Abstract
Plant organ size and shape are major agronomic traits that depend on cell division and expansion, which are both regulated by complex gene networks. In several eudicot species belonging to the rosid clade, organ growth is controlled by a repressor complex consisting of PEAPOD (PPD) and KINASE-INDUCIBLE DOMAIN INTERACTING (KIX) proteins. The role of these proteins in asterids, which together with the rosids constitute most of the core eudicot species, is unknown. We used Clustered Regularly Interspaced Short Palindromic Repeats–CRISPR-associated protein 9 genome editing to target SlKIX8 and SlKIX9 in the asterid model species tomato (Solanum lycopersicum) and analyzed loss-of-function phenotypes. Loss-of-function of SlKIX8 and SlKIX9 led to the production of enlarged, dome-shaped leaves and these leaves exhibited increased expression of putative Solanum lycopersicum PPD (SlPPD target genes. Unexpectedly, kix8 kix9 mutants carried enlarged fruits with increased pericarp thickness due to cell expansion. At the molecular level, protein interaction assays indicated that SlKIX8 and SlKIX9 act as adaptors between the SlPPD and SlTOPLESS co-repressor proteins. Our results show that KIX8 and KIX9 are regulators of organ growth in asterids and can be used in strategies to improve important traits in produce such as thickness of the fruit flesh.
Two transcriptional repressors negatively regulate organ growth in tomato, with loss-of-function lines producing enlarged fruits due to enlarged cells in the fruit flesh.
Introduction
Plants come in all shapes and sizes, yet these agronomically important traits are remarkably uniform within a given species or variety. Not surprisingly, cell division and cell expansion, the underlying processes of organ development, are under tight genetic control (Gonzalez et al., 2012; Hepworth and Lenhard, 2014; Kalve et al., 2014; Vercruysse et al., 2020). The different phases of leaf development, for instance, are regulated by complex gene networks (Gonzalez et al., 2012; Hepworth and Lenhard, 2014; Vercruysse et al., 2020). Leaf development consists of the emergence of a leaf primordium from the shoot apical meristem, followed by a period of primary cell division that transitions into a cell expansion phase, and a simultaneous phase of meristemoid division. In Arabidopsis (Arabidopsis thaliana) leaves, the shape of the primary cell cycle arrest front, which moves from tip to base as cells cease to divide, was reported to be regulated by the transcriptional regulator PEAPOD2 (AtPPD2; Baekelandt et al., 2018). Through its interaction with the adaptor protein NOVEL INTERACTOR OF JAZ (AtNINJA; Supplemental Figure S1A), AtPPD2 can recruit the transcriptional co-repressor TOPLESS (AtTPL) and, thereby, control leaf flatness (Baekelandt et al., 2018). In addition, AtPPD2 forms a transcriptional repressor complex with KINASE-INDUCIBLE DOMAIN INTERACTING 8 (AtKIX8)/AtKIX9 (Supplemental Figure S1A; Gonzalez et al., 2015), which can also recruit AtTPL, to limit the number of self-renewing asymmetric divisions that stem cell-like meristemoids can undergo before differentiating into stomatal guard cells (White, 2006; Gonzalez et al., 2015). This way, the AtPPD2–AtKIX8/AtKIX9 repressor complex restricts leaf growth, thereby controlling both leaf shape and size (White, 2006; Gonzalez et al., 2015). The functionalities of AtKIX8 and AtKIX9 are, thus, required for the repressive activity of AtPPD2 (Gonzalez et al., 2015). Consequently, double kix8-kix9 Arabidopsis knockout plants display increased transcript levels of AtPPD2 target genes and enlarged, dome-shaped leaves because of a prolonged period of meristemoid division, similar to Arabidopsis ami-ppd plants overexpressing an artificial microRNA targeting AtPPD1 and AtPPD2 (Gonzalez et al., 2015).
In Arabidopsis, the AtKIX8 and AtKIX9 proteins are known to interact with both AtPPD1 and AtPPD2 through their distinguishing N-terminal PPD domain (Supplemental Figure S1A; Bai et al., 2011; Gonzalez et al., 2015). Together with AtTIFY8 and the JASMONATE ZIM DOMAIN (AtJAZ) proteins, AtPPD1 and AtPPD2 belong to class II of the TIFY protein family (Vanholme et al., 2007; Bai et al., 2011) and are characterized by the presence of a conserved TIF[F/Y]XG motif. This motif resides within the ZINC-FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM (ZIM) domain that mediates the interaction of class II TIFY proteins with AtNINJA (Supplemental Figure S1A; Chini et al., 2009; Chung and Howe, 2009; Pauwels et al., 2010; Baekelandt et al., 2018). All class II proteins, except AtTIFY8, contain a C-terminal Jas domain (Vanholme et al., 2007; Bai et al., 2011). The Jas domain found in AtPPD proteins, however, is divergent from the Jas consensus motif in AtJAZ proteins (Bai et al., 2011) that mediates the interaction of AtJAZ proteins with transcription factors such as AtMYC2 and the F-box protein CORONATINE INSENSITIVE 1 (AtCOI1; Chini et al., 2007; Thines et al., 2007).
In addition to AtKIX8 and AtKIX9, nine other proteins that contain a KIX domain have been described in Arabidopsis (Thakur et al., 2013). Both in plant and nonplant species, such as yeast and humans, the KIX protein family includes HISTONE ACETYLTRANFERASE (HAT) proteins and Mediator subunits (Thakur et al., 2013; Kumar et al., 2018) that are known to function as co-activators through the interaction of their KIX domain with the transactivation domain of transcription factors (Thakur et al., 2014; Kumar et al., 2018). AtKIX8/AtKIX9 and their orthologs, however, are specific to plants and show, except for their N-terminal KIX domain, no similarity to these HAT and Mediator co-activators (Thakur et al., 2013). Instead, they contain an ETHYLENE RESPONSE FACTOR-ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif that allows them to recruit the co-repressor AtTPL (Kagale et al., 2010; Causier et al., 2012; Gonzalez et al., 2015). Through their KIX domain, AtKIX8 and AtKIX9 can simultaneously interact with the transcriptional repressor AtPPD2 (Gonzalez et al., 2015). Hence, AtKIX8/AtKIX9 forms a molecular bridge between AtPPD2 and AtTPL (Supplemental Figure S1A) and, because of that, AtPPD2 can act as a negative transcriptional regulator (Gonzalez et al., 2015).
In Arabidopsis, the activity of the AtPPD–AtKIX repressor complex is regulated by the F-box protein STERILE APETALA (AtSAP; Wang et al., 2016; Li et al., 2018). Interaction of the repressor complex with AtSAP results in the proteasomal degradation of both AtKIX and AtPPD proteins (Supplemental Figure S1A; Wang et al., 2016; Li et al., 2018). In accordance with these observations, AtSAP overexpression plants produce enlarged rosettes composed of enlarged and dome-shaped leaves (Wang et al., 2016).
Orthologs of the AtPPD, AtKIX8/9, and AtSAP proteins were found in members of both eudicot and monocot species, but appear to be absent from Poaceae species (grasses), suggesting that the PPD–KIX–SAP module was lost in the grass lineage (Gonzalez et al., 2015; Wang et al., 2016). It has been suggested that this might reflect the absence of self-renewing meristemoids in the stomatal lineage of grasses (Liu et al., 2009; Vatén and Bergmann, 2012; Gonzalez et al., 2015; Wang et al., 2016). Several eudicot members, in which orthologs of the AtPPD or AtKIX genes were mutated or downregulated, including Medicago truncatula, soybean (Glycine max), blackgram (Vigna mungo), and pea (Pisum sativum), produced enlarged leaves (Ge et al., 2016; Naito et al., 2017; Kanazashi et al., 2018; Li et al., 2019). Overexpression of AtSAP orthologs in M. truncatula, poplar (Populus trichocarpa), and cucumber (Cucumis sativus L.) increased leaf size as well (Yordanov et al., 2017; Yang et al., 2018; Yin et al., 2020). Next to enlarged leaves, increases in the size of other organs, such as stipules, flowers, fruits, and seeds, were also observed for several of the abovementioned mutants (Ge et al., 2016; Naito et al., 2017; Kanazashi et al., 2018; Yang et al., 2018; Li et al., 2019; Yin et al., 2020). Control of organ growth by the PPD–KIX repressor complex, together with its post-translational regulation by the F-box protein SAP, thus, seems to be conserved among distinct eudicot species (Schneider et al., 2021). However, differential developmental stages might be targeted by the repressor complex depending on the species. In M. truncatula plants in which an AtPPD ortholog was mutated, for instance, a prolonged period of primary cell division was reported to be responsible for the increase in organ size (Ge et al., 2016), whereas the enlarged leaf phenotype of Arabidopsis kix8-kix9 and ami-ppd mutants was associated with an extended duration of meristemoid division (Gonzalez et al., 2015). The role of conserved regulators can thus vary considerably between different species, illustrating that the translation of knowledge on transcriptional regulators between species is not always straightforward (Nelissen et al., 2014; Kajala et al., 2020). All of the aforementioned eudicot species, in which the function of the PPD–KIX–SAP module was described, belong to the rosid clade. Together with the asterids, the rosids constitute most of the core eudicot species (Supplemental Figure S1B) and their most recent common ancestor existed over 100 million years ago (Wikström et al., 2001). Whether the orthologs of PPD, KIX, and SAP proteins function as regulators of organ growth in asterid members is still unknown.
Here, we report a role for SlKIX8 and SlKIX9 in the regulation of organ growth in the asterid model species tomato (Solanum lycopersicum). We used protein interaction assays in yeast to demonstrate that the tomato orthologs of AtKIX8 and AtKIX9 function as SlTPL adaptor proteins for the SlPPD proteins. Next, we used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated protein 9 (CRISPR–Cas9) genome editing to simultaneously knock out SlKIX8 and SlKIX9 in the cultivar Micro-Tom. Double kix8 kix9 tomato knockout lines produced enlarged, dome-shaped leaves and displayed increased expression of genes orthologous to AtPPD2 target genes. Finally, we demonstrated that kix8 kix9 and single kix8 tomato mutants carried larger fruits with increased pericarp thickness, both important agronomic traits for fruit crops, resulting from the production of larger cells.
Results
SlKIX8 and SlKIX9 are SlTPL adaptors for SlPPD proteins
To identify the tomato orthologs of the AtKIX8 and AtKIX9 proteins, BLASTP was used. The tomato orthologs of the AtPPD proteins have been described previously (Chini et al., 2017). The SlKIX and SlPPD proteins display a similar domain structure as their Arabidopsis counterparts (Supplemental Figure S2). Amplification of the coding sequences of SlKIX and SlPPD genes revealed alternative splicing for SlKIX9, SlPPD1, and SlPPD2 (Supplemental Figure S2, B–D). Based on an alternative splicing model for AtKIX9 reported by The Arabidopsis Information Resource, we hypothesized that retention of the second SlKIX9 intron leads to the use of a downstream start codon, generating a splice variant that lacks the N-terminal KIX domain (Supplemental Figure S2B). The splice variants of SlPPD1 and SlPPD2 display retention of the Jas intron and part of the Jas intron, respectively, which is located between the two exons encoding the Jas domain (Supplemental Figure S2, C and D). These alternative splicing events are proposed to generate premature stop codons (Supplemental Figure S2, C and D), and consequently truncated SlPPD proteins, as was previously shown for AtPPD1 and AtPPD2 (Li et al., 2016).
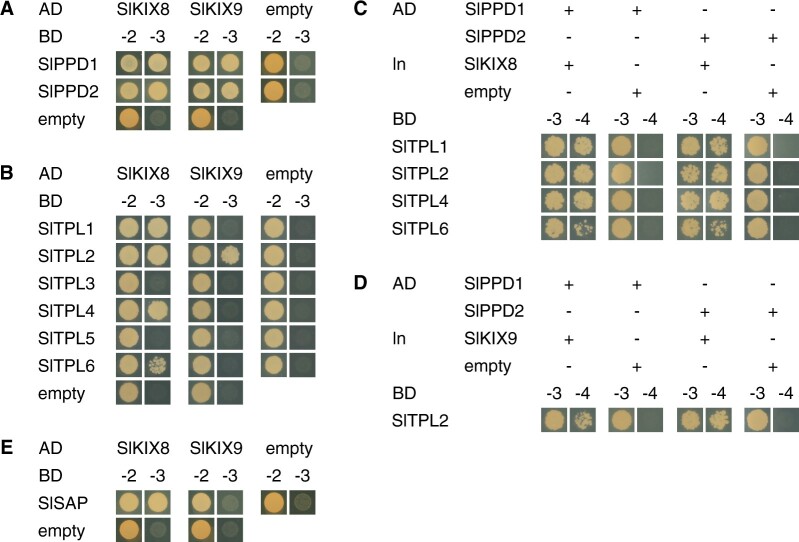
Previously, the interactions between KIX and PPD proteins from Arabidopsis and pea were analyzed in vivo and in detail in yeast (Gonzalez et al., 2015; Li et al., 2019). To determine whether SlKIX8, SlKIX9, SlPPD1, and SlPPD2 are part of a similar protein complex, we performed yeast two-hybrid (Y2H) assays. For these assays, the splice variants with the most complete coding sequence (shown in Supplemental Figure S2) were used. In the case of the SlKIX proteins, these possessed the KIX domain, which was shown to be essential for mediating the interaction between KIX and PPD proteins from Arabidopsis and pea (Gonzalez et al., 2015; Li et al., 2019). Direct interaction between the SlKIX and SlPPD proteins could be observed (Figure 1A). Next, we evaluated whether the SlKIX proteins were able to interact with SlTPL1 (Figure 1B), which is the most closely related tomato ortholog of the Arabidopsis co-repressor AtTPL (Hao et al., 2014). As only SlKIX8 was capable of interacting with SlTPL1 in the Y2H assays (Figure 1B), we also assessed the interaction between the SlKIX proteins and the five additional SlTPL proteins that were reported in tomato (Figure 1B). In addition to SlTPL1, SlKIX8 also interacted with SlTPL2, SlTPL4, and SlTPL6, whereas SlKIX9 could solely interact with SlTPL2 (Figure 1B). By means of yeast three-hybrid (Y3H) assays, we subsequently demonstrated that the SlKIX proteins can form a molecular bridge between these SlTPL proteins and the SlPPD proteins (Figure 1, C and D). In Arabidopsis, both AtKIX8 and AtKIX9 were reported to interact with the F-box protein AtSAP, resulting in their post-translational degradation (Li et al., 2018). However, we only observed interaction between SlKIX8 and the tomato ortholog of AtSAP (Figure 1E). Taken together, our results suggest that in tomato, SlKIX8 and SlKIX9 function as SlTPL adaptors for the SlPPD proteins, similar to their orthologs in Arabidopsis.
Figure 1.
SlKIX8 and SlKIX9 are SlTPL adaptors for SlPPD proteins. A and B, Y2H interaction analysis of SlKIX8 and SlKIX9 with SlPPD (A) and SlTPL (B) proteins. Yeast transformants expressing bait (BD) and prey (AD) proteins were dropped on control medium lacking Leu and Trp (−2) or selective medium additionally lacking His (−3). C and D, Y3H interaction analysis to test the bridging capacity of SlKIX8 (C) and SlKIX9 (D) to mediate the SlPPD–SlTPL interaction. Yeast transformants expressing bait (BD), bridge (ln), and prey (AD) proteins were dropped on control medium lacking Leu, Trp, and Ura (−3) or selective medium additionally lacking His (−4). E, Y2H interaction analysis of SlKIX proteins with SlSAP. Yeast transformants expressing bait (BD) and prey (AD) proteins were dropped on control medium lacking Leu and Trp (−2) or selective medium additionally lacking His (−3). Empty vectors were used in all control assays. AD, activation domain; BD, binding domain; ln, linker.
CRISPR–Cas9 genome editing of SlKIX8 and SlKIX9 leads to enlarged dome-shaped leaves
To investigate the in planta role of SlKIX8 and SlKIX9, double kix8 kix9 loss-of-function mutants (cultivar Micro-Tom) were generated using CRISPR–Cas9 genome editing (Figure 2A). A rippled, dome-shaped leaf phenotype could already be observed in regenerated double kix8 kix9 tomato knockout (T0) plants (Supplemental Figure S3). Likewise, the progeny of two independent T1 plants mono or biallelic for out-of-frame mutations at both the SlKIX8 and SlKIX9 loci (Supplemental Figure S4, A and B), displayed dome-shaped leaves with uneven leaf laminae (Figure 2, B and C). The main shoot length of these double kix8 kix9 mutants was reduced compared with that of wild-type plants (Supplemental Table S1). Single kix8 mutants (Supplemental Figure S4, A and B), obtained by pollinating the kix8 kix9#1 (T1) line with wild-type pollen, exhibited an intermediate leaf phenotype (Figure 2, B and C), whereas single kix9 plants (Supplemental Figure S4, A and B) did not show any visible phenotype (Figure 2, B and C) as was noted for Arabidopsis kix8 and kix9 single mutants (Gonzalez et al., 2015). These observations suggest that SlKIX8 and SlKIX9 may have partially redundant roles in regulating tomato leaf growth.
Figure 2.
CRISPR–Cas9 genome editing of tomato SlKIX8 and SlKIX9 causes a rippled, dome-shaped leaf phenotype. A, Schematic representation of SlKIX8 and SlKIX9 with location of the CRISPR–Cas9 cleavage sites. Dark gray boxes represent exons. Cas9 cleavage sites for gRNAs are indicated with arrowheads. B and C, Representative wild-type, kix8 kix9#1, kix8 kix9#2, kix8, and kix9 plants grown in soil for 1 month under 16:8 h photoperiods with daytime and nighttime temperatures of 26–29°C and 18–20°C, respectively, were photographed from the top (B) and the front (C).
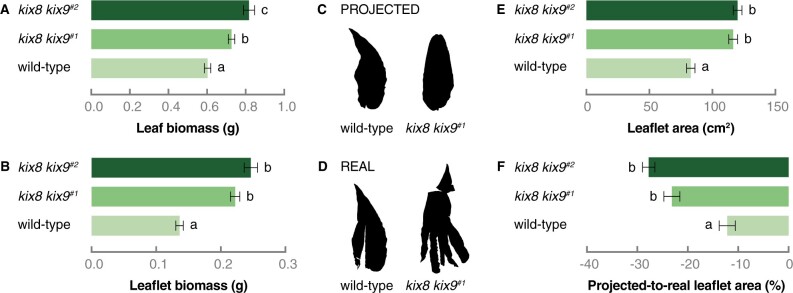
Given that the leaf shape phenotype was most pronounced for double kix8 kix9 tomato mutants, phenotypical analyses were performed on leaf eight and compared with those of the corresponding wild-type leaf. First, leaf fresh weight was determined, which was ∼30% higher for kix8 kix9 leaves compared with wild-type leaves (Figure 3A). Likewise, the fresh weights of the terminal leaflets of these kix8 kix9 leaves were increased by ∼40% compared with those of wild-type leaves (Figure 3B). Next, the area of terminal leaflets was measured before (projected area) and after (real area) terminal leaflets were cut to flatten them (Figure 3, C and D; Baekelandt et al., 2018). After flattening the terminal leaflets, those of kix8 kix9 mutants displayed an area that was ∼40% larger than corresponding wild-type leaflets (Figure 3E). In addition, the decrease in projected-to-real terminal leaflet area was ∼2 times bigger for kix8 kix9 plants compared with wild-type plants (Figure 3F), demonstrating the alteration in kix8 kix9 leaflet shape. These measurements, thus, substantiate the enlarged, dome-shaped leaf phenotype of double kix8 kix9 tomato knockout plants.
Figure 3.
Tomato kix8 kix9 plants produce enlarged, dome-shaped leaves. A and B, Biomass of leaf eight (from the top) (A) and its terminal leaflet (B). The eighth leaf (from the top) was harvested from plants grown in soil for 2 months under 16:8 h photoperiods with daytime and nighttime temperatures of 26–29°C and 18–20°C, respectively. Bars represent mean biomass relative to the mean of wild-type biomass values. Error bars denote standard error (n = 31–40). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters). C and D, The terminal leaflet area was measured before (projected, C) and after (real, D) the terminal leaflet of the eighth leaf was cut to flatten it. E−F, Area (E) and projected-to-real area (F) of the terminal leaflet of the eighth leaf. Bars represent mean area relative to the mean of wild-type area values. Error bars denote standard error (n = 31–40). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters).
Orthologs of AtPPD2 target genes are upregulated in leaves of tomato kix8 kix9 mutants
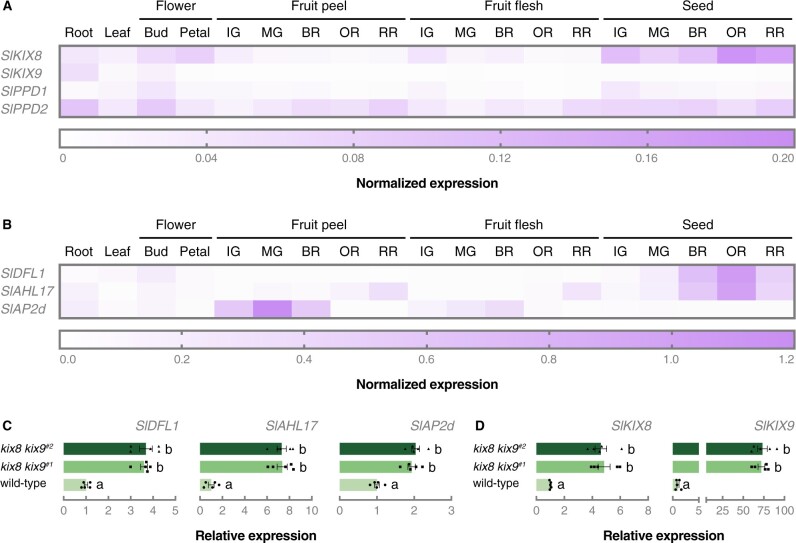
To gain further insight into the function of SlKIX8 and SlKIX9 in tomato plants, we made use of public transcriptome data (cultivar Micro-Tom; Zouine et al., 2017) to investigate the gene expression patterns of SlKIX8, SlKIX9, SlPPD1, and SlPPD2 in different tissues and throughout distinct developmental stages. A survey of these publicly available transcriptome data revealed that SlKIX8 was lowly expressed in all examined tissues and that SlKIX9 expression was (almost) absent in most tissues (Figure 4A;Supplemental Table S2). In all investigated tissues, the transcript level of SlPPD2 was higher than that of SlPPD1 (Figure 4A; Supplemental Table S2). Next, we looked up the gene expression patterns of the putative tomato orthologs of Arabidopsis DWARF IN LIGHT 1 (AtDFL1), AT-HOOK MOTIF CONTAINING NUCLEAR LOCALISED PROTEIN 17 (AtAHL17), and SCHLAFMUTZE (AtSMZ), which were top-ranked in the list of differentially expressed genes in Arabidopsis ami-ppd leaves, strongly upregulated in Arabidopsis kix8-kix9 leaves, and identified as direct AtPPD2 target genes using chromatin affinity purification (Gonzalez et al., 2015). Expression of all three tomato genes, SlDFL1, SlAHL17, and APETALA 2d (SlAP2d), was confirmed in tomato leaves (Figure 4B; Supplemental Table S2). To verify the potential differential expression of these genes in tomato kix8 kix9 mutants compared with wild-type plants, we performed a reverse transcription-quantitative PCR (RT-qPCR) analysis on the terminal leaflet of growing leaves and found that the transcription of all three genes was significantly upregulated in kix8 kix9 mutants (Figure 4C), while this was not the case in single kix8 and kix9 mutants (Supplemental Figure S5). Furthermore, the expression of SlKIX8 and SlKIX9 was increased in kix8 kix9 plants compared with wild-type plants (Figure 4D), suggesting negative feedback of the SlPPD–SlKIX complex on the expression of SlKIX8 and SlKIX9. Our findings indicate that SlKIX8 and SlKIX9 are required for the repression of tomato genes orthologous to at least three AtPPD2 target genes.
Figure 4.
SlKIX8 and SlKIX9 are required for the repression of putative SlPPD target genes. A and B, Normalized expression profiles of SlKIX8, SlKIX9, SlPPD1, SlPPD2 (A), SlDFL1, SlAHL17, and SlAP2d (B) in different tomato organs and developmental stages (cultivar Micro-Tom). Expression data were obtained from TomExpress (Zouine et al., 2017) and can be found in Supplemental Table S1. C and D, Relative expression of SlDFL1, SlAHL17, SlAP2d (C), SlKIX8, and SlKIX9 (D) in terminal leaflets of not fully developed leaves analyzed by RT-qPCR. For SlKIX8 and SlKIX9, primers allow amplification of edited alleles. The terminal leaflet from the second leaf (from the top) was harvested from plants grown in soil for 3 weeks under 16:8-h photoperiods with daytime and nighttime temperatures of 26–29°C and 18–20°C, respectively. Bars represent mean expression relative to the mean of wild-type expression values. Error bars denote standard error (n = 5). Individual wild-type (filled circle), kix8 kix9#1 (filled square), and kix8 kix9#2 (filled triangle) values are shown. Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters). IG, immature green; MG, mature green; BR, breaker; OR, orange; RR, red ripe.
Tomato kix8 kix9 mutants produce enlarged fruits due to increased cell expansion
In multiple eudicot species that belong to the rosid order of Fabales, orthologs of the KIX and PPD proteins have been reported to negatively regulate seed pod size (Ge et al., 2016; Naito et al., 2017; Kanazashi et al., 2018; Li et al., 2019). Moreover, the cucumber ortholog of the F-box protein AtSAP was shown to positively regulate fruit size (Yang et al., 2018). To examine whether the SlKIX proteins might also have a role in determining fruit size in the asterid model species tomato, we investigated if the development of reproductive organs was affected in tomato plants in which SlKIX8 and/or SlKIX9 function was disturbed.
Fruits that developed on inflorescences of the main shoot were harvested from each genotype when the ratio of ripe to unripe fruits was 65%–85%, since we noted a significant delay in flowering time for kix8 kix9 mutants compared with wild-type plants (Supplemental Figure S6). The fresh weight of individual ripe tomatoes produced by kix8 kix9 and kix8 plants was increased by ∼15% and 30%, respectively, compared with those produced by wild-type plants (Table 1 andFigure 5A). Cutting along the equatorial plane revealed that kix8 kix9 and kix8 fruits displayed an approximate increase of 50% in pericarp thickness compared with wild-type fruits, while no change was observed for kix9 fruits (Table 1 andFigure 5B). Red fruit total biomass per plant was increased for kix8 and kix9 but not kix8 kix9 mutants compared with wild-type plants (Table 1). We noted that the number of red fruits per plant remained similar for kix8 plants and increased for kix9 plants while it was lower for kix8 kix9 plants than for wild-type plants (Table 1), the latter possibly resulting from an increase in flower abortion ratio within inflorescences (Table 1). To better estimate the effect of SlKIX loss-of-function on fruit yield, we performed a second experiment for which we harvested fruits not only from inflorescences on the main shoot but also on the axillary shoots from kix8 kix9 and wild-type plants. Whereas the number of inflorescences on the main shoot was unaffected in kix8 kix9 mutants compared to wild-type plants, the number of inflorescences on the axillary shoots was reduced by ∼50% (Supplemental Table S3), indicating a delay in axillary fruit development. Although the higher biomass of kix8 kix9 fruits accompanied by increased pericarp thickness compared with wild-type fruits was confirmed (Supplemental Table S3), the reduced axillary branching reduced total fruit yield per plant by ∼30% (Supplemental Table S3). In addition, we observed that kix8 kix9 fruits contained ∼45% less seeds than wild-type fruits, though seed size was unaffected (Supplemental Table S3).
Table 1.
Tomato kix8 and kix8 kix9 plants produce bigger fruits with increased pericarp thickness
| Parameters | Wild-Type | kix8 | kix9 | kix8 kix9#1 | kix8 kix9#2 |
|---|---|---|---|---|---|
| Inflorescence parameters | |||||
| Number of inflorescences | 4.00 ± 0.17abc | 4.44 ± 0.24ab | 4.77 ± 0.22b | 3.78 ± 0.15ac | 3.44 ± 0.18c |
| Number of flowers per inflorescence | 6.15 ± 0.67ab | 6.01 ± 0.33ab | 6.74 ± 0.42a | 5.77 ± 0.38b | 5.53 ± 0.32b |
| Number of pollinated flowers per inflorescence | 5.70 ± 0.70a | 4.13 ± 0.66b | 6.67 ± 0.41a | 4.53 ± 0.55b | 4.48 ± 0.55b |
| Pollinated/total number of flowers per inflorescence (%) | 93.13 ± 3.65ab | 69.03 ± 10.01c | 99.04 ± 1.21b | 77.63 ± 7.49cd | 80.96 ± 8.12ad |
| Fruit parameters | |||||
| Red fruit biomass (g) | 4.40 ± 0.65ab | 5.78 ± 0.57c | 3.63 ± 0.41b | 5.00 ± 0.56ac | 5.12 ± 0.59ac |
| Red fruit pericarp thickness (mm) | 2.67 ± 0.08a | 4.08 ± 0.16b | 2.65 ± 0.13a | 3.97 ± 0.08b | 4.16 ± 0.13b |
| Red fruit pericarp thickness/radius (%) | 21.44 ± 0.67a | 31.78 ± 1.31b | 22.59 ± 0.85a | 32.07 ± 0.84b | 32.28 ± 0.90b |
| Fruit yield parameters | |||||
| Number of red fruits | 16.56 ± 1.97a | 15.44 ± 0.97a | 26.67 ± 1.96b | 13.56 ± 0.60a | 12.44 ± 0.75a |
| Red fruit yield (g) | 69.85 ± 6.25ab | 88.71 ± 4.64ac | 97.00 ± 6.97c | 67.09 ± 2.18b | 62.51 ± 3.63b |
Plants were grown in soil for 3.5–4.5 months under 15:9 photoperiods with daytime and nighttime temperatures of 25–29°C and 17–19°C, respectively. Inflorescence parameters were measured for the main shoot and red fruits that developed on these inflorescences were harvested from each genotype when the ratio of ripe to unripe fruits was 65–85%. Data are mean ± standard error (n = 7–9). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters).
Figure 5.
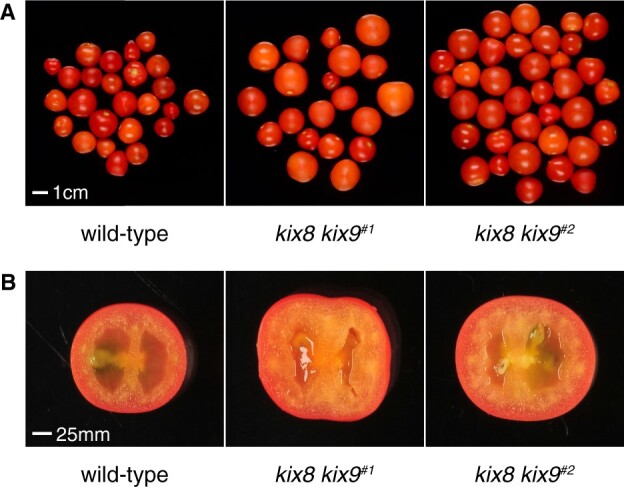
Tomato kix8 kix9 plants produce enlarged fruits that display increased pericarp thickness. A, Representative red ripe fruits produced by wild-type, kix8 kix9#1, and kix8 kix9#2 plants. B, Equatorial sections of representative red ripe fruits produced by wild-type, kix8 kix9#1, and kix8 kix9#2 plants. Plants were grown in soil under 16:8 h photoperiods with daytime and nighttime temperatures of 26–29°C and 18–20°C, respectively.
To determine the influence of an altered sink–source relationship between kix8 kix9, kix8, kix9, and wild-type plants, a follow-up experiment was performed in which fruit production was restricted. To do so, only the first two inflorescences on the main shoot, each carrying a maximum of six fruits, were kept per plant and growth parameters were documented from ovary stage until red ripe stage. Even though kix8 kix9 and kix8 mutants carried ovaries that were ∼35% smaller than wild-type plants (Table 2), final red fruit biomass was unaffected (Table 2). Moreover, the increase in pericarp thickness was confirmed in these restricted conditions for kix8 kix9 and kix8 fruits from 15 d postanthesis (DPA) onwards, with ripe kix8 kix9 and kix8 fruits harboring a pericarp that was ∼20% thicker than wild-type fruits (Table 2). To explore the cellular cause of this change in pericarp size, the number of cell layers and cell sizes were quantified. At 30 DPA, the number of cell layers across the pericarp was similar in wild-type, kix8, kix9, and kix8 kix9 fruits (Figure 6A). The average pericarp cell area, however, was increased in all mutant fruits compared with wild-type fruits (Figure 6, B and C). The increased cell area resulted from the appearance of very large cells within the pericarp of kix8, kix9, and kix8 kix9 plants and a decreased proportion of the smallest cells in kix8 and kix8 kix9 compared to wild-type plants (Figure 6D). Altogether, these data demonstrate that knocking out SlKIX8 on its own or SlKIX8 together with SlKIX9 results in the production of enlarged tomato fruits with increased pericarp thickness, suggesting that SlKIX8 and/or SlKIX9 are involved in the regulation of tomato fruit growth.
Table 2.
Pericarp thickness of fruits from tomato kix8 and kix8 kix9 plants grown in restricted conditions is increased
| Parameters | Wild-Type | kix8 | kix9 | kix8 kix9#1 |
|---|---|---|---|---|
| Ovary parameters | ||||
| Ovary diameter (mm) | 1.60 ± 0.02a | 1.13 ± 0.01b | 1.31 ± 0.04c | 0.95 ± 0.02d |
| Ovary height (mm) | 1.50 ± 0.05a | 1.17 ± 0.04b | 1.46 ± 0.05a | 0.95 ± 0.08b |
| Fruit parameters | ||||
| Biomass (g) | ||||
| 5 DPA fruit | 0.05 ± 0.01a | 0.21 ± 0.02a | 0.10 ± 0.03a | 0.04 ± 0.01a |
| 10 DPA fruit | 0.86 ± 0.06ab | 1.10 ± 0.09a | 0.68 ± 0.11b | 0.65 ± 0.03b |
| 15 DPA fruit | 2.72 ± 0.36a | 2.40 ± 0.09a | 2.00 ± 0.35a | 1.80 ± 0.06a |
| 30 DPA fruit | 4.77 ± 0.67a | 6.31 ± 0.16a | 5.15 ± 0.14a | 6.50 ± 0.76a |
| Red fruit | 6.76 ± 0.27a | 6.80 ± 0.16a | 4.73 ± 0.16b | 6.52 ± 0.31a |
| Pericarp thickness (mm) | ||||
| 5 DPA fruit | 0.54 ± 0.02ab | 0.61 ± 0.03ab | 0.72 ± 0.06a | 0.50 ± 0.04b |
| 10 DPA fruit | 1.25 ± 0.09a | 1.54 ± 0.04b | 1.26 ± 0.02a | 1.41 ± 0.05ab |
| 15 DPA fruit | 2.15 ± 0.11ab | 2.24 ± 0.05ab | 1.90 ± 0.12a | 2.35 ± 0.02b |
| 30 DPA fruit | 2.50 ± 0.04a | 3.54 ± 0.15b | 2.54 ± 0.13a | 3.81 ± 0.16b |
| Red fruit | 3.21 ± 0.06a | 3.97 ± 0.03b | 2.86 ± 0.06a | 4.20 ± 0.17b |
| Pericarp thickness/radius (%) | ||||
| 5 DPA fruit | 21.95 ± 1.06a | 20.29 ± 0.69a | 22.95 ± 1.40a | 22.86 ± 0.50a |
| 10 DPA fruit | 19.47 ± 1.02a | 21.63 ± 0.59ab | 22.11 ± 1.41ab | 23.93 ± 0.36b |
| 15 DPA fruit | 22.68 ± 1.46a | 23.86 ± 0.28a | 22.54 ± 0.50a | 27.99 ± 0.50b |
| 30 DPA fruit | 21.08 ± 0.72a | 26.98 ± 1.26b | 22.27 ± 1.12a | 29.50 ± 0.09b |
| Red fruit | 24.99 ± 1.20a | 30.45 ± 0.43b | 25.38 ± 0.39a | 33.93 ± 0.53c |
Plants were grown in soil for 3–4.5 months under 15:9 photoperiods with daytime and nighttime temperatures of 25–29°C and 17–19°C, respectively. Fruit production was restricted by keeping only the first two inflorescences, each carrying a maximum of six fruits, on the main shoot. Data are mean ± standard error (n = 3). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters).
Figure 6.
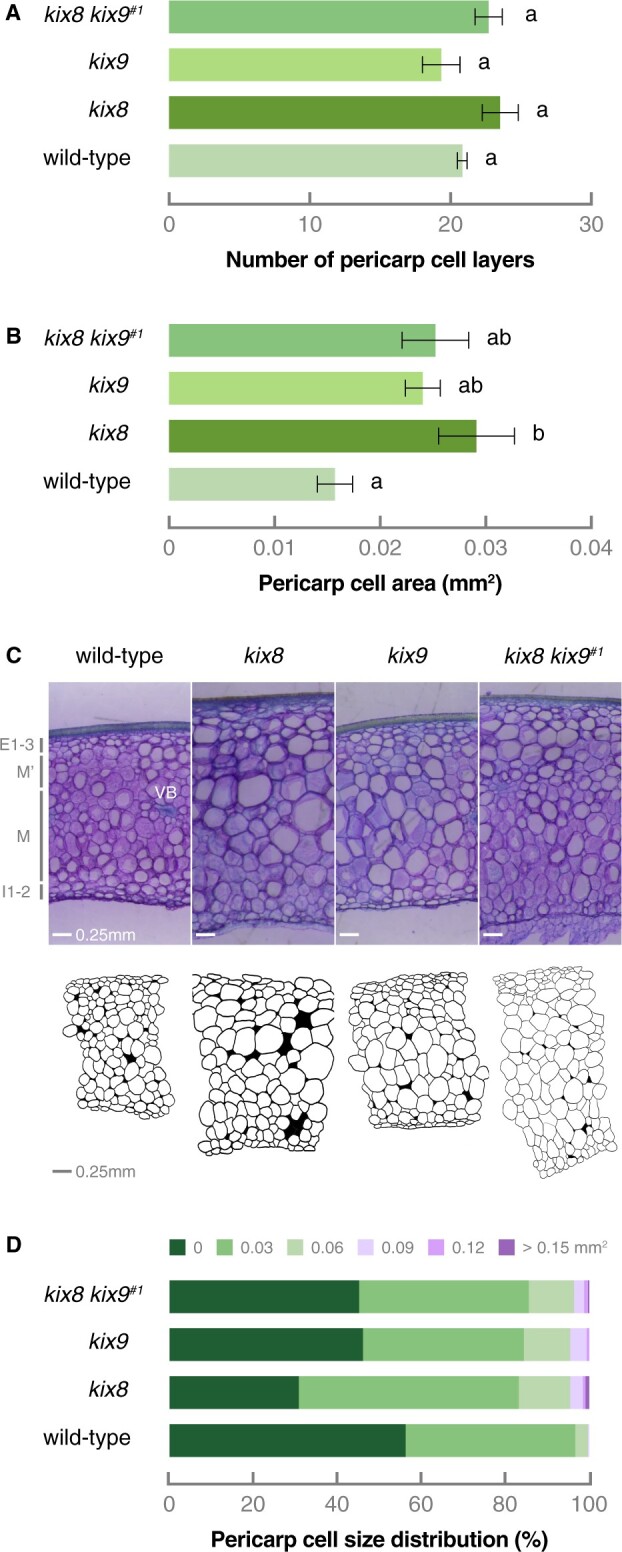
Increased pericarp thickness of kix8 and kix8 kix9 fruits results from the production of larger cells. A, Number of pericarp cell layers in wild-type, kix8, kix9, and kix8 kix9#1 fruit. B, Pericarp cell area in wild-type, kix8, kix9, and kix8 kix9#1 fruit. C, Microtome pericarp sections and drawings of a representative wild-type, kix8, kix9, and kix8 kix9#1 fruit. D, Pericarp cell size distribution in wild-type, kix8, kix9, and kix8 kix9#1 fruit. Plants were grown in soil under 15:9-h photoperiods with daytime and nighttime temperatures of 25–29°C and 17–19°C, respectively. Fruit production was restricted by keeping only the first two inflorescences, each carrying a maximum of six fruits, on the main shoot. Fruit was harvested at 30 DPA. Data are mean ± standard error (n = 2–4). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05; indicated by different letters). E, outer epidermis layer; M, mesocarp layer; I, inner epidermis layer; VB, vascular bundle.
Discussion
KIX8 and KIX9 are regulators of leaf growth in distinct eudicot species
In Arabidopsis, the asymmetric cell division of meristemoids and leaf growth are restricted by a transcriptional repressor complex in which the co-repressor AtTPL is recruited to AtPPD2 by AtKIX8/AtKIX9 (White, 2006; Gonzalez et al., 2015). Members of this repressor complex were shown to regulate leaf size and shape in a variety of species that belong to different orders of the rosids (Gonzalez et al., 2015; Ge et al., 2016; Naito et al., 2017; Kanazashi et al., 2018; Li et al., 2019), suggesting that the repressor complex is a conserved regulator of leaf growth among rosid eudicots (Schneider et al., 2021). Here, we demonstrate that the tomato orthologs of AtKIX8 and AtKIX9 act as SlTPL adaptors for SlPPD proteins and, thereby, regulate leaf growth in tomato plants. Tomato is a model species of the asterid clade that also includes tobacco (Nicotiana tabacum), carrot (Daucus carota), and sunflower (Helianthus annuus). In the rosid species Arabidopsis and pea, the interaction between KIX and PPD proteins is described to occur through the N-terminal KIX and PPD domain, respectively (Gonzalez et al., 2015; Li et al., 2019). This is likely to be the case in tomato as well, in which the SlKIX and SlPPD proteins display a similar domain structure. The interaction between tomato SlKIX8/SlKIX9 and the SlTPL co-repressors is expected to occur via the EAR motif present in the SlKIX proteins (Kagale et al., 2010; Causier et al., 2012), as was shown for their Arabidopsis and pea orthologs (Gonzalez et al., 2015; Li et al., 2019).
Tomato kix8 kix9 plants exhibited an enlarged, dome-shaped leaf phenotype, similar to the phenotype observed in Arabidopsis kix8-kix9 and ami-ppd mutants (Gonzalez et al., 2015). Moreover, terminal leaflets of young tomato kix8 kix9 leaves displayed increased expression of three putative SlPPD target genes, SlDFL1, SlAHL17, and SlAP2d, of which the orthologs were found to be directly bound by AtPPD2 and were strongly upregulated in Arabidopsis kix8-kix9 and ami-ppd leaves (Gonzalez et al., 2015). Another direct target gene of AtPPD2, ASYMMETRIC LEAVES1 (AtAS1), was shown to be involved in adaxial/abaxial leaf patterning (Byrne et al., 2000). Arabidopsis plants constitutively expressing AtAS1 displayed dome-shaped leaves (Husbands et al., 2015) reminiscent of Arabidopsis kix8-kix9 and ami-ppd mutants, suggesting that adaxial/abaxial leaf polarity might be affected in these mutants. Furthermore, the dome-shaped leaf phenotype might result from a convex-shaped primary cell cycle arrest front, as was already observed for Arabidopsis ami-ppd and ninja mutants (Baekelandt et al., 2018). The SlPPD–SlKIX complex might control similar molecular processes during leaf development as in Arabidopsis (White, 2006; Gonzalez et al., 2015). Taken together, we can conclude that both in rosid and asterid species, KIX8 and KIX9 assist PPD proteins in repressing distinct downstream target genes and in regulating leaf size and shape.
Partial redundancy of SlKIX8 and SlKIX9
In Arabidopsis, AtKIX8 and AtKIX9 were reported to have partially redundant roles in controlling leaf growth (Gonzalez et al., 2015). The intermediate and absent leaf phenotype of tomato single kix8 and kix9 mutants, respectively, compared with the markedly enlarged, dome-shaped leaf phenotype of tomato kix8 kix9 plants, suggests partial redundancy of SlKIX8 and SlKIX9 in tomato leaf development as well. In fruits, we observed stronger phenotypes for single kix8 than kix9 mutants. In line with these phenotypes, SlKIX9 expression is (almost) absent in most tomato tissues, whereas SlKIX8 displays an overall higher expression level in the examined tissues. In kix8 kix9 tomato leaflets, however, the transcript levels of not only SlKIX8 but also SlKIX9 were increased compared with wild-type leaflets, suggesting negative feedback of the SlPPD–SlKIX complex on the expression of both SlKIX8 and SlKIX9. In yeast cells, interaction with SlTPL2 was observed for both SlKIX8 and SlKIX9, but SlKIX8 could additionally interact with SlTPL1, SlTPL4, SlTPL6, and SlSAP. A previous study showed that from the six SlTPL genes, SlTPL1 had the highest overall expression in the examined tissues and developmental stages, while SlTPL2 was expressed at a much lesser extent (Hao et al., 2014). The expression of SlTPL4 dominated in ripening fruit and SlTPL6 transcripts were almost absent in all investigated tissues (Hao et al., 2014). Furthermore, SlTPL6 was suggested to have lost its functionality (Hao et al., 2014) and, therefore, calls the biological relevance of the interaction between SlKIX8 and SlTPL6 into question. To further explore this, it could be relevant to investigate the tissue-specific interactions between SlKIX and SlTPL proteins in planta. All in all, these data indicate that SlKIX8 and SlKIX9 are functionally redundant, but that SlKIX8 might play a predominant role in the regulation of leaf and fruit development.
SlKIX8 and SlKIX9 are negative regulators of fruit growth
Like any other plant organ, fruit grows by means of cell division and cell expansion. After fertilization, tomato ovary growth starts with a short period of cell proliferation followed by a longer cell expansion phase, resulting in a massive expansion of the pericarp (or fruit flesh) in particular (Xiao et al., 2009). Fruit ripening commences after fruit growth is finalized. Here, we report that SlKIX8 and/or SlKIX9 act as negative regulators of fruit growth, as simultaneously knocking out SlKIX8 and SlKIX9 by CRISRP–Cas9 genome editing results in the production of enlarged tomato fruits with increased pericarp thickness. We found that SlKIX8 loss-of-function was sufficient to trigger an increase in tomato fruit size that was associated with an increase in pericarp thickness. A stronger effect on growth-related phenotypes by downregulation of kix8 compared to that of kix9 was also observed for other plant species (Gonzalez et al., 2015, Nguyen et al., 2021). Surprisingly, the increase in tomato pericarp thickness was associated with an increased proportion of larger cells, whereas an increase in cell division was noted in the leaves of Arabidopsis kix8 kix9 mutants. However, these Arabidopsis mutants produce larger seeds resulting from both increased cell proliferation and cell elongation (Liu et al., 2020). These findings suggest that KIX8 and KIX9 might regulate multiple cellular processes, possibly through interacting with tissue-specific transcriptional regulators allowing them to target different sets of genes depending on the organ they are expressed.
In line with our findings, several rosid eudicot species, in which the KIX or PPD genes were either mutated or downregulated, displayed increased seed pod size (Ge et al., 2016; Kanazashi et al., 2018; Li et al., 2019). Ge et al. (2016) attributed the larger size of pods produced by M. truncatula plants in which an AtPPD ortholog was mutated to a prolonged period of cell division, similar to what they observed in developing leaves. Complete loss-of-function and severe downregulation of PPD genes in soybean and blackgram, respectively, led to a strong increase in seed size but was accompanied by a drastic reduction in seed number, thereby negatively impacting total yield (Naito et al., 2017; Kanazashi et al., 2018). Although we also observed a drastic reduction in seed number, a change in seed size was not observed in the tomato kix8 kix9 mutants. Orthologs of AtSAP, an F-box protein that regulates the stability of the AtPPD–AtKIX complex, are positive regulators of seed pod and flower size in M. truncatula, of fruit and flower size in cucumber and of flower size in pink shepherd’s-purse (Capsella rubella; Sicard et al., 2016; Yang et al., 2018; Yin et al., 2020).
Here, we show that tomato kix8 kix9 loss-of-function lines produce enlarged fruits, which was among the main selection criteria for nearly all fruit crops during domestication and still is today (Pickersgill, 2007). Many of the alleles selected during domestication are not severe gain- or loss-of-function alleles, but are the result of mutations residing in cis-regulatory elements (CREs) that led to spatiotemporal expression changes of genes involved in crop development (Doebley et al., 2006; Meyer and Purugganan, 2013; Swinnen et al., 2016). These CRE alterations were likely favored over severe gain- or loss-of-function mutations, which would have been accompanied by undesirable pleiotropic effects (Swinnen et al., 2016). The tomato kix8 kix9 mutants displayed reduced axillary branching, increased flower abortion, and delayed flowering, all undesirable traits for breeding, negatively impacting fruit yield. The delay in flowering time may be explained by the upregulation of SlAP2d in kix8 kix9 leaves. This putative floral repressor gene is an ortholog of AtSMZ, which encodes a protein that counteracts the activity of CONSTANS, a promoter of flowering, by repressing the expression of multiple flowering time regulators including FLOWERING TIME in Arabidopsis leaves (Mathieu et al., 2009). Using gene editing to engineer novel SlKIX8 and SlKIX9 alleles with altered protein–protein interactions or modifying CREs in the promoter regions of SlKIX8 and SlKIX9 to downregulate their expression specifically during fruit growth might present promising breeding strategies (Swinnen et al., 2016; Rodríguez-Leal et al., 2017, Nguyen et al., 2021).
Materials and methods
Ortholog identification
Tomato protein orthologs of AtKIX8 and AtKIX9 were identified through a BLASTP search in the National Center for Biotechnology Information GenBank protein database. Tomato protein orthologs of AtSAP, AtDFL1, AtAHL17, and AtSMZ were retrieved from the comparative genomics resource PLAZA 4.0 Dicots (http://bioinformatics.psb.ugent.be/plaza/; Van Bel et al., 2018).
DNA constructs
Y2H and Y3H constructs
For Y2H and Y3H assays, the coding sequence of tomato SlKIX8, SlKIX9, SlPPD1, SlPPD2, and SlSAP was PCR-amplified with the primers listed in Supplemental Table S4 and recombined in a Gateway donor vector (Invitrogen, Waltham, MA, USA). Gateway donor vectors containing the coding sequence of tomato SlTPL1–6 were obtained from Hao et al. (2014). Subsequently, Gateway LR reactions (Invitrogen) were performed with pGAD424gate and pGBT9gate, generating bait and prey constructs, respectively. Alternatively, MultiSite Gateway LR reactions (Invitrogen) were performed with pMG426 (Nagels Durand et al., 2012) to express a third protein of interest, driven by the GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE promoter and C-terminally fused to the SV40 NLS-3xFLAG-6xHis tag.
CRISPR–Cas9 constructs
To select CRISPR–Cas9 guide RNA (gRNA) target sites, CRISPR-P (http://crispr.hzau.edu.cn/CRISPR/; Lei et al., 2014) was used. We selected a gRNA target site in the first exon of SlKIX8, whereas for SlKIX9, we selected a gRNA target site in the third exon downstream of a start codon that could act as an alternative transcription start site (Supplemental Figure S2B). The CRISPR–Cas9 construct was cloned as previously described (Fauser et al., 2014; Ritter et al., 2017; Pauwels et al., 2018). Briefly, for each gRNA target site, two complementary oligonucleotides with 4-bp overhangs (Supplemental Table S4) were annealed and inserted by a Golden Gate reaction with BpiI (Thermo Scientific, Waltham, MA, USA) and T4 DNA ligase (Thermo Scientific) in a Gateway entry vector. As Gateway entry vectors, pMR217 (L1-R5) and pMR218 (L5-L2; Ritter et al., 2017) were used. Next, a MultiSite Gateway LR reaction (Invitrogen) was used to recombine two gRNA modules with pDe-Cas9-Km (Ritter et al., 2017).
Y2H and Y3H assays
Y2H and Y3H assays were performed as described previously (Cuéllar Pérez et al., 2013). Briefly, for Y2H assays, the Saccharomyces cerevisiae PJ69-4A yeast strain was co-transformed with bait and prey constructs using the polyethylene glycol/lithium acetate method. Transformants were selected on Synthetic Defined (SD) medium lacking Leu and Trp (−2) (Clontech, Mountain View, CA, USA). Three individual transformants were grown overnight in liquid SD (−2) medium and 10-fold dilutions of these cultures were dropped on SD control (−2) and selective medium additionally lacking His (−3) (Clontech). Empty vectors were used as negative controls. Yeast plates were allowed to grow for 2 d at 30°C before interaction was scored. Y3H assays were performed in the same way, but with different SD media compositions. For transformant selection and culturing in control media, SD medium lacking Leu, Trp, and Ura (−3) was used, whereas selective media additionally lacked His (−4) (Clontech).
Plant material and growth conditions
Tomato (S. lycopersicum) wild-type and CRISPR–Cas9 mutant seeds (cultivar Micro-Tom) were sown in soil. Experiments for data in Figures 2–5, Supplemental Figures S3, S5 and S6, and Supplemental Tables S1–S3 were carried out in VIB-UGent (Ghent, Belgium). Plants were grown under long-day photoperiods (16:8 h). Daytime and nighttime temperatures were 26–29°C and 18–20°C, respectively. Inflorescence and flower production, and thus fruit production, was not restricted. Experiments for data in Figure 6 and Tables 1 and 2 were carried out in INRAE (Bordeaux, France). Plants were grown under long-day photoperiods (15:9 h). Daytime and nighttime temperatures were 25–29°C and 17–19°C, respectively. Fruit production was either unrestricted or restricted by keeping only the first two inflorescences, each carrying a maximum of six flowers, on the main shoot.
Tomato plant transformation
Binary constructs were introduced in competent Agrobacterium tumefaciens (strain EHA105) cells using electroporation and transformed into tomato (cultivar Micro-Tom) using the cotyledon transformation method as reported previously (Gonzalez et al., 2007) with the following modifications. Cotyledon pieces from 1-week-old seedlings were incubated for 24 h in the dark at 25°C on solid Murashige and Skoog (MS) medium (pH 5.7) containing 4.4 g L−1 of MS supplemented with vitamins (Duchefa), 20 g L−1 of sucrose, 0.2 g L−1 of KH2PO4, 1 mg L−1 of thiamine, 0.2 mM of acetosyringone, 0.2 mg L−1 of 2,4-dichlorophenoxyacetic acid, and 0.1 mg L−1 of kinetin. Next, the cotyledon pieces were soaked in an A. tumefaciens (strain EHA105) bacterial suspension culture (0.05–0.10 OD600) containing the binary vector for 25 min while shaking. Cotyledon pieces were dried on sterile tissue paper and placed back on the aforementioned solid MS medium for 48 h in the dark at 25°C. Cotyledon pieces were washed once with liquid MS medium (pH 5.7) containing 4.4 g L−1 of MS supplemented with vitamins (Duchefa, Haarlem, Netherlands), 20 g L−1 of sucrose, 0.2 g L−1 of KH2PO4, and 1 mg L−1 of thiamine, and once with sterile water. Cotyledon pieces were dried on sterile tissue paper and placed on solid MS medium (pH 5.7) containing 4.4 g L−1 of MS supplemented with vitamins (Duchefa), 30 g L−1 of sucrose, 1 mL L−1 of 1000X Nitsch vitamin stock (for 100 mL: 0.005 g of biotin, 0.2 g of glycine, 10 g of myo-inositol, 0.5 g of nicotinic acid, 0.05 g of pyridoxine HCl, and 0.05 g of thiamine HCl), 0.5 g L−1 of folic acid, 2 mg L−1 of zeatin riboside, 100 mg L−1 of kanamycin, 25 mg L−1 of melatonin, and 300 mg L−1 of timentin and put in a 25°C controlled photoperiodic growth chamber (16:8 h photoperiods). The medium was refreshed every 14 d until regenerated shoots appeared. These shoots were placed on solid MS medium (pH 5.7) containing 2.2 g L−1 of MS, 10 g L−1 of sucrose, 1 mL L−1 of 1000X Nitsch vitamin stock, 0.5 g L−1 of folic acid, 100 mg L−1 of kanamycin, and 150 mg L−1 of timentin until their acclimatization in the greenhouse.
Identification of CRISPR–Cas9 mutants
Plant genotyping
CRISPR–Cas9 mutants were identified as described previously (Swinnen et al., 2020). Genomic DNA was prepared from homogenized leaf tissue using extraction buffer (pH 9.5) containing 0.1 M of tris(hydroxymethyl)aminomethane (Tris)–HCl, 0.25 M of KCl, and 0.01 M of ethylenediaminetetraacetic acid (EDTA). The mixture was incubated at 95°C for 10 min and cooled at 4°C for 5 min. After addition of 3% (w/v) bovine serum albumine (BSA), the supernatant was used as a template in a standard PCR reaction using GoTaq (Promega, Madison, WI, USA) with Cas9-specific primers (to select primary plant transformant (T0) lines in which the T-DNA was present or plant T1 lines in which the T-DNA was absent) or with primers to amplify a gRNA target region (Supplemental Table S4). PCR amplicons containing a gRNA target site were purified using HighPrep PCR reagent (MAGBIO, Gaithersburg, MD, USA). After Sanger sequencing of the purified PCR amplicons with an amplification primer located ∼200 bp from the Cas9 cleavage site, quantitative sequence trace data were decomposed using Tracking Indels by DEcomposition (https://www.deskgen.com/landing/tide.html#/tide) or Inference of CRISPR Editing Analysis Tool (https://ice.synthego.com/#/).
Plant ploidy level analysis
Diploid CRISPR–Cas mutants (T0) were identified using flow cytometry. Leaf material (1.0 cm2) was chopped in 200 μL of chilled CyStain UV Precise P Nuclei Extraction Buffer (Sysmex) for 2 min using a razor blade. The suspension was filtered through a 50-μm nylon filter and 800 μL of chilled CyStain UV Precise P Staining Buffer (Sysmex, Hyogo, Japan) was added to the isolated nuclei. The DNA content of 5,000–10,000 nuclei was measured using a CyFlow Space flow cytometer (Sysmex) and analyzed with FloMax software (Sysmex).
Phenotypic analyses
Plant growth parameter analysis
Primary shoot, main shoot, and internode length of 4-month-old CRISPR–Cas mutant (T2) and wild-type plants were measured. Per genotype, 12 biological replicates (plants) were collected.
Leaf growth parameter analysis
The eighth leaf (from the top) from 2-month-old CRISPR–Cas mutant (T2) and wild-type plants was harvested for leaf growth parameter analysis. Per genotype, 31–40 biological replicates (leaves) were collected. A digital balance was used to measure the biomass/fresh weight of leaves and their terminal leaflets. Pictures of terminal leaflets were taken before (projected) and after (real), and cutting the leaves to flatten them. Projected and real leaflet area was measured using ImageJ (https://imagej.nih.gov/ij/).
Flowering time analysis
Flowering time of CRISPR–Cas mutant (T2) and wild-type plants was quantified by counting the number of true leaves that were produced before initiation of the primary inflorescence (Soyk et al., 2017). Flowering time was measured for 15–16 plants per genotype.
Inflorescence parameter analysis
Inflorescence and (pollinated) flower number of 3.5– to 4.5-month-old CRISPR–Cas mutant (T2) and wild-type plants was quantified. Per genotype, 12 biological replicates (plants) were collected.
Ovary growth parameter analysis
For ovary diameter and height measurements, ovaries from CRISPR–Cas mutant (T2) and wild-type plants were harvested at anthesis. Ovary diameter was determined by averaging the maximum and minimum diameter of the equatorial axis. Per genotype, three biological replicates (ovaries) were collected.
Fruit growth parameter analysis
For fruit biomass, pericarp thickness, and yield measurements, fruits at distinct developmental and ripening stages (5 DPA-red ripe) were harvested from CRISPR–Cas mutant (T2) and wild-type plants. Pericarp thickness was measured by taking scans of equatorial fruit sections and using Tomato Analyzer (version 4.0; Brewer et al., 2006). Per genotype and developmental stage, 12 biological replicates (fruits from 12 individual plants; fruit production unrestricted; VIB-UGent), 7–9 biological replicates (fruits from 12 individual plants; fruit production unrestricted; INRAE), or 3 biological replicates (fruits from 3 batches of 3 plants; fruit production restricted; INRAE) were collected.
For cell layer number quantification and cell area measurements, fruits were harvested at 30 DPA and pericarp was fixed in a solution of FAA (18v EtOH 70% v/v, 1v acetic acid, and 1v formaldehyde). Pericarp sections with a thickness of 100 µm were made with a microtome and imaged. The number of cell layers and individual cell area were quantified using ImageJ (https://imagej.nih.gov/ij/). Per genotype, 2–4 biological replicates (fruit production restricted; INRAE) were collected.
Seed parameter analysis
For seed number and size analyses, seeds were harvested from red ripe fruits produced by CRISPR–Cas mutant (T2) and wild-type plants. Seed area was measured using ImageJ (https://imagej.nih.gov/ij/). Per genotype, 12 biological replicates were collected, constituting seeds from red ripe fruits from 12 individual plants.
Statistical analysis
For all phenotypic analyses, statistical significance was determined by analysis of variance (ANOVA) followed by Tukey’s post-hoc analysis (P < 0.05).
Gene expression analysis by RT-qPCR
The terminal leaflet of the second leaf (from the top) from 3-week-old CRISPR–Cas mutant (T2) and wild-type plants were harvested by flash freezing in liquid nitrogen and ground using the Mixer Mill 300 (Retch). Per genotype, five biological replicates, each consisting of a single terminal leaflet was collected. Messenger RNA was extracted from homogenized tissue as described in (Townsley et al., 2015) with the following modifications. Tissue was lysed using 800 µL of lysate binding buffer (LBB) containing 100 mM of Tris–HCl (pH 7.5), 500 mM of LiCl, 10 mM of EDTA (pH 8.0), 1% w/v of sodium dodecyl sulfate (SDS), 5 mM of dithiothreitol, 15 μL mL−1 of Antifoam A, and 5 μL mL−1 of 2-mercaptoethanol, and the mixture was incubated for 10 min at room temperature. Messenger RNA was separated from 200 µL of lysate using 1 µL of 12.5 µM of 5′-biotinylated polyT oligonucleotide (5′-biotin-ACAGGACATTCGTCGCTTCCTTTTTTTTTTTTTTTTTTTT-3′) and the mixture was incubated for 10 min. Next, captured messenger RNA was isolated from the lysate by adding 20 µL of LBB-washed streptavidin-coated magnetic beads (New England Biolabs, Ipswich, MA, USA) and incubated for 10 min at room temperature. Samples were placed on a MagWell Magnetic Separator 96 (EdgeBio, San Jose, CA, USA) and washed with 200 µL of washing buffer A (10 mM of Tris–HCl [pH 7.5], 150 mM of LiCl, 1 mM of EDTA [pH 8.0], 0.1% w/v of SDS), washing buffer B (10 M of Tris–HCl [pH 7.5], 150 mM of LiCl, 1 mM of EDTA [pH 8.0]), and low-salt buffer (20 mM of Tris–HCl [pH 7.5], 150 mM of NaCl, 1 mM of EDTA [pH 8.0]), all pre-chilled on ice. Elution of messenger RNA was done by adding 20 µL of 10 mM of Tris–HCl (pH 8.0) with 1 mM of 2-mercaptoethanol followed by incubation of the mixture at 80°C for 2 min.
First-strand complementary DNA was synthesized from 20 µL of messenger RNA eluate by qScript cDNA Synthesis Kit (Quantabio, Beverly, MA, USA). RT-qPCR reactions were performed with a LightCycler 480 System (Roche, Basel, Switzerland) using Fast SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA) and primers (Supplemental Table S4) designed by QuantPrime (https://www.quantprime.de/; Arvidsson et al., 2008). Gene expression levels were quantified relative to CLATHRIN ADAPTOR COMPLEXES MEDIUM SUBUNIT (SlCAC) and TAP42-INTERACTING PROTEIN (SlTIP41) using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Statistical significance was determined by ANOVA followed by Tukey’s post-hoc analysis (P < 0.05).
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank/Solgenomics data libraries under the following accession numbers: SlKIX8 (Solyc07g008100), SlKIX9 (Solyc08g059700), SlPPD1 (Solyc06g084120), SlPPD2 (Solyc09g065630), SlSAP (Solyc05g041220), SlDFL1 (Solyc07g063850), SlAHL17 (Solyc04g076220), SlAP2d (Solyc11g072600), SlCAC (Solyc08g006960), and SlTIP41 (Solyc10g049850).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. A conserved repressor complex regulates leaf growth in distinct eudicot species.
Supplemental Figure S2. Splice variants of SlKIX8, SlKIX9, SlPPD1, and SlPPD2.
Supplemental Figure S3. Regenerated tomato kix8 kix9 plants display a rippled, dome-shaped leaf phenotype.
Supplemental Figure S4. CRISPR–Cas9 mutations in double kix8 kix9 (T1), single kix8, and single kix9 tomato knockout lines.
Supplemental Figure S5. Single kix8 and kix9 mutants do not display significant upregulation of putative SlPPD target genes.
Supplemental Figure S6. Tomato kix8 kix9 plants display a delay in flowering time.
Supplemental Table S1. Tomato kix8 kix9 plants display a reduction in plant height.
Supplemental Table S2. Normalized expression of SlKIX8, SlKIX9, SlPPD1, SlPPD2, SlDFL1, SlAHL17, and SlAP2d in different tomato organs and developmental stages (cultivar Micro-Tom) used to generate heat maps in Figure 4, A and B.
Supplemental Table S3. Tomato kix8 kix9 plants display a reduction in axillary shoot formation.
Supplemental Table S4. Oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Annick Bleys for help with preparing the manuscript and Mohamed Zouine for sharing plasmids containing the coding sequence of SlTPL1–6 with us.
Funding
This work was supported by the Research Foundation Flanders (FWO) through the projects G005312N, G004515N, and 3G038719 and a postdoctoral fellowship to L.P.
Conflict of interest statement. none declared.
G.S., J.P.M., A.B., L.P., N.G., and A.G. designed the experiments. G.S., J.P.M., A.B., R.D.C., and J.V.D. performed the experiments. G.S., J.P.M., A.B., L.P., N.G., and A.G. analyzed the data. G.S. wrote the article and J.P.M., A.B., L.P., N.G., and A.G. contributed to the writing and revised the manuscript. N.G., D.I., A.G., and L.P. supervised the project. L.P. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Laurens Pauwels (http://lapau@psb.vib-ugent.be).
References
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime - a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt A, Pauwels L, Wang ZB, Li N, De Milde L, Natran A, Vermeersch M, Li Y, Goossens A, Inzé D, et al. (2018) Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol 178: 217–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Meng Y, Huang D, Qi Y, Chen M (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 [DOI] [PubMed] [Google Scholar]
- Brewer MT, Lang L, Fujimura K, Dujmovic N, Gray S, van der Knaap E (2006) Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol 141: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Ben-Romdhane W, Hassairi A, Aboul-Soud MAM (2017) Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS One 12: e0177381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R (2009) The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Howe GA (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar Pérez A, Pauwels L, De Clercq R, Goossens A (2013) Yeast two-hybrid analysis of jasmonate signaling proteins. Methods Mol Biol 1011: 173–185 [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Ge L, Yu J, Wang H, Luth D, Bai G, Wang K, Chen R (2016) Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc Natl Acad Sci USA 113: 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A (2007) The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51: 642–655 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Pauwels L, Baekelandt A, De Milde L, Van Leene J, Besbrugge N, Heyndrickx KS, Cuéllar Pérez A, Nagels Durand A, De Clercq R, et al. (2015) A repressor protein complex regulates leaf growth in Arabidopsis. Plant Cell 27: 2273–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D (2012) Leaf size control: complex coordination of cell division and expansion. Trend Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang X, Li X, Bassa C, Mila I, Audran C, Maza E, Li Z, Bouzayen M, van der Rest B, et al. (2014) Genome-wide identification, phylogenetic analysis, expression profiling, and protein–protein interaction properties of TOPLESS gene family members in tomato. J Exp Bot 65: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth J, Lenhard M (2014) Regulation of plant lateral-organ growth by modulating cell number and size. Curr Opin Plant Biol 17: 36–42 [DOI] [PubMed] [Google Scholar]
- Husbands AY, Benkovics AH, Nogueira FT, Lodha M, Timmermans MC (2015) The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27: 3321–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajala K, Shaar-Moshe L, Mason GA, Gouran M, Rodriguez-Medina J, Kawa D, Pauluzzi G, Reynoso M, Canto-Pastor A, Lau V, et al. (2020) Innovation, conservation, and repurposing of gene function in root cell type development. Cell 184: 3333–3348.e19 [DOI] [PubMed] [Google Scholar]
- Kalve S, De Vos D, Beemster GTS (2014) Leaf development: a cellular perspective. Front Plant Sci 5: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazashi Y, Hirose A, Takahashi I, Mikami M, Endo M, Hirose S, Toki S, Kaga A, Naito K, Ishimoto M, et al. (2018) Simultaneous site-directed mutagenesis of duplicated loci in soybean using a single guide RNA. Plant Cell Rep 37: 553–563 [DOI] [PubMed] [Google Scholar]
- Kumar V, Waseem M, Dwivedi N, Maji S, Kumar A, Thakur JK (2018) KIX domain of AtMed15a, a Mediator subunit of Arabidopsis, is required for its interaction with different proteins. Plant Signal Behav 13: e1428514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li N, Liu Z, Wang Z, Ru L, Gonzalez N, Baekelandt A, Pauwels L, Goossens A, Xu R, Zu Z, et al. (2018) STERILE APETALA modulates the stability of a repressor protein complex to control organ size in Arabidopsis thaliana. PLoS Genet 14: e1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yamada M, Hang X, Ohler U, Benfey PN (2016) High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell 39: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu W, Zhuang L, Zhu Y, Wang F, Chen T, Yang J, Ambrose M, Hu Z, Weller JL, et al. (2019) BIGGER ORGANS and ELEPHANT EAR-LIKE LEAF1 control organ size and floral organ internal asymmetry in pea. J Exp Bot 70: 179–191 [DOI] [PubMed] [Google Scholar]
- Liu Z, Na L, Zhang Y, Li Y (2020) Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat Commun 11: 1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC (2009) Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Murdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD (2013) Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet 14: 840–852 [DOI] [PubMed] [Google Scholar]
- Nagels Durand A, Moses T, De Clercq R, Goossens A, Pauwels L (2012) A MultiSite GatewayTM vector set for the functional analysis of genes in the model Saccharomyces cerevisiae. BMC Mol Biol 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Takahashi Y, Chaitieng B, Hirano K, Kaga A, Takagi K, Ogiso-Tanaka E, Thavarasook C, Ishimoto M, Tomooka N (2017) Multiple organ gigantism caused by mutation in VmPPD gene in blackgram (Vigna mungo). Breed Sci 67: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Moloney M, Inzé D (2014) Translational research: from pot to plot. Plant Biotechnol J 12: 277–285 [DOI] [PubMed] [Google Scholar]
- Nguyen CX, Paddock KJ, Zhang Z, Stacey MG (2021) GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol 2: 920–934 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Cuéllar Pérez A, Chico JM, Vanden Bossche R, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, De Clercq R, Goossens J, Iñigo S, Williams C, Ron M, Britt A, Goossens A (2018) A dual sgRNA approach for functional genomics in Arabidopsis thaliana. G3 (Bethesda) 8: 2603–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill B (2007) Domestication of plants in the Americas: insights from mendelian and molecular genetics. Ann Bot 100: 925–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A, Iñigo S, Fernández-Calvo P, Heyndrickx KS, Dhondt S, Shi H, De Milde L, Vanden Bossche R, De Clercq R, Eeckhout D, et al. (2017) The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat Commun 8: 15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480.e478 [DOI] [PubMed] [Google Scholar]
- Schneider M, Gonzalez N, Pauwels L, Inzé D, Baekelandt A (2021) The PEAPOD pathway and its potential to improve crop yield. Trends Plant Sci 26: 220–236 [DOI] [PubMed] [Google Scholar]
- Sicard A, Kappel C, Lee YW, Woźniak N. J., Marona C, Stinchcombe JR, Wright SI, Lenhard M (2016) Standing genetic variation in a tissue-specific enhancer underlies selfing-syndrome evolution in Capsella. Proc Natl Acad Sci USA 113: 13911–13916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB (2017) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162–168 [DOI] [PubMed] [Google Scholar]
- Swinnen G, Goossens A, Pauwels L (2016) Lessons from domestication: targeting cis-regulatory elements for crop improvement. Trend Plant Sci 21: 506–515 [DOI] [PubMed] [Google Scholar]
- Swinnen G, Jacobs T, Pauwels L, Goossens A (2020) CRISPR-Cas-mediated gene knockout in tomato. Methods Mol Biol 2083: 321–341 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Agarwal P, Parida S, Bajaj D, Pasrija R (2013) Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol Genet Genomics 288: 329–346 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Yadav A, Yadav G (2014) Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res 42: 2112–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR (2015) BrAD-seq: breath Adapter Directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci 6: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K (2018) PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46: D1190–D1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trend Plant Sci 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Vatén A, Bergmann DC (2012) Mechanisms of stomatal development: an evolutionary view. EvoDevo 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J, Baekelandt A, Gonzalez N, Inzé D (2020) Molecular networks regulating the cell division during leaf growth in Arabidopsis. J Exp Bot 71: 2365–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li N, Jiang S, Gonzalez N, Huang X, Wang Y, Inzé D, Li Y (2016) SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat Commun 7: 11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DWR (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc Royal Soc Lond B-Biol Sci 268: 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Radovich C, Welty N, Hsu J, Li D, Meulia T, van der Knaap E (2009) Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu H, Zhao J, Pan Y, Cheng S, Lietzow CD, Wen C, Zhang X, Weng Y (2018) LITTLELEAF (LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J 95: 834–847 [DOI] [PubMed] [Google Scholar]
- Yin P, Ma Q, Wang H, Feng D, Wang X, Pei Y, Wen J, Tadege M, Niu L, Lin H (2020) SMALL LEAF AND BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol 226: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanov YS, Ma C, Yordanova E, Meilan R, Strauss SH, Busov VB (2017) BIG LEAF is a regulator of organ size and adventitious root formation in poplar. PLoS One 12: e0180527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouine M, Maza E, Djari A, Lauvernier M, Frasse P, Smouni A, Pirrello J, Bouzayen M (2017) TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J 92: 727–735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.