Abstract
The resistance of human immunodeficiency virus type 1 (HIV-1) to drugs is a major cause of antiretroviral treatment failure. We have compared direct sequencing to a line probe assay (LiPA) for the detection of drug resistance-related mutations in 197 clinical samples, and we have investigated the sequential appearance of mutations under drug pressure. For 26 patients with virological failure despite the use of two nucleoside analogues and one protease inhibitor (indinavir [n = 6], ritonavir [n = 10], and saquinavir [n = 10]), genotypic resistance assays were carried out retrospectively every 3 months for up to 2 years by using direct sequencing (TruGene; Visible Genetics) and a LiPA for detection of mutations in the reverse transcriptase (INNO-LiPA HIV-1 RT; Innogenetics) and the protease (INNO-LiPA HIV Protease, prototype version; Innogenetics) genes. Comparison of the results from both assays found rare major discrepancies (<1% of codons analyzed). INNO-LiPA detected more wild-type–mutant mixtures than sequencing but suffered from a high rate of codon hybridization failures for the reverse transcriptase. LiPA detected earlier and more frequently than sequencing the transient mixed virus population that contained I84V, which appears before V82A in the protease sequence. Mutations M461, G48V, and L90M were often transient and drug pressure related. In conclusion, direct sequencing and LiPAs give concordant results for most clinical isolates. LiPAs are more sensitive for the detection of mixed virus populations. Mutation I84V appears in minor populations in the early steps of the pathways of resistance to indinavir and ritonavir. The fact that some mutations can be found only transiently and in minor virus populations highlights the importance of a low detection limit for resistance assays.
Human immunodeficiency virus (HIV) protease (PRO) inhibitors (PIs) offer the prospect of being a durable means of suppression of viremia and a sustained increase in CD4-cell counts when used at the optimal dosage in combination with two nucleoside reverse transcriptase inhibitors (NRTIs) (1). Indeed, this highly active anti retroviral therapy (HAART) markedly reduced the rates of morbidity and mortality in HIV-infected patients (16). Unfortunately, viral drug resistance is a major cause of HAART failure. Specific patterns of mutations in the PRO gene have been associated with reduced sensitivities to different PIs under in vitro and in vivo conditions (2, 20). The fast selection of drug resistance mutations is mainly attributable to an incomplete viral replication suppression and to the error-prone nature of the reverse transcriptase (RT) (10). Short insertions within the RT and the PRO sequences have also been described (M. A. Winters, E. Kim, S. Chou, et al., Abstr. Seventh Conf. Retrovir. Opportunistic Infect., abstr. 723, 2000). Often, a significant level of phenotypic PI resistance appears only after multiple mutations have accumulated (15). Interestingly, mutant virus can keep a good replication capacity, also referred to as viral fitness, through compensatory mutations (13, 14).
Rapid and sensitive drug resistance assays have been developed for detection of RT inhibitor (RTI) resistance. Hybridization assays such as the line probe assay (LiPA) for the detection of mutations in the RT gene are accurate and reliable (4, 21, 22) but suffer from hybridization failures (12). A LiPA has recently become available for detection of key mutations that are located at the catalytic site and that confer phenotypic resistance (19). The use of genotyping has improved the virological outcomes in prospective studies (7; J. D. Baxter, D. L. Mayers, D. N. Wentworth, et al., Abstr. Sixth Conf. Retrovir. Opportunistic Infect., abstr. 8, 1999), and it has previously been shown that the detection of drug resistance by direct sequencing was associated with a poor virological outcome in retrospective studies (9, 26). On the basis of these data, tests for detection of resistance could potentially be used in the clinic to select optimal drug sequences or combinations with nonoverlapping resistance patterns, which would also preserve subsequent therapeutic options.
The objectives of our study were to compare the LiPA to DNA sequencing for the detection of mutations in the PRO and RT genes. Furthermore, the sequential appearance of mutations under selective drug pressure was investigated.
MATERIALS AND METHODS
Study design and patient population.
In a retrospective study, viruses in 197 sequential samples from 26 HIV type 1 (HIV-1)-infected patients were analyzed for drug resistance. All patients were followed at the National Department of Infectious Diseases (Centre Hospitalier de Luxembourg), were PI naive, and had a rebound of viral load despite treatment with two NRTIs (zidovudine [ZDV] and zalcitabine [ddC], n = 13; ZDV and lamivudine [3TC], n = 6; ZDV and didanosine [ddI], n = 4; 3TC and ddC, n = 2; 3TC and ddI, n = 1) and indinavir (IDV; n = 6; mean duration, 1.3 ± 0.7 years), ritonavir (RTV; n = 10; mean duration, 1.2 ± 0.8 years), or saquinavir (SQV; n = 10; mean duration, 0.9 ± 0.6 years). Half of them had had a prior exposure to NRTIs. The mean follow-up was 2.56 ± 0.65 years, with sequential samples obtained every 3 months. Samples were taken between April 1995 and February 1999. Anti-HIV treatment was changed during the follow-up period, according to the physician's choice: five patients stopped PI treatment and two did not change their PI treatments. Eleven patients switched to IDV, 6 switched to RTV, and 2 switched to nelfinavir (NFV). Resistance testing had not been used in a prospective way during the study period and thus did not alter treatment decisions. The baseline characteristics of the patients are shown in Table 1.
TABLE 1.
Baseline characteristics of patientsa
| Characteristic | Value |
|---|---|
| Age (yr [mean ± SD]) | 37.1 ± 9.98 |
| No. (%) female | 4 (15) |
| No. (%) non-Caucasian | 3 (12) |
| Risk factors for infection (no. [%] of patients) | |
| Heterosexual/homosexual | 9 (35)/15 (58) |
| IVDU/transfusion | 0/2 (8) |
| No. (%) infected with non-B HIV subtype | 4 (15) |
| Time yr [mean ± SD]) from seroconversion | 4.1 ± 3.2 |
| No. (%) at clinical stage C | 14 (54) |
| Mean ± SD viral load (log no. of RNA copies/ml) | 4.41 ± 0.90 |
| Mean ± SD CD4-cell count (no. of cells/μl) | 120 ± 113 |
| No. (%) of patients at stage 3 | 21 (81) |
| History of treatment with an NRTI | |
| No. (%) of patients | 13 (50) |
| No. of drugs (median) | 2 |
| No. of of treatment (median) | 6 |
A total of 26 patients were tested. IVDU, intravenous drug user. The date of seroconversion was calculated as the mean time between the last seronegative sample and the first seropositive sample when this period was less than 2 years (n = 12). Subtyping was performed by submission of the PRO and RT sequences to the HIV subtyping tool (located at http://www.ncbi.nlm.nih.gov/retroviruses/subtype/subtype.html) of the National Center for Biotechnology, Information, Bethesda, Md.
Nucleic acid extraction.
Viral RNA was extracted from 140 μl of EDTA-treated plasma by using the QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. RNA was eluted in 50 μl of RNase-free water and was stored at −80°C until it was used for the different assays.
LiPA.
LiPA is based on the reverse hybridization of a biotinylated PCR-amplified fragment of the HIV PRO or RT gene to short oligonucleotides probes for wild-type or mutant sequences at key resistance codons immobilized as parallel lines on nitrocellulose membrane strips (22). Streptavidin labeled with alkaline phosphatase is added and binds to previously formed hybrids. Incubation with 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium chromogen results in a purple-brown precipitate. The presence of a clearly visible line is considered a positive reaction. The mutations detected by this assay are listed in Table 2.
TABLE 2.
Identification of mutations by LiPAs (three strips)a
| Strip | Amino acid substitutions (% hybridization failures) |
|---|---|
| RT | M41L (13.8), T69D/N (12.3), K70R (12.3), L74V (19.7), M184V (19.7), L214F (17.7), T215Y/F (17.7) |
| PRO (strip 1) | D30N (2.2), M46I (5.6), G48V (5.6), I50V (0), I54V/A (0), V82A/I/F/T (7.1), I84V (7.1) |
| PRO (strip 2) | L90M (4.5) |
Samples for which PCR-amplified products were detected on agarose gels were analyzed for hybridization failures.
The amplification and hybridization conditions for the detection of RT mutations (LiPA HIV-1 RT; Innogenetics, Ghent, Belgium) have been described previously (22). Isolates with amplification failures were also tested with second-generation primers (K. DeSmet, L. Celis, R. Verhelst, et al., Abstr. Second Frankfurt Symp. Clin. Implications HIV Drug Resist., abstr. 54, 2000). For the prototype INNO-LiPA HIV Protease (Innogenetics), outer primers (K. DeSmet, L. Celis, R. Verhelst, et al., Abstr. Second Frankfurt Symp. Clin. Implications HIV Drug Resist., abstr. 50, 2000) were used on a Perkin-Elmer 9700 thermocycler under the following in-house conditions: 1.5 mM MgCl2 and 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min for 40 cycles. Nested biotinylated primers for the amplification of a fragment with PRO codons 30 to 84 were used under the following in-house conditions: 1.5 mM MgCl2 and 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min for 30 cycles, which gave a 273-bp product. In a second step, for codon 90, a fragment was amplified with nested biotinylated primers under the following in-house conditions: 1.5 mM MgCl2 and 94°C for 30 s, 45°C for 30 s, and 72°C for 30 s for 35 cycles, which resulted in an 85-bp product. Eight microliters of the nested PCR products was run on an agarose gel, and the presence of amplification products was checked after ethidium bromide staining. Hybridization conditions for PRO were similar to those for RT (22).
Sequencing of PRO and RT genes.
Complete sequencing of the PRO gene and of nucleotides 111 to 741 of the RT gene was performed by the TruGene HIV-1 assay (Visible Genetics, Toronto, Ontario, Canada), following the manufacturer's recommendations. Sequences were compared to the HIV-1 LAV 1 reference sequence (GenBank/EMBL accession number K02013). Samples that gave a sequence for RT but not for PRO were analyzed with a second-generation PRO primer (primer P2) provided by the manufacturer and now incorporated in the new version of the kit.
Data analysis and statistics.
For LiPA, amplification and hybridization failures were analyzed. For sequencing, amplification and sequencing failures were assessed. For each codon of interest, results were scored as wild type, mutant, a mixture of both, or the absence of interpretable results. Concordance was defined as the same interpretable result obtained by different methods. Minor discordances were defined as a mixed result by the first method and a homogeneous result by the second method, and major discordances were defined as a mutant result by one assay and a wild-type result by the other assay. The coprevalence of mutations was evaluated by calculating Pearson's correlation coefficients. Weighted mixed-effects linear regression was used for assessment of the association between the duration of therapy and the number of resistance mutations. All calculations were made with the statistical software package SPSS (version 9.0, 1999; SPSS, Chicago, Ill.).
Nucleotide sequence accession numbers.
All sequences were submitted to the GenBank/EMBL databases and are available under accession numbers AJ 3213, 10414-17, 10488-90, 11401-07, 401723-1977.
RESULTS
Performance of genotypic assays.
For 197 samples analyzed by the LiPA HIV-1 RT, the amplification rate per patient was 81% (17% for 24 isolates at a viral load of <500 copies/ml). Hybridization failures were common (Table 2). When compared to sequencing, hybridization failures could be explained by the following reasons: presence of codon 69-serine-serine insertions (63% of failures for codon 69) or T69N mutations (19%), M184I mutations (42%), and L74I mutations (5%). For 185 samples for which sequencing was attempted, the relative frequency of amplification was 77%. RNA could not be amplified from nine samples with less than 500 copies/ml.
For 197 samples analyzed by INNO-LiPA HIV Protease, the relative frequency of amplification for the fragment containing codons 30 to 84 was 89% (50% for 24 samples with viral loads of <500 copies/ml). Hybridization failures were uncommon (Table 2). Direct sequencing identified an unexpected M46L substitution in 75% of the samples with LiPA hybridization failures for this codon. The fragment containing codon 90 could be obtained from 84 of 86 samples (98%) previously amplified for codons 30 to 84. Of these, 5% failed to hybridize. For 185 samples analyzed by sequencing, the relative frequency of amplification was 73.5%, with amplification not achieved for any of the samples with viral loads of less than 500 copies/ml.
Detection of mixed virus populations and concordance between genotypic assays.
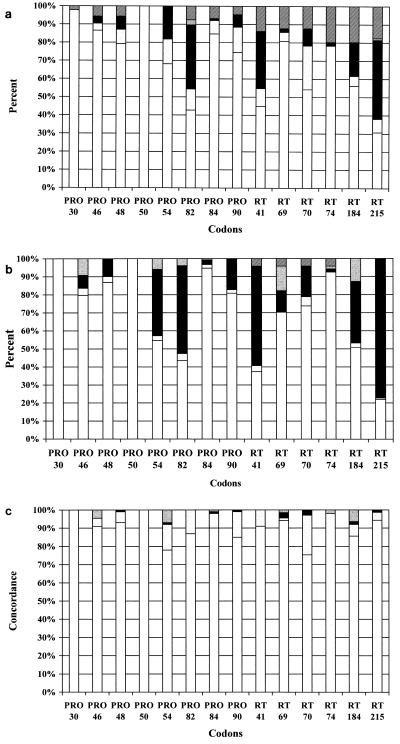
The LiPA technology detected more mixed virus populations than DNA sequencing, especially for PRO codons 54 and 90 and RT codons 41, 70, 184, and 215 (Fig. 1a and b). The results of INNO-LiPA HIV Protease and sequencing with 148 isolates were highly concordant, with identical results obtained for 87 to 100% of codons (Fig. 1c). Minor discordances were found for up to 15% of codons. Five of 887 codons (0.6%) analyzed (Fig. 1c) gave major discordances. The rates of concordance for LiPA HIV-1 RT and sequencing ranged from 91 to 98% for all codons except codons 70 and 184, which yielded more discrepancies (Fig. 1c). Major discordances were detected for 0.9% of RT codons. LiPA detected the PRO mutations V82A/F and L90M and the RT mutation K70R (four times each) as mixtures before they were detected by sequencing. However, in one case, V82A and L90M were detected earlier by sequencing.
FIG. 1.
(a) LiPA results for key positions in the PRO and RT genes. Results for wild-type (white bars), mixed wild-type and mutant (gray bars), and mutant (black bars) populations, viruses with other rare amino acid substitutions (PRO, V82F/T/I; RT, T215F) (gray bars with white dots [in PRO codon 82]), and viruses with negative results (bars with white and black lines) are shown. (b) Sequencing results for key positions in the PRO and RT genes. Results for wild-type (white bars), mixed mutant and wild-type (gray bars), and mutant (black bars) populations, viruses with other rare amino acid substitutions (PRO, M46L, I54T, V82F, and I84M, RT, T69N, codon 69 serine-serine insertion, and M184I) (gray bars with white dots [in PRO, codons 46, 54, 82, and 84; in RT, codons 69, 74, and 184]), and viruses with negative results (bars with white and black lines) are shown. (c) Comparison (percent concordance) of genotyping results obtained by LiPA and DNA sequencing. Concordance is shown as white bars, minor discrepancies are shown as gray bars, major discrepancies are shown as black bars, and unexpected amino acid substitutions are shown as gray bars with white dots (in PRO, codons 46, 54, and 84; in RT, codons 69, 74, and 184 [for RT L74I/F and M184I; for PRO, M46L, I54T, and I84M]).
Sequential appearance of PI resistance mutations.
Despite treatment failure, the viruses of six patients did not acquire key PI resistance mutations during follow-up. For both assays, the duration of PI therapy was significantly associated (P < 0.001) with the number of PI resistance mutations (for LiPA, mean ± standard error slope = 0.528 ± 0.061; for sequencing, mean ± standard error slope = 0.483 ± 0.061).
Seven patients treated with RTV or IDV transiently displayed an I84V substitution at the time of or just before the appearance of the classical V82A/F substitution. LiPA detected these transient mutations in seven samples (in two as pure populations and in five as mixed populations), and sequencing detected these transient mutations in five samples (in two as pure populations and in three as mixed populations).
For 10 patients with first-line RTV treatment, after a transient I84V in isolates from half of the patients, V82A/F gradually emerged in all of them, followed by I54V and additional mutations, e.g., K20R/M and M36I/L (n = 3) and L10I, M46I, L90M, and I93L (n = 1). Six of the patients received second-line IDV treatment. Isolates from two patients acquired an I84V mutation before the V82A/F mutation, followed by M46I/L, A71V/T, and L90M (n = 3), I93L and S37N (n = 2), and I15V, K20R, G48V, and L63P (n = 1). The G48V and A71V mutations were detected in virus from one patient with second-line RTV and SQV treatment. Viruses from three patients under first-line IDV treatment gradually developed V82A mutations; in viruses from two patients this was followed by an M46I/L mutation. In a second step, I54V/T, A71V/T, and I93L were observed. Second-line NFV treatment in one patient selected L90L/M, I54T, A71V, I77V, and I93L mutations. Of the viruses from the seven patients treated first with SQV, viruses from six developed the L90M mutation viruses from three each developed the G48V and V82A mutations; the viruses also developed some of the following mutations: L10I/V and I93L (n = 2) and K20R, M36I, S37N, R41K, L63P, A71V, and I84V (n = 1). Second-line IDV treatment selected the M46I mutation in viruses from two patients and the I54V and I93L mutations in virus from one patient.
G48V was often found as a mixed population (three of five patients) and disappeared from viruses from three of four patients after withdrawal of SQV. L90M disappeared from viruses from five of six patients after withdrawal of SQV (n = 2) and IDV (n = 2) and even after the addition of IDV to SQV (n = 1). In virus from one patient, L90M developed as a mixed population while the patient was under first-line SQV treatment, followed by additional mixed V82A and I84V substitutions. A switch to IDV resulted in the disappearance of the I84V and L90M mutations. The M46I/L mutation, mainly selected by IDV, disappeared after withdrawal of this drug in all four patients. Viruses from patients who retained their resistance mutations were the ones that had initially had more than seven PI resistance-related mutations. The D30N and I50V mutations, typical for NFV and amprenavir (APV) resistance, respectively, were not encountered in the viruses from our cohort.
Evolution of RT mutations.
Virus from a single patient had no RT mutations during follow-up. Generally, complex mutational patterns were found. Interestingly, in two patients without nonnucleoside RTI (NNRTI) treatment, the typical NNRTI resistance mutations K103N and Y181C appeared while under ZDV, 3TC, and SQV treatment in one patient, and Y181C was newly detected while under ddC, RTV, and SQV treatment in the second patient. The ZDV resistance mutation K70R was often transiently encountered as a mixed population (n = 11).
Coprevalence of PRO mutations.
PRO mutations often appeared in combinations and did not seem to be independent. The V82A/F and I54V/T mutations were associated, with good correlations (r> 0.5) in both assays. The association of M36I/L with K20R/M was not detected by LiPA because of the absence of specific probes for these positions but was found by sequencing (Table 3). Less strong but still relevant correlations were found for the A71V/T and I77V mutations (r = 0.4 to 0.5) and the E35D and M36I/L, I54V/T and A71V/T, and A71V/T and L90M mutations. The A71V/T and M36I/L mutations were inversely correlated.
TABLE 3.
Coprevalence of PRO mutations as obtained by sequencing and LiPA
| Residue | Pearson's correlation coefficienta
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L10I/V | K20R/M | E35D | M36I/L | M46I/L | G48V | I54V/T | L63P | A71V/T | I77V | V82A/F | |
| M36I/L | 0.59 | 0.41 | |||||||||
| M46I/L | 0.27 | ||||||||||
| G48V | 0.27 | ||||||||||
| I54V/T | 0.38 | ||||||||||
| L63P | −0.24 | ||||||||||
| A71V/T | 0.36 | 0.25 | 0.37 | 0.41 | 0.34 | ||||||
| I77V | −0.41 | −0.26 | 0.36 | 0.28 | 0.35 | 0.50 | |||||
| V82A/F | 0.25 | 0.26 | 0.29 | (0.19) | 0.26 | 0.64 | 0.25 | 0.37 | |||
| V82A/F | (0.19) | (0.57) | |||||||||
| I84V | 0.34 | (0.30) | (0.21) | ||||||||
| L90M | 0.25 | 0.32 | 0.46 | 0.43 | 0.25 | ||||||
Values for LiPA are given in parentheses (at a P value of 0.010).
DISCUSSION
In sequential samples from HIV-infected patients, LiPA and sequencing gave highly concordant results for the detection of resistance mutations in both PRO and RT genes. The identification of codons is more exhaustive by sequencing than by LiPA, but we showed that both LiPA are more sensitive for the detection of mixed virus populations, as previously described for LiPA HIV-1 RT (4, 12, 21). For LiPA, hybridization failures are partly explained by uncommon mutations at key codons, e.g., for the PRO I54T and M46L mutations and for the RT M184I mutation. However, due to improved primers the next generation of the LiPA HIV-1 RT is expected to have fewer failures (DeSmet et al., Abstr. Second Frankfurt Symp. Clin. Implications HIV Drug Resist., abstr. 54).
In PRO, a transient I84V mutation, mainly as a minor variant, was commonly found before the development of the V82A/F mutation for IDV and RTV resistance. Both mutations interact with the substrate at the active site. A similar phenomenon of transient detection of a mutation has been described for RT mutation K70R before the emergence of ZDV resistance. Large-scale studies have shown that I84V is also associated with APV resistance (17) (B. Schmidt, H. Walter, U. Marcus, et. al., Abstr. Seventh Conf. Retrovir. Opportunistic Infections, abstr. 726, 2000). Interestingly, the I84V mutation reappears transiently in patients receiving second-line therapy with one of two PIs, either IDV or RTV.
Second-line NFV treatment selected for the I54V/T and L90M mutations but not for the previously described D30N mutation. In IDV-treated patients, the A71V, L90M, and I93L mutations were added to the pattern of well-known IDV resistance mutations. First-line SQV treatment selected for the classical G48V and L90M mutations in association with the L10I (10, 11, 18) and I93L mutations. This confirms that, in general, secondary mutations (remote from the catalytic site) are shared between the initially used PIs.
Mixed wild-type-mutant populations of viruses were encountered in two different contexts. We have observed them as intermediate stages in resistance pathways. The detection of mixed populations can be used as an indirect tool for investigation of relative fitness. Indeed, mutations that result in less fit virus tend to disappear once the selective drug pressure stops. We observed this for most patients whose viruses had the M46I/L and G48V mutations, which are located at the upper flap region of the PRO. These mutations are associated with impaired catalytic activity and are poorly tolerated. The same phenomenon can be found for the I84V, L90M, and K70R mutations in RT. The relative fitness of a particular mutant is also enhanced in the presence of a specific PI, e.g., for L90M and SQV (11). The fitness of mutants with the I84V mutation is increased in the presence of IDV or RTV, but their fitness remains below the fitness of a virus with the L90M mutation (6, 25). This may explain the rapid emergence of the I84V mutation upon second-line IDV or RTV treatment after first-line SQV treatment. The relative fitnesses of viruses with L90M and V82A mutations are inverted for SQV and IDV (8). This explains the disappearance of the viruses with L90M mutation initially selected by SQV or IDV-SQV treatment when SQV was discontinued (n = 3 of 4 patients) and when the V82A mutation becomes predominant in a mixed population under IDV pressure (n = 1).
Our study has limitations. The use of patients with viral load rebounds to study the sequential accumulation of mutations requires a cohort different from that required for a randomized prospective trial. Sequential samples favor the investigation of intermediate stages of resistance. The high prevalence of mixed populations with mutations at some codons must be interpreted accordingly. Additionally, comparison of viruses at the intrapatient level can overestimate correlation coefficients.
In conclusion, the results of direct sequencing and LiPAs are highly concordant for samples from patients with HAART failure. LiPA is more sensitive than sequencing for the detection of mixed virus populations but results in an unacceptably high level of hybridization failures for RT. A transient mutation, I84V, often present in a minor population of viruses, is a first step toward IDV and RTV resistance. This fact highlights the importance of a low detection limit of a resistance detection assay so that important information is not missed.
ACKNOWLEDGMENTS
This study was supported by the Fondation Recherche sur le SIDA of Luxembourg, the Centre de Recherche Public-Santé (CRP-Santé) of Luxembourg, and a Glaxo Wellcome grant awarded through the Belgian Society of Infectiology and Clinical Microbiology in 1999. J. S. benefits from a Bourse-Formation Recherche (BFR97/015), Ministère de la Culture, de 1'Enseignement Supérieur et de la Recherche, Luxembourg (1997 to 2000).
REFERENCES
- 1.Collier A, Coombs R W, Schoenfeld D A, Bassett R L, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichman R C, Hooper C, Corey L. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. N Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 2.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 4.Descamps D, Calvez V, Collin G, Cécille A, Apetrei C, Damond F, Katlama C, Matheron S, Huraux J M, Brun-Vézinet F. Line probe assay for detection of human immunodeficiency virus type 1 mutations conferring resistance to nucleoside inhibitors of reverse transcriptase: comparison with sequence analysis. J Clin Microbiol. 1998;36:2143–2145. doi: 10.1128/jcm.36.7.2143-2145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devereux H L, Youle M, Johnson M A, Loveday C. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS. 1999;13:123–127. [PubMed] [Google Scholar]
- 6.Dulioust A, Paulous S, Guillemot L, Delavalle A M, Boué F, Clavel F. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J Virol. 1999;73:850–854. doi: 10.1128/jvi.73.1.850-854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher C A B, Schapiro J M, Dellamonica P. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 8.Gulnik S V, Suvorov L I, Liu B. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan P R, Hertogs K, Verbiest W, Pauwels R, Larder B, Kemp S, Bloor S, Yip B, Hogg R, Alexander C, Montaner J S G. Baseline HIV drug resistance profile predicts response to ritonavir/saquinavir protease inhibitor therapy in a community setting. AIDS. 1999;13:1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen H, Hänggi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen H, Yasargil K, Winslow D L. Characterization of human immunodeficiency virus type 1 mutants with decreased susceptibility to proteinase inhibitor RO 31-8959. Virology. 1995;206:527–534. doi: 10.1016/s0042-6822(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 12.Koch N, Yahi N, Colson P, Fantani J, Tamalet C. Genetic polymorphism near HIV-1 reverse transcriptase resistance-associated codons is a major obstacle for the line probe assay as an alternative method to sequence analysis. J Virol Methods. 1999;80:25–31. doi: 10.1016/s0166-0934(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 13.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 14.Lech W J, Wang G, Lang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molla A, Korneyeva M, Gao Q, Vasavandonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 16.Mouton Y, Alfandari S, Valette M, Cartier F, Dellamonica P, Humbert G, Lang J M, Massip P, Mechali D, Leclercq P, Modai J, Portier H. Impact of protease inhibitors on AIDS-defining events and hospitalizations in 10 French AIDS reference centres. Federation Nationale des Centres de Lutte contre le SIDA. AIDS. 1997;11:F101–F105. doi: 10.1097/00002030-199712000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Race E, Dam E, Obry V, Paulous S, Clavel F. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS. 1999;13:2061–2068. doi: 10.1097/00002030-199910220-00008. [DOI] [PubMed] [Google Scholar]
- 18.Schapiro J M, Winters M A, Lawrence J, Merrigan T C. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS. 1999;13:359–365. doi: 10.1097/00002030-199902250-00008. [DOI] [PubMed] [Google Scholar]
- 19.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:129–137. [Google Scholar]
- 20.Schmit J C, Ruiz L, Clotet B, Raventos A, Tor J, Leonard J, Desmyter J, DeClercq E, Vandamme A M. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538) AIDS. 1996;10:995–999. doi: 10.1097/00002030-199610090-00010. [DOI] [PubMed] [Google Scholar]
- 21.Schmit J C, Ruiz L, Stuyver L, VanLaethem K, Vanderlinden I, Puig T, Rossau R, Desmyter J, Clercq E D, Clotet B, Vandamme A M. Comparison of the LiPA HIV-1 RT test, selective PCR and direct solid phase sequencing for the detection of HIV-1 drug resistance mutations. J Virol Methods. 1998;73:77–82. doi: 10.1016/s0166-0934(98)00043-3. [DOI] [PubMed] [Google Scholar]
- 22.Stuyver L, Wyseur A, Rombout A, Iouwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhofstede C, VanWanzeele F, VanDerGucht B, DeCabooter N, Plum J. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS. 1999;13:2541–2546. doi: 10.1097/00002030-199912240-00007. [DOI] [PubMed] [Google Scholar]
- 24.Wainberg M A, Drosopoulos W C, Salomon H. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 25.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolopa A R, Shafer R W, Warford A, Montoya J G, Hsu P, Katzenstein D, Merigan T C, Efron B. HIV genotypic resistance patterns predicts response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]