brilliant Blue Staining. The colored bands were cut out separately and sent for LC–MS/MS LTQ Orbitrap Elite analysis. The WT protein was used as a blank control for identifying nonspecific interactions.
For the yeast two-hybrid assay, the full-length MdAMR1L1 cDNA was cloned into the prey vector pGADT7 using the BamHI (GGATCC) and SalI (GTCGAC) enzyme sites, and the MdGMP1 cDNA was cloned into the bait vector pGBKT7 using the BamHI (GGATCC) and SalI (GTCGAC) enzyme sites, which was then transformed into Y2H Gold yeast cells using the lithium acetate method. The specific experimental procedure was performed in reference to the Matchmaker® Gold Yeast Two-Hybrid System User Manual (Clontech).
For the Bi-FC assay, full-length MdGMP1 and MdAMR1L1 without their stop codons were cloned into the Bi-FC vector containing either the N- or C-terminal fragment of YFP (pEarleyGate201-YN/pEarleyGate202-YN and pEarleyGate201-YC/pEarleyGate202-YC) and co-expressed in the leaves of N. benthamiana for 3 d. Reconstituted YFP signals indicating a positive protein interaction were detected on ZEISS LSM 900 microscope with Airy scan 2. YFP was excited at 514 nm, and the fluorescence emissions were detected at 520–556 nm.
All primers used are shown in Supplemental Table S1.
Cell-free and protein degradation assays
The expression and purification of MdGMP1-His were performed as previously described (Li et al., 2016b) with slight modification. The CDS of MdGMP1 without the stop codon was cloned into the BamHI (GGATCC) and XhoI (CTCGAG) sites of the pET32a vector. The construct was transformed into Escherichia coli BL21 (DE3) competent cells. Isopropyl-β-d-thiogalactoside was added at a final concentration of 0.5 mM to induce protein expression.
Cell-free and protein degradation assays were carried out as previously described (Xia et al., 2020) with slight modifications. Total proteins were extracted from 1-month-old MdAMR1L1-OE apple seedling leaves. One milligram of total protein was mixed with 1 mg of MdGMP1-His purified protein for 0 h, 2 h, 4 h, or 6 h at room temperature. To further confirm that MdGMP1 degradation is mediated by ubiquitination, 50 µM MG132 was added. The products were subjected to immunoblotting with anti-HIS/Actin (CWBIO).
All primers used are shown in Supplemental Table S1.
Transcription factor–DNA interaction validation
For the dual luciferase assay, the CDS of MdERF98 without the stop codon was cloned into the pGreenII 62-SK vector using the related EcoRI (GAATTC) and XhoI (CTCGAG) enzyme sites to generate an effector construct. A reporter construct was generated using various fragments of the MdGMP1 promoter cloned upstream of the LUC reporter gene in the pGreenII 0800-LUC vector using the SalI (GTCGAC) and BamHI (GGATCC) enzyme sites. The reporter vector and the effector vector were, respectively, transformed into Agrobacterium strain GV3101, and N. benthamiana leaves were used for co-infiltration. Relative LUC activity was calculated as the ratio of LUC/Ren (Firefly luciferase/Renilla luciferase). The Luc/Ren ratios were calculated from three biological replicates.
For the EMSA assays, we first obtained purified MdERF98-His protein, as described above. The 5′-biotin end-labeled double-stranded DNA probes (Bioprobe and Mutant probe) were prepared by annealing complementary oligonucleotides. The sequence of the biotin-labeled bioprobe was 5′-AAAAAATCAAATACCGACTTGGTCACGGCA-3′, and the sequence of the biotin-labeled mutant probe was 5′-AAAAAATCAAATAAAAAATTGGTCACGGCA-3′. EMSA was performed using the LightShift® Chemiluminescent EMSA Kit (Cat. no.20148, Thermo Scientific).
For the ChIP-PCR (Chromatin immunoprecipitation-PCR) assays, the CDS of MdERF98 without the stop codon was cloned into the PacI (TTAATTAA) and AscI (GGCGCGCC) sites downstream of a GFP tag in pMDC83 under the control of the CaMV 35S promoter to generate the CaMV35S-MdERF98-GFP construct. The recombinant construct was transformed into apple calli, and ChIP assays were performed as described (Li et al., 2016b) with slight modifications. An anti-GFP antibody (Proteintech) was used for ChIP. The amount of immunoprecipitated chromatin was determined by qPCR.
All primers used are shown in Supplemental Table S1.
GUS staining observation and activity analysis
A reporter construct was generated using the promoter sequence of MdERF98 (1692 bp) cloned upstream of the GUS reporter gene in the binary vector pBI121 using the BamHI (GGATCC) and XhoI (CTCGAG) enzyme sites. For the transient expression assay, the reporter vector was transformed into Agrobacterium strain GV3101. “WangLin” apple fruit calli were subjected to infiltration. After growth in the dark for 3 d, the infected fruit calli were used to analyze GUS staining and activity. GUS histochemical staining and GUS activity analysis were performed according to the experimental method of Zhang et al. (2017). All primers used are shown in Supplemental Table S1.
Bioinformatics analysis
For phylogenetic analysis, the full-length amino acid sequences of AMR1-Like genes from Arabidopsis, Malus, and Solanum lycopersicum from the NCBI protein database (http://www.ncbi.nlm.nih.gov/guide/) were aligned using the integrated MUSCLE alignment program in Molecular Evolutionary Genetics Analysis (MEGA5) with default parameters. Phylogenetic analysis was performed via the Neighbor-Joining method, using MEGA6 software and bootstrap tests replicated 1,000 times (Tamura et al., 2013). The amino acid sequence of the MdAMR1L1/2 gene was submit to the website (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for analysis of conserved domains. The sequence alignment of MdAMR1L1/2 with human (SKP2 and FBX4) and Arabidopsis (AtAMR1, ORE9, SLEEPY1, FKF1, SON1, and UFO) F-box domains were aligned using DNAMAN.
Statistical analysis
All data were analyzed via IBM SPSS Statistics 16.0 and graphed with Sigma Plot 12.5 software. Data were analyzed using independent t tests or one-way analysis of variance (ANOVA) test with a significance level accepted at P < 0.05. The bar represents the ±se (standard error) of three repeated experiments, and the lowercase letters/asterisks denote results that were significantly different from those in the control (P < 0.05).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MdAMR1L1 (MD15G1235900), MdAMR1L2 (MD15G1235700), MdGMP1 (MD01G1139900), MdERF98 (MD16G1216500), and AtAMR1 (At1G65770).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Bioinformatics analysis of MdAMR1L1/2.
Supplemental Figure S2 . MdAMR1L1 expression is negatively associated with Asc levels in different developmental stages of fruit and different tissues in apple.
Supplemental Figure S3 . Effects of light on MdAMR1L1 expression and Asc levels in apple leaves.
Supplemental Figure S4 . Identification of MdAMR1L transgenic apple lines.
Supplemental Figure S5 . Overexpression of MdAMR1L1 inhibits plant growth but and has no effect on photosynthesis in leaves of transgenic apple lines.
Supplemental Figure S6 . DEGs produced in MdAMR1L1 overexpression transgenic apple lines.
Supplemental Figure S7 . Overexpression of MdAMR1L1 promotes the expression of Asc synthesis genes, but has no effect on the expression of Asc regeneration-related genes in leaves of MdAMR1L1 overexpression transgenic apple lines.
Supplemental Figure S8 . MdAMR1L inhibits the accumulation of Asc, but has no effect on the expression of Asc synthesis genes in leaves of MdAMR1L-OE transgenic Arabidopsis lines.
Supplemental Figure S9 . MdGMP1 protein levels are positively associated with the Asc levels in apple.
Supplemental Figure S10 . MdGMP1 positively regulates the biosynthesis of Asc.
Supplemental Figure S11 . MG132 enhanced MdGMP1 protein stability and Asc levels in apple leaves.
Supplemental Figure S12 . Analysis of transcription factor MdERF98.
Supplemental Figure S13 . MdAMR1L1 overexpression promotes the transcription of MdERF98 gene in leaves of transgenic apple lines.
Supplemental Figure S14 . MdERF98 overexpression promotes the biosynthesis of Asc in apple calli.
Supplemental Figure S15 . MdERF98 silencing inhibits the biosynthesis of Asc in apple leaves.
Supplemental Figure S16 . A high Asc level decreases the transcript level of MdGMP1 in leaves of WT and MdAMR1L1-overexpression transgenic lines.
Supplemental Figure S17 . MdMIOX4 expression levels are consistent with MdERF98.
Supplemental Table S1 . List of primers.
Supplementary Data Excel File S1 . mRNA expression by genes involved in D-mannose/L-galactose pathway and Asc synthetic regulation in shoot tips, mature leaves, and developed fruit of apple, based on RPKM values from RNA-seq.
Supplementary Data Excel File S2 . Different expression genes in 1OL-2, 1OL-4 and 1OL-5 lines overexpressing MdAMR1L1 compared with WT based on RNA-seq.
Supplementary Material
Acknowledgments
We thank Dr Jing Zhang, Miss Jing Zhao, and Miss Minrong Luo (Horticulture Science Research Center, Northwest A&F University, Yangling, China) for providing professional technical assistance with LC–MS/MS analysis and tissue culture.
Funding
This work was supported by the Program for the National Key Research and Development Program (2018YFD1000200), the National Natural Science Foundation of China (No. 31672128), The Chinese Universities Scientific Fund (2452020007), and the Earmarked Fund for the China Agriculture Research System (CARS-28).
Conflict of interest statement. None declared.
M.J.L. and F.W.M.: conceived and supervised this study; S.Y.M., H.X.L., L.W., B.Y.L. and Z.Y.W.: performed the experiments; S.Y.M. and H.X.L.: performed the bioinformatics analysis; S.Y.M and H.X.L.: wrote the manuscript; M.J.L, B.Q.M, and S.Y.M.: discussed the study and revised the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) are: Mingjun Li (limingjun@nwsuaf.edu.cn) and Fengwang Ma (fwm64@nwsuaf.edu.cn).
References
- Badejo AA, Tanaka N, Esaka M (2008) Analysis of GDP-D-mannose pyrophosphorylasegene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol 49: 126–132 [DOI] [PubMed] [Google Scholar]
- Broad RC, Bonneau JP, Hellens RP, Johnson AAT (2020) Manipulation of ascorbate biosynthetic, recycling, and regulatory pathways for improved abiotic stresstolerance in plants. Int J Mol Sci 21: 1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley S, Laing W (2016) The regulation of ascorbate biosynthesis. Curr Opin Plant Biol 33: 15–22 [DOI] [PubMed] [Google Scholar]
- Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, Wright M, MacRae E, Gleave A, Laing W (2009) Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J Exp Bot 60: 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Ma F (2004) Diurnal operation of the xanthophyll cycle and the antioxidant system in apple peel. J Am Soc Hort jSci 129: 313–320 [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96: 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Pallanca JE, Last RL, Smirnoff N (1997) L-ascorbic acid metabolism in the ascorbate-deficient Arabidopsis mutant vtc1. Plant Physiol 115: 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai HY, Li WR, Han GF, Yang Y, Ma Y, Li H, Zhang ZH (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci Hortic-Amst 164: 202–208 [Google Scholar]
- Fenech M, Amorim-Silva V, Esteban del Valle A, Arnaud D, Ruiz-Lopez N, Castillo AG, Smirnoff N, Botella MA (2021) The role of GDP-L-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol 185: 1574–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22: 437–438 [Google Scholar]
- Hancock RD, Viola R (2005) Biosynthesis and catabolism of L‐ascorbic acid in plants. Crit Rev Plant Sci 24:167–188 [Google Scholar]
- Hu T, Ye J, Tao P, Li H, Zhang J, Zhang Y, Ye Z (2016) Tomato HD-Zip I transcription factor, SlHZ24, modulates Asc accumulation through positively regulating the D-mannose/L-galactose pathway. Plant J 85: 16–29 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Shigeoka S (2008) Recent advances in Ascbiosynthesis and the physiological significance of Asc peroxidase in photosynthesizing organisms. Biosci Biotechnol Biochem 72: 1143–1154 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Yu S, Jung SH, Lee B, Suh MC (2019) The F-box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell 31: 2223–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. (2015) .An upstream open reading frame is essential for feedback regulation of Asc biosynthesis in Arabidopsis. Plant Cell 27: 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Ma FW, Zhang M, Pu F (2008) Distribution and metabolism of ascorbic acid in apple fruits (Malus domestica Borkh cv. Gala) Plant Sci 174: 606–612 [Google Scholar]
- Li MJ, Chen XS, Wang PP, Ma FW (2011) Ascorbic acid accumulation and expression of genes involved in its biosynthesis and recycling in developing apple fruit. J Am Soc Hortic Sci 136: 231–238 [Google Scholar]
- Li MJ, Li DX, Feng FJ, Zhang S, Ma FW, Cheng LL (2016a) Proteomic analysis reveals dynamic regulation of fruit development and sugar and acid accumulation in apple. J Exp Bot 67: 5145–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016b) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li XJ, Ye J, Munir S, Yang T, Chen WF, Liu GZ, Zheng W, Zhang YY (2019) Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato. Int J Mol Sci 20: 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schellhorn H E (2007) New developments and novel therapeutic perspectives for vitamin C. J Nutr 137: 2171–2184 [DOI] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL (2004) Myo-inositol oxygenase offers a possible entry point into plant Asc biosynthesis. Plant Physiol 134: 200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellidou I, Chagné D, Laing WA, Keulemans J, Davey MW (2012)Allelic variation in paralogs of GDP-L-galactose phosphorylase is a majordeterminant of vitamin C concentrations in apple fruit. Plant Physiol 160: 1613–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellidou I, Kanellis AK (2017) Genetic control of ascorbic acid biosynthesis and recycling in horticultural crops. Front Chem 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda T, Yabuta Y, Rapolu M, Motoki T, Takeda T, Yoshimura K, Ishikawa T, Shigeoka S (2004) Feedback inhibition of spinach L-galactose dehydrogenase by L-ascorbate. Plant Cell Physiol 45:1271–1279 [DOI] [PubMed] [Google Scholar]
- Pauly M, Porchia A, Olsen CE, Nunan KJ, Scheller HV (2000) Enzymatic synthesis and purification of uridine diphosphob-L-arabinopyranose, a substrate for the biosynthesis of plant polysaccharides. Anal Biochem 278: 69–73 [DOI] [PubMed] [Google Scholar]
- Sawake S, Tajima N, Mortimer JC, Lao JM, Ishikawa T, Yu XL, Yamanashi Y, Yoshimi Y, Kawai-Yamada M, Dupree P, et al. (2015) KONJAC1 and 2 are key factors for GDP-mannose generation and affect L-ascorbic acid and glucomannan biosynthesis in Arabidopsis. Plant Cell 27: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, Huo LQ, Che RM, Ma FW (2018)Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J 16: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipsk A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R (2013) Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Feng C, Zhai ZF, Peng X, Wang YY, Sun YT, Li J, Shen XS, Xiao YQ, Zhu SJ, et al. (2020) The Apple microR171i-scarecrow-like proteins26.1 module enhances drought stress tolerance by integrating ascorbic acid metabolism. Plant Physiol 184: 194–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393: 365–369 [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van MM (2003) GDP-mannose 3',5'-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278: 47483–47490 [DOI] [PubMed] [Google Scholar]
- Xia FN, Zeng BQ, Liu HS, Hua Q, Xie LJ, Yu LJ, Chen QF, Li JF, Chen YQ, Jiang LW, et al. (2020) SINAT E3 ubiquitin ligases mediate FREE1 and VPS23A degradation to modulate abscisic acid signalling. Plant Cell 32: 3290–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Mieda T, Rapolu M, Nakamura A, Motoki T, Maruta T, Yoshimura K, Ishikawa T, Shigeoka S (2007) Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. J Exp Bot 58: 2661–2671 [DOI] [PubMed] [Google Scholar]
- Ye J, Li WF, Ai G, Li CX, Liu GZ, Chen WF, Wang B, Wang WQ, Lu YG, Zhang JH, et al. (2019) Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLoS Genet 15: e1008149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Nakane T, Kume S, Shiomi Y, Maruta T, Ishikawa T, Shigeoka S (2014) Transient expression analysis revealed the importance of VTC2 expression level in light/dark regulation of ascorbate biosynthesis in Arabidopsis. Biosci Biotechnol Biochem 78: 60–66 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Yu JQ, Wang JH, Hu DG, Hao YJ (2017) Functional characterization of MdMYB73 reveals its involvement in cold stress response in apple calli and Arabidopsis. J Integr Agr 16: 2215–2221 [Google Scholar]
- Zhang W, Lorence A, Gruszewski HA, Chevone BI, Nessler CL (2009) AMRL1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol 150: 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Zhang R, Huang R (2012) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhu LC, Li BY, Wu LM, Li HX, Wang ZY, Wei XY, Ma BQ, Zhang YF, Ma FW, Ruan YL, et al. (2021) MdERDL6-mediated glucose efflux to the cytosol promotes sugar accumulation in the vacuole through up-regulating TSTs in apple and tomato. PNAS 118: e2022788118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.