Abstract
UV RESISTANCE LOCUS 8 (UVR8) mediates photomorphogenic responses and acclimation to UV-B radiation by regulating the transcription of a series of transcription factors (TFs). However, the origin and evolution of UVR8-mediated signaling pathways remain largely unknown. In this study, we investigated the origin and evolution of the major components of the UVR8-mediated signaling pathway (UVR8, REPRESSOR OF UV-B PHOTOMORPHOGENESIS [RUP], BRI1-EMS-SUPPRESSOR1 [BES1], BES1-INTERACTING MYC-LIKE 1 (BIM1), WRKY DNA-BINDING PROTEIN 36 (WRKY36), MYB DOMAIN PROTEIN 73/77/13 [MYB73/MYB77/MYB13], and PHYTOCHROME INTERACTING FACTOR 4/5 [PIF4 and PIF5]) using comparative genomics and phylogenetic approaches. We showed that the central regulator UVR8 presented a conservative evolutionary route during plant evolution, and the evolutionary history of downstream negative regulators and TFs was different from that of green plant phylogeny. The canonical UVR8-CONSTITUTIVELY PHOTOMORPHOGENIC 1(COP1)/SUPPRESSOR OF PHYA-105 (SPA)-ELONGATED HYPOCOTYL 5 (HY5)-RUP signaling pathway originated in chlorophytes and conferred green algae the additional ability to cope with UV-B radiation. Moreover, the emergence of multiple UVR8-mediated signaling pathways in charophytes laid the foundations for the cross-talk between UV-B signals and endogenous hormone responses. Importantly, we observed signatures that reflect plant adaptations to high UV-B irradiance in subaerial/terrestrial environments, including positive selection in UVR8 and RUPs and increased copy number of some vital TFs. These results revealed that green plants not only experienced adaptive modifications in the canonical UVR8-COP1/SPA-HY5-RUP signaling pathway, but also diversified their UV-B signal transduction mechanisms through increasing cross-talk with other pathways, such as those associated with brassinosteroids and auxin. This study greatly expands our understanding of molecular evolution and adaptive mechanisms underlying plant UV-B acclimation.
Multiple UV RESISTANCE LOCUS 8-mediated signaling pathways originated in chlorophytes or charophytes, facilitating the UV-B acclimation of plants in subaerial/terrestrial environments.
Introduction
The high-energy UV-B radiation (280–315 nm) in sunlight has a wide range of detrimental effects on organisms. Many ancient photosynthetic organisms (such as diatoms, cyanobacteria, and green algae) have evolved the capacity to synthesize sunscreen to protect themselves against UV-B damage (Rozema et al., 2002; Balskus and Walsh, 2010). In addition to the adverse effects of high UV-B irradiance, it has been reported that low-level UV-B can mediate photomorphogenesis and stress responses in Arabidopsis (Arabidopsis thaliana) and other plant species, such as inhibition of hypocotyl elongation and accumulation of flavonoids and anthocyanins (Lee, 2016; Jenkins, 2017; Podolec et al., 2021). During the conquest of land, green plants have evolved complex mechanisms to appropriately respond to drastic changes of UV-B irradiance (Fernández et al., 2016; Han et al., 2019). Although a series of downstream transcription factors (TFs) and target genes responsive to UV-B radiation have been extensively studied in A. thaliana (Huang et al., 2014; Liang et al., 2018; Yang et al., 2018; Qian et al., 2020; Tavridou et al., 2020; Yang et al., 2020), the origin and evolution of the UV RESISTANCE LOCUS 8 (UVR8)-mediated signaling pathway in green plants remain largely unclear.
UVR8 has been characterized as the exclusive UV-B photoreceptor in green plants, and plays a vital role in UV-B perception and signaling (Rizzini et al., 2011; Liang et al., 2019). UVR8 was first identified in A. thaliana, and its orthologs were subsequently identified in other green plants, including chlorophytes, charophytes, bryophytes, and other angiosperms, except for gymnosperms (Favory et al., 2009; Tilbrook et al., 2016; Soriano et al., 2018; Han et al., 2019). Moreover, experimental analyses in green algae, moss, and liverwort revealed the functional conservation of UVR8 in UV-B perception and signal transduction (Tilbrook et al., 2016; Soriano et al., 2018). All the identified UVR8 possessed highly conserved functional domains: the RCC1 domain, the Gly-Trp-Arg-His-Thr (“GWRHT”) motif, and the Val-Pro (“VP”) core in the C27 domain (Wu et al., 2012; Jenkins, 2014a, 2014b). Three “GWRHT” motifs in UVR8 formed a tryptophan triad (W233, W285, and W337), and W233 and W285 were the main UV-B sensor (Christie et al., 2012; Podolec et al., 2021). There have been two different types of UVR8-mediated signaling pathway: (1) Without UV-B irradiation, UVR8 forms a homodimer in cytoplasm maintained by interactions of charged amino acids across the dimer interaction surface (Figure 1, A; O Hara and Jenkins, 2012; Liang et al., 2019). UV-B radiation triggers the dissociation of UVR8 homodimer, and the active UVR8 monomer inhibits the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), thereby stabilizing COP1 target TFs (ELONGATED HYPOCOTYL 5 [HY5]) that in turn promote UV-B-induced gene expression changes (Figure 1, B; Rizzini et al., 2011; Liang et al., 2019; Podolec et al., 2021). The monomeric UVR8 can be regulated by REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2, which facilitate the redimerization of UVR8 to complete the UVR8 photocycle (Gruber et al., 2010; Ren et al., 2019). (2) Current studies have shown that UVR8 can directly/indirectly interacts with specific TFs and transduce UV-B signal, such as WRKY DNA-BINDING PROTEIN 36 (WRKY36), BRI1-EMS-SUPPRESSOR1 (BES1), BES1-INTERACTING MYC-LIKE 1 (BIM1), MYB DOMAIN PROTEIN 73/77 (MYB73/MYB77), MYB DOMAIN PROTEIN 13 (MYB13), and PHYTOCHROME INTERACTING FACTOR 4/5 (PIF4 and PIF5) (Figure 1, B; Liang et al., 2018; Yang et al., 2018; Sharma et al., 2019; Qian et al., 2020; Tavridou et al., 2020; Yang et al., 2020). For instance, active UVR8 monomer interacts with WRKY36, and removes it from HY5, inducing the expression of HY5 (Yang et al., 2018). BES1 and BIM1 are two TFs in the brassinosteroids (BRs) signaling cascade, and UVR8 interacts with them to regulate the BR-induced genes (Liang et al., 2018). UVR8 inhibits lateral root development by interacting with MYB73/MYB77 in a UV-B-dependent manner (Yang et al., 2020). UVR8 also interacts with MYB13 to induce the expression of genes related to auxin response and flavonoid biosynthesis (Qian et al., 2020). It has been reported that UVR8 triggers the degradation of PIF4 and PIF5 to regulate hypocotyl growth (Sharma et al., 2019; Tavridou et al., 2020). Although these more recently discovered UV-B-mediated signaling pathways have been extensively characterized in A. thaliana at functional level, their origin and evolution in plant still need further investigations.
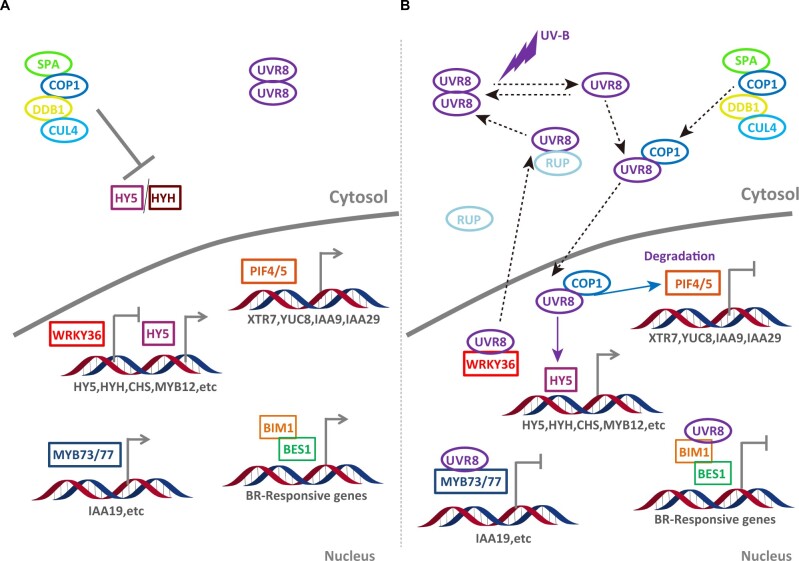
Figure 1.
Schematic summary of UVR8-mediated signaling pathways related to UV-B responses and photomorphogenesis. A, Without UV-B irradiation, UVR8 forms a homodimer in cytoplasm. B, UV-B radiation triggers the dissociation of UVR8 homodimer, and the UVR8 monomers are capable to interact with COP1, which can, in turn, regulate the expression of HY5. UVR8 regulates numerous potential transcriptional factors (WRKY36, BES1, BIM1, MYB73, MYB77, PIF4, and PIF5) to mediate the UV-B responses and photomorphogenesis. The negative feedback regulators RUP1 and RUP2 facilitate redimerization of UVR8 to complete the UV-B photocycle.
Green plants comprise the early-branching chlorophytes and streptophytes, and streptophytes are divided into two groups: streptophytes algae (charophytes) and land plants (embryophytes) (Leliaert et al., 2012; de Vries et al., 2016). It is documented that land plants originated from Zygnematophyceae (one clade of charophytes) (Zhong et al., 2013, 2014; Leebens-Mack et al., 2019; Su et al., 2021). Most chlorophytan green algae lived in water where UV-B doses are filtered, whereas subaerial/terrestrial charophytes and land plants are directly exposed to high doses of UV-B. Previous studies have showed that all chlorophytan green algae contained single-copy UVR8 orthologs, while some bryophytes, monocots, and eudicots possessed two or more copies (Fernández et al., 2016; Tilbrook et al., 2016; Han et al., 2019). The drastic changes in UV-B radiation might drive the evolution of UVR8-mediated signaling pathways in different lineages of green plants, and the changes in copy number might be an adaptive mechanism under heterogeneous evolutionary scenarios. Thus, a better understanding of the origin and evolution of UVR8-mediated signaling pathways would provide insights into the plant terrestrialization.
The conservation of structure and function of UVR8-COP1/SPA-HY5 complex has been widely studied, but the origin and evolution of other critical components (such as RUP, BES1, BIM1, WRKY36, MYB73, and MYB77) remained unclear. In this study, we performed extensive searches for orthologs of the vital components of UVR8-mediated signaling pathway using genomic and transcriptomic data of Archaeplastida. Multispecies comparative genomic, phylogenetic and positive selection analyses were conducted to investigate the gene gain/loss events and molecular adaptations in UVR8-mediated signaling pathway. Our studies provide comprehensive views of the evolutionary history of UVR8-mediated signaling pathway, and advance our understanding of the UV-B-responsive mechanisms during the plant evolution.
Results and discussion
The evolutionary conservation of UVR8
UV-B penetration in water is lower than that in terrestrial environments, and thus plants may co-evolve with ambient UV-B levels during the transition from aquatic to land. UVR8 is a plant-specific UV-B photoreceptor that can perceive low fluence rates of UV-B radiation to mediate plant metabolism and induce the photomorphogenic response (Huang et al., 2014; Jenkins, 2014a, 2014b). UVR8 uses the conserved tryptophans (W233, W285, and W337 in AtUVR8) in three “GWRHT” motifs to sense and transduce UV-B signal, rather than using typical chromophore (Wu et al., 2012). We conducted similarity searches to identify homologs of UVR8, and then reconstructed the phylogenetic tree of UVR8 homologs to systemically identify UVR8 orthologs. Homologs are genes that shared sequence similarities, and orthologs are genes in different species that have evolved from a common ancestral gene via speciation, suggesting high possibility of similar function retention of orthologs during evolution. Based on the similarity searches and phylogenetic analyses, we identified UVR8 orthologs containing the conserved “GWRHT” motif in major clades of green plants (i.e. chlorophytes, charophytes, bryophytes, lycophytes, ferns, gymnosperms, and angiosperms; Figure 2). No UVR8 orthologs were found in red algae and glaucophyte. Consistent with previous studies, our results also supported the notion that UVR8 was a specific photoreceptor in green plants (Fernández et al., 2016; Han et al., 2019).
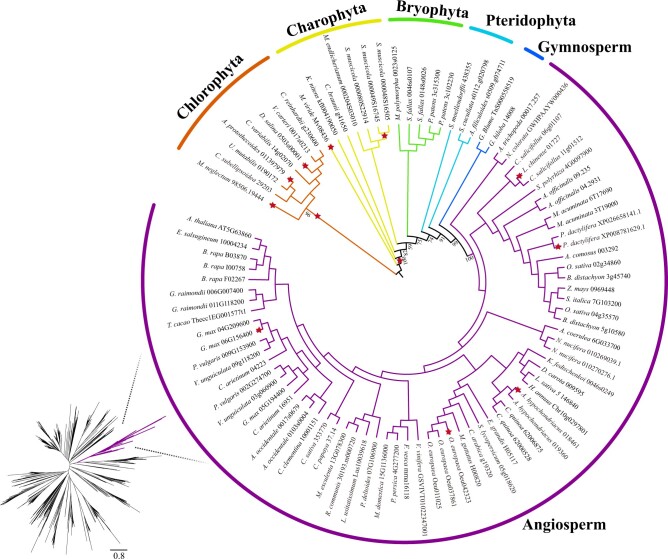
Figure 2.
Phylogenetic tree of plant UVR8 orthologs. The maximum-likelihood tree of UVR8 homologs was constructed using IQTREE with the best-fitting model (JTT + G4). The ML tree of UVR8 homologs (left) and the monophyletic group of plant UVR8 orthologs (right) were shown. Nodes under positive selection (aBSREL of Hyphy) were labeled by red asterisks.
We further inspected the phylogenetic tree of UVR8 orthologs, and found that it followed the widely accepted green plant phylogeny (Figure 2). Our phylogenetic results strongly supported that the earliest diverging clade of UVR8 was chlorophytan green algae, and the charophyte UVR8 formed a paraphyletic assemblage, covering four clades of charophyte algae (Figure 2). Our study reported that UVR8 orthologs with all conservative domains (the RCC1 domain, the “GWRHT” motif and the “VP” core) were identified in the genomes of ginkgo tree (Ginkgo biloba) and gnetophyte (Gnetum montanum) from gymnosperms. In terms of copy number of UVR8 orthologs, it was evolutionary conserved as single-copy gene across most clades of green plants (Figure 2 andSupplemental Figure S1). In addition, we noticed exceptions that Spirogloea muscicola from charophytes, Physcomitrium patens and Sphagnum fallax from mosses, and several angiosperms (e.g. rice [Oryza sativa], cotton (Gossypium raimondii), and Brassica rapa) possessed multiple copies of UVR8 genes. Interestingly, these species all experienced whole-genome duplications (WGDs) (Cheng et al., 2019; Gao et al., 2020; Wu et al., 2020). WGD events, followed by gene loss and retention, have been recognized as a vital force that drives the adaptations of plants to dramatic environmental changes (Jiao et al., 2011; De Smet and Van de Peer, 2012; Fox et al., 2020). For instance, P. patens possessed two copies of UVR8 genes, which enhances UV-B tolerance compared to A. thaliana (Wolf et al., 2010). In contrast, the marine angiosperm, Zostera marina, has lost UVR8 due to its shallow coastal soft bottom environments (almost no UV-B radiation; Olsen et al., 2016). Therefore, we presumed that multiple copies of UVR8 in land plants might result from WGD events, and their gain or loss likely reflected the adaptations of plants to complex environments.
The domain analyses showed that several amino acids were highly conserved in the C27 region (in A. thaliana residues 397-423) of UVR8 proteins among different plant species (Supplemental Figure S2). Previous studies reported that UVR8 could interact with downstream components (e.g. COP1, RUP1, and RUP2) via the “VP” core in the C27 domain (Cloix et al., 2012; Yin et al., 2015). Deletion of the C27 region from full-length UVR8 would lead to defects in UV-B signaling, and binding of RUP proteins to C27 was critical for accelerating the redimerization of UVR8 to its homodimeric ground state (Cloix et al., 2012; Heijde and Ulm, 2013). We further found that the C-terminal ends (the last 17 amino acids, C17) also had a few conserved amino acids (Supplemental Figure S2). The C17 of UVR8 could attenuate the binding between C27 and COP1, therefore inhibiting UV-B signaling (Lin et al., 2020). Based on evolutionary patterns of the C27 and C17 region of UVR8 in green plants, our results revealed the structural conservation of UVR8 in UV-B acclimation.
The origin and evolution of RUPs
The RUP proteins are members of the WD40-repeat proteins, and they only contain the WD40-repeats domain and a short N-terminal extension with unknown function (van Nocker and Ludwig, 2003; Gruber et al., 2010). Although RUPs have sequence similarity to the WD40 domain of COP1 (Figure 3, A), they showed unexpected functional divergence. COP1 is the key regulator of plant photomorphogenesis triggered by light, and RUPs function as a negative feedback regulator of UV-B signaling cascade, balancing the UV-B defense (Yin et al., 2015; Liao et al., 2020). To better understand the evolutionary history of RUP, we reconstructed the phylogenetic tree of the RUP orthologs (Figure 3, B). The phylogenetic analyses supported that RUP orthologs first arose in chlorophytan green algae and pervasively existed in most of green plants, such as chlorophytes, bryophytes, lycophytes, ferns, gymnosperms, and angiosperms (Figure 3, B). The structural analyses demonstrated that RUPs retained most of the conserved domains and motifs during the plant evolution, and only a slight increase of the conserved motifs between chlorophytes and streptophytes were observed (Figure 3, B). It was reported that UVR8-COP1/SPAs-HY5 complex originated in chlorophytes and served as the most ancient UV-B signaling module in green plants (Supplementa Figures S3–S5l ; Han et al., 2019). Not only RUPs promote UVR8 re-dimerization in UV-B signaling, but also they function as substrate receptors in CUL4-DDB1-based E3 ubiquitin ligase complexes, serving as a crucial molecular brake of UV-B signaling by modulating HY5 protein levels (Ren et al., 2019; Podolec et al., 2021). Thus, we assumed that RUP might originate in chlorophytes, probably for some primitive functions. Compared with chlorophytes, charophyte algae were the earliest diverging lineages of streptophytes, and have adapted the subaerial/terrestrial habitat (Cheng et al., 2019). Considering the negative feedback regulations of RUP in UV-B signaling cascade, the increase of motifs might facilitate the sophisticated regulation of UV-B response during transition from aquatic to terrestrial environments of green plants.
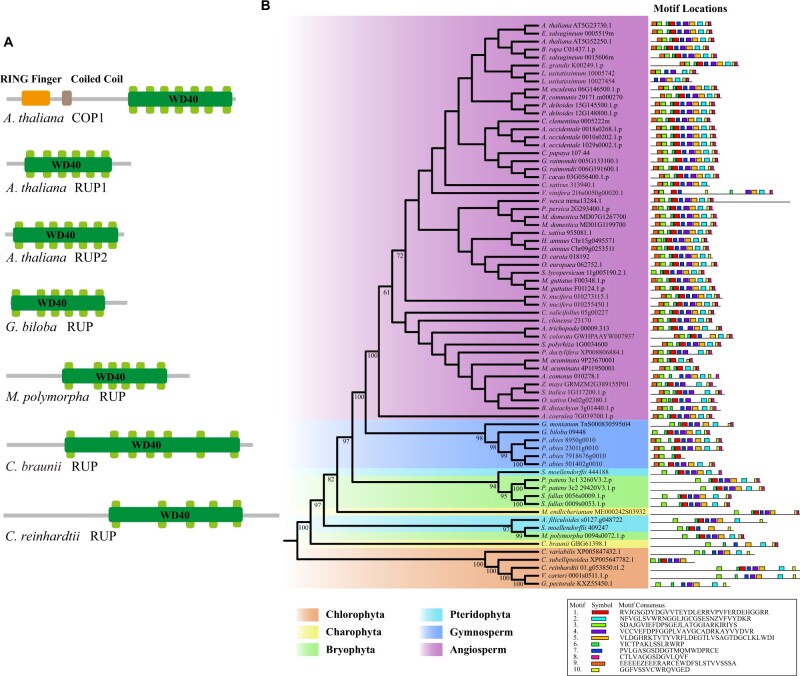
Figure 3.
The phylogenetic relationship and structural comparisons of RUP in green plants. A, The comparison of COP1 and RUPs domains among A. thaliana, G. biloba, M. polymorpha, C. braunii, and C. reinhardtii. B, The phylogenetic tree and corresponding conserved motifs in green plants.
Compared with the species tree of Viridiplantae, the phylogenetic tree of RUP showed slight differences in early-diverging streptophytic lineages (Figure 3, B), reflecting the complex evolutionary scenario. RUP orthologs from charophytes, bryophytes, and pteridophytes together formed a paraphyletic group, and the gymnosperms formed a monophyletic group (Figure 3, B). Interestingly, compared with the conservation of UVR8 orthologs within charophytes, only two RUP orthologs in five charophyte genomes were identified (Mesotaenium endlicherianum and Chara braunii), implying the gene loss of RUPs in charophytes (Figure 3, B). Moreover, we found increased copy number of RUP within the bryophytes, gymnosperms, and angiosperms (Figure 3, B). For instance, both chlorophytes and charophytes only had single-copy RUP orthologs, whereas the RUPs in Selaginella moellendorffii, P. patens, S. fallax, Picea Abies, and A. thaliana were two or more copies (Figure 3, B). Arabidopsis RUP1 and RUP2 duplicated after the splitting of Brassicaceae, and had six and seven WD40 domains, respectively (Figure 3, B). Although it has been found that RUP1 and RUP2 had functional redundancy in mediating UVR8 redimerization, the RUP2 likely played the dominant role, in line with RUP1 and RUP2 protein levels (Liao et al., 2020; Podolec et al., 2021). It is also well documented that Arabidopsis rup1 rup2 double mutant appeared slow UVR8 redimerization and an exaggerated UV-B photomorphogenic phenotype, such as dwarfism and short hypocotyl phenotype, resulting in better acclimation and protection against UV-B stress (Gruber et al., 2010; Liao et al., 2020). We, therefore, assumed that the loss of RUP in some pioneers of land plants (e.g. charophytes) might be a radical evolutionary innovation for their subaerial/terrestrial habitats, contributing to their adaptations to the increased UV-B radiation. Meanwhile, the pervasive existence and duplications of RUPs in seed plants probably reflected its vital roles in balancing UV-B resistance and plant developments.
The adaptive evolution of UVR8 and RUPs
During the transition from deep sea to shallow water, and from aquatic to terrestrial environments, UV radiation is a crucial environmental stress for green plants. When plants invaded the shallow water environments, it began to expose to increased UV-B radiation, and colonization in land envisaged the increased level and persistent exposure of UV-B (Kenrick and Crane, 1997; Maberly, 2014). Environmental selective pressures, such as temperature and UV-B radiation, have driven the evolution of sensing and signaling transduction pathways associated stress response in plants (Hauser et al., 2011).
In order to test the presence of positive selection on UVR8 and RUP, we conducted the branch-site random effects likelihood test (BSREL in Hyphy) to detect evidence of positive selection on each branch of the phylogeny (Pond and Muse, 2005; Smith et al., 2015). We found significant signatures that some nonsynonymous substitutions in UVR8 were subject to positive selection on the branches leading to the ancestor of chlorophytes, Monoraphidium neglectum, Dunaliella salina, and Mesostigma viride (Figure 2). We further performed the branch-site test using PAML to identify positively selected sites (PSSs) in several crucial nodes during the plant evolution (Yang, 2007), including the ancestral branch of land plants, and the ancestral branch of chlorophyte. We identified several PSSs of UVR8 along the ancestral branch of chlorophyte (e.g. Cys132, Trp144, Met176, and Thr236; Figure 4, A–D and Supplemental Table S1). Evidently, it was documented that the mutation of Trp144 to Phe (UVR8W144F) did not influence the interaction between UVR8 and COP1 in yeast, due to the formation of hydrophobic interactions through these aromatic residues (O’Hara and Jenkins, 2012). Importantly, the mutation of Trp285 to Phe (UVR8W285F) could alter the spectral sensitivity of UVR8, and the UVR8W285F mutant could weakly absorb UV-C instead of UV-B, due to the shorter absorption wavelength of phenylalanine than tryptophan (Christie et al., 2012). Thus, we assumed that the substitution at this residue (UVR8W144F) in the ancestral branch of chlorophyte might result from high levels of UV-C radiation, due to low concentration of ozone in atmosphere at the Archean Earth (Cnossen et al., 2007). In addition, the PSS Thr236 was adjacent to Trp233 that served as the UV-B chromophore (Christie et al., 2012), likely involved in UV-B perception and exciton coupling.
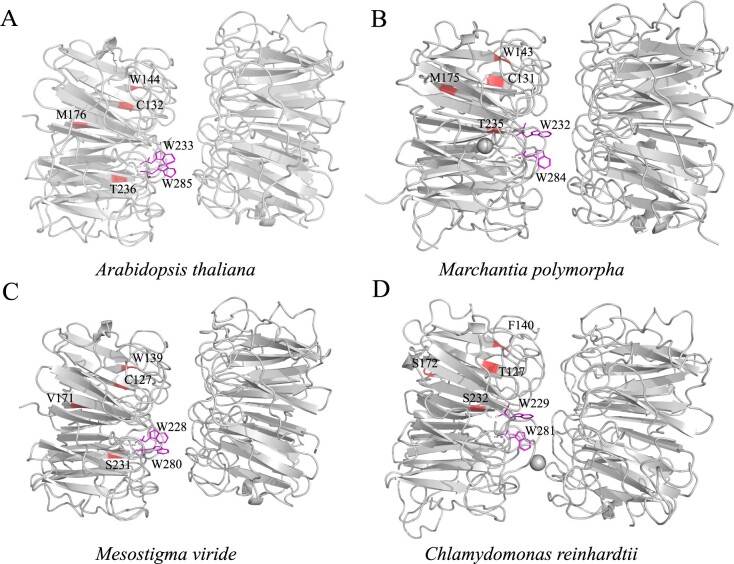
Figure 4.
Spatial distribution of PSSs on the predicted 3D structure of UVR8 proteins. A, Arabidopsis thaliana; B, M. polymorpha; C, M. viride; and D, C. reinhardtii. PSSs (Cys132, Trp144, Met176, and Thr236) were labeled in A. thaliana and the corresponding positions in C. reinhardtii, M. viride, and M. polymorpha UVR8 proteins. Trp285 and Trp233 were the main UV-B sensor, and Cys132, Trp144, Met176, and Thr236 were PSSs corresponding to the positions in homologous proteins of A. thaliana.
We also detected evidence of positive selection on RUP on the ancestral branch of chlorophyte (Supplemental Table S1). These PSSs were located in the WD40 domains of RUP proteins, likely related to the stability and status transitions of UVR8. The switching of UVR8 between homodimeric and monomeric was essential for UV-B signal transduction, and further experiments were needed to characterize the functions of these PSSs. Overall, we identified evidence of positive selection on UVR8 and RUPs leading to the ancestral branch of chlorophytes, and these PSSs were in or near the important domains of these proteins. These observations probably reflected the adaptive modifications of UVR8 and RUPs during plant terrestrialization, driven by the changes of environmental UV-B.
The evolution of the regulatory network mediated by UVR8
Not only could UVR8 mediate the UVR8-COP1/SPA-HY5 signaling pathway in the UV-B acclimation of green plants, but it also participated in other UV-B signaling pathways associated with plant growth and developments. To deepen our understanding of the complex UV-B signaling and regulatory network mediated by UVR8, we performed comprehensive similarity searches and phylogenetic analyses of other crucial components of the UVR8-mediated signaling pathways.
BES1/BIM1
BRs are kinds of endogenous plant hormones, and play important roles in the regulation of growth, developments, and environmental stress responses (Nolan et al., 2017). It has been shown that UVR8 could physically interact with BES1 and BIM1 to represses growth-associated genes that are under the control of BRs, contributing to the inhibition of hypocotyl elongation under UV-B (Liang et al., 2018). Our phylogenetic analyses suggested that BES1 originated in charophytes, and the motifs remained relatively conservative during the plant evolution (Figure 5, A and Supplemental Figure S6). In terms of BIM1, we found that BIM1 originated in chlorophytes and presented slightly different evolutionary relationships compared with the widely accepted phylogeny of green plants (Figure 5, B and Supplemental Figure S7). It was reported that the basic helix-loop-helix (bHLH) domain of BIM1 had a weak interaction with UVR8, while the bHLH domain combined with either the C terminus or N terminus of BIM1 together had a strong interaction with UVR8 in Arabidopsis (Liang et al., 2018). The domain and motif analyses indicated that BIM1 orthologs in chlorophytes only contain the bHLH domain, and the BIM1 orthologs in charophytes gradually gained other auxiliary motifs (Figure 5, B and Supplemental Figure S7). Importantly, recent genomic studies identified BES1 and BIM1 orthologs involved in BRs biosynthesis and signaling in charophytes (Cheng et al., 2019; Jiao et al., 2020). Our results supported that the UVR8-BES1/BIM1 signaling pathway likely originated in charophyte lineages, and might have experienced key structural innovations during the plant evolution.
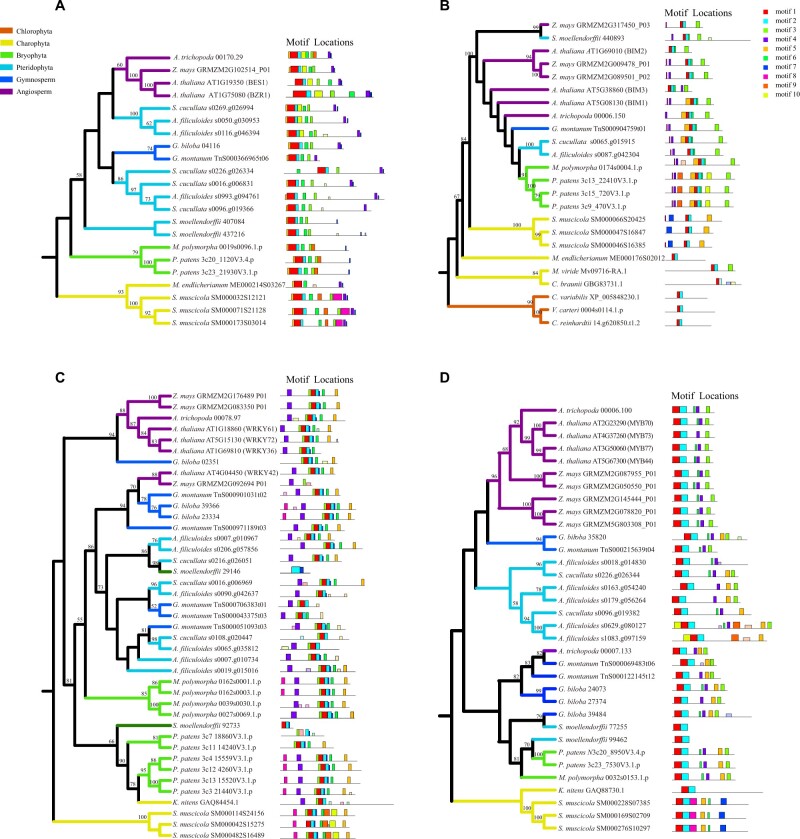
Figure 5.
The phylogenetic relationship and structural comparisons of downstream TFs in representative green plants. A, BES1 orthologs; B, BIM1 orthologs; C, WRKY36 orthologs; and D, MYB73/77 orthologs. Conserved motifs were shown against the phylogenetic tree. Bootstrap values >50 were indicated. The 10 distinct MEME-motifs were displayed in different colored boxes.
WRKY36
The WRKY superfamily has essential functions in various physiological and biological processes, especially biotic/abiotic stress responses and developments (Ülker and Somssich, 2004; Birkenbihl et al., 2017). WRKY36 could repress the transcription of HY5, and UVR8 could physically interact with WRKY36 in nuclei to promote the expression of HY5 by repressing the DNA-binding activity of WRKY36 (Yang et al., 2018). The phylogenetic tree of WRKY36 demonstrated that it originated in charophytes and existed in most clades of land plants (e.g. bryophytes, lycophytes, ferns, gymnosperms, and angiosperms; Figure 5, C). The C terminal DNA-binding domain (BD) of WRKY36 (amino acid 191-388) could directly interact with the C terminal of UVR8 (amino acid 397-440; Yang et al., 2018). Our domain and motif analyses showed that these WRKY36 orthologs of land plants contained the motifs 1, 2, 3, 6, and 7 in the C terminus, indicating the functional conservation of WRKY36 in land plants (Figure 5, C). In addition, we observed the evidence of duplication events in WRKY36 before the divergence of angiosperms (Supplemental Figure S8), probably resulting from widespread WGDs of angiosperms.
MYB73/MYB77 and MYB13
MYB is a large TF family in plants, and members of this family are essential regulators in biotic and abiotic stress response, metabolisms, and developments (Xing et al., 2019; Sun et al., 2020). Previous experimental analyses proved that UVR8 could interact with MYB73/MYB77 in a UV-B-dependent manner, and therefore directly represses the transcription of their target auxin-responsive genes, integrating light and auxin signaling and inhibiting lateral root development (Yang et al., 2020). MYB13 could also interact with the photoactivated UVR8 to regulate auxin response and flavonoid biosynthesis, inducing cotyledon expansion and UV-B acclimation (Qian et al., 2020). Our phylogenetic analyses indicated that MYB73/77 originated in charophytes (Klebsormidium nitens and S. muscicola), and MYB77/73 experienced duplication events during the divergence of Brassicaceae, resulting in the new members MYB70 and MYB40 (Figure 5, D and Supplemental Figure S9). We also discovered that MYB13 only existed in seed plants (Supplemental Figure S10).
The N terminus of MYB73/77 (amino acids 1-120, including MYB domains) was proved to be crucial for interaction with both the N and C terminus of UVR8 (Yang et al., 2020). The auxiliary motifs out of this domain were also functionally important. The mutation of these motifs could significantly influence the function of MYB (Millard et al., 2019; Jiang and Rao, 2020). Our domain and motif analyses demonstrated that all MYB73/77 had conserved MYB domains, but the auxiliary motifs of MYB73/77 from charophytes were different from that of land plants (Figure 5, D). Previous studies discovered different expression patterns and functions of closely related MYB TFs between charophytes and land plants (Higo et al., 2018; Jiang and Rao, 2020). These phylogenetic and structural analyses implied that the UVR8-MYB73/77 signaling pathway originated in charophytes, and experienced functional innovations during the plant terrestrialization.
PIFs
PIFs belong to the bHLH family TFs, and they can repress light responses and promote hypocotyl elongation, particularly the members PIF4 and PIF5 (Hornitschek et al., 2012; Pfeiffer et al., 2014; Shi et al., 2018). It was reported that PIF4 and PIF5 could be degraded in a UVR8-dependent pathway via the ubiquitin–proteasome system, resulting in the inhibition of hypocotyl growth (Sharma et al., 2019; Tavridou et al., 2020). All PIFs contained a phytochrome B-binding motif, and only PIF1 and PIF3 had an additional phytochrome A-binding (APA) motif (Leivar and Quail, 2011). We identified PIFs orthologs in charophyte lineages, and the APA motif (motif 4) was also found in C. braunii and S. muscicola (Supplemental Figure S11). The PIF-mediated light-dependent transcriptional regulation has been identified in moss (Marchantia polymorpha and P. patens), and the APA motif was an indispensable functional domain (Inoue et al., 2016; Possart et al., 2017). Consistent with previous studies, we also found APA motif in PIFs from moss (M. polymorpha and P. patens) and tracheophytes (Supplemental Figure S12). Thus, our results suggested that the PIF-mediated regulations originated in charophytes, and were conserved in land plants.
Co-expression analyses and protein interaction assays
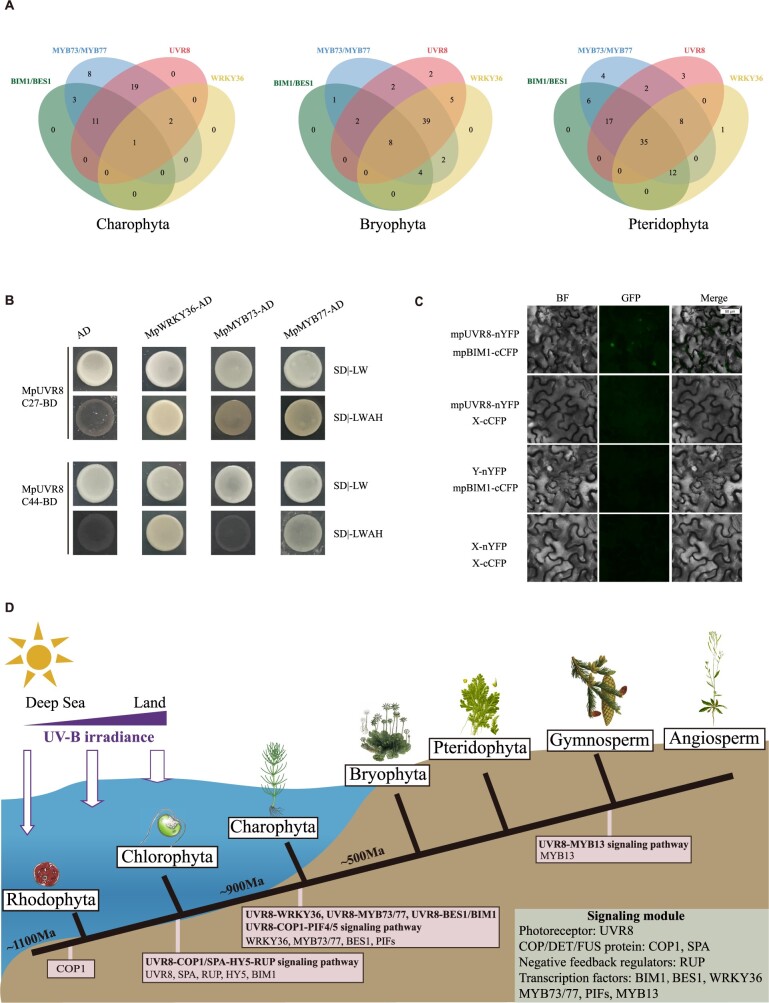
We further performed the orthologs searches in 48 charophyte transcriptomes to determine whether genes from the same pathway were all present in all species. We found 33 and 15 putatively expressed UVR8 and BES1/BIM1 in charophyte transcriptomes, respectively (Figure 6, A). UVR8 and BES1/BIM1 orthologs could be simultaneously identified in 12 charophyte transcriptomes (Figure 6, A). We also identified three putatively expressed WRKY36 and 44 putatively expressed MYB73/77 in charophyte transcriptomes. UVR8-WRKY36 and UVR8-MYB73/77 orthologs were identified at the same time in three and 33 charophyte transcriptomes (Figure 6, A). As gene expression usually presented spatial–temporal expression model, transcriptome data may not recover all the genomic information of an organism. Still, the co-expression analyses based on the transcriptome data provide evidences supporting the existence of UVR8 and TFs (BES1, BIM1, MYB73/77, and WRKY36) in charophytes. In addition, the identification of putatively expressed UVR8, WRKY36, MYB73/77, and BES1/BIM1 orthologs in several bryophytes and ferns transcriptomes implied the conservation of these pathways in early land plants (Figure 6, A).
Figure 6.
Origin and evolution of the UVR8-mediated signaling pathway. A, Co-expression analyses based on the 1KP transcriptomes. Venn diagrams of the distribution of UVR8, WRKY36, BES1, BIM1, and MYB73/73 orthologs in 48 charophytes, 69 bryophytes, and 96 ferns transcriptomes. B, Y2H assays showing the interactions between the C-terminal domain of UVR8 and WRKY36 and MYB73/MYB77. C, BiFC assays showing the interactions between BIM1 and UVR8. D, Origin and evolution of UVR8 signaling pathway in Archaeplastida. During the transition from deep sea to shallow water, and from aquatic to terrestrial environments, UV light was a crucial environmental signal for green plants. Red algae live in the deep sea where UV-B was entirely filtered. The evolution of UVR8 signaling pathway (UVR8, COP1, SPA, HY5, and RUP) in chlorophytes facilitated green algae to adapt shallow water environment. The origination of the transcriptional regulatory network (PIFs, WRKY36, BES1, BIM1, MYB73, and MYB73) in charophytes conferred freshwater algae the abilities to colonize the water surface. After plants conquered the land, UVR8 signaling pathway contributed to adapt to high and variable doses of UV-B irradiance.
We conducted the yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays to verify the interactions between UVR8 and the downstream TFs (BES1, BIM1, MYB73/77, and WRKY36). The interactions between the C-terminal domain of UVR8 and MYB73/77, the WRKY36 were identified in the Y2H assays (Figure 6, B), and BiFC assays further showed that the UVR8 had a direct interaction with BIM1 (Figure 6, C). Although we did not observe interactions in all TFs, both in vivo and in vitro assays provided substantial evidences of protein interactions between UVR8 and most TFs in early land plants. These experimental results largely confirmed that these recently discovered UVR8-mediated signaling pathways have existed at least in early land plants. Notably, consistent with our comparative analyses, previous studies have demonstrated that considerable genes essential for land plants were present in the common ancestor of charophytes and land plants (Cheng et al., 2019). Therefore, we hypothesize that these UVR8-mediated signaling pathways likely originated in charophytes, and our results provide potential directions for future experimental studies.
Hints for plant evolution and terrestrialization
During the evolution from deep sea to shallow water, and from aquatic to terrestrial environments, pioneers of land plants were exposed to complex environmental UV-B radiation (high and variable). Green plants evolved multiple UV-B signaling mechanisms, and conferred the capacity for rapid response and adaptations to changing environmental UV-B radiation. Our comparative genomic and phylogenetic analyses indicated that the orthologs of the canonical UVR8-COP1/SPA-RUP signaling pathway (UVR8, RUP, COP1, SPA, and HY5) originated in chlorophytes (Figure 6, D). It is worth mentioning that the origin of these orthologs does not necessarily indicates the presence of UVR8-COP1/SPA-RUP signaling pathway, and the proteins encoded by these orthologous genes need direct or indirect interactions to complete the signal transductions. There were documented instances of interactions between UVR8 and COP1, SPA in green algae (Tilbrook et al., 2016; Xu et al., 2021). Thus, we suggested that the UVR8-COP1/SPA-RUP signaling pathway was likely to originate in chlorophytes. Moreover, UVR8 experienced a conservative evolutionary route during the plant evolution, and the evolutionary history of RUP was different from the evolution of green plants. We also identified that UVR8 and RUPs experienced significant positive selection in the ancestral branch of chlorophytes. Adaptive modifications might influence the physiological properties and status transitions of UVR8, switching from homodimeric to monomeric ground state. Green plants have encountered the increased UV-B radiation during the colonization of shallow water and land, and the environmental stress likely has shaped the amino acid substitutions on these related proteins, eventually achieving acclimation.
Our multispecies genome-wide analyses identified that several emerging UVR8-regulated TFs originated in charophytes and were conserved in land plants (Figure 5, A–D). TFs could merge environmental and complicated endogenous signal, which contribute to adaptations of plants to drastic environmental changes (Jing and Lin, 2020). UVR8 not only mediates different hormone signals via COP1 and HY5, but also directly interacts with other TFs to quickly regulate the endogenous hormone signaling pathway (Liang et al., 2018; Yang et al., 2018). In A. thaliana, UVR8 could dissociate into the monomeric form and accumulate in the nucleus under UV-B irradiation, and the interactions between UVR8 and downstream TFs in the nucleus represented important mechanisms of early UV-B signal transduction. For instance, The UVR8-WRKY36-HY5 signaling pathway was independent from UVR8-COP1/SPA-HY5, and it provided an alternative mechanism to response to the constant and increasing UV-B irradiation (Yang et al., 2018). In contrast, plant hormones played a central role in regulating growth, developments, and defense against abiotic stresses, and the cross-talk between UV-B signal and endogeneous hormonal signal was another crucial mechanism to transduce environmental signals. Our phylogenetic analyses discovered that the components of UVR8-BES1/BIM1 and UVR8-MYB73/MYB77 signaling pathway originated in charophytes, and our experimental assays identified evidence of protein interactions in early-branching land plants (M. polymorpha; Figure 6, B and C and Supplemental Figures S6, S7, S9). By combining the motif analyses and experimental assays, we therefore suggested that the UVR8-BES1/BIM1 and UVR8-MYB73/MYB77 signaling pathway originated in charophytes (Figure 6, D). In terms of MYB13, our analyses supported that UVR8-MYB13 signaling pathway emerged in the common ancestor of angiosperms and gymnosperms. It was reported that MYB13 predominantly expressed in the cotyledons, positively regulating UV-B-induced cotyledon expansion and stress acclimation, thus further experiments are essential to verify its functions in gymnosperms. As the only known TF with indirect interaction with UVR8 in UV-B signaling pathway, the PIF-mediated signaling pathway was proposed to originate in charophytes. Binding of UV-B-activated UVR8 to COP1 could disrupt the stabilization of PIF4/5, participating in multiple responses to environmental changes, such as the shade avoidance response and thermomorphogenesis. Our results provide clues about the evolution of PIF-mediated signaling pathway, as well as the complex adaptive strategies of plants on land.
The mechanisms underlying UVR8 signaling and its outcomes (from UV-B signal transduction to gene expression changes and physiological UV-B responses) remain obscure. The cross-talk between environmental UV-B signal and hormonal signal (BR and auxin) have been mainly studied in A. thaliana, whereas their functions in green algae and early land plants are rarely documented. Integrative studies of the evolution of the UVR8 signaling pathways and cross-talks in green algae and early land plants are crucial for understanding the molecular basis of UV-B response and plant terrestrialization. Overall, our study indicates that the integrated UVR8-COP1/SPA-RUP signaling pathway that originated in chlorophytes ensures UV-B signal transduction and physiological response. The interactions between UVR8 and downstream TFs presumably originate from charophytes, which largely expand the repertoires of UV-B signaling pathway and likely facilitate the adaptations of plants to the variable UV-B radiation in subaerial/terrestrial environments.
Materials and methods
Identification of UVR8 and RUP orthologs in plant genomes
We selected 79 plant genomes from main Archaeplastida clades, including one glaucophyte, four rhodophytes, 12 chlorophytes, 5 charophytes, 3 bryophytes, 1 lycophyte, 1 fern, 3 gymnosperms, and 48 angiosperms (Supplemental Table S2). In order to identify the orthologs of UVR8 and RUP from these plant genomes, we employed both the similarity searches and phylogenetic analyses. First, the Arabidopsis (A. thaliana) UVR8 and RUP protein sequences (UVR8: AT5G63860, RUP1: AT5G52250, and RUP2: AT5G23730) were used as queries to perform BLASTP searches, and filtered with an E-value threshold of 10−5 (Altschul et al., 1997). The amino acid sequences with BLAST hits were further searched against the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd) to check the “RCC1” domain repeats and “WD40” domain, respectively. Second, the filtered protein sequences of UVR8 and RUP were separately aligned using MAFFT version 7, and then trimmed by trimAL version 1.3 with −gt = 0.03 (Capella-Gutiérrez et al., 2009; Katoh and Standley, 2013). The multiple sequence alignments (MSAs) were manually inspected, and the maximum likelihood phylogenetic tree was reconstructed using IQ-TREE version 1.6.1 (Nguyen et al., 2015). The best-fitting model was determined by ModelFinder, and branch supports were obtained using the ultrafast bootstrap approach with 1,000 replicates (Minh et al., 2013; Kalyaanamoorthy et al., 2017). The monophyletic group containing AtUVR8 and AtRUP were separately selected as candidates UVR8 and RUP orthologs. Third, we further performed functional domain analyses to check the “GWRHT” motifs and the WD40 repeat number using InterProScan version 5.24 and MEME suit version 5.0.3 (http://meme-suite.org/doc/download.html). Only the candidate orthologs having the functional domain and conserved motifs were characterized as true orthologs and used for further phylogenetic analyses.
Phylogenetic reconstruction and selective pressure analyses of UVR8 and RUP
The identified orthologs of UVR8 and RUP were separately aligned with MAFFT (Katoh and Standley, 2013). The MSAs were trimmed by trimAL with −gt = 0.03 and manually inspected (Capella-Gutiérrez et al., 2009). We further reconstructed the phylogenetic tree of the orthologs using the above-mentioned pipeline. To identify the evidence of positive Darwinian selection, the branch-site random-effect likelihood (BranchSiteREL) model in HyPhy version 2.1.2 was applied to detect episodic diversifying selection (Pond and Muse, 2005). The BranchSiteREL model could identify signs of positive selection in each branch of the phylogeny, without making priori assumptions (“foreground” and “background” branches). We also employed the branch-site models implemented in Codeml from PAML package version 4.9c to identify PSS (Yang, 2007). Model A (allows sites to be under positive selection; model = 2, Nsites = 2, fixed omega = 0, omega = 2) is compared with the null model A1 (sites may evolve neutrally or under purifying selection; model = 2, Nsites = 2, fixed omega = 1, omega = 1). As branch-site model only allows single foreground branch in each test, we separately set ancestral branch of chlorophytes and ancestral branch of land plants as foreground branch. The significance of models was determined by the likelihood ratio test with a χ2 distribution at a threshold of P values < 0.05. Ancestral sequences were reconstructed using ML method (RateAncestor = 1). Bayes empirical Bayes analysis was applied to statistically identify the PSSs on a specified branch with posterior probabilities ≥0.95 (Yang et al., 2005). The 3D structures of UVR8 proteins of Chlamydomonas reinhardtii, M. viride, M. polymorpha, and A. thaliana were retrieved from SWISS-MODEL (https://swissmodel.expasy.org), and protein structures were visualized using PyMOL (http://pymol.org).
TFs identification and phylogenetic analyses
The identification of each TF was based on the aforementioned approaches of similarity search combined with phylogenetic analyses, and the identified TFs were strictly filtered using the functional domain and conserved motif information. The queries used in BLASTP searches and functional domain information were shown in Supplemental Table S3. Sequences with blast hits (E-value < 1e−5) and functional domains were retained for further phylogenetic analyses (Altschul et al., 1997). These retained sequences were aligned using MAFFT (Katoh and Standley, 2013), and trimmed by trimAL with manually inspected (Capella-Gutiérrez et al., 2009). We reconstructed phylogenetic trees of homologs, and the monophyletic group containing Arabidopsis BES1, BIM1, WRKY36, MYB73/MYB77, MYB13, PIFs, and HY5/HYH proteins was treated as orthologs. Finally, phylogenetic trees of orthologs were reconstructed using the above-mentioned pipeline.
Co-expression analyses of the TFs
We separately selected 48, 69, and 96 transcriptomic data of charophytes, bryophytes, and ferns from the 1KP project (www.onekp.com) for the following co-expression analyses. We separately used Arabidopsis UVR8, BES1, BIM1, WRKY36, MYB73, and MYB77 proteins sequences as queries to perform BLASTP searches with an E-value threshold of 10−5 (Altschul et al., 1997). All protein sequences with the corresponding annotations in NCBI Conserved Domain Database were used for the following phylogenetic analyses. Then, we reconstructed phylogenetic trees to identify orthologs of BES1, BIM1, WRKY36, and MYB73/MYB77 based on transcriptomic data.
Y2H and BiFC assays
To further verify the protein interactions between UVR8 and related TFs (BES1, BIM1, MYB73/77, and WRKY36), we separately performed the Y2H and BiFC assays. The full-length coding sequences, C44 region and C27 region of UVR8 from M. polymorpha were synthesized and fused in-frame to the GAL4 DNA-BD of the bait vector pGBKT7. The coding sequences of BES1, BIM1, WRKY36, and MYB73/MYB77 were separately synthesized and cloned into pGADT7 vectors. The clones were co-transformed into yeast strain AH109 according to a standard yeast transformation protocol (Clontech, Beijing, China). Protein–protein interactions were characterized from the yeast transformants that were streaked onto SD/-Trp-Leu-His-Ade dropout plates. Each Y2H assay was independently repeated 3 times.
The BiFC assay followed the protocol with slight modifications (Bai et al., 2007; Ma et al., 2016), UVR8 (the full-length coding sequences, C44 region and C27 region), BES1, BIM1, WRKY36, and MYB73/MYB77 from M. polymorpha were fused to N-terminus of YFP or C-terminus of CFP, transformed to Agrobacterium strain GV3101. Overnight cultures of Agrobacteria were collected by centrifugation, resuspended in MES buffer to 0.6 OD600, mixed with GV3101 expressing pSoup-P19, and incubated at room temperature for 2 h before infiltration. Agrobacteria suspension in a 1-mL syringe (without the metal needle) was carefully press-infiltrated manually onto healthy leaves of 3-week-old Nicotiana benthamiana. Plants were left under white light (long-day conditions) for 3 d after infiltration.
Data availability statement
MSAs and phylogenetic trees used in this study are available in the FigShare repository: https://figshare.com/s/f5669bdaa79f7b3dae97.
Accession numbers
Arabidopsis protein sequence data can be found in the Arabidopsis Information Resource (https://www.arabidopsis.org/): UVR8: AT5G63860, COP1: AT2G32950, SPAs (SPA1-4: AT2G46340, AT4G11110, AT3G15354, AT1G53090), BES1: AT1G19350, BIM1: AT5G08130, WRKY36: AT1G69810, MYB73: AT4G37260, MYB77: AT3G50060, MYB13: AT1G06180, PIF1: AT2G20180, PIF3: AT1G09530, PIF4: AT2G43010, PIF5: AT3G59060, HY5: AT5G11260, and HYH: AT3G17609.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Heat map of ortholog numbers of UVR8-mediated signaling pathway in Archaeplastida.
Supplemental Figure S2. The alignments of the C terminus of UVR8 proteins in representative green plants.
Supplemental Figure S3. Phylogenetic tree of COP1.
Supplemental Figure S4. Phylogenetic tree of SPA.
Supplemental Figure S5. Phylogenetic tree of HY5.
Supplemental Figure S6. Phylogenetic tree of BES1.
Supplemental Figure S7. Phylogenetic tree of BIM1.
Supplemental Figure S8. Phylogenetic tree of WRKY36.
Supplemental Figure S9. Phylogenetic tree of MYB73/77.
Supplemental Figure S10. Phylogenetic tree of MYB13.
Supplemental Figure S11. Phylogenetic tree of PIFs.
Supplemental Figure S12. Comparisons of protein domain and conserved amino acid motif among PIFs in representative green plants.
Supplemental Table S1. Positive selection analyses of UVR8 and RUP by branch-site model.
Supplemental Table S2. The information of plant genomes used in this study.
Supplemental Table S3. The information of queries used in the similarity searches.
Supplementary Material
Acknowledgments
We thank Ziqiang Zhu and Guanzhu Han for helpful discussions, the Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry, and the Priority Academic Program Development of Jiangsu Higher Education Institutions for technical support. We also thank the editor and anonymous reviewers for their helpful suggestions.
Funding
This work was supported by the National Natural Science Foundation of China (31970229, 32122010, and 32100178).
Conflict of interest statement. Authors declare no conflict of interest.
B.Z. designed the research. Z.Z., C.X., S.Z., C.S., and H.C. performed the analyses. Z.Z. and C.X. drafted the manuscript. H.L. and B.Z. revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Bojian Zhong (bjzhong@gmail.com).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Zhang L, Gampala SS, Zhu S, Song W, Chong K, Wang Z (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balskus EP, Walsh CT (2010) The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 329: 1653–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Roccaro M, Somssich IE (2017) Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early mamp-triggered immunity. Plant Cell 29: 20–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y (2019) Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179: 1057–1067 [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen I, Sanz-Forcada J, Favata F, Witasse O, Zegers T, Arnold NF (2007) Habitat of early life: solar X-ray and UV radiation at Earth's surface 4-3.5 billion years ago. J Geophys Res 112: E02008 [Google Scholar]
- De Smet R, Van de Peer Y (2012) Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15: 168–176 [DOI] [PubMed] [Google Scholar]
- de Vries J, Stanton A, Archibald JM, Gould SB (2016) Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci 21: 467–476 [DOI] [PubMed] [Google Scholar]
- Favory J, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández MB, Tossi V, Lamattina L, Cassia R (2016) A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front Plant Sci 7: 1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Soltis DE, Soltis PS, Ashman T, Van de Peer Y (2020) Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol 30: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Chen M, Li X, Liang Y, Zhang D, Wood AJ, Oliver MJ, Zhang J (2020) Ancestral gene duplications in mosses characterized by integrated phylogenomic analyses. J Syst Evol 10.1111/jse.12683 [DOI] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chang X, Zhang Z, Chen H, He H, Zhong B, Deng XW (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol Plant 12: 847–862 [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA 110: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo A, Kawashima T, Borg M, Zhao M, López-Vidriero I, Sakayama H, Montgomery SA, Sekimoto H, Hackenberg D, Shimamura M (2018) Transcription factor DUO1 generated by neo-functionalization is associated with evolution of sperm differentiation in plants. Nat Commun 9: 5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Huang X, Yang P, Ouyang X, Chen L, Deng XW (2014) Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. Plos Genet 10: e1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Nishihama R, Kataoka H, Hosaka M, Manabe R, Nomoto M, Tada Y, Ishizaki K, Kohchi T (2016) Phytochrome signaling is mediated by phytochrome interacting factor in the liverwort Marchantia polymorpha. Plant Cell 28: 1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (2014a) Structure and function of the UV-B photoreceptor UVR8. Curr Opin Struc Biol 29: 52–57 [DOI] [PubMed] [Google Scholar]
- Jenkins GI (2014b) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (2017) Photomorphogenic responses to ultraviolet-b light. Plant Cell Environ 40: 2544–2557 [DOI] [PubMed] [Google Scholar]
- Jiang C, Rao G (2020) Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol 183: 637–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao C, Sørensen I, Sun X, Sun H, Behar H, Alseekh S, Philippe G, Palacio Lopez K, Sun L, Reed R (2020) The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell 181: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Jing Y, Lin R (2020) Transcriptional regulatory network of the light signaling pathways. New Phytol 227: 683–697 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Lee J (2016) UV-B signal transduction pathway in Arabidopsis. J Plant Biol 59: 223–230 [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O (2012) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31: 1–46 [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44: 512–523 [DOI] [PubMed] [Google Scholar]
- Liang T, Yang Y, Liu H (2019) Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol 221: 1247–1252 [DOI] [PubMed] [Google Scholar]
- Liao X, Liu W, Yang H, Jenkins GI (2020) A dynamic model of UVR8 photoreceptor signalling in UV-B-acclimated Arabidopsis. New Phytol 227: 857–866 [DOI] [PubMed] [Google Scholar]
- Lin L, Dong H, Yang G, Yin R (2020) The C-terminal 17 amino acids of the photoreceptor UVR8 is involved in the fine-tuning of UV-B signaling. J Integr Plant Biol 62: 1327–1340 [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maberly SC (2014) The fitness of the environments of air and water for photosynthesis, growth, reproduction and dispersal of photoautotrophs: an evolutionary and biogeochemical perspective. Aquat Bot 118: 4–13 [Google Scholar]
- Millard PS, Kragelund BB, Burow M (2019) R2R3 MYB transcription factors—functions outside the DNA-binding domain. Trends Plant Sci 24: 934–946 [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30: 1188–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Chen J, Yin Y (2017) Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem J 474: 2641–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O Hara A, Jenkins GI (2012) In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell 24: 3755–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin Y, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F (2016) The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Demarsy E, Ulm R (2021) Perception and signaling of ultraviolet-b radiation in plants. Annu Rev Plant Biol 72: 793–822 [DOI] [PubMed] [Google Scholar]
- Pond SLK, Muse SV (2005) Hyphy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679 [DOI] [PubMed] [Google Scholar]
- Possart A, Xu T, Paik I, Hanke S, Keim S, Hermann H, Wolf L, Hiß M, Becker C, Huq E (2017) Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 29: 310–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Chen Z, Liu Q, Mao W, Chen Y, Tian W, Liu Y, Han J, Ouyang X, Huang X (2020) Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses. Mol Plant 13: 777–792 [DOI] [PubMed] [Google Scholar]
- Ren H, Han J, Yang P, Mao W, Liu X, Qiu L, Qian C, Liu Y, Chen Z, Ouyang X (2019) Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci USA 116: 4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rozema J, Björn LO, Bornman JF, Gaberščik A, Häder DP, Trošt T, Germ M, Klisch M, Gröniger A, Sinha RP (2002) The role of UV-B radiation in aquatic and terrestrial ecosystems-an experimental and functional analysis of the evolution of UV-absorbing compounds. J Photochem Photobiol B 66: 2–12 [DOI] [PubMed] [Google Scholar]
- Sharma A, Sharma B, Hayes S, Kerner K, Hoecker U, Jenkins GI, Franklin KA (2019) UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat Commun 10: 4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lyu M, Luo Y, Liu S, Li Y, He H, Wei N, Deng XW, Zhong S (2018) Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc Natl Acad Sci USA 115: 6482–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL (2015) Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol 32: 1342–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano G, Cloix C, Heilmann M, Núñez-Olivera E, Martínez-Abaigar J, Jenkins GI (2018) Evolutionary conservation of structure and function of the UVR8 photoreceptor from the liverwort Marchantia polymorpha and the moss Physcomitrella patens. New Phytol 217: 151–162 [DOI] [PubMed] [Google Scholar]
- Su D, Yang L, Shi X, Ma X, Zhou X, Hedges SB, Zhong B (2021) Large-scale phylogenomic analyses reveal the monophyly of bryophytes and neoproterozoic origin of land plants. Mol Biol Evol 38: 3332–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhao J, Li X, Li Y (2020) E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol 227: 455–472 [DOI] [PubMed] [Google Scholar]
- Tavridou E, Pireyre M, Ulm R (2020) Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J 101: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L, Rizzini L, Stracke R, Ulm R, Rensing SA (2010) The molecular and physiological responses of Physcomitrella patens to ultraviolet-b radiation. Plant Physiol 153: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J (2012) Structural basis of ultraviolet-b perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y (2020) Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant 13: 59–71 [DOI] [PubMed] [Google Scholar]
- Xing C, Liu Y, Zhao L, Zhang S, Huang X (2019) A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ 42: 832–845 [DOI] [PubMed] [Google Scholar]
- Xu C, Chang X, Hou Z, Zhang Z, Zhu Z, Zhong B (2021) The origin of SPA reveals the divergence and convergence of light signaling in Archaeplastida. Mol Phylogenet Evol 161: 107175. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Chen P, Liang T, Li X, Liu H (2020) UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39: e101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Yin R, Arongaus AB, Binkert M, Ulm R (2015) Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27: 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Liu L, Yan Z, Penny D (2013) Origin of land plants using the multispecies coalescent model. Trends Plant Sci 18: 492–495 [DOI] [PubMed] [Google Scholar]
- Zhong B, Xi Z, Goremykin VV, Fong R, Mclenachan PA, Novis PM, Davis CC, Penny D (2014) Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol Biol Evol 31: 177–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MSAs and phylogenetic trees used in this study are available in the FigShare repository: https://figshare.com/s/f5669bdaa79f7b3dae97.