Abstract
Plant TRANS-ACTING SIRNA3 (TAS3)-derived short interfering RNAs (siRNAs) include tasiR-AUXIN RESPONSE FACTORs (ARFs), which are functionally conserved in targeting ARF genes, and a set of non-tasiR-ARF siRNAs, which have rarely been studied. In this study, TAS3 siRNAs were systematically characterized in rice (Oryza sativa). Small RNA sequencing results showed that an overwhelming majority of TAS3 siRNAs belong to the non-tasiR-ARF group, while tasiR-ARFs occupy a diminutive fraction. Phylogenetic analysis of TAS3 genes across dicot and monocot plants revealed that the siRNA-generating regions were highly conserved in grass species, especially in the Oryzoideae. Target genes were identified for not only tasiR-ARFs but also non-tasiR-ARF siRNAs by analyzing rice Parallel Analysis of RNA Ends datasets, and some of these siRNA–target interactions were experimentally confirmed using tas3 mutants generated by genome editing. Consistent with the de-repression of target genes, phenotypic alterations were observed for mutants in three TAS3 loci in comparison to wild-type rice. The regulatory role of ribosomes in the TAS3 siRNA–target interactions was further revealed by the fact that TAS3 siRNA-mediated target cleavage, in particular tasiR-ARFs targeting ARF2/3/14/15, occurred extensively in rice polysome samples. Altogether, our study sheds light into TAS3 genes in plants and expands our knowledge about rice TAS3 siRNA–target interactions.
A population of rice short interfering RNAs derived from TRANS-ACTING SIRNA3 loci direct target mRNA cleavage, revealing a broader role of TRANS-ACTING SIRNA3 genes in rice growth and stress responses.
Introduction
In plants, small RNAs have emerged as core players in almost all biological processes by regulating the expression of target genes through transcript cleavage and/or translational repression at the post-transcriptional level, or transcriptionally via directing DNA methylation (Chen, 2009; Rogers and Chen, 2013; Baulcombe, 2004; Fang and Qi, 2016; Martinez and Köhler, 2017; Tang and Chu, 2017; Yu et al., 2017). Two major types of small RNAs exist in plants, microRNAs (miRNAs) and short interfering RNAs (siRNAs), which differ in their biogenesis and modes of action. miRNAs are produced from their own MIR genes, which are mainly located in intergenic regions and transcribed into single-stranded miRNA precursors with imperfectly self-complementary hairpin structures. Many miRNAs are conserved in sequences and functions across plant species. In contrast, siRNAs could be derived from transgenes, viruses, transposons, aberrant RNAs as well as non-coding RNAs (Allen and Howell, 2010; Fei et al., 2013; Wu et al., 2020). In general, siRNAs are not characterized by unique sequences or target genes. However, there is a class of siRNAs that are unique in that they target a small number of targets in trans and is thus termed trans-acting siRNAs (ta-siRNAs) (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Axtell et al., 2006).
To date, eight TRANS-ACTING SIRNA (TAS) loci belonging to four families (TAS1–TAS4) have been shown to generate ta-siRNAs in Arabidopsis thaliana (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa, 2005; Axtell et al., 2006; Howell et al., 2007), but only the TRANS-ACTING SIRNA3 (TAS3) family is present in monocots (Williams et al., 2005; Heisel et al., 2008). ta-siRNA biogenesis is initiated by miRNA-triggered cleavage of noncoding TAS transcripts (Fei et al., 2013; Liu et al., 2020). Then the cleaved TAS transcripts are bound by the RNA-binding protein SUPPRESSOR OF GENE SILENCING 3 (SGS3) and converted into double-stranded RNAs (dsRNAs) through RNA-dependent RNA polymerase 6 (RDR6), and the dsRNAs are further processed by DICER-LIKE (DCL) proteins to generate a cluster of phased siRNAs (phasiRNAs) of 21- or 24 nt in length (Chen, 2009; Zhang et al., 2019). DCL4 is the major producer of 21-nt phasiRNAs, among which ta-siRNAs are a special subgroup that acts in trans (Peragine et al., 2004; Vazquez et al., 2004; Xie et al., 2005; Yoshikawa et al., 2005).
There are two modes of ta-siRNA biogenesis, known as the “one-hit” or “two-hit” mechanism (Fei et al., 2013; Zhang et al., 2019). In the one-hit model, TAS RNA precursors, such as TAS1, TAS2, and TAS4, only have a single miRNA binding site, and the cleavage at the site triggers the production of phasiRNAs from the fragment 3′ to (or downstream of) the target site (Allen et al., 2005; Fei et al., 2013; Liu et al., 2020). However, TAS3 is different, as its transcript bears two target sites of miR390, generating ta-siRNAs via the so-called two-hit mechanism (Axtell et al., 2006; Howell et al., 2007). Briefly, the miR390–Argonaute7 (AGO7) complex cleaves TAS3 at the 3′-site, but not at the 5′-site in Arabidopsis due to the presence of a central mismatch (Axtell et al., 2006; Howell et al., 2007; Montgomery et al., 2008; Allen and Howell, 2010). SiRNAs are produced in a 21-nt register from the 3′-miR390 cleavage site and are defined as “D1, D2, D3…,” with the orientation being indicated by the suffix “+” for the positive (the original transcript) strand, or “–” for the negative (the RDR-synthesized) strand (Allen et al., 2005; Allen and Howell, 2010; De Felippes et al., 2017).
The most well-studied TAS3 siRNAs are tasiR-ARFs, which are widely conserved across plant species and target several ARF genes (Allen et al., 2005; Williams et al., 2005). The miR390/tasiR-ARFs module regulates many developmental processes, such as lateral root growth, leaf patterning, and developmental timing in Arabidopsis, rice, maize, and even moss (Adenot et al., 2006; Fahlgren et al., 2006; Hunter et al., 2006; Marin et al., 2010; Cho et al., 2012; Dotto et al., 2014; Hobecker et al., 2017; Ding et al., 2020), indicating a highly conserved regulatory role of the tasiR-ARFs pathway (Shen et al., 2009; De Felippes et al., 2017; Xia et al., 2017). By integrating auxin signaling, the miR390/tasiR-ARFs module also regulates plant responses to environmental stresses, such as lateral root growth under salt stress in poplar (He et al., 2018), Helianthus tuberosus (Wen et al., 2020), and cotton (Yin et al., 2017), and nodule development in legumes (Hobecker et al., 2017). However, it still remains unknown whether tasiR-ARFs have non-ARF targets, or whether non-tasiR-ARF TAS3 siRNAs have targets or biological functions. Although tasiR-ARF biogenesis is highly conserved across angiosperms (Xia et al., 2017), it is striking that loss-of-function mir390, ago7, rdr6, and dcl4 mutants in rice and maize show much more severe developmental defects than those in Arabidopsis, some even fail to initiate the shoot apical meristem during embryogenesis (Itoh et al., 2000; Peragine et al., 2004; Xie et al., 2005; Nagasaki et al., 2007; Cuperus et al., 2010; Bi et al., 2020). The weak leaf patterning phenotypes of the tasiR-ARF biogenesis pathway mutants in Arabidopsis are in part due to genetic redundancy (Husbands et al., 2015). In addition to plant development, miR390 was found to act in response to multiple stresses, including cold, oxygen, salt, ultraviolet, and cadmium, etc. in rice, Arabidopsis, Medicago truncatula, and so on (Liu and Zhang, 2012; Zhou et al., 2012a, 2012b; Ding et al., 2016; Dmitriev et al., 2017; Yin et al., 2017; Lu et al., 2018; Hou et al., 2019). To date, it remains an open question as to what underlies the unusual importance of TAS3 siRNAs in monocot meristem development and stress responses. There could be two scenarios. First, the TAS3-tasiR-ARFs pathway may play a wider role in monocots than that in Arabidopsis. Lack of genetic redundancy could be part of this scenario. Second, the TAS3 loci may produce additional non-tasiR-ARF siRNAs that would target genes other than ARFs in monocots. In fact, some additional siRNAs (non-tasiR-ARFs) from TAS3 loci have been reported in Cassava and longan (Xia et al., 2014; Lin et al., 2015). Although these are not monocots, the findings suggest that the second scenario is possible. The functions of the siRNAs are as yet unknown.
In this study, we systemically investigated the origin, evolution, target genes, and potential biological functions of TAS3 siRNAs in rice (Oryza sativa, cv Nipponbare). We show that tasiR-ARFs-mediated target cleavage only occurs on polysomes in rice by analyzing rice Parallel Analysis of RNA Ends (PARE) datasets. Besides the conserved targets ARF2/3/14/15, we identified two additional targets of tasiR-ARFs involved in heavy metal responses. In addition, we found more TAS3-derived phased siRNAs that may also act in trans to direct cleavage of dozens of target transcripts, particularly on polysomes. Some of the identified targets were confirmed via expression analysis in mutants at three TAS3 loci created via clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) genome editing. Many of these targets are involved in plant growth and stress responses, implying that TAS3 siRNAs might be important for plants to adapt to their environment.
Results
A large pool of phasiRNAs is produced from TAS3 loci in rice
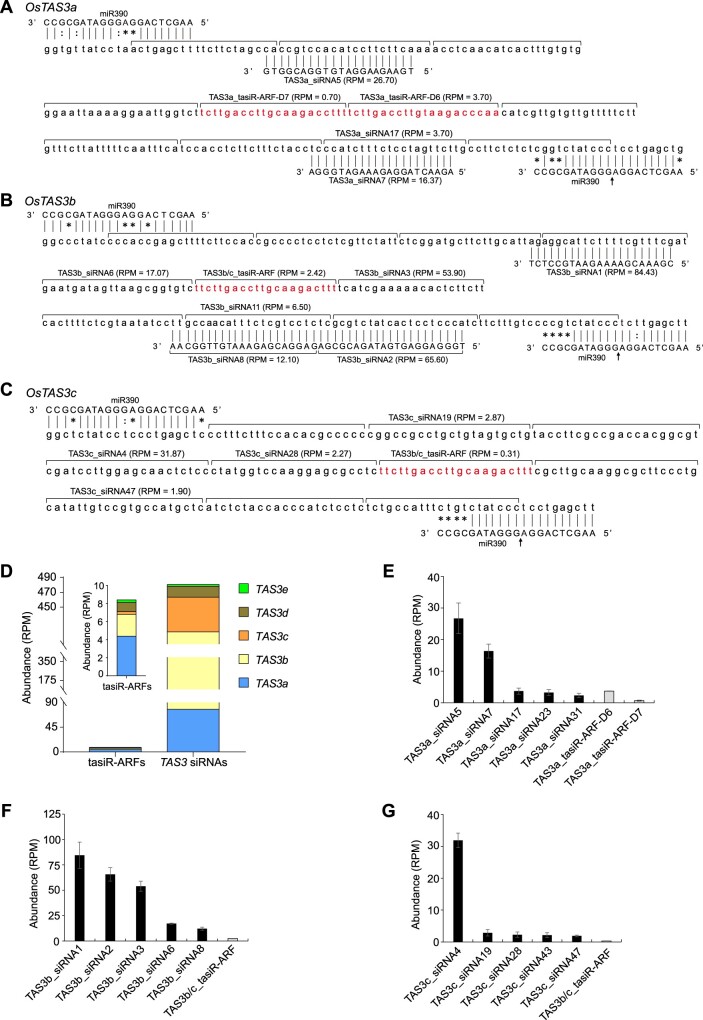
Based on the analyses of base-pairing with miR390 and the presence of both 5′- and 3′-target sites, five TAS3 loci have been identified in rice, OsTAS3a on chromosome 3, OsTAS3b on chromosome 2, OsTAS3c on chromosome 4, OsTAS3d on chromosome 1, and OsTAS3e on chromosome 5 (Supplemental Table S1; Lu et al., 2008; Allen and Howell, 2010; Wang et al., 2010b; Song et al., 2012). tasiR-ARFs produced from these five loci vary. First, the number of tasiR-ARFs is different: OsTAS3a, OsTAS3e, and OsTAS3d all give rise to two tasiR-ARFs adjacent to each other, while TAS3b and TAS3c generate a single tasiR-ARF with identical sequence. Second, the positions from which tasiR-ARFs are generated differ, with the positions being D6 (+) and D7 (+) for OsTAS3a, D6 (+) for OsTAS3b, D5 (+) for OsTAS3c, D7 (+) and D8 (+) for both OsTAS3d and OsTAS3e (Figure 1, A–C; Supplemental Figure S1, A and S1, B). Compared with those from Arabidopsis, tasiR-ARFs produced by OsTAS3a/b/c/d/e showed 1–6 nucleotide mismatches, suggesting a high degree of sequence conservation between rice and Arabidopsis (Supplemental Figure S1C).
Figure 1.
Sequences of TAS3 genes and abundance of TAS3 siRNAs in rice. A–C, Sequences of TAS3a (A), TAS3b (B), and TAS3c (C) transcripts between the two miR390 recognition sites. The phased siRNAs are indicated by brackets, and the register of the phased siRNAs is determined by cleavage (arrow) at the 3′-miR390 recognition site. The sequences of rice tasiR-ARFs, which are adopted from tasiRNAdb (https://bioinfo.jit.edu.cn/tasiRNADatabase), are indicated in red. The siRNAs with high abundance are labeled, and their mean abundance in the small RNA libraries is displayed in parentheses. D, Cumulative abundance of tasiR-ARFs and TAS3 siRNAs from the five TAS3 loci in rice. “RPM” is short for “reads per million mapped reads.” E–G, Abundance of tasiR-ARFs and top-ranked non-tasiR-ARF TAS3 siRNAs generated from TAS3a (E), TAS3b (F), and TAS3c (G) transcripts in rice. Each analysis includes three independent replicates. The abundance of siRNAs is displayed as mean ± standard deviation.
To profile TAS3 siRNAs, we sequenced small RNA libraries from three biological replicates of 1-week-old wild-type (WT) rice (Supplemental Table S2). Over 2.8 million genome-matched sequences for each library were obtained (Supplemental Table S3). Moreover, the biological repeats were highly reproducible (Supplemental Figure S1D). After normalization, siRNAs with the length of 18- to 24-nt were mapped to the TAS3 loci. The largest proportion of the identified siRNAs was 21-nt in length (35.5%), with 22-nt, 20-nt, and 19-nt classes accounting for 21.9%, 13.2%, and 11.0%, respectively, of the total TAS3 siRNA pool (Supplemental Table S4). For the identification of TAS3 siRNAs, we set the cutoff to reads per million mapped (RPM) greater than or equal to 0.1 in at least two samples out of all samples (including WT and three tas3 mutants, each with three biological replicates), and a total of 674 21-nt siRNAs were retained and named according to their locus of origin and abundance ranking. For example, the siRNA with the highest abundance was from TAS3b and named TAS3b_siRNA1, and the fourth abundant siRNA was from TAS3c and named TAS3c_siRNA4 (Supplemental Table S5). Most tasiR-ARFs were from TAS3a and TAS3b, comprising 52.4% and 32.5%, respectively, of the total tasiR-ARF pool. The remaining 15.1% were from other loci, including TAS3d tasiR-ARF-D7 (10.3%), TAS3d tasiR-ARF-D8 (1.2%), TAS3e tasiR-ARF-D7 (2.4%), and TAS3e tasiR-ARF-D8 (1.2%) (Figure 1D). Surprisingly, the abundance of tasiR-ARFs was extremely low, constituting only about 2% of all 21-nt siRNAs from TAS3 loci (Figure 1D). In contrast, the abundance of non-tasiR-ARF siRNA species was overwhelmingly higher than that of the tasiR-ARFs from the same locus (Figure 1, E–G; Supplemental Figure S1, E–K).
The average abundance (RPM) of tasiR-ARFs is about 1, therefore, we set RPM = 1 as the cutoff to define high and low abundance TAS3 siRNAs. TAS3 siRNAs with RPM greater than or equal to 1 were classified as high abundance siRNAs and those with RPM below 1 were classified as low abundance siRNAs; for example, TAS3b_siRNA70 is the top-ranked low abundance siRNA (Supplemental Table S5). Importantly, these TAS3 siRNAs were arranged in phase and possessed the 2-nt 3′-overhang in the small RNA duplexes. TAS3 siRNAs from the positive strand were almost in perfect phase to the 3′-miR390 site, suggesting that these phased siRNAs were produced together with tasiR-ARFs (Figure 1, A–C; Supplemental Figure S1, A and B). But there are a few exceptions, which may be caused by imprecise processing or trimming after processing. We analyzed the source of the 21-nt TAS3 siRNAs and found that 70.5% were derived from TAS3b, 16.2% from TAS3a, 9.7% from TAS3c, and 3.6% from TAS3e and TAS3d (Figure 1D). Thus, most TAS3 siRNAs including tasiR-ARFs and non-tasiARF siRNAs were from TAS3a and TAS3b. Since siRNAs from TAS3a/b/c accounted for >95% of the total TAS3 siRNA pool, we focused on the characterization of these siRNAs hereafter. Considering their large amount, we were curious about the evolutionary conservation and molecular functions, particularly of the high abundance TAS3 siRNAs.
TAS3 siRNAs are conserved in grasses
The miR390-TAS3-ARF pathway is a highly conserved and functionally important regulatory circuit in land plants (Allen et al., 2005; Xia et al., 2017), although TAS3 genes only comprise a small gene family in plants such as one TAS3 member in Marchantia polymorpha, three in Arabidopsis, and five in rice (Xia et al., 2017). Using plant genome information deposited in national center for biotechnology information (NCBI) and plaBiPD dataBase (plaBiPD), we analyzed the evolution of TAS3 genes in monocot and dicot plants.
In total, we identified 289 TAS3 genes, including 26 TAS3 genes from 10 dicots and 263 TAS3 genes from 50 monocots (Supplemental Table S6). Numerous studies have shown that TAS3 sequences are highly conserved in the miR390 recognition sites and tasiR-ARF sequences (Allen et al., 2005; Axtell et al., 2006; Krasnikova et al., 2013; Xia et al., 2017). We were wondering whether the regions generating non-tasiR-ARF TAS3 siRNAs, especially the ones of high abundance, were also conserved. As rice is a monocot, we first investigated the phylogenetic conservation of TAS3 genes among 14 monocots (Supplemental Figure S2A). Phylogenetic analysis revealed that these predicted TAS3 genes belonged to one of two clades, one containing rice TAS3a, TAS3d, and TAS3e and the other containing rice TAS3b and TAS3c. Then, TAS3 sequences from each clade were selected for sequence alignment. The results showed that only the sequences of tasiR-ARFs and the recognition sites of miR390 were highly conserved and this was true for the group of TAS3a, TAS3d, and TAS3e as well as that of TAS3b and TAS3c (Supplemental Figure S2B). The sequence conservation analysis of TAS3 genes of monocots was in accordance with previous studies (Allen et al., 2005; Axtell et al., 2006; Finet et al., 2013; Krasnikova et al., 2013; Lin et al., 2015; Xia et al., 2017).
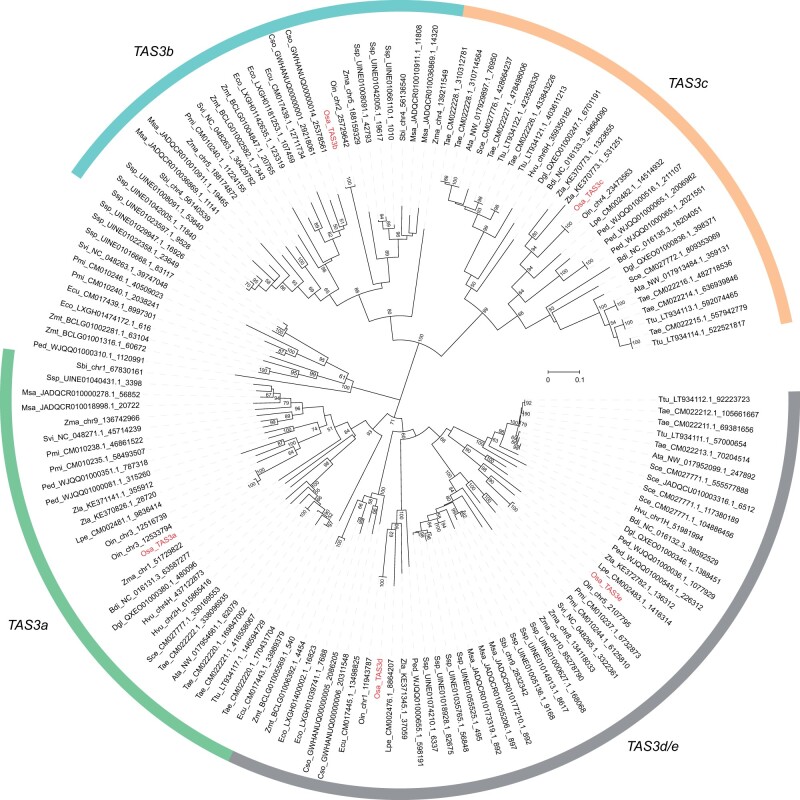
To investigate whether non-tasiR-ARF TAS3 siRNAs are conserved in more closely related species, we analyzed TAS3 genes in grasses. Totally, 141 predicted TAS3 genes were obtained from 21 grass species. Phylogenetic analysis showed that these TAS3 genes belonged to five separate clades, each represented by one of the five OsTAS3 genes. Therefore, we assigned names of the TAS3 genes in the same clade according to the corresponding rice TAS3 genes. Like the phylogeny in monocots, TAS3a/d/e clades were grouped into a single large clade, while TAS3b and TAS3c clades were grouped into another large clade (Figure 2).
Figure 2.
Phylogenetic analysis of TAS3 genes in the grass family. The neighbor-joining tree was generated by MEGA with 1,000 bootstraps, and the supporting values higher than 50% are indicated at the corresponding nodes. The scale bar represents nucleotide substitution rates. Rice TAS3 genes are labeled in red in the phylogenetic tree, and used to classify all grass TAS3 genes into five clades: TAS3a, TAS3b, TAS3c, TAS3d/e, and unclassified genes. The species used in the phylogenetic tree are Aegilops tauschii (Ata), Brachypodium distachyon (Bdi), Cleistogenes songorica (Cso), Dactylis glomerata (Dgl), Eleusine coracana (Eco), Eragrostis curvula (Ecu), Leersia perrieri (Lpe), Miscanthus sacchariflorus (Msa), Panicum miliaceum (Pmi), Phyllostachys edulis (Ped), Saccharum spontaneum (Ssp), Secale cereale (Sce), Setaria viridis (Svi), Triticum aestivum (Tae), Triticum turgidum (Ttu), Zizania latifolia (Zla), Zoysia matrella (Zmt), Oryza sativa (Osa), Oryza indica (Oin), Sorghum bicolor (Sbi), Zea mays (Zma), and Hordeum vulgare (Hvu).
Alignments of TAS3 sequences within the same clade showed that, in addition to the known sites (tasiR-ARFs and miR390 recognition sites), some siRNAs also showed sequence conservation, for instance, TAS3a_siRNA5, TAS3a_siRNA7, and TAS3a_siRNA108 from TAS3a; TAS3b_siRNA2, TAS3b_siRNA3, and TAS3b_siRNA44 from TAS3b; as well as TAS3c_siRNA135, TAS3c_siRNA175, and TAS3c_siRNA281 from TAS3c (Supplemental Figure S2C). Besides, two siRNAs, TAS3d_siRNA194 and TAS3e_siRNA523 from TAS3d and TAS3e, were also conserved (Supplemental Figure S2C). Furthermore, we performed phylogenetic analysis and sequence alignments for 69 predicted TAS3 sequences from 15 Oryza species. The results indicated that the presumed TAS3 genes were divided into five clades, which we named according to the rice genes (Lu et al., 2008; Zhu et al., 2008; Song et al., 2012; Supplemental Figure S2D). The sequence alignments showed that TAS3 sequences were highly conserved in Oryzoideae (Supplemental Figure S2E). Based on the above results, it can be concluded that some TAS3 siRNAs were conserved in grasses. The conservation of TAS3 siRNAs in Gramineae implies functional relevance.
TAS3 siRNAs are associated with polysomes
In plants, miRNAs and siRNAs repress target gene expression through either transcript cleavage or translational repression at the post-transcriptional level, or through DNA methylation at the transcriptional level (Chen, 2009; Rogers and Chen, 2013; Baulcombe, 2004; Fang and Qi, 2016; Yu et al., 2017; Jiang et al., 2020). An mRNA undergoing translation usually associates with multiple ribosomes to form polyribosome complexes (polysomes), and the number of ribosomes per transcript reflects the efficiency of translation (Kawaguchi et al., 2004; Wang et al., 2019b). Generally, the polysome association of miRNAs or AGO proteins may reflect the translational repression activity of miRNAs. In fact, miRNAs have been shown to guide the cleavage of target mRNAs on polysomes in Arabidopsis (Li et al., 2016), rice and maize (Yang et al., 2021). Therefore, we sought to determine whether TAS3 siRNAs are associated with polysomes.
We determined whether TAS3-derived siRNAs are associated with polysomes by analyzing published rice small RNA sequencing libraries from total extracts (Total), total polysomes (TPs), and membrane-bound polysomes (MBP) (Yang et al., 2021). Interestingly, TAS3 siRNAs were found not only in total extracts but also in the polysome fractions (Supplemental Figure S3, A–E). Enrichment in the MBP fraction was noted for siRNAs from TAS3a, TAS3b, and TAS3e (Supplemental Figure S3, A, B and E). However, the abundance of siRNAs from TAS3c and TAS3d in total extracts was significantly higher than that in MBP and TP (Supplemental Figure S3, C and D). We then analyzed the distribution of high abundance siRNAs and tasiR-ARFs (Figure 3, A–C; Supplemental Figure S3, F–L and Supplemental Table S7) in these three fractions. As expected, the subcellular localization of the siRNAs varied. For example, TAS3a_siRNA5 and TAS3a_siRNA31 were significantly enriched in TP and MBP (Figure 3A), whereas TAS3b_siRNA1 and TAS3c_siRNA4 were greatly enriched in Total (Figure 3, B and C). Similarly, tasiR-ARFs were found in all three fractions, but the abundance was significantly lower than that of the highly abundance siRNAs (Figure 3, A–C; Supplemental Figure S3, F–L and Supplemental Table S7). These results suggested that some TAS3 siRNAs were associated with polysomes and may mediate translational repression.
Figure 3.
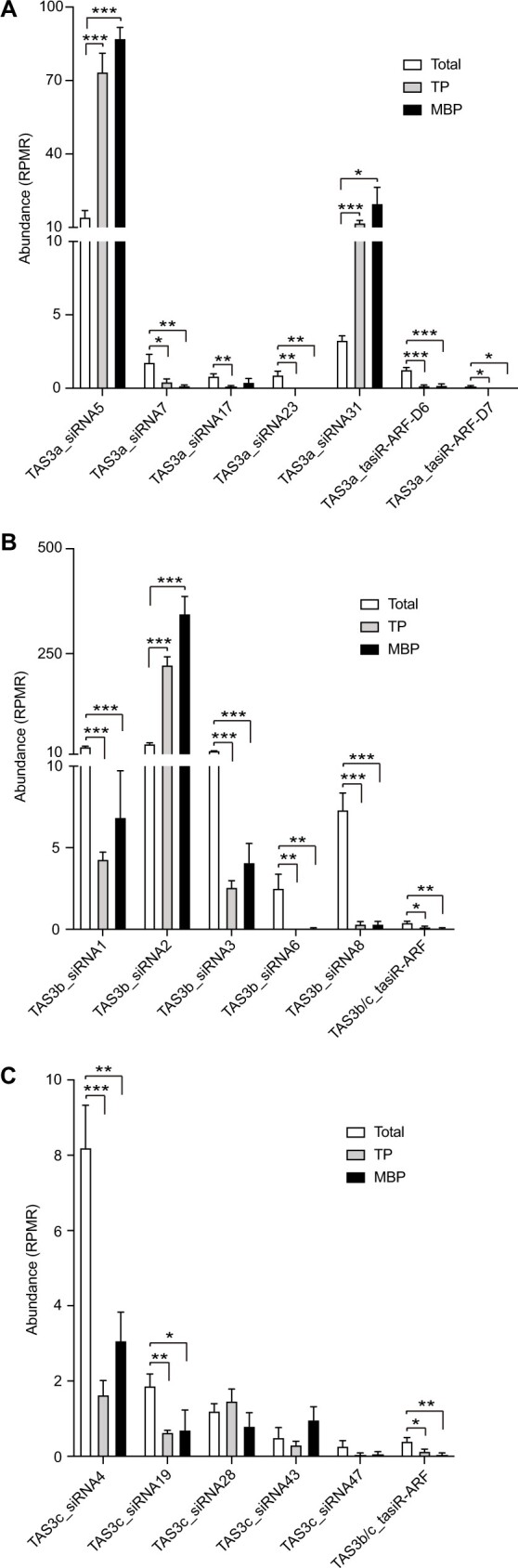
TAS3 siRNAs are associated with polysomes in rice. A–C, Abundance of tasiR-ARFs and top-ranking, highly abundant siRNAs from TAS3a (A), TAS3b (B), TAS3c (C) transcripts in Total, TP, and MBP samples. In A–C, significant differences as compared to Total are denoted by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively, with the P-values being derived from unpaired two-tailed t test. siRNA abundance is displayed as mean ± standard deviation of three biological repeats. “RPMR” is short for “reads per million mapped rRNA fragments.”
tasiR-ARFs target ARFs and additional protein-coding genes
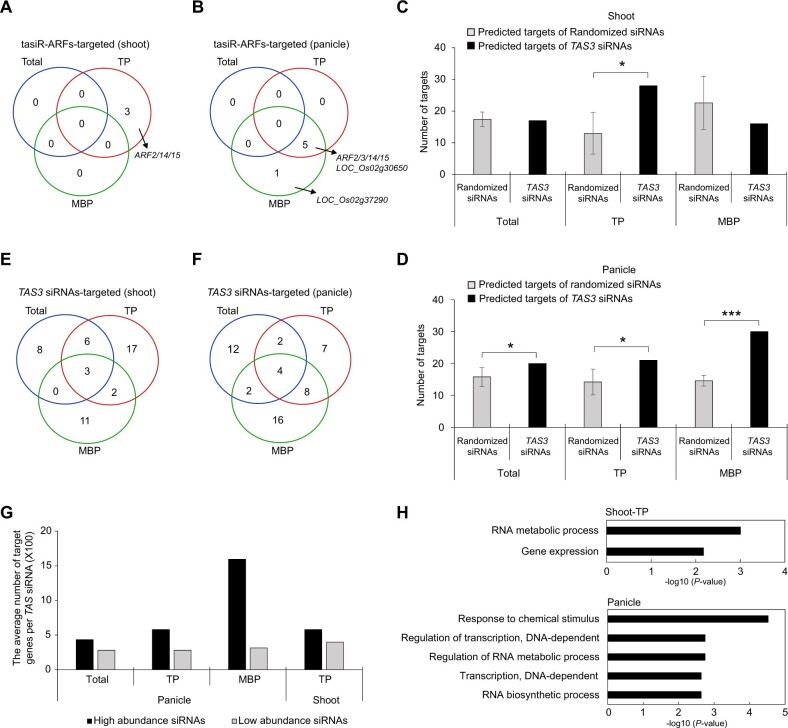
The ARFs are in a gene family ranging in numbers from 10 to more than 50 (Ha et al., 2013). Arabidopsis ARFs are divided into three clades, ARF5/6/7/8, ARF1/2/3/4/9, and ARF10/16/17, which can be traced back to the origin of land plants (Finet et al., 2013). In rice, ARF2/3/14/15 genes were identified as targets of tasiR-ARFs (Liu et al., 2007; Zhu et al., 2008; Wang et al., 2010a) because these four ARFs were of the highest sequence similarities (>80%) to Arabidopsis ARF3 and they harbor two tasiR-ARF recognition sites (Wang et al., 2007). Here, by analyzing our PARE libraries of Total, TP, and MBP fractions from rice seedling shoots and immature panicles (Yang et al., 2021) and retaining only the PARE hits present in at least two biological replicates, we identified a total of six target genes of tasiR-ARFs (Degradome category = 0; P ≤0.05; Figure 4, A and B; Supplemental Figure S4, A and B; Supplemental Table S8). ARF2/3/14/15 were among the identified targets. Moreover, the cleavage of target transcripts by tasiR-ARFs occurred only in TP and MBP, but not in Total (Figure 4, A and B; Supplemental Table S8). For instance, tasiR-ARF-targeted cleavage of ARF2 (LOC_Os01g48060) showed a better PARE signal in TP than those in Total and MBP (Supplemental Figure S4C).
Figure 4.
Identification of TAS3 siRNA target genes in rice. A and B, Numbers of tasiR-ARF target genes as determined by PARE analysis for input (Total), TP, and MBP samples from rice shoots (A) and panicles (B). C and D, Numbers of predicted targets of randomized siRNAs and TAS3 siRNAs in Total, TP, and MBP samples from shoots (C) and panicles (D). E and F, Numbers of TAS3 siRNA target genes as determined by PARE analysis for Total, TP, and MBP samples from rice shoots (E) and panicles (F). G, Summary for TAS3 siRNA targets in panicle Total, TP, and MBP and shoot TP samples. The black and gray rectangles represent target transcripts cleaved by TAS3 siRNAs with RPM ≥1 and TAS3 siRNAs with RPM <1, respectively. H, Enriched GO terms for TAS3 siRNA target genes in panicle Total, TP, and MBP and shoot TP samples as determined with the online tool agriGO v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/). The cutoff parameters for the identification of target genes were “degradomic category 0” and “P-value ≤0.05.” In (C and D), significant differences as compared to the randomized group are denoted by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively, with the P-values being derived from one-sample test (95% confidence interval). Number of target genes is displayed as mean ± standard deviation of five biological repeats.
Analysis of PARE datasets also uncovered non-ARF targets of tasiR-ARFs. tasiR-ARFs targeted two genes with heavy metal-related functions, LOC_Os02g30650 and LOC_Os02g37290 (Figure 4B; Supplemental Figure S4B and Supplemental Table S8), in rice immature panicles, indicating that tasiR-ARFs may play a role in heavy metal responses in rice panicles. For instance, LOC_Os02g37290, also named HPP03, belongs to the heavy metal-associated plant proteins (HPP) and the heavy metal-associated isoprenylated plant proteins gene family with roles in metal detoxification (Abreu‐Neto et al., 2013; Khan et al., 2019). Note that tasiR-ARFs from different TAS3 loci showed 1- to 6-nt differences (Supplemental Figure S1C). The tasiR-ARFs with the highest degree of complementarity to the targets were assigned as the regulatory tasiR-ARF in the PARE analysis. It is also noteworthy that PARE signals were not uniformly strong in the Total, TP, and MBP fractions. For example, the targeting of LOC_Os02g30650 by TAS3a_tasiARF-D6 was only obvious in the TP and MBP fractions (Supplemental Figure S4D). This might be why this gene was not previously identified as a tasiR-ARF target. In addition, for some targets, the PARE signals showed tissue-specific patterns, for instance, ARF2 (LOC_Os01g48060) was targeted by TAS3b/c tasiR-ARF in both TP and MBP fractions in rice panicles, but was only targeted in the TP fraction in rice shoots (Figure 4, A and B; Supplemental Figure S4, A and S4, C; Supplemental Table S8). These examples highlight the importance of observing small RNA–target relationships in polysome fractions.
Non-tasiR-ARF TAS3 siRNAs have potential targets in rice
We sought to determine whether non-tasiR-ARF siRNAs from TAS3 loci have endogenous targets. As non-tasiR-ARF siRNAs are numerous, it is possible that some siRNAs match a 5′-monophosphate RNA end in the PARE dataset by chance. To confidently identify targets of non-tasiR-ARF siRNAs, we implemented the following criteria. First, we only considered the best PARE hits (Degradome category = 0; P ≤0.05, as defined by the cleavage site being the only PARE signal along the transcript) as putative targets. Next, we only considered PARE hits that were present in at least two biological replicates. Finally, we generated five sets of randomized sequences from the input TAS3 non-tasiR-ARF siRNAs and used these as controls to represent the extent of fortuitous small RNA–mRNA pairing. The generation of random siRNA sequences was based on the original sequences of TAS3 siRNAs. Nucleotides from TAS3 siRNAs at each position of the 21-nt sequence were mixed and used for the generation of nucleotides at the corresponding position of randomized siRNAs (Supplemental Figure S4E). A total of five sets of randomized sequences were generated and used as controls in the PARE analysis. All the randomized datasets and TAS3 siRNAs have the same nucleotide composition but totally different sequences. The PARE datasets from Total, TP, and MBP in shoots and panicles were separately analyzed. In Total and MBP samples from shoots, the numbers of predicted targets were similar between the 674 21-nt TAS3 siRNAs and randomized siRNAs. This suggested that most of the PARE signals unlikely represented true target cleavage by an siRNA, although the results cannot exclude the presence of a small number of authentic siRNA–target interactions. Interestingly, in panicle Total, TP, and MBP and shoot TP samples, significantly higher numbers of putative targets were found for TAS3 siRNAs than randomized siRNAs (Figure 4, C and D). This indicates that TAS3 siRNAs do have targets, particularly in TP and MBP fractions, although some of the predicted targets are also likely false positives.
Given these findings, we focused on putative targets identified from panicle TP and MBP samples and shoot TP samples, and the corresponding Total samples were included for comparison. Among the 61 genes targeted by 21-nt TAS3 siRNAs in shoots, there were 17 genes in Total, 28 in TP, and 16 in MBP (Figure 4E; Supplemental Table S9). For the 71 target genes in panicles, 20 genes were in Total, 21 in TP, and 30 in MBP (Figure 4F; Supplemental Table S9). Each fraction had its own specific target transcript cleavage events; moreover, the number of target genes in TP and MBP was higher than that in Total (Figure 4, E and F). To detect the relationship between siRNA abundance and the number of target genes, we classified the siRNAs with predicted targets based on their abundance and calculated the proportion of their predicted targets in each of the three samples. Figure 4G showed that siRNAs with high abundance had more target genes than low abundance siRNAs on average, especially in panicle MBP samples.
To understand the general role of 21-nt TAS3 siRNAs in rice, we performed Gene Ontology (GO) analysis on the 71 protein-coding genes targeted by TAS3 siRNAs in panicles and the 28 in shoot TP (Figure 4H; Supplemental Table S10). The most enriched GO terms of the target genes in panicles were associated with chemical stimulus responses. In comparison, target genes in shoot TP were most enriched in RNA metabolic processes. These identified targets of TAS3-derived siRNAs implicate TAS3 siRNAs in the regulation of not only plant development but also stress responses.
We next determined the subcellular distribution of the cleavage events guided by non-tasiR-ARF TAS3 siRNAs. Most cleavage events were found in polysome fractions, although some were also found in Total (Figure 4, E and F). LOC_Os09g38520, which encodes a DOMON domain protein, was cleaved by TAS3b_siRNA45 (Supplemental Figure S4F) in Total and TP of rice shoots, and the transcription factor II F (TFIIF) gene LOC_Os06g04580 was cleaved by TAS3b_siRNA34 in MBP of rice panicles (Supplemental Figure S4G). Together with the finding that tasiR-ARFs guide the cleavage of target RNAs in polysome fractions (Figure 4, A and B; Supplemental Table S8), these results indicated that TAS3-derived siRNAs act mainly on polysomes. This is consistent with previous findings that miRNA-mediated target cleavage was widely observed on MBPs, and a large proportion of these cleavage events was MBP-unique (Yang et al., 2021).
Generation and characterization of null mutants in TAS3a, TAS3b, and TAS3c
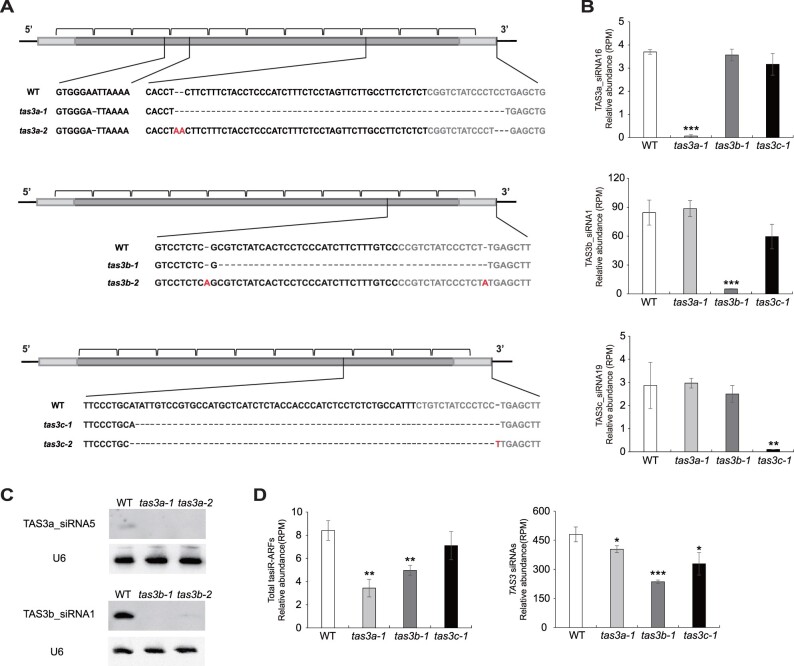
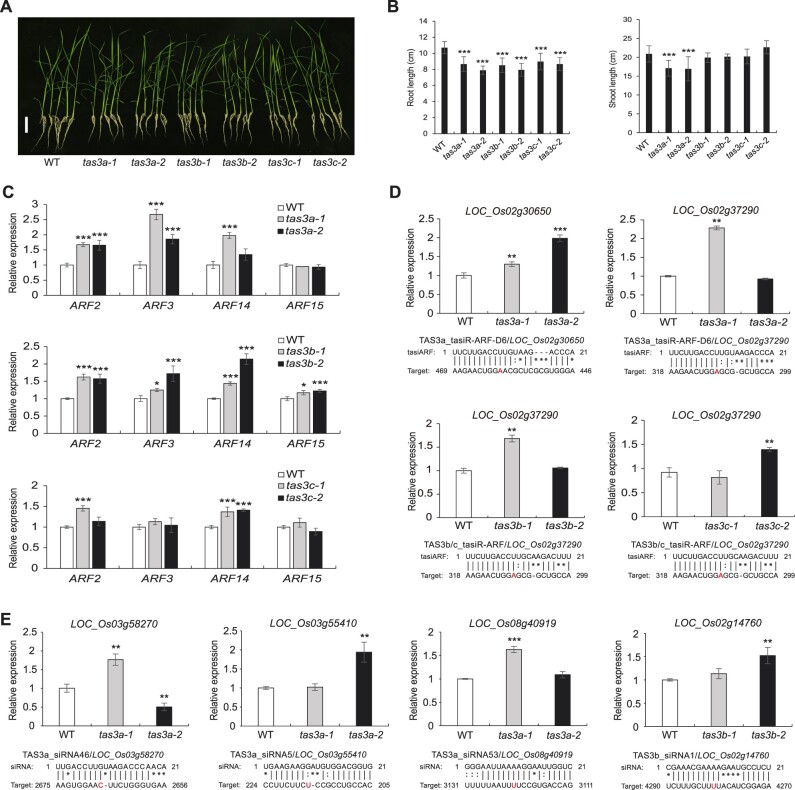
To investigate the function of TAS3 siRNAs and assess their redundancy and specificity during rice growth and development, we mutated the TAS3a-c loci by altering nucleotides within the 3′-miR390 target sites using double sgRNAs in the CRISPR/Cas9-based editing system. The three loci (TAS3a, TAS3b, and TAS3c) were selected for gene editing as they produced the majority of TAS3 siRNAs (Figure 1D). Details of mutations at each locus were as follows (Figure 5A): at the TAS3a locus, two mutants with a 60-bp deletion and a 3-bp deletion plus a 2-bp addition were designated as tas3a-1 and tas3a-2, respectively; at the TAS3b locus, two mutants with a 40-bp deletion and two individual point mutations were designated as tas3b-1 and tas3b-2, respectively; at the TAS3c locus, two mutants containing a 63-bp deletion and a 65-bp deletion were designated as tas3c-1 and tas3c-2, respectively.
Figure 5.
Generation of rice tas3a/b/c mutants by CRISPR/Cas9. A, Schematic illustration of the rice tas3a-1, tas3a-2, tas3b-1, tas3b-2, tas3c-1, and tas3c-2 mutations. The TAS3 transcripts are diagramed as rectangles with the miR390 binding sites indicated in light gray. The brackets indicate 21-nt phased siRNAs. The inserted and deleted sequences are labeled in red and represented by dotted lines, respectively. B, Abundance of three representatives TAS3 siRNAs in WT, tas3a-1, tas3b-1, and tas3c-1. Note that TAS3a_siRNA16, TAS3b_siRNA1, and TAS3c_siRNA19 are generated from TAS3a, TAS3b, and TAS3c, respectively. C, Northern blotting to detect TAS3a_siRNA5 in WT, tas3a-1 and tas3a-2, and TAS3b_siRNA1 in WT, tas3b-1 and tas3b-2. U6 was used as an internal control. D, Cumulative abundance of tasiR-ARFs (left side) and TAS3 siRNAs (right side) in WT, tas3a-1, tas3b-1, and tas3c-1. In (B) and (D), “RPM” is short for “reads per million mapped reads.” Significant differences as compared to WT are denoted by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively, with the P-values being derived from unpaired two-tailed t test. siRNA abundance is displayed as mean ± standard deviation of three biological repeats.
The tas3a-1, tas3b-1, and tas3c-1 mutants all contain truncations of the 3′-miR390 recognition site and harbor an intact 5′-miR390 binding site. Thus, these mutants offered a great opportunity to evaluate the importance of the 3′-site in ta-siRNA biogenesis. We performed small RNA sequencing with tas3a-1, tas3b-1, tas3c-1 mutants, and WT using 1-week-old shoots (Supplemental Table S2). The three biological replicates gave reproducible results (Supplemental Figure S1D). We first examined the abundance of siRNA species unique to each of the three loci. As shown in Figure 5B, the level of TAS3a_siRNA16 was dramatically reduced in tas3a-1 but was not affected in tas3b-1 or tas3c-1 (Figure 5B). Similarly, specific reduction of TAS3b_siRNA1 and TAS3c_siRNA19 were found in the corresponding tas3b-1 and tas3c-1 mutants (Figure 5B). Northern blotting was then performed to verify the accumulation of siRNAs in the mutants as determined by small RNA sequencing. TAS3a_siRNA5 was successfully detected in WT, although the band was very weak, but was not detectable in tas3a-1 and tas3a-2 (Figure 5C; Supplemental Figure S5A). Similarly, TAS3b_siRNA1 was present in WT but only showed very faint bands in tas3b-1 or tas3b-2 (Figure 5C; Supplemental Figure S5B). The abundance of siRNAs from TAS3c was too low to be detected by northern blotting. In summary, these results showed that the 3′-miR390 recognition site is crucial in the biogenesis of ta-siRNAs. In addition, the results also confirmed the production of highly abundant non-tasiR-ARF siRNAs from various TAS3 loci in a locus-specific manner.
Having shown that these tas3 mutants are likely null mutants, we went on to determine the contribution of these loci to the pool of tasiR-ARFs as well as non-tasiR-ARF siRNAs. The abundance (RPM) of total tasiR-ARFs was significantly decreased in tas3a-1 and tas3b-1 but not tas3c-1 (Figure 5D), suggesting that TAS3a and TAS3b are the main loci that produce tasiR-ARFs in seedlings. The abundance of total siRNAs was significantly reduced in all three tas3 mutants, with the reduction in tas3b-1 being the most severe (Figure 5D). Thus, TAS3a, TAS3b, and TAS3c all contribute to the TAS3 siRNA pool, with TAS3b being a major contributor. These results were also in concordance with the analyses of siRNA distribution among the five TAS3 loci (Figure 1D).
Gene regulatory and biological functions of TAS3 loci
Mutants with one of the three TAS3 loci mutated grew normally, except that root length in the two alleles of each of the three loci were shorter than WT, and shoot lengths were shorter in the two tas3a mutants (Figure 6, A and B). We analyzed the relative expression levels of ARF2/3/14/15 in dissected tissues enriched for the shoot apical meristem of WT and tas3 mutants by reverse transcription quantitative PCR (RT-qPCR). Compared with WT, the expression of ARF2 and ARF14 showed substantial upregulation in all six mutants, while that of ARF3 and ARF15 showed upregulation in some of the mutants (Figure 6C). This suggested that tasiR-ARFs from all three TAS3 loci contribute to the repression of the ARFs to some extent and that there is redundancy among the three TAS3 loci.
Figure 6.
Morphological and molecular phenotypes of rice tas3 mutants. A, Seven-day-old seedlings of WT and tas3 mutants. Scale bar = 5 cm. B, Statistical analyses of root length and plant height, the latter being defined as the distance from the shoot base to the top, of WT and different tas3 mutants. In (C–E), relative expression of various genes in WT and tas3 mutants was determined by RT-qPCR. UBQ5 was used as an internal control. Significant differences as compared to WT are denoted by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively, with the P-values being derived from unpaired two-tailed t test. The relative expression of target genes is displayed as mean ± standard deviation of three biological repeats. C, Relative expression of the tasiR-ARF targets ARF2/3/14/15. D, Relative expression of two tasiR-ARF target genes. E, Relative expression of four non-tasiR-ARF TAS3 siRNA target genes. In (D and E), the pairing between the siRNAs and the target transcripts is shown below the plots. The letters in red indicate the cleavage positions as revealed by PARE analysis.
Using these mutants, we next attempted to validate the two putative non-ARF target genes of tasiR-ARFs identified from PARE analysis. We examined the relative expression levels of the three genes in WT and the mutants by RT-qPCR. LOC_Os02g37290 encoding a heavy metal-associated domain containing protein, predicted to be targeted by tasiR-ARFs from TAS3a, TAS3b, and TAS3c, was significantly upregulated in tas3a-1, tas3b-1, and tas3c-2 (Figure 6D). Similarly, the expression of LOC_Os02g30650, which encodes a heavy metal-associated domain protein, was significantly upregulated in tas3a-1 and tas3a-2 (Figure 6D). These results suggested that the identified target genes of tasiR-ARFs are likely true targets in rice.
Finally, we sought to confirm the predicted targets of non-tasiR-ARF siRNAs using these mutants. We randomly selected several predicted target genes from the PARE data to examine the relative expression levels in WT and the corresponding mutants. For example, LOC_03g58270 was predicted to be a target of TAS3a_siRNA46, so its expression was examined in the tas3a mutants. The results showed that this gene was de-repressed in the tas3a-1 mutant (Figure 6E). Similarly, three other putative targets (LOC_Os08g40919, LOC_Os02g14760, and LOC_Os03g55410) were significantly upregulated in at least one of the mutants compared with WT (Figure 6E). However, this was not true for all tested putative targets, such as LOC_Os10g22960, LOC_03g08754, and LOC_08g35090 (Supplemental Figure S6, A–C). We performed a transient dual-luciferase (LUC) assay to further verify the effects of non-tasiR-ARF siRNAs on the expression of their potential targets. For the abovementioned four putative targets of non-tasiR-ARF siRNAs in Figure 6E, fragments containing the siRNA recognition sites were translationally fused with the LUC reporter and transformed into tas3 mutant or WT protoplasts. The results showed that the relative LUC activity, which represents the expression level of target genes, increased significantly in the tas3 protoplasts compared to WT protoplasts (Supplemental Figure S6, D and E). These results further confirmed the interactions between non-tasiR-ARF siRNAs and their targets in rice.
Discussion
In the present study, we systemically investigated the origin, evolution, target genes, and potential biological functions of TAS3-derived siRNAs in rice. Our results revealed that TAS3 loci in rice produce a large number of siRNAs with higher abundance than that of tasiR-ARFs and that the sequences of some of the non-tasiR-ARF siRNAs are conserved in grasses. In addition, we showed that tasiR-ARFs have non-ARF targets with potential roles in heavy metal responses, and that some non-tasiR-ARF siRNAs have endogenous targets.
Small RNA profiling in shoots showed that TAS3a and TAS3b are the predominant sources of TAS3-derived siRNAs in rice (Figure 1D). Phased siRNAs from TAS3 loci were composed of not only canonical tasiR-ARFs but also a large set of non-tasiR-ARF species, and the abundance of non-tasiR-ARF species was overwhelmingly greater than that of tasiR-ARFs (Figure 1D), though tasiR-ARFs are commonly considered as the only functional siRNAs in plants (Allen et al., 2005; Williams et al., 2005). This raised the question of whether the non-tasiR-ARF siRNAs regulate gene expression in rice. A first indication of their functionality is their polysome association. Our analysis of a set of published small RNA libraries prepared with rice Total, TP, and MBP samples showed that TAS3a and TAS3b siRNAs (including tasiR-ARFs and non-tasiR-ARF species) were present in TP and MBP fractions and were overrepresented in MBP in comparison to TP (Supplemental Figure S3, A–E). The MBP association of TAS3-derived siRNAs has functional implications as Arabidopsis siRNAs cause translational repression as well as phasiRNA biogenesis on polysomes, particularly MBP (Hou et al., 2016; Li et al., 2016; Bazin et al., 2017).
The polysome association of tasiR-ARFs and non-tasiR-ARF siRNAs from TAS3 prompted us to search for siRNA-guided target cleavage events in polysome fractions. This was done with PARE datasets prepared from Total, TP, and MBP fractions from rice seedling shoots and panicles (Yang et al., 2021). We observed tasiR-ARF-mediated cleavage events for ARFs in polysome fractions only (Figure 4, A and B), which demonstrated the higher sensitivity of detecting certain small RNA–target interactions in polysome fractions. In addition to the canonical target ARFs, other tasiR-ARF target genes were identified from the PARE analysis, including two heavy metal-related genes (LOC_Os02g37290 and LOC_Os02g30650). The four ARF genes and the two additional target genes were all de-repressed in tas3 mutants (Figure 6, C and D), which confirmed that they are targeted by tasiR-ARFs.
Intriguingly, this global search revealed a set of potential target genes for the non-tasiR-ARF siRNAs (Figure 4, E and F). PARE analysis determines the ends of RNAs with a 5′-monophosphate (P) and matches the ends with those that can be theoretically produced by small RNA-guided cleavage. Given the large number of TAS3-derived siRNAs, we were concerned that some siRNAs may be fortuitously matched to PARE signals although the 5′-P ends do not represent small RNA-guided cleavage events. To address this issue, we used sets of randomized sequences of the TAS3 siRNAs as controls in the PARE analysis. Indeed, we found that similar numbers of siRNA–target interactions were predicted in Total samples between randomized sets and the TAS3 siRNAs, indicating that most of the predicted interactions are not real. However, significantly higher numbers of siRNA–target interactions were found for TAS3 siRNAs as compared to the randomized controls in polysome fractions, suggesting that some TAS3 siRNAs do have targets and that they likely regulate targets on polysomes (Figure 4, C and D). A small number of the additional siRNA–target interactions were further confirmed in rice tas3 mutants, demonstrating the functionality of these noncanonical siRNAs from rice TAS3 loci (Figure 6E). The predicted target genes in panicles were mainly enriched in chemically stimulated regulatory pathways, and in shoot TP were mainly enriched in RNA metabolic process. The repression of target genes by TAS3 siRNAs could be a regulatory mechanism to enable plants to respond to environmental stresses.
Although we demonstrate that TAS3 siRNAs direct target cleavage, the biological roles of these siRNA-guided cleavage events remain to be studied. Being consistent with altered expression of target genes by tasiR-ARFs or non-tasiR-ARF species, we observed significantly shortened roots for tas3a, tas3b, and tas3c mutants as well as shorter shoots for tas3a seedlings (Figure 6, A and B). We speculate that the common phenotypes of the tas3 mutants are attributable to tasiR-ARFs, as ARF2/3/14/15 genes are generally de-repressed in the three tas3 mutants (Figure 6C). However, given the discovery that tasiR-ARFs have non-ARF targets and that non-tasiR-ARF siRNAs also have their own targets, the biological functions of the TAS3 loci are expected to be broader than that conferred by its regulation of ARFs. The tas3 mutants should be further examined to identify additional functions of these loci in the future.
Conclusion
In summary, we systemically investigated the origin, evolution, target genes, and potential biological functions of TAS3 siRNAs in rice. Small RNA sequencing results revealed that TAS3 loci produce a large set of non-tasiR-ARF siRNAs with much higher abundance than that of tasiR-ARFs. Both tasiR-ARFs and non-tasiR-ARF siRNAs were found to cleave targets on polysomes. Furthermore, some of these identified targets were experimentally confirmed by expression analysis in rice tas3 mutants created by CRISPR/Cas9 genome editing, demonstrating the functionality of tasiR-ARFs and non-tasiR-ARF siRNAs from TAS3 loci. These results expand our knowledge on rice TAS3 siRNA–target interactions and reveal a broader role of TAS3 genes in rice.
Materials and methods
Plant materials and growth conditions
WT rice (O. sativa, cv Nipponbare) or mutant rice created in the Nipponbare background was used in this study. Sterilized seeds were grown on 1/2 murashige and skoog (MS) medium (pH 5.7) with 3% (w/v) sucrose and 0.7% (w/v) agar in a growth chamber, which was set at 12 h/32°C for the light period, 12 h/28°C for the dark period, and a relative humidity of 70%. Shoot samples from 15 1-week-old seedlings were pooled together as one biological repeat, and totally three biological repeats were prepared.
CRISPR/Cas9 vector construction and rice transformation
To mutate rice TAS3a, TAS3b or TAS3c, two sgRNAs were designed and inserted into pCAMBIA1390-OsNLSCas9 in order to delete a region containing the 3′-miR390 binding site at each locus. After sequence verification of these constructs, rice transformation was carried out via an agrobacterium-mediated method at BioRun Co., Ltd. (Wuhan, China). At the T0 generation, the transformants were genotyped by PCR amplification of the TAS3 region followed by sequencing. Rice tas3 mutants, of which the 3′-miR390 target sites were truncated or altered, were selected for further analysis. Primers for construct preparation and plant genotyping are provided in Supplemental Table S11.
Construction of small RNA libraries
To construct small RNA libraries, 30 μg of total RNAs was extracted from seeding shoots using TRIzol® Reagent (Invitrogen, Waltham, MA, USA). After RNAs were resolved in a 15% (w/v) urea polyacrylamide gel electrophoresis (PAGE), gel slices containing 15–40 nt RNAs were recovered, smashed, and placed in 500 μL 0.4 M NaCl solution. The resulting mixture was agitated at 30 rpm at 4°C overnight followed by supernatant collection and small RNA recovery as described previously (Li et al., 2016). Small RNA libraries were constructed using NEB Next Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, E7300S) following the manufacturer’s instructions and sequenced on an Illumina HiSeq2500 platform with the SE50 sequencing strategy at Berry Genomics Co., Ltd. (Beijing, China).
Small RNA sequencing analysis
For raw data from small RNA sequencing libraries, removal of the 3′-adapter sequence (5′-AGATCGGAAGAGC-3′) and mapping of the remaining sequences against the “Nipponbare” V7 reference genome (http://rice.plantbiology.msu.edu/index.shtml) were performed by AUSPP (Gao et al., 2019) with the mode (-M smallRNA). Next, adaptor-trimmed reads that were aligned to each gene or miRNA were counted and normalized against total mapped reads separately for each size class ranging from 18 to 26 nt. The gene feature annotation information was obtained from Rice Genome Annotation Project 7 (http://rice.plantbiology.msu.edu/index.shtml) or miRBase release 22.1 (http://www.miRBase.org/). The normalized abundance of small RNAs for tas3 mutants was displayed as RPM (reads per million mapped reads), and statistically compared among different samples using edgeR (Robinson et al., 2010). On the other hand, the normalization of small RNAs for polysome-related analyses (Figure 3, A–C; Supplemental Figure 3, A–L) was performed by calculating the reads per million rRNA fragments value for each size class, and comparison was carried out using the R package DESeq2 (Love et al., 2014).
Northern blotting
Northern blotting was carried out for selected siRNAs according to the method described by Cai et al. (2018). After 10-min denaturation treatment, 20 µg of total RNAs were loaded into a 15% (w/v) TRIS BORATE-EDTA (TBE)–urea polyacrylamide gel and then resolved by electrophoresis at 150 V for ∼1 h. The resulting RNA samples were transferred from the gel to a Hybond NX membrane (Hybond-NX, GE Healthcare, Chicago, IL, USA) using the semi-dry method. The membrane was subjected to 90-min EDC [a1-ethyl-3-(3-dimethylaminopropyl) carbodiimide]-mediated chemical crosslinking at 65°C, and hybridized with biotin-labeled probes, which are complementary to the siRNAs or U6 (Supplemental Table S11), at 55°C overnight. After washes, the membrane was detected with the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Chemiluminescent signals were recorded with CheiScope 3300 Mini (CLINX, China). Three biological repeats were performed for each siRNA.
RT-qPCR
For RT-qPCR analysis, 25 μg of total RNAs was treated with DNase I (Invitrogen, Waltham, MA, USA), and then subjected to purification and overnight precipitation. First-strand complementary DNAs (cDNAs) were synthesized according to the manufacturer’s instructions (Invitrogen, Waltham, MA, USA). Using 5× diluted cDNAs as templates, RT-qPCR reactions were carried out in a CFX96 Real-time System (Bio-Rad, Hercules, CA, USA) with UBQ5 serving as an internal control. Data were generated with three biological replications, together with three technical replications for each biological repeat. Relative expression levels were determined with the 2−ΔΔCт method (Wang et al., 2019a). The primers used in RT-qPCR are listed in Supplemental Table S11.
Homologous gene identification
Plant genomic regions obtained from both NCBI (https://www.ncbi.nlm.nih.gov/) and plaBiPD databases (https://www.plabipd.de/) were annotated as candidate TAS3 loci by the following criteria: (1) region length ≤500 bp; (2) two miR390 binding sites with a 100- to 300-bp interval in between, and (3) generation of one or two tandem tasiR-ARFs with a 21-nt phasing register. These TAS3 genes were further assessed by manual sequence inspection.
Phylogenetic analysis
Phylogenetic analysis for TAS3 genes from various plant species was carried out using the neighbor-joining method with 1,000 bootstraps, and results were visualized with MEGA (Kumar et al., 2004). Information about sequences and accession numbers of the predicted TAS3 genes used in the phylogenetic analysis is provided in Supplemental Table S6.
Dual-LUC reporter assays
For dual-LUC reporter assays, approximately 60-bp fragments containing the target sites of putative target genes were inserted into pGreenII 0800-LUC in front of the firefly LUC to generate the pGreenII 0800-35S::target-LUC reporter constructs (Supplemental Figure S6D). The Renilla LUC under the control of the 35S promoter in pGreenII 0800-LUC was used as the internal control (Hellens et al., 2005). Rice protoplasts were isolated according to previously described methods (Bart et al., 2006). The pGreenII 0800-35S::target-LUC reporter constructs were transformed into tas3 mutant or WT protoplasts by using a PEG-mediated method (Zhang et al., 2011). After incubation in the dark for 12–16 h, LUC activities were measured by using the Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions. The relative ratio of firefly LUC to renilla LUC was calculated to represent the expression level of reporter genes. Primers used for plasmid construction are provided in Supplemental Table S11.
Statistical analysis
Each parameter was displayed as mean ± standard deviation of three independent samples, two-tailed Student’s t test and one-sample test (Figure 4, C and D) were performed for evaluating significance among different parameters at the confidence level *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively.
Data availability
The data reported in this paper have been deposited in NCBI Sequence Read Archive database (https://submit.ncbi.nlm.nih.gov/subs/sra/) with the accession no. GSE174453.
Accession numbers
Accession numbers of rice TAS3 genes are provided in Supplemental Table S1. Accession numbers of genes used for phylogenetic tree construction are provided in Supplemental Table S6.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence and abundance analysis for TAS3 genes and siRNAs in rice.
Supplemental Figure S2. Evolutionary analysis of TAS3 genes.
Supplemental Figure S3. TAS3 siRNAs are associated with polysomes in rice.
Supplemental Figure S4. Occurrence of cleavage events on target transcripts of 21-nt TAS3 siRNAs in different subcellular fractions of rice.
Supplemental Figure S5. Levels of TAS3a_siRNA5 and TAS3b_siRNA1 are reduced in tas3 and tas3b mutants, respectively.
Supplemental Figure S6. Expression analysis of putative TAS3 siRNA target genes in WT and tas3 mutants.
Supplemental Table S1. Chromosomal location of rice TAS3 loci.
Supplemental Table S2. General information for small RNA libraries constructed with WT rice and tas3 mutants.
Supplemental Table S3. The mapping information of small RNA libraries.
Supplemental Table S4. Proportion of TAS3 siRNAs of different lengths in small RNA library.
Supplemental Table S5. Sequence and abundance information of 21-nt TAS3 siRNAs used in this paper.
Supplemental Table S6. Number of TAS3 genes in plants with released genome information.
Supplemental Table S7 . Abundance of TAS3 siRNAs in Total, TP, and MBP fractions of shoots.
Supplemental Table S8. Predicted target genes of tasiR-ARFs.
Supplemental Table S9. Predicted target genes of non-tasiR-ARF TAS3 siRNAs.
Supplemental Table S10. GO terms for target genes of TAS3 siRNAs in rice shoots and panicles.
Supplemental Table S11. Probes and primers used in this study.
Supplementary Material
Acknowledgments
We thank Instrumental Analysis Center of Shenzhen University for excellent technical assistance.
X.C., L.L. (the last author), X.Y., and B.M. conceived the research. L.L.L. (the first author) and M.G. performed the experiments. L.L.L., T.L., X.Y., and L.G. analyzed the data. L.L.L., L.L., Y.Y., X.Y., and X.C. wrote the manuscript. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Lin Liu (linliu@szu.edu.cn).
Funding
This work was supported by National Key Research and Development Program of China (2019YFA0903900), Guangdong Innovation Research Team Fund (2014ZT05S078), Natural Science Foundation of Guangdong Province (2021A1515010482), and the Shenzhen Basic Research General Project (JCYJ20190808112207542).
Conflict of interest statement. The authors declare no competing interests.
References
- Abreu‐Neto JB, Turchetto‐Zolet AC, Oliveira LFV, Zanettini MHB, Margis‐Pinheiro M (2013) Heavy metal‐associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J 280: 1604–1616 [DOI] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Howell MD (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J (2017) Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA 114: E10018–E10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Fei Q, Li R, Liu B, Xia R, Char SN, Meyers BC, Yang B (2020) Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol J 18: 1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Liang C, Wang S, Hou Y, Gao L, Liu L, He W, Ma W, Mo B, Chen X (2018) The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat Commun 9: 5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SF, Coruh C, Axtell MJ (2012) miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC (2010) Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felippes FF, Marchais A, Sarazin A, Oberlin S, Voinnet O (2017) A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res 45: 5539–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Xia R, Lin Q, Gurung V, Sagawa JM, Stanley LE, Strobel M, Diggle PK, Meyers BC, Yuan YW (2020) Developmental genetics of corolla tube formation: role of the tasiRNA-ARF pathway and a conceptual model. Plant Cell 32: 3452–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ye Y, Jiang Z, Wang Y, Zhu C (2016) MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci 7: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev AA, Kudryavtseva AV, Bolsheva NL, Zyablitsin AV, Rozhmina TA, Kishlyan NV, Krasnov GS, Speranskaya AS, Krinitsina AA, Sadritdinova AF, et al. (2017) miR319, miR390, and miR393 are involved in aluminum response in flax (Linum usitatissimum L.). Biomed Res Int 2017: 4975146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto MC, Petsch KA, Aukerman MJ, Beatty M, Hammell M, Timmermans MC (2014) Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet 10: e1004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Fang X, Qi Y (2016) RNAi in plants: an argonaute-centered view. Plant Cell 28: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F (2013) Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol 30: 45–56 [DOI] [PubMed] [Google Scholar]
- Gao L, Wu C, Liu L (2019) AUSPP: a universal short-read pre-processing package. J Bioinform Comput Biol 17: 1950037. [DOI] [PubMed] [Google Scholar]
- Ha CV, Le DT, Nishiyama R, Watanabe Y, Sulieman S, Tran UT, Mochida K, Dong NV, Yamaguchi-Shinozaki K, Shinozaki K, et al. (2013) The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Res 20: 511–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Xu C, Fu X, Shen Y, Guo L, Leng M, Luo K (2018) The microRNA390/TRANS-ACTING SHORT INTERFERING RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol 177: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Liang WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel SE, Zhang Y, Allen E, Guo L, Reynolds TL, Yang X, Kovalic D, Roberts JK (2008) Characterization of unique small RNA populations from rice grain. PLoS One 3: e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobecker KV, Reynoso MA, Bustos-Sanmamed P, Wen J, Mysore KS, Crespi M, Blanco FA, Zanetti ME (2017) The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol 174: 2469–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Jiang F, Zheng X, Wu Z (2019) Identification and analysis of oxygen responsive microRNAs in the root of wild tomato (S. habrochaites). BMC Plant Biol 19: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou CY, Lee WC, Chou HC, Chen AP, Chou SJ, Chen HM (2016) Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-Nava M, Poethig SR (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands AY, Benkovics AH, Nogueira FTS, Lodha M, Timmermansa MCP (2015) The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27: 3321–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh JI, Kitano H, Matsuoka M, Nagato Y (2000) Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12: 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Lian B, Liu C, Fu Z, Shen Y, Cheng Z, Qi Y (2020) 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat Commun 11: 5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Khan IU, Rono JK, Zhang BQ, Liu XS, Wang MQ, Wang LL, Wu XC, Chen X, Cao HW, Yang ZM (2019) Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotox Environ Safe 175: 8–18 [DOI] [PubMed] [Google Scholar]
- Krasnikova MS, Goryunov DV, Troitsky AV, Solovyev AG, Ozerova LV, Morozov SY (2013) Peculiar evolutionary history of miR390-guided TAS3-like genes in land plants. Sci World J 2013: 924153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Li S, Le B, Ma X, Li S, You C, Yu Y, Zhang B, Liu L, Gao L, Shi T, et al. (2016) Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lin L, Lai R, Liu W, Chen Y, Zhang Z, Xu-Han X, Lai Z (2015) MicroRNA390-directed TAS3 cleavage leads to the production of tasiRNA-ARF3/4 during somatic embryogenesis in Dimocarpus longan Lour. Front Plant Sci 6: 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, Liu C, Cui X, Zhao X, Fang J, Xu W, Zhang H, Wang X, et al. (2007) Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang H (2012) Molecular identification and analysis of arsenite stress-responsive miRNAs in rice. J Agric Food Chem 60: 6524–6536 [DOI] [PubMed] [Google Scholar]
- Liu Y, Teng C, Xia R, Meyers BC (2020) Phased secondary small interfering RNAs (phasiRNAs) in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32: 3059–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Feng Z, Liu X, Bian L, Xie H, Zhang C, Mysore KS, Liang J (2018) MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol 18: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. (2008) Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. (2008) Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Köhler C (2017) Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr Opin Plant Biol 36: 22–28 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wang S, Chen H, Zhu QH, Helliwell C, Fan L (2009) Molecular phylogeny of miR390-guided trans-acting siRNA genes (TAS3) in the grass family. Plant Syst Evol 283: 125–132 [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, et al. (2012) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69: 462–474 [DOI] [PubMed] [Google Scholar]
- Tang J, Chu C (2017) MicroRNAs in crop improvement: fine-tuners for complex traits. Nat Plants 3: 17077. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Wang J, Gao X, Li L, Shi X, Zhang J, Shi Z (2010a) Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J Exp Bot 61: 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Quan L, Li S, You C, Zhang Y, Gao L, Zeng L, Liu L, Qi Y, Mo B, et al. (2019a) The PROTEIN PHOSPHATASE4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell 31: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li S, Zhao Y, You C, Le B, Gong Z, Mo B, Xia Y, Chen X (2019b) NAD(+)-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc Natl Acad Sci USA 116: 12094–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen D, Bo S, Chen H, Zheng J, Zhu QH, Cai D, Helliwell C, Fan L (2010b) Sequence variation and selection of small RNAs in domesticated rice. BMC Evol Biol 10: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen FL, Yue Y, He TF, Gao XM, Zhou ZS, Long XH (2020) Identification of miR390-TAS3-ARF pathway in response to salt stress in Helianthus tuberosus L. Gene 738: 144460. [DOI] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA 102: 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Li B, Iwakawa HO, Pan Y, Tang X, Ling-Hu Q, Liu Y, Sheng S, Feng L, Zhang H, et al. (2020) Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581: 89–93 [DOI] [PubMed] [Google Scholar]
- Xia J, Zeng C, Chen Z, Zhang K, Chen X, Zhou Y, Song S, Lu C, Yang R, Yang Z, et al. (2014) Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in Cassava. BMC Genomics 15: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Xu J, Meyers BC (2017) The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 29: 1232–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, You C, Wang X, Gao L, Mo B, Liu L, Chen X (2021) Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes. Genome Biol 22: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Han X, Li Y, Wang J, Wang D, Wang S, Fu X, Ye W (2017) Comparative analysis of cotton small RNAs and their target genes in response to salt stress. Genes 8: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jia T, Chen X (2017) The ‘how’ and ‘where’ of plant microRNAs. New Phytol 216: 1002–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su JB, Duan S, Ao Y, Dai JR, Liu J, Wang P, Li YG, Liu B, Feng DR, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Teotia S, Tang J, Tang G (2019) Perspectives on microRNAs and phased small interfering RNAs in maize (Zea mays L.): functions and big impact on agronomic traits enhancement. Plants 8: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZS, Song JB, Yang ZM (2012a) Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J Exp Bot 63: 4597–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2012b) Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35: 86–99 [DOI] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in NCBI Sequence Read Archive database (https://submit.ncbi.nlm.nih.gov/subs/sra/) with the accession no. GSE174453.
Accession numbers
Accession numbers of rice TAS3 genes are provided in Supplemental Table S1. Accession numbers of genes used for phylogenetic tree construction are provided in Supplemental Table S6.