Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by progressive upper and lower motor neuron (LMN) loss. As ALS and other neurodegenerative diseases share genetic risk factors, we performed whole-exome sequencing in ALS patients focusing our analysis on genes implicated in neurodegeneration. Thus, variants in the DHTKD1 gene encoding dehydrogenase E1 and transketolase domain containing 1 previously linked to 2-aminoadipic and 2-oxoadipic aciduria, Charcot-Marie-Tooth (CMT) disease type 2, and spinal muscular atrophy (SMA) were identified. In two independent European ALS cohorts (n = 643 cases), 10 sporadic cases of 225 (4.4%) predominantly sporadic patients of cohort 1, and 12 familial ALS patients of 418 (2.9%) ALS families of cohort 2 harbored 14 different rare heterozygous DHTKD1 variants predicted to be deleterious. Four DHTKD1 variants were previously described pathogenic variants, seven were recurrent, and eight were located in the E1_dh dehydrogenase domain. Nonsense variants located in the E1_dh domain were significantly more prevalent in ALS patients versus controls. The phenotype of ALS patients carrying DHTKD1 variants partially overlapped with CMT and SMA by presence of sensory impairment and a higher frequency of LMN-predominant cases. Our results argue towards rare heterozygous DHTKD1 variants as potential contributors to ALS phenotype and, possibly, pathogenesis.
Keywords: amyotrophic lateral sclerosis, DHTKD1, neurodegeneration, Charcot-Marie-Tooth disease type 2, 2-aminoadipic and 2-oxoadipic aciduria, lower motor neuron, whole-exome sequencing
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder characterized by upper and lower motor neuron loss, commonly leading to death due to respiratory paralysis within three to five years after disease onset [1]. ALS is an umbrella term covering a large spectrum of different phenotypes with distinct clinical presentations. Different ALS subtypes can be distinguished depending on the extent of bulbar/spinal upper and lower motor neuron affection with different clinical and prognostic characteristics related to gender and age [2]. A contribution of genetic factors to ALS pathogenesis was suggested by the observation that 5–10% of ALS patients have a familial predisposition, and that relatives of apparently sporadic ALS patients, representing the vast majority of cases, have an increased ALS risk [3]. Three decades of research have implicated at least 25 genes [1] and more than 120 variants in ALS risk (https://alsod.ac.uk/ accessed on 21 October 2021). Additionally, certain genetic aberrations have been reported to modify the ALS phenotype, suggesting that genetic factors also shape the clinical presentation of ALS [4]. Moreover, some of the genes identified in ALS patients are also causative for other neurodegenerative disorders, such as C9orf72 and TARDBP for frontotemporal dementia (FTD) and Parkinson’s disease, and SPG7 and SPG11 for hereditary spastic paraplegia, indicating shared pathomechanisms [4,5,6,7]. In fact, it appears as though most, if not all ALS genes may be pleiotropic [8].
Accordingly, in this study, in search of genes implicated in neurodegeneration as potential novel ALS-associated genes, we identified rare heterozygous DHTKD1 variants in ALS patients. Pathogenic variants in DHTKD1 encoding dehydrogenase E1 and transketolase domain containing 1 were previously described in patients with autosomal recessive 2-aminoadipic and 2-oxoadipic aciduria (AMOXAD), a rare metabolic disorder also characterized by hypotonia, delayed psychomotor development and seizures [9,10], autosomal dominant Charcot-Marie-Tooth disease type 2 (CMT2) [11], and autosomal recessive infantile-onset spinal muscular atrophy (SMA) with cognitive delay [12]. Here, we aimed at investigating the frequency as well as genotype–phenotype correlations of rare DHTKD1 variants in two independent ALS cohorts.
2. Materials and Methods
2.1. Patients
Cohort 1 consisted of 225 unrelated ALS patients of central European ethnicity (133 males, 92 females; 8 with familial ALS, 217 with sporadic ALS) recruited at the motor neuron disease clinic of the Department of Neurology at Hannover Medical School, Hannover, Germany. All patients were examined by a neurologist specialized in motor neuron diseases and subdivided into one of eight ALS subtypes (upper motor neuron (UMN)-dominant ALS, bulbar phenotype, flail arm syndrome, flail leg syndrome, respiratory phenotype, progressive muscular atrophy (PMA), lower motor neuron (LMN)-dominant ALS, and classic (Charcot) ALS) [2]. Extensive clinical workup including magnetic resonance imaging, cerebrospinal fluid analysis, electromyography (EMG), and nerve conduction studies (NCS) was performed at first examination. UMN-dominant ALS was defined by clinically predominant UMN signs at disease onset but development of clinically progressive LMN signs within 2–4 years, thereby distinguishing these cases from primary lateral sclerosis (PLS), according to the recently renewed diagnostic criteria for PLS [13]. While PLS can be considered as part of the ALS spectrum [14], cases of probable or definite PLS [13] were not included into our patient cohort. In cases with absence of UMN signs, the diagnosis of PMA was made after careful exclusion of disease mimics, including search for conduction blocks by extensive NCS and cerebrospinal fluid analysis to rule out immune-mediated neuropathies, genetic testing for deletions or point mutations in the SMN1 gene or expansion of the CAG repeat in the androgen receptor gene, and/or muscle biopsies [2]. Disease progression was measured using the Revised ALS Functional Rating Scale (ALSFRS-R) [15], the progression rate was calculated as previously described [16].
Cohort 2 consisted of 486 familial ALS patients from 418 unrelated families collected in Germany and Sweden [17], thus carrying a comparable genetic background as cohort 1.
2.2. Whole-Exome and Targeted DHTKD1 Sequencing
In cohort 1, extraction of genomic DNA was performed from whole blood using the QIAamp DNA Blood Maxi Kit (Qiagen, Hilden, Germany). Whole-exome sequencing (WES) on leukocyte DNA of 46 ALS patients and 148 control individuals not affected by a neurologic disease was performed using the Agilent SureSelect Human All Exon v4 or v5 + UTR Target Enrichment System (both Agilent Technologies, Santa Clara, CA, USA) or the IDT xGen Exome Research Panel (Integrated DNA Technologies, Coralville, IA, USA) on an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) or a MGISEQ2000 (MGI Tech, Shenzhen, Guangdong, China) sequencing platform. All samples were sequenced to a mean target coverage of >50×. Sequencing data were aligned to the human reference genome GRCh37/hg19 and analyzed using our in-house workflow and a candidate gene-based strategy with regard to neurodegeneration (Supplementary Table S1) using CLC Genomics Workbench 20 and Clinical Insight Interpret 8.0 (both Qiagen). Targeted sequencing of all coding exons and adjacent splice site regions (±15 base pairs into intron) of the DHTKD1 gene (NG_033248.1) was done on leukocyte DNA of 179 additional ALS patients by conventional chain termination protocols on a 3130XL Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) (oligonucleotide sequences are given in Supplementary Table S2). The identified variants were prioritized based on their minor allele frequency (MAF) extracted from the Genome Aggregation Database (gnomAD, v.2.1.1, https://gnomad.broadinstitute.org accessed on 21 October 2021) and in silico prediction of their pathogenicity using MutationTaster (http://www.mutationtaster.org accessed on 21 October 2021), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/ accessed on 21 October 2021), SIFT and PROVEAN (http://provean.jcvi.org/ accessed on 21 October 2021). DHTKD1 variants were classified according to the guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) [18].
In cohort 2, DHTKD1 variants were extracted from a WES dataset generated as previously described [17], although only DHTKD1 variants from patients without a pathogenic variant in a known ALS gene were used in this study.
Nucleotide numbering of the identified variants reflects the nucleotide position in the coding sequence of human DHTKD1 mRNA (NM_018706.6).
2.3. Metabolic Analysis of Urine and Plasma of DHTKD1 Variant Carrier VALS164
The 2-aminoadipate and 2-oxoadipate levels in urine and plasma of DHTKD1 variant carrier VALS164 were determined by the GCMS Laboratory, Heidelberg, Germany. Urine analysis was performed as previously described [19,20] with some modifications. A urine volume equivalent to 1 µmol creatinine was acidified with hydrochloric acid and extracted twice with ethyl acetate. After removal of the solvent, the residue was derivatized with N-methyl-N-trimethylsilylheptafluorobutyramide (Macherey-Nagel, Düren, Germany). The resulting trimethylsilyl derivatives were analyzed using the single quadrupole mass spectrometer DSQ II (Thermo Fisher Scientific) coupled to the gas chromatograph TRACE GC (Thermo Fisher Scientific). The mass spectrometer was run in the full scan mode (m/z 50 to m/z 650) with electron impact ionization. Gas chromatographic separation was achieved on a capillary column (DB-5MS, 30 m × 0.25 mm; film thickness: 0.25 µm; J&W Scientific, Folsom, CA, USA) using helium as a carrier gas. A volume of 1 µL of the derivatized sample was injected in splitless mode. GC temperature parameters were 80 °C for 2 min, ramp 50 °C per minute to 150 °C, ramp 10 °C per minute to 300 °C. Injector temperature was set to 260 °C and interface temperature to 260 °C. The specific mass m/z 484 was used for quantification of 2-oxoadipate.
The 2-aminoadipate levels were determined in urine and plasma by the EZ:faast amino acid GCMS analysis kit (Phenomenex, Torrance, CA, USA) with some modifications. Based on cation-exchange solid-phase extraction, 2-aminoadipate was measured as a chloroformate derivative by gas chromatography mass spectrometry. The system consisted of a gas chromatograph 7820A coupled to the mass spectrometer 5977 Inert MSD (Agilent Technologies). The capillary column used was DB-5MS, 30 m × 0.25 mm; film thickness: 0.25 µm (J&W Scientific). The temperature programming of the gas chromatograph started at an initial temperature of 60 °C and was increased to 290 °C at 7 °C/min. After a hold time of 2 min, the temperature was further increased to 300 °C at 10 °C/min. Helium was used as carrier gas in the constant pressure mode. The temperature of the MSD transfer line was 290 °C. Inlet was operated using the splitless mode with a temperature of 280 °C. Injection volume was 1 µL of the derivatized sample. Quantification ion for 2-aminoadipate was mass m/z 244.
2.4. Histological Analysis of a Muscle Biopsy from DHTKD1 Variant Carrier VALS054
A muscle biopsy of patient VALS054 obtained for differential diagnostic purposes during the initial workup was processed according to standard histological techniques. Sections were stained with hematoxylin-eosin, nonspecific esterase, modified Gomori trichrome and combined cytochrome c oxidase and succinate dehydrogenase (COX/SDH).
2.5. Statistical Analysis
Statistical analysis was done using MATLAB and Statistics Toolbox Release 2018b (The MathWorks, Natick, MA, USA). Student’s t-test, Fisher’s exact test or Mann–Whitney U test were used, as applicable; a p-value < 0.05 was considered statistically significant.
3. Results
3.1. Genetic Analysis of Two Independent Cohorts of ALS Patients
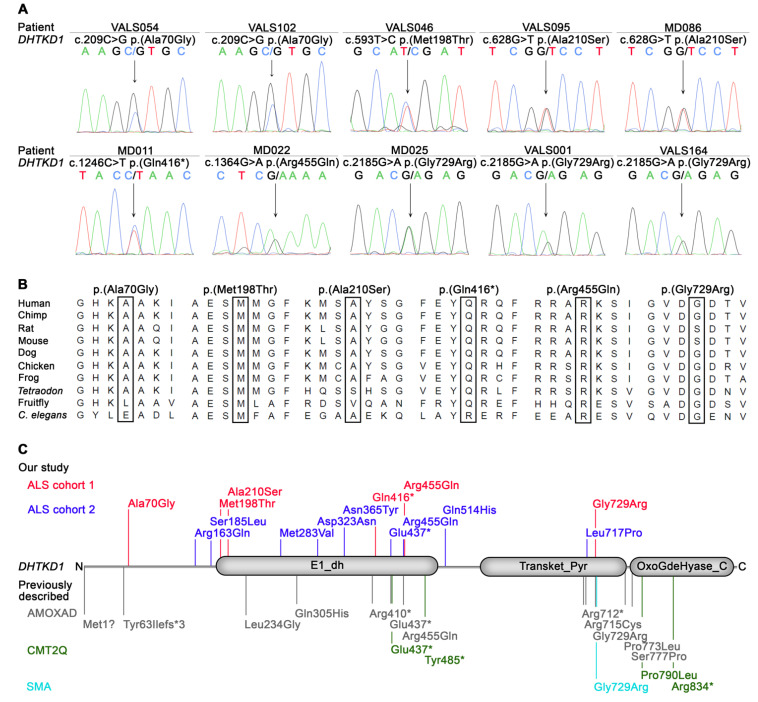
To identify novel genes potentially associated with ALS, a pilot WES analysis was performed on 27 genetically unsolved ALS patients of cohort 1. A candidate gene-based strategy was applied prioritizing rare (MAF ≤ 0.5%), non-silent variants not present in in-house controls and predicted to be deleterious in 694 genes associated with neurodegeneration, revealing the DHTKD1 gene as the only gene harboring such variants in two patients (Supplementary Table S1). In one patient, a heterozygous nonsense variant, DHTKD1:c.1246C>T p.(Gln416*), predicted to truncate the encoded protein within the first dehydrogenase domain (E1_dh; Figure 1) was detected. Another patient was found to carry a heterozygous missense variant, DHTKD1:c.1364G>A p.(Arg455Gln) (Figure 1, Table 1), previously reported in patients with AMOXAD [10]. As two ALS patients carried rare DHTKD1 variants in our pilot study and the identified aberrations were loss-of-function and known pathogenic variants, we aimed to determine the frequency of rare DHTKD1 variants in all 225 patients of cohort 1. Whole-exome and targeted sequencing revealed four additional DHTKD1 variants, i.e., c.209C>G p.(Ala70Gly), c.593T>C p.(Met198Thr), c.628G>T p.(Ala210Ser), and c.2185G>A p.(Gly729Arg), in eight of 198 further ALS patients (Table 1). Interestingly, the c.2185G>A p.(Gly729Arg) variant, previously described in AMOXAD [9] and SMA [12], was detected in three patients, significantly more frequently than in the gnomAD control data set (3/225 ALS patients of cohort 1 versus 154/59,472 controls; p = 0.022, two-sided Fisher’s exact test). In total, six different rare non-silent DHTKD1 variants predicted to be deleterious were identified in 10 sporadic ALS cases of 225 (4.4%) patients of cohort 1. All were confirmed to be heterozygous by Sanger sequencing (Figure 1A) and affected highly or moderately conserved amino acid residues (Figure 1B).
Figure 1.
DHTKD1 variants identified in ALS patients of cohort 1 (A,B) and cohort 2 in this study, and described in other disorders previously (C). (A) Electropherograms showing the six different heterozygous rare (MAF ≤ 0.5%) DHTKD1 variants predicted to be deleterious by at least one in silico prediction tool, i.e., MutationTaster, PolyPhen-2, SIFT, or PROVEAN, detected in leukocyte DNA of 10 sporadic ALS patients of cohort 1 (affected nucleotides are highlighted by an arrow). The c.209C>G p.(Ala70Gly) and c.628G>T p.(Ala210Ser) variants were detected twice, the c.2185G>A p.(Gly729Arg) variant in three ALS patients. (B) DHTKD1 variants identified in ALS patients of cohort 1 affect highly or moderately conserved amino acids, according to Alamut Visual 2.15 (Interactive Biosoftware, Rouen, France). (C) Schematic representation of rare DHTKD1 variants described in ALS patients of this study, i.e., sporadic ALS patients of cohort 1 (red) and familial ALS patients of cohort 2 (blue), and, previously, in the following other diseases: 2-aminoadipic and 2-oxoadipic aciduria (AMOXAD) [9,10,22], Charcot-Marie-Tooth disease type 2Q (CMT2Q) [11,21,23,24], and infantile-onset spinal muscular atrophy (SMA) [12]. The DHTKD1 protein contains three functional domains according to the Pfam database (http://pfam.xfam.org/protein/Q96HY7 accessed on 21 October 2021): (1) dehydrogenase E1 component (E1_dh), (2) transketolase, pyrimidine binding domain (Transket_Pyr), and (3) 2-oxoglutarate dehydrogenase, C-terminal (OxoGdeHyase_C).
Table 1.
Rare heterozygous non-silent DHTKD1 variants predicted to be deleterious identified in sporadic ALS patients of cohort 1 (n = 225 ALS patients).
| Patient ID | Chromosomal Position a | Exon | Nucleotide Change b | Amino Acid Change b | Reference SNP | MAF (gnomAD Controls c) |
Prediction | Previously Reported in |
ACMG/AMP Criteria d | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation Taster | PolyPhen-2 | SIFT | PROVEAN | |||||||||
| VALS054 VALS102 |
10:12123525 | 2 | c.209C>G | p.(Ala70Gly) | rs34644609 | 0.003700 | Disease causing | Benign | Tolerated | Neutral | - | Uncertain significance |
| VALS046 | 10:12129604 | 4 | c.593T>C | p.(Met198Thr) | - | - | Disease causing | Probably damaging | Damaging | Deleterious | - | Uncertain significance (PM2_moderate; PP3_supporting) |
| VALS095 MD086 |
10:12129639 | 4 | c.628G>T | p.(Ala210Ser) | rs146741810 | 0.002554 | Disease causing | Benign | Tolerated | Neutral | - | Uncertain significance (BP4_supporting) |
| MD011 | 10:12136158 | 7 | c.1246C>T | p.(Gln416*) | rs200722918 | 0.00004988 | Disease causing | - | - | - | - | Pathogenic (PVS1_very strong; PM1_moderate; PP3_supporting) |
| MD022 | 10:12139688 | 8 | c.1364G>A | p.(Arg455Gln) | rs142068634 | 0.0001394 | Disease causing | Probably damaging | Damaging | Deleterious | AMOXAD e (AR) | Pathogenic (PS1_strong; PS3_strong; PM1_moderate; PP3_supporting) |
| MD025 VALS001 VALS164 |
10:12154929 | 13 | c.2185G>A | p.(Gly729Arg) | rs117225135 | 0.001295 | Disease causing | Possibly damaging | Damaging | Deleterious | AMOXAD f (AR), SMA g (AR) |
Pathogenic (PS1_strong; PS3_strong; PS4_strong; PM1_moderate; PP3_supporting) |
Abbreviations: AMOXAD: 2-aminoadipic and 2-oxoadipic aciduria; AR: autosomal recessive; MAF: minor allele frequency; SNP: single nucleotide polymorphism; SMA: spinal muscular atrophy. a According to GRCh37/hg19. b According to DHTKD1 transcript NM_018706.6 and DHTKD1 protein NP_061176.4. c Genome Aggregation Database v2.1.1 controls. d [18]. e [10]. f [9]. g [12].
Next, we analyzed rare DHTKD1 variants and their frequency in familial ALS (cohort 2). In cohort 2, nine rare heterozygous DHTKD1 variants predicted to be deleterious were found in 12 unrelated familial ALS patients of 418 (2.9%) ALS families (Table 2), consisting of eight missense and one nonsense variant. The c.488G>A p.(Arg163Gln), c.847A>G p.(Met283Val) and c.1542G>T p.(Gln514His) variants were detected in two patients each. Two of the variants, i.e., c.1542G>T p.(Gln514His) and c.2150T>C p.(Leu717Pro), were not found in gnomAD database v2.1.1 controls. Five of the variants were located within the first dehydrogenase domain (E1_dh) of the DHTKD1 protein, including the nonsense variant, c.1309G>T p.(Glu437*), predicted to truncate the protein within this domain (Figure 1C). The c.1309G>T p.(Glu437*) variant was previously described in AMOXAD [10] and CMT2Q [21]. The c.1364G>A p.(Arg455Gln) missense variant, previously reported in cases of AMOXAD [10], was identified in cohort 1 and cohort 2 (Figure 1C).
Table 2.
Rare heterozygous non-silent DHTKD1 variants predicted to be deleterious identified in familial ALS patients of cohort 2 (n = 418 ALS families).
| Patient ID | Chromosomal Position a |
Exon | Nucleotide Change b | Amino Acid Change b | Reference SNP | MAF (gnomAD Controls c) |
Prediction | Previously Reported in | ACMG/AMP Criteria d | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation Taster |
PolyPhen-2 | SIFT | PROVEAN | |||||||||
| C2-Pt1 C2-Pt2 |
10:12126716 | 3 | c.488G>A | p.(Arg163Gln) | rs78189904 | 0.0006568 | Disease causing | Benign | Tolerated | Neutral | Eosinophilic esophagitis (AD) e | Uncertain significance (PS1_strong; BP4_supporting) |
| C2-Pt3 | 10:12129565 | 4 | c.554C>T | p.(Ser185Leu) | rs149544379 | - | Disease causing | Benign | Damaging | Neutral | - | Uncertain significance |
| C2-Pt4 C2-Pt5 |
10:12131114 | 5 | c.847A>G | p.(Met283Val) | rs145337285 | 0.0004113 | Disease causing | Benign | Tolerated | Deleterious | - | Uncertain significance (PM1_moderate) |
| C2–Pt6 | 10:12131234 | 5 | c.967G>A | p.(Asp323Asn) | rs529235889 | 0.00001832 | Disease causing | Possibly damaging | Damaging | Deleterious | - | Uncertain significance (PM1_moderate; PP3_supporting) |
| C2-Pt7 | 10:12133617 | 6 | c.1093A>T | p.(Asn365Tyr) | rs747758630 | 0.00001828 | Disease causing | Probably damaging | Damaging | Deleterious | - | Uncertain significance (PM1_moderate; PP3_supporting) |
| C2-Pt8 | 10:12136221 | 7 | c.1309G>T | p.(Glu437*) | rs138884194 | 0.00004988 | Disease causing | - | - | - | CMT2Q f (AD), AMOXAD g (AR) | Pathogenic (PVS1_very strong; PS1_strong; PM1_moderate; PP3_supporting) |
| C2-Pt9 | 10:12139688 | 8 | c.1364G>A | p.(Arg455Gln) | rs142068634 | 0.0001394 | Disease causing | Probably damaging | Damaging | Deleterious | AMOXAD g (AR) | Pathogenic (PS1_strong; PS3_strong; PM1_moderate; PP3_supporting) |
| C2-Pt10 C2-Pt11 |
10:12139866 | 8 | c.1542G>T | p.(Gln514His) | - | - | Disease causing | Benign | Damaging | Deleterious | - | Uncertain significance (PP3_supporting) |
| C2-Pt12 | 10:12150010 | 12 | c.2150T>C | p.(Leu717Pro) | - | - | Disease causing | Probablydamaging | Damaging | Deleterious | - | Uncertain significance (PM1_moderate; PP3_supporting) |
Abbreviations: AD: autosomal dominant; AMOXAD: 2-aminoadipic and 2-oxoadipic aciduria; AR: autosomal recessive; CMT2Q: Charcot-Marie-Tooth disease type 2Q; fALS: familial ALS; MAF: minor allele frequency; SNP: single nucleotide polymorphism. a According to GRCh37/hg19. b According to DHTKD1 transcript NM_018706.6 and DHTKD1 protein NP_061176.4. c Genome Aggregation Database v2.1.1 controls. d [18]. e [25]. f [21]. g [10].
Of the 14 different DHTKD1 variants identified in both cohorts, eight were located in the first dehydrogenase domain of the DHTKD1 protein. Notably, loss-of-function variants located in the first dehydrogenase domain of DHTKD1 were significantly more frequent in ALS patients of cohorts 1 and 2 than in gnomAD v2.1.1 controls (2/643 unrelated ALS cases of cohorts 1 and 2 versus 25/51,060 controls; p = 0.0442, two-sided Fisher’s exact test).
3.2. Clinical Characteristics of Sporadic ALS Patients Carrying DHTKD1 Variants
Sporadic ALS patients with DHTKD1 variants of cohort 1 (n = 10) were available for detailed clinical and electrophysiological phenotyping (Supplementary Table S3). DHTKD1 variant carriers were almost exclusively from Germany, had a median age of disease onset of 71 years (range: 49–73 years), and the site of onset was mainly spinal (8/10) and less frequently bulbar (2/10). The most frequent symptoms at disease onset were deterioration of fine motor skills or weakness of the hand (6/10) followed by walking difficulties (2/10) or speech difficulties (2/10).
In all sporadic ALS patients carrying DHTKD1 variants, EMG revealed disseminated signs of acute and chronic denervation. NCS were available in 9/10 DHTKD1 variant carriers, showing axonal and demyelinating motor neuropathy in 4/9 cases (VALS054, VALS102, VALS095, VALS001), axonal motor neuropathy in 5/9 cases (VALS046, MD011, MD022, VALS164, MD025), and additional sensory neuropathy in 4/9 cases (axonal and demyelinating in VALS054 and VALS095, axonal in MD022 and MD025) with matching clinical signs including pallhypaesthesia, hypaesthesia or reduced sharp/blunt differentiation. Two further DHTKD1 variant carriers (MD011, VALS164) presented with sensory impairment but normal sensory NCS. No possible underlying causes (e.g., diabetes, alcohol abuse, or chemotherapy) were present in five of the six cases with sensory impairment, whereas patient VALS054 was diagnosed with diabetes mellitus type II (Supplementary Table S3).
To test for signs of AMOXAD in patient VALS164 showing heterozygosity for the c.2185G>A p.(Gly729Arg) variant previously described in AMOXAD patients compound heterozygous for this and another variant [9], the only carrier of an AMOXAD-associated variant from cohort 1 who was still alive, urine and plasma samples of patient VALS164 were subjected to metabolic analysis of 2-aminoadipate and 2-oxoadipate levels. The urine concentration of 2-aminoadipate was close to the upper reference limit, while 2-oxoadipate was hardly detectable in patient VALS164 (Table 3).
Table 3.
Metabolite levels in sporadic ALS patient VALS164 carrying the DHTKD1:c.2185G>A p.(Gly729Arg) variant.
| Metabolite | Urine (mmol/mol Creatinine) | Plasma (µmol/L) |
|---|---|---|
| 2-aminoadipate | 5.5 (upper limit: 8) | 1.9 (upper limit: 6) |
| 2-oxoadipate | 0.2 (upper limit: 25) | Not detectable |
Metabolite levels and upper limits were determined by the GCMS Laboratory, Heidelberg, Germany.
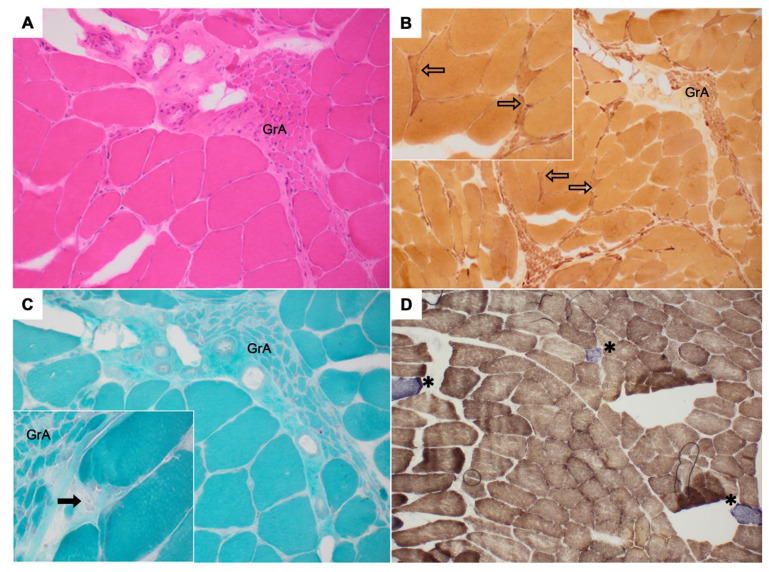
Patient VALS054 was initially diagnosed with PMA, and a muscle biopsy of the left biceps brachii was obtained for the differential diagnosis of myopathy/myositis. Neuropathological evaluation revealed a distinctive pattern of neurogenic damage including grouped atrophy, single muscle fiber atrophy, and single muscle fiber necrosis upon hematoxylin-eosin, nonspecific esterase, and modified Gomori trichrome staining. In mitochondrial-targeted staining, combined COX/SDH staining displayed reduced or absent COX activity in single muscle fibers (Figure 2).
Figure 2.
Muscle biopsy of ALS patient VALS054 of cohort 1 diagnosed with the PMA subtype carrying the DHTKD1:c.209C>G p.(Ala70Gly) variant. Muscle sections were stained using (A) hematoxylin-eosin, (B) nonspecific esterase, (C) modified Gomori trichrome, and (D) combined cytochrome c oxidase and succinate dehydrogenase (COX/SDH). Histopathological analysis revealed a distinctive pattern of neurogenic damage including groups of atrophic muscle fibers (GrA), single atrophic muscle fibers (unfilled arrows), and scattered muscle fiber necrosis (filled arrows). In the COX/SDH stain, up to six fibers per cross section were COX negative (blue fibers denoted by asterisks), suggesting mitochondrial dysfunction.
In direct comparison of DHTKD1 variant carriers to non-carriers in cohort 1, no significant differences in demographic or individual clinical parameters were identified (Table 4). However, when grouping the ALS subtypes PMA and LMN-dominant ALS together, DHTKD1 variant carriers more frequently belonged to this grouped subtype with lower motor neuron involvement (2/10, 20% versus 8/215, 3.72% in non-carriers, p = 0.066, two-sided Fisher’s exact test), although the difference was not statistically significant. Additionally, there was a trend towards a higher ALSFRS-R progression rate indicating faster disease progression in DHTKD1 variant carriers compared to non-variant carriers (median ALSFRS-R progression rate/month: 1.17 versus 0.55; p = 0.068, Mann–Whitney U test). Furthermore, patients carrying a DHTKD1 variant more frequently showed severe axonal damage (mean compound motor action potential < 1 mV in median nerve) at initial diagnosis compared to non-variant carriers (3/9, 33.3% versus 14/150, 9.3%; p = 0.057, two-sided Fisher’s exact test), although not quite statistically significant.
Table 4.
Clinical and electrophysiological characteristics of ALS patients from cohort 1 comparing DHTKD1 variant carriers and non-carriers.
| Characteristics |
DHTKD1 Variant Carriers Number (%), Mean ± SD or Median (Range) |
DHTKD1 Non-Variant Carriers Number (%), Mean ± SD or Median (Range) |
p-Value * | Number of Patients with Available Data (Carriers/ Non-Carriers) |
|---|---|---|---|---|
| Sex | ||||
| Male | 6 (60) | 127 (59.1) | 1.0 | 10/215 |
| Age at onset (years) | 71 (49–73) | 63 (25–84) | 0.272 | 10/212 |
| Disease duration (years) a | 2.50 (1.58–9.08) | 2.42 (0.25–19.25) | 0.992 | 10/212 |
| Site of onset | ||||
| Bulbar | 2 (20) | 62 (28.8) | 0.729 | 10/215 |
| Spinal | 8 (80) | 153 (71.2) | 0.729 | 10/215 |
| ALS subtype | ||||
| Classic (Charcot’s) ALS | 6 (60) | 143 (66.5) | 0.737 | 10/215 |
| Bulbar | 2 (20) | 31 (14.4) | 0.644 | 10/215 |
| UMN | 0 (0) | 3 (1.40) | 1.0 | 10/215 |
| Flail arm | 0 (0) | 24 (11.16) | 0.605 | 10/215 |
| Flail leg | 0 (0) | 4 (1.86) | 1.0 | 10/215 |
| Respiratory | 0 (0) | 2 (0.93) | 1.0 | 10/215 |
| PMA | 1 (10) | 3 (1.40) | 0.167 | 10/215 |
| LMN | 1 (10) | 5 (2.33) | 0.241 | 10/215 |
| Grouped PMA and LMN | 2 (20) | 8 (3.72) | 0.066 | 10/215 |
| Disease progression | ||||
| Time to wheelchair (years) | 1.58 (0.92–8.42) | 1.92 (0.25–16.0) | 0.911 | 7/108 |
| Time to NIV (years) | 1.04 ± 0.21 | 2.46 ± 1.45 | 0.130 | 2/37 |
| Time to PEG (years) | 1.17 (1.17–1.42) | 1.83 (0.25–7.58) | 0.160 | 3/51 |
| Weight loss (kg/month) | 0.53 (1.67–0) | 0.25 (6.33–0) | 0.532 | 10/208 |
| ALSFRS-R progression rate/month b | 1.17 (2.17–0.24) | 0.55 (7.0–0.04) | 0.068 | 9/188 |
| Nerve conduction study | ||||
| CMAP in median nerve (mV) c | 3.28 ± 2.74 | 4.44 ± 2.73 | 0.22 | 9/150 |
| Severe axonal damage (mean CMAP <1mV in median nerve) | 3 (33.3) | 14 (9.3) | 0.057 | 9/150 |
| Sensory neuropathy (NCS and/or clinical) | 6 (60) | 85 (39.5) | 0.332 | 10/215 |
Abbreviations: ALS: amyotrophic lateral sclerosis; ALSFRS-R: revised amyotrophic lateral sclerosis functional rating scale; CMAP: compound muscle action potential; LMN: lower motor neuron; NCS: nerve conduction study; NIV: non-invasive ventilation; PEG: percutaneous endoscopic gastrostomy; PMA: progressive muscular atrophy; SD: standard deviation; UMN: upper motor neuron. * Significant difference at p < 0.05. Comparisons between DHTKD1 variant carriers and non-variant carriers were made using the two tailed Fisher’s exact test for dichotomous variables, Mann–Whitney U test or Student’s t-test for continuous variables. a Until last follow-up (VALS054, VALS164), total invasive ventilation (MD086) or death (MD011, MD022, MD025, VALS001, VALS046, VALS095, and VALS102) b ALSFRS-R progression rate/month: high numbers indicate fast progression, c CMAP in-house standard for median nerve > 7 mV.
4. Discussion
Since ALS is known for its heterogeneous genetic architecture [26], we aimed at identifying additional genetic factors that may contribute to or modify the phenotype of ALS patients. DHTKD1 was identified as an interesting candidate gene by WES and a candidate-based approach using a list of genes associated with neurodegeneration (n = 694 genes) in a pilot study of ALS patients of cohort 1. Altogether, in cohort 1, 4.4% of cases, i.e., 10 sporadic ALS patients, and in cohort 2, 2.9% of families, i.e., 12 familial ALS patients, were found to carry rare heterozygous DHTKD1 variants. Collectively, the analysis of both ALS cohorts (n = 643 unrelated cases) yielded the identification of 14 different rare DHTKD1 variants in ALS.
DHTKD1 is a nuclear gene encoding dehydrogenase E1 and transketolase domain containing 1, a protein involved in the final degradative pathway of L-lysine that is critical for mitochondrial metabolism [9,27]. Accordingly, the diminished DHTKD1 expression in vitro resulted in mitochondrial dysfunction and increased production of reactive oxygen species [11,25,28]. There is evidence that the alteration of mitochondrial function occurs early and contributes to the pathogenesis of neurodegenerative diseases including ALS and other neuromuscular disorders [29,30]. Consistently, a number of genetic aberrations causing ALS, FTD, and CMT sensorimotor axonal neuropathy were shown to compromise mitochondrial function, e.g., the GGGGCC repeat expansion in C9orf72 [31], VCP (valosin containing protein) mutations [32], CHCHD10 (coiled-coil-helix-coiled-coil-helix domain containing 10) mutations [33,34], and DYNC1H1 (dynein cytoplasmic 1 heavy chain 1) mutations [35,36]. Based on these findings, it is conceivable that rare DHTKD1 variants that affect mitochondrial metabolism may increase ALS risk and be contributors to the ALS phenotype.
Of the 14 different rare DHTKD1 variants identified here in 643 unrelated ALS cases, all were predicted to be deleterious according to at least one of four prediction tools, four variants were pathogenic according to the ACMG guidelines [18], eight variants were located in the E1_dh dehydrogenase domain of the DHTKD1 protein, two of which were loss-of-function variants predicted to truncate this important functional domain, and seven variants were detected more than once, providing in silico evidence for the pathogenicity of most of the DHTKD1 variants described here. Four DHTKD1 variants detected in our ALS cohorts were previously described in patients with disorders affecting the nervous system, that is autosomal recessive AMOXAD, a metabolic condition characterized by elevated 2-aminoadipate and 2-oxoadipate levels in urine and/or plasma and varying neurological symptoms [9,10], autosomal recessive infantile-onset SMA with cognitive delay [12] and autosomal dominant CMT disease type 2Q (CMT2Q) [21], two neuromuscular disorders, as well as in patients with autosomal dominant eosinophilic esophagitis, sometimes accompanied by muscle weakness [25]. Functional in vitro analyses were previously performed on two of the DHTKD1 variants identified here and support their pathogenicity. Leandro et al. reported that mutant DHTKD1 harboring the c.1364G>A p.(Arg455Gln) variant was less soluble and enzymatically inactive [37]. Similarly, DHTKD1 containing the c.2185G>A p.(Gly729Arg) variant displayed decreased catalytic efficiency for NADH production when assembled into the 2-oxoadipate dehydrogenase complex (OADHc), of which DHTKD1 is a component [38]. Conversely, recent in vitro data suggest that deficient DHTKD1 activity may be partially compensated by oxoglutarate dehydrogenase (OGDH) [39], arguing against a very severe effect of DHTKD1 variants.
In vivo evidence for variant pathogenicity in our study came from a metabolite analysis in ALS patient VALS164 carrying the DHTKD1:c.2185G>A p.(Gly729Arg) variant, an AMOXAD disease-causing mutation [9]. Urine levels of 2-aminoadipate, elevated in AMOXAD patients with biallelic DHTKD1 mutations [9,10,22], were close to the upper reference limit in patient VALS164, although carrying a heterozygous DHTKD1 variant only. A muscle biopsy performed in ALS patient VALS054 diagnosed with the PMA subtype and carrying the DHTKD1:c.209C>G p.(Ala70Gly) variant showed the expected pattern of neurogenic atrophy. Additionally, reduced or missing COX activity was observed in single muscle fibers reflecting heterogeneously distributed mitochondrial dysfunction, potentially related to the ALS phenotype [40,41] and the identified genotype. However, partial COX deficiency may also be associated with other processes, such as ageing. Taken together, epidemiological, in silico, in vitro, and in vivo evidence support a pathogenic effect of 12 of the 14 rare DHTKD1 variants identified here.
Nonsense variants located in the E1_dh dehydrogenase domain of DHTKD1 were significantly more prevalent in ALS patients of cohorts 1 and 2 of this study compared to controls. Such nonsense variants are predicted to result in a truncated DHTKD1 protein devoid of intact functional domains (Figure 1C). One of these variants, DHTKD1:c.1309G>T p.(Glu437*), was previously described in a patient with CMT2, a hereditary motor and sensory axonal neuropathy [21]. A different DHTKD1 nonsense variant in the same domain, c.1455T>G (p.Tyr485*), was reported in a Chinese pedigree with CMT2 and symmetric muscle wasting, predominant weakness of the distal parts of the lower limbs, decreased or absent deep tendon reflexes, and mild to moderate deep sensory impairment [11]. These data suggest a genetic link between ALS and CMT2, and a phenotypic overlap may be expected in patients harboring variants in the same gene. Consistently, the majority (6/10, 60%) of ALS patients of cohort 1 carrying DHTKD1 variants suffered from sensory impairment and/or sensorimotor neuropathy, which is normally not a part of the strictly motor neuron phenotype of ALS, whereby an age-dependent effect cannot be excluded in our patients. Moreover, DHTKD1 variant carriers of ALS cohort 1 were more likely to have predominant LMN involvement than non-variant carriers. Our findings are in line with the fact that a DHTKD1 variant detected in three ALS patients of cohort 1 was previously described in infantile-onset SMA, another motor neuron disorder characterized by muscular atrophy and weakness due to LMN degeneration, whereby the variant was heterozygous in late-onset ALS and homozygous in infantile-onset SMA [12]. Dhtkd1-deficient mice are characterized by progressive weakness and atrophy in the distal limbs with motor and sensory dysfunction aggravated by age, accompanied by decreased nerve conduction velocity [42]. Similarly, nerve conduction studies showed more severe axonal damage in ALS patients with rare DHTKD1 variants compared to non-variant carriers, although these results only reached borderline significance. Taken together, our data provide evidence that rare DHTKD1 variants may modify the ALS phenotype and, possibly, contribute to ALS risk.
In our study, DHTKD1 variants were identified in both sporadic and familial ALS cases. This is in line with recent literature questioning the utility of distinguishing between familial and sporadic ALS for clinical or even genetic counseling purposes. Pathogenic variants in ALS-related genes have not only been detected in familial but also in apparently sporadic ALS, and, as in familial ALS, the ALS risk is increased in relatives of apparently sporadic ALS patients [3]. Despite the observed similarities of DHTKD1 variant carriers here, their phenotype does show some heterogeneity. This may be explained by additional genetic and environmental factors in our ALS patients carrying DHTKD1 variants. Oligogenic inheritance, environmental and lifestyle factors, e.g., smoking and extensive physical exercise, as well as age are discussed to play a role in ALS pathogenesis [26,43,44].
5. Conclusions
Considering that (i) DHTKD1 variants and diminished DHTKD1 expression resulting in reduced enzymatic activity can affect mitochondrial function [9,11,25,28], which is known to be involved in ALS pathogenesis [29,30], (ii) a sensorimotor axonal neuropathy, i.e., CMT2, and a motor neuron disease, i.e., SMA, have been associated with DHTKD1 variants [11,12,21], (iii) variants in CMT-associated genes have been identified in ALS patients and vice versa in earlier reports, e.g., CHCHD10 [33,34], DYNC1H1 [36,45], or VCP [46,47], and (iv) this study reports rare heterozygous DHTKD1 variants in two independent ALS cohorts in 4.4% of cases or 2.9% of families, respectively, with data supporting the pathogenicity of most identified variants, and describes similarities in the ALS phenotype of some DHTKD1 variant carriers, we propose that DHTKD1 variants may contribute to and modify the ALS phenotype.
Acknowledgments
The authors wish to thank the patients and their families for participating in this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13010084/s1, Table S1: Strategy used for the analysis of whole-exome sequencing data of 27 ALS patients of cohort 1, Table S2: PCR primers used for amplification and sequencing of DHTKD1, Table S3: Clinical and electrophysiological characteristics of sporadic ALS patients of cohort 1 carrying rare heterozygous DHTKD1 variants.
Author Contributions
Conceptualization, A.O., S.P. and R.G.W.; data curation, A.O., I.G., H.M., M.W., K.M. and P.M.A.; formal analysis, A.O., I.G., H.M., K.M., F.F., C.-D.L., P.M.A., J.H.W., F.B., S.P. and R.G.W.; funding acquisition, A.O., H.M., M.W., P.M.A., A.C.L., J.H.W., S.P. and R.G.W. investigation, I.G., K.M., F.F., C.-D.L. and G.S.; resources, A.O., O.S.-K., P.M.A., A.C.L., J.H.W. and S.P.; supervision, A.O., H.M., P.M.A., A.C.L., J.H.W., F.B., S.P. and R.G.W.; visualization, I.G. and F.F.; writing—original draft, A.O. and I.G.; writing—review and editing, A.O., I.G., H.M., F.F., C.-D.L., G.S., P.M.A., J.H.W., F.B., S.P. and R.G.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the following: Petermax Müller-Stiftung (grant to S.P. and R.G.W.); the PRACTIS–Clinician Scientist Program of Hannover Medical School, funded by the Deutsche Forschungsgemeinschaft (DFG) (grant no. ME3696/3–1 to A.O.); the Else Kröner-Fresenius-Stiftung (scholarship to M.W., grant to S.P. and R.G.W. within the KlinStrucMed Program of Hannover Medical School); and the DFG (grant no. MA9606/1-1 to H.M.). The work of K.M., A.C.L., and J.H.W. was funded by the German Society for Patients with Neuromuscular Diseases (Deutsche Gesellschaft für Muskelkranke, DGM) and the German Federal Ministry of Education and Research (BMBF): STRENGTH project and German ALS network (MND-NET). The work of P.M.A. was funded by the Swedish Brain Foundation (grants no. 2016-0303, 2018-0310, 2020-0353), the Swedish Research Council (grants no. 2012-3167, 2017-03100), the Knut and Alice Wallenberg Foundation (grants no. 2012.0091, 2014.0305, 2020.0232), the Ulla-Carin Lindquist Foundation, King Gustaf V:s and Queen Victoria’s Freemason’s Foundation.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki (WMA, 1964), and approved by the Ethics Committee of Hannover Medical School, Hannover, Germany (protocol code 6269, date of approval: 16.08.2012) and Ulm University, Ulm, Germany (protocol code 19/12), and by the Research Ethical Committee (FEK) in Umeå, Sweden (Um dnr 94-135, date of approval: 15.09.1994, with later amendments in 1998, 2003, 2014, 2017, and 2018).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study prior to participation.
Conflicts of Interest
A.O. received honoraria from Biogen; O.S.-K. received honoraria from the German Society for Patients with Neuromuscular Diseases (Deutsche Gesellschaft für Muskelkranke, DGM), Novartis, Biogen, Biermann Verlag, MK + S—Medizin, Kommunikation & Service, and the Jain Foundation outside of this work; F.F. received research funding from iOmx Therapeutics; P.M.A. is consultant to Biogen, Roche, Avrion, Regeneron, and Orphazyme; clinical trial site investigator for Biogen, Alexion, Sanofi, AL-S Pharma, Amylyx, and Orphazyme; since 1993 director of the ALS-FTD genetic laboratory at Umeå University Hospital that performs clinical and research genetic testing; member of the ClinGen ALS Gene Curation Expert Panel; S.P. received honoraria from Biogen, Cytokinetics, Desitin Pharma, Novartis, Roche, and Teva. The other authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brown R.H., Jr., Al-Chalabi A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017;377:1602. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 2.Chiò A., Calvo A., Moglia C., Mazzini L., Mora G. Phenotypic heterogeneity of amyotrophic lateral sclerosis: A population based study. J. Neurol. Neurosurg. Psychiatry. 2011;82:740–746. doi: 10.1136/jnnp.2010.235952. [DOI] [PubMed] [Google Scholar]
- 3.Chiò A., Battistini S., Calvo A., Caponnetto C., Conforti F.L., Corbo M., Giannini F., Mandrioli J., Mora G., Sabatelli M., et al. Genetic counselling in ALS: Facts, uncertainties and clinical suggestions. J. Neurol. Neurosurg. Psychiatry. 2014;85:478–485. doi: 10.1136/jnnp-2013-305546. [DOI] [PubMed] [Google Scholar]
- 4.Li H.F., Wu Z.Y. Genotype-phenotype correlations of amyotrophic lateral sclerosis. Transl. Neurodegener. 2016;5:3. doi: 10.1186/s40035-016-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estevez-Fraga C., Magrinelli F., Hensman Moss D., Mulroy E., Di Lazzaro G., Latorre A., Mackenzie M., Houlden H., Tabrizi S.J., Bhatia K.P. Expanding the Spectrum of Movement Disorders Associated with C9orf72 Hexanucleotide Expansions. Neurol. Genet. 2021;7:e575. doi: 10.1212/NXG.0000000000000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osmanovic A., Widjaja M., Förster A., Weder J., Wattjes M.P., Lange I., Sarikidi A., Auber B., Raab P., Christians A., et al. SPG7 mutations in amyotrophic lateral sclerosis: A genetic link to hereditary spastic paraplegia. J. Neurol. 2020;267:2732–2743. doi: 10.1007/s00415-020-09861-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rayaprolu S., Fujioka S., Traynor S., Soto-Ortolaza A.I., Petrucelli L., Dickson D.W., Rademakers R., Boylan K.B., Graff-Radford N.R., Uitti R.J., et al. TARDBP mutations in Parkinson’s disease. Parkinsonism Relat. Disord. 2013;19:312–315. doi: 10.1016/j.parkreldis.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen P.M., Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: What do we really know? . Nat. Rev. Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 9.Danhauser K., Sauer S.W., Haack T.B., Wieland T., Staufner C., Graf E., Zschocke J., Strom T.M., Traub T., Okun J.G., et al. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am. J. Hum. Genet. 2012;91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen J., Te Brinke H., Wanders R.J., Knegt A.C., Oussoren E., Hoogeboom A.J., Ruijter G.J., Becker D., Schwab K.O., Franke I., et al. Genetic basis of alpha-aminoadipic and alpha-ketoadipic aciduria. J. Inherit. Metab. Dis. 2015;38:873–879. doi: 10.1007/s10545-015-9841-9. [DOI] [PubMed] [Google Scholar]
- 11.Xu W.Y., Gu M.M., Sun L.H., Guo W.T., Zhu H.B., Ma J.F., Yuan W.T., Kuang Y., Ji B.J., Wu X.L., et al. A nonsense mutation in DHTKD1 causes Charcot-Marie-Tooth disease type 2 in a large Chinese pedigree. Am. J. Hum. Genet. 2012;91:1088–1094. doi: 10.1016/j.ajhg.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karakaya M., Storbeck M., Strathmann E.A., Delle Vedove A., Hölker I., Altmueller J., Naghiyeva L., Schmitz-Steinkrüger L., Vezyroglou K., Motameny S., et al. Targeted sequencing with expanded gene profile enables high diagnostic yield in non-5q-spinal muscular atrophies. Hum. Mutat. 2018;39:1284–1298. doi: 10.1002/humu.23560. [DOI] [PubMed] [Google Scholar]
- 13.Turner M.R., Barohn R.J., Corcia P., Fink J.K., Harms M.B., Kiernan M.C., Ravits J., Silani V., Simmons Z., Statland J., et al. Primary lateral sclerosis: Consensus diagnostic criteria. J. Neurol. Neurosurg. Psychiatry. 2020;91:373–377. doi: 10.1136/jnnp-2019-322541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amico E., Pasmantier M., Lee Y.W., Weimer L., Mitsumoto H. Clinical evolution of pure upper motor neuron disease/dysfunction (PUMMD) Muscle Nerve. 2013;47:28–32. doi: 10.1002/mus.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cedarbaum J.M., Stambler N., Malta E., Fuller C., Hilt D., Thurmond B., Nakanishi A. The ALSFRS-R: A revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III) J. Neurol. Sci. 1999;169:13–21. doi: 10.1016/S0022-510X(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 16.Labra J., Menon P., Byth K., Morrison S., Vucic S. Rate of disease progression: A prognostic biomarker in ALS. J. Neurol. Neurosurg. Psychiatry. 2016;87:628–632. doi: 10.1136/jnnp-2015-310998. [DOI] [PubMed] [Google Scholar]
- 17.Yilmaz R., Müller K., Brenner D., Volk A.E., Borck G., Hermann A., Meitinger T., Strom T.M., Danzer K.M., Ludolph A.C., et al. SQSTM1/p62 variants in 486 patients with familial ALS from Germany and Sweden. Neurobiol. Aging. 2020;87:139.e9–139.e15. doi: 10.1016/j.neurobiolaging.2019.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweetman L. Organic acid analysis. In: Hommes F.A., editor. Techniques in Diagnostic Human Biochemical Genetics: A Laboratory Manual. Wiley-Liss; New York, NY, USA: 1991. pp. 143–176. [Google Scholar]
- 20.Hoffmann G., Aramaki S., Blum-Hoffmann E., Nyhan W.L., Sweetman L. Quantitative analysis for organic acids in biological samples: Batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin. Chem. 1989;35:587–595. doi: 10.1093/clinchem/35.4.587. [DOI] [PubMed] [Google Scholar]
- 21.Dohrn M.F., Glockle N., Mulahasanovic L., Heller C., Mohr J., Bauer C., Riesch E., Becker A., Battke F., Hortnagel K., et al. Frequent genes in rare diseases: Panel-based next generation sequencing to disclose causal mutations in hereditary neuropathies. J. Neurochem. 2017;143:507–522. doi: 10.1111/jnc.14217. [DOI] [PubMed] [Google Scholar]
- 22.Stiles A.R., Venturoni L., Mucci G., Elbalalesy N., Woontner M., Goodman S., Abdenur J.E. New Cases of DHTKD1 Mutations in Patients with 2-Ketoadipic Aciduria. JIMD Rep. 2016;25:15–19. doi: 10.1007/8904_2015_462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M., et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. J. Am. Med. Assoc. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z.H., Chen Z.T., Zhou R.L., Wang Y.Z. A Chinese pedigree with a novel mutation in GJB1 gene and a rare variation in DHTKD1 gene for diverse Charcot-Marie-Tooth diseases. Mol. Med. Rep. 2019;19:4484–4490. doi: 10.3892/mmr.2019.10058. [DOI] [PubMed] [Google Scholar]
- 25.Sherrill J.D., Kc K., Wang X., Wen T., Chamberlin A., Stucke E.M., Collins M.H., Abonia J.P., Peng Y., Wu Q., et al. Whole-exome sequencing uncovers oxidoreductases DHTKD1 and OGDHL as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight. 2018:3. doi: 10.1172/jci.insight.99922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volk A.E., Weishaupt J.H., Andersen P.M., Ludolph A.C., Kubisch C. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Med. Genet. 2018;30:252–258. doi: 10.1007/s11825-018-0185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemeria N.S., Gerfen G., Nareddy P.R., Yang L., Zhang X., Szostak M., Jordan F. The mitochondrial 2-oxoadipate and 2-oxoglutarate dehydrogenase complexes share their E2 and E3 components for their function and both generate reactive oxygen species. Free Radic. Biol. Med. 2018;115:136–145. doi: 10.1016/j.freeradbiomed.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Xu W., Zhu H., Gu M., Luo Q., Ding J., Yao Y., Chen F., Wang Z. DHTKD1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. 2013;587:3587–3592. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 29.Patten S.A., Armstrong G.A., Lissouba A., Kabashi E., Parker J.A., Drapeau P. Fishing for causes and cures of motor neuron disorders. Dis. Model. Mech. 2014;7:799–809. doi: 10.1242/dmm.015719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith E.F., Shaw P.J., De Vos K.J. The role of mitochondria in amyotrophic lateral sclerosis. Neurosci. Lett. 2019;710:132933. doi: 10.1016/j.neulet.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Gonzalez R., Lu Y., Gendron T.F., Karydas A., Tran H., Yang D., Petrucelli L., Miller B.L., Almeida S., Gao F.B. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim N.C., Tresse E., Kolaitis R.M., Molliex A., Thomas R.E., Alami N.H., Wang B., Joshi A., Smith R.B., Ritson G.P., et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auranen M., Ylikallio E., Shcherbii M., Paetau A., Kiuru-Enari S., Toppila J.P., Tyynismaa H. CHCHD10 variant p. (Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol. Genet. 2015;1:e1. doi: 10.1212/nxg.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannwarth S., Ait-El-Mkadem S., Chaussenot A., Genin E.C., Lacas-Gervais S., Fragaki K., Berg-Alonso L., Kageyama Y., Serre V., Moore D.G., et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eschbach J., Sinniger J., Bouitbir J., Fergani A., Schlagowski A.I., Zoll J., Geny B., René F., Larmet Y., Marion V., et al. Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol. Dis. 2013;58:220–230. doi: 10.1016/j.nbd.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mentis A.F., Vlachakis D., Papakonstantinou E., Zaganas I., Patrinos G.P., Chrousos G.P., Dardiotis E. A novel variant in DYNC1H1 could contribute to human amyotrophic lateral sclerosis-frontotemporal dementia spectrum. Cold Spring Harb. Mol. Case Stud. 2021:mcs-a006096. doi: 10.1101/mcs.a006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leandro J., Khamrui S., Wang H., Suebsuwong C., Nemeria N.S., Huynh K., Moustakim M., Secor C., Wang M., Dodatko T., et al. Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex. ACS Chem. Biol. 2020;15:2041–2047. doi: 10.1021/acschembio.0c00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Nemeria N.S., Leandro J., Houten S., Lazarus M., Gerfen G., Ozohanics O., Ambrus A., Nagy B., Brukh R., et al. Structure-function analyses of the G729R 2-oxoadipate dehydrogenase genetic variant associated with a disorder of l-lysine metabolism. J. Biol. Chem. 2020;295:8078–8095. doi: 10.1074/jbc.RA120.012761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leandro J., Dodatko T., Aten J., Nemeria N.S., Zhang X., Jordan F., Hendrickson R.C., Sanchez R., Yu C., DeVita R.J., et al. DHTKD1 and OGDH display substrate overlap in cultured cells and form a hybrid 2-oxo acid dehydrogenase complex in vivo. Hum. Mol. Genet. 2020;29:1168–1179. doi: 10.1093/hmg/ddaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appel S.H. Is ALS a systemic disorder? Evidence from muscle mitochondria. Exp. Neurol. 2006;198:1–3. doi: 10.1016/j.expneurol.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Crugnola V., Lamperti C., Lucchini V., Ronchi D., Peverelli L., Prelle A., Sciacco M., Bordoni A., Fassone E., Fortunato F., et al. Mitochondrial respiratory chain dysfunction in muscle from patients with amyotrophic lateral sclerosis. Arch. Neurol. 2010;67:849–854. doi: 10.1001/archneurol.2010.128. [DOI] [PubMed] [Google Scholar]
- 42.Xu W.Y., Zhu H., Shen Y., Wan Y.H., Tu X.D., Wu W.T., Tang L., Zhang H.X., Lu S.Y., Jin X.L., et al. DHTKD1 Deficiency Causes Charcot-Marie-Tooth Disease in Mice. Mol. Cell. Biol. 2018:38. doi: 10.1128/MCB.00085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Chalabi A., Hardiman O. The epidemiology of ALS: A conspiracy of genes, environment and time. Nat. Rev. Neurol. 2013;9:617–628. doi: 10.1038/nrneurol.2013.203. [DOI] [PubMed] [Google Scholar]
- 44.van Blitterswijk M., van Es M.A., Hennekam E.A., Dooijes D., van Rheenen W., Medic J., Bourque P.R., Schelhaas H.J., van der Kooi A.J., de Visser M., et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21:3776–3784. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- 45.Weedon M.N., Hastings R., Caswell R., Xie W., Paszkiewicz K., Antoniadi T., Williams M., King C., Greenhalgh L., Newbury-Ecob R., et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2011;89:308–312. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez M.A., Feely S.M., Speziani F., Strickland A.V., Danzi M., Bacon C., Lee Y., Chou T.F., Blanton S.H., Weihl C.C., et al. A novel mutation in VCP causes Charcot-Marie-Tooth Type 2 disease. Brain. 2014;137:2897–2902. doi: 10.1093/brain/awu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.