Abstract
The emergence of drug-resistant viral variants is the inevitable consequence of incomplete suppression of human immunodeficiency virus type 1 (HIV-1) replication during treatment with antiretroviral drugs. Sequencing to determine the resistance profiles of these variants has become increasingly important in the clinical management of HIV-1 patients, both in the initial design of a therapeutic plan and in selecting a salvage regimen. Here we have developed a pyrosequencing assay for the rapid characterization of resistance to HIV-1 protease inhibitors (PIs). Twelve pyrosequencing primers were designed and were evaluated on the MN strain and on viral DNA from peripheral blood mononuclear cells from eight untreated HIV-1-infected individuals. The method had a limit of detection of 20 to 25% for minor sequence variants. Pattern recognition (i.e., comparing actual sequence data with expected wild-type and mutant sequence patterns) simplified the identification of minor sequence variants. This real-time pyrosequencing method was applied in a longitudinal study monitoring the development of PI resistance in plasma samples obtained from four patients over a 2 1/2-year period. Pyrosequencing identified eight primary PI resistance mutations as well as several secondary mutations. This sequencing approach allows parallel analysis of 96 reactions in 1 h, facilitating the monitoring of drug resistance in eight patients simultaneously and, in combination with viral load analysis, should be a useful tool in the future to monitor HIV-1 during therapy.
One of the greatest impediments to the long-term success of human immunodeficiency virus (HIV) therapy is the emergence of drug-resistant viral variants. Although treatment failure is a complex phenomenon, viral resistance is a major issue affecting 30 to 50% of all individuals under highly active antiretroviral therapy (HAART) (34). The rapid turnover of the virus population, which has a half-life of 1 to 2 days, coupled with the high mutation rate, provides an ideal setting for development of antiviral drug resistance (21). Therefore if viral replication is not completely suppressed during therapy, drug-resistant variants are selected for through the elimination of the drug-susceptible variants from the replicating viral pool (5). A mutant that has even a low degree of resistance can rapidly overgrow an existing wild-type viral population and thereby increase its probability of acquiring additional mutations that lead to higher degrees of resistance.
The most recent recommendations of the International AIDS Society state that resistance emergence is highly predictive of loss of antiretroviral activity (17). Resistance testing may therefore reduce antiretroviral cost and toxicity by identifying drugs that are likely to be less effective. This should improve patient treatment by predicting drug failure and assisting with the choice of initial therapy when drug resistance is suspected or with the choice of alternative treatment in the setting of treatment failure (14).
There are 175 HIV type 1 (HIV-1) drug resistance mutations, of which 88 occur in reverse transcriptase (RT), 52 in protease, 34 in the envelope gene, and 1 in integrase (20). Resistance testing generally focuses on the two main targets for current antiretroviral therapy i.e., the RT- (1,680-bp) and protease-encoding (297-bp) domains of the pol gene. While single mutations in RT can confer high-level resistance to some drugs (e.g., lamivudine and nevirapine), multiple mutations are required for development of significant resistance to several other drugs, including all currently licensed protease inhibitors (PI) (6, 17, 27). At least 33 amino acids in protease (involving 52 mutations) have been identified as potential contributors to phenotypic resistance, with high-level resistance measurable when one or two primary mutations develop in combination with a number of secondary mutations (6, 17, 27). Most primary mutations involve amino acids in or close to the active site of protease, while secondary mutations are generally observed outside of this domain but have a significant role in conferring resistance (12, 35).
Viral load analysis has provided a direct tool for monitoring treatment response (26), and the relationship between change in virus load and treatment benefit has been analyzed in several randomized, controlled clinical trials (8, 28). Failure to achieve a significant reduction of viral RNA in plasma after a few weeks of therapy or an increase in viral load during the course of treatment may be attributed to a number of factors such as poor adherence, inadequate drug absorption, or drug resistance (9, 15, 18, 25). Genotypic and phenotypic analysis of the virus should indicate whether drug failure is linked to drug resistance and perhaps also allow determination of which drugs are involved in resistance.
An increase in the understanding of the effects of resistance mutations on viral drug susceptibility and the fact that phenotypic assays are costly and time-consuming have led to an increasing use of genotypic drug resistance testing methods (1, 11). Genotypic analyses rely mostly on DNA sequencing via gel electrophoresis (with the capacity to analyze up to 96 samples, although in practice with this type of heterogeneous material usually only 48 samples are sequenced per run), but despite advances in standard sequencing methods this remains a time-consuming and laborious procedure. A line probe hybridization assay that interrogates a certain number of codons of the RT gene and a sequencing-by-hybridization approach using an oligonucleotide array have also been described (19, 31). Pyrosequencing, an alternative technique for sequencing DNA, was recently described (29). This non-gel-based sequencing technology is based on the iterative incorporation of specific nucleotides during primer-directed polymerase extension (Fig. 1). Using a four-enzyme mixture, the method relies on the luminometric detection of pyrophosphate that is released upon nucleotide incorporation, with each light signal generated being proportional to the number of nucleotides incorporated.
FIG. 1.
Schematic diagram of pyrosequencing. The reaction mixture consists of single-stranded DNA with an annealed primer, DNA polymerase, ATP sulfurylase, luciferase, and apyrase. The four nucleotide bases are added to the mixture in a defined order, e.g., A, C, G, and T. If the added nucleotide forms a base pair (in this case, two Ts base pair to the template), the DNA polymerase incorporates the nucleotide and consequently pyrophosphate (PPi) is released. The released pyrophosphate is converted to ATP by ATP sulfurylase, and luciferase uses this ATP to generate detectable light. This light is proportional to the number of nucleotides incorporated and is detected in real time. The pyrosequencing raw data are displayed simultaneously, and in this example the sequence generated reads ATCTT. The height of the signal is proportional to the number of nucleotides incorporated. Excess quantities of the added nucleotide are degraded by apyrase. If the nucleotide does not form a base pair with the DNA template, it is not incorporated by the polymerase and no light is produced. Apyrase then rapidly degrades the nucleotide.
Here we investigate the use of pyrosequencing for the detection of drug resistance mutations in the HIV-1 protease gene. We have focused on the most common mutations as stated by the International AIDS Society (17) (i.e., the primary resistance mutations at codons 30, 46, 48, 50, 82, 84, and 90 as well as 11 secondary mutations), but we also describe the sequencing of the remaining 15 amino acid positions implicated in drug resistance (Los Alamos HIV Drug Resistance website [http://hiv-web.lanl.gov] and Table 1).
TABLE 1.
Pyrosequencing primers for the HIV-1 protease gene and the codons that each primer sequences
| Primer | Nucleotide sequence (5′–3′) | Genomic locationa | Sequencing direction | Codons sequencedb | Codons involved in drug resistancec |
|---|---|---|---|---|---|
| 1 | CCCTCARATCACTCTTTGGC | 2275–2294 | Forward | 8–18 | 8, 10, 16 |
| 2 | TACTGTATYATCWGCYCCTGT | 2271–2251 | Reverse | 14–25 | 20, 23, 24 |
| 3 | CTATTAGAYACAGGRGCWGA | 2342–2361 | Forward | 30–35 | 30 |
| 4 | TTTCCATYTYCCTGGYAAA | 2404–2386 | Reverse | 32–36 | 32, 33, 36 |
| 5 | AAACCTCCAATTCCCCCTAT | 2433–2414 | Reverse | 37–46 | 45, 46 |
| 6d | TTRCCAGGAARATGGAIRCCAAA | 2387–2409 | Forward | 46–57 | 46, 47, 48, 50, 52 |
| 7 | ATAGGGGGAATTGGAGGTTT | 2414–2433 | Forward | 54–60 | 54, 55, 57, 60 |
| 8 | TTTATCAARGTAARACARTATGA | 2432–2454 | Forward | 61–68 | 63 |
| 9 | CAATTATGTTGACAGGTGTAGGTCC | 2531–2507 | Reverse | 67–77 | 69, 71, 73, 75, 77 |
| 10 | TGRGTCAACAIRTTTCTTCCA | 2550–2530 | Reverse | 75–84 | 81, 82, 84 |
| 11 | TACACCTRYCAACRTAATTGG | 2512–2532 | Forward | 87–94 | 88, 90, 91 |
| 12 | TTGGAAGAAAYITGWTGACYCA | 2529–2550 | Forward | 93–99 | 97 |
Genetic location (in nucleotides) refers to the MN sequence (accession no. AF075719).
Codons 1 to 7, 26 to 29, 85, and 86 were not sequenced with this set of primers.
The seven codons primarily involved in resistance are in boldface.
The G variant of primer 6 was designed as pyrosequencing failed in a viral DNA sample because of an A-C mismatch at the underlined A.
MATERIALS AND METHODS
Samples and DNA and RNA extraction. (i) HIV-1MN.
The MN strain of HIV-1 (a subtype-B strain) was used for initial evaluation and optimization of the pyrosequencing method. Peripheral blood mononuclear cells (PBMC) from a healthy blood donor were stimulated with phytohemagglutinin-P for 3 days and then infected by HIV-1MN. After 1 week, a crude cell lysate was prepared by incubating the virus-infected cells at 37°C for 12 h in 200 μl of lysis buffer (10 mM Tris-HCl [pH 9.0], 1 mM EDTA, 0.5% NP-40, 0.5% Tween 20, and 300 μg of proteinase K/ml) at a concentration of 107 cells/ml as previously described (24). The proteinase was inactivated at 95°C for 10 min, and the lysate containing the viral DNA was used directly as the template in the PCR.
(ii) Viral DNA.
Further evaluation was carried out on viral DNA obtained from crude cell lysates of uncultured PBMC from eight HIV-1-infected individuals (samples A to H) (24). These individuals had recently been diagnosed as HIV-1 infected and had never received antiretroviral therapy. They were included in this study because the protease and RT of their virus had already been sequenced on both the automated laser fluorescence (ALF) and Applied Biosystems (ABI) sequencer platforms (see below). DNA was extracted by incubating 2 × 106 cells in lysis buffer as described above, and the lysate was used directly in the PCR.
(iii) Patient samples.
Finally, the sequencing method was evaluated on viral RNA prepared from plasma samples from HIV-1-infected individuals, i.e., clinically relevant samples. Four HAART-treated patients monitored at the Department of Dermatovenereology, Södersjukhuset, Stockholm, Sweden, were retrospectively selected for detailed study based on the following criteria: (i) virological signs of treatment failure (i.e., plasma HIV-1 RNA levels > 1,000 copies/ml), (ii) at least one routine genotypic HIV resistance assay showing the presence of mutations associated with PI resistance, and (iii) availability of suitable frozen plasma samples. Data on the clinical history, antiretroviral treatment history, development of plasma HIV-1 RNA levels, and the genetic resistance profile were obtained from the patient records. Plasma HIV-1 RNA levels were determined by the standard or ultrasensitive Amplicor HIV-1 monitor assay (Roche Diagnostic Systems, Branchburg, N.J.). For simplicity, in this study 500 HIV-1 RNA copies/ml was considered the lower limit of detection of both assays. The genotypic HIV-1 resistance assays were performed by Professional Genetics Laboratories AB, Uppsala, Sweden. From each patient, one plasma sample obtained before the start of PI-containing treatment and four plasma samples obtained during the course of treatment were selected. These samples were retrieved from storage at −70°C, and viral RNA was extracted from 200 μl of plasma using the Nuclisens RNA extraction kit (NASBA Diagnostics, Organon Teknika, Boxtel, The Netherlands). RNA was eluted in 50 μl of elution buffer according to the recommendations of the manufacturer, and cDNA synthesis was performed on 8 μl of eluted RNA at 37°C for 60 min using the First-Strand cDNA synthesis kit (Amersham Pharmacia Biotech, Uppsala, Sweden). Fifteen out of the 20 plasma samples yielded a PCR product following RNA extraction, cDNA synthesis, and nested PCR and thus could be sequenced.
PCR.
Outer PCR was carried out using 0.1 μM concentrations of primers JA199 (5′-GAA AGG AAG GAC ACC AAA TGA AAG A-3′; nucleotides 2050 to 2075 on the MN reference sequence; accession no. AF075719) and JA202 (5′-GCC ATT GTT TAA CTT TTG GGCCAT C-3′; nucleotides 2646 to 2621 on the above MN reference sequence) in a 50-μl reaction volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR was performed using a temperature profile of 94°C for 5 min, 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Inner PCR was performed on 2.5 μl of the outer PCR product with 0.1 μM concentrations of primers JA200 (5′-CAG AGC CAA CAG CCC CAC CAG AAG A-3′; nucleotides 2158 to 2182 on the MN reference sequence) and JA201 (5′-CAT CCA TTC CTG GCT TTTA ATT TTA C-3′; nucleotides 2625 to 2600 on the MN reference sequence) as described for the outer PCR.
Sanger sequencing. (i) Viral DNA.
The 467-bp PCR products were subjected to direct Sanger sequencing using the Thermo Sequenase fluorescently labeled primer cycle sequencing kit (Amersham Pharmacia Biotech) and were analyzed on 6% polyacrylamide gels on an ALF sequencing machine (Amersham Pharmacia Biotech). Sequencing on the ABI 310 automated sequencer (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) was also carried out using Big Dye terminators. To confirm the presence of mixed variants, some of the HIV-1 viral DNA samples were A/T cloned into the (pGEM-T vector (Promega, Madison, Wis.) and sequenced using DYEnamic ET terminator chemistry on the MegaBACE platform (Amersham Pharmacia Biotech).
(ii) Patient samples.
PCR products were directly cycle sequenced using Big Dye terminator chemistry on the ABI 377 platform (Perkin-Elmer Applied Biosystems).
Pyrosequencing. (i) Design of primers for pyrosequencing.
Twelve primers were designed to anneal adjacent to codons that are involved in drug resistance. Due to the heterogeneity of HIV-1, degenerate primers were designed (Table 1) based on an alignment of 65 HIV-1 pol gene sequences that were obtained from the Los Alamos HIV Sequence Database (http://hiv-web.lanl.gov). Generally primers were designed with 8-fold (or less) degeneracy (primer 12 was an exception, with 16-fold degeneracy) with inosine used in some primers. In one case an alternate primer (nucleotides 2283 to 2302 on the above MN reference strain) to primer 1 was used to eliminate the sequencing of a cytosine stretch prior to codon 10 (see Fig. 6A, codon 10).
FIG. 6.
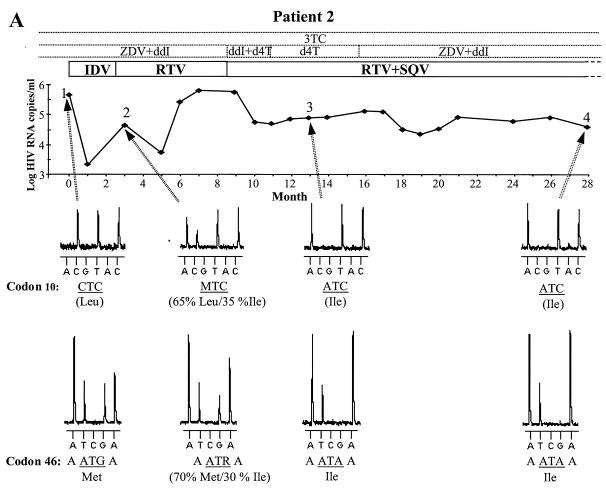
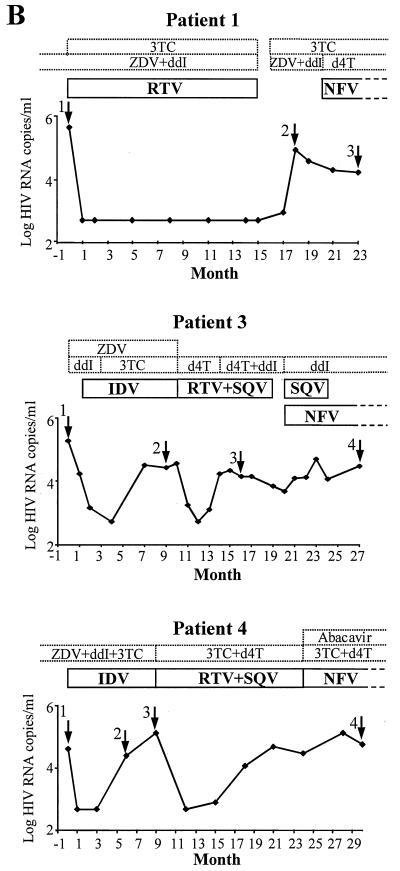
Graphs showing changes in HIV RNA levels and treatment in four patients. Arrows indicate samples from time points 1 to 4 that were subjected to pyrosequencing; dotted and solid boxes indicate NRTI and PI treatments, respectively. Open boxes illustrate that the patients were receiving these drugs before and/or after this 2 1/2-year period. ZDV, zidovudine; ddI, didanosine; 3TC, lamivudine; d4T, stavudine. (A) HIV RNA levels in patient 2. Pyrosequencing data show the development of drug resistance at codons 10 and 46. The amino acids involved in drug resistance are shown beneath the DNA sequence, with the approximate proportions of mixed variants at time point 2 indicated. The mixed bases follow the IUB code (M is A or C; R is A or G). (B) HIV RNA levels in patients 1, 3 and 4.
(ii) Preparation of template.
Nested PCR was carried out (as described above) to generate a template for pyrosequencing. The 467-bp amplicon was checked by agarose gel electrophoresis prior to its preparation for pyrosequencing. To allow for immobilization of the PCR product on streptavidin beads and preparation of single-stranded DNA, a biotinylated inner PCR primer was used. A biotinylated JA201 or JA200 primer was used in the PCR depending on whether forward or reverse pyrosequencing primers were used. The biotinylated inner PCR product (50 μl) was immobilized onto 200 μg of streptavidin-coated superparamagnetic beads (Dynabeads M280; Dynal, Oslo, Norway) in 30 μl of BW buffer (10 mM Tris-HCl [pH 7.5], 2 M NaCl, 1 mM EDTA, 0.1% Tween 20) at 43°C for 15 min. Single-stranded DNA was obtained by incubating the beads with the immobilized PCR product in 20 μl of 0.1 M NaOH for 5 min. The immobilized strand was resuspended in annealing buffer (10 mM Tris-acetate [pH 7.75] 2 mM magnesium acetate) containing 2 pmol of sequencing primer in a total volume of 10 μl. In this study, only the immobilized strand was used for pyrosequencing. The single-stranded preparation was automated and performed in a 96-well format using robotics (Magnetic BioSolutions AB, Stockholm, Sweden), with the procedure taking 40 min for 96 samples. The robotic work station consists of a 12-tip pipette head, a Peltier heating and cooling position for a microtiter plate, and positions for reagents, tips, and waste. The beads are selectively captured inside the tips by a magnet, which facilitates washing and exchange of buffers. Primer annealing was performed by incubation at 94°C for 1 min and 63°C for 1 min, with subsequent cooling to room temperature. Thirty microliters of H2O and 0.5 μg of single-stranded DNA binding protein (Amersham Pharmacia Biotech) were added to the single-stranded DNA template before sequencing.
(iii) Pyrosequencing.
Real-time pyrosequencing was performed at 28°C in a total volume of 50 μl in an automated 96-well pyrosequencer using PSQ SNP 96 enzymes and substrates (Pyrosequencing AB, Uppsala, Sweden) with cyclic dispension of the nucleotides. The base calling of the pyrograms, pattern recognition, and assignment into amino acid sequences were performed manually.
RESULTS
Pyrosequencing primers.
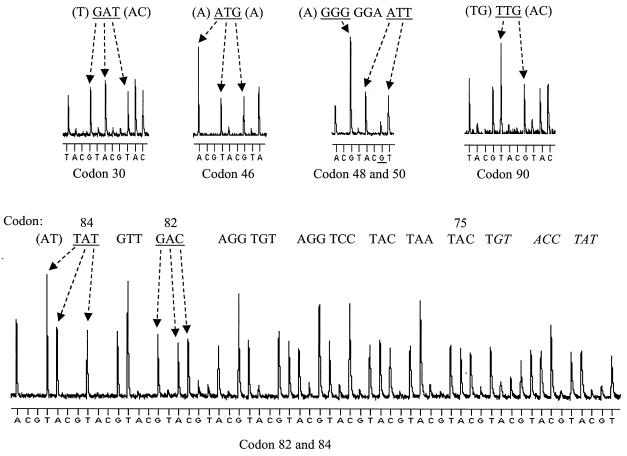
To carry out pyrosequencing (Fig. 1), 12 primers that hybridized along the length of the protease gene adjacent to codons involved in drug resistance were designed (Table 1). These primers had a certain degree of degeneracy to cover reported sequence variations in this region. Initial evaluation was carried out on the HIV-1MN strain with the 12 primers sequencing 263 bases of the protease gene (297 bp), allowing the 33 amino acid positions implicated in drug resistance to be sequenced (Table 1) (20). The results of sequencing seven codons (codons 30, 46, 48, 50, 82, 84, and 90) in the protease gene that are primarily involved in drug resistance are illustrated in Fig. 2. A slowly decreasing signal for iterative cycles of nucleotide addition was observed, resulting in an average read length of approximately 26 bases for each primer. There were some examples of ambiguities in the pyrosequencing of MN due both to “noncalls” of the last base of a homopolymeric stretch (>3 bases) and negative frameshifts (i.e., incomplete nucleotide incorporation by the DNA polymerase) after these stretches (e.g., a small G frameshift in codon 50 in Fig. 2). As described below, the ambiguities appear consistently, which allows for pattern discrimination between wild-type sequences and altered sequences.
FIG. 2.
Pyrosequencing the seven codons in the protease gene that are primarily involved in drug resistance. The sequences of these codons (underlined) and their surrounding nucleotides are displayed above the pyrosequencing data. A negative G frameshift (underlined) is observed after sequencing codon 48. Codons 82 and 84 were sequenced (in the reverse direction) using primer 10, and the complete sequence shows that readability is maintained over 33 nucleotides, i.e., codons 84 to 75 (indicated above the sequence). Ambiguous sequence data are in italics. Note that the peaks between the individual codons cannot be directly compared, as the raw data are not to scale.
Identification of polymorphic nucleotide positions.
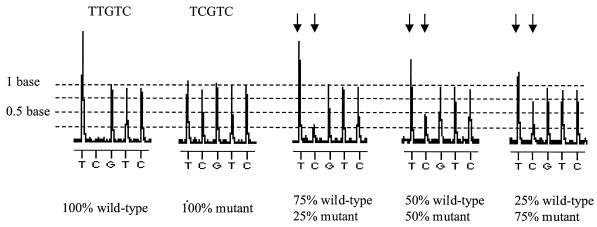
The ability to detect the presence of minor variants within the virus population is a problem with all genotypic screening methods. To establish the limit of detection of mixed variants by pyrosequencing, PCR products generated from a wild-type clone (MN) and a mutant clone were mixed at different ratios and subjected to pyrosequencing with primer 1. Pyrosequencing allowed detection of each clone when it was present at a relative concentration of 25% of the total population (Fig. 3), which is comparable to conventional sequencing strategies (23, 30).
FIG. 3.
Pyrosequencing data on defined mixtures of wild-type (MN) and mutant clones. The wild-type clone has the sequence TTGTC, while the mutant has the sequence TCGTC. Arrows correspond to positions where different ratios of the two templates give rise to different signal levels. These peaks are proportional to the percentage of each clone present in the mixture.
The performance of HIV-1 pyrosequencing was then evaluated by analyzing the sequence of the protease gene on viral DNA derived from eight patient PBMC samples (samples A to H). Since these patients were treatment naïve, they were not expected to display mutations associated with resistance to PIs. The obtained sequences correlated well with Sanger dideoxy sequencing, with a few discrepancies observed, which are described below.
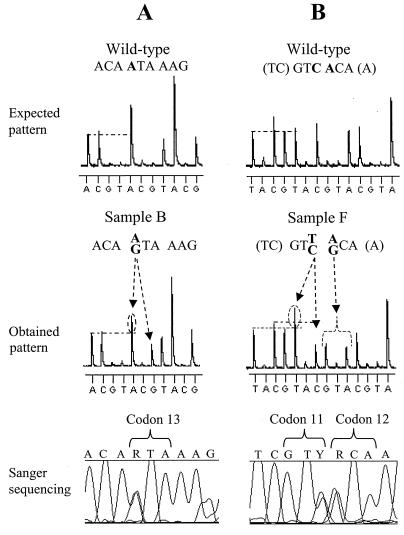
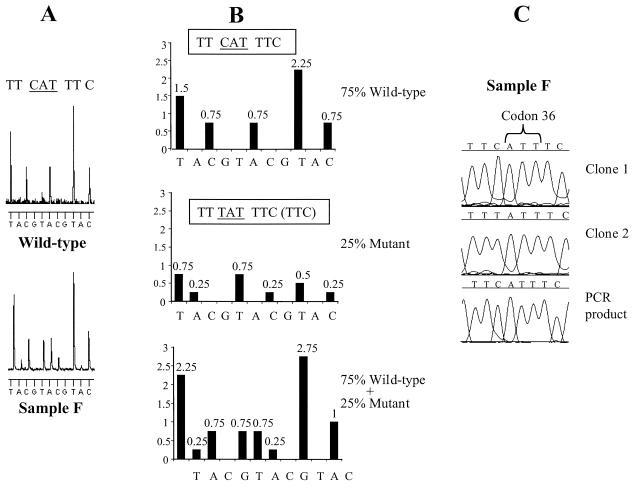
Figure 4 shows the detection of mixed variants (i.e., sequence polymorphisms) in samples B and F. Analysis of pyrosequencing raw data suggested a 40% A–60% G mixture at codon 13 (RTA) in sample B (Fig. 4A) and a 50% T–50% C and 50% A–50% G mixtures at codons 11 (GTY) and 12 (RCA), respectively, in sample F (Fig. 4B). Sanger sequencing confirmed these results. In one case, pyrosequencing identified a minor variant present in 25% of the wild-type population which conventional sequencing (on both the ALF and ABI platforms) failed to detect (Fig. 5). Due to the nature of pyrosequencing, mutations can easily be spotted by searching for changes in the pyrosequencing pattern. In this case, pyrosequencing codon 36 in sample F gave an unexpected pattern (i.e., a pattern different from that of the wild-type sample) (Fig. 5A). Analysis of this pattern suggested that mixed variants were present, and a predicted pyrosequencing pattern of a 75%–25% mixed population was constructed (Fig. 5B). Comparison of this predicted pattern with the pyrosequencing raw data reveals that indeed 25% of the sequences have the mutation TAT. Cloning this sample and sequencing multiple clones found TAT in 20% of the clones (Fig. 5C, clone 2). Since this codon was sequenced in the reverse direction, the minor variant represents a Met36→Ile mutation. This sequence variation is reported as a secondary PI resistance mutation but is also relatively common in untreated Swedish patients (2).
FIG. 4.
Pyrosequencing polymorphic positions in HIV-1 DNA populations derived from PBMC from treatment-naïve HIV-1-infected patients. (A) Comparison of the expected pattern generated from a wild-type sequence with the observed pattern (using primer 1) reveals the presence of mixed variants in sample B. Detailed analysis of the observed pattern reveals that one A peak (arrow) is approximately 40% larger than the first A peak and that the subsequent G peak is 60% of a normal G peak. Therefore the sequence of codon 13 in sample B is RTA. Direct Sanger sequencing of the PCR product shows a 50% mixture of A and G at codon 13. (B) Comparison of the expected pattern generated from a wild-type sequence with the observed pattern (using primer 1) reveals the presence of mixed variants in sample F. Detailed analysis of the observed pattern reveals that the second T peak (arrow) is 50% larger than the first T peak and that the next C peak is 50% of a normal C. The sequence of codon 11 in sample F is therefore GTY. The next base is also ambiguous in that G and A peaks which are approximately 50% of normal G and A peaks appear. The sequence of codon 12 in sample F is therefore RCA. Direct Sanger sequencing of the PCR product shows approximately 50% mixtures of T and C and A and G at codons 11 and 12, respectively. The mixed bases follow the International Union of Biochemistry (IUB) code (R is A or G, and Y is T or C).
FIG. 5.
Detection of a minor virus variant by pyrosequencing which is not detected by Sanger sequencing. (A) Pyrosequencing patterns observed with primer 4 on a wild-type sample and sample F. The pattern generated for the wild type was also expected for sample F. (B) Predicted pyrosequencing pattern of sample F if composed of mixed variants (75% wild type [CAT] and 25% mutant [TAT]). (C) Sanger sequencing on sample F. Clone 1, sequencing of a clone with the sequence CAT at codon 36; clone 2, sequencing of a clone with the sequence TAT at codon 36; PCR product, direct sequencing of the PCR product revealing the sequence CAT for codon 36 with no peak representing the minor T variant observed.
Various discrepancies between the ALF data and the pyrosequencing data were obtained in the analysis of sample E. The sequence of codon 10 is CTM according to ALF (60% A and 40% C) and ABI data (80% A and 20% C). However, the initial pyrosequencing of this codon called the sequence CTA. Pyrosequencing a new PCR product (product 2) called CTC at this position, with a small A peak appearing, which corresponds to a 15 to 20% mixture of the A variant. The direct Sanger sequencing of these two particular PCR products showed a CTA (PCR product 1) or a CTC (PCR product 2) at this position depending on which PCR product was sequenced. Cloning and sequencing PCR product 2 (eight clones sequenced) revealed a CTC in seven clones and a CTA in one clone, corresponding to the 15 to 20% mixture of the CTA variant seen when pyrosequencing PCR product 2. The ambiguous base calling suggests that viral DNA copy numbers in the sample were low, with results depending on whether CTA or CTC variants were overrepresented in the input viral DNA for the individual PCRs (i.e., a sampling artifact).
A final discrepancy is in sample H at codon 88; ABI data call MAT (70% A and 30% C) for this codon, while pyrosequencing consistently calls AAT. In agreement with pyrosequencing, we could see no C at this position when ALF sequencing was performed. Sampling artifacts cannot be excluded, as the sequences were derived from different PCR products, but this discrepancy is unlikely to be due to the presence of mixed variants, as cloning this sample did not reveal any minor variant (14 clones were all AAT).
Development of PI resistance in patients.
Pyrosequencing was used to monitor the development of drug resistance mutations in plasma samples from four patients who developed resistance to various PIs. Relevant information about the patients is presented in Table 2. All four patients had a PI added to a failing treatment regimen of two or more nucleoside analogue RT inhibitors (NRTIs). In only one case (patient 1) was a new NRTI (lamivudine) added when the PI treatment was initiated. Thus, in practice three of the four patients were given PI monotherapy, which helps to explain the poor outcome of treatment. It should be noted that the study was retrospective; today care would have been taken to exchange as many as possible of the failing NRTIs when the PI treatment was started. Samples were obtained prior to initiation of PI-containing therapy and at approximately 2-month intervals over a 2 1/2-year period with selected samples sequenced (Fig. 6). The nucleotide substitutions, discovered by pyrosequencing, in codons that have been implicated in PI resistance are listed in Table 3. There were no discrepancies between the pyrosequencing and the Sanger sequencing data for these codons.
TABLE 2.
Clinical data on patients 1 to 4
| Parameter | Data for patient:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Clinical data at start of HAART | ||||
| Sex | Male | Male | Male | Male |
| Age (yrs) | 42 | 46 | 34 | 30 |
| Time since diagnosis of HIV-1 infection (mos) | 76 | 108 | 37 | 12 |
| Prior AIDS diagnosis | No | No | No | Yes |
| CD4 cell count | 100 | 100 | 100 | 170 |
| Log plasma HIV-1 RNA copies/ml | 5.7 | 5.7 | 4.2 | 4.6 |
| Prior nucleoside analogue (NA) treatment (mos) | 32 | 56 | 33 | 9 |
| New NA (at least one) at start of HAART | Yes | No | No | No |
| PI at start of HAART | RTV | IDV | IDV | IDV |
| Clinical data at end of follow-up | ||||
| Length of follow-up after start of HAART (mos) | 23 | 28 | 27 | 30 |
| Time with no or partial HAART (days) | 127 | 0 | 28 | 0 |
| New AIDS-defining events or death | No | No | No | No |
| CD4 cell count | 370 | 330 | 280 | 150 |
| Log plasma HIV-1 RNA copies/ml | 4.3 | 4.6 | 4.5 | 4.8 |
TABLE 3.
Development of drug resistance mutations in patients 1 to 4 over a 2 1/2-year period
| Patient | Amino acida (codon) at time point:
|
PI implicatedb | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 1 | Asp30 (GAT) | Asp30 (GAT) | Asn30 (AAT) | 1°, NFV | |
| 2 | Leu10 (CTC) | Leu/Ile10 (MTC) | Ile10 (ATC) | Ile10 (ATC) | 2°, IDV-SQV |
| Leu24 (TTA) | Leu24 (TTA) | Ile24 (ATA) | Ile24 (ATA) | 2°, IDV | |
| Met46 (ATG) | Met/Ile46 (ATR) | Ile46 (ATA) | Ile46 (ATA) | 1°, IDV; 2°, RTV | |
| Gly48 (GGG) | Gly48 (GGG) | Val48 (GTG) | Val48 (GTG) | 1°, SQV | |
| Ala71 (GCT) | Ala71 (GCT) | Val71 (GTT) | Val71 (GTT) | 2°, IDV-RTV-SQV | |
| Val77 (GTA) | Val/Ile77 (RTA) | Ile77 (ATA) | Ile77 (ATA) | 2°, IDV-RTV-SQV | |
| Val82 (GTC) | Val82 (GTC) | Ala82 (GCC) | Ala82 (GCC) | 1°, IDV-RTV; 2°, SQV | |
| Ile84 (ATA) | Ile84 (ATA) | Val84 (GTA) | Val84 (GTA) | 1°, RTV; 2°, SQV | |
| 3 | Leu/Ile10 (MTC) | Ile10 (ATC) | Ile10 (ATC) | Ile10 (ATC) | 2°, IDV |
| Met46 (ATG) | Met46 (ATG) | Ile46 (ATA) | Ile46 (ATA) | 1°, IDV; 2°, RTV | |
| Gly73 (GGT) | Gly73 (GGT) | Ser73 (AGT) | Ser73 (AGT) | 2°, IDV-SQV | |
| Leu90 (TTG) | Leu90 (TTG) | Met90 (ATG) | Met90 (ATG) | 1°, SQV | |
| 4 | Leu90 (TTG) | Leu90 (TTG) | Leu90 (TTG) | Met90 (ATG) | 1°, SQV |
Resistance-associated amino acids are in boldface. Where two amino acids are listed (with their abbreviations connected by a shill), both amino acids were found at the position.
1°, primary; 2°, secondary.
Figure 6A illustrates the PI regimen and plasma HIV-1 RNA levels of patient 2 alongside pyrosequencing data for two codons implicated in PI resistance (i.e., codons 10 and 46). In this patient, the plasma HIV-1 RNA levels dropped drastically by 2.5 log units following initiation of indinavir (IDV) therapy, but after 2 months they had rebounded by 1.5 log units. Sequencing the plasma sample obtained at time point 2 revealed the presence of a mixture of wild-type and mutant sequences at both codons 10 and 46 (Fig. 6A and Table 3). At time points 3 and 4, only the mutant variants were detected at these two codons. The mutation Met46→Ile is a primary resistance mutation for IDV and, along with Leu10→Ile, probably caused resistance to IDV resulting in the increase in plasma viral levels observed at time point 2. Treatment was changed to ritonavir (RTV), and later saquinavir (SQV) was added. During this therapy, the plasma virus levels remained high and several additional PI resistance mutations developed, including primary resistance mutations for IDV-RTV (Val82→Ala) and SQV (Gly48→Val) (Table 3).
Figure 6B shows the viral load data for patients 1, 3, and 4 and the samples that were subjected to pyrosequencing. In patient 1, only the characteristic primary resistance mutation for nelfinavir (NFV) (Asp30→Asn) was observed 3 months after NFV treatment was initiated (Table 3). In patient 3, primary resistance mutations for IDV (Met46→Ile) and SQV (Leu90→Met) were observed after 9 months of IDV treatment followed by 9 months of RTV-SQV combination therapy. The secondary resistance mutation (Leu10→Ile) was present as a minor variant in the PI-naïve sample and probably represents a naturally occurring sequence polymorphism in this patient. However, upon PI selection pressure, the Ile10 variant became dominant in the virus population. In patient 4, a primary mutation which may have resulted in SQV resistance (Leu90→Met) is observed at time point 4 (Table 3). It is somewhat surprising that IDV treatment failure occurred in patients 3 and 4 without evidence of any primary IDV resistance mutations when viral RNA levels rebounded. This could indicate problems with treatment adherence or drug absorption.
DISCUSSION
An international expert panel recently stated that HIV-1 resistance testing is recommended to help guide the choice of new regimens after treatment failure and for guiding therapy for pregnant women and should be considered in treatment-naïve patients (17). To facilitate such large-scale sequencing, we have developed a pyrosequencing method for genotypic resistance analysis of relevant regions of the protease gene. This sequencing technique allows rapid real-time determination of 20 to 30 bases of a target sequence and is performed in an automated microtiter-based instrument allowing parallel analysis of 96 sequencing reactions, facilitating the monitoring of drug resistance simultaneously in eight patients. Each round of nucleotide dispensation takes approximately 1 min, and thus the sequence of the codons involved in drug resistance can be determined in 1 h (excluding time for sample preparation and data evaluation). To detect drug resistance mutations, pyrograms can easily be compared with those from a previous time point or a wild-type reference pattern (the comparison was performed manually in this study), but further development is needed before an automated genotyping method based on pyrosequencing is ready for large-scale clinical use.
Here we have designed a set of primers for the highly variable protease gene of HIV-1, which facilitates the analysis of the 33 amino acid positions that are involved in both primary and secondary drug resistance mutations. Primary mutations directly reduce the binding affinity of the drug to HIV protease (13) and, since each inhibitor has a different structure, primary mutations (with the exception of those conferring resistance to RTV and IDV) are generally distinct for a given inhibitor. In contrast, secondary mutations are common to other PIs and are located outside of the active site towards the surface of the enzyme. They usually appear after the primary mutation and often compensate for the deleterious effect of mutations in the active site, and as a result resistant variants often display significant cross-resistance to several inhibitors of the same class (7, 33). Since the effects of different combinations of mutations and polymorphisms have not all been elucidated, different interpretations of PI resistance and cross-resistance occur. However, the existence of two or more key mutations (e.g., Leu10→Ile and Leu90→Met) is likely to confer broad cross-resistance to most currently available PI classes (16, 17).
We identified a total of eight primary and six secondary mutations in a longitudinal study of four patients. The virus harbored by patient 2 had four primary mutations and appeared to be resistant to IDV, RTV, and SQV. The two classes of RT inhibitors (nucleoside analogues and the nonnucleoside analogues) also have a number of characteristic mutations that confer resistance (32), so this screening method could be applied to both the RT and the protease genes in a routine setting. Preliminary data indicate that pyrosequencing can be applied to a large template (1.2 kb) encompassing the protease-encoding region and most of the RT-encoding region (to codon 250), although lower signals are obtained for longer templates due to the immobilization capacity of the magnetic beads used in the single-strand separation. However, since codons for approximately 250 amino acid positions (by extrapolation, necessitating 30 primers) would need to be interrogated in the RT gene, pyrosequencing would need to be adapted to a 384 microtiter plate format to increase throughput if both the protease and RT genes are to be sequenced. In such applications the possibility of encountering primer-template mismatches also exists. However, primer-template mismatches were generally well tolerated in this study, as there were only three cases where mismatches affected extension. In one case, the primer was redesigned, which allowed the pyrosequencing of a viral DNA sample (primer 6, Table 1). Surprisingly, T-G and A-C mismatches were the least-tolerated mismatches, contrary to previous reports on various primer-template mismatches (22).
The detection of minor variants in the virus population is an important issue in the context of HIV-1 genotyping as it allows the identification of resistant variants before they become the dominant population (5). The various sequencing approaches have previously been shown to be highly variable, but in general, laboratories performing good-quality sequencing should be able to pick up a mutant present at a relative concentration of 25% of the total population (10, 30). Here we could easily pick up a mutant when it comprised 50% of the population, with the limit of detection lying somewhere between 20 and 25%. An interesting example is where pyrosequencing picked up a drug-resistant mutant, present at 25% of the wild-type population, which was not detected by conventional sequencing. The nature of pyrosequencing is such that, if a mutant is present, an irregular pattern will be generated. Thus it is easy to differentiate between wild-type and resistant sequences by comparing pyrograms (pattern recognition) to score mutations. Pyrosequencing can also be applied to plasmid clones; this facilitates clonal analysis, identifying minor variants present in the population.
Various discrepancies between the Sanger sequences and the pyrosequencing data occurred in the analysis of viral DNA samples. Since different PCR products from the same sample gave rise to different sequences, these discrepancies are likely to be due to differential sampling of the virus variants present in the sample. As expected, these discrepancies were usually observed in samples with low HIV-1 copy numbers, which is probably due to the higher statistical chance of amplifying one or a few individual HIV-1 genomes rather than a representative sample of the entire virus population.
During clinical trials, the management of HIV-1-infected patients relies significantly on determination of viral load as a marker for therapeutic effect and the potential emergence of resistant forms of the virus (36). Currently it is recommended that a minimum of two HIV-1 RNA measurements less than 2 weeks apart be obtained before initiating or changing therapy (3). In general, HIV RNA should be monitored monthly until the goal of therapy is reached and every 2 to 3 months thereafter (4). Failure to achieve the target level of less than 50 copies/ml or observance of an increase in viral load during the course of treatment suggests problems with poor adherence, inadequate drug absorption, or drug resistance (18, 25). In most cases, treatment failure is closely linked to emergence of drug-resistant virus variants (9, 15). Genotyping to determine resistance profiles in such patients has been shown to be beneficial in selecting a salvage regimen, and sequencing results have therefore become important in the clinical care of HIV-1-infected patients (1, 11). Hence, current recommendations of anti-HIV-1 therapy state that the selection and monitoring of patients should include both viral load and comprehensive genotypic analysis, and in light of this recommendation, this new non-gel-based sequencing technology should be useful in the future to monitor drug resistance in a routine setting.
ACKNOWLEDGMENTS
We thank Gisela Sitbon of Professional Genetics Laboratories AB, Uppsala, Sweden, for help with protease sequences and Anders Holmberg for assistance with the robotics. We acknowledge Pyrosequencing AB, Uppsala, Sweden, for supply of pyrosequencing reagents.
This work was supported by grants from NUTEK (Swedish National Board for Industrial and Technical Development), TFR (Swedish Research Council for Engineering Sciences), and the Swedish Medical Research Council.
REFERENCES
- 1.Baxter J D, Mayers D L, Wentworth D N, Neaton J D, Hoover M L, Winters M A, Mannheimer S B, Thompson M A, Abrams D I, Brizz B J, Ioannidis J P, Merigan T C. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–F93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- 2.Birk M, Sonnerborg A. Variations in HIV-1 pol gene associated with reduced sensitivity to antiretroviral drugs in treatment-naive patients. AIDS. 1998;12:2369–2375. doi: 10.1097/00002030-199818000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Brambilla D, Reichelderfer P S, Bremer J W, Shapiro D E, Hershow R C, Katzenstein D A, Hammer S M, Jackson B, Collier A C, Sperling R S, Fowler M G, Coombs R W. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. The Women Infant Transmission Study Clinics. Virology Quality Assurance Program. AIDS. 1999;13:2269–2279. doi: 10.1097/00002030-199911120-00009. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter C C, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society-USA Panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 5.Condra J H. Resistance to HIV protease inhibitors. Haemophilia. 1998;4:610–615. doi: 10.1046/j.1365-2516.1998.440610.x. [DOI] [PubMed] [Google Scholar]
- 6.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 8.Coombs R W, Welles S L, Hooper C, Reichelderfer P S, D'Aquila R T, Japour A J, Johnson V A, Kuritzkes D R, Richman D D, Kwok S, Todd J, Jackson J B, DeGruttola V, Crumpacker C S, Kahn J. Association of plasma human immunodeficiency virus type 1 RNA level with risk of clinical progression in patients with advanced HIV-1 infection. J Infect Dis. 1996;174:704–712. doi: 10.1093/infdis/174.4.704. [DOI] [PubMed] [Google Scholar]
- 9.Cozzi Lepri A, Sabin C A, Staszewski S, Hertogs K, Muller A, Rabenau H, Phillips A N, Miller V. Resistance profiles in patients with viral rebound on potent antiretroviral therapy. J Infect Dis. 2000;181:1143–1147. doi: 10.1086/315301. [DOI] [PubMed] [Google Scholar]
- 10.Demeter L M, D'Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard T M, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working Group. AIDS Clinical Trials Group. J Virol Methods. 1998;75:93–104. doi: 10.1016/s0166-0934(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 11.Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher C A, Schapiro J M, Dellamonica P. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 12.Greer J, Erickson J W, Baldwin J J, Varney M D. Application of the three-dimensional structures of protein target molecules in structure-based drug design. J Med Chem. 1994;37:1035–1054. doi: 10.1021/jm00034a001. [DOI] [PubMed] [Google Scholar]
- 13.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 14.Hammer S M, Yeni P. Antiretroviral therapy: where are we? AIDS. 1998;12(Suppl. A):S181–8. [PubMed] [Google Scholar]
- 15.Harrigan P R, Hertogs K, Verbiest W, Pauwels R, Larder B, Kemp S, Bloor S, Yip B, Hogg R, Alexander C, Montaner J S. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS. 1999;13:1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 16.Hertogs K, Bloor S, Kemp S D, van den Eynde C, Alcorn T M, Pauwels R, van Houtte M, Staszewski S, Miller V, Larder B A. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS. 2000;14:1203–1210. doi: 10.1097/00002030-200006160-00018. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch M S, Brun-Vezinet F, D'Aquila R T, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 19.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken C L, Foley B, Hahn B, Korber B, McCutchan F, Marx P A, Mellors J W, Mullins J I, Sodroski J, Wolinksy S. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1999. [Google Scholar]
- 21.Kuritzkes D R. Clinical significance of drug resistance in HIV-1 infection. AIDS. 1996;10(Suppl. 5):S27–S31. doi: 10.1097/00002030-199612005-00005. [DOI] [PubMed] [Google Scholar]
- 22.Kwok S, Kellogg D E, McKinney N, Spasic D, Goda L, Levenson C, Sninsky J J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyo E M, Uhlen M. Analysis of heterogeneous viral populations by direct DNA sequencing. BioTechniques. 1993;15:120–127. [PubMed] [Google Scholar]
- 24.Leitner T, Korovina G, Marquina S, Smolskaya T, Albert J. Molecular epidemiology and MT-2 cell tropism of Russian HIV type 1 variant. AIDS Res Hum Retroviruses. 1996;12:1595–1603. doi: 10.1089/aid.1996.12.1595. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzi P, Yerly S, Abderrakim K, Fathi M, Rutschmann O T, von Overbeck J, Leduc D, Perrin L, Hirschel B. Toxicity, efficacy, plasma drug concentrations and protease mutations in patients with advanced HIV infection treated with ritonavir plus saquinavir. Swiss HIV Cohort study. AIDS. 1997;11:F95–F99. doi: 10.1097/00002030-199712000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 27.Molla A, Granneman G R, Sun E, Kempf D J. Recent developments in HIV protease inhibitor therapy. Antivir Res. 1998;39:1–23. doi: 10.1016/s0166-3542(98)00011-4. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien W A, Hartigan P M, Daar E S, Simberkoff M S, Hamilton J D. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann Intern Med. 1997;126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 29.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 30.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winters M A, Baxter J D, Mayers D L, Wentworth D N, Hoover M L, Neaton J D, Merigan T C. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antivir Ther. 2000;5:57–63. [PubMed] [Google Scholar]
- 33.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wit F W, van Leeuwen R, Weverling G J, Jurriaans S, Nauta K, Steingrover R, Schuijtemaker J, Eyssen X, Fortuin D, Weeda M, de Wolf F, Reiss P, Danner S A, Lange J M. Outcome and predictors of failure of highly active antiretroviral therapy: one-year follow-up of a cohort of human immunodeficiency virus type 1-infected persons. J Infect Dis. 1999;179:790–798. doi: 10.1086/314675. [DOI] [PubMed] [Google Scholar]
- 35.Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 36.Young B, Kuritzkes D R. Viral kinetics: implications for treatment. AIDS. 1999;13(Suppl. 1):S11–S7. [PubMed] [Google Scholar]