Abstract
Water deficit is one of the main challenges for apple (Malus × domestica) growth and productivity. Breeding drought-tolerant cultivars depends on a thorough understanding of the drought responses of apple trees. Here, we identified the zinc-finger protein B-BOX 7/CONSTANS-LIKE 9 (MdBBX7/MdCOL9), which plays a positive role in apple drought tolerance. The overexpression of MdBBX7 enhanced drought tolerance, whereas knocking down MdBBX7 expression reduced it. Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis identified one cis-element of MdBBX7, CCTTG, as well as its known binding motif, the T/G box. ChIP-seq and RNA-seq identified 1,197 direct targets of MdBBX7, including ETHYLENE RESPONSE FACTOR (ERF1), EARLY RESPONSIVE TO DEHYDRATION 15 (ERD15), and GOLDEN2-LIKE 1 (GLK1) and these were further verified by ChIP-qPCR and electronic mobility shift assays. Yeast two-hybrid screen identified an interacting protein of MdBBX7, RING-type E3 ligase MYB30-INTERACTING E3 LIGASE 1 (MIEL1). Further examination revealed that MdMIEL1 could mediate the ubiquitination and degradation of MdBBX7 by the 26S proteasome pathway. Genetic interaction analysis suggested that MdMIEL1 acts as an upstream factor of MdBBX7. In addition, MdMIEL1 was a negative regulator of the apple drought stress response. Taken together, our results illustrate the molecular mechanisms by which the MdMIEL1–MdBBX7 module influences the response of apple to drought stress.
A ubiquitin E3 ligase, MdMIEL1, mediates drought tolerance through the ubiquitination and degradation of MdBBX7 regulating its downstream gene activation in response to drought stress.
Introduction
With its attractive appearance and pleasant taste, apple (Malus × domestica) has become an important component of the human diet. Global apple production is distributed primarily in temperate latitudes of Europe, North and South America, Asia, and Oceania. In 2018, apple production reached 86,142,200 tonnes from 4,904,304 harvested areas (FAO, 2020). Although the apple industry develops year after year, apple yields face threats from abiotic and biotic stress (Xie et al., 2018; Geng et al., 2018; Zhou et al., 2018; Li et al., 2020). Water scarcity is one of the main challenges that limit apple fruit quality and quantity (Liao et al., 2016). To date, many studies have reported useful methods for enhancing apple drought tolerance, including grafting onto drought-tolerant rootstocks, developing drought-resistant varieties by traditional crossbreeding and molecular breeding, and spraying growth regulators (Li et al., 2012, 2020; Sun et al., 2013; Wang et al., 2013a; Adams et al., 2018; Foster et al., 2018). Due to the long juvenile phase of apple trees, biotechnology is an effective way to improve apple drought tolerance. Hence, a thorough understanding of apple responses to drought stress at the molecular level is necessary for the development of drought-tolerant cultivars.
Plants have evolved a series of responses to deal with the adverse effects of drought stress at the morphological, physiological, and molecular levels (Farooq et al., 2009; Anjum et al., 2011; Santos et al., 2019). Drought stress affects the physiological morphology of plants, leading to increased peroxide content and activating antioxidant enzymes including peroxidase (POD) and catalase (CAT; Ma et al., 2018; Sun et al., 2018). As the first tissues affected by soil water deficit, roots also alter their morphology and distribution to adapt to drought stress. Under water-limited conditions, shoot growth is inhibited, but roots can continue to elongate in acclimation to drought stress (Yamaguchi and Sharp, 2010; Sharp et al., 2004).
Plant morphological and physiological responses to drought stress have been well studied; however, the molecular mechanisms that underlie drought tolerance are more elusive and complex. Transcriptional regulation plays a central role in the control of plant development and responses to abiotic stress. The B-BOX (BBX) proteins are a subgroup of zinc-finger transcription factors with special tertiary structures that are stabilized by the binding of a zinc ion (Klug and Schwabe, 1995). BBX proteins contain one or two B-box motifs at the N terminus, and some also contain a conserved CCT (CO, COL, TOC1) domain at the C terminus (Strayer et al., 2000; Ledger et al., 2001). The B-box motif in BBX proteins contains conserved residues and mediates transcriptional regulation, protein–protein interactions, and heterodimer formation within or outside the BBX family (Gangappa and Botto, 2014; Ding et al., 2018). The CCT domain functions in protein localization, transcriptional regulation, and nuclear protein transport (Yan et al., 2011; Gendron et al., 2012). BBX proteins participate in various developmental and physiological processes, including plant photomorphogenesis, flowering, shade avoidance, hormonal signaling, thermomorphogenesis, anthocyanin accumulation, carotenoid biosynthesis, leaf senescence, and stress response (Datta et al., 2007; Crocco et al., 2010; Yan et al., 2011; Gangappa and Botto, 2014; Wang et al., 2015; Ding et al., 2018; An et al., 2019; Bai et al., 2019; Fang et al., 2019; Liu et al., 2019a, 2019b; Xiong et al., 2019). CONSTANS (CO) was the first BBX protein identified and controls the photoperiodic regulation of flowering in Arabidopsis (Putterill et al., 1995). In total, 16 CO-LIKE (COL) proteins have been identified in Arabidopsis. BBX7/COL9 contains two B-box motifs and one CCT motif, and its expression was upregulated by the circadian clock in the photoperiod pathway. The overexpression of BBX7/COL9 delayed flowering, whereas knockout mutants and co-suppression lines flowered earlier under long-day conditions. Further study revealed that BBX7/COL9 functions upstream of CO and downstream of the circadian clock (Cheng and Wang, 2005). However, the potential role of BBX7/COL9 in drought stress tolerance of Arabidopsis and other crops is largely unexplored.
Post-translational modification is an effective way to regulate transcription factor activity. The ubiquitination process, which involves E1 (ubiquitin-activating enzymes), E2 (ubiquitin-conjugating enzymes), and E3 (ubiquitin ligases), is a representative means of modulating protein turnover (Lee and Seo, 2016; Morimoto et al., 2017). Among the three ubiquitination enzymes, E3 ligase is the key factor responsible for the specificity of target proteins. MIEL1 (MYB30-INTERACTING E3 LIGASE 1) has been reported to be a RING-type (Really Interesting New Gene) E3 ligase that plays a role in ubiquitination-mediated degradation of target proteins. In Arabidopsis, MIEL1 interacts with MYB30 and mediates its degradation by ubiquitination, leading to decreased transcriptional activity and reduced responses to bacterial infection (Marino et al., 2013). In addition, MIEL1 also interacts with and ubiquitinates MYB96 and negatively regulates abscisic acid (ABA) sensitivity and cuticular wax biosynthesis (Lee and Seo, 2016; Gil et al., 2017). In apple, MdMIEL1 has been found to inhibit anthocyanin accumulation by mediating ubiquitination and degradation of MdMYB1 in apple calli (An et al., 2017). In addition, MdMIEL1 targets MdMYB308L, which is a positive regulator of cold tolerance and anthocyanin accumulation (An et al., 2020b). Nevertheless, it is unclear whether MIEL1 also participates in responses to drought stress.
In this study, we demonstrate that apple MdBBX7/MdCOL9 is a positive regulator of drought tolerance. Besides the two T/G-boxes, MdBBX7 also recognizes an element (CCTTG) for transcriptional regulation of target genes, which include ETHYLENE RESPONSE FACTOR (ERF1), EARLY RESPONSIVE TO DEHYDRATION 15 (ERD15), and GOLDEN2-LIKE 1 (GLK1). MdBBX7 physically interacts with MdMIEL1 E3 ligase and is ubiquitinated and degraded by MdMILE1. In addition, MdMIEL1 has a negative effect on drought tolerance.
Results
Apple MdBBX7/MdCOL9 is a positive regulator in response to drought stress
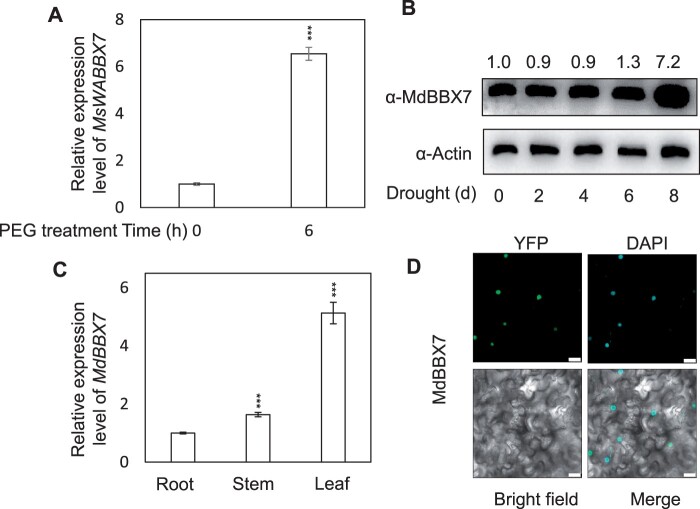
Previously, RNA-seq data showed that a gene encoding a zinc-finger transcription factor, MsBBX7, was markedly induced by simulated drought stress in the roots of Malus sieversii (Geng et al., 2019). Using the same plant material, Quantitative reverse-transcription polymerase chain reaction (RT-qPCR) analysis revealed that MsBBX7 was induced more than six-fold after PEG treatment (Figure 1A). In order to further verify whether this gene is also induced by drought in the cultivated apples (Malus × domestica), we used cultivated apples (Malus × domestica cv Golden Delicious) for further verification. Using a native antibody of MdBBX7, we found that MdBBX7 protein showed the greatest accumulation after 8 d of drought treatment in Malus × domestica (“Golden Delicious”; Figure 1B), and we speculated that MdBBX7 might participate in the apple drought response. We therefore cloned the MdBBX7 gene from Malus × domestica and found that it contained two conserved B-box motifs and one CCT motif (Supplemental Figure S1). The sequence similarity of MdBBX7 with Arabidopsis BBX7 was 57%. Phylogenic analysis showed that apple MdBBX7 proteins were widely distributed in eudicots, monocots, Lycopodiophyta, Bryophyta, Coniferophyta, Hepaticae, and Chlorophytae, suggesting its ancient origin (Supplemental Figure S2). Tissue-specific expression analysis showed that MdBBX7 was predominantly expressed in apple leaves, followed by stems and roots (Figure 1C). As a typical transcription factor, MdBBX7 was located in the nucleus (Figure 1D).
Figure 1.
Expression patterns of MdBBX7. A, Expression of MdBBX7 in response to simulated drought treatment (20% PEG) for 6 h. Root samples were collected from 2-month-old Malus sieversii for RT-qPCR. Asterisks indicated values significantly different between 0 h and 6 h. B, The protein level of MdBBX7 in response to drought stress. Three-month-old apple (Malus × domestica) plants grown in a greenhouse were treated with drought for 0, 2, 4, 6, or 8 d. C, Expression of MdBBX7 in different Malus prunifolia tissues, including roots, stems, and leaves. Asterisks indicated values significantly different from stem or leaf compared to root. D, Subcellular localization of MdBBX7. MdBBX7 was fused with a YFP for transient transformation into 4-week-old tobacco (N. benthamiana) leaves. DAPI was used as a nuclei marker. Bars = 25 μm. Data are means ± sd (n = 3). One-way ANOVA (Tukey’s test) was performed and statistically significant differences were indicated by ***P < 0.001.
To explore the biological function of MdBBX7 in the drought stress response, we generated MdBBX7 RNAi and overexpression (OE) transgenic plants by a stable transformation approach with GL-3 as the genetic background. RT-qPCR analysis showed that the relative expression level of MdBBX7 was decreased 60%–70% in the RNAi plants and upregulated three-to four-fold in the OE transgenic lines (Supplemental Figure S3, A and B). A western blot assay produced results consistent with the mRNA expression levels (Supplemental Figure S3, C and D). Under control conditions, the MdBBX7 RNAi plants were shorter than nontransgenic GL-3 controls, and the MdBBX7 OE plants were taller (Supplemental Figure S4A). Shoot dry weight displayed a similar pattern (Supplemental Figure S4B). In addition, MdBBX7 RNAi plants had shorter root lengths and lower root dry weights compared with GL-3, whereas MdBBX7 OE plants had longer root lengths and greater root dry weights (Supplemental Figure S4, C–E), suggesting a potential role of MdBBX7 in root development.
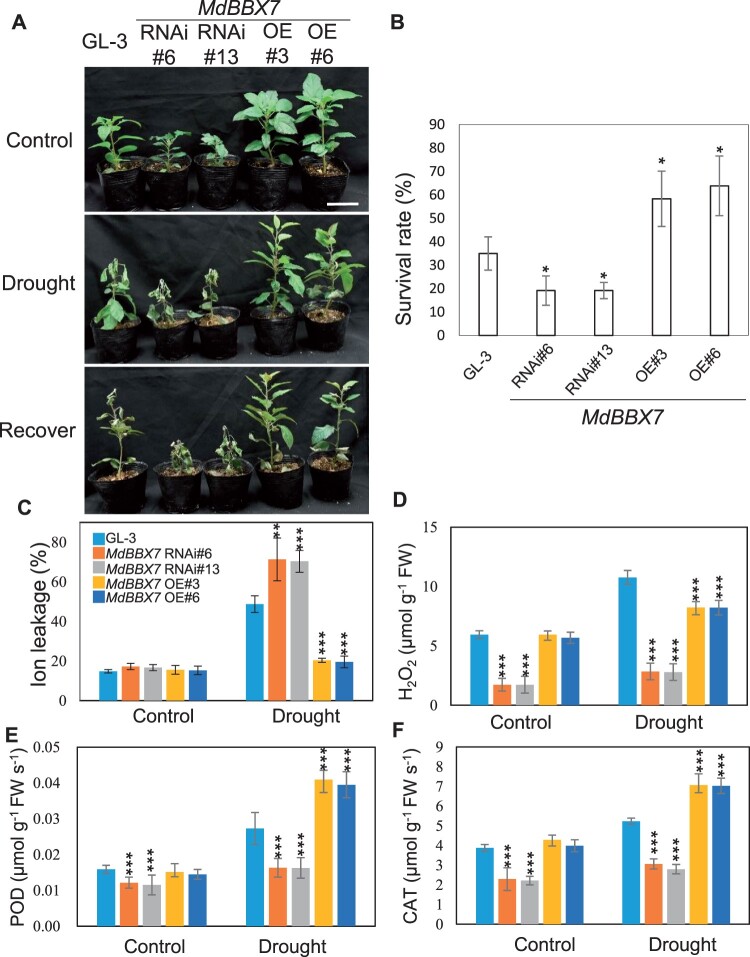
After the drought stress treatment, 35% of the GL-3 plants had survived, compared with only 20% of the MdBBX7 RNAi plants. In contrast, 59%–63% of the MdBBX7 OE plants survived the drought stress treatment (Figure 2, A and B). Consistently, membranes of the MdBBX7 RNAi leaves suffered more damage than those of GL-3 plants after drought stress treatment, as revealed by higher ion leakage levels in the MdBBX7 RNAi leaves. However, the ion leakage of MdBBX7 OE leaves was significantly lower than that of GL-3 leaves under drought. Moreover, the ion leakage of MdBBX7 OE leaves did not increase significantly after drought (19.58%) compared with control conditions (15.30%; Figure 2C;Supplemental Figure S5A). In contrast, ion leakage of MdBBX7 RNAi leaves increased from 16% to 19% under control conditions to 70% after drought stress (Figure 2C), implying that MdBBX7 RNAi plants experienced greater drought-related damage. In addition, MdBBX7 OE plants showed lower H2O2 content, and higher activities of antioxidant enzymes including POD and CAT compared with GL-3 plants in response to drought stress (Figure 2, D–F; Supplemental Figure S5, B–D). MdBBX7 RNAi plants had weaker POD and CAT activities, and lower H2O2 content under drought conditions, as compared with GL-3 plants (Figure 2, D–F; Supplemental Figure S5, B–D). ABA, the key hormone involved in drought tolerance, significantly accumulated more in MdBBX7 OE plants, whereas MdBBX7 RNAi plants accumulated less in response to drought stress, as compared with GL-3 plants (Supplemental Figure S5E).
Figure 2.
Response of MdBBX7 transgenic plants to drought. A, Morphological characteristics of GL-3 and MdBBX7 transgenic plants after drought stress. Bars = 5 cm. Three-month-old MdBBX7 RNAi and OE transgenic plants and GL-3 were withheld from water until soil volumetric water content reached to 0% and then rewatered for 7 d. B, Survival rate of the plants shown in (A). Asterisks indicated values significantly different between the transgenic lines and the GL-3. C–F, Physiological parameters of GL-3 and MdBBX7 transgenic plants after drought stress. Plants were withheld from water until soil volumetric water content reached to 5%. C, Leaf ion leakage. D, H2O2 content. E, POD activity. F, CAT activity. Asterisks indicated values significantly different between the transgenic lines and the GL-3 in each group (Control and Drought). Data are means ± sd (n = 3 in (B), 8 in (C), 5 in (D)–(F)). One-way ANOVA (Tukey’s test) was performed and statistically significant differences were indicated by *P < 0.05, **P < 0.01, or ***P < 0.001.
Taken together, the above results suggest that apple MdBBX7 is a positive regulator of drought stress tolerance.
MdBBX7 regulates the expression of drought-responsive genes
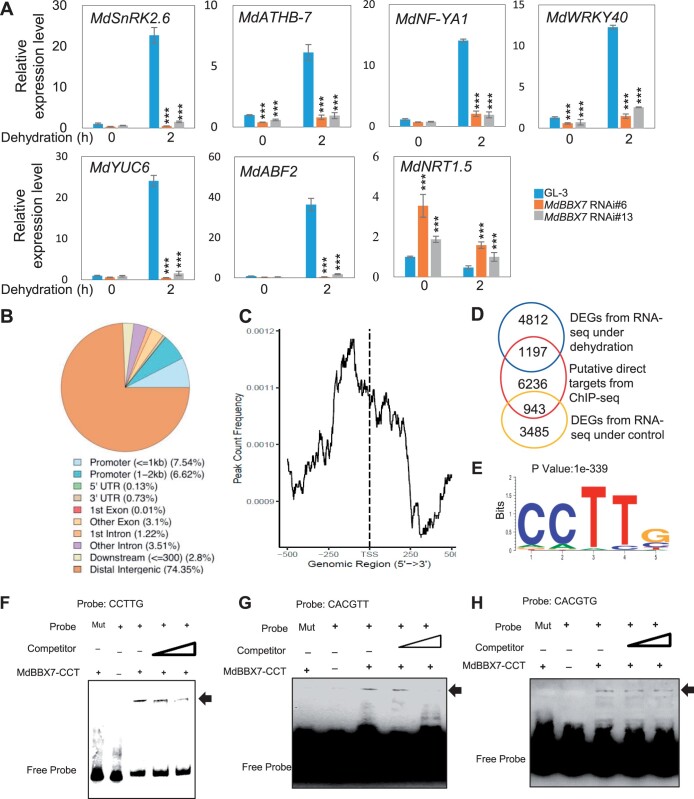
To better understand how MdBBX7 influences drought stress tolerance, we performed an RNA-seq analysis of GL-3 and MdBBX7 RNAi plants under control and drought conditions. Under control conditions, 1,862 genes were upregulated and 2,566 genes were downregulated in MdBBX7 RNAi plants, as compared with GL-3 (P < 0.05, 2.0-fold threshold; Supplemental Data Set S1). After 6 d of drought stress, 2,965 genes were upregulated in the MdBBX7 RNAi plants and 3,044 genes were downregulated compared with GL-3 (P < 0.05, 2.0-fold threshold; Supplemental Data Sets S2 and S3). Gene ontology (GO) analysis indicated that the differentially expressed genes (DEGs) were related to response to stimulus, response to oxygen-containing compound, response to abiotic stimulus, response to hormone, and response to stress (Supplemental Figure S6). KEGG enrichment analysis revealed that they were enriched in processes such as biosynthesis of secondary metabolites, plant hormone signal transduction, MAPK signaling pathway, and flavonoid biosynthesis (Supplemental Figure S7). We selected seven genes with which to verify the results of the transcriptome analysis: MdSnRK2.6, MdATHB-7, MdNF-YA1, MdWRKY40, MdYUC6, MdABF2, and MdNRT1.5. Homologs of these genes in Arabidopsis are involved in aspects of drought response, including stomatal movement, chlorophyll fluorescence, ABA accumulation, ROS production, and nitrate allocation (Söderman et al., 1996; Chen et al., 2012; Mishra et al., 2012; Li et al., 2013; Cha et al., 2015; Ding et al., 2015; Geilen and Boehmer, 2015; Wei et al., 2020). The expression of MdSnRK2.6, MdAtHB-7, MdNF-YA1, MdWRKY40, MdYUC6, and MdABF2 was downregulated in MdBBX7 RNAi plants under drought stress, whereas that of MdNRT1.5 was upregulated (Figure 3A), consistent with the RNA-seq results.
Figure 3.
Identification of MdBBX7 recognition sites. A, Expression of drought-responsive genes in MdBBX7 RNAi plants under control and dehydration conditions. B, Distribution of MdBBX7 binding regions in the apple genome. ChIP was performed using tissue-cultured GL-3 seedlings with anti-MdBBX7 antibody. C, Peak distance from the TSS of MdBBX7. The peaks were highly enriched from −250 bp to 0 from the TSS. D, Numbers of regulon detected by ChIP-seq and RNA-seq under control and dehydration conditions. DEGs were from the MdBBX7 RNAi plants detected by RNA-seq under control and dehydration conditions. E, The CCTTG element was identified from ChIP-seq analysis using MEME-ChIP. F–H, EMSA analysis showing that MdBBX7 protein bound to the element of CCTTG (F), CACGTT (G), and CACGTG (H). The C terminal region of MdBBX7 containing the CCT domain (207–410 aa) was used. Mut was presented the mutant probe. Binding sites in mutant probe were replaced with AAAAA or AAAAAA. Data are means ± sd (n = 3). Asterisks indicated values significantly different between the transgenic lines and the GL-3 in each group (0 h and 2 h). One-way ANOVA (Tukey’s test) was performed and statistically significant differences were indicated by ***P < 0.001.
MdBBX7 recognizes the T/G-box and the CCTTG element in the promoters of its targets
It has been reported that Arabidopsis BBX proteins bind to the T/G-box motif of their target gene promoters (Rajnish Khanna and Maszle, 2009; Crocco and Botto, 2013; Gangappa and Botto, 2014). Because the sequence similarity between MdBBX7 and AtBBX7 was 57% (Supplemental Figure S1), we speculated that MdBBX7 might recognize different motifs. We therefore performed a chromatin immunoprecipitation-sequencing (ChIP-seq) analysis using anti-MdBBX7 antibody produced by immunizing rabbits with polypeptides (14 aa) from MdBBX7. After analyzing the sequenced reads with Homer and MEME-ChIP, we identified 21,344 binding peaks of MdBBX7 in the apple genome (Supplemental Data Set S4), as well as one distinct and significant sequence for MdBBX7 binding, CCTTG (Figure 3, B–E). We verified the binding of MdBBX7 to this binding motif by electrophoresis mobility shift assay (EMSA) analysis. When probe CCTTG was used, the complex of MdBBX7-probe could be detected. However, when CCTTG was mutated to AAAAA, no MdBBX7-probe was present (Figure 3F). The T/G-box motifs (CACGTT/G) are known BBX recognition motifs (Xu et al., 2018; Xiong et al., 2019). Using EMSA analysis, we also verified recognition of the T/G box binding by MdBBX7 in apple (Figure 3, G–H).
Among the 23,039 peaks, approximately 22.86% were located in the genic regions of 3,185 genes, and 14.16% were located in the promoter regions of 2,239 genes (∼2 kb from the transcription start site, TSS; Figure 3B). The peaks were substantially enriched 500 bp upstream of the TSS (Figure 3C). We functionally annotated the 8,377 genes from the total peak results and performed GO and KEGG analysis. Enriched GO terms were predominantly related to organ development and metabolic processes (including hormones), such as reproductive shoot system development, flower development, regulation of hormone levels, meristem development, and phenylpropanoid metabolic process (Supplemental Figure S8). KEGG enrichment analysis revealed that these genes were enriched in diterpenoid biosynthesis (Supplemental Figure S9). We also identified 6,236 putative targets of MdBBX7 with both RNA-seq and ChIP-seq analysis (Figure 3D;Supplemental Data Set S5).
MdERF1, MdERD15, and MdGLK1 are direct targets of MdBBX7
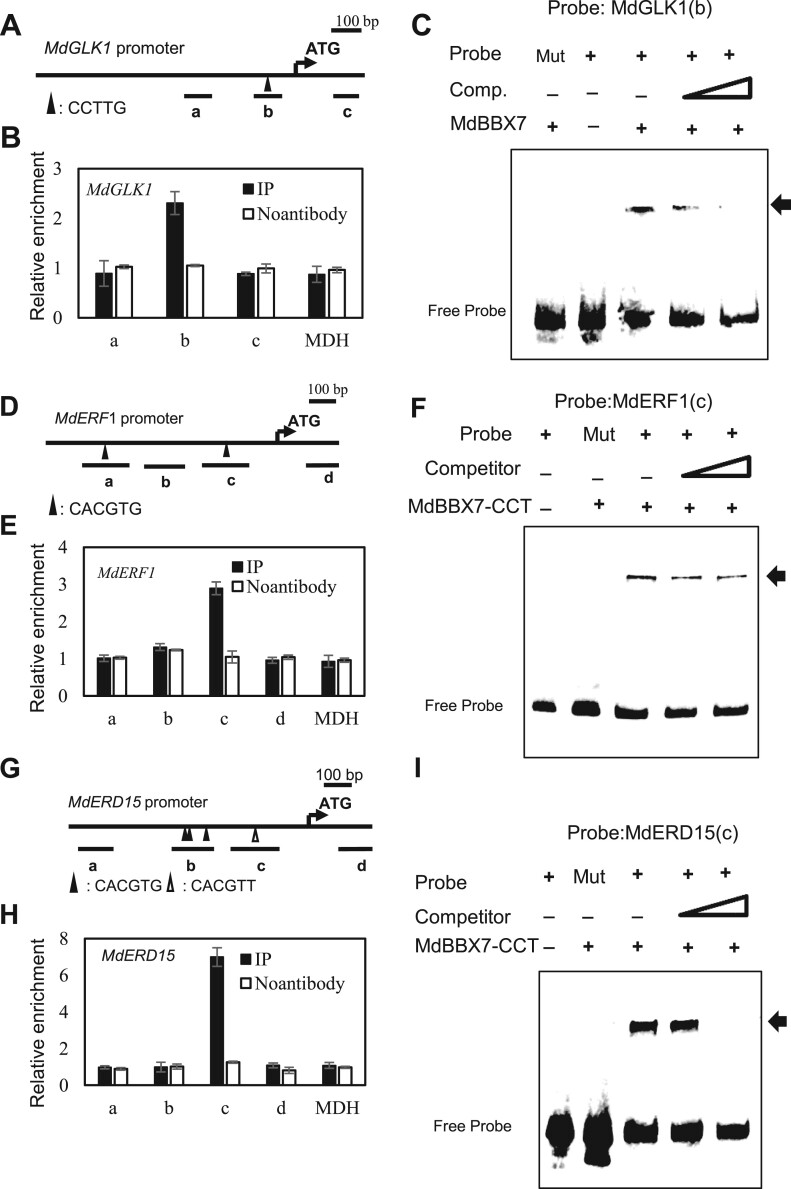
We selected several target genes from the ChIP-seq data to further verify based on three criteria: (1) The promoters contained the T/G-box or CCTTG motifs. (2) The binding peaks identified in ChIP-seq were nearby the TSS (<500 bp). (3) The genes or their homologs should play critical roles in drought tolerance. Homologs of MdERF1, MdERD15, MdGLK1, MdWRKY40, and MdCAT2 in Arabidopsis are critical regulators of drought stress tolerance (Kiyosue et al., 1994; Cheng et al.,2013; Lestari et al., 2018; Nagatoshi et al., 2016; Wang et al., 2021; He et al., 2021). We then selected MdERF1, MdERD15, MdGLK1, MdWRKY40, and MdCAT2 to confirm their identity as direct targets of MdBBX7. Using ChIP-qPCR, we found that MdBBX7 was enriched in the promoter regions of MdERF1, MdERD15, and MdGLK1(Figure 4, B, E, and H), but not MdWRKY40 or MdCAT2 (Supplemental Figure S10). Fragment b of MdGLK1 promoter contains CCTTG motif (Figure 4A). Fragment c of MdERF1 and MdGLK1 promoter contains G-box and T-box, respectively (Figure 4, D and G). Results showed that MdBBX7 could bind fragment c of MdERF1 and MdERD15, and fragment b of MdGLK1 (Figure 4). We next used EMSA to further verify the binding of MdBBX7 to these fragments. A gel shift assay revealed that MdERF1, MdERD15, and MdGLK1 were indeed direct targets of MdBBX7, as revealed by the MdBBX7-probe complex on the PAGE gel. When CCTTG or T/G box in the probes was mutated to AAAAA or AAAAAA, respectively, no MdBBX7-probe complex was observed on the gel (Figure 4, C, F, and I). We also examined whether MdGLK1, MdERF1, and MdERD15 responded to drought stress. The results showed that the expression of these three genes positively associated with that of MdBBX7 under drought conditions (Supplemental Figure S11).
Figure 4.
MdBBX7 binds to the cis-elements of CCTTG, CACGTG, or CACGTT in promoters of MdGLK1, MdERF1, and MdERD15. A, Diagram of MdGLK1 promoter regions. Fragment b contains CCTTG element. B, ChIP-qPCR analysis of MdBBX7 binding to the promoter of MdGLK1. Fragment a, c, MDH, and no antibody serve as negative controls. C, EMSA analysis showing that the MdBBX7 protein bound to the promoter of MdGLK1. (D) Diagram of MdERF1 promoter regions. Fragment a and c contain CACGTG element. E, ChIP-qPCR analysis of MdBBX7 binding to the promoter of MdERF1. Fragment b, d, MDH, and no antibody serve as negative controls. F, EMSA analysis showing that the MdBBX7 protein bound to the promoter region of MdERF1. G, Diagram of MdERD15 promoter regions. Fragment b contains CACGTG element. Fragment c contains CACGTT element. H, ChIP-qPCR analysis of MdBBX7 binding to the promoter of MdGLK1. Fragment a, d, MDH, and no antibody serve as negative controls. I, EMSA analysis showing that the MdBBX7 protein bound to the promoter region of MdERD15. The C terminal region of MdBBX7 containing the CCT domain (207–410 aa) was used. Binding sites in mutant probe were replaced with AAAAA or AAAAAA. Data are means ± sd (n = 3).
MdBBX7 interacts with a ubiquitin E3 ligase, MdMIEL1, in vivo and in vitro
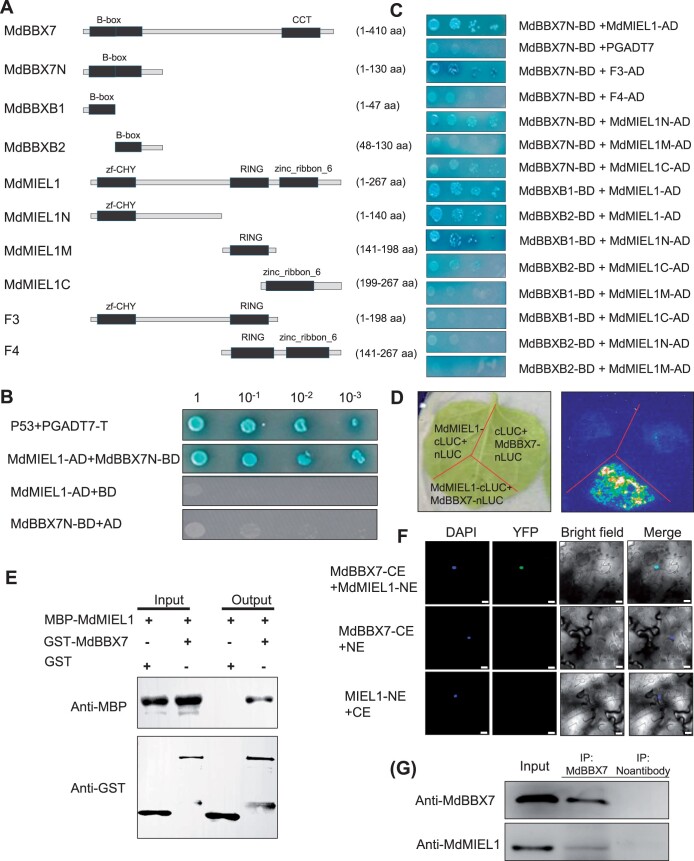
To reveal the molecular mechanism by which MdBBX7 influences drought response, a yeast-two hybrid (Y2H) screen was performed. Because MdBBX7 has self-activation activity (Supplemental Figure S12) and the B-box motifs are often responsible for protein–protein interactions, we used a deleted MdBBX7 (1–130 aa) that contained only the two B-box motifs and lacked self-activation activity (Supplemental Figure S13). Eight potential interacting proteins of MdBBX7 were identified, including the ubiquitin E3 ligase, MYB30-interacting E3 ligase 1 (MdMIEL1) (Supplemental Data S1). Y2H analysis verified this interaction (Figure 5, A–C). To identify the regions responsible for the interaction between MdBBX7 and MdMIEL1, we constructed a series of truncated MdBBX7 and MdMIEL1 proteins. Using Y2H analysis, we found that the two B-box motifs in MdBBX7 were required for the interaction between MdBBX7 and MdMIEL1 (Figure 5C). The GST pull-down assay, bimolecular fluorescence complementation (BiFC) analysis, split-luciferase assay, and co-immunoprecipitation (Co-IP) assay further confirmed the interaction between MdBBX7 and MdMIEL1 in vitro and in vivo (Figure 5, D–G).
Figure 5.
MdBBX7 interacts with MdMIEL1 in vivo and in vitro. A, Schematic diagrams of MdBBX7 deletions and MdMIEL1 deletions. F3 contains MdMIEL1 aa 1–198, whereas F4 contains MdMIEL1 aa 141–267. B, Y2H assays showing the domains responsible for the interaction between MdBBX7 and MdMIEL1. Yeast cells were grown on SD–Ade–His–Leu–Trp and subjected to x-gal assay. C, Interaction of MdBBX7N (N terminus of MdBBX7 containing the two B-box motifs) with full-length MdMIEL1 by Y2H analysis. D, Firefly luciferase complementation imaging assay for the MdBBX7 and MdMIEL1 interaction in tobacco (N. benthamiana). E, Pull-down assay analyzing the interaction of MdBBX7-N214 with MdMIEL1 in vitro. MBP-MdMIEL1 and GST-MdBBX7-N214 proteins expressed in E. coli were purified and used in this analysis. F, BiFC assay showing that MdBBX7 and MdMIEL1 interact in tobacco. Bars = 25 μm. G, In vivo interaction of MdBBX7 and MdMIEL1 detected by Co-IP assay. Proteins were extracted from 4-week-old MdBBX7 OE plants leaves, and immunoprecipitated with anti-MdBBX7 antibody. Western blotting was performed with anti-MdBBX7 or anti-MdMIEL1.
MdMIEL1 ubiquitinates MdBBX7 and mediates its degradation
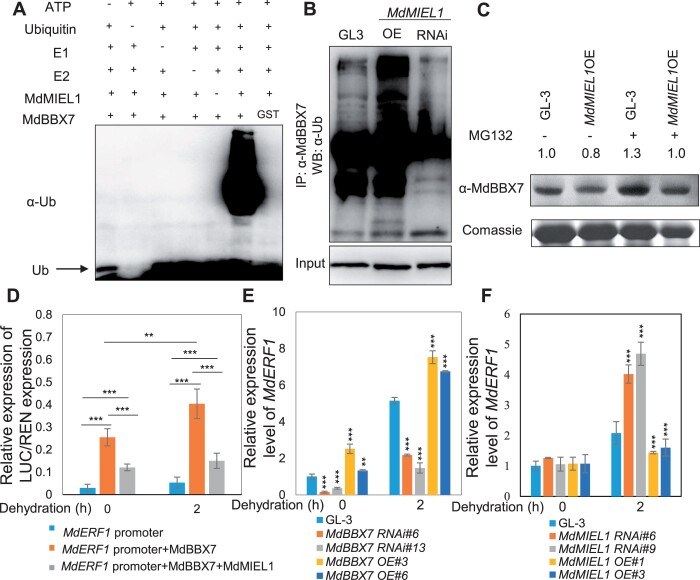
MdMIEL1 is a ubiquitin E3 ligase that contains conserved motifs, including a typical zinc-ribbon finger, a CHY zinc finger, and a RING-finger (Supplemental Figure S13). MdMIEL1 shares high sequence similarity (81%) with Arabidopsis MdMIEL1 (Supplemental Figure S13). Previous reports have shown that MdMIEL1 functions in the ubiquitination and degradation of its interacting proteins (An et al., 2017). The physical interaction of MdBBX7 and MdMIEL1 prompted us to speculate that MdMIEL1 might ubiquitinate and degrade MdBBX7. Using recombinant proteins of MdMIEL1 and MdBBX7, we performed an in vitro ubiquitination assay and found that MdMIEL1 could ubiquitinate MdBBX7 in the presence of ubiquitin E1, E2, and ATP (Figure 6A). To further verify the ubiquitination of MdBBX7 by MdMIEL1 in vivo, we generated stable MdMIEL1 transgenic plants with either increased expression (OE) or reduced expression (RNAi) of MIEL1 (Supplemental Figure S14) and performed an in vivo ubiquitination assay. Total proteins were extracted from plants and MdBBX7 protein was immunoprecipitated with anti-MdBBX7 antibody, then anti-ubiquitin antibody was used for western blot detection. Results showed that MdBBX7 had a higher ubiquitination level in MdMIEL1 OE plants than in GL-3 plants, as revealed by the presence of high-molecular-weight polypeptide bands. However, MdBBX7 had a lower ubiquitination level in MdMIEL1 RNAi plants, as shown by fewer corresponding polypeptide bands (Figure 6B). These results further suggest that MdMIEL1 mediates the ubiquitination of MdBBX7.
Figure 6.
MdMIEL1 ubiquitinates and degrades MdBBX7, and inhibits its transcriptional activity. A, MdMIEL1 ubiquitinated MdBBX7 in vitro. The ubiquitination status of MdBBX7 was detected in the presence of ubiquitin E1, E2, and MdMIEL1 using an anti-ubiquitin antibody. B, MdMIEL1 ubiquitinated MdBBX7 in vivo. Total protein was extracted from GL-3, MdBBX7 OE, and MdBBX7 RNAi transgenic plants and immunoprecipitated with anti-MdBBX7 antibody, and was subjected to western blot analysis with anti-ubiquitin. WB, western blot. C, The MdBBX7 protein was degraded by MdMIEL1 via the 26S proteasome pathway. Four-week-old tissue-cultured GL-3 and MdMIEL1 OE transgenic plants were treated with or without the proteasome inhibitor MG132 for 12 h. The proteins were extracted from GL-3, MdMIEL1 OE transgenic plants and immunoblotted with anti-MdBBX7 antibody. D, Relative luciferase activity from the dual luciferase reporter assays in N. benthamiana leaves. Pro35S::REN was used as an internal control. Quantification was performed by normalizing firefly luciferase activity to that of renilla luciferase. E, Expression of MdERF1 in MdBBX7 RNAi and OE plants in response to drought stress. F, Expression of MdERF1 in MdMIEL1 RNAi and OE plants in response to drought stress. Data are means ± sd (n = 5 in (D), 3 in (E) and (F)). Asterisks indicated values significantly different between the transgenic lines and the GL-3 in each group (0 h and 2 h) or the line indicated. One-way ANOVA (Tukey’s test) was performed, and statistically significant differences are indicated by **P < 0.01, or ***P < 0.001.
We measured the protein level of MdBBX7 in GL-3 and MdMIEL1 transgenic plants. Western blot analysis showed that MdBBX7 was less abundant in MdMIEL1 OE plants, suggesting a degradation of MdBBX7 by MdMIEL1 (Figure 6C). The degradation of MdBBX7 was accelerated in MdMIEL1 OE plants compared with GL-3 plants; however, MdBBX7 stability was increased by the application of MG132 (Figure 6C). These results indicate that MdBBX7 protein is degraded by MdMIEL1 through the 26S proteasome pathway.
Similar to MdBBX7, MdMIEL1 was localized in the nuclei (Supplemental Figure S15A). The mRNA level of MdMIEL1 decreased by drought treatment (Supplemental Figure S15B). Moreover, the accumulation of MdMIEL1 protein decreased in response to drought stress. After 6 d of drought treatment, almost no MdMIEL1 protein was detected (Supplemental Figure S15C), a pattern opposite to that of MdBBX7 expression under drought stress.
MdMIEL1 inhibits the DNA binding activity of MdBBX7
Because MdMIEL1 interacted with and degraded MdBBX7—and MdBBX7 directly regulated target gene expression, we next asked whether MdMIEL1 affected the transcriptional activity of MdBBX7. We performed transient dual-luciferase assays with the LUC reporter gene driven by the MdERF1 promoter. LUC activity analysis suggested that MdBBX7 could directly activate expression of the MdERF1 promoter. However, when MdMIEL1 was co-expressed with MdBBX7, the activation of the MdERF1 promoter by MdBBX7 was significantly decreased (Figure 6D), indicating that MdMIEL1 inhibits the transcriptional activity of MdBBX7. In addition, RT-qPCR analysis showed that MdERF1 expression was downregulated under drought in MdBBX7 RNAi plants and upregulated in MdBBX7 OE plants (Figure 6E). In contrast, MdERF1 expression was elevated in MdMIEL1 RNAi plants but reduced in MdMIEL1 OE plants (Figure 6F). The transcriptional regulation of MdERF1 by MdBBX7 and MdMIEL1 was consistent with the dual-luciferase results, further indicating that MdBBX7 activates MdERF1 expression and MdMIEL1 negatively regulates MdERF1, probably by degrading MdBBX7.
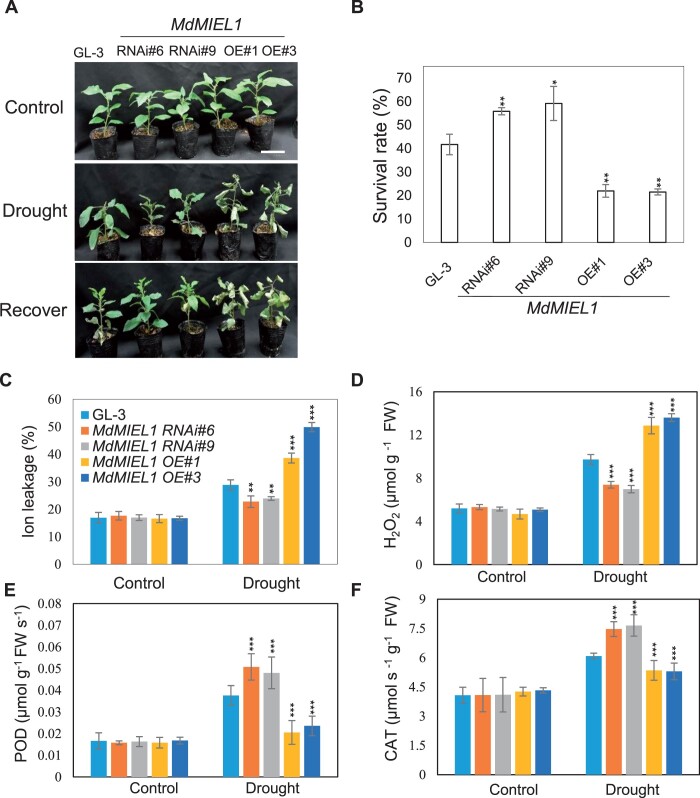
MdMIEL1 negatively regulates apple drought stress
To examine the biological function of MdMIEL1 in response to drought stress, we assessed the drought tolerance of the MdMIEL1 OE and RNAi transgenic plants. After drought stress followed by re-watering, 40% of GL-3 plants survived compared with only 22% of MdMIEL1 OE plants. In comparison, 56%–59% of the MdMIEL1 RNAi plants survived the drought stress (Figure 7, A and B). Under drought stress, the leaves of MdMIEL1 OE plants displayed the greatest cell membrane damage, followed by GL-3 and MdMIEL1 RNAi plants (Figure 7C). In addition, MdMIEL1 RNAi plants had higher activities of POD and CAT, leading to less H2O2 than GL-3 plants under drought stress conditions. However, MdMIEL1 OE plants had lower POD and CAT activities, thereby more H2O2 than GL-3 plants under drought stress (Figure 7, D–F). Finally, MdMIEL1 RNAi plants accumulated more ABA while MdMIEL1 OE plants had less in response to drought stress when comparing with GL-3 plants (Supplemental Figure S16). These results indicated that MdMIEL1 negatively regulates apple drought stress tolerance.
Figure 7.
Drought resistance of MdMIEL1 transgenic plants. A, Morphological characteristics of GL-3 and MdMIEL1 transgenic plants after drought stress. Bars = 5 cm. Three-month-old MdMIEL1 RNAi, OE transgenic plants, and GL-3 plants were withheld water until relative soil volumetric water content reached to 0%, and then rewatered for 7 d. B, Survival rate of the plants shown in (A). Asterisks indicated values significantly different between the transgenic lines and the GL-3. C–F, The physiological parameters of GL-3 and MdMIEL1 transgenic plants after drought stress. Plants were withheld from water until soil volumetric water content reached to 5%. C, Leaf ion leakage. D, H2O2 content. (E) POD activity. (F) CAT activity. Data are means ± sd (n = 3 in (B), 8 in (C), 5 in (D)–(F)). Asterisks indicated values significantly different between the transgenic lines and the GL-3 in each group (Control and Drought). One-way ANOVA (Tukey’s test) was performed and statistically significant differences were indicated by **P < 0.01 or ***P < 0.001.
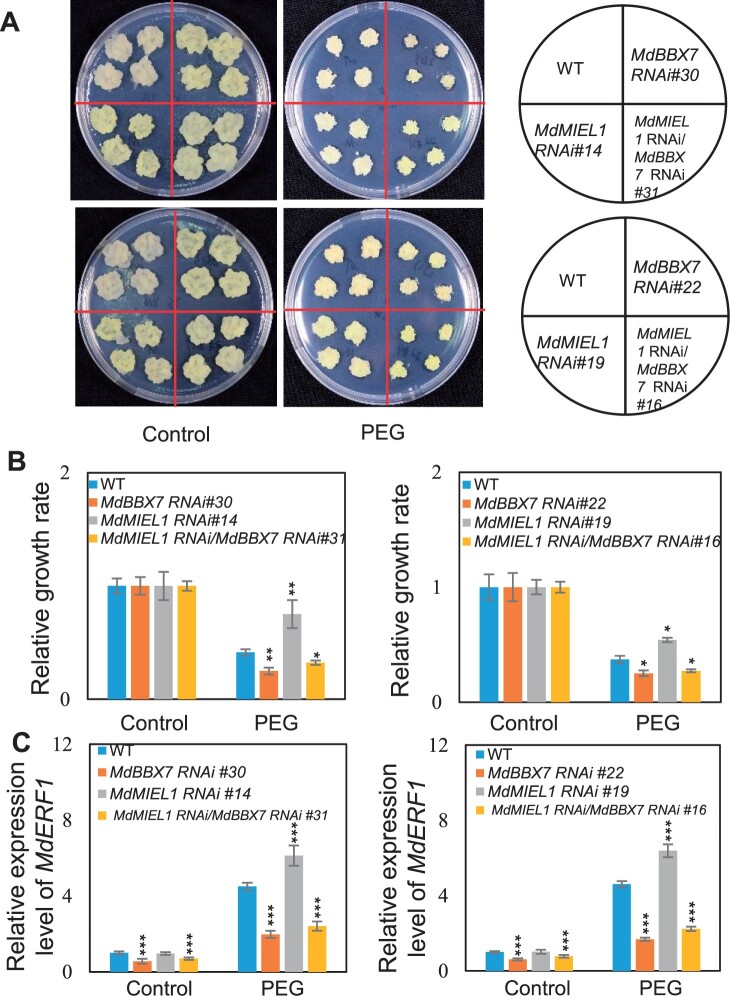
MdMIEL1 acts as an upstream factor of MdBBX7 to negatively regulate apple drought response
Our data showed that MdMIEL1 negatively regulates apple drought stress response (Figure 7), whereas MdBBX7 positively regulate drought response by positively regulate MdERF1 (Figure 2). To further investigate the relationship between MdBBX7 and MdMIEL1 in regulating apple drought response, we generated transgenic calli with reduced expression of MdBBX7 or MdMIEL1. We also knocked down the expression of MdMIEL1 in the background of MdBBX7 RNAi transgenic calli (Supplemental Figure S17). We then treated these transgenic calli and the wild-type (WT) with PEG6000 for 2 weeks. We observed that under low water potential conditions (−0.7 MPa), MdMIEL1 RNAi calli had higher relative growth rate than the WT. However, MdBBX7 RNAi calli exhibited lower growth rate (Figure 8, A and B). Under PEG treatment, reduced expression of MdMIEL1 in MdBBX7 RNAi transgenic apple calli showed similar phenotype as MdBBX7 RNAi transgenic calli. We also examined the expression of MdERF1 in the transgenic calli under PEG conditions, and noticed that reduced expression of MdMIEL1 in the MdBBX7 RNAi calli slightly increased the expression of MdERF1, compared to the MdBBX7 RNAi calli (Figure 8C), suggesting MdMIEL1 regulates MdERF1 expression by mediating MdBBX7. We also observed similar pattern for the expression of MdERD15 and MdGLK1 under PEG conditions (Supplemental Figure S18), further supporting the notion that MdBBX7 is a downstream target of MdMIEL1. These results also imply that MdMIEL1 acts upstream of MdBBX7 in regulating apple drought stress response.
Figure 8.
Genetic analysis of MdMIEL1 and MdBBX7. A, Morphological characteristics of the WT, MdMIEL1 RNAi, MdBBX7 RNAi, and MdMIEL1 RNAi/MdBBX7 RNAi transgenic apple calli after PEG treatment. B, The relative growth rate of apple calli under PEG treatment. C, Relative expression level of MdERF1 in the WT and transgenic apple calli under control and PEG conditions. MdMIEL1 RNAi/MdBBX7 RNAi, knocking down MdMIEL1 in the background of MdBBX7 RNAi calli. Equal weight of WT and transgenic calli were grown on MS or MS supplemented with PEG (−0.7 MPa) for 2 weeks. Data are means ± sd (n = 5 in (B), 3 in (C)). Asterisks indicated values significantly different between the transgenic lines and the WT in each group (Control and PEG). One-way ANOVA (Tukey’s test) was performed, and statistically significant differences are indicated by *P < 0.05, **P < 0.01, or ***P < 0.001.
Discussion
Water deficit is a great challenge for agricultural production worldwide. Plant growth, development, and distribution are greatly affected by drought stress, resulting in decreased yields and economic losses (Kim et al., 2017; Kumar et al., 2019). Apple is a perennial woody plant that requires adequate irrigation during its development (Sun et al., 2018; Li et al., 2020). The development of drought-tolerant cultivars for arid and semi-arid areas is urgently needed, necessitating a thorough understanding of the molecular mechanisms by which apple responds to drought stress. In this study, we identified a zinc-finger protein, MdBBX7, as a positive regulator of drought response. Molecular approaches identified its interacting protein MdMIEL1, an ubiquitin E3 ligase. MdMIEL1 can ubiquitinate and degrade MdBBX7, thereby inhibiting its transcriptional activity. Under drought stress, MdBBX7 protein accumulated, whereas MdMIEL1 was degraded. Using ChIP-seq and RNA-seq analysis, we identified direct and indirect targets of MdBBX7, including MdERF1, a homolog of the drought positive regulator ERF1. More importantly, we identified another recognition motif of MdBBX7 in addition to the classical T/G-box.
Previous studies have shown that the BBX proteins play important roles in plant abiotic and biotic stress tolerance (Yan et al., 2011; Wang et al., 2013b; Gangappa and Botto, 2014; Liu et al., 2019b; Xiong et al., 2019; Zhang et al., 2020). BBX18 negatively regulates thermotolerance by modulating a set of heat shock-responsive genes in Arabidopsis, and BBX19 negatively regulates drought tolerance in chrysanthemum through an ABA-dependent pathway (Wang et al., 2013b; Ding et al., 2018; Xu et al., 2020). Sweet potato IbBBX24 enhances fusarium wilt resistance by promoting the jasmonic acid pathway (Zhang et al., 2020). In apple, recent findings reveal a role of BBX proteins in anthocyanin biosynthesis (Bai et al., 2014; An et al., 2019; Plunkett et al., 2019). The overexpression of MdCOL11/MdBBX22 enhanced the accumulation of anthocyanin (Bai et al., 2014), whereas MdBBX37 inhibited anthocyanin biosynthesis and promoted hypocotyl elongation by negatively regulating light signaling (An et al., 2020a). In addition, MdBBX1 enhanced the activation of MdMYB10 and MdbHLH3 on the promoter of MdDFR, an anthocyanin biosynthesis gene (Plunkett et al., 2019). However, the roles of MdBBXs in stress tolerance are largely unknown. In our study, we identified MdBBX7 as a drought positive regulator (Figure 2). Compared with GL-3, fewer MdBBX7 RNAi transgenic plants survived under short-term drought stress, whereas more MdBBX7 OE plants survived. MdBBX7 OE plants displayed enhanced antioxidant enzyme activities and less H2O2 (Figure 2). On the contrary, under drought stress, MdBBX7 RNAi transgenic plants had weaker antioxidant enzyme activities. BBX proteins are involved in plant development. Decreasing the level of BBX protein leads to the dwarf phenotype of plants (Crocco and Botto, 2013). By comparing the relative data in plants before and after drought, the influence of development on drought tolerance can be excluded (Zhou et al., 2020). Considering that the MdBBX7 transgenic plants showed developmental phenotype on plant height, we also calculated the relative level of ion leakage, H2O2 content, and activities of POD and CAT. Results showed that MdBBX7 RNAi plants showed higher relative ion leakage and H2O2 content, and lower relative activities of POD and CAT, as compared with GL-3 plants after drought treatment. By contrast, MdBBX7 OE plants displayed higher relative activities of POD and CAT, and lower relative ion leakage and H2O2 content (Supplemental Figure S5, B–E). These data indicate that developmental phenotype may not contribute to drought tolerance of MdBBX7 transgenic plants and further suggest that MdBBX7 is a positive regulator of drought stress tolerance.
Drought stress leads to the accumulation of the reactive oxygen species, which can cause the oxidative damage in plants (Scandalios, 1993; Farooq et al., 2009). Activating antioxidant enzymes including POD and SOD is the most important way to avoid the oxidative damage of plants (Maribel et al., 1998; Ma et al., 2018). GO enrichment analysis revealed that 394 DEGs under drought conditions in the MdBBX7 RNAi genes were related to “the response to oxygen-containing compound” (Supplemental Figure S6), suggesting that MdBBX7 can affect the expression of genes responding to oxidative stress. It was possible that MdBBX7 facilitated activities of antioxidant enzymes by regulating such DEGs.
As zinc-finger transcription factors, BBX proteins play important roles in transcriptional regulation by recognizing the T/G-box in the promoter of their target genes (Yan et al., 2011; Xu et al., 2018; Zhang et al., 2020). Tomato SlBBX20 can bind to the promoter of PHYTOENE SYNTHASE 1, a key enzyme in carotenoid biosynthesis, and activate its expression (Xiong et al., 2019). Sweet potato IbBBX24 activates IbMYC2 expression by binding to T/G-box elements in its promoter (Zhang et al., 2020). BBX18 regulates heat-stress-responsive gene expression, and BBX10 can modulate the transcript levels of ABA- and stress-related genes (Wang et al., 2013b; Ding et al., 2018; Liu et al., 2019b). Because the sequence similarity between BBX7 and MdBBX7 was only 57%, we speculated that the binding motif of MdBBX differed from the classical T/G box, and we therefore performed a ChIP-seq analysis. After immunoprecipitation, sequencing, and analysis, we identified one significant motif, CCTTG (Figure 3, E and F). We also identified the T/G-box, although it was not significant. Using EMSA analysis, we confirmed that MdBBX not only binds to the T/G-box but also to the CCTTG motif (Figure 3, G and H). Therefore, we identified another recognition site of MdBBX7. However, whether this motif can be recognized by other BBX proteins in apple or other plant species requires further investigation. We noticed that the enriched region of sweet potato IbBBX24 in the promoter of IbJAZ10 did not contain the classic T/G box, but CCTTG motif (Zhang et al., 2020). Hence, it is possible that IbBBX24 recognizes CCTTG motif in sweet potato.
Among the potential MdBBX7 target genes identified by ChIP-seq, we verified MdBBX7 binding to the promoters of MdERF1, MdERD15, and MdGLK1. MdERF1 was shown to be a drought positive regulator in previous studies (Cheng et al., 2013; Xing et al., 2017). Here, MdERF1 expression was reduced in MdBBX7 RNAi plants but elevated in MdBBX7 OE plants (Figure 6E). Hence, it is possible that MdBBX7 functions in drought tolerance through its regulation of MdERF1. By RNA-seq analysis, we also identified 6,009 DEGs regulated by MdBBX7 in response to drought. Among these genes, drought positive regulators such as MdSnRK2.6, MdAtHB-7, MdNF-YA1, MdWRKY40, MdYUC6, and MdABF2 were positively regulated by MdBBX7 in response to drought stress, and the drought negative regulator MdNRT1.5 was negatively regulated by MdBBX7 (Figure 3A;Supplemental Data Set S2). Among the DEGs, 1,197 were also identified in the ChIP-seq data (Figure 3D). Therefore, MdBBX7 may regulate drought tolerance by transcriptional regulation of its direct and indirect targets.
Previous studies suggest that not only the B-box domain but also the CCT domain play roles in transcriptional regulation (Gangappa and Botto, 2014). BBX21 binds to the known T/G-box elements with its second B-box, and BBX19 lost this biological function without its first B-box (Wang et al., 2014, 2015; Xu et al., 2018). The CCT domain of the flowering time regulator CO not only functions in binding to target promoters but also has the ability to regulate gene expression (Tiwari et al., 2010). In our study, we found that the N terminus but not the C terminus of MdBBX7 bound directly to the T/G-box elements and the CCTTG motif (Figure 3), confirming the regulatory function of the CCT domain of BBX proteins. Using Y2H screening, we identified interacting proteins of MdBBX7. To identify the region of MdBBX7 responsible for protein interaction, we created a series of MdBBX7 protein deletions and found that the two B-box motifs were responsible for the interaction between MdBBX7 and MdMIEL1 (Figure 5), consistent with previous findings that the B-box mainly functions in protein–protein interaction.
BBX proteins have been reported to interact with ubiquitin E3 ligase, affecting their stability. COP1, a well-known E3 ligase that degrades its interacting proteins by the 26S proteasome pathway, can associate with BBX21, BBX22, BBX4, BBX20, and BBX24 (Crocco et al., 2010; Chang and Wu, 2011; Yan et al., 2011; Wei et al., 2016; Heng et al., 2019). As the core component that regulates photomorphogenesis, COP1 associates with BBX proteins and participates in various photomorphogenic responses, including flowering, circadian regulation, and hypocotyl growth (Crocco et al., 2010; Chang and Wu, 2011; Yan et al., 2011; Wang et al., 2015). In apple, we found that MdBBX7 interacted with MdMIEL1 in vitro and in vivo (Figure 5). MdMIEL1 has previously been identified as a RING-type E3 ligase. Here, we found that MdMIEL1 could mediate ubiquitination and degradation of MdBBX7 by the 26S proteasome pathway (Figure 6). When MdBBX7 was co-expressed with MdMIEL1, the transcriptional activity of MdBBX7 was reduced (Figure 6, D–F). In addition, MdMIEL1 was a negative regulator of drought tolerance (Figure 7;Supplemental Figure S16). We also noticed that plants maintained the MdBBX7 protein under control conditions (Figure 1B), indicating that other proteins that stabilize MdBBX7 may exist.
In summary, we identified the roles of a zinc-finger protein, MdBBX7, in response to drought and characterized its molecular mechanisms (Figure 9). Our findings provide information on the molecular mechanisms by which apple responds to drought stress and offer candidate genes for the improvement of apple drought tolerance by molecular tools in the future.
Figure 9.
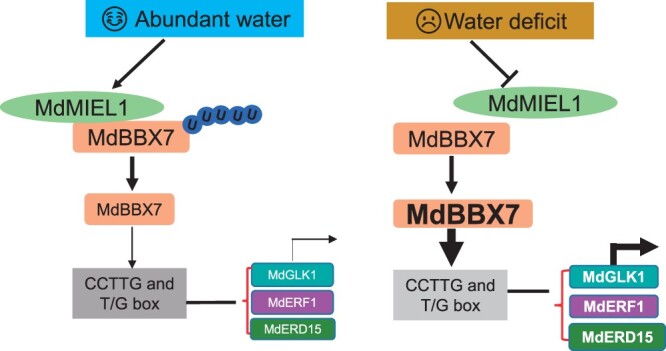
A proposed model of MdMIEL1–MdBBX7 module in apple drought stress response. When soil water is abundant, MdMIEL1 is accumulated. Accumulated MdMIEL1 then interacts with MdBBX7 and mediates the ubiquitination and degradation of MdBBX7 via the 26S protease pathway, thereby leading to the suppressed expression of target genes including MdERF1, MdGLK1, and MdERD15. Under drought stress conditions, MdMIEL1 is repressed, thereby resulting in accumulated MdBBX7. The accumulated MdBBX7 activates the expression of its downstream genes including MdERF1, MdGLK1, and MdERD15 by binding to the CCTTG or T/G box elements.
Materials and methods
Plant materials and growth conditions
Leaves of 1-year-old “Golden Delicious” (Malus × domestica) grown in a greenhouse were used to extract total RNA for gene cloning. GL-3 which has high regeneration capacity isolated from progenies of “Royal Gala” (Malus × domestica) was used for genetic transformation (Dai et al., 2013). Four-week-old tissue-cultured GL-3 plants were used for apple (Malus × domestica) stable transformation as previously reported (Xie et al., 2018). Tissue-cultured GL-3 plants were grown on MS medium (4.43-g·L-1 MS salts, 30-g·L-1 sucrose, 0.2-mg·L-1 6-BA, 0.2-mg·L-1 IAA, 7.5-g·L-1 agar, pH 6.0), and subcultured every 4 weeks. Four-week-old transgenic plants and GL-3 were rooted on MS medium (2.23-g·L-1 MS salts, 20-g·L-1 sucrose, 7.5-g·L-1 agar, 0.5-mg·L-1 IAA, 0.5-mg·L-1 IBA, pH 6.0). Fifty days later, the plants were transplanted into plant culture substrates (PINDSTRUP, Denmark) in a growth chamber with long-day conditions (16-h light/8-h dark at 25°C).
To study the tissue-specific expression, 5-year-old apple trees (Malus prunifolia), which were planted in an orchard at Northwest A&F University, Yangling, Shaanxi, China (34°16′ N, 108°4′ E) were used to collect the mature leaves, stems shoots, and roots.
Apple calli (Malus × domestica cv Orin) were grown on MS medium (4.43-g·L-1 MS salts, 30-g·L-1 sucrose, 8-g·L-1 agar, 0.4-mg·L-1 6-BA, 1.5-mg·L-1 2,4-D, pH 5.8) in darkness at 24°C. Two-week-old apple calli were used for gene transformation as described previously (Zhou et al., 2017).
Stress treatment
Two-month-old Malus sieversii grown in 1/2 Hoagland solution were treated with 20% (w/v) PEG6000 (Sigma, USA) for 0 and 6 h (Geng et al., 2019), and the roots were collected for MsBBX7 expression analysis.
Protein level of MdBBX7 and MdMIEL1 under drought stress was detected using 3-month-old “Golden Delicious” (Malus × domestica) plants. The treatment started when VWC (soil volumetric water content) reached to 43%–48%. After 8 d water withholding, four to nine mature leaves were collected and put into liquid nitrogen quickly for protein extraction. VWC was measured by TDR (FS6430, USA).
For short-term drought treatment, 3-month-old MdBBX7 and MdMIEL1 transgenic plants and GL-3 were withheld from water until VWC reached to 0, and survival rate was calculated after rewatering for 1 week. All the plants were grown in a growth chamber with long-day conditions and appropriate temperature and humidity (25°C, 50%–75% humidity).
MS medium was pre-treated with PEG (40% [w/v], SIGMA-ALDRICH, polyethylene glycol 8000, Lot#SLCF0313) for 24 h to simulate −0.7 MPa osmotic stress as described previously (Verslues et al., 2006). The WT and transgenic apple calli were transferred to MS medium or PEG medium for additional 2 weeks.
Phylogenetic tree and sequence alignment
The amino acid sequences of BBX7 from 20 plant species were downloaded from the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic analyses were conducted using MEGA version 5 and the Maximum Likelihood method with 1,000 bootstrap replicates.
Plasmid construction and genetic transformation
In order to generate overexpressing apple plants, the coding region (CDS) of MdBBX7 or MdMIEL1 was cloned into pK2WIWG2D vector. To obtain MdBBX7 RNAi transgenic plants, a 221-kb fragment was inserted into pK7WIWG2D vector. The plasmid to knock down MdMIEL1 was built with a similar method. Vectors mentioned above were individually transformed into Agrobacterium tumefaciens strain EHA105 and GL-3 plants using the Agrobacterium-mediated transformation as described by Li et al. (2020). The primers and vectors are listed in Supplemental Table S1.
Y2H screening
A Y2H screening assay was carried out according to the manufacturer’s manuals (Clontech, 630439, 630489). MdBBX7 CDS and truncated MdBBX7 were cloned into pGBKT7 individually and tested for self-activation. The fragment of MdBBX7N which contains the first 130 aa and two B-box motifs did not show self-activation was used for Y2H screening assay.
For the Y2H screening, MdBBX7N was cloned into pGBKT7. The cDNA library was prepared with apple leaves and roots. For the point-to-point Y2H assay, MdBBX7N was further divided into several fragments. MdMIEL1 CDS and truncated MdMIEL1 were individually cloned into pGADT7. MdBBX7N and its truncated forms were individually cloned into pGBKT7. MdBBX7N (130 aa)-pGBKT7 or truncated MdBBX7N-pGBKT7 and MIEL1-pGADT7 or truncated MIEL1-pGADT7 were co-transformed into yeast (Saccharomyces cerevisiae) strain Y2H Gold.
All positive clones were selected on SD–Leu–Trp, and then on SD–Leu–Trp–His–Ade + x-α-gal plates for growth observation and the x-α-gal assay.
The primers and vectors used are listed in Supplemental Table S1.
RNA extraction, RNA-seq, and RT-qPCR analysis
Mature leaves of 3-month-old MdBBX7 RNAi transgenic plants and GL-3 were collected and airdried for 0 or 2 h, each condition consists of three valid biological replicates (leaves from three trees was pooled as one biological replicate, and three biological replicates were used.). Total RNA was extracted by a CTAB method as described previously (Xie et al., 2018). mRNA was isolated and used to construct a library and then subjected for sequencing using the Illumina HiSeq platform by Novogene (Beijing, China). Clean sequences were mapped to the latest Malus × domestica reference genome from NCBI by HISAT2. Differential gene expressions were analyzed by DESeq2 with threshold of adjust P values below 0.05 and the absolute value of log2 (Fold change) above 1.
For RT-qPCR analysis, first strand cDNA was synthesized according to the manufacturer’s instructions of the RevertAid First Strand cDNA synthesis kit (Thermo Scientific, USA). The RT-qPCR was performed in a total 20-μL reaction containing GoTaq qPCR Master Mix (Promega, USA). The primers and vectors used are listed in Supplemental Table S1.
ChIP-seq and ChIP-qPCR
ChIP-seq assay was conducted as previously reported (Xie et al., 2018). Briefly, 4-month-old tissue-cultured GL-3 plants were used for crosslinking and then immunoprecipitated with anti-MdBBX7 polyclonal antibody produced from rabbit (GenScript, Nanjing, China). The recovered DNA was subjected to paired-end sequencing (150 bp) by an Illumina TruSeq platform (Novogene, Beijing, China). The clean reads were aligned to the apple reference genome from NCBI by Bowtie2. A control sample was added for background subtraction in peak calling using MACS2. With findMotifGenome.pl of HOMER, the enriched motifs were produced.
Recovered DNA was used for ChIP-qPCR analysis. qPCR primers were designed in the regions containing the MdBBX7 binding sites in promoters of MdERF1, MdGLK1, and MdERD15. Malate dehydrogenase and no antibody served as the negative controls. The primers and vectors used are listed in Supplemental Table S1.
EMSA
MdBBX7-CCT, which contains the last 204 aa (207–410 aa) in the C terminus of MdBBX7, was cloned into pMAL-c5X (MBP). The fusion protein of MdBBX7-C204 was induced by 0.3-mM IPTG in E. coli, and purified by maltose. The probes were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). EMSA was performed as previously described (Xie et al., 2018) using a LightShift Chemiluminescent EMSA Kit (20148, Thermo Scientific, USA).
Pull-down assay
MBP-MdMIEL1 and GST-MdBBX7-N214 fusion proteins were induced by IPTG in E. coli and purified with Amylose resin (E8021S, New England Biolabs, USA) and Pierce Glutathione Spin Columns (16105, Thermo Scientific, USA) respectively. MdBBX7-N214 contains the first 214 aa in the N terminus of MdBBX7. Pierce GST Protein Interaction Pull-Down Kit (21516, Thermo Scientific, USA) was used for pull-down assay by following the manufacturer’s protocols. The purified proteins were then detected by anti-MBP antibody (2396S, Cell Signaling Technology, USA) and anti-GST antibody (M20007L, Abmart Shanghai Co., Ltd. China).
Split-luciferase assay
CDS of MdBBX7 and MdMIEL1 were cloned into pCAMBIA1300-nLuc and pCAMBIA1300-cLuc, respectively. These constructs were then transformed into Agrobacterium strain C58C1 and co-infiltrated with 35S:p19 to infiltrate 4-week-old Nicotiana benthamiana leaves. The infiltrated leaves were placed in a growth chamber for additional 3 d under 16 h-light/8 h-dark at 25°C. Luciferase expression signal was captured as previous described (Ishitani et al., 1997) with Lumazone Pylon 2048B (Princeton, USA).
BiFC assay
The CDS of MdBBX7 and MdMIEL1 were introduced into pSPYCE and pSPYNE, respectively. The MdBBX7-pSPYCE and MdMIEL1-pSPYNE were transformed into C58C1 and co-infiltrated with 35S:p19 into 4-week-old N. benthamiana leaves. Three days later, the fluorescence signal was detected by confocal microscopy (FV1200 Olympus, Japan). An argon laser excitation of 488 nm (intensity 1%) was examined at the wavelength of 492–537 nm to detect yellow fluorescent protein (YFP) signal. 4′,6-Diamidino-2-phenylindole (DAPI) was used to detect nuclei under an excitation line of 405 nm (intensity 20%) and signals were detected at the wavelength of 416–466 nm.
Co-IP assay
Leaves of 4-week-old MdBBX7 OE plants were collected and pulverized with liquid nitrogen. Total proteins were extracted with extraction buffer (50-mM Tris-HCL, 150-mM NaCl, 5-mM EDTA, 1% NP-40, 10% glycerol, 1-mM PMSF, 5-mM DTT, and 1× Halt protease inhibitor cocktail [Fisher Scientific]) and then incubated with anti-MdBBX7 polyclonal antibody produced from rabbit (GenScript, Nanjing, China) for overnight. The immunocomplexes were collected by adding protein A/G agarose beads (Thermo Fisher) and were washed with immunoprecipitation buffer (50-mM Tris-HCl, pH 8.0, 150-mM NaCl, 2-mM EDTA, 1-mM DTT, 10% glycerol, 0.15% Triton X-100, 1-mM PMSF, and 1× Halt protease inhibitor cocktail [Fisher Scientific]). The pellet (immunocomplexes with beads) was resuspended in 1× SDS–PAGE loading buffer. Eluted proteins were analyzed by immunoblotting using anti-MdBBX7 antibody or anti-MdMIEL1 antibody.
Dual-luciferase assay
CDS of MdBBX7 and MdMIEL1 were cloned into pGreen62-SK vector, respectively. The promoter fragments of MdERF1 were introduced into pGreen0800-LUC vector, respectively. The recombinant vectors were transformed into Agrobacterium strain C58C1 and co-infiltrated with 35S:p19 into 4-week-old N. benthamiana leaves. The samples were collected 3 d later for luminescence detection with a detection kit (Promega, Madison, WI, USA).
In vitro and in vivo ubiquitination
For the in vitro ubiquitination assay, the fusion proteins of GST-MdBBX7–N214 and MBP-MdMIEL1 were incubated with ATP (A1852, Sigma, USA), Ubiquitin (U-115, Boston BioChem), E1 (E-304, Boston BioChem), and E2 (E2-622, Boston BioChem) at 30°C for 6 h. The purified GST protein was severed as a negative reference.
To perform the in vivo ubiquitination assay, total proteins were extracted from MdBBX7 OE, MdBBX7 RNAi transgenic plants, and GL-3 plants by extraction buffer (50-mM Tris-HCL, 150-mM NaCl, 5-mM EDTA, 1% NP-40, 10% glycerol, 1-mM PMSF, 5-mM DTT). Then the proteins were immunoprecipitated with anti-MdBBX7 polyclonal antibody (GenScript, Nanjing, China) and immunoblotted with antibody against ubiquitin (Cell Signaling Technology, USA). Immunoblotting with anti-Actin (Cell Signaling Technology, #9715) antibody was used as an input control.
Protein stability assay
Four-week-old MdBBX7 OE and RNAi transgenic plants and GL-3were pre-treated with DMSO or 50-μM MG132 for 12 h. Proteins were extracted and then detected with anti-MdBBX7 antibody and anti-H3 antibody. In addition, the fusion protein of MdBBX7 (GST-MdBBX7-N214) and the proteins from MdMIEL1 OE transgenic plants and GL-3 were incubated together with DMSO or 50-μM MG132 for 0, 45, 90, and 180 min. Immunoblotting was performed used anti-GST antibody to detect the protein level of MdBBX7 and anti-Actin antibody (M20009L, Abmart Shanghai Co., Ltd., China) to serve as a loading control.
Subcellular localization
The coding sequences of MdBBX7 and MdMIEL1 were cloned into pEearleyGate104, respectively, to generate YFP-MdBBX7 and YFP-MdMIEL1. The fusion proteins were transiently transformed into 4-week-old N. benthamiana leaves. The fluorescence signal detecting was consistent with BiFC assay.
Ion leakage analysis
Ion leakage assay was performed when VWC reached to 5% under drought stress. Briefly, mature leaves from middle parts of the stems were detached and cut into leaf discs (diameter = 8 mm). The leaf discs were soaked in deionized water for 12 h before measurement of conductivity, and data were recorded as R1. Then the leaf discs were boiled for 30 min and conductivity was measured and recorded as R2. Electrolyte leakage was calculated as the percentage of R1 to R2.TGCQ
Measurement of ABA
For endogenous ABA content determination, frozen leaves were weighed and pulverized with liquid nitrogen. The extraction steps were carried out as described previously (Xie et al., 2020). ABA content was determined using a UPLC–MS/MS system (QTRAP 5500 LC/MS/MS, USA) and a Shimadzu LC-30AD UPLC system (Tokyo, Japan).
Detection of enzyme activity of POD and CAT, and H2O2 content
The activities of POD and CAT were detected as described previously (Sun et al., 2018) with minor modifications. Leaf samples were weighed (0.05 g) and extracted with 800-μL extraction buffer (100-mM phosphate buffer, 1-mM EDTA, 0.1% Triton-X-100, and 1% PVP). The extracted proteins were used for the detection of enzyme activities with microplate reader (Victor Nivo, USA).
Measurement of H2O2 content was performed according to the manufacturer’s manual (Comin, China).
Statistical analysis
Data are reported as the means ± sd. Statistical significance was determined by one-way ANOVA (Tukey’s test) analysis using SPSS (version 21.0, USA). Variations were considered significant if P < 0.05, 0.01, or 0.001.
Data availability
The RNA-seq data have been deposited to the NCBI with the dataset identifier PRJNA627355 and PRJNA645515.
Accession numbers
Sequence data from this article can be found in the NCBI data libraries under accession numbers MdBBX7 (NM_001293833.1), MdMIEL1 (XM_008342616.3), MdERF1 (XM_008390485.3), MdERD15 (XM_008358143.3), MdGLK1 (XM_008383362.3).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Protein alignment of BBX7 from apple and Arabidopsis.
Supplemental Figure S2 . Phylogenic tree of BBX7 from different plant species.
Supplemental Figure S3 . Generation of MdBBX7 RNAi or OE transgenic plants.
Supplemental Figure S4 . Physiological parameter of GL-3 and MdBBX7 transgenic plants.
Supplemental Figure S5 . Response of MdBBX7 transgenic plants to drought.
Supplemental Figure S6 . The enriched GO terms for DEGs in MdBBX7 RNAi plants under drought stress condition.
Supplemental Figure S7 . KEGG enrichments for DEGs in MdBBX7 RNAi plants under drought stress conditions.
Supplemental Figure S8 . The enriched GO terms for putative targets of MdBBX7 revealed from the ChIP-seq analysis.
Supplemental Figure S9 . KEGG enrichments for putative targets of MdBBX7 revealed from ChIP-seq analysis.
Supplemental Figure S10 . The ChIP-qPCR analysis of MdWRKY40 and MdCAT2.
Supplemental Figure S11 . The transcript level of MdBBX7, MdGLK1, MdERF1, and MdERD15 in response to drought stress.
Supplemental Figure S12 . Self-activation test of MdBBX7 protein.
Supplemental Figure S13 . Protein alignment of MIEL1 in apple and Arabidopsis.
Supplemental Figure S14 . Identification of MdMIEL1 RNAi or OE transgenic plants.
Supplemental Figure S15 . Response of MdMIEL1 to drought stress and its subcellular localization.
Supplemental Figure S16 . ABA content of MdMIEL1 OE, MdMIEL1 RNAi, and GL-3 under drought stress.
Supplemental Figure S17 . Identification of MdBBX7 RNAi, MdMIEL1 RNAi, or MdMIEL1 RNAi/MdBBX7 RNAi transgenic apple calli.
Supplemental Figure S18 . Relative expression levels of MdERD15 and MdGLK1 in the WT and transgenic apple calli under control and PEG conditions.
Supplemental Data S1 . Y2H library screening results of MdBBX7.
Supplemental Table S1 . Primers and vectors used in this study.
Supplemental Data Set S1. Differentially expressed genes in MdBBX7 RNAi plants under control conditions.
Supplemental Data Set S2. Differentially expressed genes in MdBBX7 RNAi plants under dehydration conditions.
Supplemental Data Set S3. Dehydration responsive genes in GL-3.
Supplemental Data Set S4. Binding peaks of MdBBX7 by using ChIP-seq analysis.
Supplemental Data Set S5. The putative targets identified by ChIP-seq and RNA-seq analysis.
Supplementary Material
Acknowledgments
We thank Dr Zhihong Zhang from Shenyang Agricultural University for providing tissue-cultured GL-3 plants.
Funding
This work was supported by the National Natural Science Foundation of China 31572106 and 31622049.
Conflict of interest statement. The authors declare no potential competing interests.
Q.G. designed the project. P.C., F.Z., X.L., W.S., M.Y., T.L., and C.B. performed the experiments. Q.G., P.C., J.H., Z.F., X.L., S.Z., and F.M. analyzed the data. X.L. and P.C. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Qingmei Guan (qguan@nwafu.edu.cn).
References
- Adams SW, Lordan J, Fazio G, Bugbee B, Francescatto P, Robinson TL, Black B (2018) Effect of scion and graft type on transpiration, hydraulic resistance and xylem hormone profile of apples grafted on Geneva®41 and M.9-NICTM29 rootstocks. Sci Hortic 227:213–222 [Google Scholar]
- An JP, Liu X, Li HH, You CX, Wang XF, Hao YJ (2017) Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol 58:1953–1962 [DOI] [PubMed] [Google Scholar]
- An JP, Wang XF, Espley RV, Lin-Wang K, Bi SQ, You CX, Hao YJ (2020a) An apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol 61:130–143 [DOI] [PubMed] [Google Scholar]
- An JP, Wang XF, Zhang XW, Bi SQ, You CX, Hao YJ (2019) MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol J 17:2231–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JP, Wang XF, Zhang XW, Xu HF, Bi SQ, You CX, Hao YJ (2020b) An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J 18:337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum SA, Xie XY, Wang LC, Saleem MF, Lei W (2011) Morphological, physiological and biochemical responses of plants to drought stress. Afr J Agric Res 6:2026–2032 [Google Scholar]
- Bai S, Saito T, Honda C, Hatsuyama Y, Ito A, Moriguchi T (2014) An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240:1051–1062 [DOI] [PubMed] [Google Scholar]
- Bai S, Tao R, Tang Y, Yin L, Ma Y, Ni J, Yan X, Yang Q, Wu Z, Zeng Y (2019) BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol J 17:1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Kim WY, Kang SB, Kim JI, Baek D, Jung IJ, Kim MR, Li N, Kim HJ, Nakajima M, et al. (2015) A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat Commun 6:8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CSJ, Wu MSH (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156:228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM (2012) Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol 159:1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XF, Wang ZY (2005) Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J 43:758–768 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Botto JF (2013) BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531:44–52 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Holm M, Yanovsky MJ, Botto JF (2010) AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J 64:551–562 [DOI] [PubMed] [Google Scholar]
- Dai HY, Li WR, Han GF, Yang Y, Ma Y, Li H, Zhang ZH (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci Hortic 164:202–208 [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19:3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wang S, Song ZT, Jiang YP, Han JJ, Lu SJ, Li L, Liu JX (2018) Two B-Box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep 25:1718–1728.e1714 [DOI] [PubMed] [Google Scholar]
- Ding S, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27:3228–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Dong Y, Yue X, Hu J, Jiang S, Xu H, Wang Y, Su M, Zhang J, Zhang Z (2019) The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ 42:2090–2104 [DOI] [PubMed] [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA (2009) Plant drought stress: effects, mechanisms and management. Agron Sustain Dev 29:185–212 [Google Scholar]
- Foster TM, Ledger SE, Janssen BJ, Luo Z, Drummond RSM, Tomes S, Karunairetnam S, Waite CN, Funnell KA, van Hooijdonk BM, et al. (2018) Expression of MdCCD7 in the scion determines the extent of sylleptic branching and the primary shoot growth rate of apple trees. J Exp Bot 69:2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19:460–470 [DOI] [PubMed] [Google Scholar]
- Geilen K, Boehmer M (2015) Dynamic subnuclear relocalisation of WRKY40 in response to Abscisic acid in Arabidopsis thaliana. Sci Rep 5:13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109:3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng DL, Chen PX, Shen XX, Zhang Y, Li XW, Jiang LJ, Xie YP, Niu CD, Zhang J, Huang XH, et al. (2018) MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol 178:1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng DL, Lu LY, Yan MJ, Shen XX, Jiang LJ, Li HY, Wang LP, Yan Y, Xu JD, Li CY, et al. (2019) Physiological and transcriptomic analyses of roots from Malus sieversii under drought stress. J Integr Agric 18:1280–1294 [Google Scholar]
- Gil HL, Kim J, Chung MS, Joon PS (2017) The MIEL1 E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis stems. Plant Cell Physiol 58:1249–1259 [DOI] [PubMed] [Google Scholar]
- He H, Denecker J, Van Der Kelen K, Willems P, Pottie R, Phua SY, Hannah MA, Vertommen D, Van Breusegem F, Mhamdi A (2021) The Arabidopsis mediator complex subunit 8 regulates uxidative stress responses. Plant Cell 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y, Jiang Y, Zhao X, Zhou H, Wang X, Deng XW, Xu D (2019) BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc Natl Acad Sci USA 116:26049–26056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9:1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Matsui A, Tanoi K, Kobayashi NI, Matsuda F, Habu Y, Ogawa D, Sakamoto T, Matsunaga S, et al. (2017) Acetate-mediated novel survival strategy against drought in plants. Nat Plants 26:17097. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1994) Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol 25:791–798 [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JWR (1995) Protein motifs 5. Zinc fingers. FASEB J 9:597–604 [PubMed] [Google Scholar]
- Kumar A, Nayak AK, Das BS, Panigrahi N, Dasgupta P, Mohanty S, Kumar U, Panneerselvam P, Pathak H. (2019) Effects of water deficit stress on agronomic and physiological responses of rice and greenhouse gas emission from rice soil under elevated atmospheric CO2. Sci Total Environ 650:2032–2050 [DOI] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26:15–22 [DOI] [PubMed] [Google Scholar]
- Lee H, Seo P (. 2016) The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat Commun 7:12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestari R, Rio M, Martin F, Leclercq J, Woraathasin N, Roques S, Dessailly F, Clément-Vidal A, Sanier C, Fabre D, et al. (2018) Overexpression of Hevea brasiliensis ethylene response factor HbERF-IXc5 enhances growth and tolerance to abiotic stress and affects laticifer differentiation. Plant Biotechnol J 16:322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang P, Wei Z, Liang D, Liu C, Yin L, Jia D, Fu M, Ma F (2012) The mitigation effects of exogenous melatonin on salinity‐induced stress in Malus hupehensis. J Pineal Res 53:298–306 [DOI] [PubMed] [Google Scholar]
- Li XW, Chen PX, Xie YP, Yan Y, Wang LP, Dang H, Zhang J, Xu LY, Ma FW, Guan QM (2020) Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Hortic Res 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, Zheng CC (2013) NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS One 8:e61289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Guo X, Wang Q, Wang Y, Zhao D, Yao L, Wang S, Liu G, Li T (2016) Overexpression of MsDREB6.2 results in cytokinin‐deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J 89:510–526 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen H, Ping Q, Zhang Z, Guan Z, Fang W, Chen S, Chen F, Jiang J, Zhang F (2019a) The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep 38:15–24 [DOI] [PubMed] [Google Scholar]
- Liu X, Li R, Dai Y, Yuan L, Sun Q, Zhang S, Wang X (2019b) A B-box zinc finger protein, Md BBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol Biol 99:437–447 [DOI] [PubMed] [Google Scholar]
- Ma QJ, Sun MH, Lu J, Kang H, You CX, Hao YJ (2018) An apple sucrose transporter MdSUT2.2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnol J 17:625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maribel L, Sese D, Tobita S (1998) Antioxidant responses of rice seedlings to salinity stress. Plant Sci 135:1–9 [Google Scholar]
- Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, Jauneau A, Roby D, Rivas S (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4:1476. [DOI] [PubMed] [Google Scholar]
- Mishra KB, Iannacone R, Petrozza A, Mishra A, Armentano N, La Vecchia G, Trtílek M, Cellini F, Nedbal L (2012) Engineered drought tolerance in tomato plants is reflected in chlorophyll fluorescence emission. Plant Sci 182:79–86 [DOI] [PubMed] [Google Scholar]
- Morimoto K, Ohama N, Kidokoro S, Mizoi J, Takahashi F, Todaka D, Mogami J, Sato H, Qin F, Kim J-S, et al. (2017) BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc Natl Acad Sci USA 114:E8528–E8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatoshi Y, Mitsuda N, Hayashi M, Inoue S, Okuma E, Kubo A, Murata Y, Seo M, Saji H, Kinoshita T, et al. (2016) GOLDEN 2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc Natl Acad Sci USA 113:4218–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett BJ, Henry-Kirk R, Friend A, Diack R, Helbig S, Mouhu K, Tomes S, Dare AP, Espley RV, Putterill J, et al. (2019) Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci Rep 9:17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857 [DOI] [PubMed] [Google Scholar]
- Rajnish Khanna BK, Maszle DR (2009) The Arabidopsis B-Box zinc finger family. Plant Cell 21:3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ICd, Almeida AAFD, Pirovani CP, Costa MGC, Conceição ASd, Soares Filho WDS, Coelho Filho MA, Gesteira AS (2019) Physiological, biochemical and molecular responses to drought conditions in field-grown grafted and ungrafted citrus plants. Environ Exp Bot 162:406–420 [Google Scholar]
- Scandalios JG (1993) Oxygen stress and superoxide dismutases. Plant Physiol 101:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55:2343–2351 [DOI] [PubMed] [Google Scholar]
- Söderman E, Mattsson J, Engström P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10:375–381 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289:768–771 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, Huo LQ, Che RM, Ma FW (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J 16:545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XP, Yan HL, Kang XY, Ma FW (2013) Growth, gas exchange, and water-use efficiency response of two young apple cultivars to drought stress in two scion-one rootstock grafting system. Photosynthetica 51:404–410 [Google Scholar]
- Tiwari SB, Shen Y, Chang H-C, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187:57–66 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu JH, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523–539 [DOI] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26:3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Sarmast MK, Jiang J, Dehesh K (2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27:1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sun X, Li C, Wei ZW, Liang D, Ma FW (2013a) Long‐term exogenous application of melatonin delays drought‐induced leaf senescence in apple. J Pineal Res 54:292–302 [DOI] [PubMed] [Google Scholar]
- Wang Q, Tu X, Zhang J, Chen X, Rao L (2013b) Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol Biol Rep 40:2679–2688 [DOI] [PubMed] [Google Scholar]
- Wang TJ, Huang S, Zhang A, Guo P, Liu Y, Xu C, Cong W, Liu B, Xu ZY (2021) JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol 230:567–584 [DOI] [PubMed] [Google Scholar]
- Wei CQ, Chien CW, Ai LF, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang ZY (2016) The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics 43:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Lu W, Mao L, Han X, Wei X, Zhao X, Xia M, Xu C (2020) ABF2 and MYB transcription factors regulate feruloyl transferase FHT involved in ABA-mediated wound suberization of kiwifruit. J Exp Bot 71:305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Chen PX, Yan Y, Bao CN, Li XW, Wang LP, Shen XX, Li H, Liu XF, Niu CD, et al. (2018) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol 218:201–218 [DOI] [PubMed] [Google Scholar]
- Xie Y, Bao CN, Chen PX, Cao FG, Liu XF, Geng DL, Li ZX, Li XW, Hou N, Zhi F, et al. (2020) ABA homeostasis is mediated by a feedback regulation of MdMYB88 and MdMYB124. J Exp Bot 72:592–607 [DOI] [PubMed] [Google Scholar]
- Xing L, Di Z, Yang W, Liu J, Li M, Wang X, Cui C, Wang X, Wang X, Zhang R, et al. (2017) Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front Plant Sci 8:1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Luo D, Lin A, Zhang C, Shan L, He P, Li B, Zhang Q, Hua B, Yuan Z, et al. (2019) A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol 221:279–294 [DOI] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Holm M, Deng XW (2018) The B-Box domain protein BBX21 promotes photomorphogenesis. Plant Physiol 176:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhao X, Aiwaili P, Mu X, Hong B (2020) A zinc finger protein BBX19 interacts with ABF3 to negatively affect drought tolerance in chrysanthemum. Plant J 103:1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Sharp RE (2010) Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant Cell Environ 33:590–603 [DOI] [PubMed] [Google Scholar]
- Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, Rodriguez-Franco M (2011) Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol 156:1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang Q, Zhai H, Gao S, Yang L, Wang Z, Xu Y, Huo J, Ren Z, Zhao N, et al. (2020) IbBBX24 promotes the jasmonic acid pathway and enhances fusarium wilt resistance in sweet potato. Plant Cell 32:1102–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LJ, Zhang CL, Zhang RF, Wang GL, Li YY, Hao YJ (2018) The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis. Plant Physiol 179:88–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LJ, Li YY, Zhang RF, Zhang C, Xie X, Zhao C, Hao YJ (2017) The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ 40:2068–2080 [DOI] [PubMed] [Google Scholar]
- Zhou SX, Walker RR, Edwards E (2020) Decoupled drought responses of fine-root versus leaf acquisitive traits among six Prunus hybrids. J Plant Ecol 13:304–312 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited to the NCBI with the dataset identifier PRJNA627355 and PRJNA645515.