Abstract
Highly efficient tissue repair is pivotal for surviving damage-associated stress. Plants generate callus upon injury to heal wound sites, yet regulatory mechanisms of tissue repair remain elusive. Here, we identified WUSCHEL-RELATED HOMEOBOX 13 (WOX13) as a key regulator of callus formation and organ adhesion in Arabidopsis (Arabidopsis thaliana). WOX13 belongs to an ancient subclade of the WOX family, and a previous study shows that WOX13 orthologs in the moss Physcomitrium patens (PpWOX13L) are involved in cellular reprogramming at wound sites. We found that the Arabidopsis wox13 mutant is totally defective in establishing organ reconnection upon grafting, suggesting that WOX13 is crucial for tissue repair in seed plants. WOX13 expression rapidly induced upon wounding, which was partly dependent on the activity of an AP2/ERF transcription factor, WOUND-INDUCED DEDIFFERENTIATION 1 (WIND1). WOX13 in turn directly upregulated WIND2 and WIND3 to further promote cellular reprogramming and organ regeneration. We also found that WOX13 orchestrates the transcriptional induction of cell wall-modifying enzyme genes, such as GLYCOSYL HYDROLASE 9Bs, PECTATE LYASE LIKEs and EXPANSINs. Furthermore, the chemical composition of cell wall monosaccharides was markedly different in the wox13 mutant. These data together suggest that WOX13 modifies cell wall properties, which may facilitate efficient callus formation and organ reconnection. Furthermore, we found that PpWOX13L complements the Arabidopsis wox13 mutant, suggesting that the molecular function of WOX13 is partly conserved between mosses and seed plants. This study provides key insights into the conservation and functional diversification of the WOX gene family during land plant evolution.
WUSCHEL-RELATED HOMEOBOX 13 transcriptionally induces WOUND-INDUCED DEDIFFERENTIATION genes and cell wall modifying enzyme genes that likely contribute to callus formation and organ reconnection.
Introduction
Living organisms are constantly challenged by stress arising from tissue damage, hence regenerative capacity is critical for surviving injuries. Plants can repair local tissue damage, as is observed upon amputation of root apical meristems (RAMs; Sena et al., 2009) or debarking of trees (Stobbe et al., 2002). The regenerative capacity of plants to reconnect vascular tissues at wound sites allows connections to form between scion and rootstock of different species. This procedure, called grafting, has been widely utilized in agriculture and horticulture for various purposes, such as for clonal propagation of elite cultivars or for the introduction of stress resistance to useful crops (Melnyk and Meyerowitz, 2015). Grafting progresses through multiple cellular processes, which initiate with the cohesion of assembled tissues (Jeffree and Yeoman, 1983). Cells facing the graft junction then initiate to divide and make a cell mass called callus. Callus cells experience morphological changes and undergo extensive cell expansion, which is postulated to facilitate physical contact between a scion and rootstock pair (Jeffree and Yeoman, 1983). The grafting process completes with vascular reconnection, where auxin flowing from the scion to the rootstock instructs vascular cell differentiation. Although callus formation is thought to play physiological roles during grafting, such as facilitating physical connection between the scion and rootstock and providing pluripotent cells which can differentiate into vascular cells, the exact role of callus formation in this process remains elusive.
The molecular basis of tissue repair and organ reconnection has mainly been studied using hypocotyl grafting (Melnyk et al., 2015, 2018) or tissue repair of incised floral stem (Asahina et al., 2011) in Arabidopsis (Arabidopsis thaliana). Auxin accumulation at the top part of the cut site induces the expression of a NAC family transcription factor, ANAC071, which in turn directly induces cell wall-modifying enzymes, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19 (XTH19) and XTH20 (Pitaksaringkarn et al., 2014). XTH19 and XTH20 are subsequently involved in reactivating cell proliferation important for successful graft formation (Pitaksaringkarn et al., 2014). Furthermore, ANAC071 and ANAC096 are also required for cellular reprogramming of parenchymal tissue to cambium, a pivotal step for vascular reformation (Matsuoka et al., 2021). Finally, canalization of auxin flow, which is mediated by canalization-related auxin-regulated malectin-type RLK (CAMEL) and canalization-related receptor-like kinase (CANAR) instructs vascular regeneration (Hajný et al., 2020). Another line of evidence underscores the importance of cell wall modification during grafting. Transcriptome analyses of inter-family grafting between Nicotiana benthamiana and Arabidopsis uncovered a large number of genes encoding cell wall-modifying enzymes that are transcriptionally induced upon grafting (Notaguchi et al., 2020; Kurotani et al. 2020). Among them, a cellulase gene in the GLYCOSYL HYDROLASE 9B (GH9B) clade plays pivotal roles in the establishment of grafting (Notaguchi et al., 2020). However, the gene regulatory networks underlying wound stress-induced upregulation of cell wall-modifying enzymes remain to be elucidated.
Recent efforts identified several key transcription factors regulating wound-induced cellular reprogramming and callus formation. The AP2/ERF class transcription factors WOUND-INDUCED DEDIFFERENTIATION 1–4 (WIND1–4) induce cellular de-differentiation leading to the formation of callus or somatic embryos when over-expressed in planta (Iwase et al., 2011; Ikeuchi et al., 2013). The expression of WIND1 is abruptly induced upon wounding, which in turn promotes callus formation and shoot regeneration via transcriptional upregulation of ENHANCER OF SHOOT REGENERATION 1 (ESR1; Iwase et al., 2017). Furthermore, WIND1 orchestrates various other developmental processes including vascular reconnection and the activation of defense responses upon injury (Iwase et al., 2021). Other AP2/ERFs, including PLETHORA 3 (PLT3), PLT5, PLT7, and ERF115 also contribute to callus formation at wound sites (Ikeuchi et al., 2017) and vascular regeneration (Radhakrishnan et al., 2020). Furthermore, a yeast one hybrid-based interactome analysis identified ESR1 and PLT3 as hub nodes of a gene regulatory network controlling cellular reprogramming (Ikeuchi et al., 2018). Apart from Arabidopsis, reprogramming of Physcomitrium patens leaf cells into protonema stem cells has proved to be a useful model system for identifying key genes regulating cellular reprogramming leading to stem cell re-establishment following injury. An AP2/ERF transcription factor named STEM CELL-INDUCING FACTOR 1 (STEMIN1) is functionally similar to Arabidopsis WIND1 in that STEMIN1 is sufficient to induce stem cell formation from somatic cells upon overexpression (Ishikawa et al. 2019). Another study using P. patens revealed that WUSCHEL-RELATED HOMEOBOX 13 (WOX13)-like genes regulate cellular reprogramming (Sakakibara et al., 2014). Wild-type (WT) leaf cells undergo proliferation and elongation during reprogramming into protonema stem cells, while the ppwox13ab loss-of-function mutant specifically displays a defect in cell elongation. They showed that PpWOX13L promotes cell elongation via transcriptional activation of genes encoding cell wall loosening proteins, including EXPANSINs (EXPs) and XTHs (Sakakibara et al., 2014). In Arabidopsis, WOX13 regulates replum formation during fruit development (Romera-Branchat et al., 2013), but it is reportedly not involved in the repair of the RAM in the repair of the RAM (Sakakibara et al., 2014). On the other hand, many other members of the WOX family have well-established roles regulating both the formation and maintenance of stem cells in seed plants. For instance, WUSCHEL (WUS) maintains homeostasis of the stem cell population in shoot apical meristems (SAMs; Mayer et al., 1998; Schoof et al., 2000). WUS is also indispensable for de novo establishment of SAM in tissue culture (Gordon et al., 2007; Zhang et al. 2017). Similarly, WOX5 is specifically expressed in the RAM quiescent center, where it maintains the neighboring population of columella stem cells (Sarkar et al., 2007; Pi et al., 2015) and WOX4 maintains stem cell activity in the cambium (Hirakawa et al., 2010). Additionally, WOX5, together with WOX7 and WOX14, mediate pluripotency acquisition of callus in tissue culture, which is required for shoot regeneration (Kim et al., 2018). Notably, the WUS subclade is specific to seed plants, while PpWOX13L is part of a different, ancient subclade that includes Arabidopsis WOX13 (Deveaux et al., 2008). Molecular function of WOX13 remains as the missing link in the evolution of key regulators of organ regeneration.
In this study, we identified WOX13 as a key player controlling callus formation and organ reconnection in Arabidopsis. We show that WOX13 and WIND transcription factors constitute a gene regulatory circuit that promotes cellular reprogramming. Furthermore, WOX13 directly upregulates a variety of genes encoding cell wall-modifying enzyme. Moreover, based on our finding that PpWOX13L complements the wox13 mutant in Arabidopsis, we propose that WOX13 is an evolutionarily conserved regulator of wound-induced cellular reprogramming in land plants.
Results
WOX13 promotes callus growth at wound sites
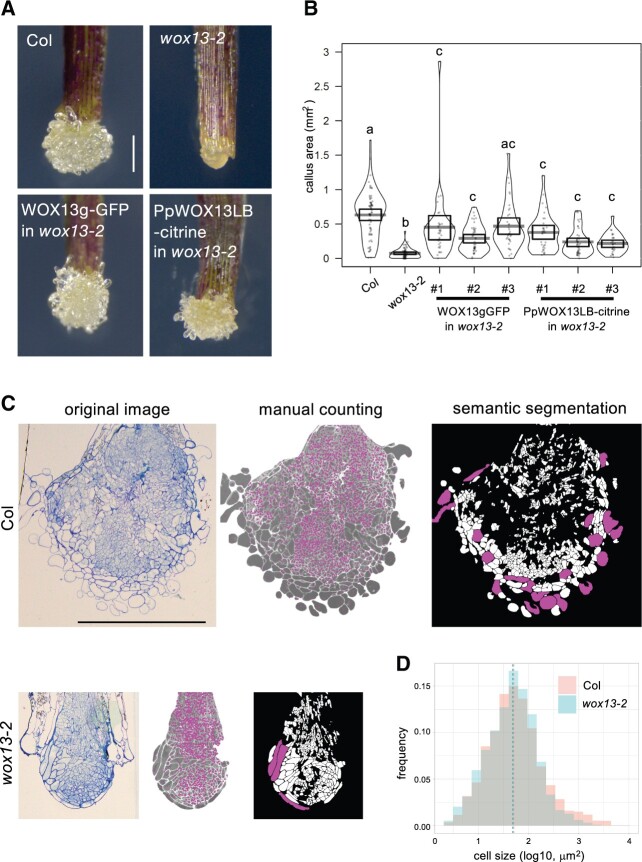
Our previous transcriptome analysis revealed that WOX13 expression is rapidly induced within 1 h after cutting hypocotyls (Supplemental Figure S1A; Ikeuchi et al., 2017). This observation prompted us to hypothesize that WOX13 may function in callus formation at wound sites. As a result, we found that a loss-of-function mutant wox13-2 (Romera-Branchat et al., 2013) displays significant reduction in callus formation efficiency in hypocotyl cutting assay at Day 4 (P = 2.713e-11, prop-test; Supplemental Figure S1B) and reduced subsequent callus growth by Day 14 (Supplemental Figure S1C). Furthermore, we found that the wox13 mutant displays even more severe defects in callus formation assays using leaf explants (Supplemental Figure 1, A and B). While WT explants form well-developed calli from petiole cut end, wox13 mutant explants generate compact calli with a significant reduction in the projection area (Figure 1B). In addition to the difference in callus size, the mutant calli display remarkable differences in tissue histology. Namely, WT generate friable calli where cells at the callus surface have loose inter-cellular adhesion. wox13 mutant calli, on the other hand, have a compact calli with smooth surface, which is implicative of tight intercellular connections (Figure 1A). To test whether the observed phenotype in wox13-2 is due to the loss of WOX13 function, we generated a translation fusion containing the WOX13 genomic fragment with GFP at the N terminus of the coding region (WOX13gGFP) and performed a complementation test. As shown in Figure 1, both the defect in callus growth and the histological feature observed in wox13-2 are complemented by expressing WOX13gGFP. Therefore, we conclude that WOX13 promotes callus formation at wound sites.
Figure 1.
WOX13 promotes callus growth at wound sites. A and B, Wound-induced callus formation assay using leaf petioles. Representative images of calli (A) and quantification of the callus projection area (B) 9 d after cutting of petioles. Gray bars in the pirateplot indicate mean value and boxes indicate 95% highest density intervals. Different letters indicate significant differences based on one-way analysis of variance (ANOVA) with Steel–Dwass test, P < 0.01. C, Semi-thin cross sections of calli, original image (left), after manual cell counting (middle), after X-net segmentation of cells (right). Magenta dots in the middle image indicate individual cells that are manually marked. White regions in the right image indicate individual cells that are automatically recognized by X-net and cells with areas larger than 1,000 µm2 are highlighted by magenta. n = 3. D, Histogram of the callus cell size. Dashed lines indicate median value. n = 3,435 (Col), n = 1,332 (wox13-2). Scale bars, 100 µm (A and C).
Given that WOX13 is known as an ancient member of the WOX family, we next investigated if the developmental function of Arabidopsis WOX13 is conserved with PpWOX13LB. We replaced the coding sequence of Arabidopsis WOX13 with PpWOX13LB-citrine in the construct containing the WOX13-genomic fragment and tested if the transgene complements the Arabidopsis wox13-2 mutant. Strikingly, PpWOX13LB partially complements the callus formation phenotype of wox13-2 with regards to both callus size and the histological features of the callus surface (Figure 1, A and B). This observation suggests that the molecular function and target genes of WOX13 orthologs are evolutionarily conserved between distantly related land plants.
WOX13 is required for the formation of highly vacuolated large cells in callus
To further identify developmental function of WOX13 in callus formation, we next asked if the reduction of callus size in wox13-2 is due to a reduction in cell number or cell size. We generated semi-thin sections at the median plane of calli and performed quantitative histological analyses. We manually counted all the cells included in the sections and found that WT calli have 2,483 cells (n = 3), whereas wox13 mutant calli have 945 cells (n = 3) for each section on average, implicating that cell proliferation is reduced in the wox13 mutant (Figure 1C). Given that the reduction in cell number in the wox13 mutant (38% relative to the WT) only accounts for the half portion of the reduction in callus area (22% relative to the WT), size of the individual cells should be affected as well. For cell size measurement, we sought to apply a computer vision approach to automatically detect cells on the microscopic images. We developed a deep-learning architecture “X-net”, which shows higher performance in cell segmentation than commonly used U-net (Fujii et al., 2021). In our X-net analysis, 46% of individual cells were identified after the elimination of incorrectly segmented cell cluster (Figure 1C). We measured the area of segmented cells and found that the median area of the cell was similar (50 µm2 in the WT; 48 µm2 in wox13). In contrast, the proportion of the large cells, whose areas are larger than 1,000 µm2, is significantly higher in the WT (5.5%) than wox13 mutant (2.3%; P = 7.264e-07, Fisher’s exact test). Hence, our data show that wox13 mutation specifically affects the formation of large cells rather than causing defects in cell expansion among all cells. (Figure 1D). We also noticed that the large cells found in WT calli are mainly located at around callus surface and they have cytological characteristics distinct from other callus cells. Smaller cells in the WT or wox13 and large cells in wox13 mutant have dense cytoplasm, whereas the large cells found in the WT are mostly occupied with central vacuoles (Figure 1C). Collectively, our histological analyses revealed that WOX13 promotes both cell proliferation and the formation of highly vacuolated large cells, which together contribute to callus growth.
WOX13 is required for the establishment of organ reconnection
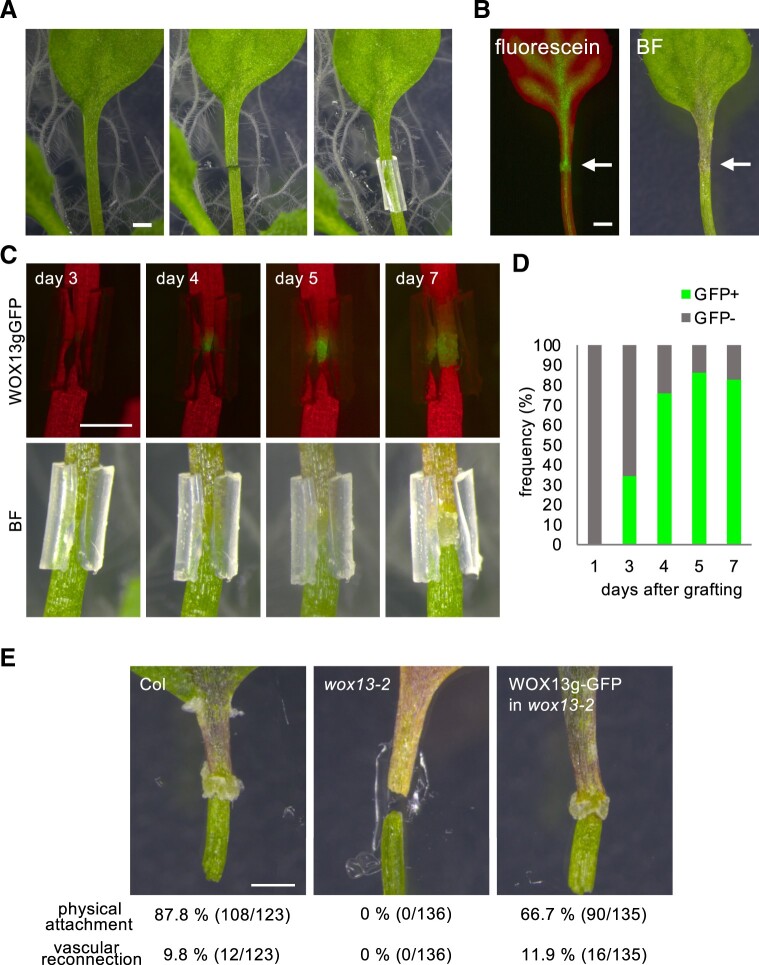
To test the potential role of WOX13 in the establishment of organ reconnection, we first set-up an experimental system to self-graft cut petioles (Figure 2A; Iwase et al., 2021). We used silicon tubes with a slit for the assembly, so that the tubes can open as callus grows and can be easily removed to assess physical re-attachment. We optimized conditions to the point where 87.8% (108/123) of WT petioles re-establish physical attachment by 13 d (Figure 2). Furthermore, 9.8% (12/123) of petioles have also established vascular connectivity as visualized by the translocation of fluorescein from roots to grafted leaf blades (Figure 2B). A large drop in the success rate of vascular connectivity as compared to the recovery of physical adhesion implies that the establishment of physical adhesion is a step within the hierarchical processes of graft establishment, as has been suggested before (Melnyk et al., 2015). Notably, we detected WOX13gGFP signal in calli formed at graft junctions. Our time-lapse observations revealed that the WOX13gGFP expression initiates at around 3–4 d after grafting, when callus cells become visible externally around the site of the graft (Figure 2, C and D). The signal attenuates as callus growth slows down after 7 d, consistent with the idea that WOX13 regulates callus development (Figure 2C). We then compared grafting efficiencies of wox13 and the WT using this experimental system. Strikingly, wox13-2 petioles are totally defective in organ adhesion (0/136), contrasting with the 87.8% success rate of WT petioles. We further confirmed that this defect in the mutant is complemented by expressing WOX13gGFP, where petioles have recovered the capacity of physical attachment to 66.7% (90/135) and the vascular connection to 11.9% (16/135), which is comparable to the WT (Figure 2E). Taken together, we conclude that WOX13 is required for organ reconnection, an essential step toward successful graft formation.
Figure 2.
WOX13 is required for the establishment of organ reconnection. A, Petiole self-grafting assay to test for reconnection of sharply cut petioles. Cut petioles are assembled and supported by silicon tubes containing a slit. B, Translocation of fluorescein dye from roots to the blade of a grafted leaf. Arrows indicate the graft junction. BF, bright field. C and D, WOX13gGFP expression detected in callus formed at a graft junction. Representative images from time-lapse observation are shown in (C) and quantification data based on the frequency of GFP+ or GFP-petiole for each graft (D, n = 29). E, Success rate of physical attachment and vascular reconnection after grafting. Scale bars, 1 mm (A, B, C, and E).
Wound-induced expression of WOX13 is partially dependent on WIND1
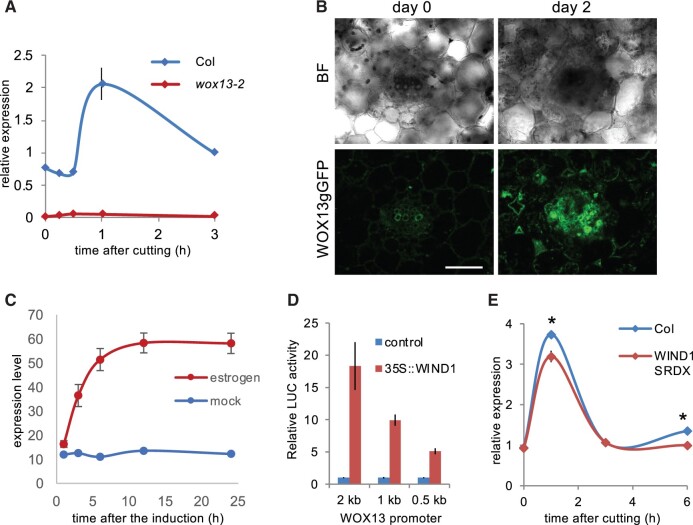
WOX13 was originally identified among wound-induced genes in a hypocotyl transcriptome data set (Ikeuchi et al., 2017). Here, we examined WOX13 expression upon cutting petioles and verified that the expression of WOX13 is transcriptionally induced within 1 h after cutting in the WT by reverse transcription quantitative polymerase chain reaction (RT-qPCR), whereas WOX13 transcripts are totally absent in wox13-2 (Figure 3A). Imaging analysis of WOX13gGFP further revealed that WOX13 expression is not detected in intact mature leaf tissues, while WOX13gGFP starts to accumulates upon cutting mainly in vascular cells (Figure 3B), which are among the major tissues where callus cells originate (Iwase et al., 2017).
Figure 3.
Wound-induced expression of WOX13 is partially dependent on WIND1. A, RT-qPCR analysis of WOX13 expression after cutting of petioles. B, Confocal images showing WOX13gGFP in a vibratome section of petiole taken from the cut end. Scale bar, 50 µm. C, Microarray analysis of WOX13 expression after WIND1 induction in XVE-WIND1 plants. D, Reporter transactivation assay used to analyze WOX13 promoter activity upon WIND1 overexpression in Arabidopsis culture cells. E, RT-qPCR analysis of WOX13 expression after cutting of petioles. Data are mean ± se (n = 3 in A, D, E and n = 4 in C, biological replicates). Asterisks indicate significant differences based on t test, P < 0.05.
Given that the transcriptional response of WOX13 resembles WIND1 in terms of the transient transcriptional induction after wounding, we next tested for potential regulatory relationships between WOX13 and WIND1. Fine time-course expression analysis showed that transcriptional activation of WIND1 initiates at around 15 min following wounding (Iwase et al., 2017), whereas WOX13 transcriptional induction starts after 30 min, slightly later than WIND1 (Figure 3A). We thus postulated that WIND1 might act as an upstream regulator of WOX13. First, we tested the effect of WIND1 on WOX13 expression using XVE-WIND1 transgenic plants harboring estradiol-inducible WIND1, in the previously published transcriptome dataset (Iwase et al., 2021). Strikingly, the transcript level of WOX13 is readily upregulated following induction of WIND1 expression via estradiol application, strongly suggesting that WIND1 is capable of inducing WOX13 expression (Figure 3C). Furthermore, we tested the regulatory role of WIND1 on WOX13 using a transactivation assay employing a luciferase reporter. As shown in Figure 3D, the 2-kb promoter of WOX13 is sufficient for the transcriptional activation upon overexpression of WIND1 in Arabidopsis culture cells. These results together suggest that WIND1 is sufficient for the transcriptional induction of WOX13. We next asked if WIND1 is required for the wound-induced activation of WOX13 in planta by checking the WOX13 expression in transgenic plants harboring a chimeric repressor of WIND1 (WIND1-SRDX) (Iwase et al., 2011). Wound-inducible expression of WOX13 is significantly reduced in WIND1-SRDX plants (Figure 3E), although WOX13 still displays wound-induced expression in the WIND1-SRDX background. Therefore, we conclude that WIND1 is sufficient to induce WOX13 transcription and is partly responsible for inducing WOX13 expression in response to wounding.
WOX13 mutation has profound impact on gene expression during callus formation
To elucidate how WOX13 affects gene expression profile changes during callus formation, we performed time-course transcriptome analysis via RNA-seq in WT and wox13 mutant. We sampled at 0, 1, 3, 6, 12, 24, and 48 h after cutting petioles, i.e. within the timeframe when we have histologically observed the onset of cell proliferation (Iwase et al., 2017). In addition, we also included samples collected after 10 d (240 h) to identify gene expression profile differences in well-developed calli. To identify overall trends in the gene expression profile among samples, we first subjected the obtained transcriptome data to principal component analysis. We found that transcriptome data from respective time point of different genotypes cluster together (Supplemental Figure S2), suggesting that the temporal trajectory has a bigger impact than genotype on shaping the overall gene expression profile. Hence, we next performed pairwise comparison of the genotypes at each time point using edgeR with thresholds false discovery rates (FDRs) <0.01 and |log2 fold change| >1. This analysis revealed that overall 557 genes are up- and 795 genes are downregulated in the wox13 mutant at least at one time point. Importantly, these differentially expressed genes include both direct targets of WOX13 and indirect downstream consequence of the misexpression of WOX13 targets.
WOX13 directly binds specific motifs within promoters
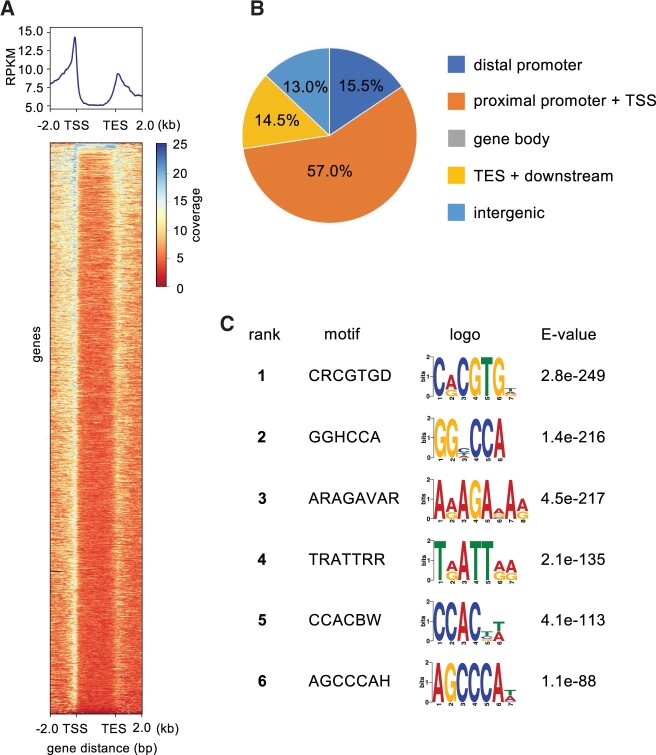
To identify WOX13 direct targets, we next investigated which genomic loci are bound by WOX13-GFP using chromatin immunoprecipitation followed by sequencing (ChIP-seq). We used the WOX13gGFP transgenic line, i.e. plants expressing this fusion protein in its native expression pattern, to identify WOX13 targets. We should note that we used intact root samples where WOX13gGFP displays strong endogenous expression, because we encountered technical difficulties performing ChIP with callus cells as a consequence of the sample properties associated with lysate from this type of tissue. We confirmed that two independent ChIP experiments gave similar results, and then we chose one representative replicate to perform peak characterization and motif analysis. Consistent with the expectation that WOX13 regulates transcription initiation of target genes, WOX13 binding sites are strongly enriched around transcription start sites (TSS), as visualized using a metagene plot (Figure 4A). Indeed, 57% of the identified peaks are found within the range of 1-kb upstream to 100-bp downstream of a TSS (Figure 4B). WOX13 binding sites are enriched around transcription end sites (TES) as well, which is sometimes observed for transcription factors (Wang et al., 2019; Favero et al., 2020). To discover candidate cis-elements bound by WOX13, identified peaks were subjected to motif analysis using DREME (Bailey, 2011). We also re-analyzed the previously published ChIP-seq data of WUS (Ma et al., 2019) and performed motif analysis side-by-side for the comparison (Supplemental Figure S3), to directly test the similarity in sequence motifs as expected from the structural conservation of the DNA binding domain. Top represented sequence motif for WOX13 was CRCGTG (first ranked), which bears striking similarity to a G-box like motif (TCACGTGA). G-box-like motif is designated as a high-confidence WUS-bound motif by multiple independent analyses, including systematic evolution of ligands by exponential enrichment (Busch et al., 2010), in vitro binding assay (Sloan et al., 2020) ChIP-chip (Busch et al., 2010), and ChIP-seq (Ma et al., 2019; Supplemental Figure S3). TRATTRR (fourth ranked) resembles a tandem repeat of TGAT or TAAT (Figure 4C), both of which the DNA binding domain of WUS specifically binds with high-affinity in vitro (Sloan et al., 2020). Other highly enriched motifs such as GGHCCA (second ranked) and ARAGAVAR (third ranked) are also similar to highly ranked motifs from the WUS motif analysis (Supplemental Figure S3). These results together indicate that the cis-elements associated with WOX13 binding that were identified via ChIP-seq resemble the DNA binding profile of WUS that has been revealed by a combination of both in vitro and in vivo approaches. Hence, this supports the idea that our WOX13gGFP ChIP-seq experiment successfully captures specific binding events of WOX13 at target sites.
Figure 4.
WOX13 directly binds specific motifs within promoters. A and B, Distribution of WOX13 binding peaks identified from WOX13 ChIP-seq data. “distal promoter” is defined as −2,000 bp to −1,000 bp relative to the TSS, “proximal promoter” is defined as −1,000 bp to + 100 bp relative to the transcription start site, and “TES + downstream” is defined as −100 bp to +1,000 bp relative to the TES (B). C, Top 6 enriched motifs in the WOX13 peaks.
WOX13 directly regulates WINDs and cell wall modifier genes
To identify genes that are directly regulated by WOX13, we next sought to combine the above-described RNA-seq and ChIP-seq datasets. For this analysis, we defined 5,582 high-confidence WOX13-bound loci, where we detected peaks in both of the replicates within genic regions ranging from −2-kb upstream of the TSS to +1-kb downstream of the TES. Among the 5,582 WOX13-bound genes, we then identified 435 directly regulated genes, including 314 that are downregulated and 135 that are upregulated in wox13 mutant (Figure 5A). Among them 14 genes are in both of the lists, because we categorized genes based on the differential expression at least at one time point. Notably, the proportion of WOX13-bound genes was significantly higher among downregulated genes (39.5%) than among upregulated genes (24.2%) (P = 6.429e-09, prop-test). These results suggest that WOX13 mainly serves as a transcriptional activator, yet it may also function as a repressor depending on the target.
Figure 5.
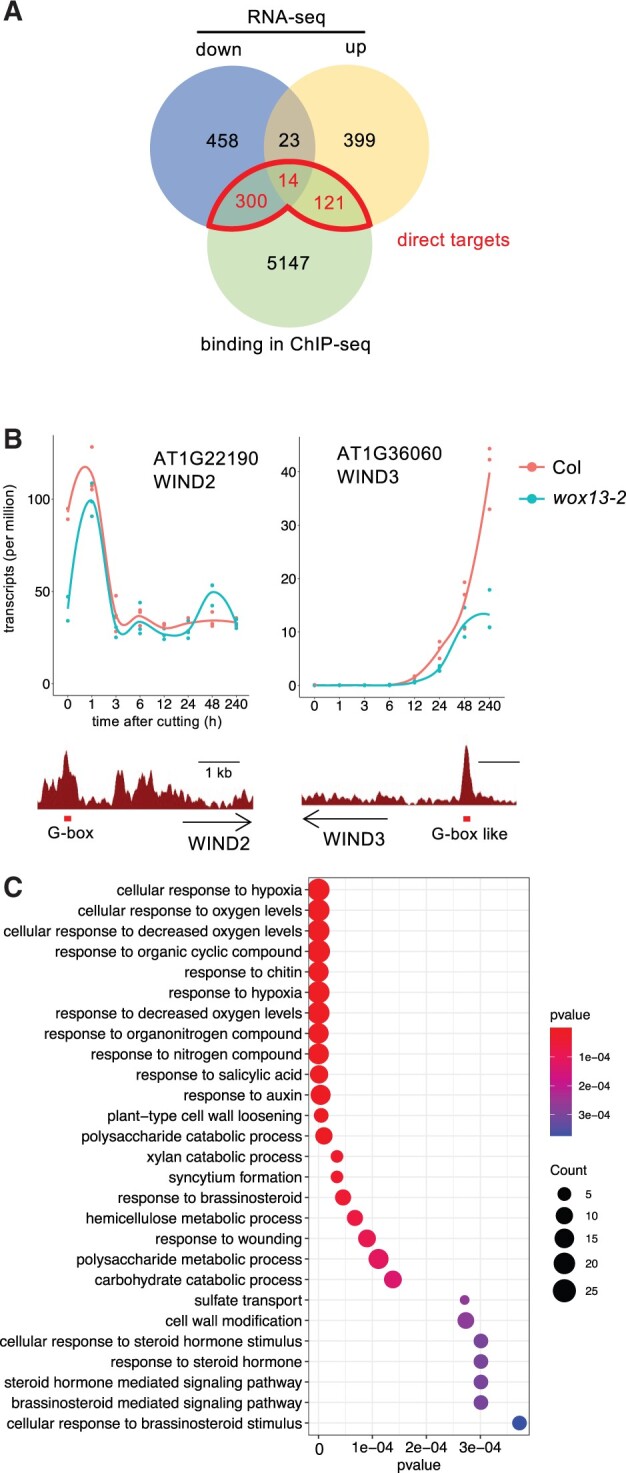
WOX13 directly regulates cellular reprogramming regulators. A, Venn diagram showing the overlap between differentially expressed genes between Col and wox13-2 and genes that are directly bound by WOX13. B, WIND2 and WIND3 are among WOX13’s direct targets. Gene expression data retrieved from RNA-seq (top part, n = 3) and WOX13 binding peaks based on ChIP-seq data (bottom part) are shown. Red boxes highlight G-box or G-box like motif found within the peaks. C, Enriched GO categories of the differentially expressed genes that are downregulated in wox13-2 and directly bound by WOX13.
To investigate the WOX13-mediated gene regulatory network, we first looked at transcription factor-encoding genes that are functionally annotated as regeneration regulators (Ikeuchi et al., 2019) and we identified WIND2 and WIND3 as WOX13 direct targets. WIND2 is rapidly induced upon wounding (Figure 5B), and the expression peak is lower in wox13 mutant (Figure 5B). WIND3 transcripts, on the other hand, slowly accumulate from 12 h onward in the WT (Figure 5B), whereas the expression level is strongly reduced in wox13-2 (Figure 5B). Both WIND2 and WIND3 have reproducible WOX13 binding peaks which contain a G-box (CACGTG) and a G-box like (TCACGTGA) motif, respectively, strongly suggesting that WOX13 directly binds these specific locations. Combined with our finding that WIND1 regulates WOX13, this result indicates that WOX13 and WIND transcription factors have a mutually dependent regulatory relationship with each other that promotes cellular reprogramming.
To further infer the biological function of WOX13, we performed gene ontology (GO) analysis on the list of direct target genes. Among various biological processes implicated by enriched GO categories, we further analyzed those that are functionally relevant to callus formation. Auxin response (4.02e−06), for instance, is important for callus formation (Ikeuchi et al., 2013). We sought to test the possibility that auxin response as visualized by DR5 reporter activity is affected in the wox13 mutant. Imaging analysis, however, showed that DR5rev:GFP signal is comparable between the WT and the wox13 mutant at petiole cut ends, suggesting that WOX13 does not affect cellular auxin response during callus formation. We next looked at GO categories associated with cell wall property, which is also relevant to callus formation (Ikeuchi et al., 2013). Functional categories such as polysaccharide catabolic process (1.01e−05) and plant cell wall loosening (5.11e−06) are among the categories highly enriched for downregulated genes (Figure 5C; Supplemental Table S1), implicating that WOX13 regulates cell wall-related genes. Among them we found a striking enrichment for GH9B family genes that encode cellulases. Four out of nine expressed (tpm > 1) GH9B genes, namely CEL1, CEL3, GH9B8, and GH9B13, are among the listed WOX13 direct target genes. The expression of GH9B13, for instance, is dramatically induced from 3 h after wounding in the WT, whereas transcriptional induction of this gene is almost completely lost in the wox13 mutant (Figure 6A). This observation strongly suggests that the wound-inducible expression of a set of GH9B genes is dependent on WOX13 activity. We also found that PECTATE LYASE LIKE (PLL) family genes involved in pectin degradation are enriched among WOX13 targets. Five out of 13 members of the expressed PLL family subfamily I genes (Sun and Van Nocker, 2010), namely PLL17, PLL18, PLL19, PLL22, and PLL26, are found among the listed genes. The expression of PLL18, for instance, rises from 3 h onward and reaches 500-fold induction at 240 h in the WT, whereas transcriptional induction of this gene is marginal in wox13 (Figure 6B). We also identified a set of EXP genes among WOX13 targets as well: EXPA4, A6, and B3 genes display a substantial reduction of wound-induced expression in the wox13 mutant (Figure 6C). These observations together suggest that WOX13 directly regulates genes involved in cell wall degradation and cell wall loosening.
Figure 6.
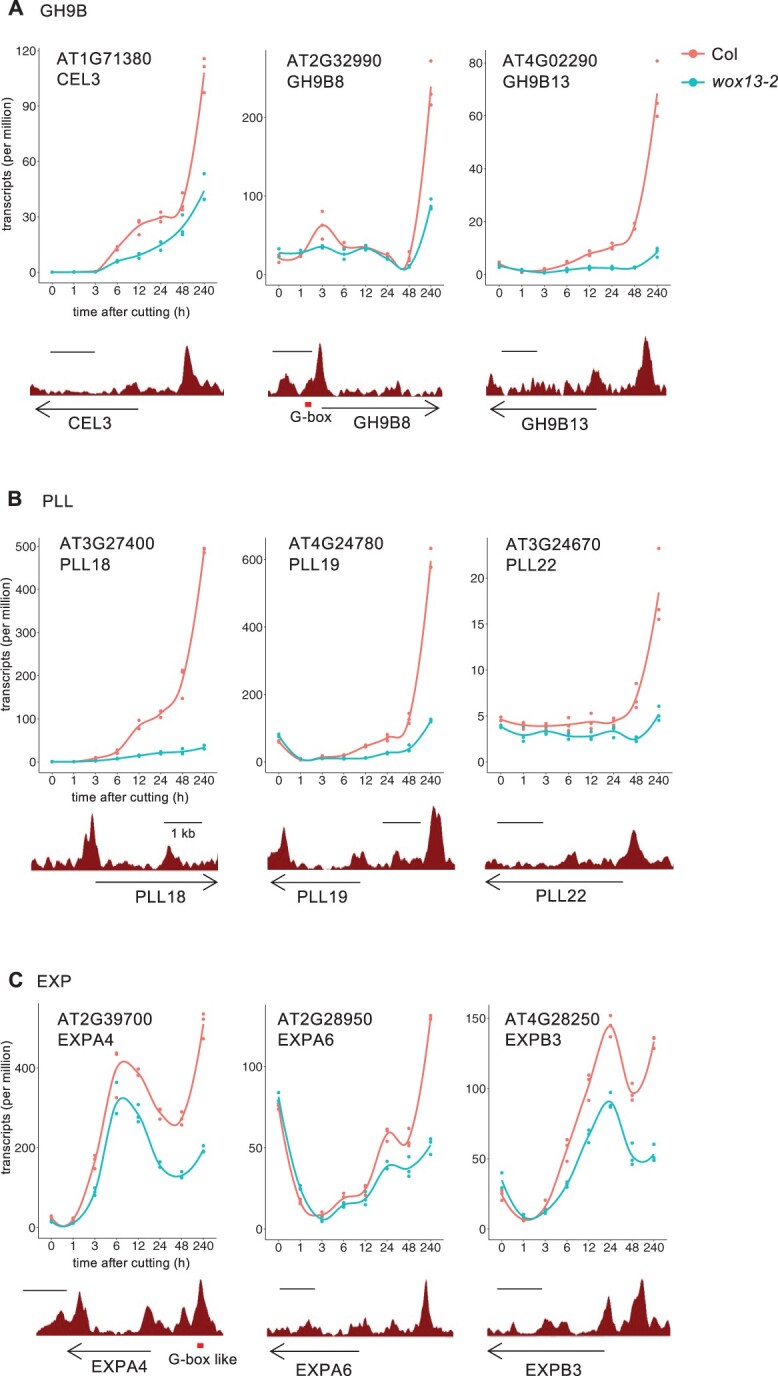
WOX13 directly upregulates cell wall modifying enzyme genes. Gene expression data retrieved from RNA-seq (top part, n = 3) and WOX13 binding data retrieved from ChIP-seq (bottom part) are shown for representative examples of WOX13 target genes in GH9B (A), PLL (B), EXP (C). Red boxes highlight G-box or G-box like motif found within the peaks.
Finally, to assess whether wox13 mutation affects cell wall composition, we next analyzed the monosaccharide composition of cell wall residue by ultra-performance liquid chromatography (UPLC) as described before (Sakamoto et al., 2015). Highly sensitive small-scale analysis allowed us to specifically analyze cell walls only from callus tissue. Such chemical analysis of callus tissue revealed that glucose (Glc) content is dramatically higher in the wox13 mutant (44.6%) in comparison to the WT (26.8%) (Figure 7A). Consistently, relative proportion of other cell wall monosaccharides is slightly reduced. These data suggest that cell walls of the wox13 mutant calli are richer in cellulose, with relative reduction in pectin and xyloglucan. Taken together, WOX13 likely modifies various aspects of cell wall properties in calli.
Figure 7.
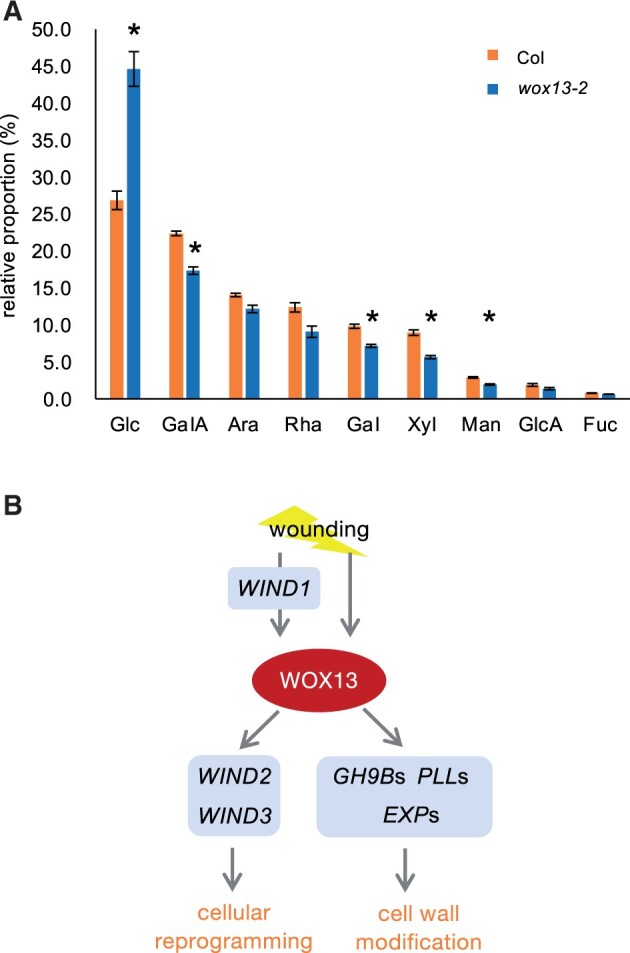
WOX13 regulates cell wall composition of callus. A, Chemical composition of cell wall monosaccharides isolated from callus. Data are mean ± se (n = 4, biological replicates). Asterisks indicate significant differences based on t test, P < 0.01. Glc, glucose; GalA, galacturonic acid; Ara, arabinose; Rha, rhamnose; Gal, galactose; Xyl, xylose; Man, mannose; GlcA, glucuronic acid; Fuc, fucose. Note that GalA, Rha and Ara are the major components of pectin. B, Schematic diagram of WOX13 function in callus formation and tissue repair.
Discussion
WOX13 as a regulator of callus formation and organ reconnection
In this study, we identified WOX13 as a regulator of callus formation at wound sites (Figure 7B). Considering that WOX13 is transcriptionally induced upon wounding and promotes callus formation in both hypocotyls and leaf petioles, it is likely that WOX13 plays general regulatory roles in various organs. Our histological analyses further revealed that WOX13 is required for both the formation of highly vacuolated cells and the promotion of cell proliferation during callus formation (Figure 1). Cellular heterogeneity within callus has been recognized for a long time (Komamine et al., 1963), yet specific cell types are poorly characterized in terms of their physiological function and specification mechanisms. Highly vacuolated cells are typically observed at the surface of wound-induced callus, yet it had been unclear whether highly expanded spherical cells are born merely as a consequence of the loss of physical pressure from surrounding cells or some genetic mechanisms actively specify this cell type. Our findings clearly support the latter hypothesis by demonstrating that wound-inducible WOX13 is required for the formation of highly vacuolated cells. We also showed that WOX13 is indispensable for the reestablishment of physical adhesion between organs upon petiole grafting (Figure 2). These findings together suggest that callus formation and organ adhesion are under the control of the common regulatory mechanism. In addition, this finding supports the previous assumption that the extensively expanded cells observed at graft junctions facilitate physical contact between the scion and rootstock (Jeffree and Yeoman, 1983). We should note, however, that wox13 mutant calli not only lack highly vacuolated cells at the surface but also display a reduction of cell mass. Further studies are required to demonstrate the importance of the specific cell type in successful establishment of the organ adhesion.
Mutually regulatory relationships among WOX13 and WINDs
Upon identification of WOX13 as a regulator of wound-induced callus formation, we sought to place WOX13 within the gene regulatory network consisting of previously identified transcriptional regulators. We found that wound-induced expression of WOX13 is partially dependent on WIND1 (Figure 3). Consistently, wox13 mutant and WIND1-SRDX display similar phenotype in compromised callus growth and reduced efficiency in organ reconnection (Figures 1 and 2; Iwase et al., 2011, 2021), further supporting the idea that WOX13 and WIND1 function in the same regulatory pathway.
In contrast to a previous report showing that ESR1 is a direct downstream target of WIND1, with wound-inducible expression of ESR1 completely lost in the WIND1-SRDX genetic background (Iwase et al., 2011), WOX13 is still transcriptionally induced upon wounding in WIND1-SRDX. Accordingly, other wound signal transducers must be involved in the transcriptional induction of WOX13 in addition to WINDs. It would be interesting to test for the potential contribution of other wound signaling pathways, such as jasmonic acid, ethylene or reactive oxygen species, to the transcriptional induction of WOX13 (Koo and Howe 2009; Marhavý et al., 2019; Ikeuchi et al., 2020).
WIND1 has its paralogs, named WIND2, WIND3, and WIND4, that also have roles in stress-induced cellular reprogramming (Iwase et al., 2011, 2021). WIND1−4 have conserved functions inducing cellular de-differentiation when overexpressed in planta, yet they display distinct temporal expression patterns upon tissue wounding. WIND1 and WIND2 are rapidly induced within 1 h after wounding, whereas WIND3 expression initiates later at around 12-h post injury (Figure 5), raising the possibility that these WIND genes rely on distinct induction mechanisms. Notably, WIND3 is under repressive control of a histone methyltransferase, POLYCOMB REPRESSIVE COMPLEX 2 (PRC2), which prevents ectopic expression under normal growth conditions (Ikeuchi et al., 2015). It is plausible that the transcriptional induction of WIND3 takes longer following perception of an inductive cue because chromatin status of this locus is usually “closed” and needs to be “opened” before transcription initiates. We identify WOX13 as a transcriptional regulator that binds directly upstream of the WIND3 locus (Figure 5B). It would be interesting to test if WOX13 is involved in the process of chromatin opening of WIND3 as described for pioneer transcription factors such as LEAFY (Jin et al., 2021).
Taken together, our data show that WIND1 induces WOX13 expression, with WOX13 in turn directly upregulating WIND2 and WIND3. These regulatory relationships may together facilitate a robust change in the gene expression profile that leads to cellular reprogramming. Future elucidation of functional relationships between WOX13 and WINDs should further unveil the mechanisms of cellular reprogramming.
Roles of WOX13 in the control of cell wall property during callus formation and graft formation
Cell wall-modifying enzyme genes display dynamic transcriptional responses during tissue repair, yet the underlying mechanism regulating the expression of these genes remained unclear. In this study, we identified WOX13 as a transcriptional regulator to orchestrate the expression of genes which are involved in cell wall degradation and loosening (Figure 6). This is in agreement with our phenotypic analysis of the wox13 mutant, because defects in cell expansion and cell–cell adhesion are potentially attributable to the changes in cell wall property. Specifically, we find that cellulase encoding genes in the GH9B clade are highly enriched among WOX13 direct targets, and indeed the wound-induced upregulation of CEL1, CEL3, GH9B8, and GH9B13 is severely compromised in the wox13 mutant (Figure 6). Recent studies showed that GH9B3/CEL3 homologs are required for successful grafting in Nicotiana benthamiana and haustrium formation in Phtheirospermum japonicum (Notaguchi et al., 2020; Kurotani et al. 2020). It would thus be interesting to test the potential role of WOX13 homologs in these experimental systems. In addition to cellulose degradation, dynamic changes in pectin deposition are observed at graft junctions (Jeffree and Yeoman, 1983). Considering that successful grafting involves de novo formation of middle lamella, the failure of the wox13 mutant in the establishment of organ attachment may be partly attributable to the reduced pectin content of callus cells as suggested from monosaccharide composition of cell walls (Figure 7A). We also observed strong wound-induced expression of several members of PLL subfamily I. Biochemical function of PLL in plants are yet to be established, yet they display high sequence similarity with bacterial pectate lyases, which cleaves pectate by β-elimination (Collmer and Keen, 1986; Sun and van Nocker, 2010). Several studies suggest potential roles of PLLs in the promotion of cell expansion (Vogel et al., 2002) and cell proliferation (Leng et al., 2017) during plant organogenesis. Hence, dramatic reduction in PLL gene expression in the wox13 mutant might be relevant to the reduced callus growth.
There are several other examples where key transcription factors mediating callus formation directly regulate the expression of cell wall modifying factors. LATERAL ORGAN BOUNDARIES DOMAIN 18 (LBD18), for instance, directly regulates EXP14 (Xu et al., 2018). LBD16 induces FAD-BD (Lee et al., 2013) and LBD29 upregulates PME2 (Xu et al., 2018). Yet, wound-induced callus and LBD16/18/29-mediated lateral root-like calli harbor distinct histological and developmental features (Ikeuchi et al., 2013). Further investigation should uncover the specific roles of WOX13 and LBDs in the regulation of cell wall modification during callus formation.
Evolution of WOX family genes in land plants
This study provides key evolutionary insights into the WOX family of transcription factors. We show that WOX13 is transcriptionally activated upon tissue injury in both the Arabidopsis hypocotyl and leaf petiole, as has been described for PpWOX13L in the P. patens leaf (Sakakibara et al., 2014). We also show that PpWOX13LB complements the Arabidopsis wox13 mutant, suggesting that the molecular function of the transcription factor to recognize specific DNA sequences is conserved between mosses and seed plants. Furthermore, a subset of target genes, such as EXPs, are commonly upregulated by WOX13 in Arabidopsis (Figure 6) and PpWOX13L in P. patens (Sakakibara et al., 2014). These striking parallels strongly suggest that WOX13 is embedded in a conserved gene regulatory network that regulates cellular reprogramming upon tissue damage. On the other hand, it is also clear that there are fundamental differences between WOX13 orthologs in these different systems. Whereas PpWOX13LB-mediated cell elongation is part of the stem cell initiation process, Arabidopsis WOX13 is involved in the formation of highly vacuolated, fairly differentiated cells in calli and at graft junctions. We suspect that cell wall loosening may be a property associated with stem cells in P. patens, while it is instead coupled with cellular differentiation in seed plants. Why cell wall loosening is oppositely associated with cellular differentiation status in mosses and seed plants poses an attractive evo-devo question. In addition, we also found functional differences between the orthologs, where Arabidopsis WOX13 is involved in the regulation of cell proliferation while P. patens WOX13 is not. WOX13 may have acquired an additional role to control cell proliferation in seed plants, although we thus far have not found any good candidates as cell cycle regulators among WOX13 direct targets (Supplemental Table S1). It is plausible that either the identified target genes may have unknown function in the regulation of cell proliferation, or some relevant targets may have been missed because we used root tissues for ChIP analyses.
The WOX family transcription factors have diversified during the course of seed plant evolution, and it is likely that modern WUS subclade members have taken over the regulatory roles of stem cell formation. WUS subclade members, including WUS in particular, have been mainly subjected to extensive functional characterization. Recent studies have identified genome-wide WUS binding sites (Ma et al., 2019) and structural as well as biochemical properties associated with WUS binding to target DNA sequences (Sloan et al., 2020). In this study, we performed ChIP-seq analysis for WOX13-GFP driven by its native promoter. By comparing the results for WOX13-GFP motif analysis with those from WUS-GFP ChIP-seq data, we find that genome-wide binding patterns are highly similar for these two transcription factors. This is consistent with sequence conservation observed for the DNA binding domain among WOX family members. Namely, key amino acid residues identified in the WUS homeodomain, such as R38, Q89, and R94 (Sloan et al., 2020) are conserved in WOX13 and PpWOX13LB. Therefore, it is plausible that WOX13 has the capacity to bind common targets with WUS, either cooperatively or competitively. Future studies are still required to untangle the functional relationships between ancient and derived WOX family members in the control of cellular reprogramming and organ regeneration.
Materials and methods
Plant materials and growth conditions
The WT Arabidopsis described in this study is Columbia (Col-0). The wox13-2 mutant was originally Ler background and was backcrossed to Col-0 three times (Romera-Branchat et al., 2013). WIND1-SRDX and XVE-WIND1 are in the Col-0 background (Iwase et al., 2011). We used DR5rev:GFP (Benková et al., 2003) as a auxin response reporter. Plants were grown on Murashige–Skoog (MS) medium containing 0.6% (w/v) gelzan and 1% (w/v) sucrose under constant light at 22°C unless otherwise described.
Generation of transgenic plants
To generate the WOX13 genomic fragment fused with GFP, 5-kb genomic fragment of WOX13 was amplified and subcloned into pDONR221 by Gateway BP cloning (Invitrogen). After verifying the full sequence of the amplified fragment, a SmaI recognition site was incorporated in front of the start codon by PCR (WOX13pro-SmaI-WOX13coding). The resultant plasmid was digested by SmaI and ligated with a green fluorescent protein (GFP) fragment obtained from the pHSG399 plasmid via SmaI digestion. The resultant WOX13gGFP fragment was transferred to the binary vector pGWB501 by LR reaction. To generate PpWOX13LB, the coding region of the WOX13 genomic sequence was replaced by PpWOX13LB-citrine as follows. We first incorporated a SmaI site after the stop codon of the WOX13pro-SmaI-WOX13coding vector and then removed the WOX13 coding region by SmaI digestion. The insert was prepared by PCR amplification of PpWOX13LB-citrine from the PpWOX13LB-citrine vector (Sakakibara et al., 2014) and then ligated into the prepared destination vector. The resultant WOX13g-PpWOX13LB-citrine fragment was transferred to the binary vector pGWB501 via an LR reaction. For plant transformation, the binary vectors carrying T-DNAs were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation and the resultant bacteria were infiltrated into wox13-2 plants by the floral dip method.
Callus formation assay
Hypocotyl assay was performed as described previously (Ikeuchi et al., 2017). For petiole assay, petioles of the first pair of leaves from 14-d-old seedlings were cut and incubated on MS medium with the adaxial side of the leaves facedown. Callus images were taken at 9 d after cutting and callus size quantified using a semi-automated pipeline in ImageJ (1.5). We performed the “Crop”, “Make Inverse”, and “Fill” actions, and then converted the image to a binary image using “Make Binary”. Small holes in calli were manually filled using “Fill Holes” and then the region of interest area was measured via “Analyze Particle”. Data were visualized using pirateplot in R version 3.6.
Self-grafting of petioles
To test organ reconnection capacity, we established an experimental system to self-graft petioles as follows. Petioles from first or second foliage leaves from 16-d-old seedlings grown on half-strength MS medium were cut in the middle and re-aligned using ∼3-mm-long silicon tubes 0.5 mm in diameter. We then tested if grafted petioles are physically attached after removal of the tubes 13 d after grafting. Attached petioles were further tested for xylem connection by the translocation of 1-mM fluorescein. We applied fluorescein to roots for 3 h and looked for fluorescence within the xylem of self-grafted leaf blades using an MZ10F dissection microscope (Leica).
Histological analysis
For the analysis of GFP signal in tissue sections, cross sections of petioles were prepared using a vibratome VT1200 (Leica) and observed under the confocal laser scanning microscopy SP8 (Leica). Detailed microscopy settings are as follows: 488 nm laser with 27.5% intensity was used for the excitation and GFP fluorescent was detected with HyD with Gain 500, bandwidth 500–550 nm and 0–3.5 ns time gating (for WOX13gGFP); 488 nm laser with 1.7% intensity was used for the excitation GFP fluorescent was detected with HyD with Gain 131, bandwidth 495–548 nm (for DR5rev:GFP). For the analysis of cellular composition of callus, petiole calli were pre-fixed with 4% (w/v) formaldehyde, 2% (w/v) glutaraldehyde, and 0.05 M cacodylate for overnight, and post-fixed with 1% (w/v) osmium tetroxide in 0.05-M cacodylate for 3 h. The samples were dehydrated in a methanol dilution series, and embedded in EPON 812 resin (TAAB). The resin blocks were cut with an ultramicrotome (Leica EM-UC7) using a diamond knife (DIATOME Histo), and 1-µm thick sections were prepared. The obtained sections were stained with 0.05% (w/v) toluidine blue solution, and micrographs were taken using an upright light microscope Olympus BX51M equipped with DP26 digital camera.
Automatic cell area measurement
Segmentation of cells on tissue section images was performed using X-net as described in details in Fujii et al (2021). Briefly, this method integrates two encoders and decoders to improve the accuracy of semantic segmentation. As input dataset, individual raw images of 2448 × 1920 pixels with the resolution 0.1 pixel per micrometer were stitched to provide high-resolution whole callus tissue section images. Three sub-images representing high, middle, and low contrast regions were cropped from these images to make the ground truth dataset consisting of two classes: cells and cell walls. All the cells in the three images were manually annotated using a pen-tablet, and regions, which are neither of the cells nor the cell walls, were manually masked, so that all the remaining regions were to be considered as cell walls. We cropped the regions of 256 × 256 pixels from the three original images and used 57 regions for training and 20 regions for validation. In our data set, the trained X-net showed 45.9%, 80.8%, and 63.3% of Intersection over Union (IoU) for cell wall, cell and overall average IoU, respectively. This performance is substantially higher than the result of U-Net, which is 43.9%, 78.8%, and 61.4% (Fujii et al., 2021).
After performing segmentation by the X-net, we used Morphology as post-processing to connect disconnected cell walls. In addition, fill hole processing was used to convert small regions misassigned as “cell wall” pixels inside a cellular region to “cell” pixels. Cases where more than two cells are misannotated as single cells were excluded for the downstream analyses, by using the information of manually annotated center positions of each cell which were obtained during manual counting process of cell numbers on ImageJ (1.5). After this filtering, cell area from 46% of all cells were measured and compared between Col and wox13-2. Histogram drawing and statistical tests were performed with R version 4.0 and package ggplot2 (Wickham, 2016).
Transactivation assay
Transactivation assay was performed as described previously (Iwase et al., 2017). The 35Sp::WIND1 and 35Sp::NOS terminator vectors were used as an effector and control, respectively. The WOX13p::L-LUC vector was used as a reporter, and the 35Sp::R-LUC was used as an internal control. To construct the WOX13p:::L-LUC reporter vectors, the 2-kb, 1-kb, and 0.5-kb promoter sequence of WOX13 was amplified by PCR and cloned between ApaI and NotI sites of the ESR1pro:L-LUC vector. Particle bombardment was performed using the Biolistic PDS-1000/He system (Bio-Rad) and luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega). Arabidopsis MM2d cultured cells were used as host cells and luciferase activities were quantified using the Mithras LB940 Microplate Luminometer (Berthold Technologies).
RNA extraction
To evaluate gene expression following petiole cutting, petioles from first or second foliage leaves were sharply cut in the middle and the remaining apical half of the petioles were harvested for RNA extraction at the designated time after cutting. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) according to manufacturer’s instructions. For RNA-seq, isolated total RNA solution was subjected to on-column DNase digestion (Qiagen) to eliminate genomic DNA (gDNA) contamination.
Reverse transcription quantitative PCR
For RT-qPCR, isolated RNA was treated with gDNA eraser before reverse transcription was performed using random hexamer primers and PrimeScript reverse transcriptase (Takara). RT-qPCR was performed in technical duplicates using the THUNDERBIRD SYBR qPCR Mix (Toyobo) with the primers indicated in Supplemental Table S2. A total of 400 ng of cDNA was used as template for each sample. Relative transcript quantities were calculated using standard curves for each primer set and values normalized to an internal control gene, PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3).
Transcriptome data analysis
For the transcriptome comparison between WT and wox13 mutant, total RNA was prepared from cut petiole samples from the first pair of leaves of 14-d-old seedlings. Biological triplicates were prepared for each genotype (Col, wox13-2) and time point (0-, 1-, 3-, 6-, 12-, 24-, 48-, and 240-h post injury). Isolated total RNA was subjected to library preparation using the KAPA Stranded mRNA-Seq Kit (Kapa Biosystems) with NEBNext Multiplex Oligos for Illumina (New England Biolabs) used as adapters and Agencourt AMPure XP (Beckman Coulter) beads in place of KAPA Pure Beads. Single-end sequencing was performed using the Illumina NextSeq500 platform. Raw data files (bcl format) were converted to fastq files by bcl2fastq (Illumina). On average 92% (85%–95%) of reads were mapped to the Arabidopsis TAIR10 reference transcript model using the RSEM package (Li and Dewey, 2011) with default parameters. The samples with 6–14 million mapped reads were used for the downstream analysis. Differentially expressed transcripts between genotypes at each time points were identified using the edgeR package in R/Bioconductor with FDR <0.01 and |log2FC| >1 cutoff. GO analysis was performed using clusterProfiler package (3.18.1; Yu et al., 2012) and the obtained GO category list was simplified using “simplify” function.
ChIP
ChIP was carried out twice independently as previously reported (Rymen et al., 2019) with minor modifications. One gram of whole roots was harvested from 14-d-old WOX13gGFP in wox13-2 seedlings and frozen with liquid nitrogen. Samples were ground to a fine powder using a multibeads shocker (MB1200, Yasui Kikai) and the nuclear fraction was isolated after cross-linking for 10 min with 1% formaldehyde (Sigma) under vacuum. Chromatin was sheared at 5°C with a focused ultrasonicator (Covaris) with the following settings: duty cycle 5%, intensity 4, and cycles per burst 200 for 25 min (the first replicate) or duty cycle 10%, intensity 5, and cycles per burst 200 for 15 min (the second replicate). Sheared chromatin was immunoprecipitated using antibodies against GFP (ab290, Abcom). The isolated DNA was quantified with the Qubit dsDNA High Sensitivity Assay kit (Thermo Fisher Scientific) and 1–2 ng of DNA was used to make each ChIP-seq library. Libraries were prepared using the KAPA Hyper Prep Kit for Illumina (KK8502, KAPA Biosystems) and Illumina compatible adaptors (E7335, E7500, E7710, E7730, NEB). Libraries were pooled and 84-bp single-stranded sequences were obtained using Illumina NextSeq500 sequencer.
ChIP-seq data analysis
Obtained sequence data files (bcl format) were converted to fastq files by BaseSpace Sequence Hub (Illumina) or bcl2fastq (Illumina). Over 48% of reads were uniquely mapped to the Arabidopsis TAIR10 reference using Bowtie 1 (Langmead et al., 2009) with the setting “-m 1”. The total number of uniquely mapped reads per sample was 8–30 million. Peaks were called by comparing ChIP samples with the input using the “callpeak” command in MACS2 (Zhang et al., 2008) with the following parameters: “-g 1.2e8 -q 0.01”. Fold-enrichment bdg format peak files were generated by using the treatment pileup and control lambda output files generated from “callpeak” as inputs for the MACS2 “bdgcmp” command with the setting “-m FE”. Peaks were viewed using Integrative Genomics Viewer version 2.5.0 (Robinson et al., 2011). Motif analysis was performed using BEDTools (Quinlan and Hall, 2010) and DREME 5.1.1 (Bailey, 2011) with the default parameters. pyGenomeTracks (Lopez-Delisle et al., 2021) was used for peak visualization.
Monosaccharide composition analysis
For determination of cell wall monosaccharide composition, we exploited ultra performance liquid chromatography/p-aminobenzoic ethyl ester (UPLC-ABEE) method (Sakamoto et al., 2015), which is 10–100 times more sensitive compared with previous methods. The establishment of highly sensitive method allowed us to use extremely small amount of starting material. The callus sample 9 d after induction was fixed in 80% ethanol (EtOH). After exchanging from ethanol to methanol, the fixed callus were treated three times with methanol at 25°C for 30 min, three times with acetone at 25°C for 30 min, three times with methanol/chloroform (1:1, v/v) at 25°C for 30 min, and then dried at 65°C for 18 h. Cell wall hydrolyzation and sample preparation were performed by 1/25 downscaled method as previously described (Sakamoto and Mitsuda, 2015) and monosaccharide content in hydrolysate was determined by UPLC-ABEE method (Sakamoto et al., 2015).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: WOX13 (AT4G35550), WIND1 (AT1G78080), WIND2 (AT1G22190), WIND3 (AT1G36060), PP2AA3 (AT1G13320). Next-generation sequence data from this article can be found under the following accession numbers: RNA-seq data, DRR300300-DRR300347; ChIP-seq data, DRR300348-DRR300351.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. WOX13 promotes callus formation upon hypocotyl cutting.
Supplemental Figure S2. PCA of time-course transcriptome data.
Supplemental Figure S3. Motif analysis of WUS ChIP-seq data published in Ma et al. (2019).
Supplemental Figure S4. DR5rev:GFP signal at the petiole cut end.
Supplemental Table S1. List of WOX13 targets.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr Keiko Sakakibara (Rikkyo University) and Dr Mitsuyasu Hasebe (National Institute for Basic Biology) for sharing PpWOX13LB plasmid, Dr Michitaka Notaguchi (Nagoya University) and Dr Hiroki Tsutsui (University of Zurich) for their advice on petiole grafting experiments, Ms Mariko Ohnuma, Ms Mariko Mouri, and Ms Chika Ikeda for technical assistance in Sugimoto laboratory (RIKEN), Dr Mayuko Sato and Dr Noriko Takeda for microscopy assistance in Toyooka laboratory (RIKEN), Ms Satomi Kawai, Ms Nao Ogura, and Mr Takeshi Tsuchida for technical assistance in Ikeuchi laboratory (Niigata University).
Funding
This work was supported by JSPS KAKENHI Grant Numbers (17K15146, 17J40121, 20K06712, 20H05431, and 20H04894 to M.I.; 17H03704, 20H05911, and 20H05905 to K.S.); by The Naito Foundation to M.I.; by Takeda Science Foundation to M.I; by Shiseido Female Researcher Science Grant to M.I.; a grant from Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry to K.S.
Conflict of interest statement. None declared.
Conceptualization, M.I., A.I., and K.S.; Methodology, H.F. and K.H; Software, H.F. and K.H; Investigation, M.I., A.I., T.I., H.T., D.S.F., A.K., S.S., M.W., N.H., K.T., T.S.; Formal analysis, T.I., T.T.; Writing—original draft, M.I.; Writing—review and editing, M.I., D.S.F., N.M., and K.S.; Supervision, M.I., N.M., and K.S.; Project Administration, M.I.; Funding acquisition, M.I. and K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Momoko Ikeuchi (ikeuchi@bio.sc.niigata-u.ac.jp).
References
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev. Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Collmer A, Keen NT (1986) The role of pectic enzymes in plant pathogenesis. Ann Rev Phytopathol 24: 383–409 [Google Scholar]
- Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H, Laufs P, Moreau H, Kreis M, Lecharny A (2008) Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Biol 8: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero DS, Kawamura A, Shibata M, Takebayashi A, Jung JH, Suzuki T, Jaeger KE, Ishida T, Iwase A, Wigge PA, et al. (2020) AT-Hook transcription factors restrict petiole growth by antagonizing PIFs. Curr Biol 30: 1454–1466 [DOI] [PubMed] [Google Scholar]
- Fujii H, Tanaka H, Ikeuchi M, Hotta H (2021) X-net with different loss functions for cell image segmentation. In Proceedings of IEEE CVF Conference on CVPR Workshops, pp 3937–3800
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Hajný J, Prát T, Rydza N, Rodriguez L, Tan S, Verstraeten I, Domjan D, Mazur E, Smakowska-Luzan E, Smet W, et al. (2020) Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 370: 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 8: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A (2013) Plant callus: mechanisms of induction and repression. Plant Cell 25: 3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, Veylder LD, et al. (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1: 15089. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Lambolez A, Kojima M, Takebayashi Y, Heyman J, Watanabe S, Seo M, De Veylder L, et al. (2017) Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol 175: 1158–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Shibata M, Rymen B, Iwase A, Bågman AM, Watt L, Coleman D, Favero DS, Takahashi T, Ahnert SE, et al. (2018) A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol 59: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Favero DS, Sakamoto Y, Iwase A, Coleman D, Rymen B, Sugimoto K (2019) Molecular mechanisms of plant regeneration. Annu Rev Plant Biol 70: 377–406 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Rymen B, Sugimoto K (2020) How do plants transduce wound signals to induce tissue repair and organ regeneration? Curr Opin Plant Biol 57: 72–77 [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Morishita M, Higuchi Y, Ichikawa S, Ishikawa T, Nishiyama T, Kabeya Y, Hiwatashi Y, Kurata T, Kubo M, et al. (2019) Physcomitrella STEMIN transcription factor induces stem cell formation with epigenetic reprogramming. Nat. Plants 5: 681–690 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Iwase A, Harashima H, Ikeuchi M, Rymen B, Ohnuma M, Komaki S, Morohashi K, Kurata T, Nakata M, Ohme-Takagi M, et al. (2017) WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29: 54–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Kondo Y, Laohavisit A, Takebayashi A, Ikeuchi M, Matsuoka K, Asahina M, Mitsuda N, Shirasu K, Fukuda H, Sugimoto K (2021) WIND transcription factors orchestrate wound-induced callus formation, vascular reconnection and defense response in Arabidopsis. New Phytol. DOI: 10.1111/nph.17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree E, Yeoman MM (1983) Development of intercellular connections between opposing cells in a graft union. New Phytol 93: 491–509 [Google Scholar]
- Jin R, Klasfeld S, Zhu Y, Fernandez Garcia M, Xiao J, Han SK, Konkol A, Wagner D (2021) LEAFY is a pioneer transcription factor and licenses cell reprogrammming to floral fate. Nat Commun 12: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yang W, Forner J, Lohmann JU, Noh B, Noh Y (2018) Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J 37: e98726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komamine A, Sato M, Shimokoriyama M (1963) Physiological studies on the outgrowth of the epicotyl in Stizolobium hassjo I. Properties of the outgrowth. Bot Mag Tokyo 76: 130–137 [Google Scholar]
- Koo AJK, Howe GA (2009). The wound hormone jasmonate. Phytochemistry 70: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani K, Wakatake T, Ichihashi Y, Okayasu K, Sawai Y, Ogawa S, Cui S, Suzuki T, Shirasu K, Notaguchi M (2020) Host-parasite tissue adhesion by a secreted type of β-1,4-glucanase in the parasitic plant Phtheirospermum japonicum. Commun Biol 30: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim M, Kim NY, Lee SH, Kim J (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J 73: 212–224 [DOI] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X, et al. (2017) A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol 174: 1151–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T (2021) pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics 37: 422–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K, et al. (2019) WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Kurenda A, Siddique S, Dénervaud Tendon V, Zhou F, Holbein J, Hasan MS, Grundler FM, Farmer EE, Geldner N (2019) Single‐cell damage elicits regional, nematode‐restricting ethylene responses in roots. EMBO J 38: e100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Sato R, Matsukura Y, Kawajiri Y, Iino H, Nozawa N, Shibata K, Kondo Y, Satoh S, Asahina M (2021) Wound-inducible ANAC071 and ANAC096 transcription factors promote cambial cell formation in incised Arabidopsis flowering stems. Commun Biol 4: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Meyerowitz EM (2015) Plant grafting. Curr Biol 25: R183–188 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Schuster C, Leyser O, Meyerowitz EM (. 2015) A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol 25: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk CW, Gabel A, Hardcastle TJ, Robinson S, Miyashima S, Grosse I, Meyerowitz EM (2018) Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc Natl Acad Sci USA 115: E2447–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M, Kurotani K, Sato Y, Tabata R, Kawakatsu Y, Okayasu K, Sawai Y, Okada R, Asahina M, Ichihashi Y, et al. (2020) Cell-cell adhesion in plant grafting is facilitated by β-1,4-glucanases. Science 369: 698–702 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Pitaksaringkarn W, Matsuoka K, Asahina M, Miura K, Sage‐Ono K, Ono M, Yokoyama R, Nishitani K, Ishii T, Iwai H, Satoh S (2014) XTH 20 and XTH 19 regulated by ANAC 071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems. Plant J 80: 604–614 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan D, Shanmukhan AP, Kareem A, Aiyaz M, Varapparambathu V, Toms A, Kerstens M, Valsakumar D, Landge AN, et al. (2020) A coherent feed-forward loop drives vascular regeneration in damaged aerial organs of plants growing in a normal developmental context. Development. DOI: 10.1242/dev.185710 [DOI] [PubMed] [Google Scholar]
- Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M, Ripoll JJ, Yanofsky MF, Pelaz S (2013) The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J 73: 37–49 [DOI] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Lambolez A, Inagaki S, Takebayashi A, Iwase A, Sakamoto Y, Sako K, Favero DS, Ikeuchi M, et al. (2019) Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun Biol 2: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Reisewitz P, Aoyama T, Friedrich T, Ando S, Sato Y, Tamada Y, Nishiyama T, Hiwatashi Y, Kurata T, et al. (2014) WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 141: 1660–1670 [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Yoshida K, Sugihara S, Mitsuda N (2015) Development of a new high-throughput method to determine the composition of ten monosaccharides including 4-O-methyl glucuronic acid from plant cell walls using ultra-performance liquid chromatography. Plant Biotechnol 32: 55–63 [Google Scholar]
- Sakamoto S, Mitsuda N (2015) Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol 56: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu H-Y, Hofhuis H, Birnbaum KD (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J, Hakenjos JP, Gebert M, Ermakova O, Gumiero A, Stier G, Wild K, Sinning I, Lohmann JL (2020) Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL. Nat Commun 11: 2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, van Nocker S (2010) Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobbe H, Schmitt U, Eckstein D, Dujesiefken D (2002) Developmental stages and fine structure of surface callus formed after debarking of living lime trees (Tilia sp.). Ann Bot 89: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li S, Li Y, Xu Y, Wang Y, Zhang R, Sun W, Chen Q, Wang XJ, Li C, Zhao J (2019) MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling. Nat Plants 5: 616–625 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. New York, Springer [Google Scholar]
- Xu C, Cao H, Xu E, Zhang S, Hu Y (2018) Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol 59: 744–755 [DOI] [PubMed] [Google Scholar]
- Xu C, Cao H, Zhang Q, Wang H, Xin W, Xu E, Zhang S, Yu R, Yu D, Hu Y (2018) Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat Plants 4: 108–115 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T-Q, Lian H, Zhou C-M, Xu L, Jiao Y, Wang J-W (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.