Abstract
Brassinosteroids (BRs) play essential roles in regulating plant growth and development, however, gaps still remain in our understanding of the BR signaling network. We previously cloned a grain length quantitative trait locus qGL3, encoding a rice (Oryza sativa L.) protein phosphatase with Kelch-like repeat domain (OsPPKL1), that negatively regulates grain length and BR signaling. To further explore the BR signaling network, we performed phosphoproteomic analysis to screen qGL3-regulated downstream components. We selected a 14-3-3 protein OsGF14b from the phosphoproteomic data for further analysis. qGL3 promoted the phosphorylation of OsGF14b and induced the interaction intensity between OsGF14b and OsBZR1. In addition, phosphorylation of OsGF14b played an important role in regulating nucleocytoplasmic shuttling of OsBZR1. The serine acids (Ser258Ser259) residues of OsGF14b play an essential role in BR-mediated responses and plant development. Genetic and molecular analyses indicated that OsGF14b functions as a negative regulator in BR signaling and represses the transcriptional activation activity of OsBZR1. Collectively, these results demonstrate that qGL3 induces the phosphorylation of OsGF14b, which modulates nucleocytoplasmic shuttling and transcriptional activation activity of OsBZR1, to eventually negatively regulate BR signaling and grain length in rice.
Protein phosphatase qGL3/OsPPKL1 modulates the phosphorylation of a 14-3-3 protein, which inhibits the nuclear function of transcription factor OsBZR1 to repress brassinosteroid signaling in rice.
Introduction
Rice (Oryza sativa L.) is one of the most important food crops worldwide and increasing grain yield is a major goal of rice breeding. Plant architecture, which is a crucial factor for grain yield in rice, is determined by plant height, leaf angle, tiller number, panicle morphology, and grain size. Brassinosteroids (BRs) regulate various biological processes in Arabidopsis (Arabidopsis thaliana), including seed germination, stomata formation, vascular differentiation, plant architecture, flowering, male fertility, ovule integument growth, root growth, and senescence (Clouse and Sasse, 1998; Jiang et al., 2013; Chen et al., 2019; Jia et al., 2020). BRs affect many agricultural traits and have been widely used to increase yields in various crops since the 1980s (Divi and Krishna, 2009; Zhang et al., 2014; Nolan et al., 2020). Therefore, manipulating BR biosynthesis and signaling in rice to modify plant architecture represents a feasible approach for improving yields (Tong and Chu, 2018).
The BR signaling pathway in rice is majorly mediated by protein phosphorylation and dephosphorylation. Several components in this pathway have been characterized using molecular approaches. Oryza sativa BRASSINOSTEROID-INSENSITIVE1 (OsBRI1) (Nakamura et al., 2006) and O. sativa BRI1-ASSOCIATED RECEPTOR KINASE1 (OsBAK1) (Li et al., 2009) form a transmembrane BR receptor complex. Oryza sativa BR-SIGNALING KINASE3 (OsBSK3) is a direct substrate of OsBRI1, and the phosphorylation of OsBSK3 by OsBRI1 disrupts the interaction between its TPR and kinase domains (Zhang et al., 2016). In addition, O. sativa GLYCOGEN SYNTHASE KINASES (OsGSK1, OsGSK2, and OsGSK3) (Koh et al., 2007; Tong et al., 2012; Gao et al., 2019), which are homologs of Arabidopsis thaliana BR INSENSITIVE2 (BIN2) (He et al., 2002; Li and Nam 2002), repress BR signaling by negatively regulating the levels of proteins including O. sativa BRASSINAZOLE RESISTANT1 (OsBZR1) (Bai et al., 2007) and DWARF AND LOW-TILLERING (DLT) (Tong et al., 2009). The bri1 SUPPRESSOR1 (BSU1) in Arabidopsis was the first functionally characterized protein phosphatase with Kelch-like domains (PPKL) shown to be a positive effector of BR signaling (Kim et al., 2009). Overexpressing BSU1 suppressed the BR-insensitive phenotype of the bri1 mutant and led to the accumulation of dephosphorylated BES1/BZR1 (Kim et al., 2009). qGL3/OsPPKL1 encodes a protein phosphatase that is the rice homolog of BSU1. qGL3 negatively regulates BR signaling via a combination of dephosphorylation and proteasome-mediated degradation of OsGSK3, which phosphorylates OsBZR1, resulting in the degradation of OsBZR1 in the cytoplasm (Gao et al., 2019).
Interestingly, several components of the BR signaling pathway in rice lack their homologs in Arabidopsis, such as LEAF AND TILLER ANGLE INCREASED CONTROLLER (LIC), DLT, ENHANCED LEAF INCLINATION AND TILLER NUMBER1 (ELT1), O. sativa TAIHU DWARF1 (OsTUD1, a U-box E3 ubiquitin ligase also known as ERECT LEAF1 [ELF1]), O. sativa RELATED TO ABI3/VP1 RAV-LIKE1 (OsRAVL1), O. sativa BRASSINOSTEROID UPREGULATED 1-LIKE1 (OsBUL1), SHORT GRAIN1 (SG1), OsWRKY53, and GRAIN WIDTH5 (GW5). This observation indicates that the BR signaling pathways in rice and Arabidopsis have some species-specific components (Tanaka et al., 2009; Tong et al., 2009; Il Je et al., 2010; Nakagawa et al., 2012; Zhang et al., 2012a; Hu et al., 2013; Liu et al., 2017; Tian et al., 2017; Yang et al., 2017).
Among grain size-associated quantitative trait loci (QTLs) cloned in rice, GS2 (or GL2) is the first QTL that was clearly shown to be involved in BR signaling. GS2/GL2 encodes OsGRF4 and belongs to a transcription factor gene family whose members are targeted by the microRNA396 (miR396). The GS2/GL2 alleles contain mutations in the miR396 targeting sequence and thus show elevated expression (Hu et al., 2015; Che et al., 2016; Duan et al., 2016). We previously cloned a major grain-length QTL qGL3 encoding a protein phosphatase with Kelch-like repeat domains (OsPPKL1). N411, a japonica rice variety with extra-large grains, contains a rare qgl3 allele that causes its long-grain phenotype (Zhang et al., 2012b). The qGL3 protein in N411 contains two amino acid substitutions compared with qGL3 in the variety 9311: an aspartate-to-glutamate substitution in the second Kelch domain and a histidine-to-tyrosine substitution. qGL3 directly interacts with OsGSK3 to dephosphorylate and stabilize this protein (Gao et al., 2019). The near-isogenic line NILqgl3 (carrying the qGL3N411 allele in the 9311 background) exhibited increased grain length compared with 9311 (Zhang et al., 2012b). Treatment with brassinolide (BL), a highly active BR, significantly increased lamina joint bending and coleoptile length in m-qgl3 and NILqgl3, implying that the negative role is abolished in N411 and the BR signaling is therefore induced. Moreover, the two amino acid substitutions in qGL3N411 make its phosphatase inactive (Gao et al., 2019). It was also found that GL3.1, encoded by an allele of qGL3, modulates grain length by dephosphorylating the cell-cycle protein Cyclin-T1;3 (Qi et al., 2012).
In a previous study, we clarified the role of qGL3, which negatively regulates BR signaling and grain length by regulating the phosphorylation level and stability of OsGSK3, modulating OsBZR1 phosphorylation status and subcellular distribution (Gao et al., 2019). To further explore the rice BR signaling network, we performed phosphoproteomic screening using two groups of rice materials: japonica rice cultivar Dongjin and its qGL3 knockout mutant m-qgl3, and the indica rice cultivar 9311 and its near-isogenic line NILqgl3 (Zhang et al., 2012b). In this work, we focused on a 14-3-3 protein OsGF14b for further research. We found that qGL3 induces the phosphorylation of OsGF14b and the phosphorylated OsGF14b plays critical roles in the nucleocytoplasmic shuttling of OsBZR1 to negatively regulate BR signaling. Therefore, the qGL3-OsGSK3-OsBZR1 module was expanded to qGL3→OsGSK3→OsBZR1←OsGF14b←qGL3 module in this work. This research lays the foundation for addressing how the BR signal is transduced and modulates plant architecture in rice.
Results
qGL3 regulates the phosphorylation of OsGF14b to modulate BR signaling
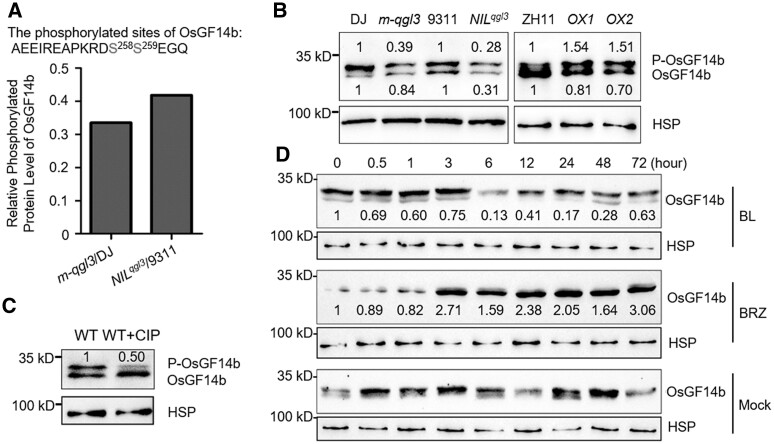
To better understand phosphorylation events in BR signaling affected by qGL3/OsPPKL1, we performed phosphoproteomic screening using young panicles (∼10 cm) from m-qgl3, NILqgl3, and their corresponding wild type (WT) lines. From these differentially expressed phosphorylated proteins, it was found that the phosphorylation status of OsGF14b was altered in both m-qgl3 and NILqgl3 (Figure 1, A). To further validate the protein level and status of OsGF14b, we performed western-blot analysis in WTs and qGL3-related plants. The phosphorylated protein bands migrate more slowly than the unmodified proteins through the gel due to the phosphate attachment. The results showed that OsGF14b has similar phosphorylation tendencies as the phosphoproteomic data indicated (Figure 1, B). Moreover, the absence of the slower-migrating bands following treatment with calf intestine alkaline phosphatase (CIP) confirmed their identity as phosphorylated proteins (Figure 1, C). Since qGL3 plays essential roles in rice BR signaling, we analyzed whether OsGF14b modulates BR signaling. First, we measured the effects of BL and brassinozole (BRZ) treatments on the protein level of OsGF14b. OsGF14b levels substantially decreased in WT plants treated with BL, by contrast, BRZ treatment increased OsGF14b accumulation (Figure 1, D).
Figure 1.
qGL3 induces the phosphorylation of OsGF14b. A, The relative phosphorylated protein level of OsGF14b in two pairs of rice materials (m-qgl3/DJ) and (NILqgl3/9311). Data are from isobaric tags for relative and absolute quantification (iTRAQ)-based phosphoproteomic analysis of two pairs of rice materials in two biological replicates. B, Comparison of the level of OsGF14b in the WT, m-qgl3 mutant, NILqgl3, and qGL3-OXs plants. For the quantitation of protein level, we first normalized to heat shock protein (HSP) (approximately 90 kDa), and then analyzed the protein level of the phosphorylated proteins and the unphosphorylated proteins. The protein level of DJ, 9311, and ZH11 was defined as 1. C, Immunoprecipitated OsGF14b protein from WT was treated with calf intestine alkaline phosphatase (CIP). For the quantitation of protein level, we first normalized to HSP, and then analyzed the protein level of the phosphorylated proteins. The phosphorylated protein level of WT was defined as 1. D, Induction of OsGF14b protein accumulation by BL and brassinazole (BRZ) treatment. OsGF14b was detected by immunoblot assays with anti-OsGF14b antibody. Rice HSP was used as the internal control. The quantification of the BL and BRZ treatment immunoblot results was normalized to HSP, and then normalized to the mock control. The protein levels of OsGF14b in response to “0 h” were defined as “1.”
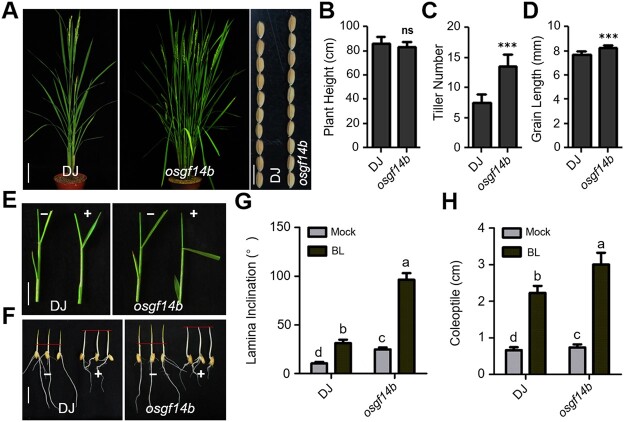
To test whether the loss of OsGF14b function may lead to an altered response to BR, we introduced T-DNA insertion mutant, osgf14b, with abolished OsGF14b protein expression (Supplemental Figure S1). Loss of function of OsGF14b led to larger angles between the leaf sheath and leaf blade compared with the WT. The tiller number was significantly higher and the grains were longer in osgf14b compared with the WT (Figure 2, A–D). Then we analyzed the sensitivity of the mutants to BL using a lamina inclination assay. As expected, osgf14b plants were more sensitive to BL treatment than WT plants (Figure 2, E and G). In addition, we used a coleoptile growth experiment to quantify the BR sensitivity of WT and mutant plants. BL treatment increased coleoptile to a significantly longer length in osgf14b seedlings compared with the WT (Figure 2, F and H). These results indicate that OsGF14b plays a negative role in BR signaling.
Figure 2.
Phenotypes of the osgf14b mutant. A, Morphology of the osgf14b mutant at the reproductive phase. Scale bar for plants, 10 cm; scale bar for grains, 1 cm. B–D, Quantification of plant height (B), tiller number (C), and grain length (D) of the osgf14b mutant compared with the WT. For quantification of plant height and tiller number, data are means ± se (n = 10). For quantification of grain length, data are means ± se (n = 20). The data were compared by Student’s t test. ***P < 0.001, and “ns” means no significance. E and F, Lamina joint inclination and coleoptile elongation response to 1 μM BL in the dark. Scale bar for lamina joint inclination, 1 cm; scale bar for coleoptile, 1 cm. The osgf14b plants showed more sensitive to BL treatment compared with WT. G and H, Statistical analyses of the lamina joint angle and coleoptile length under treatment with 1 μM BL. Values are means ± se (n = 15). Statistical analyses were performed by Duncan’s multiple range tests. The presence of the same lowercase letter denotes a non-significant difference between means (P > 0.05).
Genetic analysis of qGL3 and OsGF14b
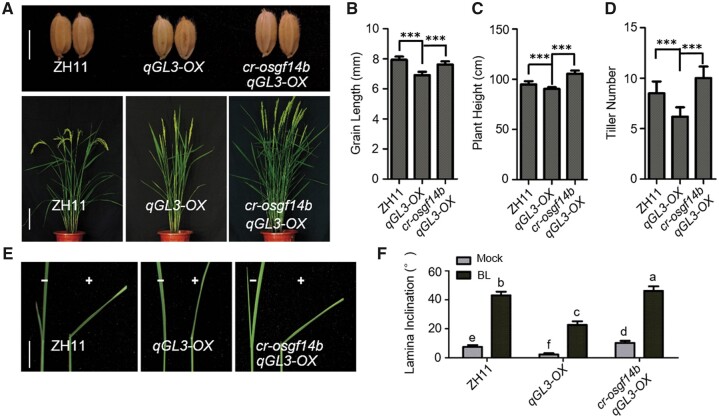
Since qGL3 induces the phosphorylation level of OsGF14b to modulate BR signaling, it is of interest to investigate their genetic relationship. We used CRISPR-Cas9 to knock-out OsGF14b in qGL3-OX plants and obtained the homozygous cr-osgf14b/qGL3-OX line. To ascertain the genotype and heritability of each line, we sequenced the target genes to confirm the genotypes (Supplemental Figure S2). Compared with qGL3-OX, cr-osgf14b/qGL3-OX increased plant height and displayed longer grain length and more tillers (Figure 3, A–D). Then we used the lamina inclination assay to quantify the BR sensitivity of the WT and transgenic plants. BL significantly decreased the lamina joint bending of qGL3-OX seedlings compared with the WT, while knocking out OsGF14b in qGL3-OX plants increased lamina inclination under BL treatment (Figure 3, E and F). These results indicate that OsGF14b may have genetic interactions with qGL3 in rice.
Figure 3.
Genetic analyses of qGL3 and OsGF14b in rice. A, Morphology of the WT, qGL3-OX, and cr-osgf14b/qGL3-OX plants at the reproductive phase. Scale bar for grains, 0.5 cm; scale bar for plants, 10 cm. B–D, Quantification of grain length (B), plant height (C), and tiller number (D) of the qGL3-OX and cr-osgf14b/qGL3-OX plants compared with the WT. For quantification of grain length, data are means ± se (n = 20). For quantification of plant height and tiller number, data are means ± se (n = 10). The data were compared by Student’s t test. ***P < 0.001. E, Lamina joint inclination response to 1 μM BL in the dark. Scale bar for lamina joint inclination, 1 cm. F, Statistical analyses of the lamina joint angle under treatment with 1 μM BL. Data are means ± se (n = 15). Statistical analyses were performed by Duncan’s multiple range tests. The presence of the same lowercase letter denotes a non-significant difference between means (P > 0.05).
Phosphorylation of OsGF14b at Ser258Ser259 induced by qGL3 promotes interaction to OsBZR1
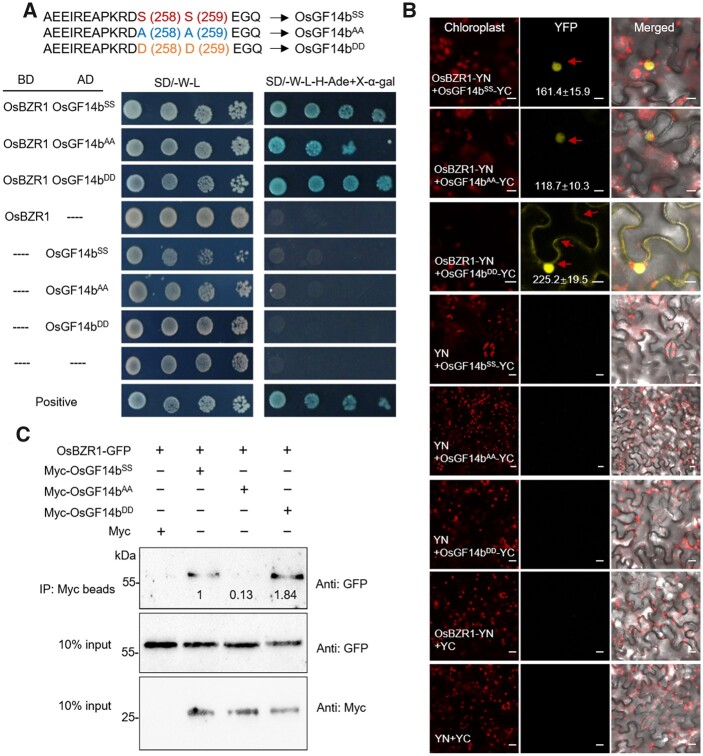
To further explore the function of OsGF14b in BR signaling pathway, we examined the interactions of OsGF14b with qGL3 and OsBZR1, the critical BR components in rice. Our data showed that only OsBZR1 interacts with OsGF14b (Figure 4, A). In the qGL3-dependent phosphoproteomic data, the phosphorylated OsGF14b decreased in both m-qgl3 and NILqgl3 plants, and the phosphorylation sites are Ser258 and Ser259 in OsGF14b protein. Phosphorylation of 14-3-3 proteins is important for their functions. To determine the phosphorylation sites of OsGF14b, we replaced these Ser (S) residues of OsGF14b with Ala (A) residue, producing OsGF14bS258AS259A (OsGF14bAA), to mimic non-phosphorylation. We also mutated Ser (S) residues with Asp (D) to generate OsGF14bDD (phosphomimic mutated form; Figure 4, A). To explore whether these mutations affect the interaction of OsGF14b and OsBZR1, we performed a yeast two-hybrid assay. The results showed that OsBZR1 interacted with all OsGF14bSS, OsGF14bAA, and OsGF14bDD in yeast cells (Figure 4, A). To analyze their interactions in vivo, in planta interactions of OsGF14b and OsBZR1 were examined in Nicotiana benthamiana leaf with bimolecular fluorescence complementation (BiFC) assays. These results showed that interaction of OsGF14bSS-OsBZR1 was detected in nucleus and OsGF14bDD-OsBZR1 was observed in both the cytosolic and nuclear fractions (Figure 4, B). Moreover, OsGF14bAA-OsBZR1 interaction was observed in nuclear, showing a reduced interaction intensity compared with OsGF14bSS-OsBZR1 interaction (Figure 4, B). In addition, we performed a coimmunoprecipitation (co-IP) assay, and observed a stronger interaction intensity between OsGF14bDD-OsBZR1 compared with OsGF14bAA-OsBZR1 interaction (Figure 4, C). OsGF14bSS still interacted with OsBZR1, but the immunoprecipitation product reduced substantially, indicating that the phosphorylation of OsGF14b affects its interaction with OsBZR1.
Figure 4.
The interaction analyses between OsGF14bSS, OsGF14bAA, OsGF14bDD, and OsBZR1. A, Yeast two-hybrid analyses of OsGF14bSS, OsGF14bAA, OsGF14bDD, and OsBZR1. B, BiFC analyses of OsGF14bSS, OsGF14bAA, OsGF14bDD, and OsBZR1 interaction. The YFP fluorescence signals and autofluorescence signals from chloroplasts were pseudo-colored as yellow and red, respectively. Scale bars, 20 μm. The fluorescent signal intensities of YFP were analyzed using Zeiss Image Browser software. Ten N. benthamiana cells per combination (OsBZR1-YN+OsGF14bSS-YC, OsBZR1-YN+OsGF14bAA-YC, OsBZR1-YN+OsGF14bDD-YC) were used to analyze data. C, Interaction between OsBZR1 and OsGF14bSS, OsGF14bAA, OsGF14bDD was detected by co-IP, respectively, in N. benthamiana. Input, extracted proteins; IP, immunoprecipitated proteins; anti-Myc, Myc antibodies; anti-GFP, GFP antibodies. For quantification of the immunoblot results, the relative protein levels of OsBZR1-GFP co-expressed with Myc-OsGF14bSS were defined as 1.
OsGF14b interaction increases cytoplasmic localization and decreases transcriptional activation activity of OsBZR1
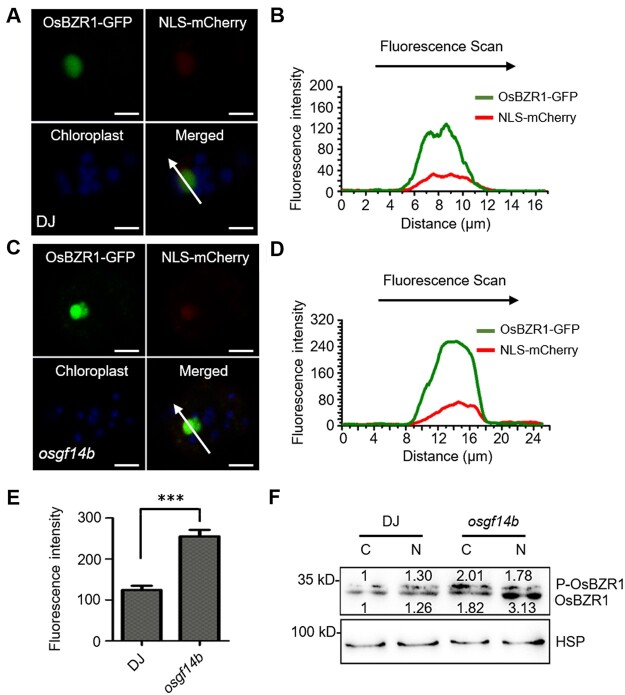
14-3-3 proteins usually regulate their target proteins by altering their stability, enzymatic activity, or subcellular localization (Wilson et al., 2016). To analyze the effects of OsGF14b on the subcellular localization of OsBZR1, we transfected OsBZR1-GFP into the WT and osgf14b mutant plants. We detected the strong OsBZR1-GFP signals in the nuclei of protoplasts from the osgf14b mutant (Figure 5, A and C). The peak of OsBZR1-GFP signal overlapped with that of NLS-mCherry (Figure 5, B and D). Analysis of the OsBZR1-GFP intensities in the nucleus revealed that GFP signals significantly increased in osgf14b mutant plants (Figure 5, E). We further analyzed the protein level and status of OsBZR1 in the nucleus and cytoplasm. Nuclear and cytoplasmic fractions were prepared from the mature leaves of the osgf14b mutant and WT plants. Immunoblotting showed that the protein level of OsBZR1 in the nucleus was largely increased in osgf14b mutant compared with DJ (Figure 5, F). These results indicate that OsGF14b influences the nuclear localization of OsBZR1.
Figure 5.
OsBZR1 subcellular distribution is modulated by OsGF14b. A and C, Subcellular localization of OsBZR1 in the protoplasts of the WT and osgf14b mutant plants. NLS-mCherry was used as a nuclear marker, the OsBZR1-GFP and NLS-mCherry signals match well in the nucleus. The GFP fluorescence signals, mCherry fluorescence signals, and autofluorescence signals from chloroplasts were pseudo-colored as green, red, and blue, respectively. Scale bars, 5 μm. B and D, The fluorescent signal was analyzed using Zeiss Image Browser software. Different signal peaks of OsBZR1-GFP were detected in the nucleus and cytoplasm. The fluorescence intensities of the regions indicated by arrows are shown. E, The statistical analysis of the fluorescence intensities. For quantification of fluorescence intensities, data are means ± se (n = 10). The data were compared by Student’s t test. ***P < 0.001. F, The localization of phosphorylated OsBZR1 was affected by OsGF14b. Proteins of the WT and osgf14b plants were separately extracted from the cytoplasm and nucleus. C and N represent the cytoplasm and nucleus, respectively.
Next, to investigate whether the interaction intensity with OsGF14b affects the subcellular localization of OsBZR1, we co-expressed OsBZR1-GFP with Myc-OsGF14bSS, Myc-OsGF14bAA, and Myc-OsGF14bDD in N. benthamiana leaf cells. OsBZR1-GFP could be detected in the cytosol in the presence of Myc-OsGF14bSS and Myc-OsGF14bDD, and in the nucleus in the presence of Myc and Myc-OsGF14bAA (Figure 6, A). Moreover, the nuclear and cytoplasmic proteins were extracted from N. benthamiana leaves. The phosphorylated OsBZR1 accumulated both in nucleus and cytoplasm in the presence of Myc-OsGF14bSS compared with that in the presence of Myc (Figure 6, B). We further investigated whether OsGF14bDD and OsGF14bAA regulate the protein status and subcellular localization of OsBZR1. Dephosphorylated OsBZR1 decreased in the presence of OsGF14bDD, and there is no obvious change when it was co-expressed with OsGF14bAA (Figure 6, B). These data suggest that qGL3 induced the phosphorylation of OsGF14b, which in turn, increases the interaction intensity between OsGF14b and OsBZR1, and maintains OsBZR1 in cytoplasm to negatively regulate BR signaling.
Figure 6.
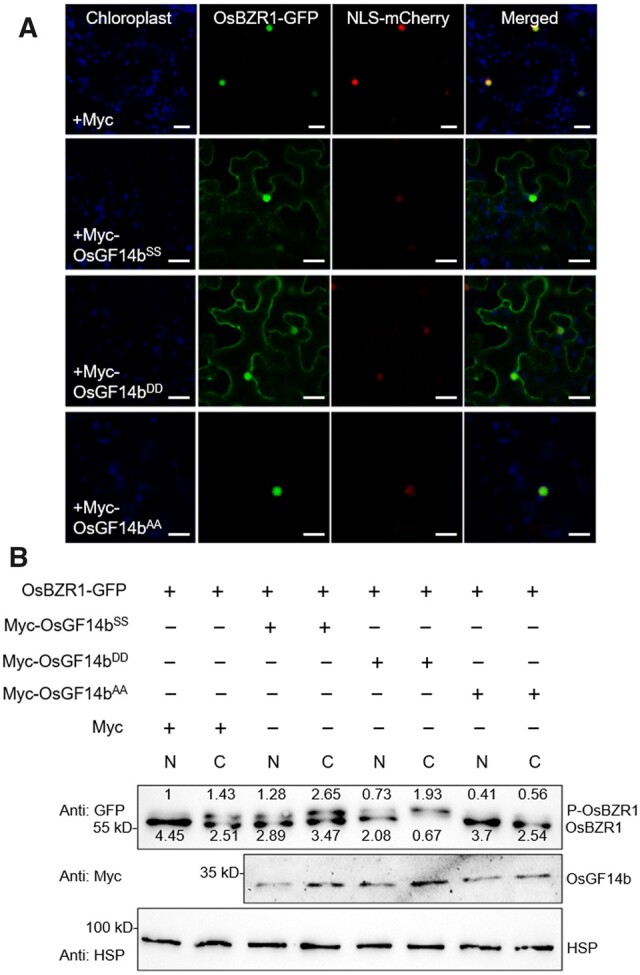
The phosphorylation of OsGF14b induces the cytoplasmic localization of OsBZR1. A, Subcellular localization of OsBZR1 in the presence of OsGF14bSS, OsGF14bDD, and OsGF14bAA. The GFP fluorescence signals, mCherry fluorescence signals, and autofluorescence signals from chloroplasts were pseudo-colored as green, red, and blue, respectively. Scale bars, 10 μm. B, Immunoblot analyses of OsBZR1 protein in the presence of OsGF14bSS, OsGF14bDD, and OsGF14bAA. The nuclear (N) and cytosol (C) proteins were separated by CeILytic PN Isolation/Extraction kit (Sigma), and OsBZR1 proteins were detected by immunoblot analyses with an anti-GFP antibody. For quantification of the immunoblot results, the relative protein levels of phosphorylated OsBZR1-GFP in cytoplasm co-expressed with Myc were defined as 1.
Furthermore, we measured the expression levels of BR biosynthetic and signaling related genes, including OsD2, O. Sativa DWARF (OsDWF), O. Sativa DWARF4 (OsDWF4), OsGSK3, OsBZR1, and O. Sativa INCREASED LAMINA INCLINATION1 (OsILI1), in the WT and cr-osgf14b plants (Supplemental Figure S2) using reverse transcription quantitative PCR (RT-qPCR). These results showed that expression level of OsD2 was downregulated in cr-osgf14b, and OsDWF, OsDWF4, OsBZR1, and OsILI1 were upregulated in cr-osgf14b plants (Supplemental Figure S3). OsBZR1 can directly bind to the promoter regions of a number of genes to regulate their expression, and OsILI1 is a direct target of OsBZR1 in rice (Zhang et al., 2009). We hypothesized that OsGF14b could regulate the transcription activation activity of OsBZR1 as the tight nuclear–cytoplasm shuttling regulation on OsBZR1 by OsGF14b. We then conducted dual-luciferase assays using rice protoplast to confirm the role of OsGF14b on OsBZR1 activity. We used the promotor of OsILI1 to drive the luciferase gene (LUC) as a reporter, and OsBZR1-GFP, OsGF14bAA-GFP, OsGF14bSS-GFP, OsGF14bDD-GFP, and GFP were used as the effectors (Figure 7, A). The OsILI1pro-LUC reporter activity was largely activated by OsBZR1-GFP. When we transformed OsBZR1-GFP together with OsGF14bSS-GFP, the LUC reporter activity was significantly reduced, and the LUC reporter activity level became much lower when incubated with OsGF14bDD-GFP. But there is no obvious change of the LUC activity when co-expressed with OsBZR1-GFP and OsGF14bAA-GFP compared with OsBZR1-GFP alone (Figure 7, B). These findings suggest that phosphorylation at Ser258 and Ser259 residues promotes nuclear export of OsBZR1 and reduces the transcriptional activation activity of OsBZR1.
Figure 7.
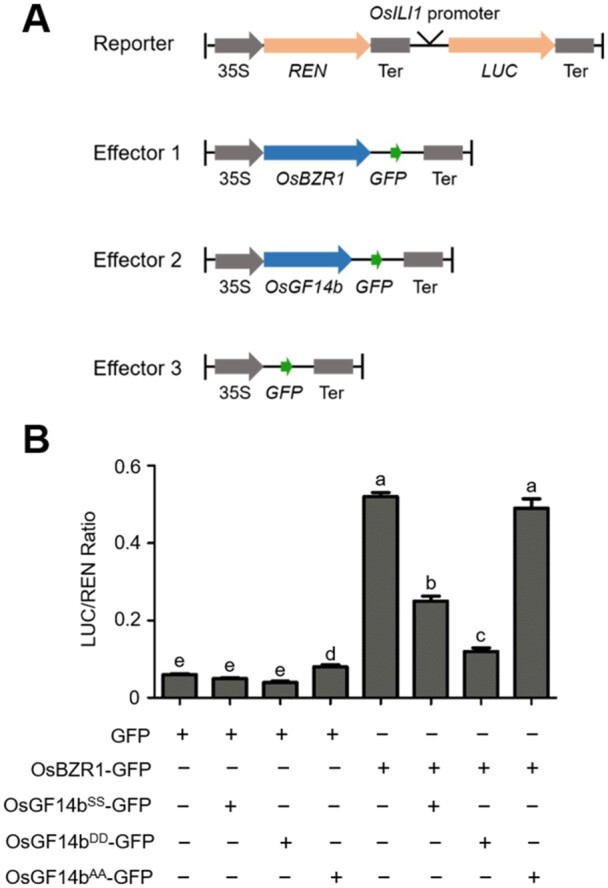
OsGF14b regulates the transcriptional activation activity of OsBZR1. A, Schematic diagrams of the dual-luciferase reporter and effector constructs. The firefly luciferase (LUC) gene driven by the OsILI1 promoter was used as the reporter. The Renilla luciferase (REN) reporter gene was controlled by the Cauliflower mosaic virus promoter (35S) and terminator (Ter). For the effectors, OsBZR1 and OsGF14b were fused with GFP. B, Transient gene expression assays in rice protoplast cells. The LUC reporter gene was co-transfected with OsBZR1-GFP, OsGF14bSS-GFP, OsGF14bDD-GFP, OsGF14bAA-GFP, or both. Data are means ± se (n = 3). Statistical analyses were performed by Duncan’s multiple range tests. The presence of the same lowercase letter denotes a non-significant difference between means (P > 0.05).
Genetic analysis of OsGF14b and OsBZR1
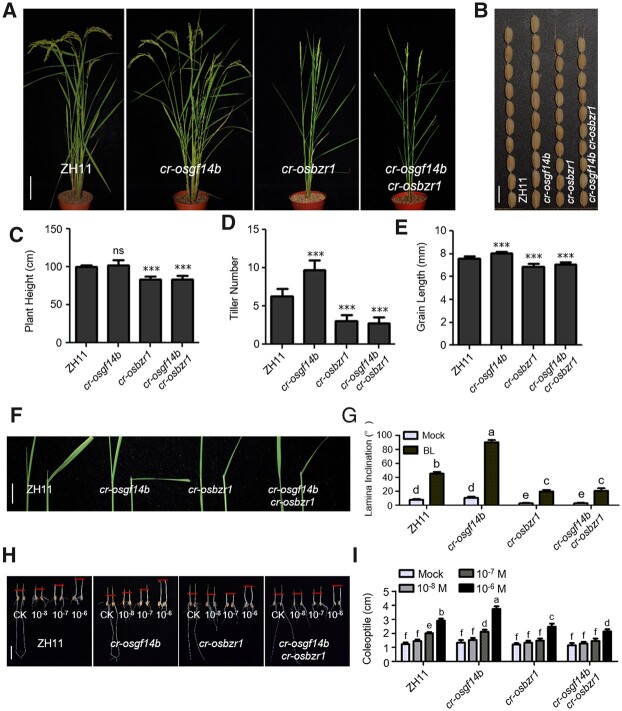
To analyze the genetic interaction of OsGF14b and OsBZR1, we used CRISPR/Cas9 system to create multiple mutants (specific target sites on each gene are shown in Supplemental Table S1; Supplemental Figure S2). In the cr-osgf14b/cr-osbzr1 double mutant, knocking out OsBZR1 suppressed the plant height of cr-osgf14b (Figure 8, A and C). The tiller number was similar in cr-osgf14b/cr-osbzr1 and cr-osbzr1 plants (Figure 8, A and D). Moreover, cr-osgf14b/cr-osbzr1 plants formed shorter grains than cr-osgf14b plants (Figure 8, B and E). These results suggest that OsBZR1 likely functions downstream of OsGF14b.
Figure 8.
Genetic analyses of OsGF14b and OsBZR1. A and B, Morphology of the WT, cr-osgf14b, cr-osbzr1, and cr-osgf14b/cr-osbzr1 plants at the reproductive phase. Scale bar for grains, 1 cm; scale bar for plants, 10 cm. C–E, Quantification of plant height (C), tiller number (D), and grain length (E) of the cr-osgf14b, cr-osbzr1, and cr-osgf14b/cr-osbzr1 plants compared with the WT. For quantification of plant height and tiller number, data are means ± se (n = 10). For quantification of grain length, data are means ± se (n = 20). The data were compared with the WT by Student’s t test. ***P < 0.001, and “ns” means no significance. F, Lamina joint inclination response to 1 μM BL in the dark. Scale bar for lamina joint inclination, 1 cm. G, Statistical analyses of the lamina joint angle under treatment with 1 μM BL. Data are means ± se (n = 15). Statistical analyses were performed by Duncan’s multiple range tests. The presence of the same lowercase letter denotes a non-significant difference between means (P > 0.05). H, Coleoptile elongation of the WT, cr-osgf14b, cr-osbzr1, and cr-osgf14b/cr-osbzr1 plants in response to BL. Scale bar for coleoptile, 2 cm. I, Quantitative comparison of coleoptile length of these plants. Data are means ± se (n = 15). Statistical analyses were performed by Duncan’s multiple range tests. The presence of the same lowercase letter denotes a non-significant difference between means (P > 0.05).
The lamina joint inclination, coleoptile length, and root growth inhibition are widely used to reflect sensitive responses to BR in rice. To further investigate the relationship between OsGF14b and OsBZR1, we examined the effects of exogenously applied BL on these plants. We found that BL significantly increased the lamina joint bending of cr-osgf14b plants compared with WT but had opposite impact on cr-osbzr1 and cr-osgf14b/cr-osbzr1 (Figure 8, F and G). As shown in Figure 8, H, BL increased the coleoptile length in cr-osgf14b plants compared with WT, but not in cr-osbzr1 and cr-osgf14b/cr-osbzr1 seedlings (Figure 8, H and I). Together with the biochemical evidences, these genetic evidences strengthened the conclusion that the BR signaling involves the actions of qGL3-OsGF14b and OsGF14b-OsBZR1.
Discussion
In previous studies, we found that the m-qgl3 mutant in DJ background and NILqgl3 containing the qgl3 allele from N411 in 9311 background display enhanced BR-signaling phenotypes, suggesting that qGL3 negatively regulates grain length and BR signaling in rice (Gao et al., 2019). Since qGL3 is a protein phosphatase, we analyzed a set of differentially phosphorylated proteins regulated by qGL3. We used two groups of materials: m-qgl3/DJ and NILqgl3/9311 for phosphoproteomics analysis. In the common proteins verified in phosphoproteomic screenings, we found a 14-3-3 protein OsGF14b showing the decreased phosphorylation level in the absence of qGL3, suggesting that qGL3 positively modulates the phosphorylation on OsGF14b.
In our previous work, qGL3 directly dephosphorylates and stabilizes OsGSK3, which negatively regulates BR signaling. Moreover, the inhibition of OsGSK3 degradation via the overexpression of qGL3 increases the level of phosphorylated OsGSK3 in the nucleus. OsGSK3 phosphorylates OsBZR1 in nucleus and the phosphorylated OsBZR1 shuttles to the cytoplasm for degradation to repress BR signaling (Gao et al., 2019). Indeed, the regulation of the nucleocytoplasmic trafficking of proteins by hormones and environmental stress has been widely reported in plants. Bai et al. (2007) supports a model that the OsGF14c interacts with phosphorylated OsBZR1 and inhibits OsBZR1 by retaining it in the cytoplasm, whereas BRs activate OsBZR1 by inducing its dephosphorylation and increasing its nuclear localization and transcriptional activity. However, how phosphorylated OsBZR1 shuttles from nucleus to the cytoplasm remains unclear. We hypothesize that OsGF14b, which was identified from qGL3-regulated phosphorylated proteins, plays an essential role in the nucleocytoplasmic shuttling of OsBZR1.
14-3-3 proteins are phosphopeptide-binding proteins that are highly conserved in all eukaryotes. These proteins interact with numerous cellular proteins in a sequence-specific and phosphorylation-dependent manner. 14-3-3 proteins have been shown to interact with large numbers of proteins in plants, including metabolic enzymes, signaling proteins, and transcription factors (DeLille et al., 2001; de Vries, 2007; Wilson et al., 2016). The main role of 14-3-3 proteins was thought to bind to and modulate the functions of phosphorylated proteins. In Arabidopsis, the essential roles for 14-3-3 proteins were also demonstrated in BR signal transduction (Gampala et al., 2007; Wang et al., 2011). The 14-3-3 proteins could interact with phosphorylated BZR1, while the mutation in 14-3-3 binding site and BIN2-phosphorylation site in BZR1 abolished BZR1 binding to 14-3-3 proteins, leading to the increased nuclear localization of BZR1 and enhanced BR signaling in Arabidopsis. In the model proposed by Wang et al. (2021), BR signaling first recruits phosphorylated BZR1 to the nucleus where it is dephosphorylated by protein phosphatase 2A (PP2A) and both BZR1 phosphorylation and dephosphorylation occur in the nucleus, where 14-3-3 proteins promote the phosphorylated BZR1 from nucleus (Wang et al., 2021). Similarly, mutation of a putative 14-3-3-binding site of OsBZR1 abolished its interaction with the 14-3-3 proteins in rice and 14-3-3 proteins inhibit OsBZR1 function at least in part by reducing its nuclear localization (Bai et al., 2007). These studies suggest that 14-3-3 proteins play essential roles in the nucleocytoplasmic shuttling of BZR1 in Arabidopsis and rice. However, how 14-3-3 protein is regulated by BR signaling and then modulates nucleocytoplasmic shuttling of BZR1 is still unclear.
We found that the phosphorylation of OsGF14b strengthened the interaction intensity with OsBZR1 and increased its localization in the cytoplasm (Figures 4–6). Most of the identified phosphorylation sites in 14-3-3 proteins locate in C terminus and we found that Ser258 and Ser259 are major phosphorylated residues of OsGF14b. As expected, the interaction of OsGF14bAA and OsBZR1 remained majorly in the nucleus (Figure 6, A). The phosphorylated OsGF14b interacts with OsBZR1 to decrease its protein level and repress the transcription activation activity of OsBZR1. In total, qGL3 induces the phosphorylation of OsGF14b and enhances the binding between phosphorylated OsGF14b and OsBZR1 to mediate the nucleocytoplasmic shuttling of OsBZR1, resulting in the repressed BR signaling.
Furthermore, identification of the protein kinase (PK) that phosphorylates OsGF14b will shed more light on the molecular mechanism of qGL3-OsGF14b-OsBZR1 module. This PK remains to be explored. It has been reported that R-X-X-S/T is a target sequence for calcium-regulated PKs (CDPKs), therefore Ser259 in R-D-S-S of OsGF14b may be phosphorylated by a CDPK (Hong et al., 2011; Khan et al., 2014). Moreover, these two serine acids are positioned in the extension of a typical SUMOylation motif, with the target Lys255. Since phosphorylation impacts SUMOylation and SUMOylation regulates nuclear distribution, abundance, and activity of proteins in various signal transduction cascades (Khan et al., 2014; Shahpasandzadeh et al., 2014), it will be of interest to investigate whether OsGF14b is SUMOylated and if this involves BR signaling.
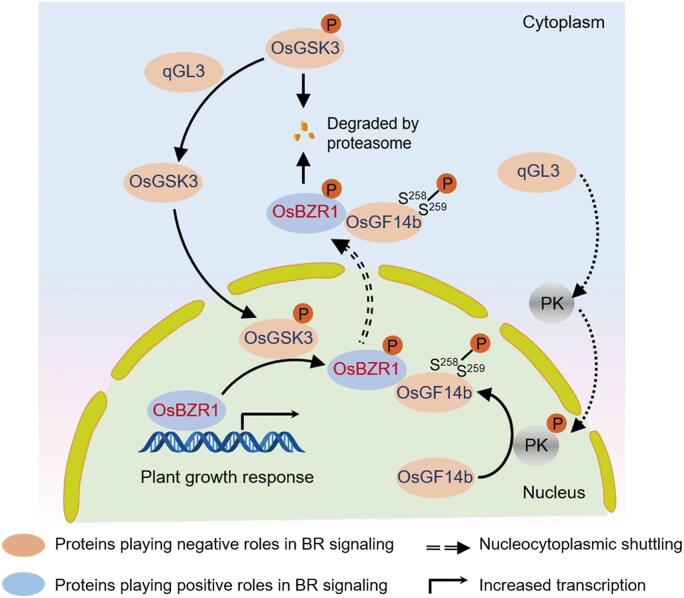
Based on these findings, we propose a model illustrating the relationship between protein level of qGL3 and intensity of BR signaling. In the absence of BR, qGL3 inhibits the degradation of OsGSK3 by dephosphorylating OsGSK3 and causes the OsGSK3 accumulation (Gao et al., 2019). Moreover, the accumulation of OsGSK3 increases the level of phosphorylated OsGSK3 in the nucleus. OsBZR1 was phosphorylated by OsGSK3 in the nucleus and promoted proteasome-mediated degradation of P-OsBZR1 and reduced BR signaling. Here, OsGF14b functions in the nucleocytoplasmic trafficking of OsBZR1. qGL3 induced the phosphorylation of OsGF14b and increased the interaction intensity of OsGF14b-OsBZR1. OsBZR1 is exported from the nucleus and is targeted for degradation in the cytoplasm with the accumulation of OsGF14b. Moreover, phosphorylated OsGF14b decreased in m-qgl3 plants, resulting in the accumulation of dephosphorylated OsBZR1 and induced BR signaling (Figure 9). Collectively, qGL3 dephosphorylates and inhibits the degradation of OsGSK3, leading to the increased protein level of P-OsBZR1. Furthermore, qGL3 induced the interaction intensity of P-OsGF14b and P-OsBZR1, thus, P-OsBZR1 shuttles to the cytoplasm for degradation. Therefore, qGL3-OsGSK3-OsBZR1 module was expanded to qGL3→OsGSK3→OsBZR1←OsGF14b←qGL3 module in this work (Figure 9).
Figure 9.
A proposed qGL3-OsGF14b-OsBZR1 module regulating BR signaling. The phosphorylation of OsGF14b is regulated by qGL3. Meanwhile, OsGF14b modulates subcellular distribution of OsBZR1. In the presence of qGL3, qGL3 inhibits the degradation of OsGSK3 by dephosphorylating OsGSK3 and causes OsGSK3 accumulation. Moreover, accumulation of OsGSK3 by overexpression of qGL3 also increases the level of phosphorylated OsGSK3 in the nucleus. OsBZR1 is phosphorylated by OsGSK3 in the nucleus and promotes proteasome-mediated degradation of P-OsBZR1 and reduced BR signaling. On the other hand, qGL3 induces phosphorylation of OsGF14b and increases the interaction intensity of OsGF14b-OsBZR1. OsBZR1 is exported from the nucleus and targeted for degradation in the cytoplasm with the accumulation of OsGF14b. Moreover, phosphorylated OsGF14b decreases in m-qgl3 plants, resulting in the accumulation of dephosphorylated OsBZR1 and induced BR signaling. The serine acids (Ser258Ser259) residues of OsGF14b play an essential role in BR-mediated responses and plant development. The “P” in orange circles indicates phosphorylation.
BRs regulate many essential agronomic traits in rice, including grain size, plant height, lamina joint bending, tiller number, flowering, and stress tolerance, highlighting the great potential for utilization of BRs to improve plant architectures and yields (Zhang et al., 2014; Zuo and Li 2014; Nolan et al., 2020). For the breeding application of OsGF14b in rice, knocking out OsGF14b in rice increased grain length and tiller number, implying its breeding potential. Thus, studies of the functional mechanisms of BRs in crop plants could facilitate the use of BR-related traits in the field. Gain- or loss-of functions of these BR signaling genes including qGL3, OsGSK3, OsGF14b, and OsBZR1 or their combinations may result in the improvement of plant architecture and increase of grain length and tiller number, suggesting their potential application values in enhancing rice production.
Materials and methods
Plant materials
Rice (O. sativa L.) varieties 9311 (xian/indica), Dongjin (DJ, geng/japonica), and Zhonghua 11 (ZH11, geng/japonica) were used as the WTs. The T-DNA insertion mutants in the DJ backgrounds were obtained from Kyung Hee University, Korea (http://cbi.khu.ac.kr/RISD_DB.html). The constructs used for gene editing OsGF14b and OsBZR1 via CRISPR/Cas9 were designed as previously described (Gao et al., 2019). All constructs were confirmed by sequencing. The edited targets are listed in Supplemental Table S1. NILqgl3 was developed in the genetic background of indica variety 9311 and the qgl3 allele is from the japonica N411 background (Zhang et al., 2012b). To evaluate tiller number and plant height, 10 plants were examined before harvesting. For each line, the lengths of 20 filled grains were measured with an electronic digital caliper (WSEEN, Hangzhou, China).
BL and BRZ treatments
The WTs, osgf14b mutant, qGL3-OX, cr-osgf14b/qGL3-OX, cr-osgf14b, cr-osbzr1, and cr-osgf14b/cr-osbzr1 were grown in the field or greenhouse under a 30°C/25°C day/night cycle. BL and BRZ (Sigma, USA) were dissolved in DMSO to suitable concentrations as stock solutions. The lamina joint inclination assay and excised leaf segment assay were performed as described previously (Gao et al., 2019). Thirty plants were used per test. Leaf angle was measured with ImageJ software. For the coleoptile growth experiments, 30 rice seeds were grown on 0.3% (w/v) agar medium with or without BL at 30°C under continuous darkness for 5 d. To measure protein levels, BL and BRZ treatments were performed using WT plants. The leaves of 2-week-old WT seedlings were cut into 0.5 cm segments and immersed in 1 μM BL and 1 μM BRZ containing 0.1% (v/v) Triton X-100 for various periods of time. An equal volume of solvent (DMSO) was used as the mock treatment.
RNA extraction and RT-qPCR
Total RNA was extracted with Trizol reagent (Takara, Japan) according to the manufacturer’s instruction. First-strand cDNA was synthesized using HiScript II Q RT SuperMix for RT-qPCR (+gDNA wiper) Kit (Vazyme, Nanjing, China). The cDNA was used as a template in a 10-μL PCR amplification. For RT-qPCR, SYBR Green I was added to the reaction and amplified on a Roche 480 real-time PCR detection system. The transcript data were calculated by Roche’s software and the relative expression level was calculated by 2−ΔΔCt. Each experiment was performed with three replicates. The primers for RT-qPCR are listed in Supplemental Table S1.
Yeast two-hybrid assays
For the yeast two-hybrid assays, OsBZR1 were cloned into the pGBKT7 vector, and OsGF14b were cloned into the pGADT7 vector, resulting in OsBZR1-BD and OsGF14b-AD, respectively. The reporter gene assay was performed following the manufacturer’s instructions (Clontech, USA). The yeast cells were cultured on SD/-W-L and SD/-W-L-H-A medium containing X-α-gal at 30°C for 3 d in the dark. SD/-W-L is yeast culture medium without tryptophan and leucine. SD/-W-L-H-A is culture medium without tryptophan, leucine, histidine, and adenine. The PCR primers used for the yeast two-hybrid assays are listed in Supplemental Table S1.
Bimolecular fluorescence complementation
For BiFC analysis, OsBZR1 was cloned into the pSPYNE173 vector, and OsGF14b was cloned into the pSPYCE (M) vector. The plasmids were transformed into Agrobacterium tumefaciens strain EHA105. Agrobacterium tumefaciens cells containing each construct were prepared and mixed to an OD600 of 0.5:0.5. YFP fluorescence was visualized under a confocal laser-scanning microscope at 40–48 h after infiltration. YFP fluorescence was observed with a confocal laser scanning microscope (LSM780, Carl Zeiss, Jena, Germany). The YFP fluorescence was excited with a 488-nm laser and emission spectra was collected between 520 and 540 nm for YFP. The PCR primers used for the BiFC assays are listed in Supplemental Table S1.
Co-IP assay
Myc-OsGF14b and OsBZR1-GFP were transformed in Agrobacterium strain EHA105 and injected into N. benthamiana leaves. Total proteins were extracted from the samples with lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2 mM DTT, 0.5% NP-40, protease inhibitor cocktail [P9599; Sigma]), incubated with Anti-Myc Affinity Gel (Sigma) for 2–3 h, and washed three times with buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2 mM DTT, 0.1% [v/v] NP-40). After adding 2× SDS loading buffer to the tubes, the samples were denatured at 95°C for 10 min. OsBZR1-GFP was detected with anti-GFP antibody and Myc-OsGF14b was detected with anti-Myc antibody (ab32072, Abcam).
Transient transcription dual-luciferase assay
Full-length coding sequence of OsBZR1, OsGF14b (S/A/D) was cloned into pAN580, and effectors were generated. The promoter regions (upstream of the ATG) of OsILI1 were amplified and cloned into pGreenII 0800-LUC vector and used as reporter. The resulting effector and reporter constructs were cotransfected into rice protoplasts. The Renilla luciferase (REN) gene directed by 35S promoter in the pGreenII 0800-LUC vector was used as an internal control. Firefly LUC and REN activities were measured with a Dual-Luciferase reporter assay kit using a GloMax 20/20 luminometer (Promega). The LUC activity was normalized to REN activity and LUC/REN ratios were calculated. For each plasmid combination, six independent transformations were performed.
Immunoblotting
To detect OsGF14b protein, anti-OsGF14b was prepared by immunizing rabbits with purified GST-OsGF14b fusion proteins (Abclonal, Wuhan, China). The detection of HSP was used as a control for equal loading. Young leaf tissue was ground into a powder in liquid nitrogen and suspended in lysis buffer (100 mM Tris–HCl pH 7.5, 300 mM NaCl, 2 mM EDTA pH 8.0, 1% [v/v] Triton X-100, 10% [v/v] glycerol, and 1× phosphatase inhibitor cocktail and 1× protease inhibitor cocktail). The total extracts were centrifuged at 15,000 g for 30 min at 4°C; the supernatant (containing total proteins) was analyzed by immunoblotting with the appropriate antibodies. For data analysis, protein levels were calculated using Tanon Image Software. The immunoblot results from plants subjected to BL and BRZ treatment were normalized to HSP.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: qGL3, Os03g44500; OsGSK3, Os02g14130; OsBZR1, Os07g39220; OsGF14b, Os04g38870; OsILI1, Os04g54900; OsD2, Os01g10040; OsDWF4, Os03g12660; OsDWF, Os03g40540. The phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner Repository with the dataset identifier PXD025494.
Supplemental data
Supplemental Figure S1. Molecular characterization of the osgf14b mutant.
Supplemental Figure S2. The sequences of the target genes.
Supplemental Figure S3. Expression patterns of BR biosynthesis and signaling-related genes in the seedlings of the WT and cr-osgf14b plants.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the supports from Jiangsu Collaborative Innovation Center for Modern Crop Production and Cyrus Tang Seed Innovation Center, Nanjing Agricultural University.
Funding
This work was supported by the National Key Research and Development Program (2016YFD0100400), the Natural Science Foundation of China (32071918, 32000227), and Youth Science Foundation of Jiangsu Province (SBK2019040714).
Conflict of interest statement. None declared.
J.H. and X.G. conceived the original research plans, analyzed the data, and wrote the manuscript. X.G. performed most of the experiments. J.Z., G.C., H.D., J.L., R.W., Y.W., J.Y., and W.Z. conducted some experiments. J.H. and H.Z. supervised the project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Ji Huang (huangji@njau.edu.cn).
References
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104:13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che RH, Tong HN, Shi BH, Liu YQ, Fang SR, Liu DP, Xiao YH, Hu B, Liu LC, Wang HR, et al. (2016) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2 [DOI] [PubMed] [Google Scholar]
- Chen WY, Lv MH, Wang YZ, Wang PA, Cui YW, Li MZ, Wang RA, Gou XP, Li J (2019) BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat Commun 10:4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49:427–451 [DOI] [PubMed] [Google Scholar]
- de Vries SC (2007) 14-3-3 proteins in plant brassinosteroid signaling. Dev Cell 13:162–164 [DOI] [PubMed] [Google Scholar]
- DeLille JM, Sehnke PC, Ferl RJ (2001) The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol 126:35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Krishna P (2009) Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol 26:131–136 [DOI] [PubMed] [Google Scholar]
- Duan PG, Ni S, Wang JM, Zhang BL, Xu R, Wang YX, Chen HQ, Zhu XD, Li YH (2016) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang WQ, Deng ZP, Bai MY, Guan SH, Lalonde S, Sun Y, Gendron JM, et al. (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13:177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XY, Zhang JQ, Zhang XJ, Zhou J, Jiang ZS, Huang P, Tang ZB, Bao YM, Cheng JP, Tang HJ, et al. (2019) Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31:1077–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang YL, Li JM, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99:10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JY, Chae MJ, Lee IS, Lee YN, Nam MH, Kim DY, Byun MO, Yoon IS (2011) Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72:27–36 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang YX, Fang YX, Zeng LJ, Xu J, Yu HP, Shi ZY, Pan JJ, Zhang D, Kang SJ, et al. (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465 [DOI] [PubMed] [Google Scholar]
- Hu XM, Qian Q, Xu T, Zhang YE, Dong GJ, Gao T, Xie Q, Xue YB (2013) The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G alpha subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet 9:e1003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il Je B, Piao HL, Park SJ, Park SH, Kim CM, Xuan YH, Park SH, Huang J, Do Choi Y, An G, et al. (2010) RAV-like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22:1777–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Chen LG, Yin G, Yang X, Gao Z, Guo Y, Sun Y, Tang W (2020) Brassinosteroids regulate outer ovule integument growth in part via the control of INNER NO OUTER by BRASSINOZOLE-RESISTANT family transcription factors. J Integr Plant Biol 62:1093–1111 [DOI] [PubMed] [Google Scholar]
- Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH (2013) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162:1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Unterholzner SJ, Chen TT, Eremina M, Wurzinger B, Bachmair A, Teige M, Sieberer T, Isono E, et al. (2014) Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat Commun 5:4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan SH, Sun Y, Deng ZP, Tang WQ, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11:1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, Lee SC, Kim MK, Koh JH, Lee S, An G, Choe S, Kim SR (2007) T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65:453–466 [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7:791–806 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301 [DOI] [PubMed] [Google Scholar]
- Liu JF, Chen J, Zheng XM, Wu FQ, Lin QB, Heng YQ, Tian P, Cheng ZJ, Yu XW, Zhou KN, et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Tanaka A, Tanabata T, Ohtake M, Fujioka S, Nakamura H, Ichikawa H, Mori M (2012) SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol 158:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan TM, Vukasinovic N, Liu D, Russinova E, Yin Y (2020) Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32:295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22:1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahpasandzadeh H, Popova B, Kleinknecht A, Fraser PE, Outeiro TF, Braus GH (2014) Interplay between sumoylation and phosphorylation for protection against alpha-synuclein inclusions. J Biol Chem 289:31224–31240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, et al. (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151:669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Li X, Zhou W, Ren Y, Wang Z, Liu Z, Tang J, Tong H, Fang J, Bu Q (2017) Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol 175:1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong HN, Chu CC (2018) Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci 23:1016–1028 [DOI] [PubMed] [Google Scholar]
- Tong HN, Jin Y, Liu WB, Li F, Fang J, Yin YH, Qian Q, Zhu LH, Chu CC (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816 [DOI] [PubMed] [Google Scholar]
- Tong HN, Liu LC, Jin Y, Du L, Yin YH, Qian Q, Zhu LH, Chu CC (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24:2562–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang C, Zhang C, Wang N, Lu D, Wang J, Zhang S, Wang ZX, Ma H, Wang X (2011) Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell 21:825–834 [DOI] [PubMed] [Google Scholar]
- Wang RJ, Wang RX, Liu MM, Yuan WW, Zhao ZY, Liu XQ, Peng YM, Yang XR, Sun Y, Tang WQ (2021) Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc Natl Acad Sci USA 118:e2101838118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Swatek KN, Thelen JJ (2016) Regulation of the regulators: Post-translational modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front Plant Sci 7:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BJ, Lin WH, Fu FF, Xu ZH, Xue HW (2017) Receptor-like protein ELT1 promotes brassinosteroid signaling through interacting with and suppressing the endocytosis-mediated degradation of receptor BRI1. Cell Res 27:1182–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BW, Wang XL, Zhao ZY, Wang RJ, Huang XH, Zhu YL, Yuan L, Wang YC, Xu XD, Burlingame AL, et al. (2016) OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol 170:1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Bai MY, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33:683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu YY, Guo SY, Zhu JY, Huan G, Liu HH, Wang L, Luo GZ, Wang XJ, Chong K (2012a) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8:651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu JX, Zhu JY, Wang H, Zhang ZG, Wang WF, Sun Y, Zhao J, Sun XH, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21:3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Wang JF, Huang J, Lan HX, Wang CL, Yin CF, Wu YY, Tang HJ, Qian Q, Li JY, et al. (2012b) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109:21534–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Li JY (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48:99–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.