Abstract
Despite recent progress in our understanding of graft union formation, we still know little about the cellular events underlying the grafting process. This is partially due to the difficulty of reliably targeting the graft interface in electron microscopy to study its ultrastructure and three-dimensional architecture. To overcome this technological bottleneck, we developed a correlative light electron microscopy (CLEM) approach to study the graft interface with high ultrastructural resolution. Grafting hypocotyls of Arabidopsis thaliana lines expressing yellow FP or monomeric red FP in the endoplasmic reticulum (ER) allowed efficient targeting of the grafting interface for examination under light and electron microscopy. To explore the potential of our method to study sub-cellular events at the graft interface, we focused on the formation of secondary plasmodesmata (PD) between the grafted partners. We showed that four classes of PD were formed at the interface and that PD introgression into the cell wall was initiated equally by both partners. Moreover, the success of PD formation appeared not systematic with a third of PD not spanning the cell wall entirely. Characterizing the ultrastructural characteristics of these incomplete PD gives us insights into the process of secondary PD biogenesis. We found that the establishment of successful symplastic connections between the scion and rootstock occurred predominantly in the presence of thin cell walls and ER–plasma membrane tethering. The resolution reached in this work shows that our CLEM method advances the study of biological processes requiring the combination of light and electron microscopy.
A correlative light electron microscopy method provides access to the ultrastructural level at the graft interface and allows focusing on the 3-D architecture of plasmodesmata between grafted partners.
Introduction
Grafting plants permits us to select and combine different rootstock and scion phenotypes, such as root pathogen resistance and fruit quality traits, independently of genetic introgression (Mudge, 2009). Nevertheless, graft compatibility is an essential prerequisite (Flaishman et al., 2008; Chen et al., 2017). As such, the study of graft union formation and the characterization of the mechanisms involved are important for the breeding of many horticultural crops (Kyriacou et al., 2017). In addition, grafting is also used in scientific studies to explore long-distance movement of molecules and to graft-inoculate pathogens (Ragni et al., 2011; Liang et al., 2012; Bidadi et al., 2014; Zhang et al., 2014; Simone et al., 2016; Ota et al., 2020).
Grafting involves cutting the scion and rootstock, and maintaining the cut surfaces in close proximity. During grafting, as opposed to wounding, all physical cell communications are interrupted by cutting at the grafting point. To survive, both grafted partners have to interact together quickly and effectively to establish new connections. During graft union formation the cells at the interface collapse, a necrotic layer forms, and the scion and rootstock adhere to each other (Sala et al., 2019). Cells then proliferate at the cut surfaces to form a callus, some of these callus cells are thought to differentiate into phloem and xylem that connect the vasculature of the scion and rootstock (Melnyk et al., 2015). Recent progress has been made in understanding of graft union formation by using the Arabidopsis thaliana hypocotyl grafting model (Matsuoka et al., 2016; Sala et al., 2019; Tsutsui et al., 2020). In this model, the developmental framework and transcriptomic changes have been excellently documented (Melnyk et al., 2015, 2018). The establishment of a vascular continuum begins by phloem reconnection at 3 d after grafting (DAG) and then, xylem reconnection occurs from 5 to 7 DAG (Melnyk et al., 2015). New vascular tissue formation is initiated above, and then below the graft interface indicating that the scion could be the leader of this cellular differentiation processes. At the graft interface, new differentiated vascular cells expand and divide to bypass the wounded regions, permitting the vascular reconnection between both partners (Melnyk et al., 2015).
While long-distance signaling is ensured by the vascular tissues, cell-to-cell communication is mediated via nanometric channels called plasmodesmata (PD) (Li et al., 2021b). These channels span the cell wall and participate in developmental processes through the trafficking of cytosolic and membrane molecules between neighboring cells (Cantrill et al., 1999; Lucas and Lee, 2004; Benitez-alfonso et al., 2009; Stonebloom et al., 2009; Xu et al., 2011; Furuta et al., 2012). PD are composed of a central cylindrical element, called the desmotubule (Tilney et al., 1991; Ding et al., 1992). The desmotubule, localized exactly at the cell-to-cell interface, is a specialized constricted domain of endoplasmic reticulum (ER) shared between both cells. Inside the PD, the desmotubule interacts physically with a particular plasma membrane domain, which is in continuity with the plasma membranes of neighboring cells (Grison et al., 2015; Nicolas et al., 2017; Brault et al., 2019). The space between the desmotubule and the plasma membrane, called the cytoplasmic sleeve, is not visible in very young PD (PD of type I, i.e. PD with no discernible cytoplasmic sleeve), but is larger in more mature PD (PD of type II, i.e. PD with visible space between the desmotubule and the plasma membrane; Nicolas et al., 2017). Another nomenclature used to describe PD, primary and secondary PD, distinguishes PD based on their biogenesis (Ehlers and Kollmann, 2001). Primary PD are formed during cell division by trapping ER strands in a neo cell wall during its formation (Hepler, 1982; Nicolas et al., 2017), whereas secondary PD are formed in pre-existing cell walls by an unknown mechanism.
The studies of Kollmann and Glockmann (Kollmann et al., 1985; Kollmann and Glockmann, 1985, 1991) on the heterograft faba bean (Vicia faba)/sunflower (Helianthus annuus), where callus cells possess species-specific ultrastructural differences, demonstrated the existence of PD at the graft interface. Because the scion and rootstock cells belong to different cell lineages, PD at the graft interface are necessarily secondary. Unfortunately, this approach is limited to studying very different species grafted together, so possibly characterizes grafts with a certain degree of incompatibility (at best only 50% of V. faba/H. annuus grafts survived 4–6 weeks) and does not necessarily give insights into the development of compatible grafts. In addition, relying on species-specific ultrastructural differences does not allow the unambiguous identification of every cell studied because of the intrinsic ultrastructural cell heterogeneity occurring between different cells.
Horizontal-DNA transfers have been found at the graft interface of Nicotiana tabacum grafts (Stegemann and Bock, 2009; Stegemann et al., 2012; Hertle et al., 2021), suggesting that large openings form at the graft interface. A recent study showed that callus cells of the graft interface could have cell wall holes of approximately 1.5 µm in diameter, which could allow DNA and organelle transfers (Hertle et al., 2021). Despite an attempt made to track plastids with live-imaging to show plastid exchanges through these holes, the absence of a complete three-dimensional (3-D) view of the observed cells and of a single plastid tracking method means that the authors could not unequivocally show that movements of a plastid observed around a cell wall correspond to the same plastid going through the cell wall (Hertle et al., 2021). Therefore, direct evidence of the movement of organelles between the scion and rootstock of grafted plants has yet to be found.
In order to gain insights into the ultrastructural events underpinning the graft union formation, we developed a Correlative Light and Electron Microscopy (CLEM) approach on grafted hypocotyls of A. thaliana to target the graft interface. Correlative light and electron microscopy approaches have been developed for several model organisms and have been used for plant tissues (Bell et al., 2013; Marion et al., 2017; Wang and Kang, 2020). The different protocols published to date give access to the ultrastructure context of fluorescence labeling, but the ultrastructure preservation and resolution are limited to relatively low magnifications and provide only ultrastructural overviews. Here, we describe in detail a CLEM protocol to localize and study with a very high reliability the graft interface of the model plant A. thaliana. This method is based on the in-resin fluorescence CLEM methods published by Kukulski et al. (2012) that we adapted for plant samples. Combined with electron tomography (ET), it permits us to study the fine 3D ultrastructural details of our samples, up to the resolution of the bilayer of the plasma membrane. ER-localized fluorescent proteins were used to label the scion and the rootstock, and permitted us to localize the graft interface under light microscopy before revealing its ultrastructure with TEM and ET. The ability to identify the origin and the type of cells at the graft interface offers perspectives to understand cytological events involved in establishing communication between the scion and rootstock of grafted plants. We chose as a proof of concept to use this approach to answer the following questions: Are PD formed at the graft interface of homo-grafted hypocotyls? Do the PD formed at the graft interface differ in ultrastructure? What are the characteristics of successful PD biogenesis at the graft interface and how can they inform us on the mechanisms involved? Do both the scion and rootstock initiate PD biogenesis, if so, are both partners equally involved?
Results
Targeting the graft interface using correlative light electron microscopy
In order to precisely and accurately identify the graft interface of hypocotyls of A. thaliana at the cellular level, we used yellow and red intracellular FPs to tag the scion and rootstock, respectively. To do this we made use of transgenic lines of A. thaliana expressing either YFP (yellow FP) or mRFP (monomeric red FP) with an ER retention signal (HDEL_YFP or HDEL_mRFP) (Figure 1, A and B). The HDEL retention signal prevents the FPs from being secreted and retains them in the ER lumen (Pelham et al., 1988). We visualized graft interfaces of the scion/rootstock combination HDEL_YFP/HDEL_mRFP by confocal microscopy at 5 DAG; we observed a clear demarcation between YFP- or mRFP-containing cells at the graft interface (Figure 1, B). As a positive control, we used A. thaliana lines expressing a free cytosolic FP (GFP or mRFP), which are known to diffuse from cell-to-cell through PD (Oparka et al., 1999; Nicolas et al., 2017). When the lines expressing a free cytosolic FP were used as one graft partner, the cells in the vicinity of the graft interface were always labeled with both the cytosolic and ER-bound FPs (Figure 1, C and D). We concluded that, as expected, HDEL-tagged FPs cannot freely pass from one grafted partner to the other and that using the HDEL_YFP and HDEL_mRFP expressing lines permits the precise and accurate identification of the graft interface.
Figure 1.
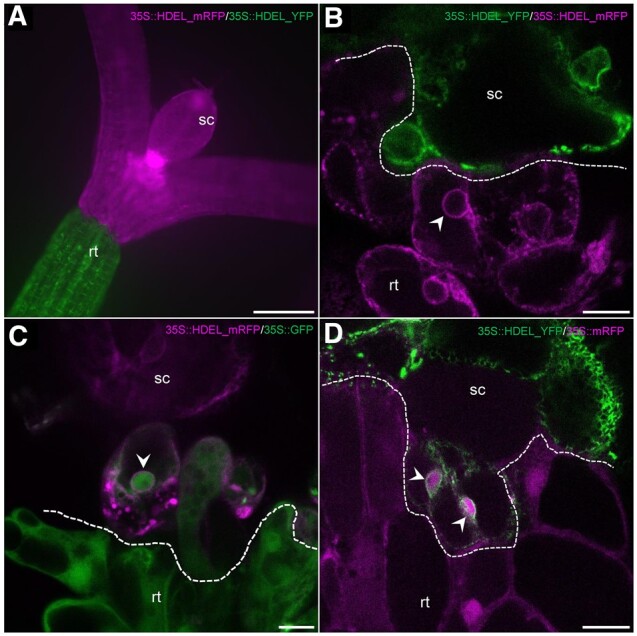
The graft interface of A. thaliana hypocotyls visualized with ER markers in the scion and rootstock. The scion (sc) and the rootstock (rt) are at the top and bottom positions, respectively. A, Large view of a graft between a scion expressing 35S::HDEL_mRFP (magenta) onto a rootstock expressing 35S::HDEL_YFP (green). B, Observation by confocal microscopy of a graft interface of 35S::HDEL_YFP (scion)/35S::HDEL_RFP (rootstock). No YFP nor RFP signal is found in the opposite partner. C, Observation by confocal microscopy of a graft interface of 35S::HDEL_mRFP (scion)/35S::GFP (rootstock). GFP and HDEL_mRFP can be observed in same cells. D, Observation by confocal microscopy of a graft interface of 35S::HDEL_YFP (scion)/35S::mRFP (rootstock). mRFP and HDEL_YFP can be observed in same cells. White arrowheads pinpoint nuclei. Dashed lines indicate graft interface. Scale bars: (A) 200 μm and (B–D) 10 μm.
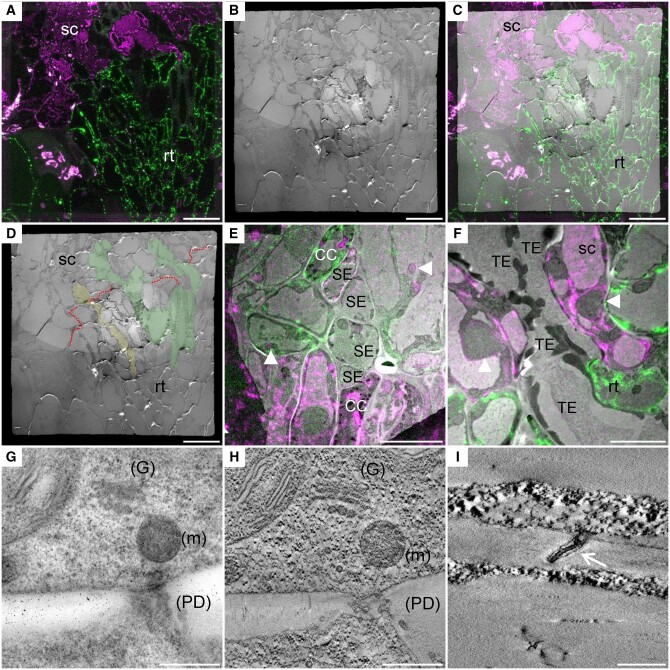
To investigate the graft interface at the ultrastructural level, we then used a CLEM approach on the grafted HDEL_YFP/HDEL_mRFP hypocotyls (Figure 2). The graft interfaces could be harvested from 3 DAG without the scion and rootstock detaching from each other. After harvesting, graft interfaces were then quickly fixed by high pressure freezing in order to ensure the biological preservation of samples closest to their native state as possible. During freeze substitution steps, the fluorescence preservation of YFP and mRFP needs to be maintained without compromising the ultrastructure preservation. To this end, fixative contrasting compounds were limited to uranyl acetate and we used limited incubation times for acetone, resin, and UV exposure. Following freeze substitution in HM20 resin, ultrathin sections of 70 nm were made and harvested on TEM grids and then visualized under confocal microscopy after mounting in water between a glass slide and a cover slip. Despite the measures taken during fixation, the YFP and mRFP fluorescence signals were too weak to discriminate with enough confidence the FP-specific signals. To overcome this technical hurdle, the thickness of ultrathin sections was increased from 70 to 150 nm to maximize the total number of FPs within a single section. Precautions were made during sample mounting to permit the complete water immersion of the section by maintaining the grid on a drop of water with a thin pin during the positioning of the cover slip. In order to improve the visualization of FPs and because the TEM grids were not perfectly flat, Z-stack acquisitions followed by maximal Z-projections were systematically performed. Overall, YFP detection required a higher excitation power and a numerical compression of the intensity levels compared with mRFP. Thus, mRFP fluorescence is less affected during TEM sample preparations than YFP (Supplemental Figure S1). We also noted that the TEM-sample-embedding process induced a shift in the chlorophyll emission spectra so that it overlaps with the mRFP spectra (Supplemental Figure S2). Despite this, cells expressing mRFP and YFP ER-markers were reliably differentiated and graft interfaces were targeted without ambiguity (Figure 2). Following confocal acquisition, the TEM observation of the same section gave access to the ultrastructural data. Correlation between the images obtained from both microscopies was based on the overlay of natural markers (the cell shapes, plastids, and nuclear envelops), there was no ambiguity in the identity and the ultrastructure of the cells except for completely differentiated tracheid and sieve elements. This is because the complete differentiation of sieve and tracheid elements results in loss or modification of ER, which was no longer labeled with HDEL-markers (Figure 2, E and F and Supplemental Figure S3). Despite this loss of labeling, the ultrastructure details permitted us to identify the vascular tissues and to study the sieve and tracheid elements continuities between both grafted partners at the graft interface (Figure 2). Magnified observations permitted us to evaluate the level of ultrastructural preservation. The cell compartmentation was conserved with notably the maintenance of the integrity of chloroplasts, nuclei, Golgi, and vesicles (Figure 2, G and H). An important technical advance, compared with previous CLEM protocols in plants (Bell et al., 2013; Marion et al., 2017; Wang and Kang, 2020), is the capacity to reveal 3-D shapes with high ultrastructural details thanks to ET. The different parts of PD, such as the bilayer of plasma membrane and the desmotubule was spatially resolved indicating the high quality of the ultrastructure preservation despite the absence of strong chemical fixative agents (Figure 2, I).
Figure 2.
CLEM as a tool to investigate the ultrastructure of the graft interface. The scion and the rootstock are, respectively, at the top and bottom positions (A–C, E, and F). A, Confocal acquisition of the graft interface of a 35S::HDEL_mRFP scion grafted onto a 35S::HDEL_YFP rootstock. B, Transmission electron micrograph tiles of the regions observed in (A). C, Correlated light electron microscopy view of the graft interface which allows the precise identification of the scion and rootstock cells. D, Thanks to correlative images, the “map” of the graft interface (red dotted line) is made and allows us to identify the scion (sc), the rootstock (rt), phloem sieve element (yellow), and xylem vessels (green). E, Correlated view of sieve elements (SE) and companion cells (CC) aligned at the interface. F, Correlated view of xylem tracheary element (TE) properly aligned at the graft interface. G, Electron micrograph of a cell wall that also shows the Golgi apparatus (G), mitochondria (m), and PD (H) its corresponding tomogram that increases the resolution and shows the preservation of the membranes of the organelles. I, Tomogram showing the PD plasma membrane (white arrow) and the desmotubule membranes. White arrowheads denote “natural landmarks,” i.e. plastids, nuclei, that were used to correlate confocal and transmission electron microscopy images. Scale bars: (A–D) 20 µm, (E) 10 µm, (F) 5 µm, (G and H) 0.5 µm, and (I) 0.2 µm.
Ultrastructure of PD at the graft interface
We took advantage of our method of CLEM and the accurate targeting of the graft interface to study the PD formed between the scion and rootstock. PD were already present from 3 DAG and every PD observed contained a desmotubule structure in their center and often had a constricted neck region where the plasma membrane and desmotubule were in very close interaction (Figure 3). We defined four classes of PDs based on their shapes (Figure 3, A). Class I PD were found in 32.4% of cases and correspond to isolated straight, simple stranded PD crossing the cell wall. The class II was characterized by tortuous PD containing in total from three to six branches with compressed neck regions. These branched PD represented 20% of observed PD. Class III PD corresponds to twinned PD, which were defined as two straight, simple stranded PD (class I) closer to each other than their average length spanning the cell wall; this class of PD was rarely observed (7.6%). Unlike classes I–III, class IV, which corresponded to the most frequently found PD at the graft interface (39.4%), showed the unique feature of not spanning the entire cell wall separating the scion and rootstock (Figure 3, E and F). These PD, called hemi-PD, were often seen forming loop-like structures with cytosolic ends opening within the same cell. On two occasions, we observed an unprecedented desmotubule with a “ball of wool”-like structure where compressed ER had rolled onto itself in a huge central cavity (Figure 3, F). We interpreted hemi-PD as incomplete attempts at secondary PD formation. A similar frequency of hemi-PD was observed on the scion and rootstock sides of the graft interface indicating that the scion and rootstock are similarly able to insert PD at the graft interface. Thus, the cell machinery for the ER maturation in desmotubules and the cell wall modifications seem to be functional in both grafted partners.
Figure 3.
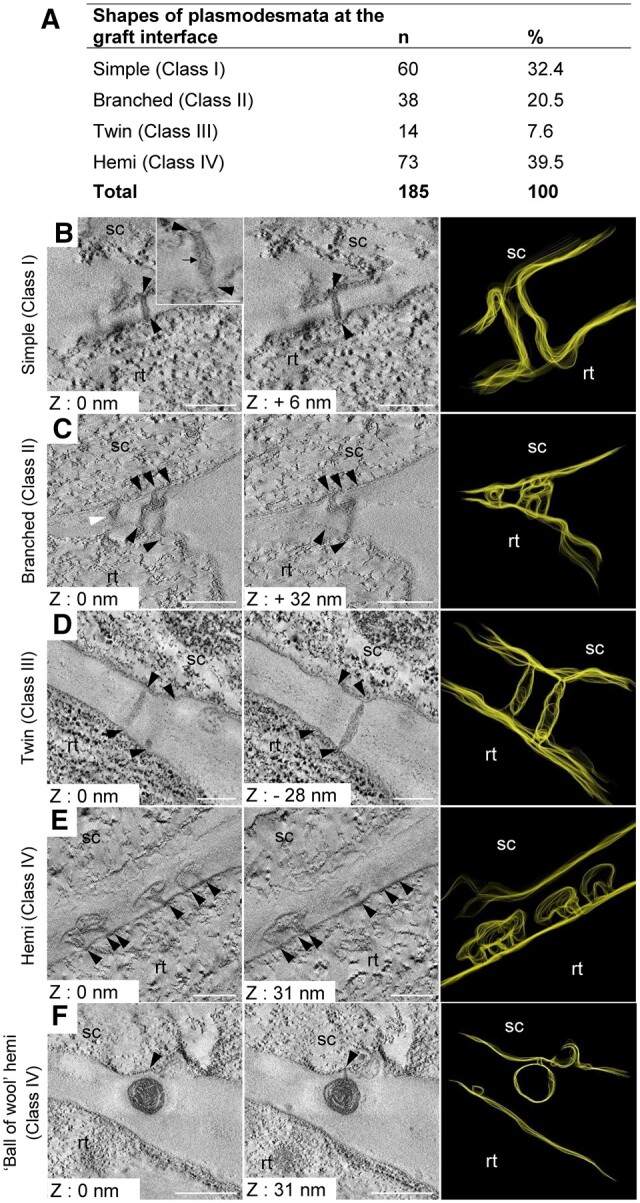
Electron tomograms and 3-D segmentation of simple, branched, and twin PD. Number and proportion of the different shapes of PD at the graft interface. B–F, The scion (sc) and the rootstock (rt) are at the top and bottom positions, respectively. Black arrowheads pinpoint the extremities of simple (B), branched (C), and twin (D) PD. For each shape of PD, two Z plans and one 3-D segmentation are presented. B, Simple PD with a visible desmotubule (black arrow). C, Branched PD with three branches on the scion side and two branches on the rootstock side, and another PD in its vicinity (B, white arrowhead). D, Twin PD characterized by the close proximity of two simple PD. E, Hemi-PD do not span the cell wall and have a large central cavity. F, Hemi-PD with a ball of wool shape have an aberrant accumulation of desmotubule in this large central cavity. Scale bars: (B–E) 0.1 µm, (F) 0.5 µm, and inset in (B) 50 nm.
In order to identify events leading to successful symplastic connections between the scion and rootstock, hemi-PD (class IV) and PD of classes I–III were compared. The measurements of the thickness of the cell wall around 184 PD (60 simple, 38 branched, 14 twin, and 72 hemi) revealed that the median of the cell wall thickness around class IV was more than two times thicker (261 nm) than around PD that crossed the cell wall (121 nm) (Figure 4). However, no significant difference was found between the classes I–III that successfully made symplastic connections between the scion and rootstock (Supplemental Figure S4). Interestingly, the median of the penetration depth of hemi-PD into the cell wall (99 nm, with a minimum of 38 and a maximum of 353 nm) was equivalent to the cell wall thickness around classes I–III (with a minimum of 40 and a maximum of 528 nm, Figure 4). This suggests that cell wall thinning could be a prerequisite for the establishment of cell-to-cell connections between the scion and the rootstock.
Figure 4.
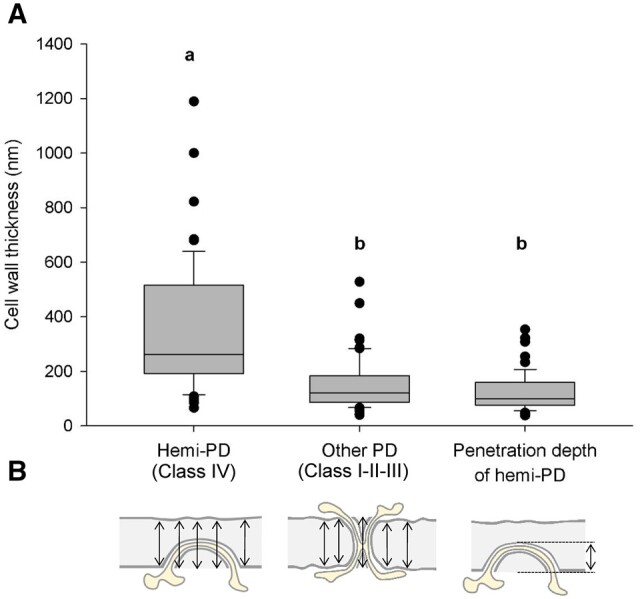
Box plots of the thickness of the cell wall around different classes of PD versus penetration depth of hemi-PD at the graft interface. A, Box plots showing the median (center line), upper and lower quartiles (box limits), and 10th and 90th percentiles (whiskers) and outliers (points). Letters indicate the results of ANOVA on ranks and Dunn’s tests (P-value < 0.05). B, Scheme illustrating the method of cell wall thickness measurements. To assess cell wall thickness around PD represented in (A), five measurements were done in the vicinity of PD and mean thickness was calculated. For the penetration depth of hemi-PD, only one measurement was taken of the maximal PD penetration into the cell wall. Data were generated using 10 biological samples in total coming from five experimental repetitions.
In order to catch PD biogenesis events, we decided to focus on regions where the cell wall is thin. ET allowed us to reveal ultrastructural details invisible using conventional 2-D TEM projections. ET showed that the cell wall thickness at the graft interface could be reduced down to 6 nm causing both plasma membranes to be extremely close together (Figure 5, A–C). Exceptionally, in one case of extreme cell wall thinning, we caught an event of organelle transfer from one cell to another through a cell wall gap (Supplemental Figure S5). In this particular graft interface, we also found ER-YFP and ER-RFP in the same cells (Supplemental Figure S6). The regions of extreme cell wall thinning occurred at sites where the plasma membrane and ER were in close contact (Figure 5, D–F), suggesting that it could be a preparation process for a PD insertion by one of the graft partners. Moreover, in regions of thin cell walls of 50 nm, we observed tortuous simple PD spanning the entire cell wall (Figure 5, G–I). This suggests that the involvement of only one partner could allow the formation of class I PD. On the contrary, in a thin cell wall of 25 nm, we observed a complex PD in a thicker cell wall spot of 113 nm (Figure 5, J–L). This complex PD had four arms on the scion side and one arm on the rootstock side, which met together at a central cavity. This complex PD associated with the local cell wall thickening could be the consequence of the meeting of two PD coming from both partners. In a neighboring cell, in a thicker cell wall of 134 nm, a hemi-PD was positioned in front of two hemi-PD coming from the other partner (Figure 5, M–O). Thus, in this case, both grafted partners inserted PD arms in the same site of the cell wall. Unfortunately, it is impossible to follow the ultrastructure developments of the same PD over time. Therefore, we cannot know whether these different arms would fuse together or not. We hypothesize that the large volume of the central cavity of hemi-PD could facilitate hemi-PD contact and fusion across the graft interface.
Figure 5.
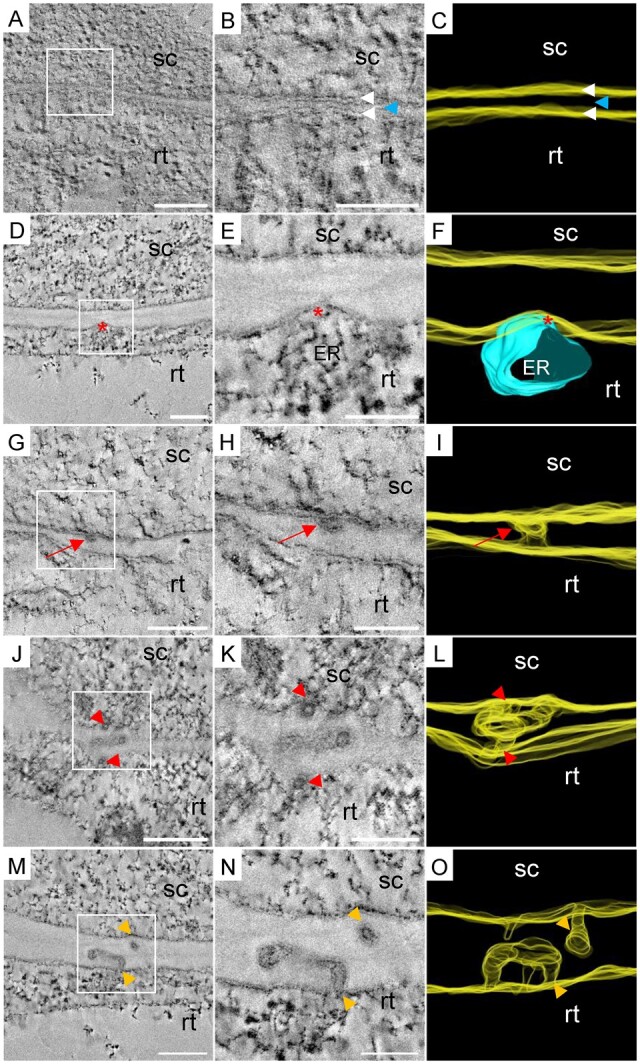
Electron tomograms and 3-D segmentations of PD biogenesis events at the graft interface. The scion (sc) and the rootstock (rt) are, respectively, at the top and bottom positions. A–C, Extreme thinning observed all along the cell wall between two grafted cells. White arrowheads indicate the bilayer of plasma membranes and the blue arrowhead indicates the residual cell wall between them. D–F, Localized thinning of the cell wall at the ER/plasma membrane contact site at the rootstock side (red asterisks). G–I, Simple PD (red arrows) observed in a thin cell wall. J–L, Complex PD in a localized thickening of a thin cell wall (entrances are depicted with red arrowheads). M–O, Hemi-PD facing each other across the graft interface (orange arrowheads). Scale bars: (A, D, G, J, M) 0.2 µm, (B, E, H, K, N) 0.1 µm.
Secondary PD formation at the graft interface seems to require cell wall remodeling with a thinning of the cell wall. In addition, the PD characteristics described here could indicate that PD could be formed by a mono-directional PD penetration in the cell wall in cases where the cell wall is thinner than 100 nm. In the case of thicker cell walls, PD formation could depend on a synchronized spatial–temporal hemi-PD penetration from both graft partners.
Discussion
Understanding the cellular events occurring during graft union formation has long been a goal for plant scientists. The emergent high-end microscopy technique, CLEM, allowed us to target unambiguously the graft interface and characterize its ultrastructure in detail. This method allowed us to characterize different events occurring at the graft interface from 3 DAG such as cell differentiation, cell wall modifications, the establishment of communication between the scion and rootstock, and especially to determine the 3D structure of secondary PD formed at the graft interface.
Thanks to the absence of cell-to-cell diffusion of the HDEL motif, fluorescent labeling of ER lumen of scions and rootstocks allowed the accurate targeting of the graft interface. During sample preparation for TEM, despite the higher brightness of YFP (44.89 units) than mRFP (12.25 units), the mRFP signal was better preserved during sample processing than YFP. Our hypothesis to explain this difference is that in the acid resin environment (pHHM20 = 5), the fluorescent state of mRFP (i.e. the deprotonated state) is better maintained than the fluorescent state of YFP due to differences of their pKa (PkamRFP = 4,5; PkaYFP = 6,9); as has been shown for an autophagic flux probe (Kaizuka et al., 2016). Therefore, choosing FPs with low pKa could be an important parameter to take into account before starting a CLEM approach. Our method allows the preservation of both fluorescence and ultrastructure with a high resolution of around 3 nm, as such this CLEM method could be used to study the ultrastructural events relevant to other biological questions.
In A. thaliana homografts, we show that secondary PD are formed at the graft interface by at least 3 DAG (the first time point we were able to study for technical reasons). We distinguished four classes of PD: simple (I), branched (II), twin (III), and hemi (IV). The latter, hemi-PD, have the characteristics of not entirely spanning the cell wall and could correspond to failed attempts of PD formation. Hemi-PD were the class the most frequently found at the graft interface; therefore, it seems that even between cells of the same species, secondary PD biogenesis at the graft interface appears not to be systematically a successful event. Hemi-PD are found in the same proportions on the scion and rootstock sides of the graft interface demonstrating that the scion and the rootstock have the same capacity to insert PD into the cell wall.
Every class of PD had a desmotubule in the central part, which could be simple, branched, or forming a loop to a “ball of wool” shape. The formation of ball of wool shaped PD could be the consequence of hemi-PD having only one cytosolic opening, through which the desmotubule is sent, without the possibility of exit. This suggests that, PD progression into the cell wall is a directed, active mechanism where the desmotubule–plasma membrane interactions observed at the entrances of PD could play a central role. In addition, the accumulation of compressed desmotubule in the central cavity shows that the desmotubule compression is, at least sometimes, independent of direct plasma membrane–ER interactions.
The idea that cell wall modifications at the graft interface are required for the correct establishment of the graft union has been suggested previously (Jeffree and Yeoman, 1983; Kollmann and Glockmann, 1991; Notaguchi et al., 2020). The thickness of cell walls of un-grafted A. thaliana hypocotyls has been described in the literature (Derbyshire et al., 2007); cell wall thickness is approximately 150–200 nm in the inner epidermis, inner and outer cortex, and in the endodermis, and approximately 430 nm in the outer epidermis. In general, we found that PD spanning the cell wall at the graft interface was found in particularly thin cell walls. In addition, extreme cell wall thinning at the graft interface was observed; in rare cases, this thinning was so excessive that it leads to the formation of gaps in the cell wall and this could explain the presence of some cells that contain both red and yellow ER and nuclei in our experiments. The idea that gaps formed between the cells of the scion and the rootstock is supported by the observation of local horizontal transfer of organelles between the scion and rootstock of grafted plants (Stegemann and Bock, 2009; Stegemann et al., 2012; Lu et al., 2017; Hertle et al., 2021). Cell wall remodeling could be essential to establishing symplastic connections between the rootstock and scion, and cell wall remodeling could be particularly concentrated at the ER–plasma membrane contact sites. In fact, our results suggest that the PD progression through the cell wall is impaired when the cell wall is insufficiently thinned. This could be because of the loss of sufficient contact between the desmotubule and the plasma membrane in the middle of cell walls that have not undergone adequate thinning. In the absence of contact between the desmotubule and the plasma membrane, the desmotubule could lose its sense of direction and could form a loop to come back to its “home-cell” (hemi-PD), or get lost in a central cavity (ball of wool shaped PD). Thus, the contact between the ER and the plasma membrane could be a key event in the process of the secondary PD biogenesis by participating in the targeted thinning of the cell wall at PD entry sites and by driving the desmotubule across the cell wall into the neighboring cell. In summary, the formation of secondary PD that span the cell wall entirely seems to require cell wall thinning and ER–plasma membrane tethering sites.
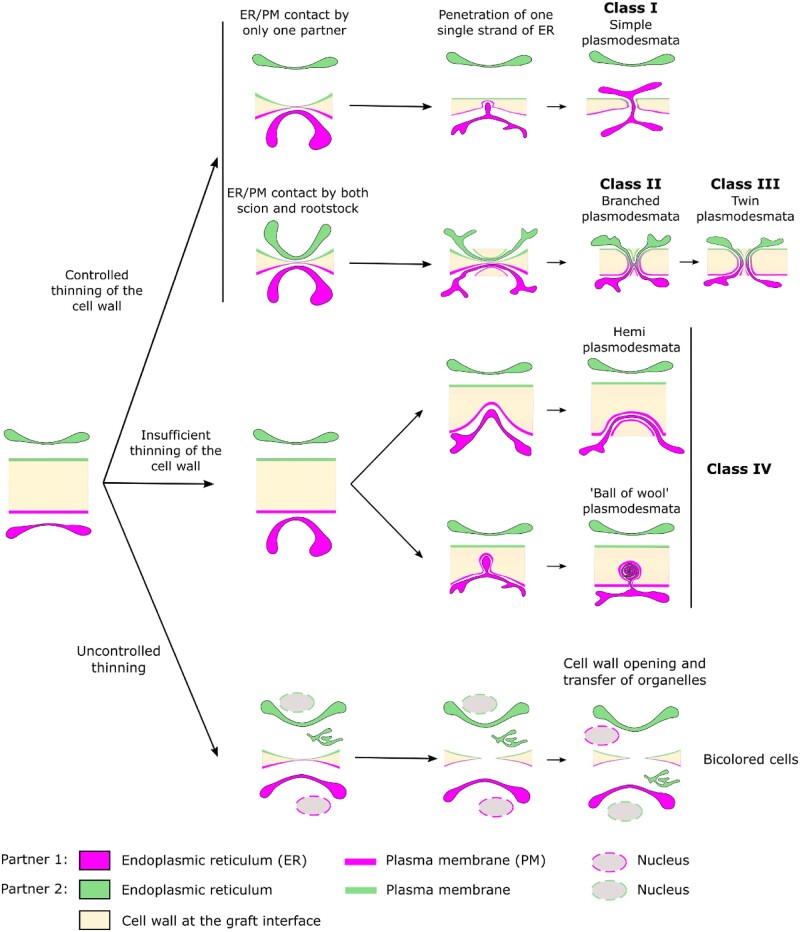
The presence of four different PD classes could be the result of several biogenesis mechanisms or different maturation states of PD illustrated in Figure 6. Our results show that in the thin cell wall at the graft interface, new secondary PD can belong to the class I, simple PD, potentially formed by a directed active progression of the desmotubule across the cell wall. These class I PD resemble de novo primary PD (Nicolas et al., 2017). The branched PD (class II) could result from the maturation of simple PD by the trapping of ER strands in continuity with the desmotubule during cell wall thickening, as seen in the maturation of primary PD (Faulkner et al., 2008). This idea is supported by the fact that newly divided cells have a majority of simple PD and that mature cells have a majority of branched PD (Oparka et al., 1999; Nicolas et al., 2017). However, the presence of branched PD at the graft interface soon after grafting is not clearly associated with a general thickening of the cell wall. Furthermore, ultrastructural examinations indicate that at the graft interface branched PD could be in some cases the result of the fusion of two well-aligned hemi-PD, rather than a maturation process. For branched PD biogenesis at the graft interface, the scion and rootstock cells would have to initiate hemi-PD opposite each other at the same moment and at the same site (with a precision around several hundred nanometers in 3D). In this case, the plasma membranes and the desmotubules of both grafting partners have to fuse in order to establish the symplastic continuity between the scion and the rootstock. This symmetric process, on either side of the cell wall at the graft interface, would require the precise spatial–temporal coordination of processes between the scion and the rootstock. This coordination could be mediated by signals produced during cell wall remodeling (for reviews: Anderson and Kieber, 2020; Duman et al., 2020) and/or by local ER–plasma membrane interactions (for review: Li et al., 2021a). The genesis of a large central cavity, by for example the formation of hemi-PD running in parallel to the cell wall, could increase the chance of fusing of hemi-PD on either side of the graft interface. If the cell wall is too thick, or the scion and rootstock are not well coordinated, desmotubule entry into the cell wall by only one of the grafted partners occurs, PD progression into the cell wall is deviated and PD evolve into a loop-form, and finally, a hemi-PD is produced. It is possible that branched PD turn into twin PD (class III) during cell wall extension as this mechanism could permit a relatively efficient increase of the number of PD (Faulkner et al., 2008).
Figure 6.
A model of secondary PD biogenesis at the graft interface. Class I PD could result from unidirectional desmotubule entry by one partner and extreme cell wall thinning. Class II PD could be the consequence of synchronized and symmetric desmotubule entry into a thinned cell wall by both the scion and the rootstock. Subsequently, during cell wall extension, Class II PD could mature into Class III PD. Class IV could correspond to de novo PD formation attempts which are incomplete due to an insufficient thinning of the cell wall at the graft interface. The existence of “ball of wool” shape PD suggests that desmotubule entry into the cell wall is an active mechanism. The presence of PD of Class IV in the same proportions in the scion and the rootstock suggest that both grafting partners are able to initiate PD biogenesis at the graft interface. In addition, bicolored cells at the graft interface could be the consequence of uncontrolled cell wall thinning leading to the formation of cell wall gaps through which nucleus and ER could transit from one partner to the other.
This work provides a view of cell-to-cell connections through PD: it suggests that desmotubules are more than structural components of PD, but could also be dynamic ER compartments actively moving through the cell wall to make PD connections from cell-to-cell. In the future, our CLEM approach could be further exploited to phenotype mutant lines of A. thaliana to identify genes involved in the formation of the graft interface as well as providing an improved understanding of secondary PD biogenesis. Thanks to the excellent preservation of both fluorescence and ultrastructural details of this CLEM method, it could also be used on other plant tissues and to answer biological questions in the future.
Materials and methods
Plant material, growth conditions, and plant grafting
Arabidopsis thaliana were grown vertically in a growth chamber on solid medium composed of half-strength Murashige and Skoog medium including vitamins (1/2 MS) and plant-Agar (8 g L−1), pH 5.8. Growth conditions were set at 22°C in a growth chamber with a 10-h photoperiod with a photosynthetic photon flux density of 120 µmol m−2 s−1. All lines 35S::HDEL_mRFP, 35S::HDEL_YFP, and 35S::GFP were previously published (Nelson et al., 2007; Lee et al., 2013; Clark et al., 2016). The 35S::TagRFP lines were produced in a Col-0 background.
For grafting, 7-d-old plants were selected and grafted using the transverse cut and butt alignment method described by Melnyk (2017). This method was slightly modified: ultra pure water plus plant-Agar (1.6 g L−1) was used instead of humid paper to maintain the moisture level required for graft union formation.
High pressure freezing and cryosubstitution
Copper carriers (100 µm deep and 1.5 mm wide, Leica Microsystems, catalog number 16707898) were filled with ultra pure water plus 20% w/v bovine serum albumin (BSA) (which functions as a cryoprotectant). Rapidly, grafts at 3 or 6 DAG were cut above and below the graft point and installed in the well of the carrier. Samples were frozen in the carriers using an EM-PACT high-pressure freezer (Leica) and then transferred at −90°C into an AFS 2 freeze-substitution device (Leica). Grafts were incubated in a cryosubstitution mix containing only uranyl acetate 0.1% w/v in pure acetone for 30 h (freeze substitution and embedding protocol adapted from Nicolas et al., 2018). Afterward the temperature was raised progressively to −50°C, at 3°C h−1. The cryosubstitution mix was removed and thoroughly washed three times with pure acetone and then three times in pure ethanol. Low temperature and the limited amount of chemical compounds help in preserving the fluorescence of FPs while preserving a sufficient staining for TEM and ET. Samples were then carefully removed from the carriers before progressive embedding in HM20 Lowicryl resin (Electron Microscopy Science): HM20 25% v/v, 50% v/v (2 h each), 75% v/v (overnight), 100% (twice for 2 h), and a last 100% for 8 h. The polymerization was done under UV light for 24 h at −50°C followed by 12 h at +20°C. Samples in resin blocks were stored at −20°C protected from light.
Microscope acquisitions and correlations
Prior to observation, sections were made with an EM UC7 ultramicrotome (Leica). For an optimal balance between resolution under the electron microscope and signal quantity under the confocal microscope, sections from 150 to 200 nm were collected on parlodion-coated 200-square mesh copper grids with thin bars (Electron Microscopy Science). Fluorescence Z-stack acquisitions were made on a Zeiss LSM 880 with excitation 488 nm (with a power from 1.5% to 26%) for YFP, and 561 nm (with a power from 1.5% to 3%) for mRFP on the spectral detector (from 414 to 691nm) with a gain set at 900. The acquisitions were achieved with a 63× apochromatic N.A 1.4 oil objective. The spectral deconvolution was performed on Zen black software by using YFP and mRFP emission spectra in HM20. The tile acquisition tool of the Zen black software was used to get the large field of view with the best resolution. Moreover, the Z-stack tool was use to acquire the maximum of photons emitted by the samples. Afterward maximal or summed Z-projections were used to improve the visualization. Transmission electron microscopy observations were carried out on a FEI TECNAI Spirit 120 kV electron microscope equipped with an Eagle 4Kx4K CCD camera. Correlation was based on natural landmarks such as cell shapes, plastids, and nucleus. Correlated images were obtained thanks to the plugin ec-Clem (Paul-Gilloteaux et al., 2017) of Icy software (De Chaumont et al., 2012).
Tomogram acquisition and reconstruction
We used the process described by Nicolas et al. (2018). Before tomogram acquisition, sections need to be coated with gold fiducials for subsequent alignment, a 1:1 mixture of 0.5% BSA w/v, and 5 nm colloidal gold solution from BBI solutions (referenced as EM-GC5 on http://bbisolutions.com) was used. Tilt series were acquired with a single tilt specimen holder (Fischione Instruments, model-2020), using the batch mode of FEI 3D Explore tomography software. Each tilt series was acquired between −65° to 65° when it was possible, with an acquisition at every degree. For dual axis, the grid was turned to 90° manually. The raw tilt series were aligned and then reconstructed using the fiducial alignment mode with the eTomo software (http://bio3d.colorado.edu/imod/). A total of 10–25 fiducials were used to accurately align all images. Reconstruction was performed using the back-projection with SIRT-like filter (10–50 iterations).
Image processing and analysis
Image analysis was carried out using the ImageJ software (https://imagej.nih.gov/ij/download.html) with the Bio-Format plugin (https://www.openmicroscopy.org/bio-formats/) to open and process the Zeiss .czi files and to open the .mrc tilt series files and tomograms. Measurement of cell wall thickness was done using ImageJ. Mosaic images from the confocal microscope were assembled with the BigStitcher plugin of Fiji. Mosaic images of TEM were assembled using Photomerge function of Adobe Photoshop. No image analysis and measurements were carried out on mosaic and correlated images, they were only used as a visualization tools.
Data analysis
The thickness of cell walls around PDs was done by measuring the cell wall thickness at five locations distributed in the vicinity of PD followed by the calculation of the mean thickness of the cell wall around each PD. Cell wall thickness was compared with the Kruskal–Wallis one way analysis of variance on ranks, to isolate the group or groups that differ from the others. The Dunn’s Method was used on all pairwise comparisons. Data analysis and box plot presentation were done using SigmaPlot, Systat Software Inc.
Supplemental data
Supplemental Figure S1. Histogram display and laser power.
Supplemental Figure S2. Emission spectra of chlorophyll, mRFP, and YFP in HM20 section of a graft interface.
Supplemental Figure S3. CLEM as a tool to investigate the ultrastructure of the graft interface.
Supplemental Figure S4. Box plots of the thickness of the cell wall near to the PD found at the graft interface of A. thaliana by class of PD.
Supplemental Figure S5. CLEM reveals cell wall gap through which organelle is moving from cell-to-cell close to the graft interface at three DAG.
Supplemental Figure S6. Bicolored cells at the graft interface of A. thaliana 35S::HDEL_YFP (scion) on 35S::HDEL_mRFP (rootstock) three DAG.
Supplementary Material
Acknowledgments
All sample preparation and imaging were done on the Pôle Imagerie du Végétale, appended to the Bordeaux Imaging Centre (http://www.bic.u-bordeaux.fr/) which is a part of the France BioImaging Infrastructure (https://france-bioimaging.org/). Claire Brehelin provided transgenic lines. We thank Christophe Garcion for the use of his plant culture room and Charles Melnyk for showing us the grafting procedure. Research support was provided by the ORIGINE project of the Plan National Dprissement du Vignoble (FranceAgriMer-22001149-00001505).
Funding
The master 2 and the Ph.D. thesis of C.C. was financed by: the grant “PEPS2015 Super-CLEM-3D”, the grant “Etude des mécanismes de l’union greffon/porte-greffe”, 50% from the INRAE department Plant biology and breeding and 50% from the Région Nouvelle Aquitaine, France. Further financial support was provided by the Starter grant “Etude des mécanismes de l’union greffon/porte-greffe” from the INRAE department Plant biology and breeding. Research support was provided by the ORIGINE project of the Plan National Deperissement du Vignoble (FranceAgriMer-22001149-00001505).The Region Aquitaine also supported the acquisition of the electron microscope (Grant No. 2011 13 04 007 PFM). The “Contrat-Plan-Etat-Region PMUSCIVI-Plateforme mutualisée en sciences du vivant 2015_2020” provided the funding for the acquisition of the confocal microscope.
Conflict of interest statement. None declared.
L.B., S.J.C., N.O., and E.B. conceived and designed the experiments. C.C. and L.B. performed the experiments, analyzed the data, and wrote the first draft of the article. All authors revised and contributed to the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is lysiane.brocard@inrae.fr.
References
- Anderson CT, Kieber JJ (2020) Dynamic construction, perception, and remodeling of plant cell walls. Annu Rev Plant Biol 71: 39–69 [DOI] [PubMed] [Google Scholar]
- Bell K, Mitchell S, Paultre D, Posch M, Oparka K (2013) Correlative imaging of fluorescent proteins in resin-embedded plant material. Plant Physiol 161: 1595–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-alfonso Y, Cilia M, San A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidadi H, Matsuoka K, Sage-Ono K, Fukushima J, Pitaksaringkarn W, Asahina M, Yamaguchi S, Sawa S, Fukuda H, Matsubayashi Y, et al. (2014) CLE6 expression recovers gibberellin deficiency to promote shoot growth in Arabidopsis. Plant J 78: 241–252 [DOI] [PubMed] [Google Scholar]
- Brault ML, Petit JD, Immel F, Nicolas WJ, Glavier M, Brocard L, Gaston A, Fouché M, Hawkins TJ, Crowet J, et al. (2019) Multiple C2 domains and transmembrane region proteins (MCTP s) tether membranes at plasmodesmata. EMBO Rep 20: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrill LC, Overall RL, Goodwin PB (1999) Cell-to-cell communication via plant endomembranes. Cell Biol Int 23: 653–661 [DOI] [PubMed] [Google Scholar]
- De Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al. (2012) Icy: An open bioimage informatics platform for extended reproducible research. Nat Methods 9: 690–696 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhao J, Hu F, Qin Y, Wang X, Hu G (2017) Transcriptome changes between compatible and incompatible graft combination of Litchi Chinensis by digital gene expression profile. Sci Rep 7: 3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G, Blilou I, Gratton E, Benfey PN, Sozzani R (2016) Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5: e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, Findlay K, McCann MC, Roberts K (2007) Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J Exp Bot 58: 2079–2089 [DOI] [PubMed] [Google Scholar]
- Ding B, Turgeon R, Parthasarathy MV (1992) Substructure of freeze-substituted plasmodesmata. Protoplasma 169: 28–41 [Google Scholar]
- Duman Z, Eliyahu A, Abu-Abied M, Sadot E (2020) The contribution of cell wall remodeling and signaling to lateral organs formation. Isr J Plant Sci 67: 110–127 [Google Scholar]
- Ehlers K, Kollmann R (2001) Primary and secondary plasmodesmata : structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Faulkner C, Akman OE, Bell K, Jeffree C, Oparka KJ (2008) Peeking into pit fields: A multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell Online 20: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaishman MA, Loginovsky K, Golobowich S, Lev-Yadun S (2008) Arabidopsis thaliana as a model system for graft union development in homografts and heterografts. J Plant Growth Regul 27: 231–239 [Google Scholar]
- Furuta K, Lichtenberger R, Helariutta Y (2012) The role of mobile small RNA species during root growth and development. Curr Opin Cell Biol 24: 211–216 [DOI] [PubMed] [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK (1982) Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111: 121–133 [Google Scholar]
- Hertle AP, Haberl B, Bock R, Bock R (2021) Horizontal genome transfer by cell-to-cell travel of whole organelles. Sci Adv 7: eabd8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree CE, Yeoman MM (1983) Development of intercellular connections between opposing cells in a graft union. New Phytol 93: 491–509 [Google Scholar]
- Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N (2016) An autophagic flux probe that releases an internal control. Mol Cell 64: 835–849 [DOI] [PubMed] [Google Scholar]
- Kollmann R, Glockmann C (1985) Studies on graft unions—I. Plasmodesmata between cells of plants belonging to different unrelated taxa. Protoplasma 124: 224–235 [Google Scholar]
- Kollmann R, Glockmann C (1991) Studies on graft unions—III. On the mechanism of secondary formation of plasmodesmata at the graft interface. Protoplasma 165: 71–85 [Google Scholar]
- Kollmann R, Yang S, Glockmann C (1985) Studies on graft unions—II. Continuous and half plasmodesmata in different regions of the graft interface. Protoplasma 126: 19–29 [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JAG (2012) Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol 111: 235–257 (doi: 10.1016/B978-0-12-416026-2.00013-3) [DOI] [PubMed] [Google Scholar]
- Kyriacou MC, Rouphael Y, Colla G, Zrenner R, Schwarz D (2017) Vegetable grafting: The implications of a growing agronomic imperative for vegetable fruit quality and nutritive value. Front Plant Sci 8: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L (2013) An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol 197: 481–489 [DOI] [PubMed] [Google Scholar]
- Li C, Qian T, He R, Wan C, Liu Y, H Yu (2021a) Endoplasmic reticulum–plasma membrane contact sites: Regulators, mechanisms, and physiological functions. Front Cell Dev Biol (doi: 10.3389/fcell.2021.627700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZP, Paterlini A, Glavier M, Bayer EM (2021b) Intercellular trafficking via plasmodesmata: molecular layers of complexity. Cell Mol Life Sci 78: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, White RG, Waterhouse PM (2012) Gene silencing in Arabidopsis spreads from the root to the shoot, through a gating barrier, by template-dependent, nonvascular, cell-to-cell movement. Plant Physiol 159: 984–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Stegemann S, Agrawal S, Karcher D, Ruf S, Bock R, Lu Y, Stegemann S, Agrawal S, Karcher D, et al. (2017) Horizontal transfer of a synthetic metabolic pathway between plant species report horizontal transfer of a synthetic metabolic pathway between plant species. Curr Biol 27: 3034–3041.e3 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Lee J-Y (2004) Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol 5: 712–726 [DOI] [PubMed] [Google Scholar]
- Marion J, Le Bars R, Satiat-Jeunemaitre B, Boulogne C (2017) Optimizing CLEM protocols for plants cells: GMA embedding and cryosections as alternatives for preservation of GFP fluorescence in Arabidopsis roots. J Struct Biol 198: 196–202 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Sugawara E, Aoki R, Takuma K, Terao-Morita M, Satoh S, Asahina M (2016) Differential cellular control by cotyledon-derived phytohormones involved in graft reunion of arabidopsis hypocotyls. Plant Cell Physiol 57: 2620–2631 [DOI] [PubMed] [Google Scholar]
- Melnyk CW (2017) Grafting with Arabidopsis. Methods Mol Biol 1497: 9–18 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Gabel A, Hardcastle TJ, Robinson S, Miyashima S (2018) Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc Natl Acad Sci USA 115: E2447–E2456 (doi: 10.1073/pnas.1718263115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk CW, Schuster C, Leyser O, Meyerowitz EM (2015) A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol 25: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge K (2009) A history of grafting. Hortic Rev 35: 437–494 [Google Scholar]
- Nelson BK, Cai X, Nebenfu A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Bayer E, Brocard L (2018) Electron tomography to study the three-dimensional structure of plasmodesmata in plant tissues–from high pressure freezing preparation to ultrathin section collection. Bio-protocol 8: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Trépout S, Gaston A, Fouché M, Cordelières FP, Oparka K, Tilsner J, Brocard L, Bayer EM (2017) Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat Plants 3: 17082 (doi: 10.1038/nplants.2017.82) [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Kurotani KI, Sato Y, Tabata R, Kawakatsu Y, Okayasu K, Sawai Y, Okada R, Asahina M, Ichihashi Y, et al. (2020) Cell-cell adhesion in plant grafting is facilitated by b-1,4-glucanases. Science 369: 698–702 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Cruz SS, Roberts I, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Ota R, Ohkubo Y, Yamashita Y, Ogawa-Ohnishi M, Matsubayashi Y (2020) Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat Commun 11: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Gilloteaux P, Heiligenstein X, Belle M, Domart MC, Larijani B, Collinson L, Raposo G, Salamero J (2017) EC-CLEM: Flexible multidimensional registration software for correlative microscopies. Nat Methods 14: 102–103 [DOI] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ (1988) Sorting of soluble ER proteins in yeast. EMBO J 7: 1757–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Nieminen K, Pacheco-Villalobos D, Sibout R, Schwechheimer C, Hardtke CS (2011) Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 23: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala K, Karcz J, Rypień A, Kurczyńska EU (2019) Unmethyl-esterified homogalacturonan and extensins seal Arabidopsis graft union. BMC Plant Biol 19: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone D, Paultre G, Gustin M-P, Molnar A, Oparka KJ. (2016) Lost in transit : Long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell 28: 2016–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Bock R (2009) Exchange of genetic material between cells in plant tissue grafts. Science 324: 649–651 [DOI] [PubMed] [Google Scholar]
- Stegemann S, Keuthe M, Greiner S, Bock R (2012) Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA 109: 2434–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Burch-smith T, Kim I, Meinke D, Mindrinos M, Zambryski P (2009) Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Cooke TJ, Connelly PS, Tilney MS (1991) The structure of plasmodesmata as revealed by plasmolysis, detergent extraction, and protease digestion. J Cell Biol 112: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Yanagisawa N, Kawakatsu Y, Ikematsu S, Sawai Y, Tabata R, Arata H, Higashiyama T, Notaguchi M (2020) Micrografting device for testing systemic signaling in Arabidopsis. Plant J 103: 918–929 [DOI] [PubMed] [Google Scholar]
- Wang P, Kang BH (2020) Correlative light and electron microscopy imaging of the plant trans-Golgi network. Methods Mol Biol 2177: 59–67 [DOI] [PubMed] [Google Scholar]
- Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PGM, Hariharan N, Kim JY, Jackson D (2011) Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Zhang W, Kollwig G, Stecyk E, Apelt F, Dirks R, Kragler F (2014) Graft-transmissible movement of inverted-repeat-induced siRNA signals into flowers. Plant J 80: 106–121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.