Abstract
Stunted growth in saline conditions is a signature phenotype of the Arabidopsis SALT OVERLY SENSITIVE mutants (sos1-5) affected in pathways regulating the salt stress response. One of the mutants isolated, sos4, encodes a kinase that phosphorylates pyridoxal (PL), a B6 vitamer, forming the important coenzyme pyridoxal 5′-phosphate (PLP). Here, we show that sos4-1 and more recently isolated alleles are deficient in phosphorylated B6 vitamers including PLP. This deficit is concomitant with a lowered PL level. Ionomic profiling of plants under standard laboratory conditions (without salt stress) reveals that sos4 mutants are perturbed in mineral nutrient homeostasis, with a hyperaccumulation of transition metal micronutrients particularly in the root, accounting for stress sensitivity. This is coincident with the accumulation of reactive oxygen species, as well as enhanced lignification and suberization of the endodermis, although the Casparian strip is intact and functional. Further, micrografting shows that SOS4 activity in the shoot is necessary for proper root development. Growth under very low light alleviates the impairments, including salt sensitivity, suggesting that SOS4 is important for developmental processes under moderate light intensities. Our study provides a basis for the integration of SOS4 derived B6 vitamers into plant health and fitness.
Phosphorylated B6 vitamer content is decreased in salt overly sensitive4 mutant alleles, causing shoot and root developmental defects that are alleviated by growth under very low light.
Introduction
Vitamin B6 is a water-soluble vitamin essential for all living organisms. The term vitamin B6 collectively refers to a family of compounds that have a pyridine ring in common but differ in their 4ʹ as well as their 5ʹ moieties, comprising the alcohol pyridoxine (PN), the amine pyridoxamine (PM), the aldehyde pyridoxal (PL), and their 5′ phosphorylated forms (PNP, PMP, and PLP) (Fitzpatrick et al., 2007). Among these compounds, PLP acts as a co-enzyme in over 284 enzymatic reactions involved in a variety of processes (Colinas et al., 2016). The different forms of vitamin B6 have also been touted as potent antioxidants and are associated with numerous environmental stress responses, both biotic and abiotic (Bilski et al., 2000; Titiz et al., 2006; Havaux et al., 2009; Asensi-Fabado and Munné-Bosch, 2010; Zhang et al., 2015; Dell’Aglio et al., 2017). PLP may also have non-coenzyme functions in carbohydrate metabolism, acting as a proton donor to facilitate starch and glycogen phosphorylase reactions and is reported to inhibit export of triose phosphates from chloroplasts (Rueschhoff et al., 2013).
Biosynthesis de novo of vitamin B6 takes place only in microorganisms and plants, thus animals must acquire it from their diet. Forms of vitamin B6 that can be taken up and functionally utilized are termed vitamers. Most organisms (including those that can biosynthesize the vitamin de novo) have enzymes that can interconvert the different B6 vitamers—sometimes referred to as “vitamin B6 salvage” enzymes. In humans, the kinase which can phosphorylate the non-phosphorylated vitamers (PN, PL, PM) and the oxidase that converts PMP/PNP to PLP are very well characterized and have received a lot of attention as they have been implicated in several diseases (Di Salvo et al., 2012; Wilson et al., 2019). However, the importance of these enzymes in plants has been somewhat neglected in comparison. Recently, it has been demonstrated that balancing of B6 vitamers through the salvage pathway is important for plant fitness in relation to nitrogen metabolism (Colinas et al., 2016). In particular, perturbation of homeostasis in Arabidopsis (Arabidopsis thaliana) by knocking out the PMP/PNP oxidase (PDX3) leads to a dependence on ammonium as a source of nitrogen and pdx3 knock-out mutant plants were constitutively activated for defense, thereby negatively impacting growth (Colinas et al., 2016).
Interestingly, the kinase that phosphorylates the non-phosphorylated vitamers went unrecognized in plants until it was retrieved in an Arabidopsis screen for salt-hypersensitive mutants and was thus named SALT OVERLY SENSITIVE4 (SOS4) (Shi et al., 2002; Shi and Zhu, 2002). Under sodium chloride stress, sos4-1 mutants are unable to maintain the Na+ to K+ balance and are characterized by a low K+ and high Na+ content compared with wild type plants (Shi et al., 2002). However, the connection between SOS4 and salt sensitivity remained largely unexplained, although PLP is known to regulate P2X receptor ion channels in animal cells (Thériault et al., 2014). Curiously, a later study reported that PLP content is increased in sos4-1 (González et al., 2007), although the biochemical activity of intact SOS4 is phosphorylation of PL (Lum et al., 2002). More recently, an allele of sos4-1 was isolated in a screen for nitric oxide (NO) hypersensitivity based on impaired growth in the presence of the NO donor, sodium nitroprusside (SNP), and was annotated as sensitive to nitric oxide1 (sno1) (Xia et al., 2014). Intriguingly, an elevated PLP content in sno1 was also reported using the same methodology as for sos4-1 (Xia et al., 2014). However, little is known about the contribution of SOS4 to the PLP pool in the absence of abiotic stress and its role under these conditions is imperative to understand stress sensitivity.
Here, we show that SOS4 is important for plant development and health, independent of the salt-sensitive phenotype. Our data demonstrate that sos4-1 and more recently isolated alleles are deficient in phosphorylated B6 vitamers including PLP, concomitant with an increase in non-phosphorylated vitamers PM and PN but, intriguingly, a deficit in PL. Interestingly, ionomic analyses of plants grown on soil reveal that sos4 mutants are severely perturbed in mineral nutrient homeostasis with a hyperaccumulation of transition metal micronutrients particularly in the root. A deeper analysis of root growth shows that the meristem is impaired in sos4 alleles with consequential defects in general root development and differentiation. This includes enhanced lignification and suberization of the endodermis, although the Casparian strip is intact and functional, as well as accumulation of reactive oxygen species (ROS). Interestingly, micrografting demonstrates that regulation of vitamin B6 contents by SOS4 in shoots is crucial for root development. These defects can be alleviated under very low light conditions and implicate correct balancing of B6 vitamers by SOS4 in regulation of root (and shoot) development under standard laboratory conditions.
Results
SOS4 contributes to shoot and root growth and development
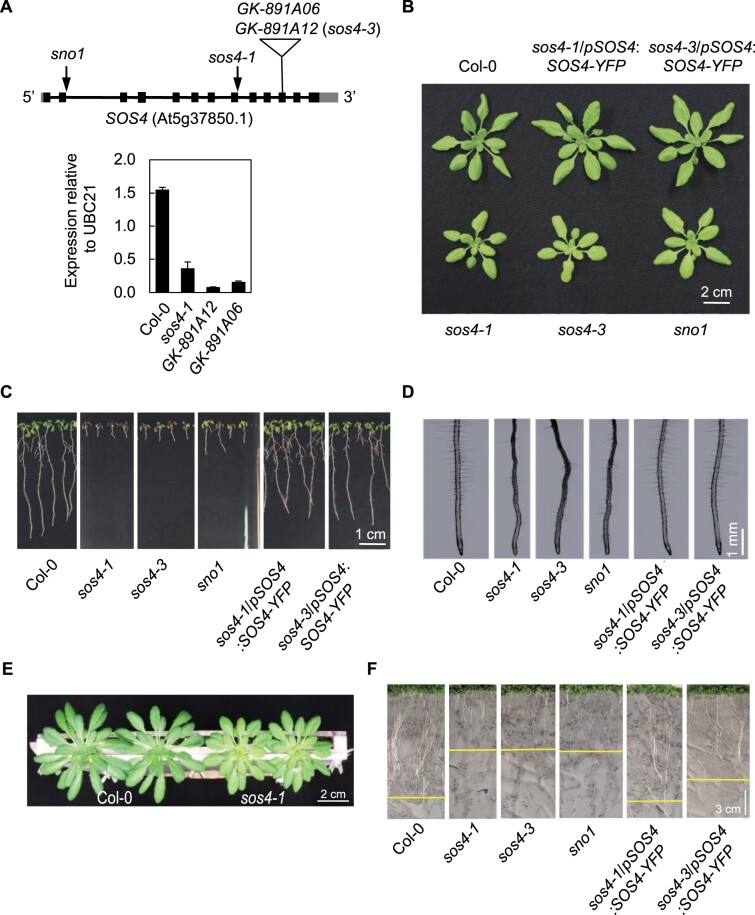
The original EMS mutant allele of the kinase that phosphorylates B6 vitamers (At5g37850) was isolated in a salt hypersensitivity screen and annotated sos4-1 (Shi et al., 2002). sos4-1 seedlings fail to form root hairs, and their overall root growth is reduced considerably (Shi et al., 2002; Shi and Zhu, 2002). A later study also demonstrated shoot growth impairment of sos4-1 under standard laboratory growth conditions (Rueschhoff et al., 2013), although the more recently isolated EMS sno1 allele was reported to behave like wild type under similar conditions (Xia et al., 2014). The majority of studies on sos4-1/sno1 were carried out on seedlings grown on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), which is primarily salt based, and moreover, in the presence of sucrose, which masks photosynthesis impairments that could potentially cause defective shoot growth of the mutants. Therefore, in this work, we initially focused on clarifying the role of SOS4/SNO1 with Arabidopsis grown on soil. We used the previously characterized sos4-1 and sno1 mutants, both of which have G to A nucleotide mutations at acceptor splicing sites, which leads to retention of intron 8 in the case of sos4-1 and intron 2 in the case of sno1 (Figure 1, A), resulting in non-functional truncated proteins (Shi et al., 2002; Xia et al., 2014). To strengthen our observations, we isolated independent T-DNA insertion mutant alleles GK-891A06 and GK-891A12 to homozygosity (Figure 1, A). PCR analysis established insertion of the T-DNA at the same position in exon 11 in both alleles and only GK-891A12 was used for further studies. Analysis of the transcript level of SOS4 expression by reverse transcription quantitative PCR (RT-qPCR) confirmed severely reduced expression (Figure 1, A) and we annotated this line sos4-3.
Figure 1.
SOS4 is important for shoot and root development. A, Upper panel: Scheme of the architecture of SOS4. Arrows indicate position of EMS mutations, the triangle shows the location of the T-DNA insertion. Lower panel: expression analysis of sos4 alleles bearing T-DNA insertions. Data are the mean of three biological replicates ± SD (each comprising ≥20). B, 3-week-old plants grown on soil in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8 h dark (at 18°C) cycles. C, 10-d-old seedlings grown on sterile culture medium under the same conditions as (B). D, Roots of 7-d-old seedlings grown on sterile culture medium under the same conditions as (B). E, 6-week-old plants grown in a rhizobox in 8-h light (120 µmol photons m−2 s−1 at 21°C), 16-h dark (at 18°C) cycles. F, 2-week-old plants in a rhizobox under the same conditions as (B).
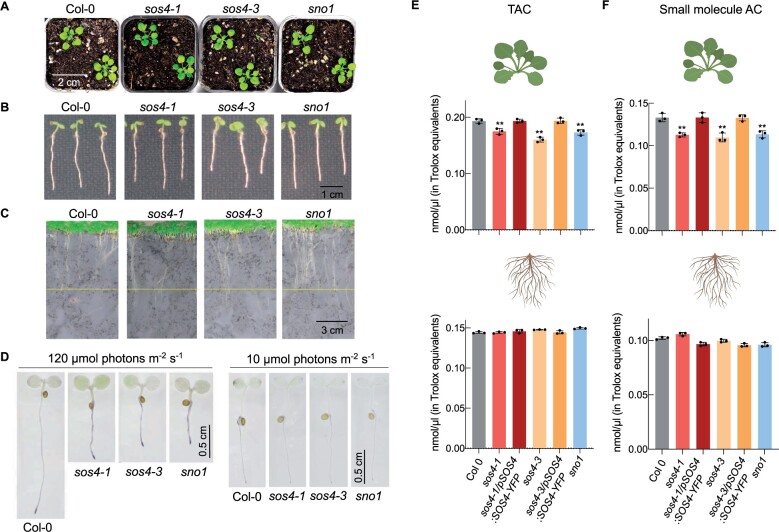
When grown on soil, the aerial parts of sos4-1, sos4-3, and sno1 are stunted in growth and exhibit pale leaf phenotypes (Figure 1, B). When grown in culture on the standard MS medium, all three mutants show reduced root growth with less root hairs, confirming previous reports (Figure 1, C and D). Shoot growth is also impaired under these conditions (Figure 1, C). In order to uncouple sos4/sno1 from their presumptive salt sensitivity, we used so-called rhizoboxes, a vertical soil culture system where roots are separated from soil by a porous nylon membrane, which allows for solute exchange and visualization of this tissue (Durand et al., 2016). Notably, this system allows for growth of plants to maturity, while protecting roots from light exposure. The sos4/sno1 mutant lines grown in this system had chlorotic leaves which is consistent with previously reported shoot defects (Rueschhoff et al., 2013) and were stunted in shoot and root growth compared with wild type plants (Figure 1, E and F). These observations suggest that salt stress is not the primary cause of the growth impairment of sos4 mutants. Importantly, transformation of sos4 mutant lines with a construct carrying the full length SOS4 gene fused to YFP at the C-terminus expressed under the control of its upstream region (pSOS4:SOS4-YFP) rescued growth to that of the wild-type and complemented the morphological phenotype (Figure 1, B, C, D, and F). This confirms that SOS4/SNO1 is responsible for the growth defects observed in the mutant lines. Taken together, these observations suggest that sos4/sno1 plants are impaired in both shoot and root growth independent of salinity stress.
SOS4 is expressed in shoots and roots
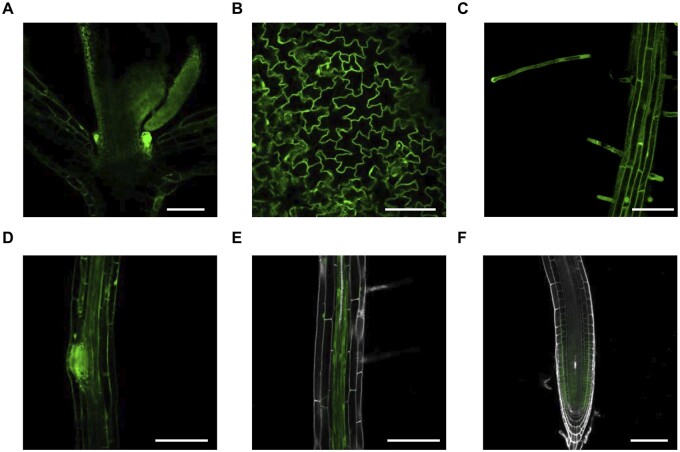
To get insight into the physiological role of SOS4, we studied its tissue expression pattern using the sos4-1/pSOS4:SOS4-YFP lines. As these transgenic lines are morphologically like wild type (Figure 1), the SOS4-YFP fusion can be considered to be functional and thus its expression pattern is biologically relevant. SOS4 was found to be expressed in both shoots and roots of sos4-1/pSOS4:SOS4-YFP seedlings (Figure 2, A–F). In young seedlings, strong expression was observed in the shoot apex in emerging organ primordia (Figure 2, A). Fluorescence was also detected in epithelial pavement cells of cotyledons (Figure 2, B). A strong signal was detected in root hairs (Figure 2, C), in emerging lateral roots (Figure 2, D), as well as in the vasculature (Figure 2, E). SOS4 was also strongly expressed in the root tip, particularly in division and elongation zones (Figure 2, F). These observations match with publicly available datasets (https://www.arabidopsis.org) and previous reports (Shi et al., 2002; Shi and Zhu, 2002). Moreover, the data are in line with the observed growth impairments of all sos4 mutants, corroborating the notion that SOS4 might play an important role in both shoots and roots.
Figure 2.
Expression pattern of SOS4. The expression pattern was visualized using 5-d-old sos4-1/pSOS4:SOS4-YFP lines grown in sterile culture under 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. In each case, the scale bar represents 100 µm and the photo shown is representative of the pattern observed. A, Shoot apex with first pair of true leaves and emerging leaf primordia. B, Cotyledon pavement cells. C, Root epithelial cells and root hairs. D, Emerging lateral roots. E, Root vasculature. F, Meristematic and elongation zones in the root tip.
sos4 alleles have a deficit in phosphorylated vitamers of B6
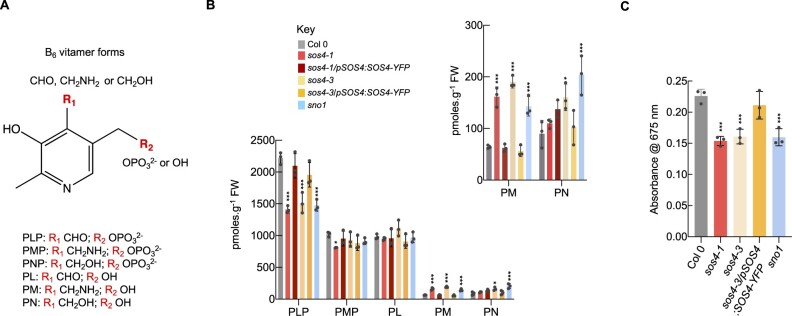
The biochemical activity of SOS4 and its homologs from all domains of life is reported to be phosphorylation of at least one of the B6 vitamers PN, PL, and PM (Ghatge et al., 2021). Therefore, in the absence of SOS4, less of the respective phosphorylated vitamers could be expected. We have previously employed an established HPLC method allowing profiling of B6 vitamers in crops and in lines that are altered either in biosynthesis de novo or the salvage pathway (Li et al., 2015; Colinas et al., 2016; Mangel et al., 2019). This method was used to examine the distribution of B6 vitamers (Figure 3, A) in shoots of the three SOS4 mutant alleles, namely sos4-1, sno1, as well as sos4-3 isolated in this study, from plants grown in pots on soil (see Supplemental Figure S1, A for a profile example). We find that all of these lines mutated in the vitamin B6 kinase SOS4 have less of the phosphorylated vitamer PLP (Figure 3, B). PMP content was not significantly different from that of the wild type among the mutant lines tested, with the exception of sos4-1, which showed a decreased PMP content (Figure 3, B). PNP content was below detection level in all tested genotypes. The mutants also exhibited an altered distribution of non-phosphorylated B6 vitamers. Specifically, there was an increase in the level of PM and to a lesser extent PN (albeit inconsistent) (Figure 3, B), although the PL content was not significantly altered in this shoot material (Figure 3, B). Considering the lowered PLP abundance, the unchanged PL level in sos4 mutants could be considered surprising. However, it has been reported previously that levels of PLP could not be readily associated with changes in the corresponding unphosphorylated vitamer, PL (Huang et al., 2011). In particular, supplementation studies with PL show that it is converted into PM or PN (Huang et al., 2011). This would explain the increases in PM and PN in sos4 mutants, while PL levels remain stable. Importantly, the vitamin B6 profiles are largely restored to that of wild type in morphologically complemented lines (sos4-1/pSOS4:SOS4-YFP and sos4-3/pSOS4:SOS4-YFP), that is the imbalance between unphosphorylated and phosphorylated vitamer levels is restored (Figure 3, B). Taking this data altogether provides very strong evidence that the biochemical activity in vivo of SOS4 is phosphorylation of B6 vitamers. Furthermore, it indicates that SOS4 makes a strong contribution to the PLP pool in plants that is not compensated for by another PLP producing enzyme such as PDX3 of the salvage pathway or the biosynthesis de novo pathway. However, in contrast to our findings it has been reported previously that PLP accumulates in sos4 mutants (González et al., 2007; Xia et al., 2014). Therefore, to verify our findings with an independent method, we assessed the PLP content of shoot material of all sos4 mutants grown on soil using a commercial assay kit (A/C Diagnostics), which employs recombinant homocysteine-α, γ-lyase (rHCYase) that is stripped of its co-enzyme PLP (apoenzyme form) and therefore catalytically inactive. Enzymatic activity is restored by reconstitution of the holoenzyme, through binding of the stripped rHCYase to PLP present in the plant extract. The approach confirmed the lowered PLP level in all analyzed sos4 mutants (Figure 3, C). In addition, we repeated the HPLC protocol that was used in previous studies that showed an increased PLP content in sos4 alleles (González et al., 2007; Xia et al., 2014) and detected a peak at a retention time of 6.4 min that is close to that of PLP (6.0 min) (Supplemental Figure S1, B). The identity of this slower retention peak is not known but it is substantially increased in the sos4 mutant alleles (Supplemental Figure S1, B). If this peak was assumed to correspond to PLP, it may explain the interpretation of increased levels of the PLP vitamer in the assessment of sos4/sno1 mutant alleles in previous studies. Of note also is that the fluorescence intensity of PLP is very low with this method (Supplemental Figure S1, B).
Figure 3.
Vitamin B6 content of shoots of plants grown in soil. A, Chemical structures of different forms of vitamin B6 (vitamers). The R1 and R2 positions vary as indicated. The individual vitamers measured here are pyridoxal 5′-phosphate (PLP), pyridoxamine 5′-phosphate (PMP), pyridoxal (PL), pyridoxamine (PM), pyridoxine (PN). Note pyridoxine 5′-phosphate was not detected in our study. B, Individual B6 vitamer content in shoots of 3-week-old plants grown on soil in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The individual vitamers measured are abbreviated as in (A). To aid visualization, PM and PN contents are shown as an inset on a smaller scale. The key indicates the color coding used for each line. The data are represented as the means of three biological repeats (≥10 plants each; plants were grown in individual pots) ± sd. Asterisks indicate significance by one-way ANOVA test (*P-value < 0.05, ***P-value < 0.001). A separate ANOVA was used for each line in comparison with the wild type. C, PLP-specific enzyme assay of shoots of 4-week-old plants grown on soil in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The data are represented as the means of three biological repeats (≥10 plants each; plants were grown in individual pots) ± sd. Asterisks indicate significance by one-way ANOVA test (***P-value < 0.001). A separate ANOVA was used for each line in comparison with the wild type.
Taken together, this data suggests that SOS4 is responsible for the phosphorylation of B6 vitamers and is an important contributor to the PLP pool in Arabidopsis, as sos4/sno1 mutant lines are deficient in PLP. Moreover, the B6 profiling shows that the lowered PLP content is not accompanied by an increase in PL, but does however coincide with elevated PM and PN.
Ion homeostasis is severely disrupted in sos4 alleles
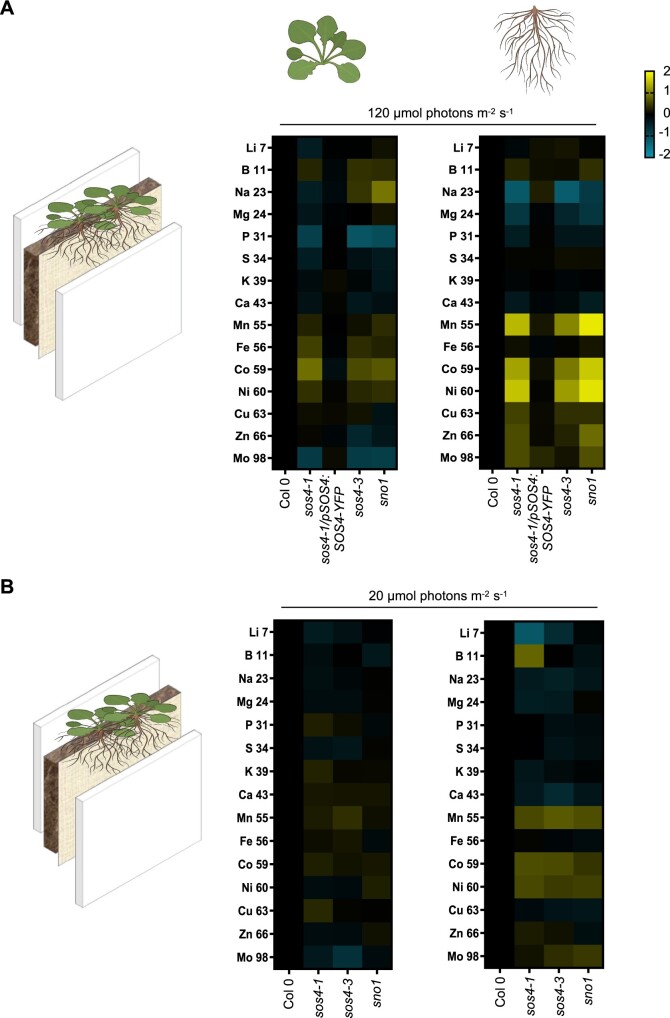
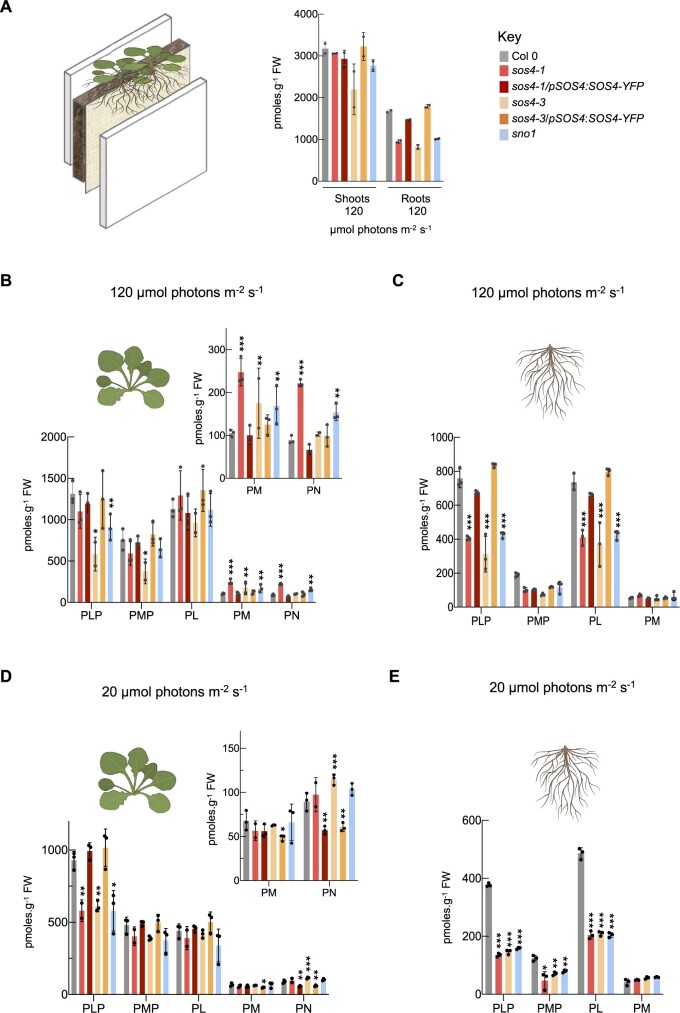
The imbalance in vitamin B6 homeostasis and the previous identification of SOS4 in a salt sensitivity screen prompted us to perform a full ionomic analysis of the mutant lines using Inductively-Coupled Plasma Mass Spectrometry (ICP-MS). In order to facilitate this analysis and to avoid the pleiotropic or indirect effects that may result from the mixed salt content in sterile culture medium, we grew the Arabidopsis lines on soil in rhizoboxes as above. In addition, the rhizobox system allows us to easily harvest both the shoot and root material for downstream molecular analyses. The levels of mineral elements were determined by ICP-MS in both shoots and roots of the three sos4 mutant alleles, the sos4-1/pSOS4:SOS4-YFP complementing line and the corresponding wild type (Supplemental Table S1). We observed a consistent lower level of phosphate and molybdenum in the shoots of the three sos4 mutant lines compared with either wild type or the complemented line sos4-1/pSOS4:SOS4-YFP (Figure 4, A, see also Supplemental Table S1). By contrast, levels of cobalt, iron, nickel, boron, and manganese were slightly elevated in shoots of the sos4 mutant lines (Figure 4, A and Supplemental Table S1). The elevated level of these latter elements in addition to copper was even more pronounced in roots of the sos4 lines, with the exception of iron and boron that were not consistently altered (Figure 4, A and Supplemental Table S1). Similar to the shoots, phosphate levels were slightly lower in roots of the mutant lines, but by contrast to shoots, molybdenum levels were not consistently changed (Figure 4, A and Supplemental Table S1). Sodium and magnesium levels were consistently lower in roots of the mutant lines but not in shoots, and potassium levels were not altered in any tissue or line compared with wild type or the complemented line (Figure 4, A and Supplemental Table S1). All of these ionic perturbations were restored to wild type levels in the sos4-1/pSOS4:SOS4-YFP complemented line (Figure 4, A and Supplemental Table S1). Therefore, disruption of SOS4 is clearly implicated in the mineral element perturbance. We propose that the accumulation of highly reactive transition metals in sos4/sno1 mutants contributes to the impaired root growth independent of salt stress.
Figure 4.
Ionomic analysis of sos4 and wild type grown in rhizoboxes under standard and low light conditions. On the left side in (A) and (B) is a scheme of the rhizobox system used. The soil and a membrane are layered between two perspex plates. Seeds are sown on the top at the membrane side and do not touch the soil. A, Ionome profile of tissues of 2-week-old plants grown in rhizoboxes in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles as measured by ICP-MS. The representation of the data is log2 (fold-change)| > 0.5. The color shades in the key indicate the extent of change of the particular element relative to wild type. B, As in (A) but from plants grown under a light intensity of 20 µmol photons m−2 s−1. The data are representative of three biological replicates (each biological repeat comprised ≥20 plants collected from three rhizoboxes).
Root morphology and differentiation is impaired in sos4 coincident with ectopic lignification and suberization
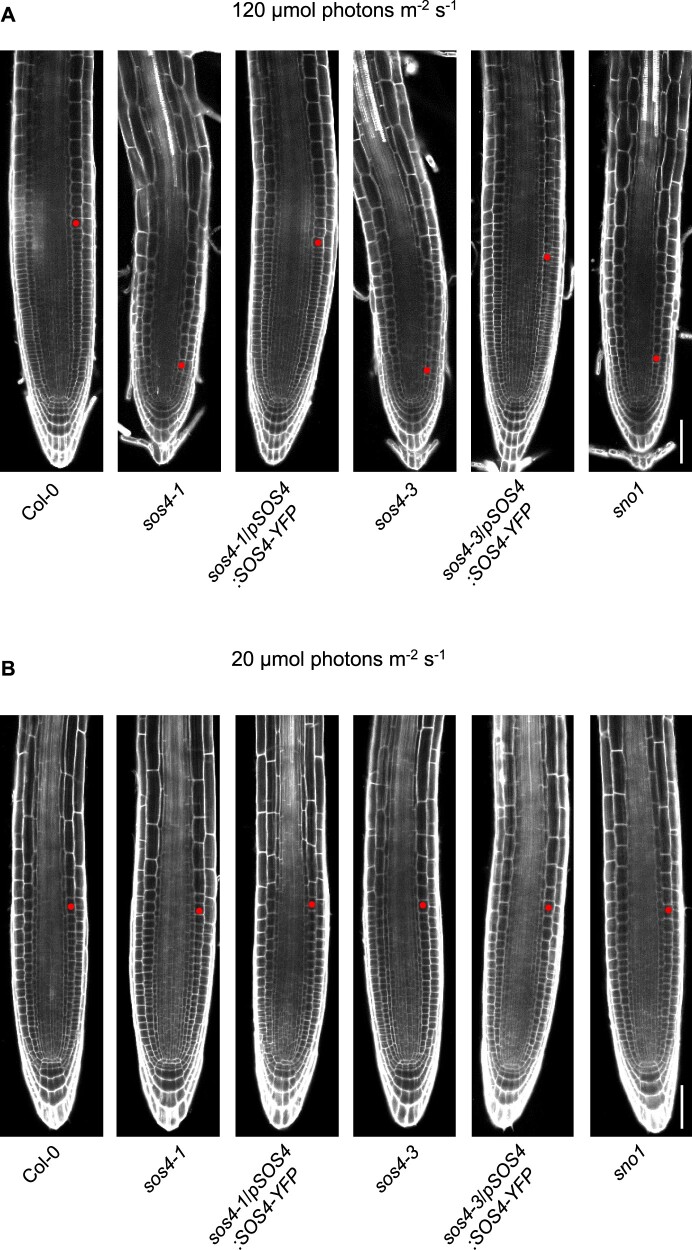
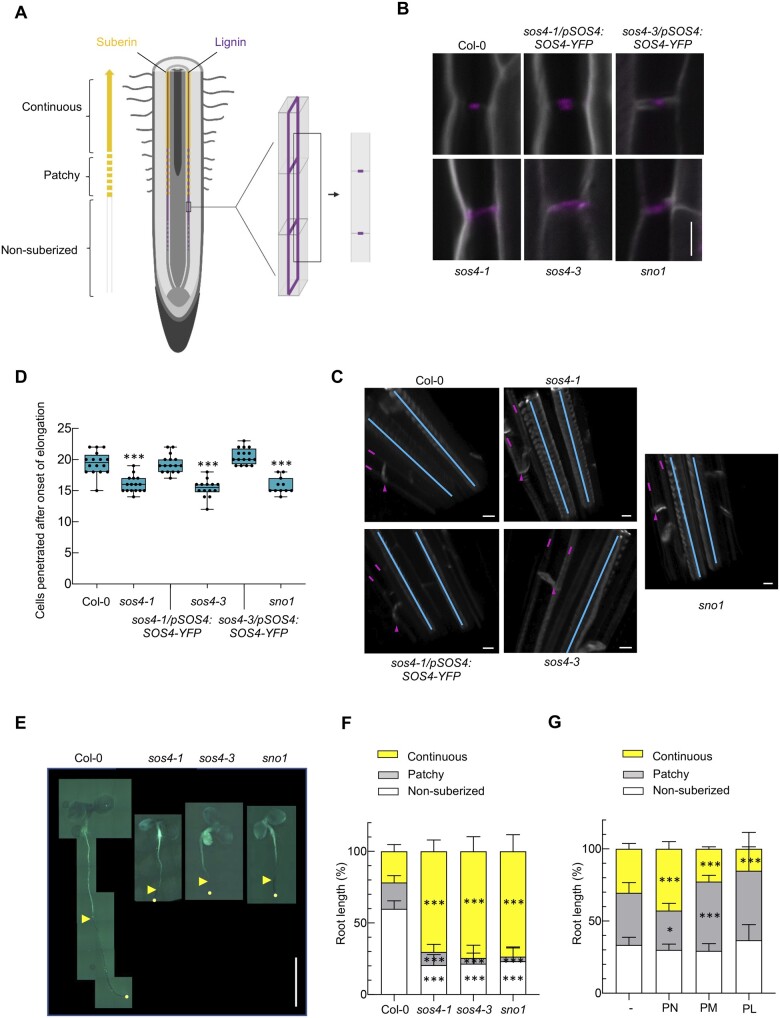
At the macroscopic level, the primary root growth defects (stunted growth, lack of root hairs) are strong in sos4 alleles (Figure 1, C, D, and F). To provide more insight at the microscopic scale and in an attempt to diagnose the underlying issues with sos4, we examined root tip morphology. In sos4, meristem size as defined by the number of cortex cells in a file extending from the quiescent center to the first elongated cell (length double the width in the median, longitudinal section) is severely reduced (Figure 5, A). These defects are completely abolished in lines carrying the sos4/pSOS4:SOS4-YFP transgene (Figure 5, A). The endodermis that surrounds the vasculature is an important bidirectional barrier that manages the flow of nutrients into the stele, as well as preventing their backflow (Barberon, 2017). This is achieved by impregnating cell walls with lignin (giving rise to the Casparian strips) and adding a layer of suberin (Doblas et al., 2017). Moreover, it has previously been reported that disturbance of ion homeostasis through external application of a combination of iron, zinc, and manganese increases suberization (Barberon et al., 2016). Thus, we examined lignin and suberin deposition in sos4 alleles. First, when viewed from the endodermal cell surface (longitudinal view, z-axis) lignin deposition is normally observed as a continuous strip (Figure 6, A), whereas a central patch is seen in the transverse view, corresponding to the Casparian strip band, and commences within the root differentiation zone (Figure 6, A). Both the transverse and longitudinal view of the root endodermis stained with Calcofluor White (cell wall) and Basic Fuchsin (lignin) demonstrated ectopic lignin deposition in sos4 alleles compared with wild type (Figure 6, B and C). Ectopic lignin deposition was not observed in either the sos4-1/pSOS4:SOS4-YFP or sos4-3/pSOS4:SOS4-YFP complemented lines (Figure 6, B and C). Notably, the ectopic deposition of lignin in the sos4 alleles was observed in both periclinal and transverse walls of the endodermal cells (analogous to a 3D brick, Figure 6, A and B). To assess for Casparian strip barrier functionality we scored the number of endodermal cells penetrated by the fluorescent cell wall dye propidium iodide (PI) after the onset of cell elongation (Fujita et al., 2020). Indeed, the dye could not penetrate into the stele after a number of endodermal cells close to that of the wild type or complemented lines, indicating that the Casparian strip is functional in sos4 (Figure 6, D). Suberin was examined using the specific stain Fluorol Yellow (Franke et al., 2005; Naseer et al., 2012) following the protocol described in Shukla et al. (2021). Two zones are distinguished in the endodermis suberization region that begins in the more mature root area after the onset of lignification—the patchy zone where individual cells are suberized in a random fashion and the zone of continuous suberization, where the entire endodermal cell layer is homogenously coated by suberin (Figure 6, A). Quantification of suberization through scoring of suberized cells within the two zones revealed an enhancement of suberization in the sos4 alleles compared with wild type (Figure 6, E and F). The zone of continuous suberization was extended and total suberization suffused almost 80% of the entire length of the root in sos4 alleles, whereas it is approximately 20% in wild type under these conditions (Figure 6, F). To relate our findings on suberin to vitamin B6, we treated wild type by external application of PN, PM, or PL. In the presence of PN suberization was enhanced, whereas it was decreased by PL (Figure 6, G). Thus, the imbalance in B6 vitamers (increase in PN in shoots that is possibly transported to roots, decrease in PL in roots (see below)) may account for the altered suberization (and lignification) in sos4. Taken together, our data show that root morphology and differentiation is impaired in sos4 coincident with ectopic lignification and suberization.
Figure 5.
Root tip morphology of sos4 and wild type under standard and low light conditions. A, PI staining of the root tip zone of 3-d-old seedlings grown on sterile culture medium in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The images are aligned to the position of the quiescent center. The scale bar represents 50 µm. The red dot marks the first cortical cell of the elongation zone defining the end of the meristem region. B, As in (A) but from seedlings grown under a light intensity of 20 µmol photons m−2 s−1. Each image is representative of n ≥ 20.
Figure 6.
Lignification and suberization in roots of sos4 compared with wild type. A, Schematic representation of the root showing differentiation, lignification, and suberization zones. Magenta represent Casparian strips, yellow lines are suberin lamellae. Schematics are indicating the position of optical sections in a 3D illustration (middle) and in a median illustration (right). B, Lignin accumulation patterns at median positions. Lignin and cellulosic cell walls of 3-d-old seedlings grown in sterile culture medium in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles are stained with Basic Fuchsin and Calcofluor White, shown in magenta and white, respectively. Images are representative of at least eight analyzed seedlings. The scale bar represents 5 µm. C, 3D views showing xylem poles (blue lines) and endodermal cells (magenta lines and arrows indicate longitudinal and transverse deposition of lignin, respectively). The scale bar represents 5 µm. D, PI penetration assay. Scoring of number of cells after the onset of cell elongation until the PI signal is excluded from the inner side of the endodermis. The number of endodermal cells was scored from the onset of cell elongation (defined as endodermal cell length being more than two times the width in the median, longitudinal section) until PI could stain the inner endodermal cell wall. For each genotype, at least 11 roots of 3-d-old seedlings grown in sterile culture medium in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles were tested and showed similar results in two independent experiments. The data represent the mean ± sd (N ≥ 11). Asterisks indicate significance by one-way ANOVA tests for P-values < 0.001. A separate ANOVA was used for each line in comparison with the wild type. Data are presented as a box plot, where the line indicates the median, the box indicates the first and third quartiles, and the whiskers indicate ± 1.5 × interquartile range, outliers are not indicated on the plot. E, Suberization pattern of 3-d-old seedlings grown in sterile culture medium in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The scale bar represents 0.5 cm. Images were digitally extracted for comparison; adjustments to brightness, contrast, or color balance were made uniformly across the entire image. F, Quantification of (E). The data represent the mean ± sd (N ≥ 17). The asterisks indicate significance by one-way ANOVA tests for P values < 0.001. A separate ANOVA was used for each line in comparison with the wild type. G, Quantification of the suberization pattern of 3-d-old wild type seedlings grown in sterile culture medium in the absence or presence of either pyridoxine (PN), pyridoxamine (PM), or pyridoxal (PL) (200 µM in each case). The data represent the mean ± sd of three biological replicates (each comprising ≥8). The asterisks indicate significance by one-way ANOVA tests (*P-value < 0.05, ***P-value < 0.001). A separate ANOVA was used for each line in comparison with the wild type.
An imbalance in shoot vitamin B6 homeostasis impairs root growth in A. thaliana
Given the strong root developmental defects in sos4 mutants and pronounced ionome alterations in the root, we next addressed shoot versus root perturbance in vitamin B6 homeostasis of rhizobox grown plants, from which we could get both tissues thus facilitating these experiments. First, we observed that the overall vitamin B6 content was considerably lower in roots than in shoots of all plant lines (Figure 7, A). At the level of individual vitamers in shoots, the phosphorylated vitamer levels are generally decreased (PLP, PMP) in sos4 lines compared with wild type, whereas the unphosphorylated vitamers PM and PN increase but PL contents are not significantly different from wild type (Figure 7, B). This is consistent with the observed B6 profile of shoots of plants grown in pots on soil (Figure 3, B). In roots, there was a consistent decrease in PLP in the sos4 lines, while PMP tended to be decreased in all transgenic lines but was not significantly different from wild type (Figure 7, C). In stark contrast to the shoots, in roots of sos4, PL was significantly decreased, whereas there was no significant change in PM (Figure 7, C) and PN could not be detected. Notably, the deficit in PLP is much more pronounced in the root (coincident with the decrease in PL) than in the shoot, suggesting that basipetal transport of PLP could be impaired in sos4 plants. Importantly, the imbalance in individual B6 vitamers in both shoots and roots was restored in the complemented lines (Figure 7, C).
Figure 7.
HPLC profiling of vitamin B6 content of plants grown in rhizoboxes. A, Left side, a scheme of the rhizobox system used. The soil and a membrane are layered between two perspex plates. Seeds are sown on the top at the membrane side and do not touch the soil. Right side, total vitamin B6 content of shoots and roots of 2-week-old plants grown in rhizoboxes in 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The key indicates the color coding used for each line. B, Individual B6 vitamer content in shoots of plants grown as in (A). The individual vitamers are pyridoxal 5′-phosphate (PLP), pyridoxamine 5′-phosphate (PMP), pyridoxal (PL), pyridoxamine (PM), and pyridoxine (PN). To aid visualization PM and PN contents are shown as an inset on a smaller scale. C, Individual B6 vitamer content in roots of plants grown as in (A). Vitamers as in (B). D, Individual B6 vitamer content in shoots of 2-week-old plants grown in rhizoboxes in 16-h light (20 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. Vitamers and inset as in (B). E, Individual B6 vitamer content in roots of plants grown as in (D). Vitamers as in (B). The data represent the mean ± sd of three biological replicates (each comprising ≥20 plants collected from three rhizoboxes). Asterisks indicate significance by one-way ANOVA test (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001). A separate ANOVA was used for each line in comparison with the wild type.
It now became important to be aware of previous studies showing that inclusion of non-phosphorylated B6 vitamers in culture plates only partially rescues root growth of sos4, if at all (Shi and Zhu, 2002). The partial rescue is facilitated by PN, whereas PL does not alleviate the phenotype. Here, we saw no restoration of root growth in sos4 alleles by external application of PL (or PN) (Supplemental Figure S2). This is different than biosynthesis de novo mutants such as pdx1.3, the mild root growth impairment of which is rescued by external application of PN or PL under similar conditions (Titiz et al., 2006 and Supplemental Figure S2), presumably due to the action of the salvage pathways. This strongly suggested that the principal defect in sos4 roots is the deficit in PLP. However, direct external application of PLP cannot be used to compensate for the deficit, as phosphorylated vitamers do not appear to be taken up and have not been reported to rescue root growth defects (Shi and Zhu, 2002). Therefore, to probe the contribution of the imbalance in B6 vitamers to the respective shoot and root phenotypes observed in sos4 alleles, we next performed a series of micrografting experiments. Micrografting of a wild type scion onto rootstocks of the sos4 alleles rescued the root growth impairments observed in the mutants (Figure 8, A and B and Supplemental Figure S3). On the other hand, micrografting of sos4 scions onto wild type root impaired wild type root growth—stunted root length and diminished root hairs (Figure 8, A and B and Supplemental Figure S3). Notably, self-grafting of sos4 scions and rootstocks demonstrated the same short root phenotype as seen with whole intact seedlings, as well as a visibly lower density of root hairs (Figure 8, A and B and Supplemental Figure S3). This implies that irregularities in the shoot of sos4 mutants are the predominant cause of the whole seedling phenotypes. Based on the observations in rhizobox grown plants, the growth impairment could be caused either by the deficit in PLP/PMP or the build-up of non-phosphorylated vitamers PN/PM in the shoot. To facilitate vitamin B6 profiling of the micrografted plants, they were transplanted onto soil. Interestingly, mutant rootstocks did not impair growth of wild type scions (Figure 8, C and Supplemental Figure S3). However, the sos4 scions grafted with wild type rootstocks were stunted in growth compared with self-grafted wild type or even wild type scions grafted to sos4 rootstock (Figure 8, C). Although growth of sos4 scions grafted to the wild type rootstocks was less impaired than that of self-grafted sos4, indicating partial rescue of the sos4 shoot phenotype by wild type rootstock (Figure 8, A and C). Vitamin B6 profiling of the aerial parts of the transplanted micrografts revealed that the B6 vitamer balance of the wild type scions was not disturbed by grafting with sos4 rootstocks (Figure 8, D and Supplemental Figure S3). In addition, grafting of the wild type rootstock could partially rescue the B6 imbalance in sos4 scions (Figure 8, D and Supplemental Figure S3).
Figure 8.
Reciprocal micrografting of sos4 lines and wild type. A, Elongation of micrografted roots at 7 d after grafting. The data are the mean ± sd of three biological replicates (each comprising ≥ 12). The scion and rootstock used are as indicated below. B, Microscope pictures of roots of 7-d-old micrografts grown on sterile culture medium under 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The scion and rootstock used are as indicated in (A). C, Photographs of micrografted seedlings from (B) 14 d after transfer to soil and grown under 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The scion and rootstock used are as indicated in (A). D, Individual B6 vitamer content in the shoots of plants grown in (C). The individual vitamers are pyridoxal 5′-phosphate (PLP), pyridoxamine 5′-phosphate (PMP), pyridoxal (PL), pyridoxamine (PM), and pyridoxine (PN). The key indicates the color coding used for each micrografted line. The data are represented as the mean ± sd of three biological replicates (each comprising ≥ 15 plants grown in individual pots). Asterisks indicate significance by one-way ANOVA test (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001). A separate ANOVA was used for each line in comparison with the wild type.
Taken together, our data imply that a shoot derived B6 vitamer imbalance impairs both shoot and root growth in sos4 mutant alleles. The data also reveal distinct functions of SOS4 in the maintenance of homeostasis of B6 vitamers between the shoot and the root. In particular, SOS4 is clearly implicated in homeostasis of PLP/PMP/PM/PN levels in shoots, while its absence in roots results in a deficit of PLP and PL that is likely derived from the imbalance in B6 vitamers of the shoot.
Growth in very low light bypasses the need for SOS4
To clarify the biological function of SOS4 further, it is important to know that vitamin B6 metabolism responds to light, and moreover, there have been several demonstrations of potent antioxidant activity of B6 vitamers in vitro (Bilski et al., 2000; Havaux et al., 2009; Asensi-Fabado and Munné-Bosch, 2010). To put this into context with the experiments here, we profiled the B6 vitamers from shoot and root material of rhizobox grown lines under very low light conditions (less accumulation of ROS) to compare to the standard conditions used (20 and 120 µmol photons m−2 s−1, respectively; Figure 7, D and E). First, the level of B6 vitamers was generally decreased in the lower light condition in comparison with that in the standard light intensity regime for all analyzed genotypes (Figure 7, D and E). Among the phosphorylated B6 vitamers measured, there was still a significant deficit in PLP and PMP content in sos4 alleles in both shoots and roots at the lower light intensity (Figure 7, D and E). Among the non-phosphorylated vitamers of shoots, the level of PM and PL was not significantly different to wild type, although PN levels were still slightly elevated (Figure 7, D and E). In roots, there was still a considerable deficit in PL levels, PM levels were unchanged, and PN could not be detected (Figure 7, E). We then hypothesized that although SOS4 is needed to regulate vitamin B6 levels it may be more important under standard light conditions, which if true then its requirement should be bypassed under the very low light conditions. Indeed, the phenotype of sos4 mutants grown either in pots on soil (Figure 9, A), or in sterile culture (Figure 9, B), or in rhizoboxes (Figure 9, C) is comparable to wild type upon growth under the lower light intensity. We did a qualitative analysis of superoxide levels (as a representative ROS) using nitroblue tetrazolium (NBT) in the sos4 alleles versus wild type under both 120 µmol photons m−2 s−1 (standard light intensity used in this study) and 20 µmol photons m−2 s−1 light intensities. Under the higher light intensity, the staining was much stronger in the sos4 alleles compared with wild type, indicating higher accumulation of superoxide in the mutant lines (Figure 9, D). On the other hand, staining intensity was equivalent across all lines under the lower light intensity (Figure 9, D). We then measured total antioxidant capacity (TAC) to generally assess ROS neutralizing power of shoots and roots of seedlings grown under the standard light regime. The analysis revealed that TAC was decreased in the shoots of sos4 alleles, while it was not altered in the roots of the mutants (Figure 9, E). B6 vitamers could contribute to cellular reducing power either by directly neutralizing ROS or through the action of PLP as a coenzyme supporting either ROS detoxifying enzymes or the generation of antioxidant chemical derivatives. To discriminate between a direct enzymatic or small molecule role, a protein mask was included in the TAC assay that prevents enzymatic Cu2+ reduction, thereby revealing the contribution of small molecule antioxidants to the total cellular reducing power. This analysis demonstrated a clear reduction in antioxidant capacity in shoots of sos4 alleles, suggesting a considerable contribution of small molecule antioxidants to the total reducing capacity (Figure 9, F).
Figure 9.
Growth of plants under low light. A, Photographs of 6-week-old plants grown on soil under 16-h light (20 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. B, Photographs of 5-d-old seedlings grown on sterile culture medium under 16-h light (20 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. C, Photographs of 2-week-old plants grown in a rhizobox under 16-h light (20 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. D, Microscope pictures of NBT staining for superoxide of 3-d-old seedlings grown on sterile culture medium under 16-h light (either 120 or 10 µmol photons m−2 s−1 as indicated and at 21°C), 8-h dark (at 18°C) cycles. E, TAC of shoots (upper panel) or roots (lower panel) of 7-d-old seedlings grown on sterile culture medium 16-h light (120 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. The data are the mean of three biological repeats ± sd (each comprising at least 50 seedlings). Asterisks indicate significance by one-way ANOVA test (**P-value < 0.01). A separate ANOVA was used for each line in comparison with the wild type. F, As in (E) but with the use of a protein mask to determine small molecule antioxidant capacity (AC).
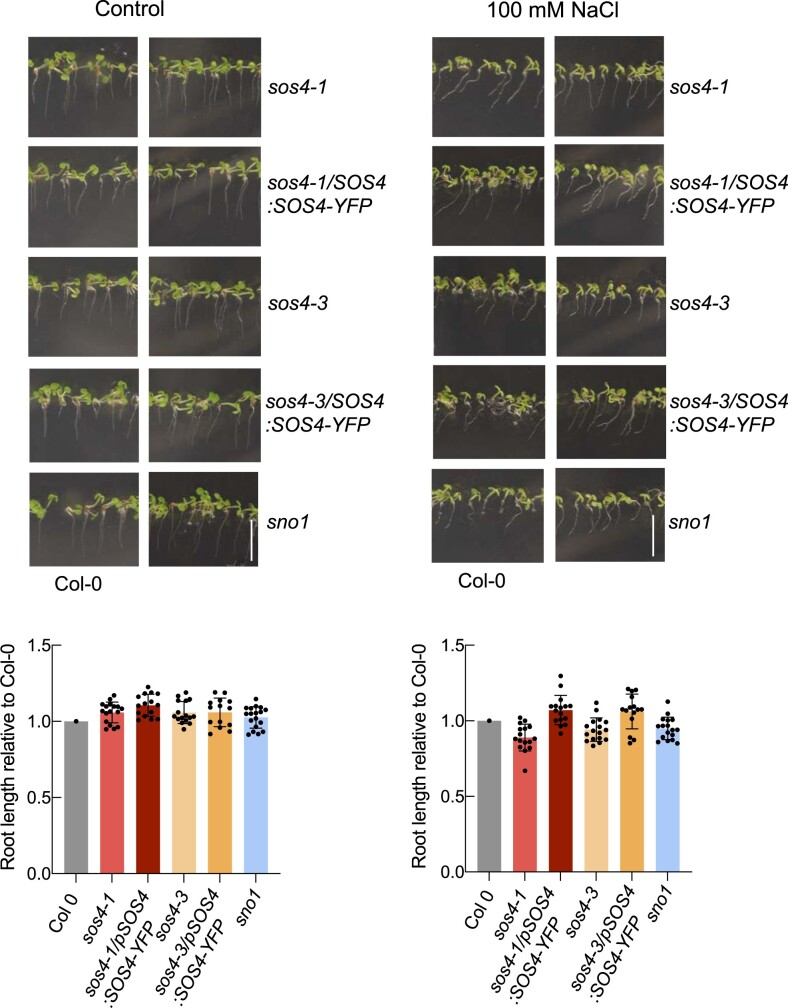
Next, given that growth of sos4 is comparable to wild type under very low light, we were prompted to analyze the ionome of these lines under such conditions. This analysis revealed that the nutrient perturbations seen under the standard light condition were alleviated in both shoots and roots under the lower light regime (compare Figure 4, A and B). In addition, we examined root tip morphology and observed that the meristem was indistinguishable from that of wild type under very low light conditions (Figure 5, B). Furthermore, as stress conditions (such as salinity) are associated with increased ROS production, we hypothesized that sos4 alleles might show elevated sensitivity to such stress due to their compromised reducing capacity under standard light conditions. To address this hypothesis, we assessed salt sensitivity of sos4 alleles by exposure to sodium chloride under the lower light conditions and found that they did not differ in their response from wild type seedlings (Figure 10).
Figure 10.
Sensitivity of sos4 mutants to salt upon growth in low light. Upper panels: Photographic images of 7-d-old seedlings grown on sterile culture medium in the absence (Control) or presence of 100 mM NaCl under 16-h light (20 µmol photons m−2 s−1 at 21°C), 8-h dark (at 18°C) cycles. Representative images of mutant and complemented lines are paired with images of Col-0 (wild type) grown on the same plate. The scale bar represents 1 cm. Lower panel: root lengths of mutant and complemented lines normalized to the mean of the root length of Col-0 growth on the same plate. The data are mean lengths ± sd of at least 15 seedlings.
Therefore, growth under very low light effectively dissipates all of the morphological and developmental defects associated with the absence of SOS4 presumably because of less oxidative load under these conditions and thereby less requirement for reducing antioxidant capacity. Thus, SOS4 is required under increasing light intensities, where its absence renders plants more susceptible to abiotic stress.
Discussion
PLP is a vital coenzyme to several life processes and is predominantly associated with transaminases that facilitate amino acid biosynthesis. There are several pathways to PLP biosynthesis comprising both de novo and salvage pathways. However, the latter have not been well characterized particularly in plants and little is known about their overall contribution to the PLP pool. Here we studied SOS4, first identified in a salt sensitivity screen in Arabidopsis almost two decades ago (Shi et al., 2002). SOS4 is a homolog of the well characterized pyridoxal kinases in other domains of life including bacteria and humans that phosphorylates non-phosphorylated B6 vitamers (notably PL but also PN and PM; Newman et al., 2006; Gonzalez-Ordenes et al., 2021). We focused our study on facets of SOS4 in the absence of salt stress and observed that sos4 mutant alleles are developmentally impaired in both shoot and root growth. The use of several methods establishes that the PLP content is reduced in both tissues as could be expected for the biochemical activity of SOS4, and likely accounts for the compromised growth and development that renders sos4 mutants unhealthy. We propose that such compromised plant fitness leaves sos4 mutants susceptible to stress and could explain its discovery in screens related to abiotic stress.
Intriguing to this study and in contrast to other PLP biosynthesis mutants such as those of the de novo pathway, developmental impairments in sos4 alleles are not rescued by supplementation with PL or PN. While rescue of biosynthesis de novo mutants by such supplementation can be explained by the compensatory kinase activity of SOS4, it emphasizes the important role of SOS4 in contributing to the PLP pool. Moreover, the PLP deficiency in sos4 alleles suggests that lack of SOS4 is not compensated for by another PLP biosynthesis pathway and that SOS4 has a unique contribution to the PLP pool. This is exemplified by the distinct morphological phenotypes of sos4 alleles not seen in other PLP biosynthesis mutants most notable of which is the strong root growth impairment. The strong impairment in root meristem size in particular may be ultimately accountable for the plethora of growth and development defects in root tissue. Here, we consider that PLP deficiency is the underlying problem. On the one hand, PLP is renowned for its coenzyme function and is necessary for the action of enzymes involved in phytohormone biosynthesis (Boycheva et al., 2015), notably auxin and ethylene both of which are essential for proper root development (Liu et al., 2017). On the other hand, PLP and the other B6 vitamers are notable antioxidants (Czégény et al., 2019) and as such the deficit of PLP and imbalance in other vitamers in roots may account for the enhancement of ROS (i.e. superoxide as measured here) levels in this tissue in sos4 alleles. The increased ROS in sos4 alleles may also be derived for the substantial increase in transition metals particularly in the root, as they can generate hydroxyl radicals through the Fenton or Haber–Weiss reactions (Merchant, 2010). Incidentally, the coincident enhancement of many of these metals in the shoot implies that the endodermal barriers are crossed. In this context, the plant root is a highly selective filter often likened to an inverted gut (Waisel and Eshel, 2002; Pfister et al., 2014; Ramakrishna and Barberon, 2019). Nutrient selectivity in roots is achieved through epithelial-like characteristics both at the outer epidermis which controls acquisition at this layer and the diffusion barriers at the endodermis which surrounds the vascular system. Radial passage through the root is achieved through cell-to-cell pathways comprising the symplastic and transmembrane routes, as well as passage through the intercellular space and cell walls that defines the apoplastic route. The endodermal diffusion barrier of the root is comprised of the lignin infused net-like structure of the Casparian strips and suberin lamellae that surround the endodermal surface. Areas where both are in place together force symplastic passage of nutrients and water from outer cell layers. Furthermore, and equally important, the endodermal barriers stop nutrients from leaking back out of the plant. In the case of sos4, the nutrient increases observed (notably for Mn, Co, Ni, Cu, Zn, Mo) may be due to overstimulation of uptake via symplastic or transcellular pathways. Yet, the block in the penetration of PI into the vasculature indicates that the Casparian strip is functional and thus that the apoplastic barrier is intact. With this in mind, the ectopic lignification and enhanced suberization at the endodermis observed in sos4 likely poses an additional problem and suggest inability to flush out elements that reach potentially toxic concentrations. Suberization in particular is hypothesized to have a strong impact not only on nutrient uptake but also nutrient efflux through non-symplastic barriers i.e. it is a bidirectional barrier that provides a checkpoint for nutrients (Barberon, 2017). Previous studies have demonstrated that enhanced suberin deposition is mediated by the antagonistic action of abscisic acid (ABA) (a positive regulator) and ethylene (a negative regulator) (Barberon et al., 2016). Although enzymes involved in ABA metabolism do not appear to require PLP as a coenzyme, ethylene biosynthesis does, as mentioned above, because the key regulatory protein 1-aminocyclopropane-1-carboxylate (ACC) synthase is a PLP dependent enzyme. Thus, the deficit of PLP in roots of sos4 alleles may in turn led to a reduction in ethylene biosynthesis or even ACC itself permitting excessive suberization and should be investigated, along with ABA content, in future studies. With regards to lignification, it has recently been shown that ROS production is localized to allow for spatially restricted lignification via the Schengen pathway, that is the Casparian strip (Fujita et al., 2020). This appears to be intact in sos4 as mentioned above. However, the enhanced ROS may trigger ectopic radical coupling of mono-lignols through the action of peroxidases and laccases independent of the precisely localized Schengen module.
While root development is severely impaired in sos4 alleles, we demonstrated two ways of rescuing these defects in this study. One is through micrografting of a wild type scion onto a sos4 root. This strongly suggests that SOS4 action in the shoot is vital for proper root development. The requirement for SOS4 in the shoot may be either to provide PLP or support the production of a molecular entity that is required for proper root development. On the one hand, the PLP made by SOS4 in the shoot may be transported to the root to support needs in this tissue—perhaps by SOS4 itself which could be corroborated by expression of SOS4 in the vascular system (Figure 2, E). The function of PLP as provided by SOS4 (in shoots or roots) may be in its role as coenzyme for enzymes involved in phytohormone biosynthesis and/or enzymes involved in scavenging ROS. Interestingly, the TAC assay reveals that ROS neutralizing power is compromised in the shoots of sos4 seedlings and implies a role in cellular reducing power. We could not detect changes in whole roots using this assay but this does not rule out that reducing power may be compromised in particular root tissues. Nonetheless, the contribution of SOS4 to reducing power could be provided through support of ROS detoxifying enzymes such as those involved in biosynthesis of cysteine or glutathione, potent cellular antioxidants, or in the manufacture of coenzymes for antioxidant enzymes, for example haem for associated peroxidases (Czégény et al., 2019). Alternatively, the B6 vitamers may be directly acting as antioxidants, as shown in earlier studies in vitro (Bilski et al., 2000; Czégény et al., 2019) and SOS4 is required to contribute to their maintenance. That the decrease in cellular reducing power is similar when we compared total and small molecule antioxidant capacity alone suggests that SOS4 contributes to small molecule antioxidant capacity. Here, we cannot deduce if that capacity is derived from B6 vitamers directly or the biosynthesis of small molecule antioxidants derived from the use of PLP as a coenzyme. Future work could strive to dissect these contributions. The second way of rescuing root development is by growth under very low light intensity and provides equally plausible hypotheses for the physiological function of SOS4. Under very low light, the deficit in PLP contents was still observed in sos4 alleles but they conform morphologically to wild type behavior and suggests that shoot and root development under these conditions is independent of SOS4. As there is less accumulation of ROS under the lower light intensity and therefore less demand for dissipating capacity, the requirement for SOS4 derived PLP might be bypassed under these conditions and development is not impaired. This suggests that the deficit in PLP is less important under very low light intensity and points to an important role either as antioxidant or its integration into ROS scavenging networks. Furthermore, light promotes PLP biosynthesis as deduced from the higher content under the higher light intensity (Figure 7) and would therefore favor the shoots as a source of PLP for the root as a sink tissue under such conditions.
Our finding that PLP is decreased in sos4 alleles contrasts with previous studies on sos4-1 and sno1 (González et al., 2007; Xia et al., 2014). In our hands, the detection of a peak close to the retention time of PLP using the previous methodology that is substantially increased in sos4 alleles may account for the earlier interpretations. While PLP content is altered in sos4 alleles, the imbalance in other B6 vitamers is also noteworthy. Particularly striking is the strong decrease in PL levels specifically in the root. Thus, SOS4 could also be seemingly implicated in PL maintenance in the root, which is either derived from phosphatase action on PLP transported from the shoot or SOS4 is associated with PL transport from the shoot. The deficit in root PL may be compounded by its conversion to PM and PN in the shoot, in the absence of SOS4, and is corroborated by the buildup of PM and PN in shoots. However, earlier studies that are corroborated here show that external application of PL does not rescue root growth of sos4-1 (Shi and Zhu, 2002; Supplemental Figure S2); therefore, it does not appear likely that the deficit in PL accounts for the root phenotype, rather it is more likely that the deficit in PLP is the predominant problem with sos4 alleles.
In summary, we conclude that SOS4 is important for vitamin B6 management in shoots and roots required for appropriate development. That SOS4 is directly accountable for morphological and chemical phenotypes reported here is substantiated by their reversion in the presence of the SOS4 transgene. The contribution of SOS4 to the PLP pool, and in particular, maintenance of root PLP levels from the shoot has an important role to play in root development. Its absence under higher light intensities has a detrimental effect on the root apical meristem, which in turn affects root development and differentiation.
Materials and methods
General plant material and growth conditions
Arabidopsis (A. thaliana) (Columbia ecotype) was used from an in-house stock. The mutant lines sos4-1 (N24930 in Col (gl1) background), GK-891A06 (N485446) and GK-891A12 (N485452, sos4-3) were obtained from the European Arabidopsis Stock Center (http://arabidopsis.info). The sos4-1 EMS mutant allele was backcrossed and segregated to remove the gl1 mutation. The sno1 EMS mutant allele was a kind gift from Yikun He (Capital Normal University, Beijing, China). Plant lines homozygous for sos4-3 were segregated based on the presence of the T-DNA insertion by PCR analysis of genomic DNA (see Supplemental Table S2 for primers used), as well as the phenotype. The genotype of each mutant allele was verified by sequencing (Microsynth AG). The expression of SOS4 in the respective lines was assessed by RT-qPCR (see below). In general, plants were grown on soil (Einheitserde, Classic Ton Kokos) in pots or in rhizoboxes (see below), or sterile culture on half-strength Murashige and Skoog (MS) medium (Duchefa, M0221) (Murashige and Skoog, 1962) containing 0.55% [w/v] agar (Duchefa, P1001) under 16-h or 8-h photoperiods as specified (60% relative humidity, 20 or 120 µmol photons m−1 s−1 generated by fluorescent lamps [Philips Master T-D Super 80 18W/180] and 22°C) followed by the corresponding hours of darkness (8 or 16 h) at 18°C and ambient CO2 to complete a diel cycle. Seeds used for sterile culture were surface sterilized and air-dried prior to plating. For salt sensitivity assessment, seedlings were grown for 7 d on sterile culture in the absence or presence of 100 mM NaCl. In all cases, seeds were stratified for 2–3 d in the dark at 4°C before transferring to a growth incubator. Seeds cultivated on soil were grown in a CLF Climatics AR-66 phytochamber, whereas a CLF Climatics CU-22L phytochamber was used for sterile cultures. In all cases, individual biological replicates were generated from pools of plants sampled from different pots or plates.
Construction of transgenic plant lines
Full-length SOS4 but excluding the stop codon was amplified from cDNA of 10-d-old seedlings using a proofreading polymerase (Stratagene) and specific oligonucleotides (Supplemental Table S2). The amplified products were cloned into the pENTR/D-TOPO vector using the pENTR/D-TOPO cloning kit (Life Technologies) according to the manufacturers’ instructions and sequenced. Subsequently, the SOS4 insert was cloned into the Gateway destination vector pB7YWG2 (Karimi et al., 2002) using LR clonase enzyme mix II (Life Technologies) to generate p35S:SOS4-YFP. A 1973 bp promoter fragment upstream of the ATG start codon was amplified from genomic DNA using primers that included SacI and SpeI restriction sites (Supplemental Table S2) and cloned into pCR2.1TOPO using the TOPO-TA cloning kit (Invitrogen). The 35S promoter fragment in p35S:SOS4-YFP was then exchanged for the upstream region of SOS4 by restriction enzyme cloning using SacI and SpeI to generate pSOS4:SOS4-YFP. The construct was introduced into Agrobacterium tumefaciens strain C58 and used to transform sos4-1 or sos4-3 A. thaliana mutant plants by the floral dip method (Clough and Bent, 1998). As the respective constructs contain the BAR gene, transformants were selected by resistance to BASTA and phenotypic complementation. Resistant plants were allowed to self-fertilize and homozygous lines were selected from the T3 generation according to their segregation ratio for BASTA resistance.
Gene expression analysis by RT-qPCR
Tissue samples were collected from 21-d-old soil grown plants under a 16-h photoperiod. RNA was extracted using the RNA NucleoSpin Plant kit (Macherey-Nagel) according to the manufacturers’ instructions. DNA was removed by an on-column DNase digest during the RNA extraction and by an additional in-solution digest using DNAse RQ1 (Promega) for 20 min according to the manufacturers’ instructions. Reverse transcription was performed using 1 μg of total RNA as a template and Superscript II (Invitrogen) according to the instructions with the following modifications: oligo (dT) 20 primer stock concentration was 50 ng/μL and 0.5 μL Superscript II enzyme was used per reaction. RT-qPCR was performed in 384 well plates on a 7900HT Fast Real-Time PCR system (Applied Biosystems) using Power SYBR Green master mix (Applied Biosystems) and the following amplification program: 10 min denaturation at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The data were analyzed using the comparative cycle threshold method (2−ΔCT) normalized to the reference gene UBC21 (At5g25760). Primers used are listed in Supplemental Table S2. Each experiment was performed with three biological and three technical replicates.
Tissue expression pattern analysis
Tissue-specific expression of SOS4 was analyzed using 5-d-old sos4-1/pSOS4:SOS4-YFP seedlings grown vertically on sterile culture plates under a 16-h photoperiod. Roots and shoots of the seedlings were observed with a 40× IRAPO objective of a confocal microscope (Leica SP8) using a white light laser with an excitation at 488 nm and emission collection at 515–535 nm (bandwidth 20 nm), intensity at 3%, gain <20%. Images were acquired with the Leica LAS-X confocal software.
Vitamin B6 analysis by HPLC
In each case, plant material was ground using glass beads and a tissue lyser (Retsch). Two volumes of 50 mM ammonium-acetate (pH 4) were added, the samples were vortexed vigorously for 10 min and centrifuged at 20,238 g for 15 min, heated to 99°C for 3 min, and centrifuged as before. The resulting supernatant was decanted and used for analysis in two runs, using 10 and 50 µL injection volumes. Separation and detection of B6 vitamers by HPLC was performed as described previously (Szydlowski et al., 2013). All standards except PNP were purchased from Sigma–Aldrich. PNP is not commercially available and was produced enzymatically using PN as a substrate and recombinant Escherichia coli PdxK (Park et al., 2004). The protocol described in González et al. (2007) was repeated during which B6 vitamers were extracted in 5% (w/v) TCA and subjected to HPLC on a C18 reverse phase column (HyperClone 5-μm BDS C18, Phenomenex). Separation was achieved using 50% solution A (50 mM phosphoric acid) and 50% solution B (solution A containing 1% acetonitrile), both at pH 3.2, at a flow rate of 1 mL min−1. Detection was carried out using an excitation wavelength of 290 nm and an emission at 395 nm. To account for any possible variations in fresh to dry mass between genotypes, the fresh to dry (incubated overnight at 50°C) mass ratio was determined and is equivalent in all lines used (Supplemental Figure S4).
Enzymatic PLP assay
Shoots of 28-d-old plants grown on soil under a 16-h photoperiod were frozen in liquid nitrogen and ground using a tissue-lyser (Retsch). The assay was performed using 100 mg of the resulting material and the enzymatic vitamin B6 assay kit (A/C Diagnostics) according to the manufacturers’ instructions.
Rhizobox design and assembly
The rhizoboxes were made with two opaque Plexiglass plates (20 cm × 20 cm × 1 cm depth with 1 cm spacers and a perforated bottom on one [back] plate) by Vienna Scientific Instruments as described in Durand et al. (2016). The rhizoboxes were filled with 500 mL of sieved soil. A nylon membrane with a 0.7-µm mesh width (Sefar Nitex 03-7/2) was placed inside the rhizoboxes on the front plate and the rhizobox was closed with pliers. Seeds were sown in a drop of 0.65% (w/v) agar, placed between the front Plexiglas plate and the nylon membrane at the top of rhizoboxes (four plants per rhizobox). The top of the rhizoboxes was sealed with cellophane for 5–7 d after sowing to maintain a high humidity and prevent seedling dehydration. Where a clear plexiglass plate was used Rhizoboxes were wrapped with opaque plastic to avoid exposing roots to light and in all cases seedlings were grown as specified under a 16-h or 8-h photoperiod and either 20 or 120 µmol photons m2 s−1. Rhizobox soil water content was kept constant by daily watering.
Root length measurement
Root length was determined using the ImageJ software https://imagej.nih.gov/.
Micrografting of A. thaliana
Micrografting was performed as previously described (Martinis et al., 2016). Specifically, seedlings were grown in sterile culture under a 16-h photoperiod using 60 µmol photons m2 s−1 and 27°C followed by 8-h dark period at 25°C. Grafting was performed under sterile conditions 4 d after germination using a stereoscope. Hypocotyls were cut using a sharp razor blade (Gillette Wilkinson). Freshly cut scions and rootstocks were moved to a new plate, aligned, and joined using 0.3 mm silicon tubing (HelixMark) that was cut in half longitudinally and transversely into segments of about 1.5 mm. Plates were then placed back in the growth chamber for 7 d before grafting efficiency was evaluated. Grafts were examined under a stereomicroscope and were considered successful if no adventitious roots could be observed.
Ionomic analysis
The ionomic analysis was performed as previously described (Lahner et al., 2003). Leaf or root material was collected from seedlings grown in rhizoboxes under a 16-h photoperiod at either 20 or 120 µmol photons m2 s−1 using plastic tweezers and a ceramic knife (Victorinox) for separation of tissues. The harvested material was rinsed with ultrapure water (18.2 MΩcm) and placed in 10 mM CaCl2 for 10 min, in order to remove metals adhering to the root surface. The material was rinsed again with ultrapure water (18.2 MΩcm) and placed into Pyrex digestion tubes. The collected material was dried at 88°C overnight. Samples were digested with 1 mL of concentrated metal grade nitric acid (Primar Plus, Fisher Chemicals). Prior to the digestion, 20 µg/L of indium (In) was added to the nitric acid as an internal standard for assessing errors in dilution, variations in sample introduction and plasma stability in the ICP-MS. The samples were then digested in dry block heaters (SCP Science; QMX Laboratories) for 4 h at 115°C. After cooling down, 0.5 mL of 30% hydrogen peroxide (Fisher Chemicals) was added to samples that were then digested for an additional 1.5 h at 115°C in a heating block. After cooling down, the digests were diluted to 10 mL with ultrapure water (18.2 MΩcm). Elemental analysis was performed using an ICP-MS (NexION 2000; Perkin-Elmer) and 23 elements were monitored comprising Li, B, Na, Mg, P, S, K, Ca, Ti, Cr, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, Rb, Sr, Pb, and Cd. To correct for variation within ICP-MS analysis runs, liquid reference material was prepared using pooled digested samples, and run after every nine samples. Sample concentrations were calculated using the external calibration method within the instrument software. Further data processing was performed in Microsoft Excel. The concentrations for all the samples were normalized to calculated weights. In brief, the dry weights of eight reference samples were measured and used to calculate the weights and then final element concentration of the remaining samples based on a heuristic algorithm, which uses the best-measure elements in these samples, the weights of the eight weighed samples, and the solution concentrations as described by Lahner et al. (2003).
Lignin deposition analysis
ClearSee-adapted cell wall staining was performed as described in Fujita et al. (2020). Specifically, 3-d-old seedlings grown vertically in sterile culture under a 16-h photoperiod were fixed in 3 mL of phosphate-buffered saline (PBS) containing 4% (w/v) paraformaldehyde for 1 h at room temperature in six-well plates and washed twice with 3 mL of PBS. Following fixation, seedlings were incubated in 3 mL of ClearSee solution with gentle shaking overnight. The solution was then exchanged to new ClearSee solution containing 0.2% (w/v) Fuchsin and 0.1% (w/v) Calcofluor White for lignin and cell wall staining, respectively, for 12 h. The dye solution was removed, the material rinsed once with fresh ClearSee solution followed by washing in ClearSee solution first for 30 min, and then overnight with gentle shaking before observation. Basic Fuschin fluorescence was detected at 561 nm excitation and emission at 570–650 nm; Calcofluor White fluorescence was detected at 405 nm excitation and emission 425–475 nm using a Leica SP8 microscope.
PI staining
The staining of root tips with PI was performed on 3-d-old seedlings grown vertically on half-strength MS plates under a 16-h photoperiod at either 20 or 120 µmol photons m2 s−1 by incubation in water containing 10 μg mL−1 PI for 10 min. After staining, seedlings were rinsed in water and mounted on microscopy slides. Cells were scored using a Leica SP8 confocal microscope equipped with a white light laser using an excitation at 488 nm and emission collection at 500–550 nm (bandwidth 50 nm), intensity at 5%, gain <20%. For the PI penetration assay, the number of endodermal cells was scored from the onset of cell elongation (defined as endodermal cell length being more than two times the width in the median, longitudinal section) until the inner wall of an endodermal cell could not be stained by PI as described in Fujita et al. (2020).
Suberin assay
Seedlings grown vertically in sterile culture under a 16-h photoperiod at either 20 or 120 µmol photons m2 s−1 for 3 d were incubated in a freshly prepared solution of Fluorol Yellow 088 (0.01% w/v, in lactic acid) at 70°C for 30 min following the protocol described in Shukla et al. (2021). After incubation seedlings were rinsed three times in water baths for 5 min each. Counter-staining was done with aniline blue (0.5% w/v, in water) at room temperature for 30 min in darkness. Samples were washed in water for 30 min and mounted on slides using 50% glycerol prior to microscope examination. Samples were kept in the dark during the whole procedure. Fluorescence was detected using a ZEISS Axio Zoom.V16 stereomicroscope with a GFP filter (excitation 450–490 nm, emission 500–550 nm). For suberin pattern determination, tiled images covering the whole seedlings in single images were taken. For high-resolution imaging of the large field of view, multiple smaller images were made as tiles and stitched. The region of interest of a sample was defined by marking the ‘tile-region’ after a quick scan of the sample at a lower resolution. At least five focal points were used to adjust the focus of the sample (for 3-d-old seedlings) along the region of interest. A 10% area of overlap was defined for alignment and stitching of tiles. Fiji (http://fiji.sc/Fiji) was used on Zen2.3 blue exported stitched tile images for quantification of suberin patterns (in mm) along the root. Suberin deposition was assessed and quantified according to the non-suberized zone, the zone of patchy suberization, and the zone of continuous suberization. The results are presented as percentage of the root length as previously done (Pfister et al., 2014).
Staining for superoxide
Staining for superoxide with NBT was performed according to Dunand et al. (2007). Three-day-old seedlings grown in sterile culture under 16-h light (either 10 or 120 µmol photons m−2 s−1 as indicated and at 21°C), 8-h dark (at 18°C) cycles were incubated in 2 mM NBT (Sigma–Aldrich) in 20 mM potassium phosphate buffer, pH 6.1, for 2 h. Images were taken using a stereomicroscope (Leica MZ16).
TAC assay
For the analysis of TAC, 7-d-old seedlings grown on sterile culture under a 16-h photoperiod (120 µmol photons m−1 s−1) were ground to a powder in liquid nitrogen and homogenized in 50% acetone (1 mL per 100 mg FW) for TAC determination using the TAC Assay Kit according to the manufacturers’ instructions in the absence and presence of the included protein mask (MAK187, Sigma–Aldrich).
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank/TAIR data libraries under AGI locus identifiers SOS4, At5g37850; UBC21, At5g25760.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of HPLC methods for assessment of B6 vitamers.
Supplemental Figure S2. Effect of external application of PN or PL on sos4 root growth.
Supplemental Figure S3. Reciprocal micrografting of sos4 lines and wild type.
Supplemental Figure S4. Determination of the dry-to-fresh mass ratio of genotypes.
Supplemental Table S1. Elemental content of lines relative to wild type.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre for seeds of sos4-1 (N24930 in Col (gl1) background), GK-891A06 (N485446) and GK-891A12 (N485452, sos4-3) and Yikun He (Capital Normal University, Beijing, China) for donating sno1. We acknowledge Michael Moulin for initial HPLC analysis aspects and Denise Touré for analysis of Arabidopsis lines during the early stages of this study (both University of Geneva). We are grateful to Vinay Shukla for sharing the suberin methodology used and Marie Barberon for critical reading of the manuscript (both University of Geneva).
Funding
Financial support is gratefully acknowledged from the Swiss National Science Foundation (Grant 31003A-141117/1 and IZLIZ3_183193) to T.B.F. as well as the University of Geneva.
Conflict of interest statement. None declared.
V.G. performed the majority of the experimental work, analyzed the data, and contributed to writing the article. M.C. contributed research tools and did some of the experimental work. E.D. performed some of the experimental work. P.F. and D.E.S. performed and analyzed the ionome. T.B.F. conceived, obtained funding, supervised the research, analyzed the data, and wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Teresa B. Fitzpatrick (theresa.fitzpatrick@unige.ch).
References
- Asensi-Fabado MA, Munné-Bosch S (2010) Vitamins in plants: Occurrence, biosynthesis and antioxidant function. Trends Plant Sci 15: 582–592 [DOI] [PubMed] [Google Scholar]
- Barberon M (2017) The endodermis as a checkpoint for nutrients. New Phytol 213: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Barberon M, Vermeer JE, De Bellis D, Wang P, Naseer S, Andersen TG, Humbel BM, Nawrath C, Takano J, Salt DE, et al. (2016) Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164: 447–459 [DOI] [PubMed] [Google Scholar]
- Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF (2000) Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol 71: 129–134 [DOI] [PubMed] [Google Scholar]
- Boycheva S, Dominguez A, Rolcik J, Boller T, Fitzpatrick TB (2015) Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol 167: 102–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colinas M, Eisenhut M, Tohge T, Pesquera M, Fernie AR, Weber AP, Fitzpatrick TB (2016) Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. Plant Cell 28: 439–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czégény G, Kőrösi L, Strid Å, Hideg É (2019) Multiple roles for vitamin B6 in plant acclimation to UV-B. Sci Rep 9: 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aglio E, Boycheva S, Fitzpatrick TB (2017) The pseudoenzyme PDX1.2 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiol 174: 2098–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvo M, Safo M, Contestabile R (2012) Biomedical aspects of pyridoxal 5′-phosphate availability. Front Biosci 4: 897–913 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Geldner N, Barberon M (2017) The endodermis, a tightly controlled barrier for nutrients. Curr Opin Plant Biol 39: 136–143 [DOI] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N (2016) Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol 170: 1460–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T (2007) Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J 407: 1–13 [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues—A typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Fujita S, De Bellis D, Edel KH, Köster P, Andersen TG, Schmid-Siegert E, Dénervaud Tendon V, Pfister A, Marhavy P, et al. (2020) SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J 39: e103894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatge MS, Al Mughram M, Omar AM, Safo MK (2021) Inborn errors in the vitamin B6 salvage enzymes associated with neonatal epileptic encephalopathy and other pathologies. Biochimie 189: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Danehower D, Daub ME (2007) Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol 145: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ordenes F, Bravo-Moraga F, Gonzalez E, Hernandez-Cabello L, Alzate-Morales J, Guixé V, Castro-Fernandez V (2021) Crystal structure and molecular dynamics simulations of a promiscuous ancestor reveal residues and an epistatic interaction involved in substrate binding and catalysis in the ATP-dependent vitamin kinase family members. Protein Sci 30: 842–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Ksas B, Szewczyk A, Rumeau D, Franck F, Caffarri S, Triantaphylidès C (2009) Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zeng H, Zhang J, Wei S, Huang L (2011) Interconversions of different forms of vitamin B6 in tobacco plants. Phytochemistry 72: 2124–2129 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L,et al. (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Li KT, Moulin M, Mangel N, Albersen M, Verhoeven-Duif NM,Ma Q, Zhang P, Fitzpatrick TB, Gruissem W, Vandershuren H (2015) Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency. Nat Biotechnol 33: 1029–1032 [DOI] [PubMed] [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K (2017) Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Mol Plant 10: 1480–1496 [DOI] [PubMed] [Google Scholar]
- Lum HK, Kwok F, Lo SC (2002) Cloning and characterization of Arabidopsis thaliana pyridoxal kinase. Planta 215: 870–879 [DOI] [PubMed] [Google Scholar]
- Mangel N, Fudge JB, Li KT, Wu TY, Tohge T, Fernie AR, Szurek B, Fitzpatrick TB, Gruissem W, Vandershuren H (2019) Enhancement of vitamin B6 levels in rice expressing Arabidopsis vitamin B6 biosynthesis de novo genes. Plant J 99: 1047–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis J, Gas-Pascual E, Szydlowski N, Crèvecoeur M, Gisler A, Bürkle L, Fitzpatrick TB (2016) Long-distance transport of thiamine (vitamin B1) is concomitant with that of polyamines. Plant Physiol 171: 542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS (2010) The elements of plant micronutrients. Plant Physiol 154: 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth of bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JA, Das SK, Sedelnikova SE, Rice DW (2006) The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parallel emergence of enzyme activity during evolution. J Mol Biol 363: 520–530 [DOI] [PubMed] [Google Scholar]
- Park J-H, Burns K, Kinsland C, Begley TP (2004) Characterization of two kinases involved in thiamin pyrophosphate and pyridoxal phosphate biosynthesis in Bacillus subtilis: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and pyridoxal kinase. J Bacteriol 186: 1571–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, Vermeer JE, Yamazaki M, Li G, Maurel C, Takano J, et al. (2014) A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. Elife 3: e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna P, Barberon M (2019) Polarized transport across root epithelia. Curr Opin Plant Biol 52: 23–29 [DOI] [PubMed] [Google Scholar]
- Rueschhoff EE, Gillikin JW, Sederoff HW, Daub ME (2013) The SOS4 pyridoxal kinase is required for maintenance of vitamin B6-mediated processes in chloroplasts. Plant Physiol Biochem 63: 281–291 [DOI] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu J-K (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhu J-K (2002) SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol 129: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V, Han JP, Cléard F, Lefebvre-Legendre L, Gully K, Flis P, Berlin A, Andersen TG, Salt DE, Nawrath C,et al. (2021) Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors. Proc Natl Acad Sci USA 118: e2101730118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N, Burkle L, Pourcel L, Moulin M, Stolz J, Fitzpatrick TB (2013) Recycling of pyridoxine (vitamin B6) by PUP1 in Arabidopsis. Plant J 75: 40–52 [DOI] [PubMed] [Google Scholar]
- Thériault O, Poulin H, Thomas GR, Friesen AD, Al-Shaqha WA, Chahine M (2014) Pyridoxal-5′-phosphate (MC-1), a vitamin B6 derivative, inhibits expressed P2X receptors. Can J Physiol Pharmacol 92: 189–196 [DOI] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N,Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48: 933–946 [DOI] [PubMed] [Google Scholar]
- Waisel Y, Eshel A (2002) Plant roots: the hidden half. Ann Bot 90 [Google Scholar]
- Wilson MP, Plecko B, Mills PB, Clayton PT (2019) Disorders affecting vitamin B. J Inherit Metab Dis 42: 629–646 [DOI] [PubMed] [Google Scholar]
- Xia J, Kong D, Xue S, Tian W, Li N, Bao F, HU Y, Du J, WangY, Pan X, et al. (2014) Nitric oxide negatively regulates AKT1-mediated potassium uptake through modulating vitamin B6 homeostasis in Arabidopsis. Proc Natl Acad Sci USA 111: 16196–16201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jin X, Ouyang Z, Li X, Liu B, Huang L, Hong Y, Zhang H, Song F, Li D (2015) Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J Plant Physiol 175: 21–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.