Abstract
SAMBA has been identified as a plant-specific regulator of the anaphase-promoting complex/cyclosome (APC/C) that controls unidirectional cell cycle progression in Arabidopsis (Arabidopsis thaliana), but so far its role has not been studied in monocots. Here, we show the association of SAMBA with the APC/C is conserved in maize (Zea mays). Two samba genome edited mutants showed growth defects, such as reduced internode length, shortened upper leaves with erect leaf architecture, and reduced leaf size due to an altered cell division rate and cell expansion, which aggravated with plant age. The two mutants differed in the severity and developmental onset of the phenotypes, because samba-1 represented a knockout allele, while translation re-initiation in samba-3 resulted in a truncated protein that was still able to interact with the APC/C and regulate its function, albeit with altered APC/C activity and efficiency. Our data are consistent with a dosage-dependent role for SAMBA to control developmental processes for which a change in growth rate is pivotal.
Designing gRNAs and interpreting the effects of genome edited mutations at the DNA level should be done with caution, because translation re-initiated can lead to truncated, functional proteins.
Introduction
A phytomer is a repetitive structural component that originates from the shoot apical meristem and that forms the basis of the plant body plan. A typical phytomer consists of a node, a leaf, an internode, and an axillary meristem. A typical characteristic of monocot leaf development is the sheathing leaf base leading to older leaves to be wrapped around younger developing leaves and internodes (Conklin et al., 2019). A mature maize leaf consists of the leaf sheath and blade, which are separated by the auricle and the ligule. Auricles are two wedge-shaped structures on either side of the midvein, which act as hinges to allow the leaf blade to project at an angle from the vertical leaf sheath axis. The size of the auricles and ligule has been shown to be a major determinant of the leaf angle (Kong et al., 2017). When the leaf becomes visible from the whorl, two key growth parameters, the leaf elongation rate (LER) or maximal growth rate and the leaf elongation duration or how long maximal growth is maintained, can be determined by daily measuring its length until it is maximal (Nelissen et al., 2013).
Both leaves and internodes grow post-embryonically through cell division and cell expansion. The contribution of cell division to the final organ size is determined by the number of dividing cells and the cell division duration and rate that is controlled by the cell cycle progression. The cell cycle is one of the most highly conserved processes among eukaryotes and is a strictly controlled process in which a cell duplicates its genome and equally distributes the genome copies between its two daughter cells. To ensure correct duplication and distribution of the genome and separation of the two daughter cells, the cell cycle has several checkpoints, which only allow progression to the next step if the previous step has been correctly completed (De Veylder et al., 2007). Ubiquitin-mediated degradation of essential checkpoint proteins is a major mechanism ensuring unidirectional progression through the cell cycle checkpoints (Eloy et al., 2015; Sharma et al., 2016), for which E3 ligases provide substrate specificity. The SKP/CUL/RBX/F-box protein complex and the anaphase-promoting complex/cyclosome (APC/C) play a major role in the regulation of cell cycle progression by ubiquitin-mediated degradation (Eloy et al., 2015). The APC/C is a large multi-subunit complex that is conserved among yeast, mammals, and plants (Lima et al., 2010; Eloy et al., 2015) and consists of a catalytic and substrate recognition module, a tetratricopeptide repeat lobe, and a platform module. The catalytic core of the APC/C comprises APC2 and APC11 (Gmachl et al., 2000; Leverson et al., 2000; Tang et al., 2001). Substrate specificity of the catalytic core is obtained through association with APC10 and the CELL DIVISION CYCLE 20 and CELL CYCLE SWITCH 52 (CCS52) families of APC/C co-activator proteins (Da Fonseca et al., 2011; Chang et al., 2015).
During the mitotic cell cycle, the APC/C has been shown to target cell cycle regulators such as CYCLINs (CYCs), CYCLIN-DEPENDENT KINASE inhibitors, and proteins involved in DNA replication (Castro et al., 2005). The ability of the APC/C to mark proteins for degradation depends on the presence of destruction signals (degrons), such as the D-box or KEN-box motifs, in its target proteins (Glotzer et al., 1991; Pfleger and Kirschner, 2000). Besides mitotic cell division, the APC/C is also involved in regulating endoreduplication, which is a different type of cell cycle consisting of several rounds of DNA synthesis without subsequent cell division (De Veylder et al., 2011). The onset of endoreduplication often coincides with the exit from the mitotic cell cycle and the onset of cell differentiation (Breuer et al., 2010). Furthermore, the endocycle may serve as a mechanism to sustain cellular expansion without cell division, as increased endocycle rates often correspond with an increased final cell size (Melaragno et al., 1993), although this is not always the case (Beemster et al., 2002; De Veylder et al., 2011). Furthermore, several plant-specific APC/C inhibitors have also been identified, such as SAMBA (Eloy et al., 2012). In Arabidopsis (Arabidopsis thaliana), SAMBA has been shown to directly interact with APC3b, and the SAMBA gene is highly expressed during early development (Eloy et al., 2012). SAMBA loss of function results in an increased cell proliferation due to a shortened cell cycle duration, leading to a larger meristem size as well as the formation of larger leaves, roots, and seeds (Eloy et al., 2012).
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein9 (CAS9) system allows to efficiently introduce double-strand DNA breaks (DSBs) to a specific target site in the genome. Nonhomologous end-joining is the main mechanism to repair these DSBs, which can introduce small insertions or deletions (indels), resulting in a shift in the open reading frame, leading to a premature stop codon in the expressed transcript. Normally, the aberrant mRNAs are degraded by nonsense-mediated decay resulting in a loss of function (Doudna and Charpentier, 2014). Recently, multiple studies further analyzed the post-transcriptional effect of indels on the target gene, showing that alternative splicing (skipping the mutated exon) and/or translation re-initiation downstream of the frameshift mutation can produce truncated functional proteins in cell lines, animals, and plants (Tang et al., 2018; Smits et al., 2019). When targeting 136 genes in human near-haploid cells using the CRISPR system, approximately one-third of the residual proteins were the result of skipping of the mutated exon or translation re-initiation (Smits et al., 2019). Also in rice, de novo alternative splicing of rice (Oryza sativa) Auxin/Indole-3-Acetic Acid 23 (OsIAA23) circumvented the premature stop and thereby preserved the wild-type (WT) phenotype in a CRISPR/Cas9 mutant (Jiang et al., 2019).
Here, we found that the interaction of SAMBA and APC/C was conserved in maize and remained stable in the cell division and cell expansion zone. CRISPR/Cas9 mutants in the maize SAMBA ortholog displayed dwarfism, erect upper leaves, reduced organ, and tissue growth, but the onset and severity of the phenotypes was altered in two alleles although the frameshift occurred at the same position. The difference in the allelic strength was caused by the formation of a truncated protein due to translation re-initiation in one allele, while no detectable protein or a very short N-terminal peptide was formed in the null allele. Phenotypic analysis demonstrated that the knockout SAMBA mutant showed an increased cell production level by a higher cell division rate but reduced cell size, and mainly affected developmental processes for which a change in growth rate is instrumental, such as ligule formation and internode elongation. The truncated SAMBA protein retained partial functionality and was still able to interact with the APC/C. In addition, the partial complementation of the null allele by the truncated protein showed that SAMBA functions in a dose-dependent manner to control the cell division rate.
Results
SAMBA interacts with the APC/C in vivo
To verify whether the maize SAMBA ortholog also interacts with the APC/C, the full-length SAMBA encoding sequence fused to the GSrhino tag (SAMBA-GS; Van Leene et al., 2015) was expressed under the control of the UBIQUITIN promoter (Coussens et al., 2012) in the maize B104 inbred line. Affinity purification followed by mass spectrometry (AP/MS) was performed on SAMBA-GS embryogenic calli, identifying orthologs of the core components of the APC/C, which were previously identified as interacting proteins with SAMBA in Arabidopsis (Eloy et al., 2012; Table 1;Supplemental Data Set S1). Furthermore, APC15b, previously not identified in Arabidopsis, was enriched in maize callus. In addition to these known components and regulators of the APC/C, 56 proteins that were significantly enriched compared to the control purifications were identified. None of these proteins were previously found to interact with SAMBA in Arabidopsis, but some were reported as putative targets of APC/C in human cells (Horn et al., 2011; Supplemental Table S1 and Supplemental Data Set S1). Among these proteins are several members of the DYNAMIN-RELATED PROTEIN (DRP) family, i.e. DRP1A, DRP1E, DRP2A, DRP2B, and DRP5B. Furthermore, CALLOSE SYNTHASE, which is involved in cell plate synthesis and is regulated by DRPs (Verma, 2001) was retrieved as a SAMBA-interacting protein.
Table 1.
Known APC subunits identified through AP/MS from maize callus, leaf division zone (DZ), and expansion zone (EZ) and in Arabidopsis using SAMBA-GSrhino as bait
| Protein ID | At ortholog | Description | In maize callus |
In maize leaf DZ | In maize leaf EZ | In Arabidopsis | |
|---|---|---|---|---|---|---|---|
| −log(p) | Difference | ||||||
| GRMZM2G157878 | AT1G32310 | SAMBA | 7.05 | 11.07 | x | x | x |
| GRMZM2G392710 | AT2G20000 | APC3b | 6.6 | 10.11 | x | x | x |
| GRMZM2G089296 | AT2G39090 | APC7 | 7.08 | 9.76 | x | x | x |
| GRMZM2G170591 | AT3G48150 | APC8a | 6.95 | 10.16 | x | x | x |
| GRMZM2G168886 | AT2G04660 | APC2 | 7.55 | 8.87 | x | x | x |
| GRMZM2G053766 | AT4G21530 | APC4 | 9.27 | 9.5 | x | x | x |
| GRMZM2G053980 | AT5G05560 | APC1 | 8.72 | 7.93 | x | x | x |
| GRMZM2G012220 | AT5G05560 | APC1 | 6.19 | 7.81 | x | x | x |
| GRMZM2G431251 | AT1G06590 | APC5 | 6.39 | 7.51 | x | x | x |
| GRMZM2G054247 | AT2G18290 | APC10a | 6.62 | 7.12 | x | x | x |
| GRMZM2G354696 | AT5G05560 | APC1 | 4.67 | 6.48 | x | x | x |
| GRMZM2G020201 | AT5G63135 | APC15b | 5.05 | 4.27 | x | x | |
| GRMZM2G064005 | AT4G11920 | CCS52A2 | 3.91 | 3.92 | x | x | x |
| GRMZM2G159427 | AT4G22910 | CCS52A1 | 4.47 | 3.46 | x | x | |
| GRMZM2G166684 | AT1G78770 | APC6b | 4.83 | 3.02 | x | x | x |
| GRMZM2G147603 | AT1G78770 | APC6a | NA | NA | x | x | |
| GRMZM5G821639 | AT5G13840 | CCS52B | NA | NA | x | ||
Protein identifications are based on significant enrichment compared to the non-SAMBA-related GIF2 and GIF3 control purifications grouped together (n = 3). Arabidopsis orthologs were determined using PLAZA 3.0 based on best-hit Interparalog families (BHIF) (Proost et al., 2015). Difference: log2 (LFQ ratio [SAMBA/GIF]) presents the log2 fold change in protein abundance in GIF samples compared to SAMBA samples. Significance of enrichment was calculated using permutation-based FDR-corrected t test (FDR = 0.01, S0 = 1). x indicates that a subunit was identified in this sample; NA = not applicable.
In addition, seven independent stable transgenic plants were obtained of which SAMBA-GS4, SAMBA-GS5, and SAMBA-GS6 showed a high accumulation of the fusion protein (Supplemental Figure S1A). Using AP/MS, we mapped the dynamics of the SAMBA-interacting proteins in the division and expansion zone of the growing SAMBA-GS5 leaf four. All APC/C components identified in the maize callus, except APC6a and CCS52B, were retrieved in the leaf tissue (Table 1). Label-free quantification (LFQ) of co-purified proteins showed that the abundance of SAMBA and of the majority of the identified APC/C subunits did not change significantly between both zones (Supplemental Figure S2). However, APC7 and even more pronounced CCS52B were found to be significantly enriched in the division zone (Supplemental Figure S2 and Supplemental Data Set S2). Furthermore, 20 out of the 56 additional proteins that were identified after affinity enrichment from callus, were also identified after affinity enrichment from leaf tissue, including DRP1A, DRP1E, DRP2A, DRP2B, and DRP5B (Supplemental Table S1). Nevertheless, none of these proteins were significantly enriched in either developmental subzone (Supplemental Data Set S2).
samba frameshift mutants are dwarfed with decreased organ size
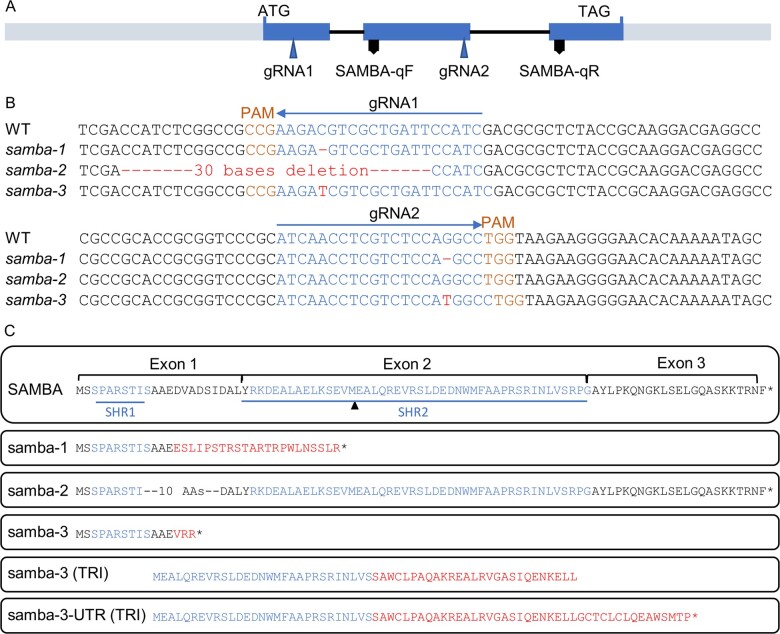
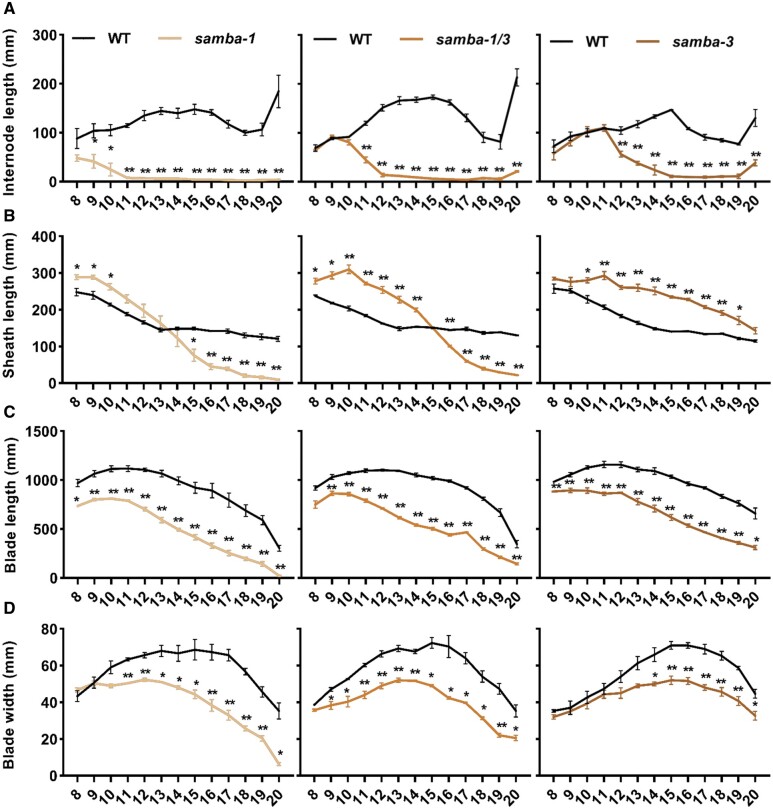
Because ectopic expression of SAMBA-GS in maize did not induce observable phenotypes in leaf and plant growth (Supplemental Figure S1, B and C), the function of SAMBA during maize development was investigated by generating mutants using CRISPR/Cas9 with a dual gRNA approach (Figure 1A). Both gRNAs had a very high Specificity Score (91% for the first gRNA and 96% for the second gRNA) in CRISPOR (Concordet and Haeussler, 2018), indicating a very low risk of off-targets. The samba-1 allele contained a single nucleotide deletion at both target sites, which caused a frameshift in the SAMBA encoding sequence, resulting in 22 missense amino acids (AAs) prior to a premature stop codon (Figure 1, B and C). The samba-2 allele contained a 30-nucleotide deletion at the first target site, which caused an in-frame deletion of 10 AAs while no mutation occurred at gRNA2 (Figure 1, B and C). The samba-3 allele contained a single T insertion at both target sites (Figure 1B), resulting in a frameshift at the same location as the samba-1 allele but with only three missense AAs prior to the premature stop codon (Figure 1C). The T0 samba mutants were backcrossed with B104 and the T1 heterozygote plants without Cas9 were self-pollinated to obtain the T2 generation. The T3 plants obtained by self-pollinating the T2 heterozygotes were further analyzed. We compared the phenotypes of the samba T3 plants 70 d after sowing with those of their WT segregating siblings. The phenotype of samba-2 was similar to the WT (Figure 2A), while samba-1 mutants were severely dwarfed (plant height was reduced by 79.7%; Figure 2, A and B; Supplemental Table S2), which was due to a significantly decreased internode length from internode 9 onward (Figure 3A). The average mature cell length of samba-1 internode 9 was significantly decreased by 70.8% (Supplemental Figure S3A). The samba-3 plants displayed a semi-dwarfed phenotype (plant height was decreased by 64.7%) because the internodes from internode 12 onward were significantly reduced (Figures 2, A and B and 3, A; Supplemental Table S2). The average mature cell length of samba-3 internode 12 was significantly decreased by 28.9% (Supplemental Figure S3B). Both samba-1 and samba-3 plants transitioned from vegetative development into reproductive development but with limited to no anthesis, and despite the presence of silking, seed production was unsuccessful even in reciprocal crosses with the WT. Leaf development was also affected in the samba mutants. The sheath length of lower and higher leaves was significantly increased and significantly decreased, respectively, in samba-1, whereas all leaf sheath lengths were significantly increased in samba-3 (Figure 3B). The failure of internode elongation at the later formed phytomers restricted the growth of leaf sheath and blade in samba-1. Furthermore, both blade length and width were significantly reduced in samba-1 and samba-3; for the leaf width this was significant (P < 0.05) earlier in samba-1 than in samba-3 (Figure 3, C and D). The average mature leaf 12 cell length was significantly reduced by 31.3% and 14.5% in samba-1 and samba-3, respectively, compared with their WT (Supplemental Figure S3, C and D). Also, the transition between the leaf sheath and lamina was affected in the samba-1 and samba-3 alleles. The ligule was elongated and elliptical in shape and the auricle size was extremely decreased in both mutants, resulting in an erect leaf architecture (Figure 2C;Supplemental Figure S4). The samba-1 leaves showed this ligule phenotype from leaf 8 onward, whereas the samba-3 ligule phenotype was visible from leaf 10 onward (Figure 2C). Furthermore, the length of the root system did not differ significantly compared with the WT, but visually, the samba mutants showed a decreased root density (Supplemental Figure S5). To conclude, the samba-1 and samba-3 mutants showed similar growth phenotypes, but the onset of the phenotypes differed between the two alleles. The mutant phenotypes could be complemented to the WT upon introgression of a SAMBA overexpression line, SAMBA-GS5, into both samba lines (Figure 4), which confirmed that the growth defects were caused by the samba-1 and samba-3 mutations.
Figure 1.
samba mutants obtained through CRISPR/Cas9 gene editing. A, Structural representation of the maize SAMBA gene showing the target sites of the two gRNAs. B, The gRNA sequences (blue), PAM (orange), and mutation sites (red) of samba alleles. C, The amino acid sequences of SAMBA WT and putative samba mutant isoforms from translation re-initiation (TRI). SAMBA homology region 1 (SHR1) and SHR2, as defined by Eloy et al. (2012) are marked in green and the missense AAs are indicated in red. SAMBA-qF and SAMBA-qR indicate the position of the forward and reverse primers, respectively, used for SAMBA RT-qPCR. Asterisk represents a stop codon.
Figure 2.
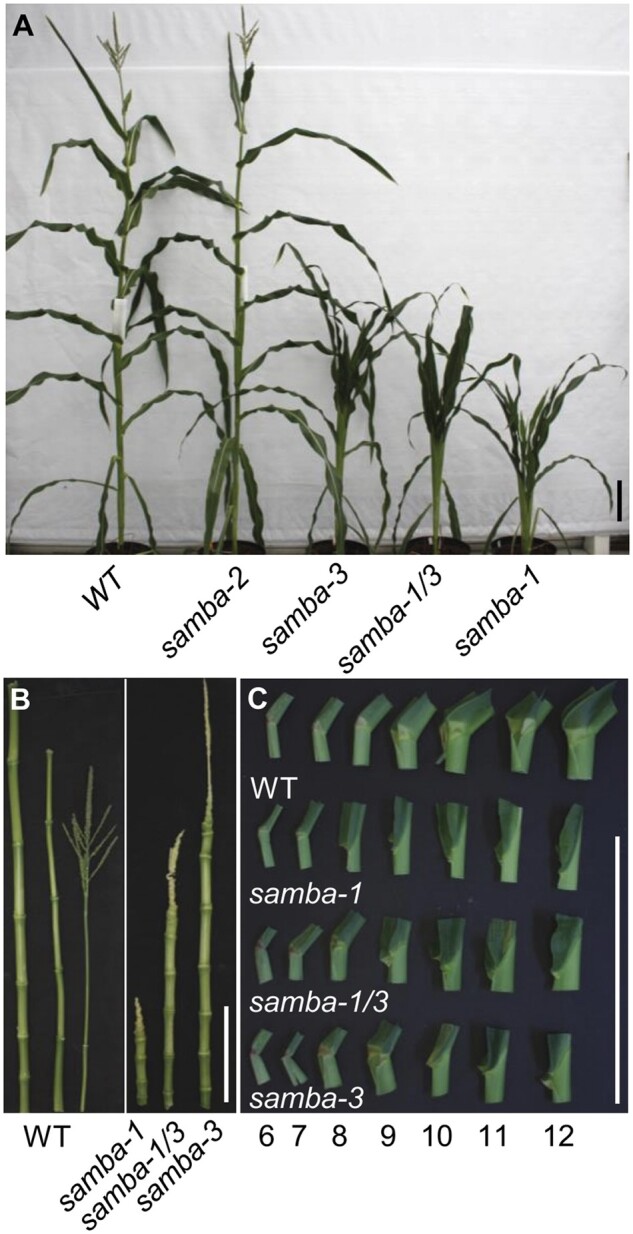
Overview of the phenotypes of the WT and samba mutant plants. A, WT and samba mutant plants grown in the greenhouse 70 d after sowing. The internodes (B) and ligule morphology (C) of WT plant and samba mutants from leaf 6 to leaf 12. Scale bars = 20 cm.
Figure 3.
Quantification of the phenotypes of phenotypes of the WT and samba mutant plants. Quantification of internode (A), leaf sheath (B) and blade (C) length and blade width (D) of samba mutants in comparison to the WT. Error bars represent standard error and significant differences were determined using Student’s t test: *P < 0.05, **P < 0.01 (n ≥ 3).
Figure 4.
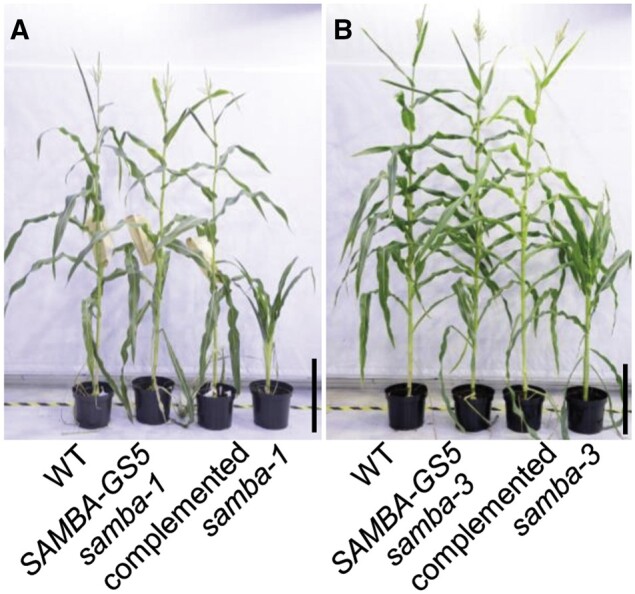
Complementation of the samba mutant plants by introducing SAMBA-GS5. Plant phenotype of the WT, SAMBA-GS5, samba complemented plants and samba-1 (A), or samba-3 (B). Bars = 50 cm.
Both samba alleles appeared to have a different strength and onset of the phenotypes, and to analyze if this was due to a dosage effect, the heterozygous samba-1 and samba-3 mutants were crossed to generate the samba-1/3 bi-allelic mutant. The bi-allelic samba-1/3 mutant showed the significantly shortened internode length above internode 11. The fact that internodes above the fifteenth internode barely elongated showed that the phenotype aggravated over time (Figure 3A). Moreover, the elongated ligule was present at leaf 9, while the elliptical ligule was visible on leaf 8 and leaf 10 for samba-1 and samba-3, respectively (Figure 2C). These data show that the samba-1/3 mutant showed the typical samba growth defects such as reduced plant height, shortened upper internode length, increased leaf sheath length, smaller and erect leaves, and fewer roots compared to the WT, but the onset of the phenotypes was intermediate to that of the single mutants (Figures 2 and 3; Supplemental Figures S4 and S5).
samba mutants have an accelerated cell cycle and decreased cell size
To investigate the cellular effect of the samba-1 mutation, the epidermal layer of internodes and mature leaves was imaged. Both WT and samba-1 internode and leaf epidermal cells were organized in cell files parallel with the vertical internode axis or leaf midvein, but the cells of samba-1 were smaller than those of the WT (Supplemental Figure S6, A and B). Stomata developed normally in the samba-1 mutant (Supplemental Figure S6, C and D), but leaf blades showed an altered leaf hair development (Supplemental Figure S6F). WT leaves developed long macrohairs and bicellular microhairs, spaced by prickle hairs (Supplemental Figure S6, E and G), whereas in samba-1 mutants, shorter macrohairs were formed and a large number of enlarged prickle hairs were regularly spaced alongside the files of bulliform cells (Supplemental Figure S6, F and H). The morphology and size of the samba-1 leaf vascular bundles were similar to those of the WT, but the number of minor veins per major vein was significantly reduced by 54% (Supplemental Figure S6, I and J and Supplemental Table S3).
To gain more insights into possible defects in cell division or cell expansion, a kinematic analysis of the growing leaf four was performed in the most severe samba-1 allele and the WT (Table 2). The kinematic analysis was based on daily leaf growth measurements, a DAPI staining to determine the size of the division zone and cell length profiles along the growth zone to quantify the contributions of cell division and/or cell expansion to the growth phenotype. The fourth leaf of samba-1 showed a moderate, but significant decrease of 6.64% (P < 0.05) in the final leaf length, while the LER was not significantly affected (Table 2). The effect of the samba-1 mutation was much more pronounced at the cellular level, where a significant increase in cell production of 13.61% (P < 0.05) was observed, while the mature cell size was reduced with 20.92% (P < 0.05). This opposite effect on cell division and cell expansion explained the rather mild effect observed for the final leaf length. The increase in cell production was caused by a significantly increased cell division rate (25.78%, P < 0.05) and a decreased cell cycle duration (35.42%, P < 0.05). The average cell cycle duration in WT leaves was 23.55 h, whereas it was reduced to 17.4 h in the samba-1 mutant leaves (Table 2). Flow cytometry on mature leaves 12 showed a similar distribution of ploidy level in the WT and samba-1 (Supplemental Figure S7, A and B), whereas the shift towards more 2C cells was significant in samba-1 developing internodes compared with those of the WT (Supplemental Figure S7, C and D).
Table 2.
Effect of samba-1 mutation on cellular parameters of leaf 4 growth
| Parameter | WT | samba-1 | P-value | % change |
|---|---|---|---|---|
| Final leaf length (mm) | 558.43 ± 5.80 | 523.67 ± 3.91 | 2.38E−05 | −6.64 |
| Mature cell size (µm) | 119.96 ± 1.15 | 99.21 ± 1.61 | 7.75E−04 | −20.92 |
| Cell production (cell h−1) | 17.40 ± 0.18 | 20.14 ± 0.38 | 0.01 | 13.61 |
| Cell division rate (cells cell−1 h−1) | 0.030 ± 0.002 | 0.040 ± 0.002 | 0.03 | 25.78 |
| Cell cycle duration (h) | 23.55 ± 1.62 | 17.40 ± 0.89 | 0.04 | −35.42 |
| Division zone size (cm) | 1.49 ± 0.10 | 1.24 ± 0.06 | 0.11 | −19.84 |
| Expansion zone size (cm) | 2.88 ± 0.32 | 1.87 ± 0.08 | 0.08 | −53.83 |
| Average size of dividing cells (µm) | 26.91 ± 0.46 | 26.37 ± 0.34 | 0.4 | −2.06 |
| Leaf elongation rate (mm·h−1) | 2.08 ± 0.04 | 2.04 ± 0.04 | 0.19 | −2.31 |
Statistics were calculated based on Student’s t test, significance P ≤ 0.05, n = 3, error indicates standard error.
The frameshift mutation in samba-3 leads to a partially functional truncated protein by translation re-initiation
To understand the differences in the severity and onset of the growth phenotypes between the different samba alleles, we analyzed the SAMBA mRNA level by reverse transcription quantitative polymerase chain reaction (RT-qPCR). In samba-1, the relative expression of SAMBA was reduced by 83% and 74% in leaf 5 and leaf 12, respectively, compared to the WT (Supplemental Figure S8A). Similarly, SAMBA expression was diminished by 57.12% and 44.4% in samba-3 for leaf 5 and leaf 12, respectively (Supplemental Figure S8B). However, the relative expression of SAMBA was also reduced by 47.7% in samba-2 leaf 5, while the samba-2 mutant did not reveal any growth defect (Supplemental Figure S8C). These data suggest that frameshifts, as well as in-frame deletions, affect the stability of the transcript and that downregulation of the SAMBA expression by approximately 50% is insufficient to evoke phenotypes.
Next, we examined SAMBA protein formation by fusing the coding sequences without stop codon of the WT (positive control), samba-1, samba-2, and samba-3 alleles to a C-terminal yellow fluorescent protein (YFP) tag and overexpressing the constructs under the control of the CaMV 35S promoter in Nicotiana benthamiana leaves. The samba-2 protein, which was expected to be about 0.96 kDa smaller than the WT because of the deletion of 10 AAs, showed a barely visible shift on Western blot (Figure 5A). The samba-3 protein expected to be about 6.31 kDa smaller than the WT and without the YFP tag because of the premature stop codon was not detected in the Western blot. The observed samba-3 protein (3.82 kDa smaller than the WT) could be explained when the protein started from the second methionine in SAMBA, of which the ATG, located at the start of exon 2, was recognized by translation re-initiation, resulting in an N-terminally truncated and C-terminally altered protein (Figure 1C). Because only cDNA containing three exons was detected in the WT and samba alleles (Supplemental Figure S9), it was concluded that the truncated protein is not due to exon skipping but rather to translation re-initiation. In samba-1, the stretch of missense AAs reaches until the second methionine, most likely prohibiting the re-initiation of the translational machinery, explaining the absence of the N-terminally truncated SAMBA protein in samba-1 on Western blot. Conversely, in samba-3 the translational machinery is released from the mRNA after building in only three missense AAs, making the second methionine more accessible for translation re-initiation (Figure 1C).
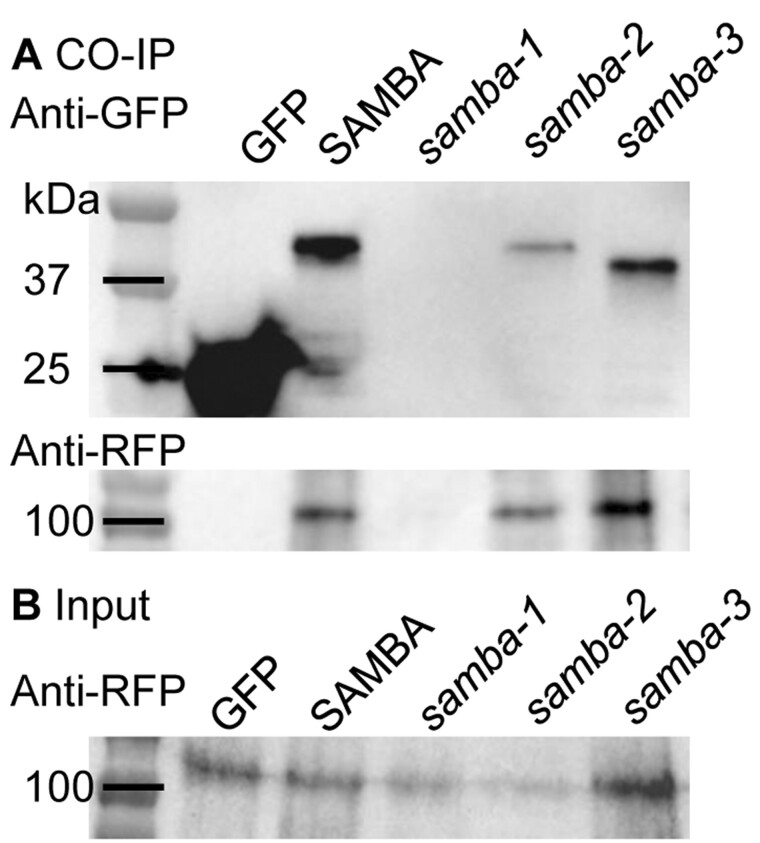
Figure 5.
Co-IP results of C-terminally YFP-tagged WT SAMBA and samba alleles with C-terminally RFP-tagged APC3. Co-IP (A) and input of the extracted whole proteins (B).
To analyze whether the mutated SAMBA proteins interact with the APC/C, a co-immunoprecipitation (Co-IP) of YFP-tagged SAMBA (WT and the three alleles) with APC3b through its RFP tag were performed in N. benthamiana leaves. Although APC3b-RFP proteins (107.05 kDa) were present in the input samples (Figure 5B), they were detected after immunoprecipitation of the WT, samba-2, and samba-3, indicating that the N-terminal and the C-terminal AAs of SAMBA were not essential for the protein interaction with the APC3b (Figure 5A).
To further investigate the functionality of SAMBA in samba-3, we performed AP/MS in maize callus using the WT and the truncated SAMBA as it occurs in the samba-3 mutant. Because of the one base insertion at the second target site in samba-3, there is no stop codon at the end of the samba-3 encoding sequence when the protein translation starts from the second methionine. The stop codon appears later in the 3′-untranslated region (UTR), causing an additional 15 AAs compared to samba-3 (Figure 1C). Therefore, we fused the full samba-3 encoding sequence and samba-3 with 3′-UTR (samba-3-UTR) to the GSrhino affinity tag next to the SAMBA WT, and ectopically expressed these three constructs in maize callus for AP/MS. All APC/C members that were identified in the AP/MS with SAMBA WT were also retrieved both for samba-3 and samba-3-UTR, except APC7, suggesting that the mutated samba-3 or samba-3-UTR proteins still interact with the APC/C (Supplemental Figure S10 and Supplemental Data Set S3). Moreover, 49 proteins were significantly enriched in the SAMBA WT compared to the samba-3 and samba-3-UTR mutants including DRP1E, while 30 proteins were significantly enriched in the samba-3 mutants including two UBIQUITIN6 proteins and an ubiquitin-specific protease. The altered enrichment in targets and UBIQUITIN6 suggests a distinct efficiency of the APC/C when bound to a mutated samba-3 (Supplemental Data Set S4). For the proteins enriched in the SAMBA WT and the samba-3 and samba-3-UTR mutants, a similar percentile, 65.4% and 63.6%, respectively, of the mutants with described phenotypes displayed growth-related phenotypes.
Discussion
In Arabidopsis, SAMBA has been identified as a negative regulator of the APC/C, because its loss of function results in increased cell proliferation (Eloy et al., 2012). Here, we validated that the maize SAMBA ortholog also interacts with APC/C. Furthermore, samba loss of function in maize samba-1 and samba-3 mutants also accelerates cell proliferation during leaf development, indicating that, also in maize, SAMBA negatively regulates cell proliferation through its interaction with APC/C. However, despite this seemingly conserved role of SAMBA, the phenotypic readout is distinct in Arabidopsis and maize mutants, which most likely results from a combination of inter-species differences. In maize, samba mutants show an increased cell production, due to a substantially faster cell cycle, resulting in significantly smaller cells. In Arabidopsis, an increased cell division was also shown to result in smaller cells. For example, the overexpression of E2F transcription factors (E2Fa, De Veylder et al., 2002 and E2Ff, Ramirez-Parra et al., 2004) or Cyclin D (CYCD, CYCD3, Menges et al., 2006 and CYCD4, Kono et al., 2007) genes in Arabidopsis stimulates cell division by promoting the G1/S transition, and reduces or does not alter the cell DNA ploidy level, leading to a reduced cell size. However, in Arabidopsis, where endoreduplication in leaves is much more pronounced than in some monocots such as maize, the cell size of the samba mutants is increased through higher ploidy levels.
Another striking difference between maize and Arabidopsis is the timing at which the phenotypic effects upon SAMBA perturbation become apparent. In Arabidopsis, the effects of SAMBA loss of function are most pronounced early during seedling development and decreased throughout further development. In maize, however, SAMBA loss-of-function seedlings develop normally and show only a moderate reduction in final leaf length, while later in development more severe reductions of both leaf and internode length are observed. This might be explained by the SAMBA expression pattern differences between Arabidopsis and maize. In Arabidopsis, SAMBA shows high expression in all tissues of germinating seedlings but its expression quickly diminishes in root tissues and becomes restricted to the hypocotyl at 8 d after stratification. Conversely, in maize, SAMBA is more stably expressed throughout the entire development (Sekhon et al., 2011). The distinct developmental regulation of SAMBA indicates that in maize, SAMBA is not only instrumental for the cell divisions in organ primordia, but also during processes where cell division plays a role throughout development. The most conceivable phenotypes upon SAMBA loss of function, being leaf and internode elongation and ligule formation, but also the less pronounced effects on leaf width, are associated with changes in the rate and/or orientation of cell divisions (Sylvester et al., 1990; McKim, 2019).
Using affinity enrichment of the SAMBA-GSrhino fusion protein from both embryogenic callus and leaf tissues, we were able to recover all APC/C subunits, including the CCS52 proteins. CCS52A proteins regulate the onset of the endocycle in Arabidopsis leaves and roots. Both loss-of-function mutants of AtCCS52As are smaller than the WT, and their leaves contain more but smaller cells, whereas the double ccs52a1/a2 mutant is lethal (Vinardell et al., 2003; Vanstraelen et al., 2009; Baloban et al., 2013). Downregulation of the rice OsCCS52A results in semi-dwarfism and smaller seeds with an endosperm defect in endoreduplication (Su'udi et al., 2012). The reduced plant height and leaf size are also observed in the rice tillering and dwarf 1 (tad1) mutant, which is an ortholog of CCS52A (Xu et al., 2012). The CCS52B protein is reported as a plant-specific APC/C activator (Tarayre et al., 2004) and the rice ccs52b mutant is semi-dwarfed and shows narrow kernels (Su'udi et al., 2012). As the phenotype of the ccs52 mutants in Arabidopsis and rice show similar growth defects as the maize samba mutant, it would be quite interesting to investigate the function of maize CCS52 genes by generating the ccs52 CRISPR knockout mutants to further decipher the functions of the maize APC/C. In addition, we identified several members of the DRP family, including DRP1A, DRP1E, DRP2A, DRP2B, and DRP5B, as SAMBA interactors. All of these proteins, except DRP5B, have been shown to localize to the cell plate during cytokinesis (Park et al., 1997; Hong et al., 2003; Fujimoto et al., 2008) and mutations in DRP1A and DRP1E result in defective cell plate assembly and cytokinesis, as well as defects in cell expansion (Kang et al., 2003). Furthermore, CALLOSE SYNTHASE was identified as maize SAMBA interactor, which is involved in cell plate synthesis and is regulated by DRP proteins (Verma, 2001). Interestingly, APC3b, which has been shown to directly interact with SAMBA in Arabidopsis, has also been found to be localized at the cell plate during the early telophase (Perez-Perez et al., 2008). In mammalian cells, DRP1 has been shown to be degraded by the APC/C in a D-box dependent way (Horn et al., 2011), and DRP2A, DRP2B, DRP5B, and CALLOSE SYNTHASE also contain D-box motifs (Verma, 2001) and could, therefore, be direct targets of the APC/C. In the samba-3 mutant, where the SAMBA SHR1 domain is lacking, but (the majority of) the SHR2 domain is present, SAMBA was still able to interact with APC3, indicating that the SHR2 domain is pivotal for SAMBA interaction with APC/C. Although SAMBA in the samba-3 mutant interacted with the APC/C, in total 49 or 30 proteins, such as DRP1E or UBIQUITIN6, were found to be significantly enriched in the SAMBA or samba-3 callus, respectively, indicating that the activity or efficiency of the APC/C are altered in samba-3. Performing AP/MS with bait proteins on callus tissue initiated from immature embryos during the early steps of maize transformation provided good insights into the interacting proteins and could help to circumvent the generation of transgenic plants (Dedecker et al., 2016).
Because of the phenotypic differences in timing and severity of seemingly similar samba-1 and samba-3 alleles, it became clear that addition or removal of one nucleotide (frameshift mutations) at the same position can make a distinction between a true knockout and the formation of a truncated, but partially functional protein. The observations that processes such as translation re-initiation and exon skipping might cause rather unexpected readouts of genome editing were already made several times for animal tissues (Tang et al., 2018; Smits et al., 2019), but remained fragmentary in plants so far. Alternative splicing was shown to be the underlying reason why CRISPR/Cas9-induced frameshifts in the rice OsIAA23 gene did not result in the expected severe phenotypes (Jiang et al., 2019). Here, we showed that besides alternative splicing, plants also can use translation re-initiation as a possibility to overcome CRISPR/Cas9-induced point mutations. The understanding that in-frame deletions as well as point mutations affect transcript stability and that plants possess inherent strategies such as alternative splicing and translation re-initiation will create an awareness in the choice of gRNAs. Often gRNAs are designed close to the translational start site with the aim to create early frameshifts and knockout, but the samba-3 allele shows that early mutations can cause translation re-initiation, which was in turn dependent on the number of missense AAs that shielded the re-initiation methionine. Taken together, our data show that caution should be taken in designing gRNAs and in interpreting the effects of mutations at the DNA level only.
Materials and methods
Growth conditions in growth chamber and greenhouse
Plants for leaf growth monitoring were grown under growth chamber conditions with controlled relative humidity (55%), temperature (24°C d/18°C night), and light intensity (170–200 μmol·m−2·s−1 photosynthetic active radiation at plant level) provided by a combination of high-pressure sodium vapor (RNP-T/LR/400W/S/230/E40; Radium) and metal halide lamps with quartz burners (HRI-BT/400W/D230/E40; Radium) in a 16-h/8-h (d/night) cycle. Plants for adult plant trait characterization were grown under controlled greenhouse conditions (26°C/22°C, 55% relative humidity, the light intensity of 180 μmol·m−2·s−1 photosynthetic active radiation, in a 16-h/8-h d/night cycle).
Generation of SAMBA constructs
GRMZM2G157878 was identified as the maize (Zea mays) SAMBA ortholog using PLAZA (Eloy et al., 2012; Proost et al., 2015). CRISPOR (Concordet and Haeussler, 2018) was used to determine possible gRNAs and their off-target scores in the coding sequence of SAMBA. Two gRNA sequences were chosen based on their location, MitSpecScore and off-target count: 35rev and 297fwd (Supplemental Data Set S6). These sequences target the coding sequence of SAMBA and do not overlap with other genes. The CRISPR construct containing these two gRNA sequences was generated according to Xing et al. (2014), with minor modifications. Briefly, the kanamycin resistance cassette in the pBUN411 destination vector was replaced with a spectinomycin resistance cassette. This vector is called pBUN411-Sp. Primers were designed that matched with the 35rev and 297fwd gRNA sequences, after which PCR was performed on the pCBC-MT1T2 plasmid, resulting in a fragment containing the desired target sites and the correct sites for ligation into the pBUN411-Sp destination vector using Golden Gate cloning (Invitrogen).
The coding sequences of SAMBA, samba-3, and samba-3-UTR flanked by AttB1/AttB2 Gateway recombination sites were amplified from B104 WT cDNA using PCR, which was then ligated into pDONR221 entry vectors using Gateway BP reactions. The SAMBA entry vector together with the pUBI-L promoter (Coussens et al., 2012) and GSrhino tag (Van Leene et al., 2015) were ligated into the pBb7m34GW destination vector (Karimi et al., 2013) to generate the pUBIL:SAMBA::GSrhino (SAMBA-GS) expression vector by Multisite Gateway recombination (Invitrogen). This vector is codon-optimized for monocots and contains a Bar selection marker for the selection of transgenic plants using phosphinotricin (Karimi et al., 2013). Expression vectors for affinity enrichment experiments in callus tissues were generated by Multisite Gateway recombination (Invitrogen) into the in-house developed pBb-G7-BBMR-Ef1a destination vector. This vector contains a Bar selection marker but also contains a pEF1a:BBM cassette, allowing co-transformation of the morphogenic factor BABY BOOM, which can enhance callus growth.
Maize transformation, genotyping, and biomass generation
Immature embryos of the maize inbred line B104 were transformed by Agrobacterium tumefaciens co-cultivation (Coussens et al., 2012). In short, immature B104 embryos were co-cultivated with A. tumefaciens for 3 d followed by 1-week growth on nonselective medium. Transformed embryogenic calli were subsequently selected on increasing concentrations of phosphinotricin. After shoot induction from the selected calli, transgenic T0 shoots were transferred to soil. At maturity, these T0 shoots were backcrossed with B104 WT, resulting in a collection of T1 seeds from multiple independent transgenic events. For the overexpression lines, this segregating population of 1:1 nontransgenic:transgenic plants was used for phenotyping and biomass generation for AP/MS. For the genome editing lines, the offspring containing the heterozygous mutant alleles that did not contain the T-DNA harboring CRISPR/Cas9 were self-crossed to identify WT plants and homozygous mutants. The segregating population was self-pollinated for phenotypic analysis in the next generation.
Screening for mutations and genotyping of samba CRISPR mutants was performed by sequencing the genomic region containing both target sites after PCR amplification (Supplemental Data Set S6). To select suitable transgenic lines for AP/MS, the collection of T1 lines was first subjected to a segregation analysis to select single-locus insertion lines. Because the high accumulation of the fusion protein forms the basis of a successful affinity purification experiment, expression analysis was performed at the protein level using antibodies specific for the GSrhino tags via western blotting. Based on this expression analysis, the transgenic lines showing the highest accumulation of the bait protein were selected to be upscaled for phenotypical analysis and AP/MS. To perform AP/MS from division zone and expansion zone in triplicate, approximately 720 seeds were sown in small pots and grown in the growth chamber. When the first leaf was fully grown, a solution for the Bar selection marker, containing 1% w/v glufosinate ammonium (Sigma), was applied to its blade. After 3 d, nontransgenic plants showed severe first leaf necrosis while transgenic plants were resistant to this treatment, allowing easy genotyping of a large number of plants. Transgenic seedlings were harvested 2 d after leaf four appearance. The fourth leaf of each seedling was dissected and the division zone (first basal cm of the leaf) and the expansion zone (cm three to four) were cut out of the leaf growth zone. Harvested division and expansion zone samples were frozen in liquid nitrogen and pooled for all harvested plants. Harvested tissues were stored at −70°C.
The SAMBA-GS5 line was crossed with samba-1 and samba-3, and the heterozygote offspring was self-pollinated to generate homozygous WT plants and samba-1 and samba-3 mutants expressing the complementation construct.
The callus was generated by slight modifications to the transformation protocol (Coussens et al., 2012). Thirty immature embryos from three different pollinated ears were transformed. Embryos were cocultivated with A. tumefaciens for 3 d and subsequently transferred to a resting medium for 1 week. Transgenic calli were selected by transferring them to selection medium I for 2 weeks, after which the calli were transferred to selection medium II and incubated for 2 weeks. Nontransgenic callus tissue, which became necrotic due to phosphinothricin selection, was removed during each transfer step. Calli were then transferred to fresh selection medium II and incubated for one additional week, after which the healthy callus tissues were harvested, frozen in liquid nitrogen, ground to a fine powder (∼10 g) using mortar and pestle, and stored at −70°C.
AP/MS
The AP/MS was performed by a slightly adjusted protocol (Nelissen et al., 2015). During protein extraction, Benzonase treatment was performed for 15 min instead of 30 min to shorten the duration of the purification protocol. The protein extracts were divided into three technical repeats, containing 65-mg protein extract each. To each technical repeat, 50 µL of home-made magnetic IgG beads, equilibrated in extraction buffer, were added and incubated for 45 min on a rotating device at 4°C. Beads were washed with 2×500 µL ice-cold extraction buffer and transferred to a fresh tube. Beads were washed again with 1×500 µL ice-cold extraction buffer, 1×500 µL ice-cold extraction buffer without detergents, and a final time with 800-µL 50-mM ammonium bicarbonate solution at pH 8. On bead digestion was performed in 50 µL of ammonium bicarbonate solution containing 4 µL of Trypsin/LysC (Promega) solution and incubated at 37°C for 3 h in a thermomixer at 1,000 rotations per minute. After 3 h, the beads were separated from the digest and the digest was spiked with an additional 2 µL of Trypsin/LysC solution and left to incubate overnight at 37°C in a thermomixer at 1,000 RPM. Samples were dried in a SpeedVac system and stored at −70°C for MS analysis.
Liquid chromatography-tandem MS analysis
The digested peptide mixtures were injected into a liquid chromatography-tandem MS system using an Ultimate 3000 RSLCnano LC (Thermo Fisher Scientific) in-line connected to a Q Exactive (SAMBA callus versus GIF control group and SAMBA leaf) or Q Exactive HF Biopharma (samba-3 mutants callus versus SAMBA WT) mass spectrometer (Thermo Fisher Scientific). The sample mixture was first bound on an in-house prepared trapping column (100-μm internal diameter [I.D.] × 20 mm, 5-μm beads C18 Reprosil-HD, Dr Maisch). For Q Exactive, peptides were separated on an in-house prepared analytical column (75-μm I.D. × 150 mm, 5-μm beads C18 Reprosil-HD, Dr Maisch) packed in the needle (PicoFrit SELF/P PicoTip emitter, PF360-75-15-N-5; New Objective). Peptides were separated through a linear gradient from 98% solvent A (0.1% formic acid in water [v/v]) to 56% solvent B (0.1% formic acid in water/acetonitrile, 20/80 [v/v]) for 30 min at a flow rate of 300 nL·min−1. Next, the column was washed for 5 min, hence reaching 99% solvent B. The operation mode of the mass spectrometer was set at the data-dependent, positive ionization mode, which automatically switches between MS and MS/MS acquisition for the five most abundant peaks in a given MS spectrum. The Q Exactive implemented source voltage was 2.8 kV and the temperature of the capillary was set at 275°C. After one MS1 scan (m/z 400–2,000, AGC target 3 × 106 ions, maximum ion injection time 80 ms) obtained at a resolution of 70,000 (at 200 m/z), up to five MS/MS scans (resolution 17,500 at 200 m/z) were obtained of the most intense ions that fitted the predefined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 60 ms, isolation window 2 Da, fixed first mass 140 m/z, spectrum data type: centroid, underfill ratio 2%, intensity threshold 1.3 × E4, exclusion of unassigned, 1, 5 to 8, >8 charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time 12 s). The higher-energy C-trap dissociation (HCD) collision energy was kept at 25% of the normalized collision energy. For Q Exactive HF Biopharma, the peptides were separated on a 250-mm Waters nanoEase M/Z HSS T3 Column, 100Å, 1.8 µm, 75-µm inner diameter (Waters Corporation) kept at a constant temperature of 50°C. Peptides were eluted by a non-linear gradient starting at 1% MS solvent B reaching 55% MS solvent B (0.1% FA in water/acetonitrile [2:8, v/v]) in 65 min, 97% MS solvent B in 70 min followed by a 5-min wash at 97% MS solvent B and re-equilibration with MS solvent A (0.1% FA in water). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 12 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375–1,500 m/z) were acquired at a resolution of 60,000 in the Orbitrap analyzer after accumulation to a target value of 3,000,000. The 12 most intense ions above a threshold value of 13,000 were isolated with a width of 1.5 m/z for fragmentation at a normalized collision energy of 30% after filling the trap at a target value of 100,000 for maximum 80 ms. MS/MS spectra (200–2,000 m/z) were acquired at a resolution of 15,000 in the Orbitrap analyzer.
Protein identification and LFQ
The raw files were processed using MaxQuant software (Tyanova et al., 2016a). The data were searched against the Z. mays B73 RefGen_v3 database (obtained from ftp://ftp.ensemblgenomes.org/pub/release-27/plants/fasta/zea_mays/pep/) using the built-in Andromeda search engine. A second database containing sequences of all possible types of contaminants that are often found with tandem affinity purification (TAP) or with proteomics experiments in general was added. These contaminants include a common repository of adventitious protein sequences, a list with proteins that are often found in proteomics experiments and are present as unavoidable contaminations of protein samples or by accident (The Global Proteome Machine, www.thegpm.org/crap/). In addition, tag sequences of many common tags and typical TAP contaminants, including sequences of resin proteins or implemented proteases, were added. For the searches, only oxidation of methionines and N acetylation of protein N termini were accepted as variable modifications. A mass tolerance for precursor (peptide) ions was set at 4.5 ppm, whereas for fragment ions this was set at 20 ppm. The used enzyme was trypsin/P, permitting two missed cleavages as well as cleavage in cases where proline followed arginine or lysine. A target-decoy approach with a false discovery rate (FDR) of 1% was used to filter protein identifications and for peptide-spectrum matches. LFQ was performed using the MaxLFQ algorithm integrated into MaxQuant (Cox et al., 2014).
Due to the high degree of protein sequence similarity between both CC52A1 and CC52A2 in Arabidopsis (86.4%) and the two maize CCS52A members (94.5%), it was impossible to determine which maize gene is the correct ortholog of which Arabidopsis gene, and to recognize the peptide in the AP/MS data. Therefore, GRMZM2G159427 was arbitrarily assigned as CCS52A1 and GRMZM2G064005 as CCS52A2, and both genes were identified as CCS52As in the AP/MS data.
Statistical analysis of the LFQ
The identification of significantly enriched proteins in the samples was performed using Perseus (Tyanova et al., 2016b). The MaxQuant output files were loaded into Perseus and LFQ intensities were log2 transformed, after which samples were grouped in bait and control, or in division zone and expansion zone. Proteins that did not contain at least two valid values in at least one group were filtered out and missing LFQ values were imputed from the normal distribution, as previously described (Smaczniak et al., 2012; Wendrich et al., 2017). A Student’s t test was performed to determine significantly enriched proteins in either developmental zone, or a permutation-based FDR correction (FDR = 0.01, S0 = 1) was applied to correct for multiple hypothesis testing. Significantly enriched proteins after affinity enrichment with SAMBA-GSrhino in callus were determined using a negative control AP/MS data set from embryogenic callus transformed with the non-SAMBA-related proteins GIF2-GSrhino and GIF3-GSrhino. Additionally, nonspecific and sticky proteins were marked in the lists, based on Supplemental Data Set S5, which is assembled from a set of control affinity purification experiments from maize leaf tissues (Besbrugge et al., 2018) and a set of proteins whose Arabidopsis orthologs were included in a previously published background list based on 543 TAP purifications over 155 different bait proteins (Van Leene et al., 2015).
RT-qPCR
Expression analysis of SAMBA was checked by RT-qPCR. Total RNA was extracted from each repeat using Trizol (Life Technologies, Invitrogen) and DNA was removed by RQ1 DNase (Promega) treatment. Preparation of cDNA was performed using the iScript cDNA synthesis kit (BioRad) according to manufacturer’s recommendations starting with 1 µg of RNA. RT-qPCR was performed with the LightCycler 480 Real-Time SYBR Green PCR System (Roche); used primers are listed in Supplemental Data Set S6. For RT-qPCR on maize samples, we used 18S RNA as the housekeeping gene.
Flow cytometry
For flow cytometry analysis, at least three leaves or stem samples per time point of a biological repeat (n = 3) were chopped with a razor blade in 200 mL of Cystatin UV Precise P Nuclei Extraction buffer (Partec), followed by the addition of 800 mL of staining buffer and filtering through a 50-mm filter. Nuclei were analyzed with the Cyflow MB flow cytometer (Partec) and the FloMax software.
Anatomical analysis
The second cm of the samba-1 mature ninth leaf blade was cut into cross slides by hand section. Segments were soaked in 100% ethanol for discoloration and then transferred into the ACA staining solution (0.5% w/v Astra blue, 0.5% w/v Chrysopidae and 0.5% w/v Acridine red solution mixed at a volume ratio of 16:1:1) for 5 min. The stained sections were washed three times, 5 min per time in water and mounted in 50% glycerol solution for imaging.
Co-IP assays
The coding sequence without stop codon of WT SAMBA, samba-1, samba-2, samba-3 and were fused with a 3′-YFP tag; that of APC3b was fused with a 3′-RFP tag. All genes were cloned behind the 35S promoter in the pBb7m34GW destination vector (Karimi et al., 2013). All these constructs, including the empty p35S::YFP vector, were introduced into A. tumefaciens cells. All transient expression assays were performed as described previously (Marin-de la Rosa et al., 2015). Nicotiana tabacum leaves were harvested 48 h after infiltration and were ground in liquid nitrogen, homogenized in the extraction buffer (50-mM Tris–Cl, pH 7.5, 1-mM EDTA, 75-mM NaCl, 0.1% Triton X-100, and protease inhibitor cocktail [Sigma]). The supernatant was incubated with the GFP-Trap agarose beads (Chromotek) for 2 h at 4°C. After washing, beads were eluted by boiling with 2× sodium dodecyl sulfate (SDS) sample buffer for 10 min. Samples were separated on 4%–20% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad) and then immunoblotted with anti-GFP and anti-RFP antibody (Abcam, 1:2,000 dilution).
Accession numbers
The accession number of SAMBA is GRMZM2G157878. The accession numbers of all interacting proteins are indicated in Supplemental Figure S2.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression and phenotypic analysis of the SAMBA-GS transgenic overexpression lines.
Supplemental Figure S2. LFQ of APC/C subunits and regulatory proteins co-purified with the SAMBA-GSrhino fusion protein using AP/MS on the division (DZ) and expansion zone (EZ) of the growing maize leaf.
Supplemental Figure S3. Average mature internode and leaf epidermal cell length of samba-1 and samba-3 mutants.
Supplemental Figure S4. Leaf 12 ligule morphology of the WT and samba-1, samba-1/3, and samba-3 mutants.
Supplemental Figure S5. Overview of the root phenotypes of the WT and samba mutant plants. Representative root systems of samba-1 (A), samba-3 (B), and samba-1/3 (C) mature plants at tasseling stage of the respective WT.
Supplemental Figure S6. Microscopic comparison of mature leaves and internodes of the WT and samba-1 mutants.
Supplemental Figure S7. Ploidy distribution of the mature leaf and internode cells in samba-1 and samba-3 mutants determined by flow cytometry.
Supplemental Figure S8. Comparison of the relative expression of SAMBA in the WT and samba mutant plants. The relative expression level of SAMBA determined by RT-qPCR in samba-1 (A), samba-3 (B), and samba-2 (C) relative to the respective segregating WT plants.
Supplemental Figure S9. The PCR for the UTRs of SAMBA in WT plants and the samba alleles on cDNA from the leaf 4 division zone.
Supplemental Figure S10. Label-free quantification of APC/C subunits and regulatory proteins co-purified with SAMBA, samba-3, and samba-3-UTR using AP/MS on the maize callus.
Supplemental Table S1. Additional proteins, besides the known SAMBA interactors listed in Table 1 that were significantly enriched after AP/MS using SAMBA-GS as bait protein from transformed embryogenic callus.
Supplemental Table S2. Plant height and root length of samba-1 and samba-3. Error is standard error and P ≤ 0.05 indicates significance.
Supplemental Table S3. Minor veins per major vein of the nine major veins in the center of the blade midrib in samba-1. Error is standard error and P ≤ 0.05 indicates significance.
Supplemental Data Set S1. Protein identifications through affinity enrichment from maize callus by using SAMBA-GSrhino as bait protein.
Supplemental Data Set S2. Protein identifications through affinity enrichment from the leaf division and expansion zone using SAMBA-GSrhino as bait protein. Error is standard error and P ≤ 0.05 indicates significance.
Supplemental Data Set S3. Protein identifications through affinity enrichment from SAMBA WT and samba-3 mutant calli using SAMBA-GSrhino as bait protein. Error is standard error and P ≤ 0.05 indicates significance.
Supplemental Data Set S4. Proteins enriched in Supplemental Data Set S3.
Supplemental Data Set S5. Lists of nonspecific and sticky proteins used for background filtering.
Supplemental Data Set S6. Nucleotide sequences of the primers used for cloning and RT-qPCR.
Supplementary Material
Acknowledgments
The authors thank Dr Bert De Rybel, Helena Arents, and Ji Li for helping with the leaf hand section and ACA staining, Dr Hannes Vanhaeren, Dr Ting Li, Ying Chen, and Reinout Laureyns for helping with the Co-IP Assay, Rudy Vanderhaeghen for callus generation, Thomas Eekhout for sharing the pBUN411-Sp vector and VIB Proteomics core facility for analyzing the maize AP/MS samples.
Funding
The research was funded by the European Research Council under the European Community’s Seventh Framework Programme [FP7/2007‐2013] under ERC grant agreement no. [339341‐AMAIZE]11 and by the “Bijzonder Onderzoeksfonds Methusalem Project” (no. BOFMET2015000201) of Ghent University. Pan Gong received a scholarship from the China Scholarship Council (CSC no. 201606320217).
Conflict of interest statement. None declared.
P.G., M.B., L.P., G.D.J., D.I., and H.N. conceived and planned the experiments. P.G., M.B., K.D., J.D.B., G.P., S.A., G.C. carried out the experiments. P.G., M.B., K.G., D.E., M.V.L., G.D.J., D.I., and H.N. contributed to the interpretation of the results. P.G., M.B., D.I., and H.N. took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Hilde Nelissen (hilde.nelissen@psb.vib-ugent.be).
References
- Baloban M, Vanstraelen M, Tarayre S, Reuzeau C, Cultrone A, Mergaert P, Kondorosi E (2013) Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol 198:1049–1059 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inze D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129:854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besbrugge N, Van Leene J, Eeckhout D, Cannoot B, Kulkarni SR, De Winne N, Persiau G, Van De Slijke E, Bontinck M, Aesaert S, et al. (2018) GS(yellow), a multifaceted tag for functional protein analysis in monocot and dicot plants. Plant Physiol 177:447–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13:654–660 [DOI] [PubMed] [Google Scholar]
- Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T (2005) The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 24:314–325 [DOI] [PubMed] [Google Scholar]
- Chang LF, Zhang ZG, Yang J, McLaughlin SH, Barford D (2015) Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Haeussler M (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46:W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PA, Strable J, Li S, Scanlon MJ (2019) On the mechanisms of development in monocot and eudicot leaves. New Phytol 221:706–724 [DOI] [PubMed] [Google Scholar]
- Coussens G, Aesaert S, Verelst W, Demeulenaere M, De Buck S, Njuguna E, Inze D, Van Lijsebettens M (2012) Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J Exp Bot 63:4263–4273 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Fonseca PCA, Kong EH, Zhang ZG, Schreiber A, Williams MA, Morris EP, Barford D (2011) Structures of APC/C-Cdh1 with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature 470:274–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Engler JD, Ormenese S, Maes S, Naudts M, Van der Schueren E, Jacqmard A, Engler G, et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. Embo J 21:1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inze D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8:655–665 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16:624–634 [DOI] [PubMed] [Google Scholar]
- Dedecker M, Van Leene J, De Winne N, Eeckhout D, Persiau G, Van De Slijke E, Cannoot B, Vercruysse L, Dumoulin L, Wojsznis N, et al. (2016) Transferring an optimized TAP-toolbox for the isolation of protein complexes to a portfolio of rice tissues. Plant Mol Biol 91:341–354 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:1077. [DOI] [PubMed] [Google Scholar]
- Eloy NB, Gonzalez N, Van Leene J, Maleux K, Vanhaeren H, De Milde L, Dhondt S, Vercruysse L, Witters E, Mercier R, et al. (2012) SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc Natl Acad Sci USA 109:13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, Lima MD, Ferreira PCG, Inze D (2015) The role of the anaphase-promoting complex/cyclosome in plant growth. Crit Rev Plant Sci 34:487–505 [Google Scholar]
- Fujimoto M, Arimura SI, Nakazono M, Tsutsumi N (2008) Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep 27:1581–1586 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349:132–138 [DOI] [PubMed] [Google Scholar]
- Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM (2000) The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci USA 97:8973–8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong ZL, Geisler-Lee CJ, Zhang ZM, Verma DPS (2003) Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol 53:297–312 [DOI] [PubMed] [Google Scholar]
- Horn SR, Thomenius MJ, Johnson ES, Freel CD, Wu JQ, Coloff JL, Yang CS, Tang WL, An J, Ilkayeva OR, et al. (2011) Regulation of mitochondrial morphology by APC/C-Cdh1-mediated control of Drp1 stability. Mol Bio Cell 22:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang MM, Hu HY, Kai J, Traw MB, Yang SH, Zhang XH (2019) Different knockout genotypes of OsIAA23 in rice using CRISPR/Cas9 generating different phenotypes. Plant Mol Biol 100:467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY (2003) Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15:899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Van Lijsebettens M, Hilson P (2013) Gateway vectors for transformation of cereals. Trends Plant Sci 18:1–4 [DOI] [PubMed] [Google Scholar]
- Kong F, Zhang T, Liu J, Heng S, Shi Q, Zhang H, Wang Z, Ge L, Li P, Lu X, et al. (2017) Regulation of leaf angle by auricle development in maize. Mol Plant 10:516–519 [DOI] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Adachi S, Nagata N, Konomi M, Nakagawa T, Uchimiya H, Umeda M (2007) The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19:1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson JD, Joazeiro CAP, Page AM, Huang HK, Hieter P, Hunter T (2000) The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Bio Cell 11:2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima MD, Eloy NB, Pegoraro C, Sagit R, Rojas C, Bretz T, Vargas L, Elofsson A, de Oliveira AC, Hemerly AS, Ferreira PCG (2010) Genomic evolution and complexity of the anaphase-promoting complex (APC) in land plants. BMC Plant Biol 10:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Gronlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PloS Genet 11: e1005337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim SM (2019) How plants grow up. J Integr Plant Biol 61:257–277 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell-size in epidermal tissue of arabidopsis. Plant Cell 5:1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JAH (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18:893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Eeckhout D, Demuynck K, Persiau G, Walton A, van Bel M, Vervoort M, Candaele J, De Block J, Aesaert S, et al. (2015) Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27:1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Coppens F, Dhondt S, Fiorani F, Beemster GT (2013) Kinematic analysis of cell division in leaves of mono- and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol Biol 959:247–264 [DOI] [PubMed] [Google Scholar]
- Park JM, Kang SG, Pih KT, Jang HJ, Piao HL, Yoon HW, Cho MJ, Hwang I (1997) A dynamin-like protein, ADL1, is present in membranes as a high-molecular-mass complex in Arabidopsis thaliana. Plant Physiol 115:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Serralbo O, Vanstraelen M, Gonzalez C, Criqui MC, Genschik P, Kondorosi E, Scheres B (2008) Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J 53:78–89 [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev 14:655–665 [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inze D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43:D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of arabidopsis cell growth and differentiation. Plant Cell 16:2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin HN, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563 [DOI] [PubMed] [Google Scholar]
- Sharma B, Joshi D, Yadav PK, Gupta AK, Bhatt TK (2016) Role of ubiquitin-mediated degradation system in plant biology. Front Plant Sci 7:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muino JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu SJ, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109:1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Ziebell F, Joberty G, Zinn N, Mueller WF, Clauder-Munster S, Eberhard D, Savitski MF, Grandi P, Jakob P, et al. (2019) Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 16:1087–1093 [DOI] [PubMed] [Google Scholar]
- Su'udi M, Cha JY, Ahn IP, Kwak YS, Woo YM, Son D (2012) Functional characterization of a B-type cell cycle switch 52 in rice (OsCCS52B). Plant Cell Tiss Org 111:101–111 [Google Scholar]
- Su'udi M, Cha JY, Jung MH, Ermawati N, Han CD, Kim MG, Woo YM, Son D (2012) Potential role of the rice OsCCS52A gene in endoreduplication. Planta 235:387–397 [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110:985–1000 [DOI] [PubMed] [Google Scholar]
- Tang JX, Chen DF, Deng SL, Li J, Li YY, Fu Z, Wang XX, Zhang Y, Chen SR, Liu YX (2018) CRISPR/Cas9-mediated genome editing induces gene knockdown by altering the pre-mRNA splicing in mice. BMC Biotechnol 18:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZY, Li B, Bharadwaj R, Zhu HH, Ozkan E, Hakala K, Deisenhofer J, Yu HT (2001) APC2 cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell 12:3839–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayre S, Vinardell JM, Cebolla A, Kondorosi A, Kondorosi E (2004) Two classes of the Cdh1-type activators of the anaphase-promoting complex in plants: novel functional domains and distinct regulation. Plant Cell 16:422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J (2016a) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protocols 11:2301. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016b) The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods 13:731. [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Vercruysse L, Dedecker M, Verkest A, Vandepoele K, et al. (2015) An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protocols 10:169–187 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106:11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DPS (2001) Cytokinesis and building of the cell plate in plants. Annu Rev Plant Phys 52:751–784 [DOI] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, Tarayre S, Roudier F, Mergaert P, Kondorosi A, et al. (2003) Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15:2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Boeren S, Moller BK, Weijers D, De Rybel B (2017) In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol Biol 1497:147–158 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang YH, Yu YC, Duan JB, Liao ZG, Xiong GS, Meng XB, Liu GF, Qian Q, Li JY (2012) Degradation of MONOCULM 1 by APC/C-TAD1 regulates rice tillering. Nat Commun 3:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.