Abstract
The first ever FDA-approved messenger RNA (mRNA) vaccines are highly protective against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1–3. However, the contribution of each dose to the generation of antibodies against SARS-CoV-2 spike (S) protein and the degree of protection against novel variants warrant further study. Here, we investigated the B cell response to the BNT162b2 vaccine by integrating B cell repertoire analysis with single-cell transcriptomics pre- and post-vaccination. The first vaccine dose elicits a recall response of IgA+ plasmablasts targeting the S subunit S2. Three weeks after the first dose, we observed an influx of minimally-mutated IgG+ memory B cells that targeted the receptor binding domain (RBD) on the S subunit S1 and likely developed from the naive B cell pool. This response was strongly boosted by the second dose and delivers potently neutralizing antibodies against SARS-CoV-2 and several of its variants.
BNT162b2 is one of the two first vaccines that are based on lipid nanoparticle delivery of modified mRNA and are dependent on the host cells for translation and expression of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein, which consists of subunits S1 and S24. S1 contains the receptor binding domain (RBD) that binds the host entry receptor angiotensin-converting enzyme 2 (ACE2) and initiates viral cell entry, while S2 mediates virus-cell membrane fusion5,6. RBD is the target of most neutralizing antibodies found in coronavirus disease 2019 (COVID-19) patients7. The cellular processes that generate potent neutralizing antibodies in response to mRNA vaccines are not fully characterized, and to what degree these antibodies protect against novel variants remains unclear.
Nine healthy individuals without prior SARS-CoV-2 infection were included in this study (Supplementary Table 1). One individual contracted COVID-19 eight weeks after the second dose. Peripheral blood B cells were investigated by droplet-based single-cell sequencing before vaccination (day 0), as well as 7–9 days (day 7), 21–23 days (day 21) and 28 days (day 28) after the first dose (Fig. 1a). The second dose was given on day 21. Additionally, SARS-CoV-2 S-specific B cells were labeled with S1-, S2- and RBD-tetramers conjugated to fluorochromes and DNA-barcodes, and sorted by fluorescence-activated cell sorting (FACS) before sequencing (Fig. 1a). 131,138 B cells were included in the global transcriptomic analysis. Dimensionality reduction by uniform manifold approximation and projection (UMAP)8 and graph-based clustering distinguished nine B cell populations present in all individuals at all timepoints, including naive B cells (Bnaive cells), unswitched and switched memory B cells (U-Bmem cells and S-Bmem cells) and plasmablast clusters (Fig. 1b, Extended Data Fig. 1a-c, Supplementary Table 2). Bnaive cells were further sub-categorized into CCR7loPTRCAPhiJUNBhi Bnaive cells (Bnaive1 cells) and CCR7hiPTRCAPloJUNBlo Bnaive cells (Bnaive2 cells) (Extended Data Fig. 1b). S-Bmem cells were divided into PTPN6loBLKloCD86lo resting switched Bmem cells (SR-Bmem cells) and PTPN6hiBLKhiCD86hi activated switched Bmem cells (SA-Bmem cells) (Extended Data Fig. 1b)9. Fitting to the cluster assignments, Bnaive cells mainly expressed low-mutation IgM, whereas Bmem cells and plasmablasts expressed other isotypes with higher frequencies of somatic hypermutation (SHM) (Extended Data Fig. 1d-e). Single-cell transcriptome sequencing on days 0, 7, 21 and 28 showed decreased frequencies of U-Bmem cells (day 0, 35.46%; day 7, 13.88%) and increased frequencies of SR-Bmem cells (day 0, 9.38%; day 7, 23.42%) on day 7, which was sustained until day 28 (Fig. 1c,d). To understand the fate of U-Bmem cells, we tracked them from day 0 to day 28 using B cell receptor (BCR) sequencing. Clonally related BCR sequences were defined by shared heavy-chain and light-chain variable genes and >70% overlap in both CDR3 regions. B cells clonally related to day 0 U-Bmem cells were identified in day 7, 21 and 28 datasets (Fig. 2a). The majority of clonal day 0 U-Bmem cells differentiated into SR-Bmem cells on days 7–28 (day 7, 46.91%; day 21, 63.21%; day 28, 66.34%)(Fig. 2a,b, Extended Data Fig. 2a,b). SHM frequencies of day 0 U-Bmem cells did not differ from clonally-related days 7–28 SR-Bmem cells (Extended Data Fig. 2c), suggesting differentiation without germinal center (GC) maturation. Only 1.98% of the B cells clonally related to day 0 U-Bmem cells developed into plasmablasts on day 28 (Fig. 2a,b, Extended Data Fig. 2a,b), suggesting that the differentiation of day 0 U-Bmem cells was separate from the plasmablast response to SARS-CoV-2. Pathway enrichment analysis revealed increased expression of B cell activation genes in U-Bmem cells between day 0 and days 7–28 (Fig. 2b, Extended Figure 2f,j). In contrast, the same genes were downregulated in SR-Bmem cells between day 0 and days 7–28 (Fig. 2c, Extended Figure 2g), although upregulation of CD83 and CD69 in SR-Bmem cells indicated that they were recently activated (Extended Data Fig. 2k)10,11. Together, these observations suggest that BNT162b2 activated U-Bmem cells and induced class-switching and differentiation into SR-Bmem cells independent of the GC.
Fig. 1: Single cell transcriptomic analysis of the B cell response to BNT162b2 vaccination.
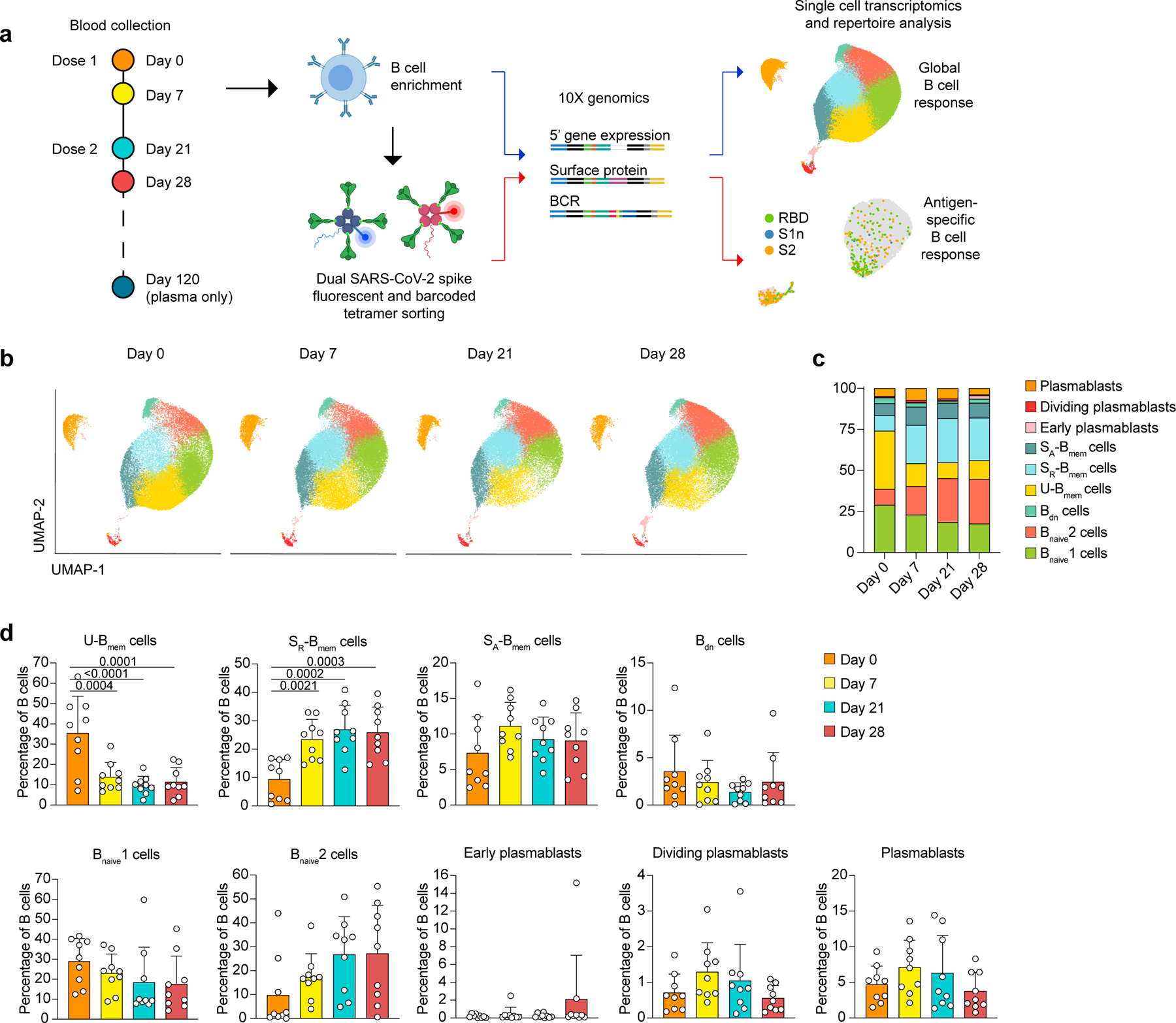
a, Outline of experimental approach. b-d, Single-cell transcriptome analysis showing, b, UMAP visualization of B cell clusters for all individuals at four timepoints. Cluster assignments are based on gene expression and cell surface expression (CITE-seq) of canonical B cell markers. c, Mean percentages of B cell clusters shown in (b). d, Individual and mean percentages of B cell clusters in (b) at four timepoints. n=9 individuals at all timepoints. Individual values, means and standard deviations are shown. Exact p-values according to two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test. Memory B cells (Bmem cells), resting switched memory B cells (SR-Bmem cells), activated switched memory B cells (SA-Bmem cells), double-negative B cells (Bdn cells), naive B cells (Bnaive cells).
Fig. 2: Unswitched memory B cell differentiation and expansion of IgG+ plasmablasts in response to vaccination.
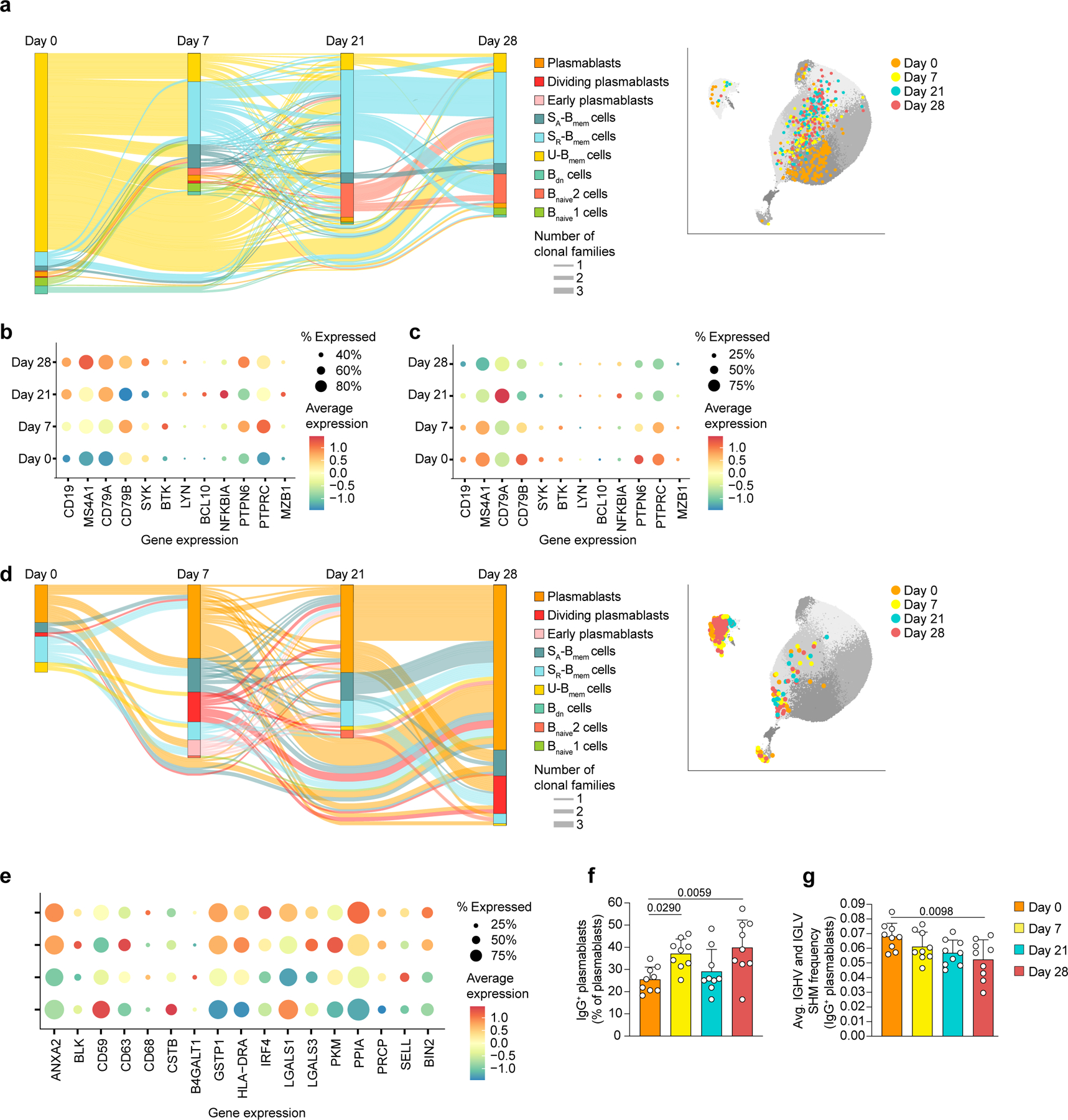
a, Single-cell BCR repertoire data, showing trajectories of B cell clonal families across timepoints as alluvial plot (left) and UMAP projection of clonal B cells (right) that are related to U-Bmem cells at day 0 of n=9 participants b-c, Single-cell transcriptomic sequencing data showing expression of genes involved in B cell activation and BCR signaling in b, U-Bmem cells and c, SR-Bmem cells. d, Single-cell BCR repertoire data, showing trajectories of B cell clonal families across timepoints as alluvial plot (left) and UMAP projection of clonal B cells (right) that are related to plasmablasts at day 28 of n=9 participants. e-g, Single-cell transcriptomic sequencing data showing e, expression of genes involved in B cell activation in plasmablasts, f, percentage of IgG+ plasmablasts in all plasmablasts per individual (n=9), at four timepoints, and g, average SHM frequency of IgG+ plasmablasts per individual (n=9) at four timepoints. a,d, Clonal families included have ≥3 members at ≥2 timepoints. Line thickness represents the number of clonal families. f,g, Individual values, means and standard deviations are shown. Exact p-values according to two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test. Memory B cells (Bmem cells), resting switched memory B cells (SR-Bmem cells), activated switched memory B cells (SA-Bmem cells), double-negative B cells (Bdn cells), naive B cells (Bnaive cells), immunoglobulin heavy-V gene (IGHV), immunoglobulin lightV gene (IGLV), somatic hypermutation (SHM).
U-Bmem cells are thought to develop independently of the GC and possess a polyreactive repertoire for rapid B cell responses12. The observed separation of clonal U-Bmem cells from the plasmablast lineages indicates that U-Bmem cells were likely not a major source of S-antigen-specific antibodies. Plasmablasts are short-lived activated B cells that expand in response to antigen stimulation and secrete large amounts of antibodies13. To identify B cell populations directly involved in the antigen-specific response, we focused on clonally related B cell expansions that included plasmablasts at day 28. Based on shared BCR sequences, the majority of B cells clonally related to day 28 plasmablasts were found in the plasmablast compartment at days 0–21 (day 0, 53.57%; day 7, 52.75%; day 21, 66.67%) (Fig. 2d, Extended Data Fig. 2d,e). Considering the short life span of plasmablasts14, this indicates continued recruitment from either Bnaive cell or Bmem cell pools. Genes associated with leukocyte activation and protein processing were enriched in plasmablasts on days 7–28 over day 0 (Fig. 2e, Extended Data Fig. 2h), consistent with their function as antibody producers. Genes associated with SARS-CoV-2 infection were enriched in plasmablasts on day 21 over day 0 (Extended Data Fig. 2h), suggesting that recognition of viral proteins triggers similar pathways in vaccination and infection.
IgA+ plasmablasts are prevalent in peripheral blood during health, while IgG+ plasmablasts increase during systemic infection and vaccination15,16. Frequencies of IgG+ plasmablast and in particular IgG1+ plasmablast increased between days 0 and 7 and between days 21 and 28 in response to each dose (Fig. 2f, Extended Data Fig. 3a-c). The average SHM frequency in IgG+ plasmablasts decreased from day 0 to day 28 (Fig. 2g, Extended Data Fig. 3d-m). These observations show an influx of minimally-mutated IgG+ plasmablasts developed in response to vaccination, likely originating from Bnaive cell pool.
To further characterize the S-specific B cell response to vaccination, we labeled B cells with fluorescent S1-, S2- and RBD-tetramers, each with a unique barcode, and FACS-sorted antigen-tetramer-specific and non-specific IgA+, IgG+ and IgM-plasmablasts and S-Bmem cells (Extended Data Fig. 4a). Antigen-specificities were determined by their barcode after demultiplexing (Extended Data Fig. 5a-e). S-specific IgA+ and IgG+ plasmablasts expanded at days 7 and 28 (day 0, 0.22%; day 7, 4.58%; day 28, 1.37%)(Fig. 3a, Extended Data Fig. 4b-c). The S-specific IgA+ and IgG+ S-Bmem cells response increased from day 0 to day 28 (day 0, 0.08%; day 28, 0.395%) (Fig. 3a, Extended Data Fig. 4b,e). After day 7, the S-specific B cell response shifted from an IgA+ to an IgG+response (Fig. 3a, Extended Data Fig. 4d,f).
Fig. 3: Vaccination induces an IgA+ anti-S2 response on day 7 followed by an IgG+ anti-RBD response on days 21 and 28.
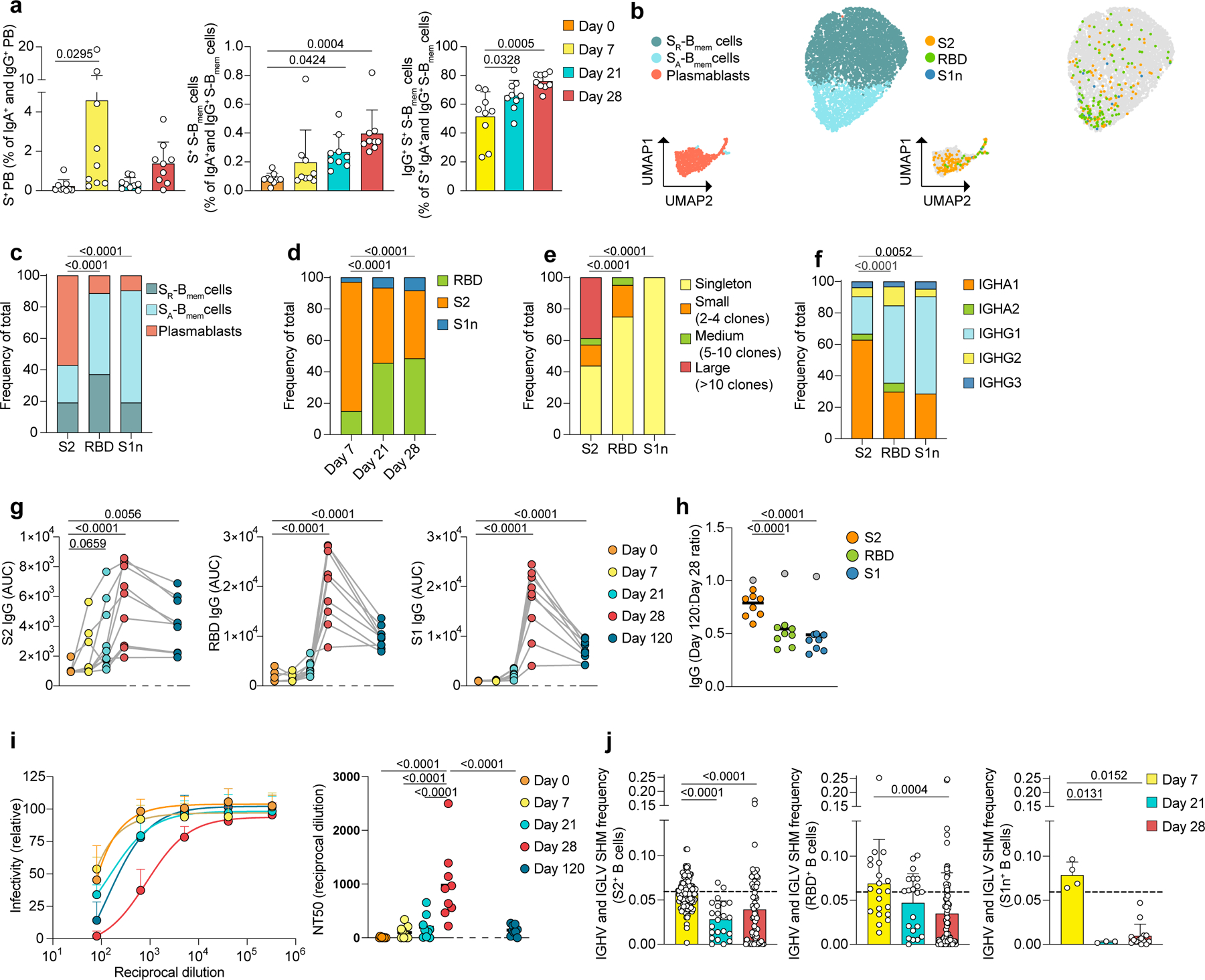
a, Flow cytometry data, showing percentages of S-specific plasmablasts in all IgG+ and IgA+ plasmablasts (left), S-specific S-Bmem cells in all IgG+ and IgA+ S-Bmem cells (center) and IgG+ S-specific S-Bmem cells in all S-specific IgG+ and IgA+ S-Bmem cells (right) Individual data points are averages of two independent experiments, including n=9 participants at four timepoints. b-f, Single-cell transcriptome and BCR repertoire sequencing data of S-specific and non-specific sorted B cells showing b, UMAP visualization with cluster assignments of SR-Bmem cells, SA-Bmem cells and plasmablasts (left) and antigen-specificity to S2, RBD, and S1n (right). c-f, Proportions of cells shown in (b), separated by c, antigen and cluster, d, day and antigen, e, antigen and clonality and f, antigen and immunoglobulin class. g, Plasma IgG levels against S2 (left), RBD (center), and S1 (right) for n=9 individuals at five timepoints. Area under the curve (AUC) for plasma dilutions are shown as individual data points. h, Ratio of IgG levels of day 120 AUC to day 28 AUC for antigens S2, RBD, and S1. Grey dots represents the vaccinated participant who contracted COVID-19 two weeks before day 120, and is excluded from statistics. Individual data points and medians for n=9 participants are shown. i, Plasma neutralization of SARS-CoV-2 pseudovirus shown as dilution curves (left) and quantification (NT50) of neutralization titers (right) of n=9 individuals at five timepoints. j, Single-cell BCR repertoire sequencing data showing IGHV and IGLV gene SHM frequencies of B cells specific for S2 (n=210 cells, left), RBD (n=124 cells, center), and S1n (n= 24 cells, right) at three timepoints post vaccination. Dashed line indicates the average mutation frequency of sorted non-specific B cells. Individual data points represent single B cells from n=9 individuals. a, j, Individual values, means and standard deviations are shown. a, g-i, Exact p-values according to two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test, c-f, chi-square test, j, two-tailed Kruskal Wallis test followed by Dunnett’s multiple comparison test. Plasmablasts (PB), Memory B cells (Bmem cells), resting switched memory B cells (SR-Bmem cells), activated switched memory B cells (SA-Bmem cells), S-specific (S+), immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Antibody responses against distinct epitopes contribute differently to immune protection17. We investigated how the antibody response to each S subunit varies within B cell subsets and over time. UMAP-analysis indicated that S2-specific B cells were predominantly plasmablasts and accounted for 82.14% of all S-specific B cells at day 7 (Fig. 3b-d). In contrast, the majority of B cells specific to RBD and specific to S1, but not RBD (S1n) were SA-Bmem cells (Fig. 3b,c, Extended Data Fig. 4g). RBD-specific and S1n-specific B cells increased from less than 20% on day 7 to over 50% on day 28 of all S-specific B cells (Fig. 3b-d). The S2-specific response was highly clonal and dominated by IgA1+ plasmablasts, while RBD-specific and S1n-specific SA-Bmem cells used predominantly IgG1 and were less clonally expanded than S2 plasmablasts (Fig. 3e,f). The rapid onset of the S2-specific response and the delayed S1n- and RBD-specific response indicated that the S2-specific B cell response is a secondary response, while the S1n and RBD-specific B cell response is a primary response.
The early S2-specific plasmablast response corresponded with S2-specific plasma antibody titers, which rose quickly from day 0 to day 7 and plateaued around day 21. RBD-specific and S1-specific IgG and IgA levels were low until day 21 and increased at day 28 in response to the second dose (Fig. 3g, Extended Data Fig. 6a-b). Accordingly, day 28 plasma neutralized the SARS-CoV-2 Wuhan-Hu-1 pseudovirus more potently than day 0–21 plasma (Fig. 3i). Notably, by day 120, RBD-specific and S1-specific IgG levels decreased by 52%, whereas S2-specific IgG levels only decreased by 23.61% (excluding the participant who contracted COVID-19 two weeks before day 120) (Fig. 3g,h). This suggests less effective induction of RBD-specific long-lived plasma cells upon recruitment from Bnaive cells.
The participant with the lowest RBD-specific IgG and IgA levels contracted COVID-19 eight weeks after the second vaccination (Extended Data Fig. 6a-d). While this participant produced high-affinity neutralizing B cells (Fig. 4f,g), antibody levels were not protective. In addition, the participant’s antibody levels did not increase by day 120, two weeks after the COVID-19 infection (Extended Data Fig. 6a-c), suggesting that the low antibody levels were not due to the effectiveness of the mRNA vaccine.
Fig. 4: Neutralization of SARS-CoV-2 pseudovirus and variants by BNT162b2-induced antibodies.
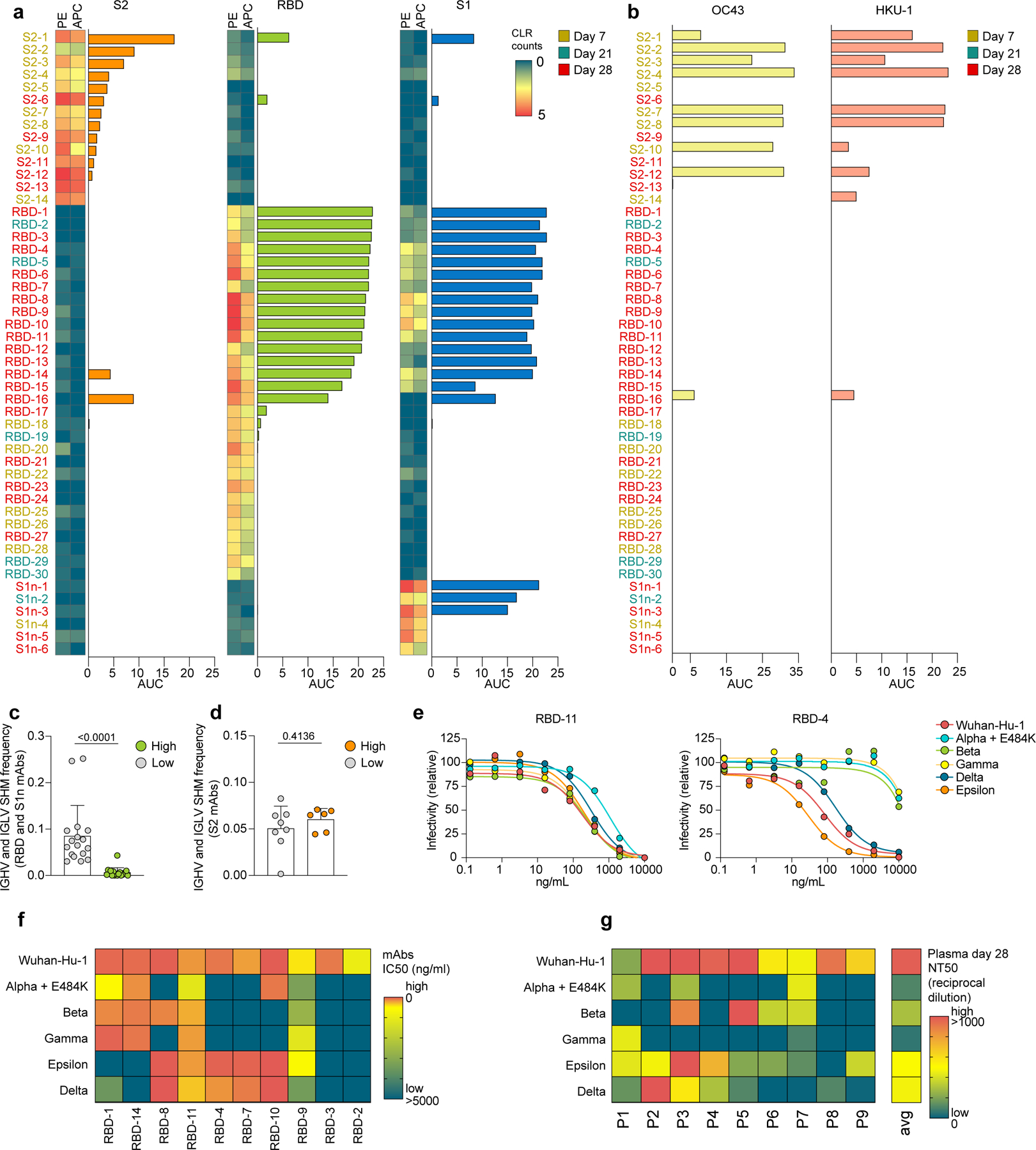
a-b, Single-cell sequencing data indicating abundance of barcoded and fluorochrome-tagged antigen-tetramers for each sorted B cell (two-column heatmaps, one barcode corresponding to PE and APC, respectively), paired with ELISA data of the corresponding expressed mAbs (bar graphs). Reactivity against S2, RBD, and S1, are shown. Barcoded tetramer binding data are represented as centered log ratio transformed (CLR) counts. b, ELISA data showing reactivity of the same mAbs as (a) against OC43 and HKU1 spike. a,b, ELISA data are shown as area under the curve (AUC) of plasma dilutions. Threshold at 0 was set to the average binding to bovine serum albumin (BSA) plus its three-fold standard deviation. c,d, Single-cell BCR repertoire sequencing data showing IGHV and IGLV SHM frequencies of c, RBD- and S1-specific B cells (high, n=17 cells; low, n=18 cells), and d, S2-specific B cells (high, n=8 cells; low, n=6 cells). High binding mAbs are defined by the data shown in (a) as positive for both PE and APC barcode tetramer binding and ELISA AUC >3. Individual values, means and standard deviations are shown, exact p-values according to unpaired two-tailed t-test. e-g, Neutralization of SARS-CoV-2 pseudovirus Wuhan-Hu-1 and variants alpha-epsilon are shown e, as dilution curves for antibodies RBD-11 (left) and RBD-4 (right) and f-g, as heatmaps indicating f, IC50 (ng/ml) for each mAb and g, NT50 (reciprocal dilution) of day 28 plasma for each of the n=9 participants. Immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Neutralizing antibodies from COVID-19 patients show characteristically low mutation rates18, indicating recruitment of Bnaive cells to the GC in response to a novel antigen. We found that on day 7, when the majority of antigen-specific Bmem cells and plasmablasts was S2-specific (Fig. 3d), mutation frequencies of antigen-specific Bmem cells and plasmablasts were similar to control-sorted Bmem cells and plasmablasts without specificity to S-antigens (Fig. 3j, Extended Data Fig. 7a-c). In contrast, mutation frequencies of antigen-specific Bmem cells and plasmablasts on days 21 and 28, when the majority of B cells reacted to S1 and RBD, were significantly lower than on day 7 (Fig. 3d,j, Extended Data Fig. 7a-c). Thus, the SHM analysis indicated that S2-specific response was a recall response from Bmem, whereas the delayed S1n- and RBD-specific response was a primary response from Bnaive cells.
To further characterize the primary and secondary B cell response, we expressed 50 representative BCR sequences from S-antigen-specific B cells — 14 S2-specific, 30 S1- and RBD-specific and 6 S1n-specific, as recombinant monoclonal antibodies (mAbs) (Supplementary Table 3). ELISA and bio-layer interferometry indicated that of the selected mAbs, 8 potently bound S2, 15 bound RBD and 3 bound S1n (Fig. 4a, Extended Data Fig. 8). When tested against S protein of 4 other human-pathogenic coronaviruses (229E, NL63, OC43 and HKU1), nine and eight of the twelve S2-reactive mAbs cross-reacted with betacoronaviruses OC43 and HKU-1 S protein, respectively, but none with the alphacoronaviruses 229E and NL63 (Fig. 4b). Of the RBD-specific mAbs, only the polyreactive mAb RBD-16 (Extended Data Fig. 9), showed cross-reactivity to OC43 and HKU-1 S protein (Fig. 4b). These observations are in line with a higher conservation of S2 than S1 across coronavirus species19,20.
mAbs that bound strongly to S1 by ELISA and bio-layer interferometry (high binders) harbored low SHM frequencies in comparison to mAbs that bound S1-specific antigen-tetramers but showed little or no reactivity to S antigens by ELISA (low binders) (Fig. 4c, Extended Data Fig. 7d). In contrast, SHM frequencies in S2-specific mAbs did not differ significantly between high and low binders (Fig. 4d, Extended Data Fig. 7e). mAbs with the highest affinity for S2 were derived from day 7 plasmablasts, while mAbs with the highest affinity for S1 (including RBD) originated from S-Bmem cells from days 21 and 28 (Fig. 4a). While a correlation between increased affinity and low SHM frequency is counter-intuitive, it is characteristic for the B cell repertoire in COVID-19 patients21,22, and likely caused by the reduced structural overlap of the S1 protein to other pathogens, requiring recruitment from the Bnaive cell pool.
The emergence of novel SARS-CoV-2 variants could jeopardize vaccine efficacy. RBD mutations contribute significantly to immune escape23. After testing neutralization efficacy of all RBD-specific mAbs to the Wuhan-Hu-1 pseudovirus (Extended Data Fig. 10), we selected the ten best Wuhan-Hu-1-neutralizing RBD-specific mAbs and tested their neutralization of five SARS-CoV-2 variants –– alpha+E484K (B.1.1.7+E484K; N501Y, E484K), beta (B.1.351; N501Y, K417N), gamma (P.1; N501Y, K417T), delta (B.1.617.2; L452R, T478K) and epsilon (B.1.429; L452R) (Supplementary Table 4). Each variant was neutralized by at least four of the ten mAbs (Fig. 4e,f, Extended Data Fig. 10), indicating at least partial protection. Most antibodies neutralized either alpha, beta and gamma (N501Y) or delta and epsilon (L452R) (Fig. 4f), indicating that neutralization was dependent on RBD mutations shared between the variants. Two antibodies were broadly neutralizing (Fig. 4f, Extended Data Fig. 10). Additionally, we tested neutralization efficacy in plasma of the nine participants (Fig. 4g). Overall, plasma neutralization potency was highest against Wuhan-Hu-1, followed by epsilon and delta, and lowest against alpha, beta and gamma. Together, despite low plasma neutralization potency against variants in some participants, we detected B cells with potently neutralizing BCRs against variants in the same participants.
Here we provided a detailed characterization of the B cell response to the BNT162b2 mRNA vaccine on a single-cell level. Parsing the S1-specific and S2-specific responses provides important insights into why a second dose is vital for protection. Our results indicate that the first dose activated a non-neutralizing recall response that initially targets epitopes in the S2 protein subunit, conserved across human-pathogenic betacoronaviruses19,20, while the second dose boosted the neutralizing B cell responses to S1 including RBD.
The first dose an IgA-dominant plasmablast response against S2 with high SHM, which was cross-reactive to the betacoronaviruses OC43 and HKU1. This is consistent with a recall response of mucosal Bmem cells to prior pulmonary coronavirus infections. The first dose conveys a degree of protection against COVID-193, and this initial S2-specific response likely contributes to it. S2-specific antibodies can neutralize SARS-CoV-2 by inhibiting virus-cell membrane fusion and boosting anti-viral T cell immunity24–26
After the S2-specific response on day 7, the frequency of minimally-mutated SA-Bmem cells increased on days 21 and 28. The SA-Bmem cells at these later timepoints mostly targeted S1, including RBD, and their low SHM frequency indicates a primary B cell response. As shown for B cell responses in influenza vaccine settings, blocking of a dominant epitope by early antibodies (anti-S2) can facilitate the development of later antibodies against another epitope (anti-S1)27. We observed that low mutation frequency corresponded to high affinity against RBD. High BCR affinity and a naive phenotype foster preferential recruitment into GC28–30 and high affinity also promotes release from the GC as plasmablasts, plasma cells or Bmem cells31. High affinity in minimally mutated BCRs could therefore limit GC maturation to a relatively short time frame. mRNA vaccines against SARS-CoV-2 induce robust and prolonged GC reactions, with plasmablasts and S-Bmem cells persisting in GC for over three months32,33. However, despite the prolonged GC reaction, the more rapid decrease of antibody titers against RBD and S1 than against S2 suggests less efficient development of long-lived plasma cells upon recruitment from Bnaive cells34.
Plasma neutralization assays indicated a significant degree of immune escape of SARS-CoV-2 variants. However, several selected mAbs from the same participants were neutralizing, indicating that even individuals with low neutralizing titers can raise a recall memory response after infection with SARS-CoV-2 variant. Together, our study provides a detailed characterization of the blood B cell response to the BNT162b2 mRNA vaccine. Our data emphasize the importance of the second dose in inducing generation of RBD-specific antibodies that contribute to neutralization of SARS-CoV-2 variants.
Methods
Study design, sample collection, and storage
All studies were approved by the Institutional Review Board of Stanford University (IRB-3780), and the studies complied with the relevant ethical regulations. All participants provided written informed consent before participating in the study. Nine healthy individuals were enrolled in the study (Supplementary Table 1). All individuals had undergone routine RT-PCR testing for SARS-CoV-2 infection prior to study. None of the participants had been previously diagnosed with SARS-CoV-2 infection. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications35–37. Blood samples were collected in heparin tubes (BD) at four different timepoints including pre-vaccination (day 0, D0), 7- to 9 days post initial vaccination (day 7, D7), on the day of and prior to the second dose 21- to 23 days post initial vaccination, day 21, D21), and 28 to 30 days after initial and 7–9 days after second vaccination (day 28, D28). Plasma samples were obtained after centrifugation, and stored at −80˚C. Peripheral Blood Mononuclear Cells (PBMCs) were obtained by density gradient centrifugation over Ficoll PLUS media (Cytiva) and stored in cell freezing media (Thermo Fisher Scientific). PBMCs were aliquoted and stored until use at −80°C.
Generation of barcoded fluorescent antigen-tetramers
Recombinant Avi-tag biotinylated SARS-CoV-2 S2 protein (Acro Biosystems, S2N-C52E8–25ug), SARS-CoV-2 RBD (Acro, SPD-C82E9–25ug), and SARS-CoV-2 S1 (Acro, S1N-C82E8–25ug) were mixed with barcoded, fluorescently labeled streptavidin (Biolegend) at a 4 to 1 molar ratios for 45 minutes while rotating. Excess biotin was added to saturate all streptavidin binding sites.
Flow cytometry, cell sorting, and 10X sample preparation
PBMCs were thawed at 37C°, treated for 15min with DNAse and washed in complete RPMI. PBMCs were enriched for B cells using the EasySep Human Pan-B Cell Enrichment Kit (Stem Cell Technologies) according to the manufacturer instructions. B cell samples without antigen enrichment were stained with CD19, IgD, CD27, CD38 TotalSeq-C antibodies (0.5µg per 1,000,000 cells) (all Biolegend). For antigen-sorted B cell samples, cells were stained with the following fluorescently labeled antibodies according to standard protocols: CD19 (1:100 dilution), CD20 (1:300 dilution), CD38 (1:100 dilution) (all BD Biosciences), CD3 (1:60 dilution) , CD27 (1:100 dilution) , IgM (1:100 dilution) , IgD (1:100 dilution) , (all BioLegend), IgA (1:100 dilution) (Miltenyi Biotec), Sytox blue (1:1000 dilution) (Thermo Fisher Scientific), and S-antigen-tetramers. Additionally, sorted samples were labeled with TotalSeq-C hashtag 1–9 antibodies (0.5µg per 1,000,000 cells) (Biolegend) for demultiplexing individual samples in downstream analysis. Single cells were sorted with a FACSAria II cell sorter (BD Biosciences) into cooled 1.5 ml tubes (BioRad). FACS data was collected with the BD FACSDiva (v.8.0) software. FlowJo Version 10.7.1 (BD Biosciences) was used for flow cytometry data analysis. Flow cytometry experiments were performed with n=9 biological replicates (study participants), and the experiment was performed twice.
Droplet-based single-cell sequencing
Using a Single Cell 5′ Library and Gel Bead Kit v1.1(10X Genomics, 1000165) and Next GEM Chip G Single Cell Kit (10X Genomics, 1000120), the cell suspension was loaded onto a Chromium single cell controller (10X Genomics) to generate single-cell gel beads in the emulsion (GEMs) according to the manufacturer’s protocol. Briefly, approximately 8,000 cells were added to each channel and approximately 4,000 target cells were recovered. Captured cells were lysed and the released RNA was barcoded through reverse transcription in individual GEMs. 5’ Gene expression (GEX) libraries, Single Cell V(D)J libraries (1000016), and Cell surface protein libraries were constructed according to manufacturer protocols. Library quality was assessed using a 2200 TapeStation (Agilent). The libraries were sequenced using an Illumina Novaseq6000 sequencer with a paired-end 150-bp (PE150) reading strategy (Novogene).
Single cell RNA-seq data processing
Raw gene expression and cell surface matrices were generated for each sample by the Cell Ranger Pipeline (v.6.0.1) coupled with human reference version GRCh38. Briefly, gene expression analyses of single cells were conducted using the R package Seurat (v4.0.2) to perform data scaling, transformation, clustering, dimensionality reduction, differential expression analyses and most visualization38. The count matrix was filtered to remove cells with >10% of mitochondrial genes or low gene counts (<600 for enriched B cells, <200 for sorted B cells). The normalized data were integrated into one Seurat data file using the IntegrateData function. Principal component analysis was performed using variable genes. We compared the ranking of principal components (PCs) with the percentage of variance to determine the number of first ranked PCs to use for Uniform Manifold Approximation And Projection (UMAP)8 to reduce the integrated dataset into two dimensions. Afterwards, the same number of first ranked PCs were used to construct a shared nearest-neighbor graph (SNN), and this SNN used to cluster the cells. For sorted cells, all sorted cells (antigen-specific and non-specific) were used in the UMAP projection. Contaminant cells (non-B cells) were removed and the data was normalized, integrated, and clustered with only the B cells. Specific B cell clusters were identified using canonical B cell markers9(Extended Data Fig. 1).
Identification of differentially expressed genes (DEG) and functional enrichment
We performed differential gene expression testing using the FindMarkers function in Seurat with Wilcoxon rank sum test, and the Benjamini–Hochberg method was used to adjust the p-values for multiple hypothesis testing. DEGs were filtered using a minimum log2(fold change) of 0.25 and a maximum FDR value of 0.05. Pathway analysis for the DEGs was conducted using the Metascape39.
VDJ Sequence analysis
BCR VDJ regions were generated for each sample using the Cell Ranger Pipeline (v.6.0.1). BCR sequences were then filtered to include cells that have one light and one heavy chain per cell. Consensus sequences were aligned to germline variable-chain immunoglobulin sequences with IMGT HighV-QUEST v1.8.340. Clonal families were defined based on sharing the same heavy and light chain V and J genes with >70% amino acid identity in heavy and light chain CDR3s. Mutations were identified by aligning the nucleotide sequence to germline variable-chain immunoglobulin sequences with IMGT HighV-QUEST. To calculate the mutation frequency, we divided the number of mutations (silent and non-silent) by the length of the V-gene.
Recombinant monoclonal antibody (mAb) production
Heavy chain and light chain variable sequences were codon optimized and cloned into in-house vectors, containing human IgG1 constant region or kappa or lambda constant regions, respectively. Expi293F cells were transfected with heavy chain and light chain plasmids using FectoPro (Polyplus transfection). Media was harvested after seven days, and mAbs were purified with AmMag Protein A magnetic beads (Genscript). Antibody concentrations were measured with a nanodrop spectrophotometer (Thermo Fisher Scientific) and human IgG quantitation ELISAs (Bethyl Laboratories).
ELISA
For protein ELISAs, MaxiSorp 384-well plates (Thermo Fisher Scientific) were coated with 1 µg/ml recombinant SARS-CoV-2 S2 protein (Acro, S2N-C52H5), SARS-CoV-2 RBD (Acro, SPD-C52H3), or SARS-CoV-2 S1 (Acro, S1N-C52H3), HCoV-OC43 Spike protein (Sino Biological, 40607-V08B), HCoV-HKU1 Spike protein (Sino Biological, 40606-V08B), HCoV-229E Spike protein (Sino Biological, 40605-V08B), HCoV-NL63 Spike protein (Sino Biological, 40604-V08B), 10µg/ml LPS (Sigma), 10µg/ml Calf Thymus DNA (Invitrogen), 5µg/ml of Insulin (Sigma), or 2µg/ml of flagellin (Invivogen) in carbonate-bicarbonate buffer at 4˚C overnight. Plates were washed 6 times with PBST (PBS + 0.1% Tween20) after each step. The plates were blocked with blocking buffer (PBS + 1% BSA) for 1 hour at room temperature. Human plasma was serially diluted, and added for 1 hour at room temperature. Human mAbs were added at concentrations of 10µg/ml and three 10-fold serial dilutions, and incubated overnight at 4°C. Secondary HRP-conjugated antibodies goat anti-human IgG (Bethyl Laboratories) or HRP-conjugated goat anti-human IgA (Bethyl Laboratories) were applied for 1h at RT, and plates were developed with TMB substrate (Thermo Fisher Scientific), and stopped with 2N sulfuric acid. On each plate, four dilutions of positive control plasma and secondary only controls were run. Additionally, BSA only plate was run in parallel to each antigen. Plates were read on a GloMax Explorer Microplate Reader (Promega). ELISA assays were performed at least 2 times, in duplicate or triplicate.
Bio-Layer Interferometry
mAbs interactions with S2, RBD, and S1 protein were measured on an Octet Red96e (Fortebio / Sartorius). Association and dissociation curves were measured with mAbs bound to anti-human IgG Fc Capture (AHC) sensors at 20 nM and antigens in solution at 0, 16.7, 50, 150, and 450 nM in 1x kinetic buffer (Fortebio / Sartorius). BLI analysis software (Fortebio/Sartorius, version 7.1) was used for data processing and analysis. Buffer controls were subtracted from antigen values and curves were fitted globally for each group, consisting of all concentrations of the same ligand. Association and dissociation curves and constants as well as KD values for each antibody were reported and graphed with GraphPad Prism. Bio-layer interferometry assays were performed 1–2 times, in triplicate.
Cell culture
Expi293F cells were cultured in 33% Expi293 Expression Medium (Gibco) and 67% Freestyle293 Expression Medium (Gibco). HeLa-ACE2 were kindly provided by of Dennis Burton41 and were cultured in Eagle’s Minimum Essential Medium (ATCC, 30–2003) with 10% heat-inactivated FBS (Corning) and 100 U/ml of penicillin–streptomycin (Gibco). Lenti X 293T cells (Takara Bio) were cultured in DMEM (ATCC, 30–2002) with 10% heat-inactivated FBS (Corning) and 100 U/ml of penicillin–streptomycin (Gibco).
Generation SARS-CoV-2 spike pseudotyped lentiviral particles
Pseudotyped lentiviral particles were generated as previously described42. Briefly, LentiX 293T cells were seeded in 10cm plates. After 24 hours, cells were transfected using Fugene transfection reagent (Promega) with pHAGE-CMV-Luc2-IRES-ZsGreen-W, lentiviral helper plasmids (HDM-Hgpm2, HDM-tat1b, pRC-CMV-Rev1b), and wildtype or variant SARS-CoV-2 spike plasmids (parent plasmids publicly available from Jesse Bloom lab). After 48 to 60 hours, viral supernatants were collected and spun at 1000 x g for 10m to remove cell debris. The lentiviral supernatants were concentrated using LentiX concentrator (Takara) according to the manufacturer’s instructions. The lentiviral pellets were resuspended at 20-fold viral increase in EMEM media and stored at −80°C. Virus was titrated on HeLa-ACE2 cells.
Viral inhibition assays
Neutralization assays were performed as previously described42. Briefly, eight-fold serially diluted plasma starting at 1:80 from vaccinated individuals or five-fold serially diluted monoclonal antibodies starting at a concentration of 10µg/ml were incubated with SARS-CoV-2 pseudotyped virus for 1 hour at 37°C. The mixture was added to HeLa-ACE2 cells plated the prior day. After ~50 hours post-infection, luciferase activity was measured on a GloMax Explorer Microplate Reader (Promega). Neutralization assays were performed 1–2 times, in triplicate.
Statistics and software
GraphPad Prism version 9.1.0 and R version 4.0.3 were used for statistical analyses. Statistical tests used and significance levels are indicated in the respective methods section or in the figure legends. Normal distribution was assumed for the nine biological replicates where Student’s t test and ANOVA was performed. Graphical illustrations were created with BioRender.
Materials Availability
Materials generated in this study will be made available on request and may require a material transfer agreement.
Data Availability Statements
The data have been deposited in the Sequence Read Archive linked to BioProject accession number PRJNA775994. All other data needed to evaluate the conclusions in the paper are present in the paper or the Source Data files.
Extended Data
Extended Data Fig. 1. Classification of UMAP B cell clusters.
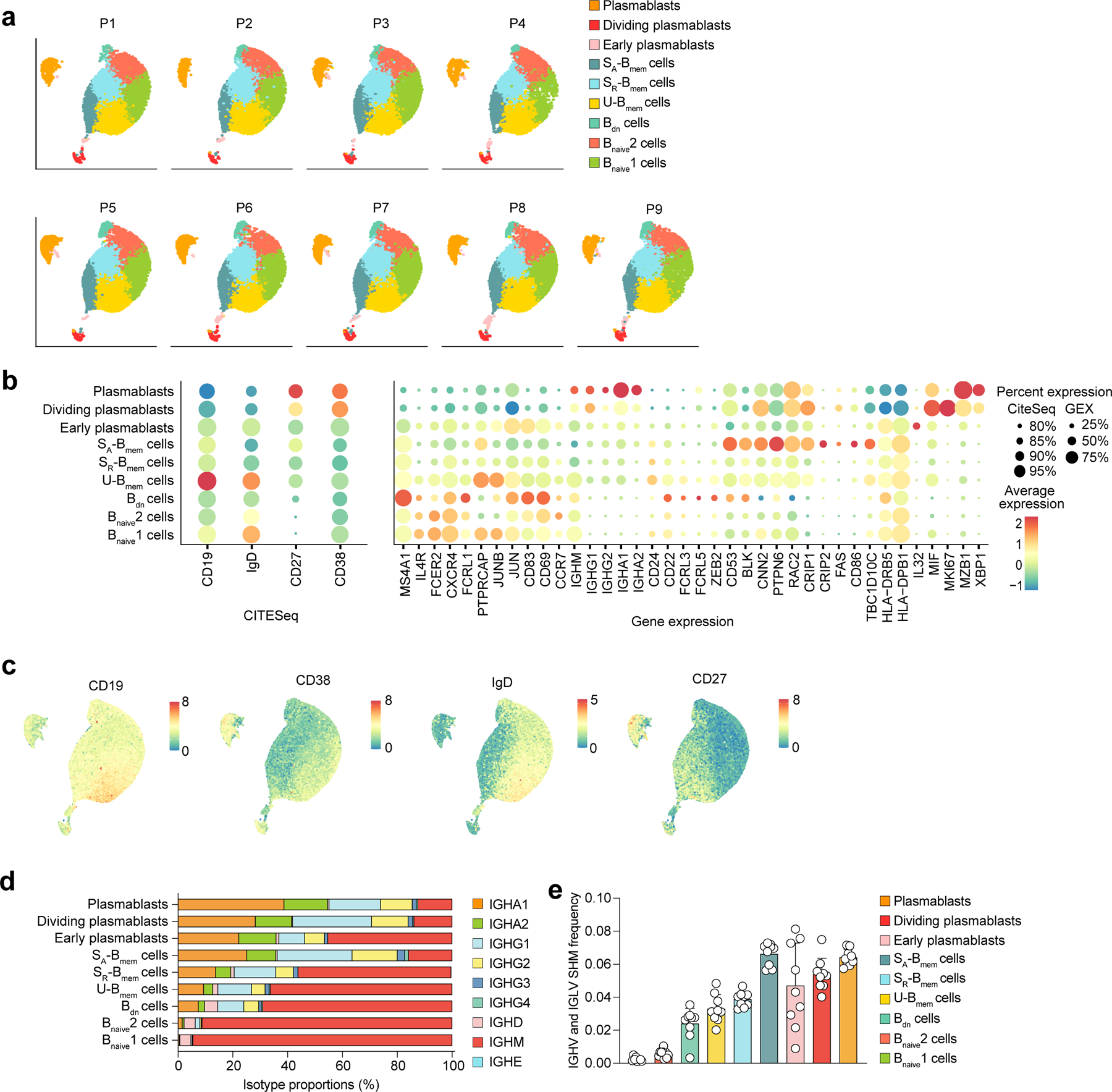
a-c, Single-cell transcriptome and CITESeq sequencing data showing a, the distribution of B cell clusters for each participant included in the study (n=9). b, gene and cell surface expression of CITESeq markers (left) and sets of differentially expressed genes (right) for each B cell cluster, represented as heatmaps (average expression levels) and dot plots (percent expression). c, UMAP visualization with overlaid CITESeq data showing expression of cell surface markers CD19, CD38, IgD, and CD27. d, e, Single-cell BCR repertoire sequencing data showing d, average percentages of immunoglobulin classes in each B cell cluster in n=9 participants across all timepoints, and e, average IGHV and IGLV SHM frequency in n=9 individuals in the respective nine different B cell clusters. Individual values, means, and standard deviations are shown. Memory B cells (Bmem cells), double-negative B cells (Bdn cells), naive B cells (Bnaive cells), immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Extended Data Fig. 2. Clonal and transcriptomic B cell responses to mRNA vaccination.
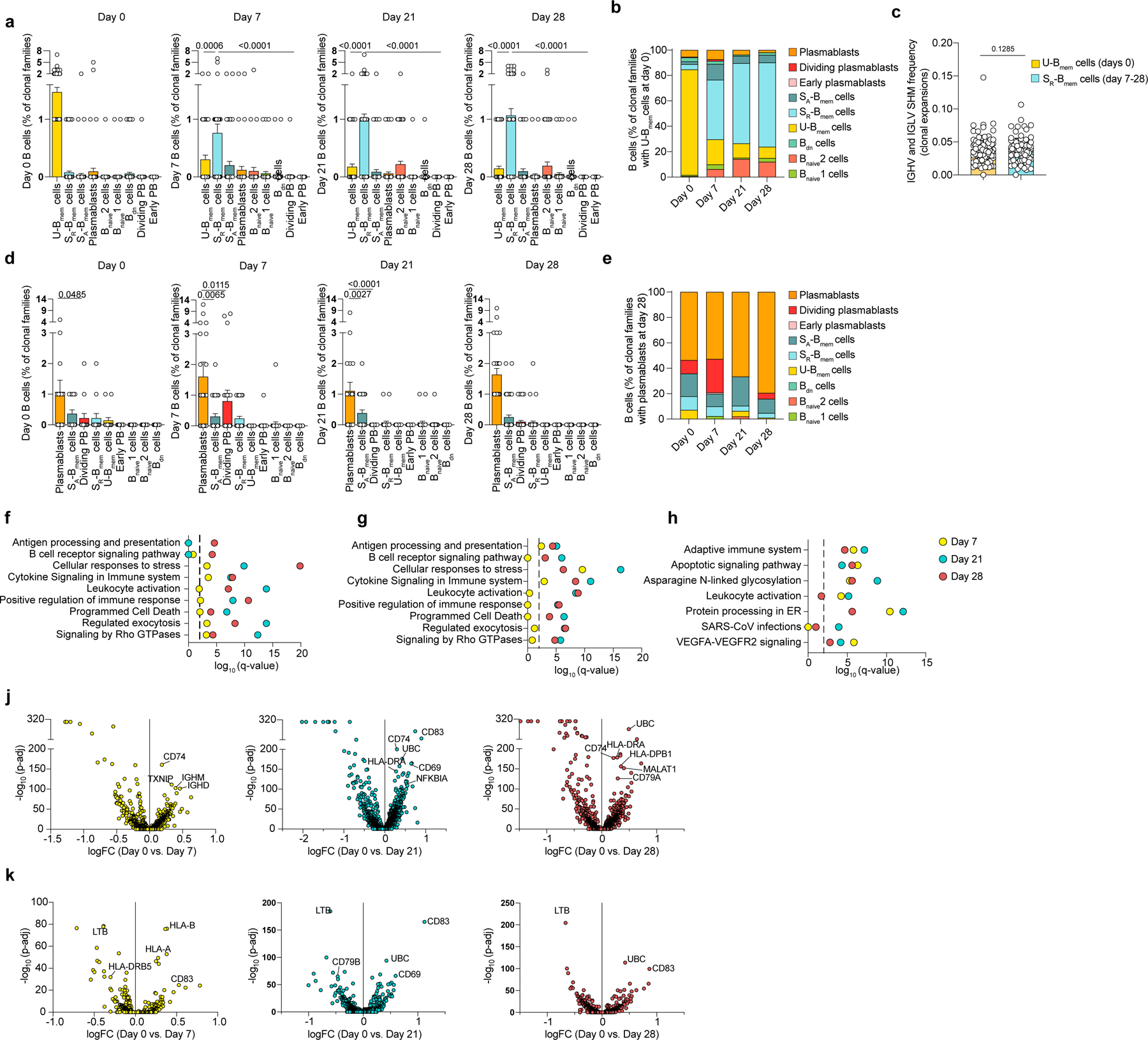
a-k, Single-cell transcriptome and BCR repertoire sequencing data showing a, cells corresponding to clonal families shown in (Fig. 2a). Number of cells in each B cell cluster for each clonal family (n=106 clonal families) at all four timepoints. Statistics are not calculated for day 0 because clonal families are selected to contain at least one clone at day 0 in the U-Bmem cell cluster. b, Proportions of cells (n=476 cells) in each cluster from clonal families containing U-Bmem cells at day 0 for all four timepoints. c, Average SHM frequency of clonal U-Bmem cell families at day 0 and clonal SR-Bmem cell families at days 7–28. Each dot represents the average of each mutation frequency for one clonal family. d, Cells corresponding to the clonal families shown in (Fig. 2d). Number of cells in each clonal family (n=52 clonal families) in each B cell cluster at all four timepoints. Statistics are not calculated for D28 because clonal families are selected to contain at least one clone at D28 in the plasmablast cluster. e, Proportions of cells (n= 274 cells) in each cluster from clonal families with a plasmablasts at D28 for all four timepoints. f-h, Gene enrichment signatures of differentially expressed genes via Metascape in f, U-Bmem cells, g, SR-Bmem cells and h, plasmablasts for day 7, day 21 and day 28 versus day 0. Dashed line represents q < 0.01. j,k, Volcano plots of differentially expressed genes in j, U-Bmem cells and k, SR-Bmem cells at timepoints day 7 (left), day 21 (center), and day 28 (right) relative to day 0. a, d, Individual values with mean and standard errors of the mean are shown. c, Individual values with mean and standard deviations are shown. Exact p-values according to a,d, two-tailed Kruskal-Wallis test, followed by Dunnett’s multiple comparison test and c, paired two-tailed t test. Memory B cells (Bmem cells), resting switched memory B cells (SR-Bmem cells), activated switched memory B cells (SA-Bmem cells), double-negative B cells (Bdn cells), naive B cells (Bnaive cells), immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Extended Data Fig. 3. Plasmablast characteristics in response to mRNA vaccination.
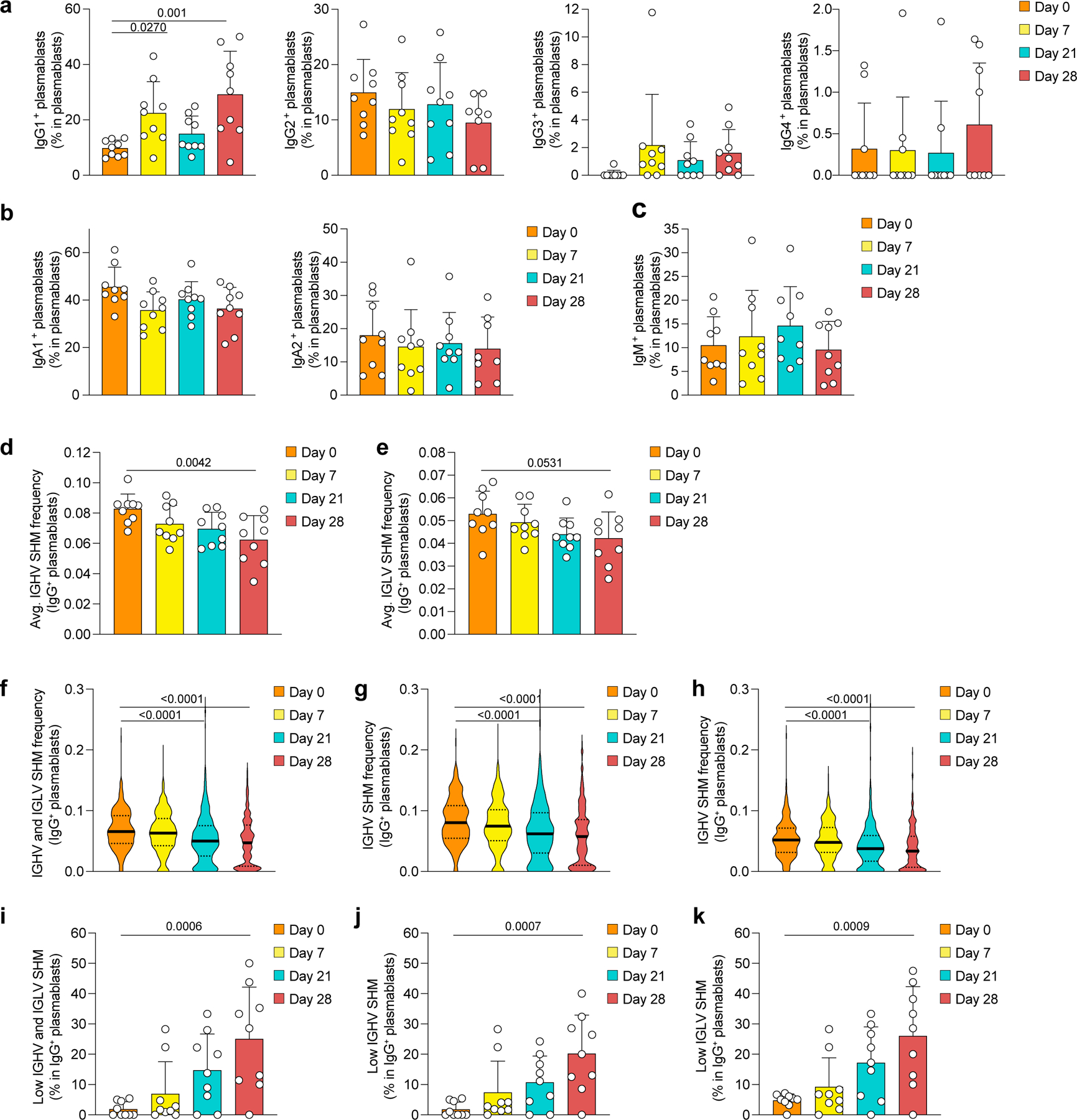
a-k, Single-cell transcriptome and BCR repertoire sequencing data of the global (non-sorted) B cell dataset showing proportions of a, IgG subclasses, b, IgA subclasses and c, IgM in total plasmablasts in n=9 participants. d,e, Average SHM frequency in IgG+ plasmablasts for d, IGHV and e, IGLV in n=9 participants. f-h, Distribution of SHM frequencies in single plasmablasts from all n=9 individuals, separated by f, IGHV and IGLV, g, IGHV, and h, IGLV. i-k, Proportion of low SHM frequency (≤ 0.01) IgG+ plasmablasts in all plasmablasts, separated by i, IGHV and IGLV, j, IGHV and k, IGLV for n=9 individuals. a-e, i-k, Individual values, means, and standard deviations are shown. f-h, Violin plots with medians and interquartile ranges are shown. Exact p-values according to a-e, i-k, two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test, and f-h, Kruskal-Wallis test followed by Dunnett’s multiple comparison test. Immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Extended Data Fig. 4. Characterization of tetramer-sorted S-antigen-specific switched BMem and plasmablasts.
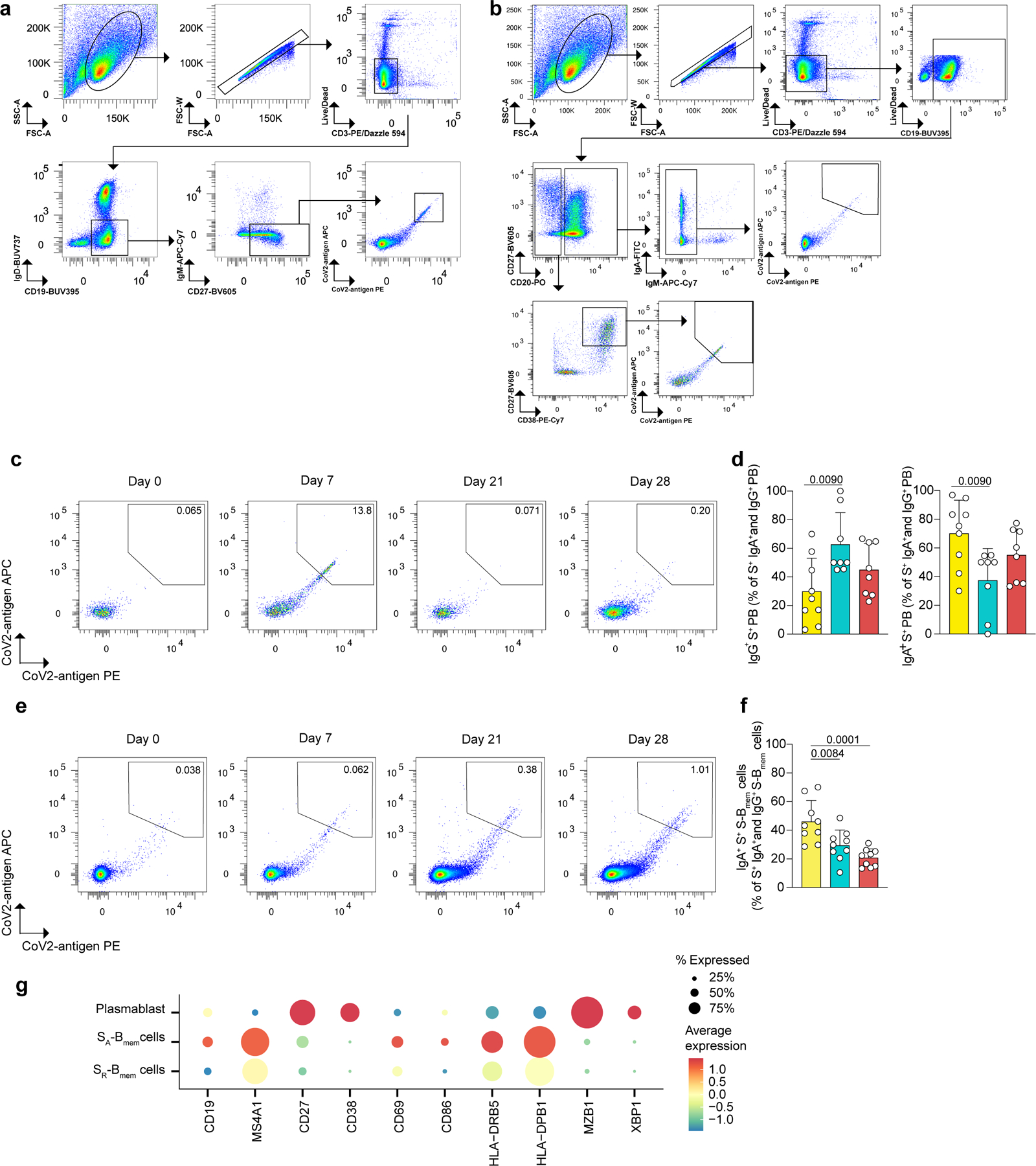
a-f, Flow cytometry data showing a, the gating strategy for cell sorting of live singlet S-Bmem cells and plasmablasts based on marker expression: CD3-, CD19+, IgD-, IgM-, CD27+. Cells were stained with two antigen-tetramers (PE labeled and APC labeled) for each subunit of S (S1, S2, and RBD), respectively. Tetramer-double-positive cells were sorted and tetramer-double-negative cells were sorted as controls. b, Gating strategy for flow cytometry analysis of S-Bmem cells and plasmablasts. c, Representative flow cytometry analyses showing two-dimensional antigen-tetramer staining of S-specific plasmablasts. d, Proportion of IgG+ (left) and IgA+ (right) S-specific plasmablasts in all IgG+ and IgA+ S-specific plasmablasts at the indicated timepoints post vaccination. e, Representative flow cytometry analyses showing two-dimensional antigen-tetramer staining of S-specific S-Bmem cells. f, Proportion of IgA+ S-specific S-Bmem cells in IgG+ and IgA+ S-specific S-Bmem cells at the indicated timepoints post vaccination. g, Single cell transcriptome sequencing data showing marker genes characteristic for the respective clusters shown in (Fig. 3d). d, f, individual values, means, and standard deviations are shown for n=9 participants. Exact p-values according to two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test. Memory B cells (Bmem cells), switched memory B cells (S-Bmem cells), plasmablasts (PB), S-specific (S+).
Extended Data Fig. 5. Specific binding of S-antigen-tetramers to sorted B cells.
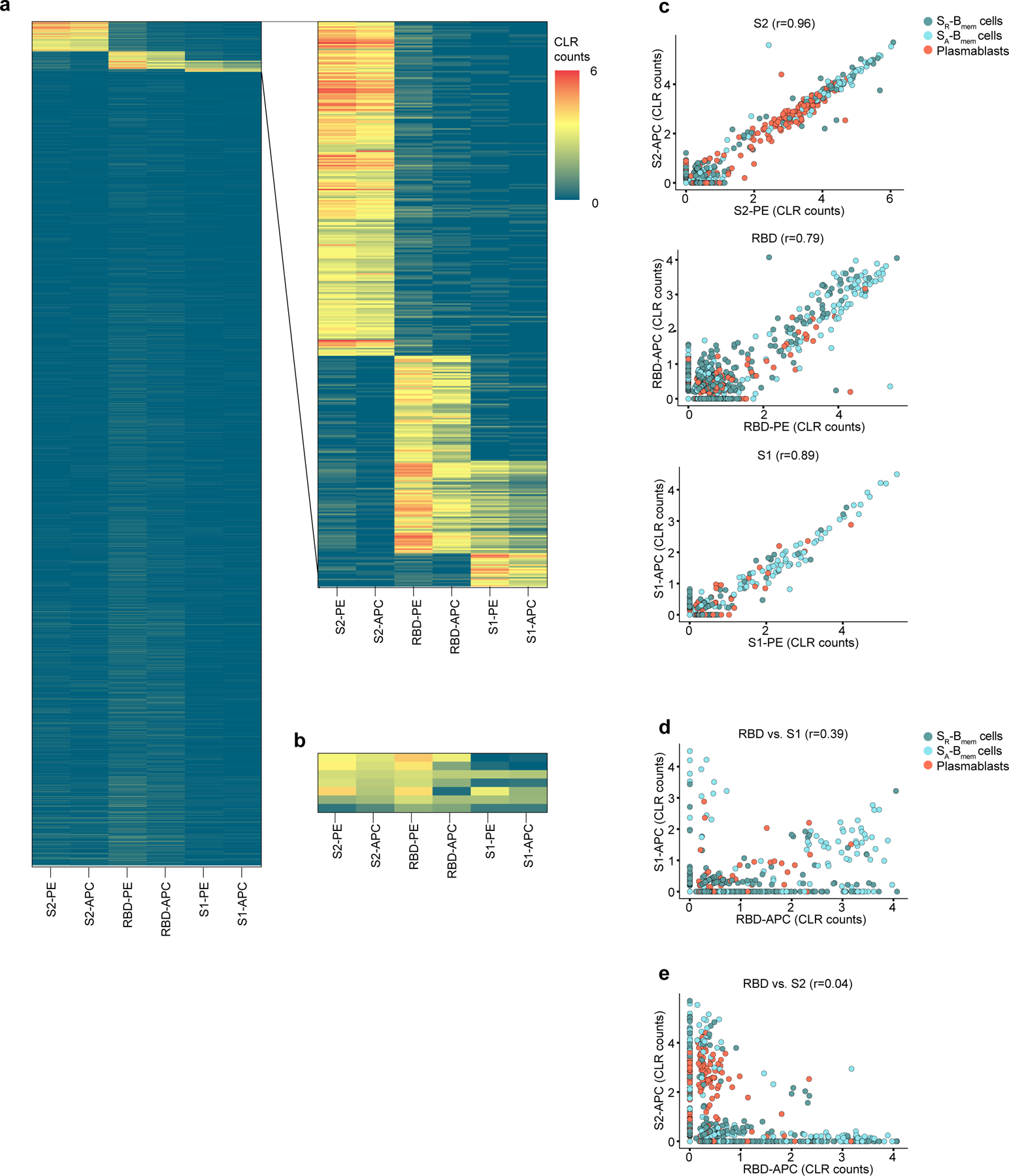
a-e, Single-cell CITESeq sequencing data of antigen-tetramer barcodes identified in the de-multiplexed data set. Antigen-tetramer positive and negative CD3-, CD19+, IgD-, IgM-, CD27+ B cells were mixed during the sort and distinguished during analysis. a, Heatmap of centered log ratio transformed counts (CLR) of barcoded PE and APC tetramers with S2, RBD, and S1 for all sorted cells. Inset magnifies the antigen-specific cells used for downstream analyses. b, Heatmap of CLR counts of polyreactive cells that were removed from downstream analyses. c, Correlation of barcoded PE and APC tetramers for S2 (top), RBD (middle) and S1 (bottom). d, Correlation of barcoded APC tetramers of S1 and RBD. e, Correlation of barcoded APC tetramers of S2 and RBD. c-e, r: Pearson’s correlation coefficient
Extended Data Fig. 6. Plasma reactivity to S protein subunits.
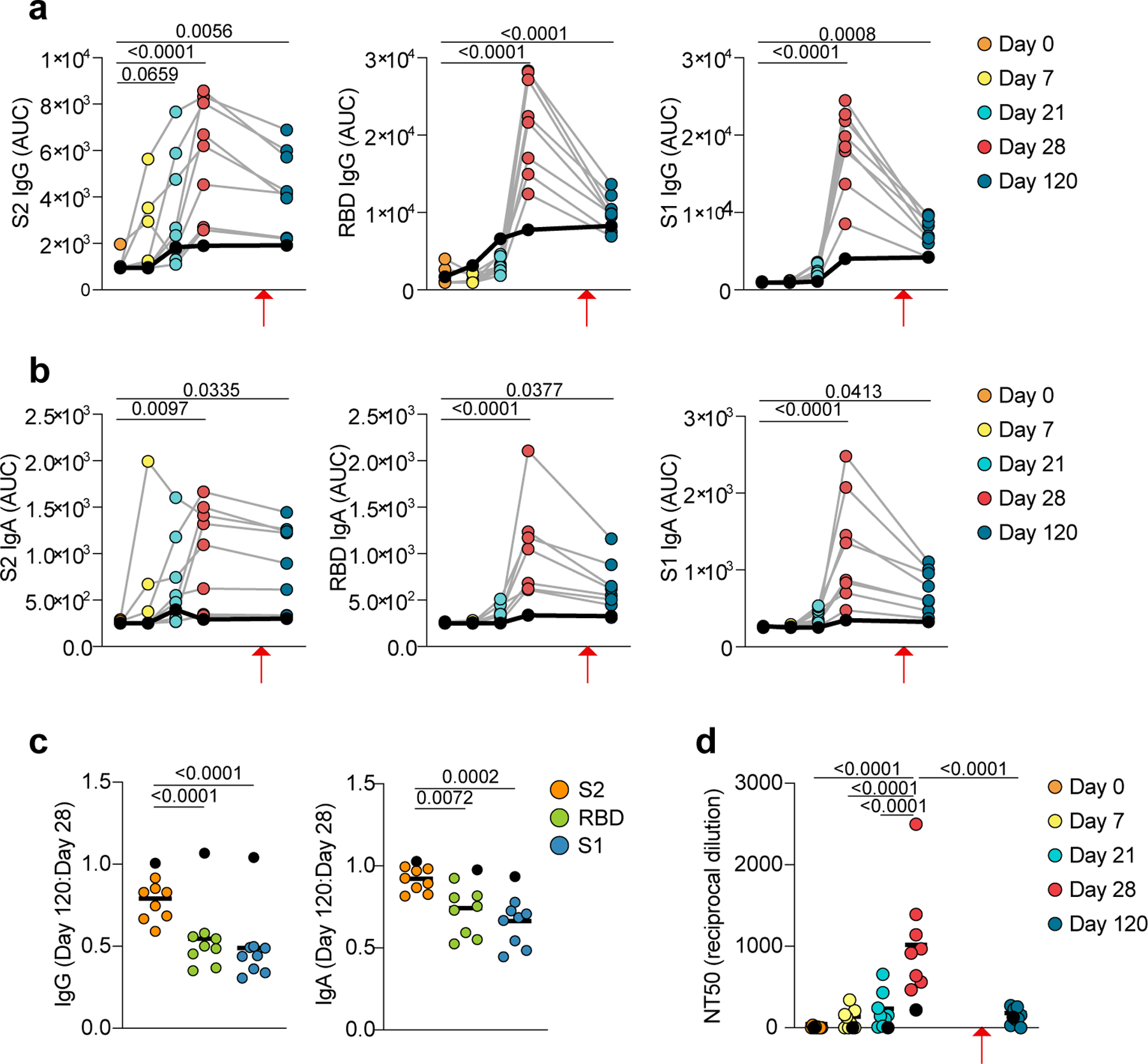
a-c, ELISA data showing plasma reactivity against S-antigen subunits. a, IgG and b, IgA levels against S2 (left), RBD (center) and S1 (right). Area under the curve (AUC) of plasma dilutions is shown for n=9 participants at five timepoints. c, Ratio of IgG titers (left) and IgA titers (right) of day 120 AUC to day 28 AUC for S2, RBD, and S1. d, Plasma neutralization of SARS-CoV-2 pseudovirus. Quantification (NT50) of neutralization titers is shown for n=9 participants at five timepoints. Individual data points represent single B cells from n=9 individuals. a, b, individual values are shown. c,d, Individual values and medians are shown. a-d, Exact p-values according to two-tailed one-way ANOVA, followed by Dunnett’s multiple comparison test. P1 contracted COVID-19 at day 105, eight weeks after the second vaccination dose and two weeks before day 120 (red arrows). P1 is denoted by black dots and black lines. P1 is excluded from statistics in (c).
Extended Data Fig. 7. Somatic hypermutation frequencies of SARS-CoV-2 S-specific B cells.
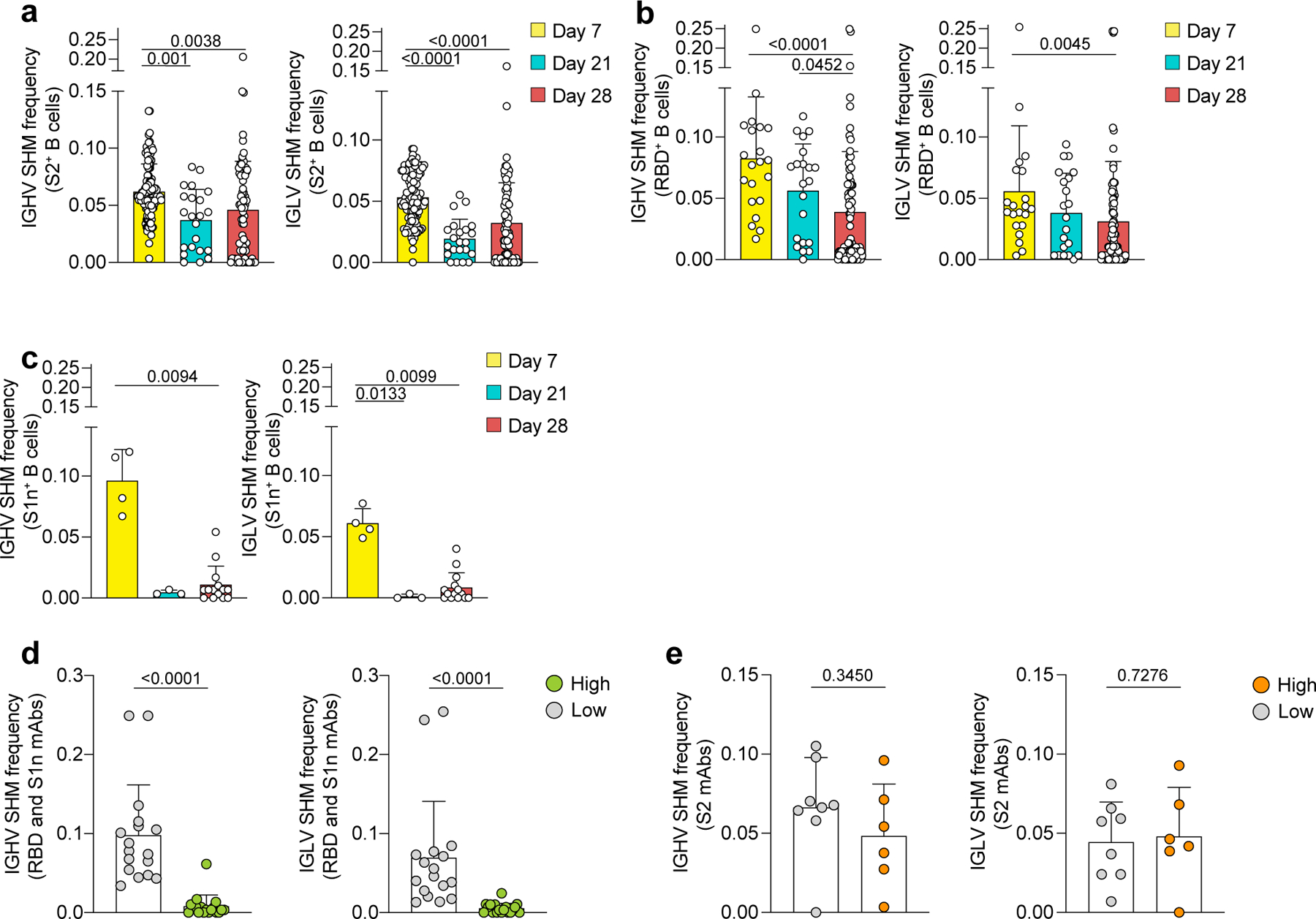
a-e, Single-cell BCR repertoire sequencing data showing SHM frequencies for IGHV (left) and IGLV (right) of B cells specific for a, S2 (n=210 cells), b, RBD (n=124 cells) and c, S1n (n=21 cells), as determined by barcoded S-antigen-tetramers at three timepoints post vaccination. d-e, SHM frequencies of B cells that were recombinantly expressed as mAbs specific for d, S1 (high, n=17 cells; low, n=18 cells) and e, S2 (high, n=8 cells; low, n=6 cells), corresponding to (Fig. 4a). High binding mAbs are defined by the data shown in (Fig. 4a) as positive for both PE and APC barcode tetramer binding and ELISA AUC >3. Individual values, means and standard deviations are shown. Exact p-values according to two-tailed one-way Kruskal Wallis test followed by Dunnett’s multiple comparison test. Immunoglobulin heavy-V gene (IGHV), immunoglobulin light-V gene (IGLV), somatic hypermutation (SHM).
Extended Data Fig. 8. Binding affinities of RBD-, S1n-, and S2-pecific mAbs.
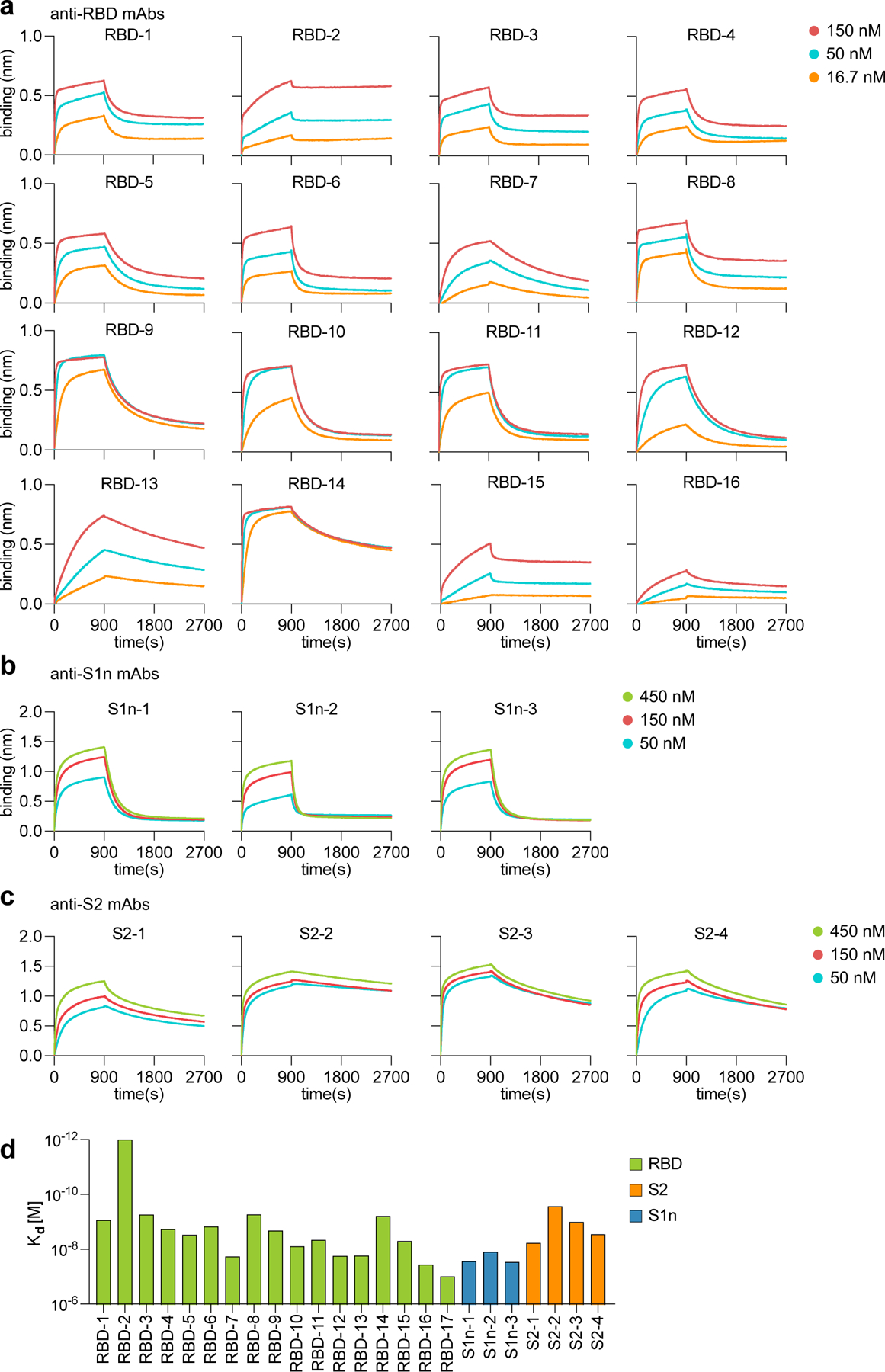
a-d, Bio-layer interferometry data showing mAb-antigen interaction affinities for a, RBD, b, S1n, and c, S2. Binding curves are shown for the indicated concentrations of antigen. d, Kd values of antibodies calculated from the binding curves shown in (a-c).
Extended Data Fig. 9. Polyreactivity of recombinantly expressed antibodies.
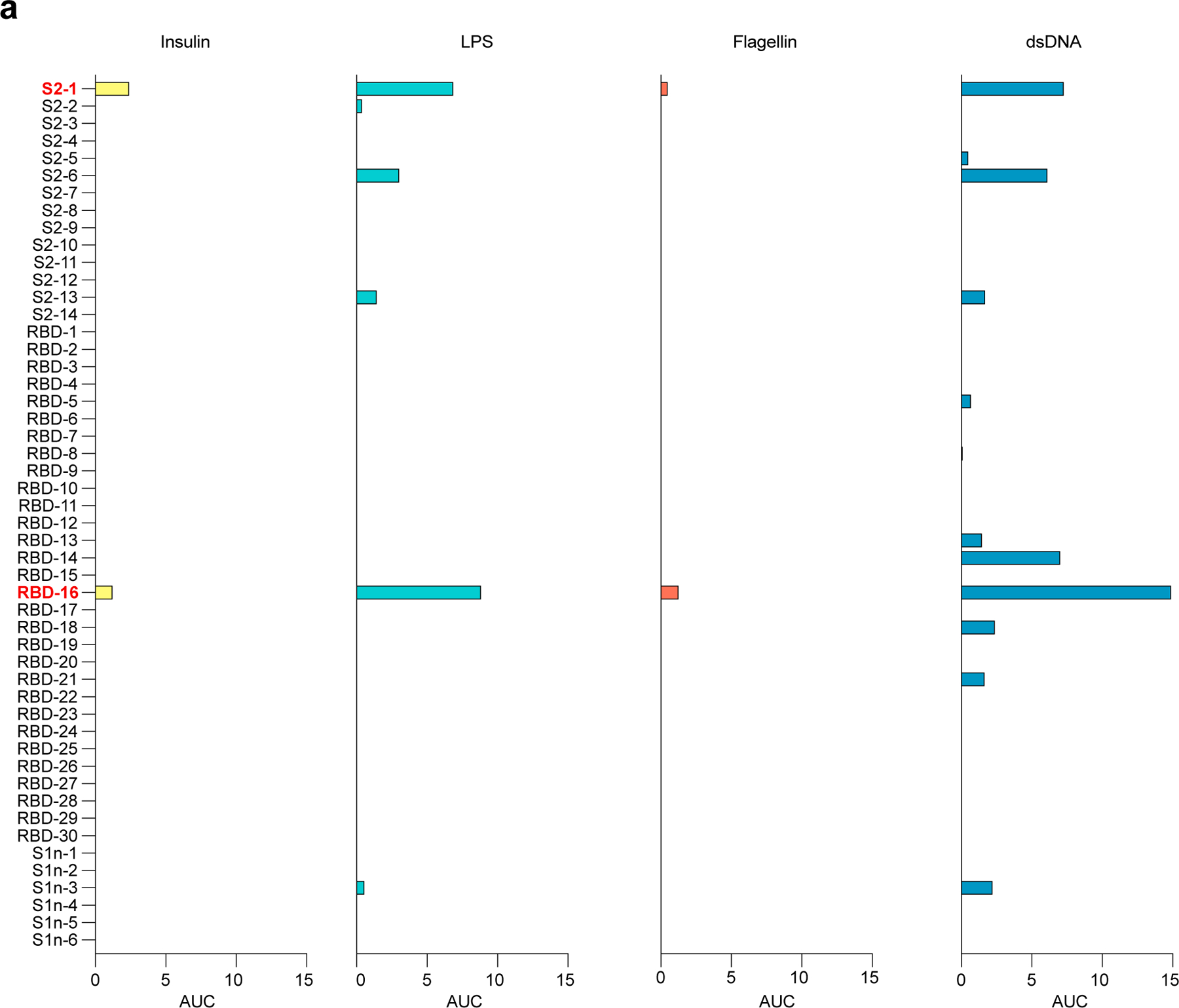
a, ELISA data showing reactivity of recombinant mAbs corresponding to (Fig. 4a,b) against human insulin, LPS, Flagellin and dsDNA, represented as area under the curve (AUC) of serial dilutions. Threshold at 0 was set to the average binding to BSA plus its three-fold standard deviation. Lipopolysaccharide (LPS), double-stranded DNA (dsDNA), bovine serum albumin (BSA).
Extended Data Fig. 10. Neutralization of SARS-CoV-2 Wuhan-Hu-1 pseudovirus and variants by mAbs from vaccinated participants.
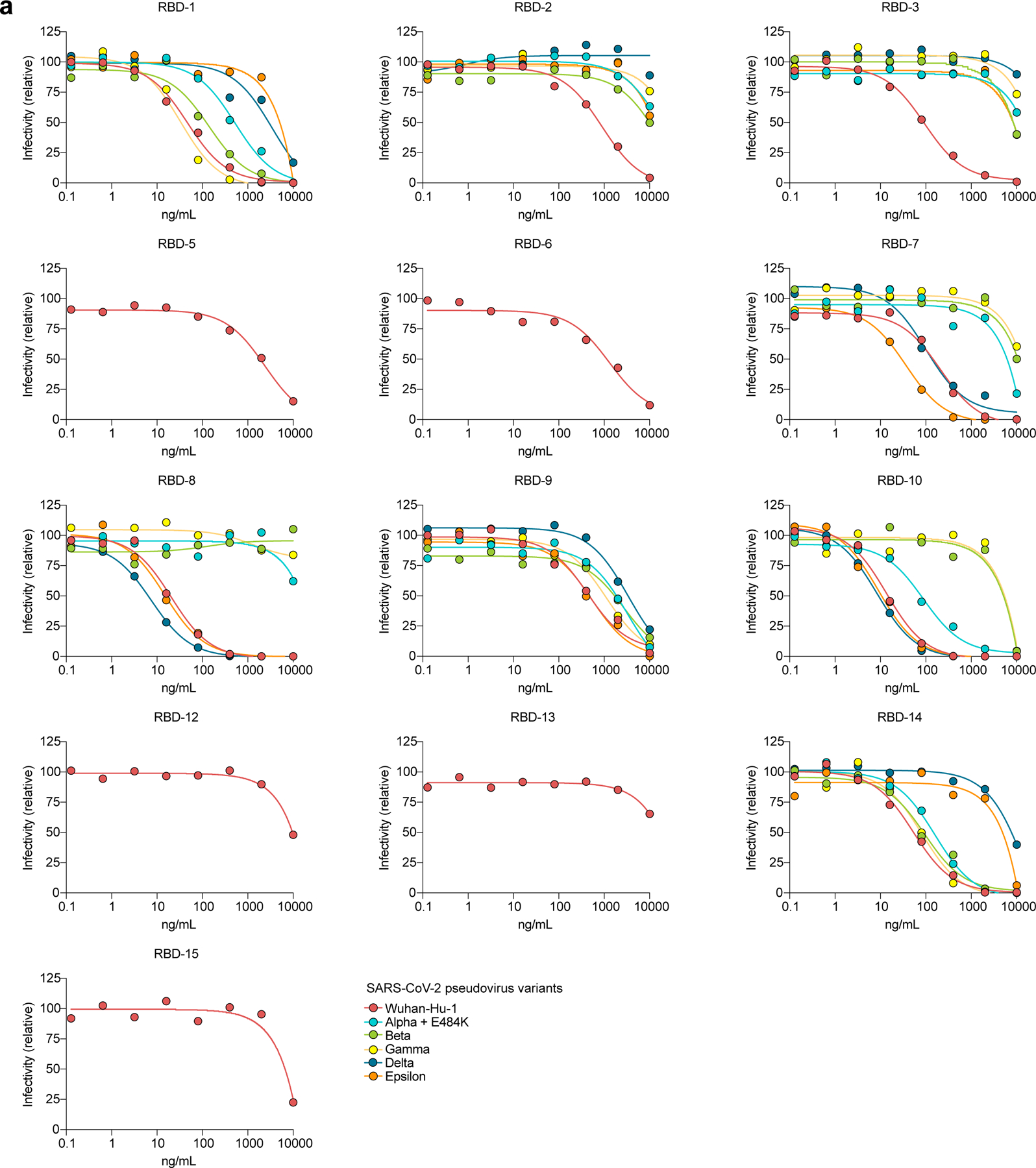
a, Neutralization of SARS-CoV-2 pseudovirus Wuhan-Hu-1 and variants alpha to epsilon are shown as dilution curves for mAbs corresponding to the indicated mAbs in (Fig. 4a).
Supplementary Material
Acknowledgements:
We thank all study participants who devoted time to our research. We thank D. Burton at Scripps Research Institute for providing the ACE2-HeLa cells. We thank M. Baker at Stanford University for key discussions. This work was supported by Foundation for the NIH training grant (T32 AI007290–35) and National Science Foundation Graduate Research Fellowship (DGE-1656518) to R.C.B; the German Research Foundation (DFG, LA3657/1) to T.V.L.; Foundation for the National Institutes of Health R01 AR078268, U19 AI057229, and U01 AI101981 to W.H.R.
Footnotes
Competing Interests Statement:
W.H.R. is a Founder, member of the Board of Directors, and consultant to Atreca, Inc. The remaining authors declare no competing interests.
References
- 1.Amanat F et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell (2021) doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Röltgen K et al. mRNA vaccination compared to infection elicits an IgG-predominant response with greater SARS-CoV-2 specificity and similar decrease in variant spike recognition. medRxiv (2021) doi: 10.1101/2021.04.05.21254952. [DOI] [Google Scholar]
- 3.Polack FP et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel AB et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Wrapp D et al. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. bioRxiv (2020) doi: 10.1101/2020.02.11.944462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Yang C, Xu X-F, Xu W & Liu S-W Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin 41, 1141–1149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccoli L et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol (2018) doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 9.Sanz I et al. Challenges and Opportunities for Consistent Classification of Human B Cell and Plasma Cell Populations. Front. Immunol 10, 2458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krzyzak L et al. CD83 Modulates B Cell Activation and Germinal Center Responses. J. Immunol 196, 3581–3594 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Maddur MS et al. Human B cells induce dendritic cell maturation and favour Th2 polarization by inducing OX-40 ligand. Nat. Commun 5, 4092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palm A-KE & Henry C Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Front. Immunol 10, 1787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bortnick A & Allman D What is and what should always have been: long-lived plasma cells induced by T cell-independent antigens. J. Immunol 190, 5913–5918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodadadi L, Cheng Q, Radbruch A & Hiepe F The Maintenance of Memory Plasma Cells. Front. Immunol 10, 721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fink K Origin and Function of Circulating Plasmablasts during Acute Viral Infections. Front. Immunol 3, 78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei HE et al. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood 113, 2461–2469 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Krammer F The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol 19, 383–397 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Dugan HL et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladner JT et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with an endemic human CoV. bioRxiv (2020) doi: 10.1101/2020.07.27.222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Premkumar L et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreer C et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell 182, 843–854.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaebler C et al. Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greaney AJ et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi X et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah P, Canziani GA, Carter EP & Chaiken I The case for S2: The potential benefits of the S2 subunit of the SARS-CoV-2 spike protein as an immunogen in fighting the COVID-19 pandemic. Front. Immunol 12, 637651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler ES et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell vol. 184 1804–1820.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellebedy AH et al. Adjuvanted H5N1 influenza vaccine enhances both cross-reactive memory B cell and strain-specific naive B cell responses in humans. Proc. Natl. Acad. Sci. U. S. A 117, 17957–17964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuccarino-Catania GV et al. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol 15, 631–637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbott RK et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner JS et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X et al. Cbl Ubiquitin Ligases Control B Cell Exit from the Germinal-Center Reaction. Immunity 48, 530–541.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Lederer K et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity 53, 1281–1295.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner JS et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature (2021) doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisel FJ, Zuccarino-Catania GV, Chikina M & Shlomchik MJ A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity 44, 116–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 35.Wilk AJ et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med 26, 1070–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang AC et al. Publisher Correction: Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature (2021) doi: 10.1038/s41586-021-04080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Combes AJ et al. Publisher Correction: Global absence and targeting of protective immune states in severe COVID-19. Nature 596, E8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alamyar E, Duroux P, Lefranc M-P & Giudicelli V IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol 882, 569–604 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Rogers TF et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford KHD et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited in the Sequence Read Archive linked to BioProject accession number PRJNA775994. All other data needed to evaluate the conclusions in the paper are present in the paper or the Source Data files.


