Abstract
In a recent study, immunoglobulin G in human plasma was identified as a major inhibitor of diagnostic PCR (W. Abu Al-Soud, L. J. Jönsson, and P. Rådström. J. Clin. Microbiol. 38:345–350, 2000). In this study, two major PCR inhibitors in human blood cells were purified using size exclusion and anion-exchange chromatographic procedures. Based on N-terminal amino acid sequencing and electrophoretic analysis of the purified polypeptides, hemoglobin and lactoferrin were identified as PCR-inhibitor components in erythrocytes and leukocytes, respectively. When different concentrations of hemoglobin or lactoferrin were added to PCR mixtures of 25 μl containing 10 different thermostable DNA polymerases and 1 ng of Listeria monocytogenes DNA as template DNA, AmpliTaq Gold, Pwo, and Ultma were inhibited in the presence of ≤1.3 μg of hemoglobin and ≤25 ng of lactoferrin, while rTth and Tli were found to resist inhibition of at least 100 μg of hemoglobin. In addition, the quantitative effects of seven low-molecular-mass inhibitors, present in blood samples or degradation products of hemoglobin, on real-time DNA synthesis of rTth using the LightCycler Instrument were investigated. A reaction system based on a single-stranded poly(dA) template with an oligo(dT) primer annealed to the 3′ end was used. It was found that the addition of 0.25 to 0.1 mg of bile per ml, 2.5 mM CaCl2, 0.25 mM EDTA, 5 μM FeCl3, and 0.01 IU of heparin per ml reduced the fluorescence to approximately 76, 70, 46, 17, and 51%, respectively. Finally, the effects of nine amplification facilitators were studied in the presence of hemoglobin and lactoferrin. Bovine serum albumin (BSA) was the most efficient amplification facilitator, so that the addition of 0.4% (wt/vol) BSA allowed AmpliTaq Gold to amplify DNA in the presence of 20 instead of 1 μg of hemoglobin and 500 instead of 5 ng of lactoferrin. Including 0.02% (wt/vol) gp32, a single-stranded-DNA binding protein, in the reaction mixture of AmpliTaq Gold was also found to reduce the inhibitory effects of hemoglobin and lactoferrin.
Blood samples are extensively used for the PCR-based diagnosis of microbial infections and genetic diseases, as well as for forensic analysis and blood banking (14, 35, 40, 42, 50). However, when applying nucleic acid amplification techniques to blood samples, the amplification capacity can be dramatically reduced or blocked by the presence of PCR-inhibitory substances. Inhibitors in blood which have been identified are either natural components of blood, mainly heme (4) and leukocyte DNA (34), or added anticoagulants such as EDTA (51) and heparin (47). Recently, immunoglobulin G present in human plasma was identified as a major inhibitor of diagnostic PCR in blood (1). Therefore, different methods of sample preparation have been developed to remove the inhibitory effect of blood (2, 10, 26, 50, 56). Despite the various advantages of these methods, generally they (i) are time-consuming, (ii) are labor-intensive, (iii) have potential of losing target microorganism or nucleic acids during processing, (iv) are sample specific, and (v) are not suitable for automation. Thus, more understanding of the nature of PCR inhibitors present in blood and the mechanism of inhibition will be helpful for the development of more general pre-PCR treatments of blood samples.
The presence of PCR-inhibitory substances can be studied by monitoring the presence or absence of the PCR product(s) at the end of thermal cycling by gel electophoresis, dot blots, high-pressure liquid chromatography or microtiter, plate-based, calorimetric assay (25, 33, 44, 45). The quantitative effect of inhibitors on DNA synthesis can also be studied by measuring the efficiency of incorporation of radiolabeled nucleotides. Recently, thermal cyclers with real-time detection of PCR product accumulation were introduced, offering a new possibility to study amplification efficiency and/or DNA synthesis efficiency. These instruments monitor the increase in fluorescence signal using different fluorescence techniques, such as double-stranded-DNA (dsDNA) binding dyes or hybridization probes.
The aim of the present study was to identify and characterize the major inhibitors of diagnostic PCR in human blood cells using a standardized PCR assay containing the thermostable DNA polymerase AmpliTaq Gold. The effects of the major PCR inhibitors in human blood cells on 10 commercial thermostable DNA polymerases were also investigated. The quantitative effects of low-molecular-mass PCR-inhibitory components present in blood samples or degradation products of hemoglobin on real-time DNA synthesis using the LightCycler Instrument were also investigated. Finally, the ability of nine amplification facilitators to relieve the PCR inhibition by hemoglobin and lactoferrin was also studied.
MATERIALS AND METHODS
Template DNA.
DNA of Listeria monocytogenes 167 vet, which was obtained from Swedish Meats R&D, Kävlinge, Sweden, was used as target DNA in this study. Extraction of DNA was performed in accordance with a standard technique described by Sambrook et al. (45), modified by the addition of 30 U of mutanolysin (Sigma Chemical Co., St. Louis, Mo.) per ml to the lysis solution. The concentration of DNA was determined spectrophotometrically (45).
PCR assay and incubation conditions.
The total volume of the PCR mixture was 25 μl. All of the PCR mixtures contained 0.5 μM (each) primers rU8 and LM2 (31, 39) and 0.2 mM (each) deoxyribonucleoside triphosphates. Reaction buffers for the DNA polymerases were as specified by the manufacturers (Table 1). The reaction mixtures were subjected to 30 cycles consisting of heat denaturation at 94°C for 40 s, primer annealing at 53°C for 40 s, and DNA extension at 72°C for 40 s. Finally, the samples were maintained at 72°C for 7 min for the final extension of DNA. These incubation conditions were the same for all amplification reactions except those containing AmpliTaq Gold, since this polymerase requires a hot start (95°C for 10 min). Incubation was carried out in a GeneAmp 9700 (Perkin-Elmer Cetus, Norwalk, Conn). The 0.55-kb PCR product was visualized by 1.3% agarose gel electrophoresis with ethidium bromide (45). The gel was analyzed by the gel documentation system (Bio-Rad Laboratories). The results were recorded as follows: +, PCR product of high yield, ±, PCR product of low yield; or −, no PCR product.
TABLE 1.
Reaction buffers for the DNA polymerases
| DNA polymerase | Reaction buffer components |
|---|---|
| AmpliTaq Gold | 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, and 0.75 U AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus) |
| DyNAzyme II | 10 mM Tris-HCl (pH 8.8; 25°C), 1.5 mM MgCl2, 50 mM KCl, 0.1% TritonX-100, and 1 U of DyNAzyme II DNA polymerase (FINNZYMES OY, Riihitontuntie, Espoo, Finland) |
| DyNAzyme EXL | 50 mM Tris-HCl (pH 9.0; 25°C), 2.5 mM MgCl2, 15 mM (NH4)2SO4, 0.1% Triton X-100, and 1 U of DyNAzyme EXT DNA polymerase (FINNZYMES OY) |
| HotTub | 50 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, 2.5 mM MgCl2, and 0.75 U of HotTub DNA polymerase (Amersham Pharmacia Biotech) |
| Pwo | 10 mM Tris-HCl (pH 8.85; 20°C), 25 mM KCl, 5 mM (NH4)2SO4, 1.5 mM MgCl2, and 1.25 U of Pwo DNA polymerase (Roche Molecular Biochemicals) |
| rTth | 10 mM Tris-HCl (pH 8.3), 5% (vol/vol) glycerol, 0.1 M KCl, 0.05% (wt/vol) Tween 20, 0.75 mM EGTA, 2.5 mM MgCl2, and 1.25 U of rTth DNA polymerase (Perkin-Elmer Cetus) |
| Taq | 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3; 20°C), and 0.75 U of Taq DNA polymerase (Roche Molecular Biochemicals) |
| Tfl | 20 mM Tris-acetate (pH 9.0), 10 mM (NH4)2SO4, 75 mM K-acetate, 0.05% Tween 20, 2.5 mM MgSO4, and 0.5 U of Tfl DNA polymerase (Promega Corporation, Madison, Wis.) |
| Tli | 10 mM Tris-HCl (pH 9.0; 25°C), 0.1% Triton X-100, 50 mM KCl, 2.5 mM MgCl2, and 0.3 U of Tli DNA polymerase (Promega Corporation) |
| Ultma | 10 mM Tris-HCl (pH 8.8; room temperature), 10 mM KCl, 0.002% (vol/vol) Tween 20, 2.5 mM MgCl2, and 0.75 U of Ultma DNA polymerase (Perkin-Elmer Cetus) |
Preparation of blood sample.
The blood sample used was drawn from a healthy person in a quadruple blood bag (CPD; Baxter S.A., Maurpas, France). The bag was centrifuged in a cold centrifuge (Hettich, Tottlingen, Germany) at 2,800 × g for 9 min. Plasma and platelets were extracted in one bag and buffy coat and a portion of erythrocytes were extracted in another bag, using an Optipress plasma extractor (Baxter). Adsol was added to the erythrocytes. The plasma bag was recentrifuged at 1,200 × g for 7 min, plasma was extracted into an empty bag, and the concentrated platelets were suspended in 60 ml of plasma. Each blood fraction was poured into a sterile 1.5-ml Eppendorf tube, flash frozen in liquid nitrogen, and stored at −80°C. The frozen samples were thawed at room temperature before use.
Purification of PCR inhibitors in human erythrocytes by FPLC.
The ability of different fractions of blood cells to inhibit PCR was evaluated by the addition of 5 μl of the different fractions to PCR mixtures of AmpliTaq Gold containing 1 ng of L. monocytogenes DNA. The PCR inhibitors were purified by a chromatographic procedure using a fast protein liquid chromatography (FPLC) system (Amersham Pharmacia Biotech, Uppsala, Sweden) containing two model P-500 high-precision pumps, a model LCC-501 Plus liquid chromatography controller, three motor valves (MV-7 and MV-8), and a model REC 102 recorder. The elution was monitored with a UV-M II control unit (at 280 nm), and fractions were collected with a model FRAC-200 fraction collector. All of the buffers and solutions were filtered through 0.2-μm-pore-size AcroCap membrane filters (Gelman Sciences, Ann Arbor, Mich.) and degassed before use. Erythrocytes in Adsol were thawed at room temperature and diluted once with deionized water. A volume of 5.5 ml of diluted erythrocytes was mixed with approximately 3.7 ml of glass beads and vortexed at maximum speed for 5 min. The lysate of the erythrocytes was diluted twice with deionized water and centrifuged at 16,000 × g for 5 min. A 2.0-ml sample of erythrocyte lysate was injected into a Hiload 16/60 Superdex 200 prepacked gel filtration column (Amersham Pharmacia Biotech), which was equilibrated with a buffer consisting of 20 mM Tris-HCl and 100 mM NaCl (pH 7.2). Different fractions of the erythrocyte components were eluted with a buffer consisting of 20 mM Tris-HCl and 100 mM NaCl (pH 7.2) at a flow rate of 1.0 ml/min. Six fractions were collected, dialyzed overnight against 20 mM Tris-HCl (pH 8.6) by using dialysis tubing with a cutoff of 12 to 14 kDa (Spectra/Por, Houston, Tex.), and tested for their ability to inhibit the amplification capacity of AmpliTaq Gold. Only one fraction (18 ml) was found to be inhibitory to AmpliTaq Gold. This fraction was filtered through a 0.2-μm-pore-size Minisart membrane filter, injected into a Mono Q HR 5/5 anion-exchange column (Amersham Pharmacia Biotech), and eluted with 20 mM Tris-HCl (pH 8.6) and a sodium chloride gradient (0 to 0.5 M) for 30 min at a flow rate of 1 ml/min. Three fractions were collected; two of them were eluted by the NaCl gradient, while the third was collected in the flowthrough. These fractions were dialyzed overnight against 20 mM Tris-HCl (pH 8.6), and tested for their ability to inhibit the amplification capacity of AmpliTaq Gold. Only one of the two fractions eluted by the salt gradient, red in color, was highly inhibitory to AmpliTaq Gold and required 100 times dilution to remove the inhibition, while the other two fractions were not inhibitory. The concentration of hemoglobin was determined with a Sysmex K-1000 automated hematology analyzer (Sysmex Corporation of America, Long Grove, Ill.).
Purification of PCR inhibitors in human leukocytes by FPLC.
The buffy coat was thawed at room temperature. The leukocytes were isolated by a modification of a method described by Polacek et al. (37). Forty milliliters of buffy coat was drawn into 10 50-ml sterile plastic tubes containing 8 ml of 6% dextran (60 to 90 kDa) (Sigma Chemical). The contents of the tubes were mixed gently by inversion and left to stand for 1 h at 22°C. The leukocyte-rich top layer was aspirated into 50-ml sterile tubes. The cells were centrifuged at 400 × g for 10 min at 22°C, the supernatant was poured off, and each cell pellet was resuspended in 10 ml of 0.87% ammonium chloride, after which an additional 30 ml of ammonium chloride solution was added to each tube. The contents of the tubes were mixed gently and left to stand for 2 min for hypotonic lysis of the residual erythrocytes. The cells were centrifuged at 150 × g for 10 min at 22°C. The leukocytes were washed three times with 25 mM HEPES buffer–0.85% NaCl (pH 7.2) and suspended in 3 ml of the washing buffer. The leukocytes were lysed with an X-press apparatus. The leukocyte lysate was centrifuged in Eppendorf tubes at 16,000 × g for 5 min at 4°C. A 2-ml sample of cell extract was injected into a Hiload 16/60 Superdex 200 column (Amersham Pharmacia Biotech) as described above. Four fractions, of 12, 12, 9, and 15 ml, were collected and dialyzed overnight against 20 mM Tris-HCl (pH 8.6). Only one fraction of 12 ml was inhibitory, and its inhibitory effect was removed after reducing its concentration to 2% (vol/vol). This fraction was filtered and injected into a Mono Q HR 5/5 column (Amersham Pharmacia Biotech) as described above. Three fractions were collected; two of these fractions were eluted by the NaCl gradient, while the third fraction was eluted from the column after injection of 2 ml of NaCl (2 M). The three fractions were dialyzed overnight against 20 mM Tris-HCl (pH 8.6) and tested for their ability to inhibit the amplification capacity of AmpliTaq Gold. Only one of the two fractions eluted by the salt gradient was highly inhibitory to AmpliTaq Gold and required a dilution of >20-fold to remove the inhibition, while the other two fractions were not inhibitory.
PCR inhibitors in human thrombocytes.
Seventeen milliliters of thrombocyte-rich plasma was thawed at room temperature and centrifuged at 4,500 × g for 10 min. The thrombocytes were washed three times with 25 mM HEPES buffer–0.85% NaCl (pH 7.2) and suspended in 3 ml of the washing buffer. The thrombocytes were lysed with the X-press and centrifuged at 16,000 × g for 5 min. Two milliliters of the lysate of the thrombocytes was injected into a Hiload 16/60 Superdex 200 column (Amersham Pharmacia Biotech) as described above. Six fractions, of 7.5, 8, 12, 15, 13, and 18 ml, were collected and dialyzed overnight against 20 mM Tris-HCl (pH 8.6). Only one thrombocyte fraction of 7.5 ml was inhibitory to the amplification capacity of AmpliTaq Gold. This fraction was filtered and injected into a Mono Q HR 5/5 column (Amersham Pharmacia Biotech) as described above. Ten fractions were collected, dialyzed overnight against 20 mM Tris-HCl (pH 8.6), and tested for their ability to inhibit the amplification capacity of AmpliTaq Gold.
SDS-PAGE and N-terminal amino acid sequencing of the major plasma PCR inhibitor.
All the chemicals used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Bio-Rad Laboratories. The low-molecular-mass standard containing phosphorylase b (94.0 kDa), bovine serum albumin (BSA) (67.0 kDa), ovalbumin (43.0 kDa), carbonic anhydrase (30.0 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa) was supplied by Amersham Pharmacia Biotech. The erythrocyte and leukocyte PCR inhibitors were analyzed by discontinuous SDS–12% PAGE, as described by Laemmli (29), using a Mini-gel apparatus (Bio-Rad Laboratories). The protein bands were visualized with Coomassie brilliant blue R-250 or were subjected to electroblotting to polyvinylidene difluoride membranes for N-terminal sequencing, as described in the Bio-Rad Laboratories Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell instruction manual (catalog number 170–3940). The membrane was stained with Coomassie brilliant blue, and the bands with approximate molecular masses of 16 and 60 kDa, purified from erythrocytes, and of 80 kDa, purified from leukocytes, were excised and subjected to N-terminal sequencing. Edman degradation was performed by the Department of Plant Biology at the Swedish University of Agricultural Sciences (Uppsala, Sweden).
Reaction conditions in the LightCycler Instrument.
The volume of the LightCycler Instrument (Roche Molecular Biochemicals) mixture was 25 μl. All the mixtures contained 0.2 mM dTTP, a 1:10,000-diluted stock solution of SYBR Green I (Roche Molecular Biochemicals), 4 mM MgCl2, and poly(dA)-oligo(dT)12–18 (Amersham Pharmacia Biotech). The reaction mixtures for AmpliTaq Gold contained 1 × Gold buffer and 0.75 U of AmpliTaq Gold, while the reaction mixtures for rTth DNA polymerase contained 1 × chelating buffer and 1.25 U of rTth (Table 1). AmpliTaq Gold was activated by incubating the reaction mixtures of AmpliTaq Gold, without single-stranded DNA template and inhibitors, at 90°C for 40 min as recommended by the manufacturer for this type of application. The reaction mixtures were maintained at 65°C for 30 min. Ninety fluorescence measurements were taken, at 20-s intervals. The LightCycler Instrument monitored the fluorescence signal of samples online. The mean value of the fluorescence signals of three independent experiments was calculated.
Effects of PCR inhibitors on real-time DNA synthesis.
Two strategies were used to study the effects of bile salts, bilirubin, CaCl2, EDTA, FeCl3, hemin, and heparin on the synthesis of DNA by AmpliTaq Gold and rTth. The first strategy was the addition of 1 mg of bile salts per ml, 2 μg of bilirubin per ml, 10 mM CaCl2, 5 mM EDTA, 15 μM FeCl3, 0.1 μM hemin, and 0.2 IU of heparin per ml to reaction mixtures of rTth containing 3, 4, 5, 6, 7, 8, or 9 ng of poly(dA)-oligo(dT)12–18 per reaction tube. The linearity of the relationship (r2) between poly(dA)-oligo(dT) concentration and fluorescence level in the presence and absence of PCR-inhibitory substances was then calculated (r2 equals 0 when the values of the independent variable do not allow any prediction of the dependent variables, and r2 equals 1 when the values of the independent variable can perfectly predict the dependent variables). The second strategy was the addition of different concentrations of bile salts, CaCl2, EDTA, FeCl3, hemin, and heparin (namely, 0.025, 0.05, 0.1, 0.25, and 0.5 mg of bile salts per ml, 2.5, 5, 10, 15, 30, and 20 mM CaCl2; 0.25, 0.5, 1, 2.5, and 5 mM EDTA; 5, 10, 25, 50, and 100 μM FeCl3; 0.025, 0.05, 0.1, 0.25, and 0.5 μM hemin; and 0.01, 0.025, 0.05, 0.01, and 0.25 IU of heparin per ml) to the rTth and AmpliTaq Gold reaction mixtures containing 10 ng of poly(dA)-oligo(dT) per reaction tube. The results were then recorded as percent fluorescence [(fluorescence signal in the presence of inhibitor/fluorescence signal without inhibitor) ×100].
RESULTS
Purification of blood cell PCR inhibitors.
Cell lysates of erythrocytes, leukocytes, and thrombocytes were prepared and centrifuged to remove the cell debris from the cell extracts. The cell debris were resuspended in deionized water to have the same concentration as in the total cell lysates. Different concentrations of cell debris and cell extracts (20, 4, 2, 0.8, 0.4 and 0.2% [vol/vol]) were added to standardized reaction mixtures of 25 μl containing AmpliTaq Gold and 1 ng of L. monocytogenes DNA (Table 2). The water-soluble supernatants of the blood cell lysates were used for chromatographic purification of the PCR inhibitors, and the inhibitory effects of the different fractions after each purification step were tested. The cell extract of thrombocytes was divided into six main fractions using size exclusion chromatography. Only one fraction was inhibitory. This fraction was further purified using anion-exchange chromatography and collected as 10 fractions, one of which was slightly PCR inhibitory. The inhibitory effect was removed after the concentration was reduced to 4% (vol/vol). This fraction was not used for further purification of PCR-inhibitory components.
TABLE 2.
Effects of different concentrations of erythrocyte, leukocyte, and thrombocyte cell extracts and cell debrises on PCR
| Concn (%, vol/vol)a | PCR resultsb
|
|||||
|---|---|---|---|---|---|---|
| Erythrocytes
|
Leukocytes
|
Thrombocytes
|
||||
| Cell extract | Cell debris | Cell extract | Cell debris | Cell extract | Cell debris | |
| 20 | − | ± | − | + | − | − |
| 4 | − | + | − | + | − | − |
| 2 | − | + | + | + | − | − |
| 0.8 | − | + | + | + | − | + |
| 0.4 | − | + | + | + | + | + |
| 0.2 | + | + | + | + | + | + |
Percentage of different blood fractions in the PCR mixtures containing 1 ng of L. monocytogenes DNA and AmpliTaq Gold.
+, PCR product of high yield; ±, PCR product of low yield; −, no PCR product.
The cell extracts of erythrocytes and leukocytes were fractionated by size exclusion and anion-exchange chromatography. Only one erythrocyte fraction, red in color, was found to be highly inhibitory to AmpliTaq Gold, and its concentration had to be reduced to 0.2% (vol/vol) in the PCR mixture. SDS-PAGE analysis of this fraction showed three protein bands with approximate molecular masses of 16, 30, and 60 kDa (Fig. 1). The 30-kDa-molecular-mass polypeptide was suspected to be carbonic anhydrase, based on the similarity of its electrophoretic pattern to that of carbonic anhydrase. A similar band was also observed in a commercial preparation of human hemoglobin from Sigma Chemical. Furthermore, the addition of 52 μg of carbonic anhydrase was not inhibitory to AmpliTaq Gold, which excluded carbonic anhydrase as being the PCR inhibitor. The N-terminal sequence of the 60-kDa band (-DAHKSEVAHRF-) showed 100% identity with the N-terminal sequence of human albumin, which is a known amplification facilitator (27), whereas the N-terminal sequence of the 16 kDa band [-(V(LH)(LS)(PT)(PA)(DE) (KE)(KT)(NS)(VA)-] was shown to be a mixture of two polypeptides corresponding to the N-terminal sequences of human hemoglobin alpha- and beta-polypeptides. SDS-PAGE analysis of the leukocyte fraction inhibitory to AmpliTaq Gold showed one protein band with an approximate molecular mass of 80 kDa (Fig. 1). The N-terminal sequencing of this band (-GRRRRSVQW-AVSQP-) showed 100% identity with the N-terminal sequence of human lactoferrin for the residues determined (the 10th amino acid position was not determined).
FIG. 1.
SDS-PAGE of purified hemoglobin and lactoferrin in comparison with BSA, hemoglobin, and human milk lactoferrin. Electrophoretic separation was carried out by using SDS–12% PAGE. Proteins were detected with Coomassie brilliant blue. The polypeptides marked by rectangles were subjected to N-terminal sequencing. Lanes: 1, low-molecular-weight protein standards (Amersham Pharmacia Biotech); 2, BSA (Sigma Chemical); 3, human hemoglobin (Sigma Chemical); 4, purified hemoglobin; 5, human milk lactoferrin (Sigma Chemical); 6, purified lactoferrin.
The inhibitory effects of hemoglobin and lactoferrin on the ability of AmpliTaq Gold or rTth to amplify different concentrations of L. monocytogenes DNA (1 μg to 1 pg) were evaluated (Table 3). The detection limit for L. monocytogenes DNA in pure water was 10 pg of DNA for both polymerases. When 25 μg of hemoglobin was added to the PCR mixtures, the detection sensitivity of AmpliTaq Gold was reduced by more than 6 log units, while the detection sensitivity of rTth was reduced by 2 log units. Addition of 25 ng of lactoferrin reduced the detection sensitivities of both polymerases by 3 log units.
TABLE 3.
AmpliTaq Gold and rTth detection sensitivities in water in the presence of hemoglobin or lactoferrina
| L. monocytogenes concn | PCR resultb
|
|||||
|---|---|---|---|---|---|---|
| Water
|
Hemoglobin
|
Lactoferrin
|
||||
| AmpliTaq | rTth | AmpliTaq | rTth | AmpliTaq | rTth | |
| 10−6 | +, + | +, + | −, − | +, + | +, + | +, + |
| 10−7 | +, + | +, + | −, − | +, + | +, + | +, + |
| 10−8 | +, + | +, + | −, − | +, + | +, + | +, + |
| 10−9 | +, + | +, + | −, − | +, + | −, − | −, − |
| 10−10 | +, + | +, + | −, − | −, − | −, − | −, − |
| 10−11 | +, + | +, + | −, − | −, − | −, − | −, − |
| 10−12 | −, − | −, − | −, − | −, − | −, − | −, − |
Ability of AmpliTaq Gold and rTth DNA polymerases to amplify different concentrations of L. monocytogenes DNA in water, in the presence of 25 μg of hemoglobin (Sigma Chemical) or 25 ng of lactoferrin (Sigma Chemical).
Results of two independent PCRs. +, PCR product of high yield; −, no PCR product.
Effect of hemoglobin and lactoferrin on the amplification capacities of 10 DNA polymerases.
The capacities of 10 DNA polymerases to amplify 1 ng of L. monocytogenes DNA in pure water and in the presence of different concentrations of hemoglobin and lactoferrin were studied (Table 4). It was found that AmpliTaq Gold, Pwo, and Ultma were completely inhibited in the presence of 1.3 μg of hemoglobin in the PCR mixture, while DyNAzyme EXL, rTth, Tfl, and Tli DNA polymerases could amplify the specific PCR product in the presence of all concentrations of hemoglobin tested in both replicates. The polymerases DyNAzyme II, HotTub, and Taq amplified the specific product in one of two replicates in the presence of 100 μg of purified human hemoglobin. Ultma was also the DNA polymerase most sensitive to lactoferrin, amplifying the specific product in one of two replicates in the presence of 2.5 and 1 ng of human milk lactoferrin, while rTth and Tfl were the most resistant and amplified the specific product in one of two replicates in the presence of 25 ng of human milk lactoferrin.
TABLE 4.
Inhibitory effects of different concentrations of hemoglobin and lactoferrin on the amplification capacities of 10 thermostable DNA polymerases
| DNA polymerase | PCR result with the indicated concn ofa:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemoglobin (μg/reaction tube)
|
Lactoferrin (ng/reaction tube)
|
|||||||||||||
| 0 | 1 | 1.3 | 0.4 | 2 | 4 | 10 | 100 | 0 | 0.5 | 1 | 2.5 | 5 | 25 | |
| AmpliTaq Gold | +, + | ±, ± | − | −, − | − | −, − | − | −, − | +, + | +, + | +, + | +, + | +, + | −, − |
| DyNAzyme II | +, + | +, + | + | +, + | + | +, + | + | ±, − | +, + | +, + | +, + | +, + | +, + | −, − |
| DyNAzyme EXL | +, + | +, + | + | +, + | + | +, + | + | +, ± | +, + | +, + | +, + | +, + | +, ± | −, − |
| HotTub | +, + | +, + | + | +, + | + | +, + | + | +, − | +, + | +, + | +, + | +, + | +, + | −, − |
| Pwo | +, ±b | −, − | − | −, − | − | −, − | − | −, − | ±, ± | ±, ± | ±, ± | ±, ± | −, − | −, − |
| rTth | +, + | +, + | + | +, + | + | +, + | + | +, + | +, + | +, + | +, + | +, + | +, + | +, − |
| Taq | +, + | +, + | + | +, + | + | +, + | + | ±, − | +, + | +, + | +, + | +, + | +, + | −, − |
| Tfl | +, + | +, + | + | +, + | + | +, + | + | +, ± | +, + | +, + | +, + | +, + | +, + | +, − |
| Tli | +, + | +, + | + | +, + | + | +, + | + | +, + | +, + | +, + | +, + | +, + | +, + | −, − |
| Ultma | +, ± | −, − | − | −, − | − | −, − | − | −, − | +, + | +, + | +, − | ±, − | −, − | −, − |
Results of two independent PCRs. +, PCR product of high yield; ±, PCR product of low yield; −, no PCR product. Purified hemoglobin conc. and human milk lactoferrin. (Sigma Chemical) were added to the PCR mixtures of the 10 DNA polymerases containing 1 ng of L. monocytogenes DNA.
Pwo had many nonspecific products.
Quantitative inhibitory effect of ions and low-molecular weight solutes on real-time DNA synthesis.
The aim of this study was to evaluate the inhibitory effects of seven low-molecular-mass solutes, present in blood samples or degradation products of hemoglobin, on real-time DNA synthesis using the LightCycler Instrument. The DNA synthesis was measured by monitoring the increase in the fluorescence signal. The increase in fluorescence signals of AmpliTaq Gold and rTth were compared in the presence of different concentrations of poly (dA)-oligo(dT) (data not shown). The increase in the fluorescence signal of the reaction mixtures of rTth was ∼15 times higher than that of AmpliTaq Gold. To check whether this difference was due to differences in the buffers of the two polymerases, the efficiency of Taq DNA polymerase, which has a buffer content similar to that of AmpliTaq Gold, was compared with those of the other two polymerases. It was found that the real-time DNA synthesis of Taq DNA polymerase was as efficient as that of rTth (data not shown).
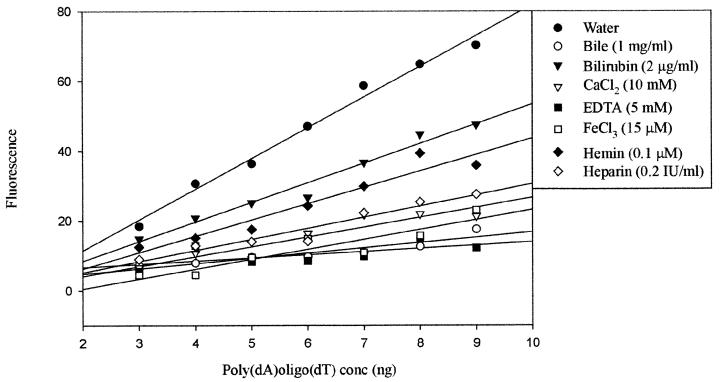
A linear relationship (r2 = 0.99) was found between the increase in concentration of template DNA in water and the increase in fluorescence (Fig. 2). When the effect of PCR inhibitors on the real-time DNA synthesis of rTth was also studied, a linear relationship were noted for bilirubin (r2 = 0.97), hemin (r2 = 0.93), and heparin (r2 = 0.94), whereas the linearity was less in the presence of bile (r2 = 0.86), CaCl2 (r2 = 0.87), FeCl3 (r2 = 0.86), and EDTA (r2 = 0.34).
FIG. 2.
Effects of seven PCR inhibitors on the real-time synthesis of rTth.
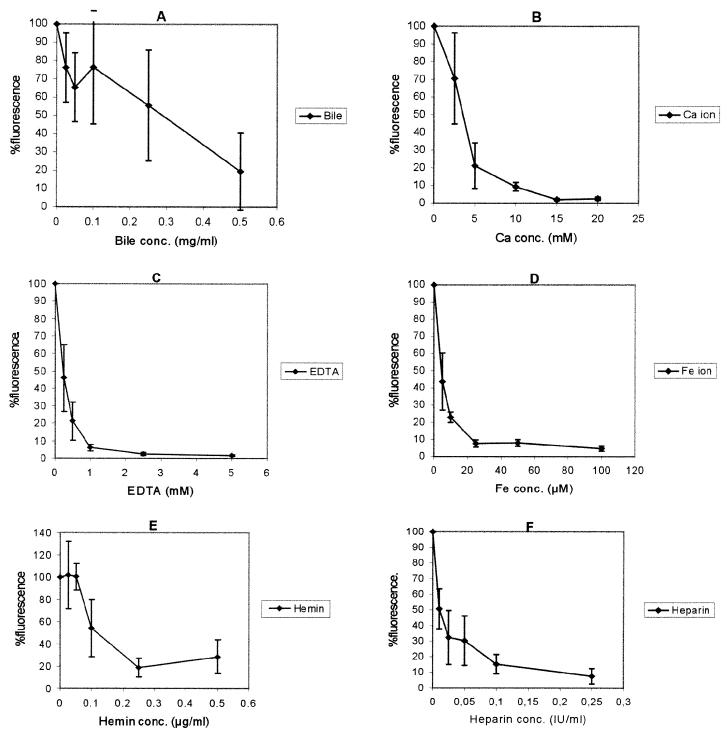
When the effects of different concentrations of bile salts, CaCl2, EDTA, FeCl3, hemin, and heparin on real-time DNA synthesis were studied, the mean of three independent fluorescence signals decreased with increasing concentrations of inhibitory substances (Fig. 3). The standard deviations were found to increase as the concentration of inhibitors decreased, except when bile and hemin were added to the reaction mixtures. Addition of 0.25 to 0.1 mg of bile per ml, 2.5 mM CaCl2, 0.25 mM EDTA, 5 μM FeCl3, 0.1 μM hemin, and 0.01 IU of heparin per ml reduced the fluorescence to approximately 76, 70, 46, 17, 54, and 51%, respectively. However, addition of 0.25 to 0.05 μg of hemin per ml was found to slightly enhance the fluorescence (to 101 to 102%).
FIG. 3.
Effects of different concentrations of bile salts, bilirubin, CaCl2, EDTA, FeCl3, hemin, and heparin on real-time DNA synthesis of rTth.
The relieving effects of nine amplification facilitators on PCR inhibition by hemoglobin and lactoferrin.
The effects of nine PCR facilitators on the amplification capacity of AmpliTaq Gold in the presence of different concentrations of hemoglobin and lactoferrin were studied (Table 5). Betaine, BSA, and gp32 were able to reduce the inhibitory effect of hemoglobin. The addition of 0.4% (wt/vol) BSA reduced the inhibitory effect of hemoglobin and allowed DNA amplification in the presence of 20 μg of hemoglobin in one of two independent replicates. Betaine (11.7% [wt/vol]) and gp32 (0.01% [wt/vol]) allowed the amplification of the specific PCR product in the presence of 10 μg of hemoglobin. The inhibitory effect of hemoglobin was not affected by the addition of the other PCR facilitators tested. Addition of 0.4% (wt/vol) BSA had the highest relieving effect on AmpliTaq Gold inhibition by lactoferrin and was found to allow DNA amplification in the presence of all amounts of lactoferrin tested (5 to 500 ng). The inhibitory effect of lactoferrin was also reduced by the addition of 0.01% (wt/vol) gp32. The relieving effect of the other amplification facilitators was slight or not observed.
TABLE 5.
Effects of nine amplification facilitators on the inhibitory effects of hemoglobin and lactoferrin on the amplification capacity of AmpliTaq Golda
| Facilitator (%, wt/vol) | PCR resultb with the indicated concn of:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemoglobin (μg/reaction tube)
|
Lactoferrin (ng/reaction tube)
|
||||||||||||||
| 0 | 1 | 1.3 | 2 | 4 | 10 | 20 | 100 | 0 | 5 | 25 | 50 | 125 | 250 | 500 | |
| Water (0) | +, + | ±, ± | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
| Betaine (11.7) | +, + | +, + | +, + | ±, ± | ±, ± | ±, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
| BSA (0.4) | +, + | +, + | +, + | +, + | +, + | +, + | +, − | −, − | +, + | +, + | +, + | +, + | +, + | +, + | +, + |
| Dextran 50 (1) | +, + | ±, − | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
| Dimethyl sulfoxide (1) | +, + | ±, − | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | +, − | −, − | −, − | −, − | −, − |
| Glycerol (5) | +, + | ±, − | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
| gp32 (0.01) | +, + | +, + | +, + | +, + | +, + | ±, ± | −, − | −, − | +, + | +, + | +, + | +, ± | −, − | −, − | −, − |
| Nonidet NP-40 (0.1) | +, + | ±, − | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | +, − | −, − | −, − | −, − | −, − |
| Polyethylene glycol 400 (1) | +, + | ±, − | −, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
| Tween 80 (0.1) | +, + | ±, ± | ±, − | −, − | −, − | −, − | −, − | −, − | +, + | +, + | −, − | −, − | −, − | −, − | −, − |
Amplification facilitators were added to reaction mixtures of AmpliTaq Gold containing 1 ng of L. monocytogenes DNA and different concentrations of purified hemoglobin and human milk lactoferrin.
Results of two independent PCRs. +, PCR product of high yield; ±, PCR product of low yield; −, no PCR product.
DISCUSSION
Several compounds in blood have been suggested to be PCR inhibitory, namely, heme (4), leukocyte DNA (34), and anticoagulants (47, 51). In a previous study, we identified immunoglobulin G as a major PCR inhibitor in human plasma (1). Hemoglobin and lactoferrin were found to be major PCR inhibitors in erythrocytes and leukocytes, respectively. Despite the fact that albumin and carbonic anhydrase were copurified with hemoglobin in the inhibitory fraction of erythrocytes, neither 30 μg of BSA nor 52 μg of carbonic anhydrase was inhibitory to AmpliTaq Gold (data not shown). Both hemoglobin and lactoferrin contain iron. Therefore, the inhibitory effects of both proteins may be related, in part, to their ability to release iron ions. When the inhibitory effect of iron was investigated, it was found that the addition of ≥25 μM FeCl3 reduced the DNA synthesis activity of rTth to <10% (Fig. 3D). PCR inhibition by hemin, a hemoglobin derivative, and its breakdown products bilirubin and bile salts were also found to be PCR inhibitory (Fig. 2 and 3A and E) (4, 5, 52). It has been suggested that heme regulates DNA polymerase activity and coordinates the synthesis of components in hemoglobin in erythroid cells by feedback inhibition (9). Hemin has been found to be inhibitory to thermolabile polymerases such as DNA polymerase from human neuroblasoma cells (7), cytoplasmic DNA polymerase from erythroid hyperplastic bone marrow cells (9) and Rauscher murine leukemia virus reverse transcriptase (49). Hemin inhibition has been found to be reversible and appears to be directed against the enzyme rather than the template (9, 49); hemin has been found to be competitive with respect to the template and a noncompetitive inhibitor with respect to nucleotides (9). Human lactoferrin is a single-polypeptide glycoprotein which binds two Fe3+ ions together with two CO32− ions (6). Lactoferrin is present in several biological fluids and has several biological functions, such as protection against infections, regulation of myelopoiesis, promotion of cellular growth, and immunostimulatory activity (21, 28, 32). Lactoferrin has been found to interact with nucleic acids (15, 19), which is another possible mechanism of PCR inhibition in addition to its ability to release iron ions. When the inhibitory effects of hemoglobin and lactoferrin on 10 thermostable DNA polymerases were investigated, AmpliTaq Gold, Pwo, and Ultma were inhibited in the presence of 1.3 μg of hemoglobin, while DyNAzyme EXL, rTth, Tfl, and Tli DNA polymerases could amplify the specific PCR product in the presence of all hemoglobin concentrations. In a previous study (3), we found that AmpliTaq Gold was highly sensitive to blood, and the addition of 0.004% (vol/vol) blood was totally inhibitory, whereas HotTub, Pwo, rTth, Tfl, and Tli DNA polymerases were found to resist 20% (vol/vol) blood. In this study, Taq DNA polymerase was found to be more resistant and Pwo was found to be more sensitive to blood. Batch-to-batch variability in the performance of Taq DNA polymerase was observed when the detection limit in the presence of blood was determined for different commercial batches (data not shown). Also, other reports have shown differences in the sensitivities of various thermostable DNA polymerases to PCR-inhibitory samples such as blood (3, 36), aqueous and vitreous fluids of the eye (53), feces (3), and phenol (24).
Addition of BSA to reaction mixtures in the LightCycler Instrument is recommended, because it coats the capillary wall and reduces the binding of SYBR Green I, DNA, oligonucleotides, and the polymerase (41, 55). On the other hand, addition of BSA can also relieve inhibition of DNA amplification (4, 27, 38). Therefore, it was necessary to exclude BSA from the reaction mixtures in order to study the effect of inhibitors on real-time DNA synthesis by AmpliTaq Gold and rTth. However, the real-time DNA synthesis by rTth was more efficient than that by AmpliTaq Gold (data not shown), a result which could be related to the molecular modification of AmpliTaq Gold, which increased its adsorption to the glass capillaries. This was supported by the ability of PCR inhibitors as well as facilitators to enhance the fluorescence signal of AmpliTaq Gold reaction mixtures through a possible coating of the capillary wall, so that the polymerase was available for DNA synthesis (data not shown). The rTth results were reproducible, and a linear relationship was obtained between the fluorescence signal and the template concentration (Fig. 2). However, the reproducibility was reduced when the inhibitors were added, which may be related to (i) the ability of the capillaries to bind different components of the reaction mixture, including inhibitors, or (ii) the binding of inhibitors to dye and/or dsDNA, reducing the number of dye molecules, which can bind to the dsDNA formed. The high linearity in the presence of bilirubin, hemin, and heparin (r2 > 0.93) indicates that they interfere with the real-time DNA synthesis by competing with the template. In a study by Byrnes et al. (9), heme was found to be a competitive inhibitor with respect to the template and a noncompetitive inhibitor with respect to nucleotides. The inhibitory effect of heparin has been suggested on the basis of an interaction between heparin and DNA, which could be mediated by Mg2+ (47). In a study by Ishii et al. (22), it was suggested that heparin inhibition of topoisomerase might be attributed to the highly sulfated, polyanionic nature of the molecules rather than a specific oligosaccharide sequence within it, and this inhibition was overcome by increasing either the substrate DNA or the enzyme concentration. Heparin was also found to copurify with DNA in traditional phenol-chloroform extraction, and the inhibition is not reversed by repeated ethanol precipitation, boiling, or pH shifts followed by gel filtration (23), which suggests a similarity between heparin and DNA. Therefore, heparin inhibition could be related to its ability to compete with target nucleic acid. Despite the fact that increasing the concentration of template reduced the inhibition by bile salts, CaCl2, and FeCl3, the reduction in linearity (r2 ∼ 0.86) indicates that their mechanisms of inhibition are different from those of bilirubin, hemin, and heparin. Increasing the negatively charged template reduced inhibition by Ca2+ and Fe3+ by binding them and preventing them from competing with Mg2+. Bile salts are polar derivatives of cholesterol and contain both polar and nonpolar regions. Bile salts have been found to be inhibitory for the amplification capacity of Taq DNA polymerase (30). In a previous study (3), the amplification capacities of nine thermostable DNA polymerases were compared in the presence of various PCR-inhibitory samples, and it was observed that Pwo and rTth were the DNA polymerases most resistant to fecal samples, which suggests that the inhibitory components in feces, such as bile salts, have a direct effect on the DNA polymerase. Magnesium ions act as cofactors of the polymerase, and their concentration affects primer annealing, the DNA melting temperature, and the polymerase activity (17). Calcium ions at a concentration of 3 mM have been found to inhibit PCR when detecting L. monocytogenes in the presence of 1.5 mM magnesium ions (8). Those authors have suggested that calcium ions inhibit PCR by competing with the magnesium ions as a cofactor for the DNA polymerase, since higher tolerance to calcium ions was observed in the presence of higher magnesium ion concentrations. The inhibition by EDTA may be related to its ability to inhibit DNA synthesis by chelating the Mg2+ necessary for the activity of DNA polymerase (43). Therefore, use of an increased magnesium ion concentration has been employed to maintain PCR activity in the presence of chelating agents.
PCR facilitators are added to relieve PCR inhibition. In this study, 0.4% BSA and 0.01% gp32 were efficient in reducing the inhibition of AmpliTaq Gold by hemoglobin and lactoferrin. However, the relieving efficiency of BSA was much more pronounced than that of gp32. The binding efficiency of albumin may explain its ability to reduce the amplification inhibition of hemoglobin and lactoferrin (13). Other studies have noted a similar relieving effect of BSA when it was added to PCR mixtures (4, 20, 27, 38, 49). The gp32 protein is a single-stranded-DNA (ssDNA) binding protein that is encoded by gene 32 of bacteriophage T4 (12). This protein has been shown to be involved in T4 DNA replication, repair, and recombination (54); to improve the accuracy of DNA replication in vitro (18, 48); and to relieve nucleic acid amplification inhibition (11, 27, 36). Betaine was efficient in reducing the inhibition only by hemoglobin. Betaine has been suggested to increase PCR specificity and product yield (16) and to increase the thermal stability of proteins (46). In conclusion, the use of DNA polymerases resistant to PCR-inhibitory components, in combination with the use of appropriate amplification facilitators, can, to some extent, eliminate the need for extensive processing of blood samples prior to diagnostic PCR.
ACKNOWLEDGMENT
This work was partially supported by the Swedish National Board for Industrial and Technical Development.
REFERENCES
- 1.Abu Al-Soud W, Jönsson L J, Rådström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J Clin Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Al-Soud W, Lantz P-G, Bäckman A, Olcén P, Rådström P. A sample preparation method which facilitates detection of bacteria in blood cultures by the polymerase chain reaction. J Microbiol Methods. 1998;32:217–224. [Google Scholar]
- 3.Abu Al-Soud W, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 5.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Purification of highly degraded DNA by gel filtration for PCR. Bio Techniques. 1994;16:235–238. [PubMed] [Google Scholar]
- 6.Anderson B F, Baker H M, Norris G E, Rice D W, Baker E N. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 A resolution. J Mol Biol. 1989;209:711–734. doi: 10.1016/0022-2836(89)90602-5. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya P, Simet I, Basu S. Differential inhibition of multiple forms of DNA polymerase alpha from IMR-32 human neuroblastoma cells. Proc Natl Acad Sci USA. 1981;78:2683–2687. doi: 10.1073/pnas.78.5.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bickley J, Short J K, McDowel D G, Parkes H C. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153–158. doi: 10.1111/j.1472-765x.1996.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 9.Byrnes J J, Downey K M, Esserman L, So A G. Mechanism of hemin inhibition of erythroid cytoplasmic DNA polymerase. Biochemistry. 1975;14:796–799. doi: 10.1021/bi00675a023. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo C, Craig O E, James N T, Sokol R J. Comparison of three DNA extraction methods on bone and blood stains up to 43 years old and amplification of three different gene sequences. J Forensic Sci. 1997;42:1126–1135. [PubMed] [Google Scholar]
- 11.Chandler D P, Wagnon C A, Bolton H., Jr Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl Environ Microbiol. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chase J W, Williams K R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 13.Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid. Nat Struct Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 14.Cursons R T, Jeyerajah E, Sleigh J W. The use of polymerase chain reaction to detect septicemia in critically ill patients. Crit Care Med. 1999;27:937–940. doi: 10.1097/00003246-199905000-00029. [DOI] [PubMed] [Google Scholar]
- 15.Fleet J C. A new role for lactoferrin: DNA binding and transcription activation. Nutr Rev. 1995;53:226–227. doi: 10.1111/j.1753-4887.1995.tb01556.x. [DOI] [PubMed] [Google Scholar]
- 16.Frackman, S., G. Kobs, D. Simpson, and D. Storts. 1998. Betaine and DMSO: enhancing agents for PCR. Promega Notes, 27.
- 17.Griffin H G, Griffin A M. PCR technology: current innovations. Boca Raton, Fla: CRC Press; 1994. [Google Scholar]
- 18.Gustafson C E, Alm R A, Trust T J. Effect of heat denaturation of target DNA on the PCR amplification. Gene. 1993;123:241–244. doi: 10.1016/0378-1119(93)90130-u. [DOI] [PubMed] [Google Scholar]
- 19.He J, Furmanski P. Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA. Nature. 1995;373:721–724. doi: 10.1038/373721a0. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi R. Simple and rapid preparation of samples for PCR. In: Erlich H A, editor. PCR technology: principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 31–38. [Google Scholar]
- 21.Ikeda M, Sugiyama K, Tanaka T, Tanaka K, Sekihara H, Shimotohno K, Kato N. Lactoferrin markedly inhibits hepatitis C virus infection in cultured human hepatocytes. Biochem Biophys Res Commun. 1998;245:549–553. doi: 10.1006/bbrc.1998.8481. [DOI] [PubMed] [Google Scholar]
- 22.Ishii K, Futaki S, Uchiyama H, Nagasawa K, Andoh T. Mechanism of inhibition of mammalian DNA topoisomerase I by heparin. Biochem J. 1987;241:111–119. doi: 10.1042/bj2410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung R, Lübecke C, Wagener C, Neumaier M. Reversal of RT-PCR inhibition observed in heparinized clinical specimens. BioTechniques. 1997;23:24–28. doi: 10.2144/97231bm03. [DOI] [PubMed] [Google Scholar]
- 24.Katcher H L, Schwartz I. A distinctive property of Tth DNA polymerase: enzymatic amplification in the presence of phenol. BioTechniques. 1994;16:84–92. [PubMed] [Google Scholar]
- 25.Katz E D, Dong M W. Rapid analysis and purification of polymerase chain reaction products by high-performance liquid chromatography. BioTechniques. 1990;8:546–555. [PubMed] [Google Scholar]
- 26.Klein A, Barsuk R, Dagan S, Nusbaum O, Shouval D, Galun E. Comparison of methods for extraction of nucleic acid from hemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J Clin Microbiol. 1997;35:1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreader C A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwata H, Yip T T, Yip C L, Tomita M, Hutchens T W. Bactericidal domain of lactoferrin: detection, quantitation, and characterization of lactoferricin in serum by SELDI affinity mass spectrometry. Biochem Biophys Res Commun. 1998;245:764–773. doi: 10.1006/bbrc.1998.8466. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lantz P-G, Matsson M, Wadström T, Rådström P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods. 1997;28:159–167. [Google Scholar]
- 31.Lantz P-G, Tjerneld F, Borch E, Hahn-Hägerdal B, Rådström P. Enhanced sensitivity in PCR detection of Listeria monocytogenes in soft cheese through use of an aqueous two-phase system as a sample preparation method. Appl Environ Microbiol. 1994;60:3416–3418. doi: 10.1128/aem.60.9.3416-3418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lonnerdal B, Iyer S. Lactoferrin: molecular structure and biological function. Annu Rev Nutr. 1995;15:93–110. doi: 10.1146/annurev.nu.15.070195.000521. [DOI] [PubMed] [Google Scholar]
- 33.Mantero G, Zonaro A, Albertini A, Bertolo P, Primi D. DNA enzyme immunoassay: general method for detecting products of polymerase chain reaction. Clin Chem. 1991;37:422–429. [PubMed] [Google Scholar]
- 34.Morata P, Queipo-Ortuno M I, de Dios Colmenero J. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J Clin Microbiol. 1998;36:2443–2446. doi: 10.1128/jcm.36.9.2443-2446.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulley J, Saar K, Hewitt G, Ruschendorf F, Phillips H, Colley A, Sillence D, Reis A, Wilson M. Gene localization for an autosomal dominant familial periodic fever to 12p13. Am J Hum Genet. 1998;62:884–889. doi: 10.1086/301793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panaccio M, Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991;19:1151. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polacek D, Byrne R E, Fless G M, Scanu A M. In vitro proteolysis of human plasma low density lipoproteins by an elastase released from human blood polymorphonuclear cells. J Biol Chem. 1986;261:2057–2063. [PubMed] [Google Scholar]
- 38.Powell H A, Gooding C M, Garrett S D, Lund B M, McKee R A. Proteinase inhibition of the detection of Listeria monocytogenes in milk using the polymerase chain reaction. Lett Appl Microbiol. 1994;18:59–61. [Google Scholar]
- 39.Rådström P, Bäckman A, Qian N, Kragsbjerg P, Pählson C, Olcén P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rautenberg P, Lubbert C, Weers W, Boetel E, Schweichler J, Zhou L, Costard-Jackle A, Kraemer-Hansen H, Harder T C. Evaluation of the AmpliSensor PCR and the SHARP signal detection system for the early prediction of symptomatic CMV infection in solid transplant recipients. J Clin Virol. 1999;13:81–94. doi: 10.1016/s1386-6532(99)00013-x. [DOI] [PubMed] [Google Scholar]
- 41.Ririe K M, Rasmussen R P, Wittwer C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 42.Robertson J S. International standardization of gene amplification technology. Biologicals. 1998;26:111–113. doi: 10.1006/biol.1998.0136. [DOI] [PubMed] [Google Scholar]
- 43.Rossen L, Nøskov P, Holmstrøm K, Rasmussen O F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solution. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 44.Saiki R K, Walsh P S, Levenson C H, Erlich H A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci USA. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Habor, N.Y: Cold Spring Habor Laboratory; 1989. [Google Scholar]
- 46.Santoro M M, Liu Y, Khan S M, Hou L X, Bolen D W. Increased thermal stability of proteins in the presence of naturally occurring osmolytes. Biochemistry. 1992;31:5278–5283. doi: 10.1021/bi00138a006. [DOI] [PubMed] [Google Scholar]
- 47.Satsangi J, Jewell D P, Welsh K, Bunce M, Bell J I. Effect of heparin on polymerase chain reaction. Lancet. 1994;343:1509–1510. doi: 10.1016/s0140-6736(94)92622-0. [DOI] [PubMed] [Google Scholar]
- 48.Topal M D, Sinha N K. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J Biol Chem. 1983;258:12274–12279. [PubMed] [Google Scholar]
- 49.Tsutsui K, Mueller G C. Hemin inhibits virion-associated reverse transcriptase of murine leukemia virus. Biochem Biophys Res Commun. 1987;149:628–634. doi: 10.1016/0006-291x(87)90414-1. [DOI] [PubMed] [Google Scholar]
- 50.Walsh P S, Metzger D A, Higuchi R. Chelex ® 100 as medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 51.Wang J T, Wang T H, Sheu J C, Lin S M, Lin J T, Chen D S. Effects of anticoagulants and storage of blood samples on efficacy of the polymerase chain reaction assay for hepatitis C virus. J Clin Microbiol. 1992;30:750–753. doi: 10.1128/jcm.30.3.750-753.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedbrauk D L, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams K R, LoPresti M B, Setoguchi M. Primary structure of the bacteriophage T4 DNA helix-destabilizing protein. J Biol Chem. 1981;256:1754–1762. [PubMed] [Google Scholar]
- 55.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. Bio Techniques. 1997;22:130–131. doi: 10.2144/97221bi01. , 134–138. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Isaacman D, Wadowsky R, Rydquist-White J, Post J, Ehrlich G. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]