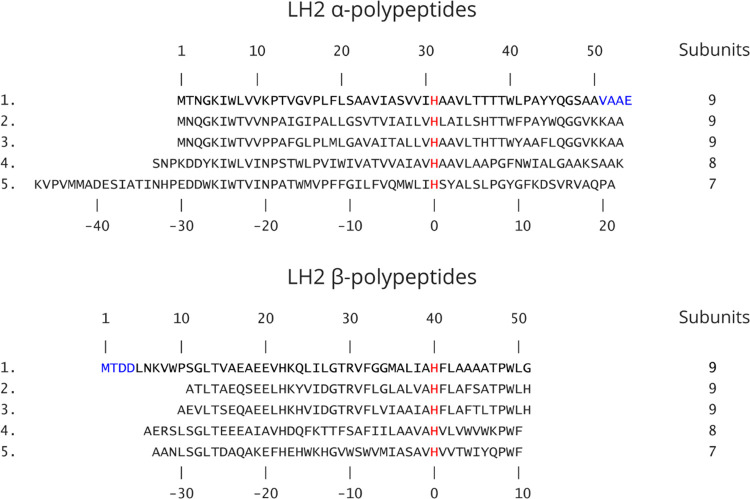
Figure 1.
Amino acid sequence alignments of α- and β-polypeptides in LH2 complexes for which a high-resolution 3D structure is available. 1, Rba. sphaeroides 2.4.1 (PDB 7PBW); 2, Rbl. acidophilus 10050 (PDB 1NKZ); 3, Rbl. acidophilus 7050 (PDB 1IJD); 4, Phs. molischianum DSM-119 (PDB 1LGH); and 5, Mch. purpuratum DSM-1591 (PDB 6ZXA). The sequences shown in black are those reported and resolved in the PDB entries; the blue characters in the α and β sequences for Rba. sphaeroides indicate those not modeled into the density map. The sequences are aligned against the central His residue (red) that coordinates the BChl a molecules forming the main ring of pigments. The top set of numbers refers to the residues in the Rba. sphaeroides LH2 complex. The lower set of numbers designates the His ligand to the B850-type BChls (red) as 0 and counts forwards or backwards from that point. This numbering system, from a central, conserved residue, allows comparison of polypeptides of differing lengths and from different bacteria. The LH2 oligomer sizes (number of αβ subunits) for different strains are indicated.