Abstract
The oxidoreductase protein disulfide isomerase A1 (PDIA1) functions as a cofactor for many transcription factors including estrogen receptor α (ERα), nuclear factor (NF)-κB, nuclear factor erythroid 2-like 2 (NRF2) and regulates the protein stability of the tumor suppressor p53. Taking this into account we hypothesized that PDIA1, by differentially modulating the gene expression of a diverse subset of genes in the ERα-positive vs. the ERα-negative breast cancer cells, might modify dissimilar pathways in the two types of breast cancer. This hypothesis was investigated using RNA-seq data from PDIA1-silenced MCF-7 (ERα-positive) and MDA-MB-231 (ERα-negative) breast cancer cells treated with either interferon γ (IFN-γ) or etoposide (ETO), and the obtained data were further analyzed using a variety of bioinformatic tools alongside clinical relevance assessment via Kaplan-Meier patient survival curves. The results highlighted the dual role of PDIA1 in suppressing carcinogenesis in the ERα(+) breast cancer patients by negatively regulating the response to reactive oxygen species (ROS) and promoting carcinogenesis by inducing cell cycle progression. In the ERα(−) breast cancer patients, PDIA1 prevented tumor development by modulating NF-κB and p53 activity and cell migration and induced breast cancer progression through control of cytokine signaling and the immune response. The findings reported in this study shed light on the differential pathways regulating carcinogenesis in ERα(+) and ERα(−) breast cancer patients and could help identify therapeutic targets selectively effective in ERα(+) vs. ERα(−) patients.
Keywords: breast cancer, protein disulfide isomerase, transcription, oxidative stress, personalized medicine
Introduction
Protein disulfide isomerases (PDIs) are involved in a wide range of biological pathways as mediators of oxidative folding in the endoplasmic reticulum (1,2), in the formation, breakage, and rearrangement of disulfide bonds (1,3), and in the regulation of apoptosis, exerting both pro-apoptotic and pro-survival effects (4,5). The PDI family member prolyl 4-hydroxylase β (P4HB, also known as PDIA1) functions as a chaperone that assists protein folding by inhibiting the aggregation of partially folded or damaged polypeptides (6). Recent findings have implicated PDIA1 in the processes of antigen processing and presentation, immunomodulation and tumor immunorecognition (4,7-9).
PDIA1 has been shown to play an important role in various stages of carcinogenesis in a wide variety of cancers, being involved in the early stages of carcinogenesis possibly by contributing to the management of misfolded and non-functional proteins (5) as well as in the evasion of the immunosurveillance (4). Evidence has been presented that PDI family members are involved in the proliferation and metastasis of brain, kidney, and lung cancers (10,11). Overexpression of PDI family members has been positively correlated with metastasis and invasion of breast cancer (12). For instance, significantly higher PDIA1 levels have been observed in axillary lymph node metastatic breast tumor compared to primary breast tumors (13). In addition, PDIA1 mRNA levels have been found to be positively correlated with malignant glioma metastasis and invasion (14) and the epithelial-mesenchymal transition (EMT) of liver cancer cells (15-17). The potential molecular mechanisms by which PDIA1 contributes to metastasis include the regulation of the hypoxia-inducible factor-1α (HIF-1α) pathway (17,18), and induction of matrix metalloproteinase-9 (MMP-9) secretion (19) or other metalloproteases such as the disintegrin and metalloprotease domain family member ADAM metallopeptidase domain 17 (ADAM17) (20).
PDIA1 is mainly localized in the endoplasmic reticulum, but cell surface and nuclear localization under certain micro-environmental conditions have also been reported (21,22). Distinct PDIA1 subcellular locations determine diverse PDIA1 signaling networks including the regulation of the cellular redox state which in turn regulates its function (2). Nuclear PDIA1 has been shown to play a role in the regulation of the transcriptional activity of redox responsive transcription factors (23,24). Indeed, PDIA1 has been shown to directly interact and function as an estrogen receptor (ER)α transcriptional cofactor (25,26). In addition, PDIA1 suppresses nuclear factor (NF)-κB transcriptional activity (27,28) and regulates p53 protein stability (29,30). These observations raise the possibility that PDIA1 regulates the crosstalk between ERα and NF-κB (31). Other redox-responsive transcription factors associated with PDIA1 include HIF-1α (18) and nuclear factor (erythroid-derived 2)-like 2 (NRF2) (32). PDIA1 is also implicated in regulating the transcription of the gene targets of thyroid hormone receptors α and β (33,34).
Several endoplasmic reticulum chaperones including PDI have been detected in the mitochondria (35) and the mitochondrial and the endoplasmic reticulum contact sites (MERC) (36). The function of the mitochondrial PDIs has not been elucidated clearly but there is evidence to suggest that PDIs regulate the exchange of Ca2+ between the endoplasmic reticulum and mitochondria, mitochondrial energy generation (37) and the immune response to tumor cells (38), suggesting that PDIs might affect carcinogenesis via multiple pathways. Given that PDIA1 protein levels are breast cancer subtype-specific (4,13) and taking into account the fact that PDIA1 functions as a co-modulator for several transcription factors including ERα (26), it was hypothesized that PDIA1 would differentially affect carcinogenesis in ERα-positive vs. ERα-negative breast cancer cells.
In the present study, PDIA1 gene expression was silenced in the ERα-positive MCF-7 and the ERα-negative MDA-MB-231 breast cancer cells and the expression of genes under conditions stimulating reactive oxygen species (ROS) generation (treatment with interferon-γ (IFN-γ) (39) or etoposide (ETO) (40) was followed using RNA-seq. Kaplan-Meier survival curves of the modulated genes detected by RNA-seq were generated to identify whether high or low expression of the upregulated or downregulated genes in the ERα-positive (MCF-7) or triple-negative breast cancer (TNBC) cells (MDA-MB-231) had positive or negative effect on the overall survival (OS) of the ERα-positive or ERα-negative breast cancer patients, respectively. The results indicated that PDIA1, in the ERα-positive breast cancer patients, suppresses carcinogenesis by negatively regulating catabolic processes of ROS. PDIA1 was found to induce carcinogenesis by affecting cell cycle progression at the G2/M checkpoint alongside spindle formation in the ERα-positive breast cancer patients. In the ERα-negative breast cancer patients, PDIA1 was found to prevent tumor development by regulating the response to NF-κB signaling, mitochondrial biogenesis, glycolysis and the process of metastasis. PDIA1 induced breast cancer progression in ERα-negative breast cancer patients by modifying the immune response, cytokine signaling and calcium homeostasis. The findings reported here shed new light on the differential pathways inducing carcinogenesis in the ERα-positive and ERα-negative breast cancer patients and could assist in the identification of selectively beneficial therapeutic targets for ERα-positive or ERα-negative patients.
Materials and methods
Cell culture
The human breast carcinoma cell lines MCF-7 (expressing ERα and wild-type p53) and MDA-MB-231 [ERα-negative, bearing mutated p53 (R280K)] were obtained from the European Collection of Cell Cultures (ECACC) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Lonza Group, Ltd.) at 37°C in a humidified atmosphere containing 5% CO2 until they reached 70% confluency. Where indicated, cells were treated with 10 ng/ml IFN-γ (Sigma-Aldrich; Merck KGaA) for 24 h or 10 µM etoposide (ETO) for 24 h (Sigma-Aldrich; Merck KGaA).
siRNA transfection
Concentration of 5 µM of the siGENOME PDIA1 siRNA and 5 µM of the siGENOME non-targeting siRNA pool was added to each well containing 2×105 cells in DMEM and incubated for 72 h according to the suppliers' instructions (Dharmacon, UK) as described previously (4). The sequences of the siRNAs used are indicated as follows: siGE-NOME PDIA1-targeting siRNA pool: ACA GGA CGG UCA UUG AUU A, GGA CGG UCA UUG AUU ACA A, CCA AGAG UGU GUC UGA CUA, and CAGAGAGGAUCACAGAGUU; siGENOME non-targeting (scramble) siRNA pool: UAG CGA CUA AAC ACA UCA A, UAA GGC UAU GAA GAG AUA C, AUG UAU UGG CCU GUA UUA G, and AUG AAC GUG AAU UGC UCA A.
Western blotting
Cellular extracts from MCF-7 and MDA-MB-231 and 30 µg of total protein per sample were loaded on a 20% precast polyacrylamide gel and transferred to a PVDF membrane. The membranes were then incubated in 5% milk in PBS-0.1% Tween-20 (v/v) with anti-P4HB anti-body (Santa Cruz Biotechnology, sc-74551) (dilution 1:500) or β-actin (Sigma-Aldrich; Merck KGaA; A1978) (dilution 1:10,000) overnight at 4°C. After incubation with secondary anti-mouse immunoglobulin G conjugated to horseradish peroxidase (GE Healthcare) (dilution 1:1,000) in 2.5% milk in PBS-0.1% Tween-20 (v/v) for 1 h at 25°C, the protein bands were visualized using the ChemiDoc MP imaging system (Bio-Rad Laboratories) The blots were quantified using ImageJ version 1.51 (National Institutes of Health).
RNA extraction
MCF-7 and MDA-MB-231 breast cancer cell lines were seeded in 6-well plates at a concentration of 3×105 cells per well for 24 h after treatment. RNA was extracted according to the RNeasy Mini Kit supplier instructions (Qiagen, UK). After collecting RNA, the samples were maintained at −80°C before sequencing carried out by the Next Generation Sequencing Facility (University of Leeds, UK).
RNA sequencing
Initial alignment of RNA transcript counts, sequence short reads, and normalization were carried out by the Next Generation Sequencing Facility (University of Leeds, UK). Sequence data in Fastq format were quality-checked using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cutadapt version 1.16 software (https://cutadapt.readthedocs.org/en/stable/) was used to trim poor quality bases (Phred quality score <20) and contaminating adapter sequences from raw reads. Reads trimmed to fewer than 30 were discarded. Reads were aligned to a human (hg38) genome reference sequences, obtained from the UCSC database (41) using the splicing-aware STAR aligner (42). STAR aligner was run with known splice junctions supplied in GTF file format, obtained from the UCSC database using Table Browser tool (43,44). The resulting alignments in BAM file format were checked for quality using QualiMap software (44) and Picard tools version 1.90 (http://picard.sourceforge.net). Picard tools also used to mark PCR/Optical duplicate alignments. BAM files were indexed using Samtools software (45) and visualised using IGV browser (46) to check for genomic DNA contamination and the presence of PCR duplicates. Bioconductor R package RSubread (47) was used to extract raw sequenced fragment counts per transcript using the RefSeq hg38 annotation dataset used by STAR during alignment. Multi-mapping read pairs were counted as a fraction of all equivalent alignments. Read count data were generated with the inclusion of reads marked as PCR/optical duplicates.
Filtration of the gene list
Significantly downregulated or upregulated genes (Benjamini-Hochberg <0.01) in the PDIA1-silenced MCF-7 and MDA-MB-231 treated with IFN-γ or ETO compared to MCF-7 and MDA-MB-231 cells expressing PDIA1 treated with IFN-γ (39) or ETO (40) were selected. To visualize the overall changes in gene expression between PDIA1-silenced and scramble-transfected MCF-7 and MDA-MB-231 cells, the log value of the change in gene expression was graphed against the normalized read count for each transcript using the plotMA function of the DeSeq2 R package (47). The read count data required for each pairwise analysis was imported into the R package DeSeq2 and the effect size of each sample's library was defined and used to normalize the read count data.
Kaplan-Meier overall survival analysis
To understand the clinical importance of the genes identified and filtered from the RNA-seq data, the Kaplan-Meier Plotter website (http://kmplot.com/analysis/) was utilized. For this purpose, Kaplan-Meier (KM) plots for ERα-positive and ERα-negative patients were generated for the genes identified to be upregulated or downregulated in the RNA-seq data (48). The following parameters were selected: Survival: OS, Split patients by: upper quartile, Follow-up threshold: 240 months, Probe set option: only JetSet best probe sets (49), ER status-IHC: ER-positive (for the genes upregulated or down regulated in MCF-7 cells) or ER negative (for the genes upregulated or downregulated in MDA-MB-231 cells) ER status-array: ER positive (for the genes upregulated or downregulated in MCF-7 cells) and ER negative (for the genes upregulated or down regulated in MDA-MB-231 cells). Log rank P-values <0.05 for the KM plots of all genes was considered statistically significant.
cBioPortal analysis
The Pearson correlation coefficient for the comparison of PDIA1 mRNA levels with those of the genes upregulated or downregulated in the PDIA1-silenced MCF-7 cells treated with etoposide (ETO) or interferon γ (IFN-γ) in breast cancer patients was explored using the cBioPortal platform (http://cbioportal.org) in combination with data provided in the Breast Cancer (SMC 2018) database using the mRNA expression z-scores relative to all sample (RNA seq TPM-168 samples) options.
Functional annotation
In order to understand the biological importance of the gene lists generated from the previous section, the GeneCards tool (50) was used to identify linked genes to the genes listed in Tables I and II. After this, Metascape tool (51) was employed. Gene lists were entered in the metascape tool using the Official Gene Symbol as the identifier and Homo sapiens as the species.
Table I.
Genes upregulated or downregulated in interferon γ (IFN-γ) or etoposide (ETO)-treated MCF-7 cells in which PDIA1 gene expression was silenced compared to PDIA1-expressing IFN-γ- or ETO-treated MCF-7 cells.
| MCF-7 cells
| |
|---|---|
| Upregulated | Downregulated |
| L1CAM | AURKA |
| MAGED1 | BUB1 |
| MDK | CCNB2 |
| PHLDA2 | CDC25A |
| PRDX2 | CDC25C |
| PTPRS | CENPF |
| PYCR1 | CKAP5 |
| RHOD | E2F8 |
| SMAD6 | ESPL1 |
| TKT | FEN1 |
| UCP2 | GTSE1 |
| JUND | |
| KIF11 | |
| KIF20A | |
| NEK2 | |
| NUSAP1 | |
| PKMYT1 | |
| RACGAP1 | |
| RANBP1 | |
| S100PBP | |
| TACC3 | |
| TRIP13 | |
| TUBB | |
| UBE2C | |
PDIA1, protein disulfide isomerase A1; AURKA, aurora kinase A; BUB1, BUB1 mitotic checkpoint serine/threonine kinase; CCNB2, cyclin B2; CDC25A, cell division cycle 25A; CDC25C, cell division cycle 25C; CENPF, centromere protein F; CKAP5, cytoskeleton associated protein 5; E2F8, E2F transcription factor 8; ESPL1, extra spindle pole bodies like 1, separase; FEN1, flap structure-specific endonuclease 1; GTSE1, G2 and S-phase expressed 1; JUND, JunD proto-oncogene; KIF11, kinesin family member 11; KIF20A, kinesin family member 20A; L1CAM, L1 cell adhesion molecule; MAGED1, MAGE family member D1; MDK, midkine; NEK2, NIMA related kinase 2; NUSAP1, nucleolar and spindle associated protein 1; PHLDA2, pleckstrin homology like domain family A member 2; PKMYT1, protein kinase, membrane associated tyrosine/threonine 1; PRDX2, peroxiredoxin 2; PTPRS, protein tyrosine phosphatase receptor type S; PYCR1, pyrroline-5-carboxylate reductase 1; RACGAP1, Rac GTPase activating protein 1; RANBP1, RAN binding protein 1; RHOD, ras homolog family member D; S100PBP, S100P binding protein; SMAD6, SMAD family member 6; TACC3, transforming acidic coiled-coil containing protein 3; TKT, transketolase; TRIP13, thyroid hormone receptor interactor 13; TUBB, tubulin β class I; UBE2C, ubiquitin conjugating enzyme E2 C; UCP2, uncoupling protein 2.
Table II.
Genes upregulated or downregulated in interferon γ (IFN-γ) or etoposide (ETO)-treated MDA-MB-231 cells in which PDIA1 gene expression was silenced compared to PDIA1 expressing IFN-γ- or ETO-treated MDA-MB-231 cells.
| MDA-MB-231 cells
| |
|---|---|
| Upregulated | Downregulated |
| HSP90B1 | CHCHD3 |
| IGFBP3 | PTPRJ |
| LAPTM5 | VMA21 |
| RNF213 | XIAP |
| SYNE1 | |
PDIA1, protein disulfide isomerase A1; CHCHD3, coiled-coil-he lix-coiled-coil-helix domain containing 3; HSP90B1, heat shock protein 90 β family member 1; IGFBP3, insulin like growth factor binding protein 3; LAPTM5, lysosomal protein transmembrane 5; PTPRJ, protein tyrosine phosphatase receptor type J; RNF213, ring finger protein 213; SYNE1, spectrin repeat containing nuclear envelope protein 1; VMA21, vacuolar ATPase assembly factor; XIAP, X-linked inhibitor of apoptosis.
Identification of repurposed drugs
Following the identification of genes important for breast cancer patient survival, the potential clinical targeting of each gene was explored via DRUGSURV (52). Genes exerting oncogenic function that were identified as statistically significant for patient OS via KM Plotter were queried in turn, and the approved drugs that targeted the genes directly or indirectly were recorded.
Results
Since PDIA1 regulates the transcriptional activity of ERα, RNA-seq was employed to investigate whether this oxidoreductase differentially affected gene expression in the ERα-positive (MCF-7) vs. the ERα-negative (MDA-MB-231) breast cancer cells. The differences in the gene expression of genes involved in the oxidative stress pathways were explored in cells treated with either IFN-γ or ETO, both of which are known to induce ROS generation (39,40).
Genes upregulated and downregulated in MCF-7 and MDA-MB-231 breast cancer cells in the absence of PDIA1 under oxidative stress conditions
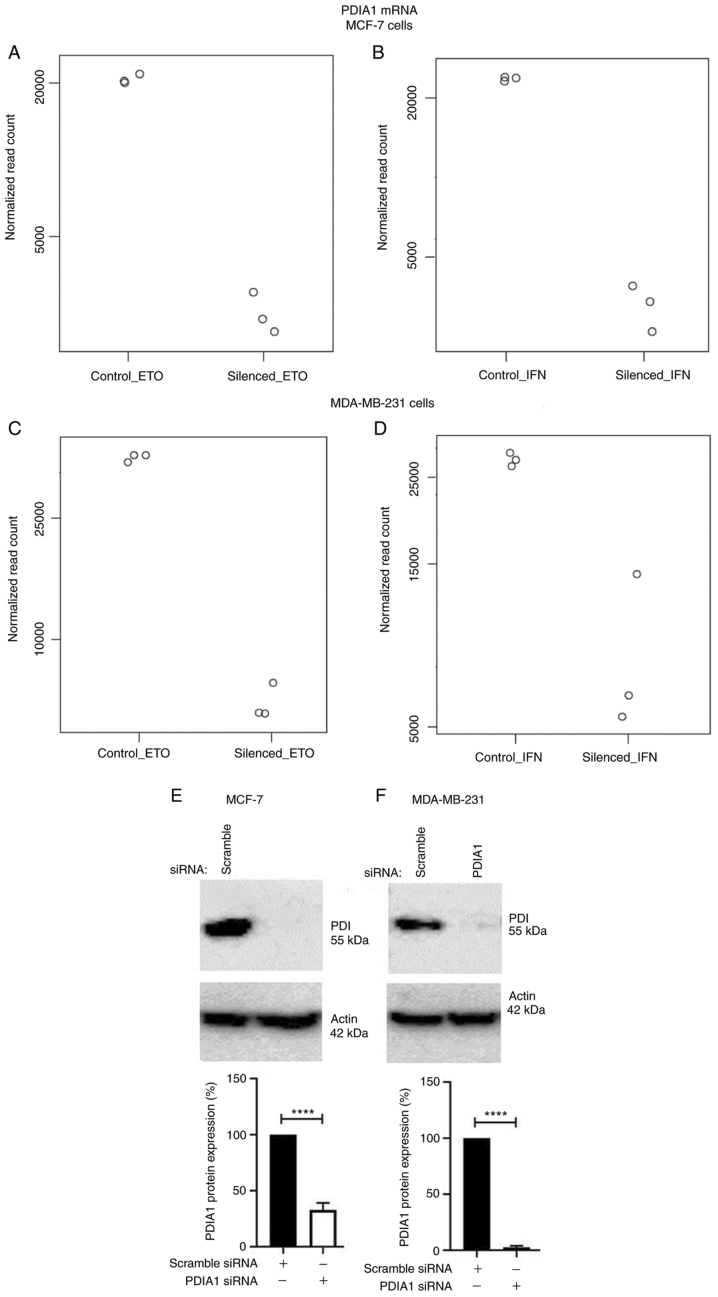
Total mRNA obtained from MCF-7 (Fig. 1A and B) or MDA-MB-231 (Fig. 1C and D) cells transfected with either scramble or specific siRNA to silence PDIA1 gene expression and treated with either ETO (Fig. 1A and C) or IFN-γ (Fig. 1B and D) was submitted to RNA-seq analysis. The transfection efficiency for the PDIA1-siRNA transfected MCF-7 cells compared to the transfection efficiency for the scramble siRNA was 67.13% (Fig. 1E). The transfection efficiency for the PDIA1-siRNA transfected MDA-MB-231 cells compared to the transfection efficiency for the scramble siRNA was 96.67% (Fig. 1F).
Figure 1.
Total mRNA obtained from MCF-7 (A and B) or MDA-MB-231 (C and D) cells transfected with either scramble or specific siRNA to silence PDIA1 gene expression and treated with either etoposide (ETO) (A and C) or interferon γ (IFN-γ) (B and D). PDIA1, protein disulfide isomerase A1 protein levels in scramble or PDIA1 siRNA transfected MCF-7 (E) or MDA-MB-231 (F) cells. The plots at the bottom of the figure indicate the transfection efficiency for the PDIA1-siRNA transfected compared to that for the scramble siRNA transfected cells. The data represent the mean of three independent experiments and the error bars represent mean ± SEM. ****P<0.001. PDIA1, protein disulfide isomerase A1.
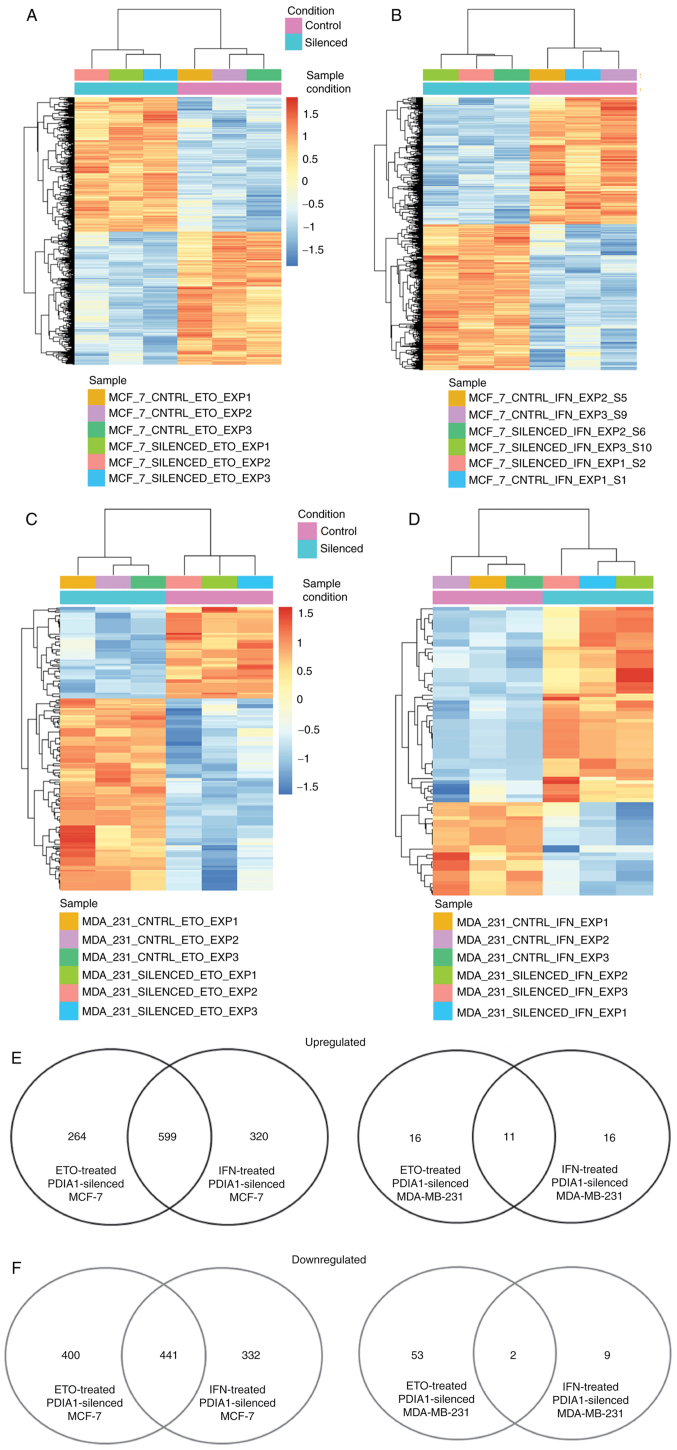
The total number of genes exhibiting statistically significant downregulation in ETO-treated MCF-7 cells in which PDIA1 gene expression had been silenced compared with ETO-treated MCF-7 cells transfected with scramble siRNA was 841, and the total number of genes upregulated under these conditions was 863 (Fig. 2A). The total number of genes exhibiting statistically significant (P-adj<0.01) downregulation in IFN-γ-treated MCF-7 cells in which PDIA1 gene expression had been silenced compared with IFN-γ-treated MCF-7 cells transfected with scramble siRNA was 773, and the total number of genes upregulated under these conditions was 919 (Fig. 2B). The total number of genes exhibiting statistically significant downregulation in ETO-treated MDA-MB-231 cells in which PDIA1 gene expression had been silenced compared with ETO-treated MDA-MB-231 cells transfected with scramble siRNA was 55, whereas the total number of genes upregulated in these cells under the same conditions was 27 (Fig. 2C). In the IFN-γ-treated MDA-MB-231 cells in which PDIA1 gene expression had been silenced compared with IFN-γ-treated MDA-MB-231 cells transfected with scramble siRNA the total number of the downregulated genes was 11, whereas the total number of genes upregulated in these cells under the same conditions was 27 (Fig. 2D). The numbers of upregulated and downregulated genes are presented in Fig. 2E and F, respectively.
Figure 2.
Genes upregulated or downregulated in PDIA1 siRNA-transfected compared to scramble transfected MCF-7 (A and B) or MDA-MB-231 (C and D) and treated with etoposide (ETO) (A and C) or interferon γ (IFN-γ) (B and D). Number of genes upregulated (E) or downregulated (F) in ETO- or IFN-γ-treated and PDIA1-silenced compared to ETO- or IFN-γ-treated and scramble-transfected MCF-7 or MDA-MB-231 breast cancer cells. PDIA1, protein disulfide isomerase A1.
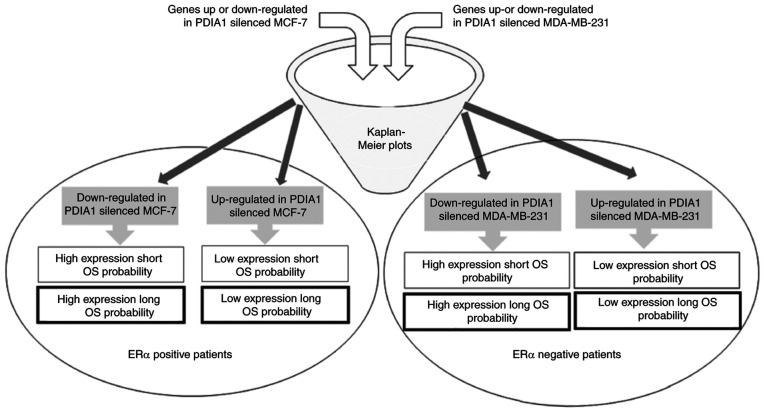
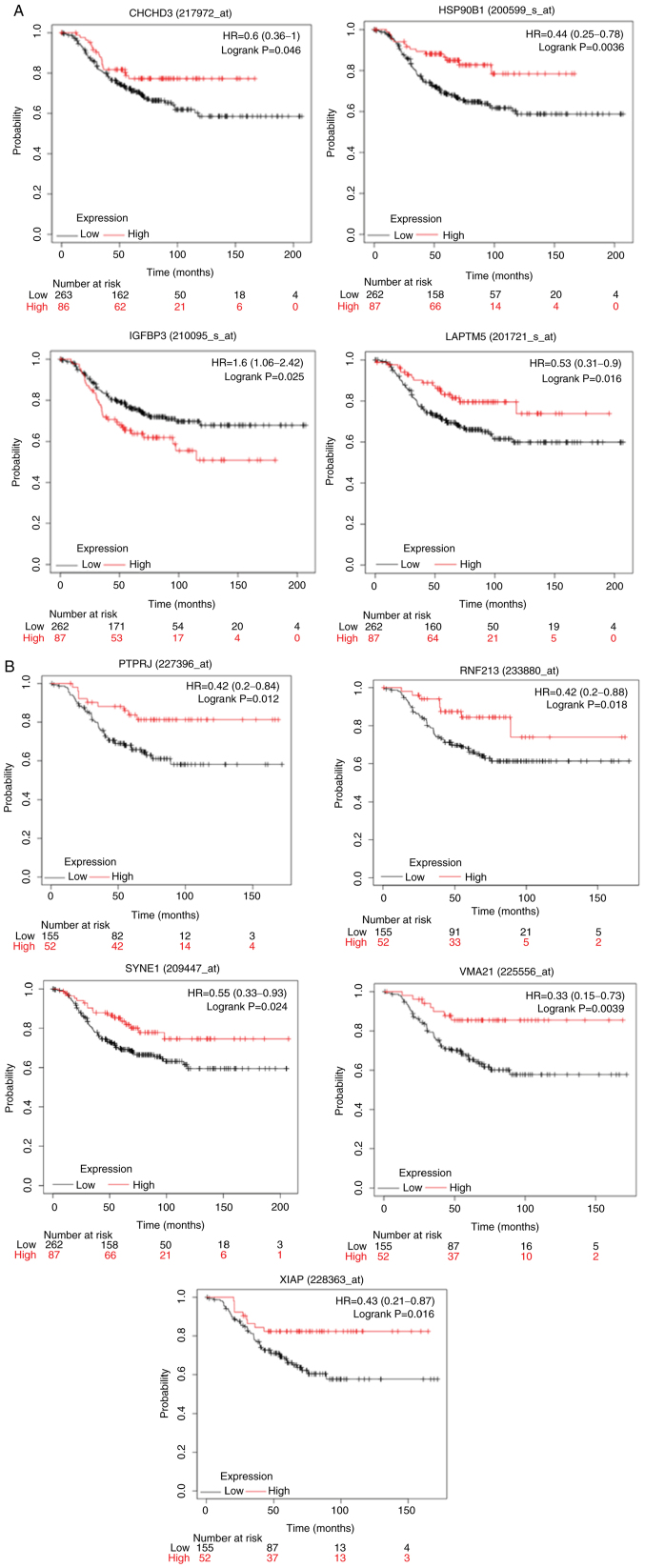
Clinical significance of the genes modulated in IFN-γ- or ETO-treated and PDIA1-silenced MCF-7 cells
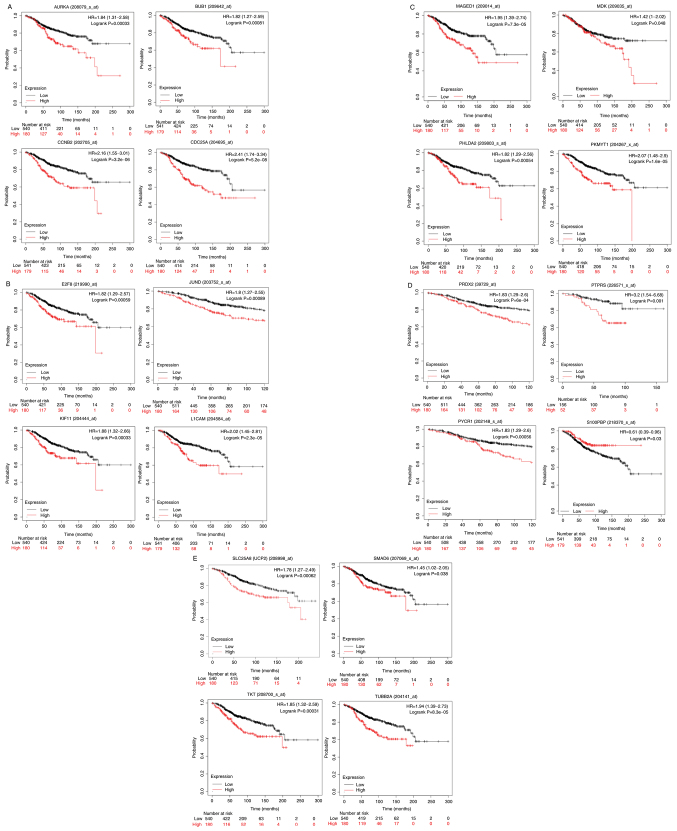
To shed light on the clinical significance of the aforementioned findings, KM curves for the upregulated or downregulated genes in the PDIA1-silenced compared with the PDIA1-expressing and either IFN-γ- or ETO-treated MCF-7 and MDA-MB-231 cells were plotted in the patients with ERα-positive and ERα-negative breast cancer, respectively, as described in the summary of the experimental design (Fig. 3). The upregulated or downregulated genes in MCF-7 cells under the conditions described above that exhibited statistically significant (P<0.05) KM results are shown in Table I. The KM curves plotted for these genes for ERα-positive patients are shown in Fig. 4A-E. High expression of aurora kinase-A (AURKA), mitotic checkpoint serine/threonine-protein kinase BUB1 (BUB1), cyclin-B2 (CCNB2), cell division cycle 25C (CDC25A) (Fig. 4A), transcription factor E2F8 (E2F8), junD proto-oncogene (JUND), kinesin family member 11 (KIF11), L1 cell adhesion molecule (L1CAM) (Fig. 4B), melanoma antigen gene D1 (MAGED1), midkine (MDK), pleckstrin homology-like domain family A member 2 (PHLDA2), membrane-associated tyrosine- and threonine kinase 1 (PKMYT1) (Fig. 4C), peroxiredoxin-2 (PRDX2), protein tyrosine phosphatase receptor type S (PTPRS), pyrroline-5-carboxylate reductase 1 (PYCR1) (Fig. 4D), uncoupling protein 2 (UCP2), SMAD family member 6 (SMAD6), transketolase (TKT) and tubulin β 2A class IIa (TUBB2A) (Fig. 4E) genes was associated with statistically significant (P<0.05) shorter OS probability in the patients with ERα-positive breast cancer. On the other hand, high expression of the S100P-binding protein (S100PBP) gene was associated with statistically significant (P<0.05) longer OS probability when comparing the 75th quartile with the 25th quartile in KM plotter in patients with ERα-positive breast cancer (Fig. 4D).
Figure 3.
Schematic presentation of the experimental design followed in the study. PDIA1, protein disulfide isomerase A1; OS, overall survival; ER, estrogen receptor.
Figure 4.
(A-E) Kaplan-Meir survival curves in ERα-positive breast cancer patients for genes upregulated or downregulated in the absence of PDIA1 in etoposide (ETO)- or interferon γ (IFN-γ)-treated MCF-7 breast cancer cells exhibiting statistically significant OS probability in patients with ERα-positive breast cancer. PDIA1, protein disulfide isomerase A1; OS, overall survival; ER, estrogen receptor.
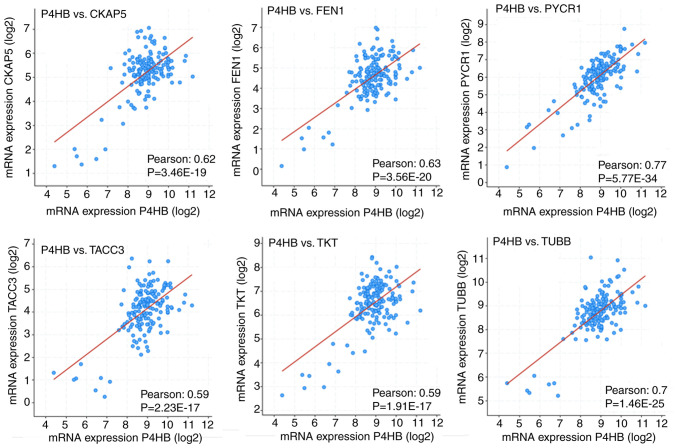
To explore whether PDIA1 gene expression in patients with ERα-positive breast cancer was correlated with the upregulated or downregulated genes in either IFN-γ- or ETO-treated PDIA1-silenced MCF-7 cells, the cBioPortal bioinformatics tool was used to investigate the Pearson correlation coefficient between PDIA1 mRNA levels and that of the genes shown in Table I. This investigation was carried out by analyzing the data provided in the Breast Cancer (SMC 2018) database using the mRNA expression z-scores relative to all samples (RNA seq TPM, 168 samples) option. Positive correlations were observed between PDIA1 gene expression and that of AURKA, CDC25A, centromere protein F (CENPF), cytoskeleton-associated protein 5 (CKAP5), E2F8, extra spindle pole bodies like 1, separase (ESPL1), flap structure-specific endonuclease 1 (FEN1), G2 and S phase-expressed protein 1 (GTSE1), JUND, KIF11, kinesin-like protein KIF20A (KIF20A), MAGED1, MDK, PKMYT1, PRDX2, PTPRS, PYCR1, RAN binding protein 1 (RANBP1), S100PBP, SMAD6, transforming acidic coiled-coil-containing protein 3 (TACC3), TKT, thyroid hormone receptor interactor 13 (TRIP13), tubulin b class 1 (TUBB), ubiquitin-conjugating enzyme E2 C (UBE2C) and UCP2 genes. In other words, in the presence of PDIA1, the expression of these genes would be upregulated, whereas in the absence of PDIA1, the expression of these genes would be downregulated in patients with ERα-positive breast cancer. Among these genes, the correlation between PDIA1 (shown as P4HB) gene expression and that of the CKAP5, FEN1, PYCR1, TACC3, TKT and TUBB genes exhibited higher Pearson correlation coefficients (Fig. 5).
Figure 5.
Pearson correlation coefficient between PDIA1 (P4HB) mRNA levels and that of CKAP5, FEN1, PYCR1, TACC3, TKT and TUBB genes. CKAP5, cytoskeleton associated protein 5; FEN1, flap structure-specific endonuclease 1; PYCR1, pyrroline-5-carboxylate reductase 1; TACC3, transforming acidic coiled-coil containing protein 3; TKT, transketolase; TUBB, tubulin β class I.
Positive correlations between PDIA1 gene expression and that of MAGED1, MDK, PRDX2, PTPRS, PYCR1, SMAD6, TKT and UCP2 genes were found, which were demonstrated to be upregulated in the PDIA1-silenced MCF-7 cells (Table I), thus indicating that in the presence of PDIA1 in patients with ERα-positive breast cancer these genes would be downregulated. According to the KM plots, the downregulation of MAGED1, MDK, PRDX2, PTPRS, PYCR1, SMAD6, TKT and UCP2 mRNA levels was associated with higher OS probability in the patients with ERα-positive breast cancer (Fig. 4). This suggested that PDIA1 can associate with MAGED1, MDK, PRDX2, PTPRS, PYCR1, SMAD6, TKT and UCP2 to induce anti-oncogenic effects in patients with ERα-positive breast cancer. To explore the pathways involved in the PDIA1-mediated anti-oncogenic effects in patients with ERα-positive breast cancer, the Metascape bioinformatics tool was employed (51). The results of this analysis showed that the pathways by which PDIA1 exerts tumor-suppressive effects include cell-substrate adhesion, regulation of growth and response to ROS (Table III).
Table III.
Gene Ontology pathways and gene networks through which PDIA1 confers anti-carcinogenic effects to patients with ERα-positive breast cancer.
| Term | Description | Hits | Log P | Log (q-value) | |
|---|---|---|---|---|---|
| 1 | GO:0031589 | Cell-substrate adhesion | SMAD6 | −6.61 | −2.26 |
| 2 | GO:0040008 | Regulation of growth | PTPRS, TKT | −3.93 | −0.48 |
| 3 | GO:0000302 | Response to reactive oxygen species | PYCR1, PRDX2, UCP2 | −3.98 | −0.48 |
SMAD6, SMAD family member 6; PTPRS, protein tyrosine phosphatase receptor type S; TKT, transketolase; PYCR1, pyrroline-5-carboxylate reductase 1; PRDX2, peroxiredoxin 2; UCP2, uncoupling protein 2.
Direct correlations between PDIA1 gene expression and that of the AURKA, CDC25A, CENPF, CKAP5, E2F8, ESPL1, FEN1, GTSE1, JUND, KIF11, KIF20A, PKMYT1, RANBP1, S100PBP, TACC3, TRIP13, TUBB and UBE2C genes were also observed, which were found to be downregulated in the PDIA1-silenced MCF-7 cells (Table I). This suggested that in the presence of PDIA1 in patients with ERα-positive breast cancer these genes would be upregulated. The KM plots indicated that high levels of these genes were associated with shorter OS in patients with ERα-breast cancer (Fig. 4A-C and E), apart from S100PBP (Fig. 4D). Therefore, the association of PDIA1 with these genes is a potential route by which PDIA1 exerts oncogenic effects in patients with ERα-positive breast cancer. The pathways affected by the networks formed by these genes were investigated using the Metascape bioinformatics tool and they included control of cell cycle transition to the mitotic phase as well as regulation of the cell cycle checkpoints (Table IV).
Table IV.
Gene Ontology pathways and gene networks through which PDIA1 confers pro-oncogenic effects to ERα-positive breast cancer patients.
| Term | Description | Hits | Log P | Log (q-value) | |
|---|---|---|---|---|---|
| 1 | GO:0010564 | Regulation of cell cycle process | AURKA, CDC25A, CENPF, CKAP5, E2F8, ESPL1, FEN1, GTSE1, KIF11, KIF20A, PKMYT1, TACC3, TRIP13, TUBB, UBE2C | −42.08 | −37.73 |
| 2 | GO:0044770 | Cell cycle phase transition | AURKA, CDC25A, CENPF, CKAP5, E2F8, ESPL1, GTSE1, PKMYT1, TACC3, TRIP13, TUBB | −37.03 | −33.15 |
| 3 | GO:0007346 | Regulation of mitotic cell cycle | AURKA, CDC25A, CENPF, CKAP5, E2F8, ESPL1, GTSE1, KIF11, PKMYT1, RANBP1, TACC3, TRIP13, TUBB, UBE2C | −25.16 | −21.80 |
| 4 | GO:0000075 | Cell cycle checkpoint | AURKA, CENPF, E2F8, GTSE1, TRIP13 | −16.50 | −13.66 |
| 5 | GO:0044839 | Cell cycle G2/M phase transition | AURKA, CDC25A, CENPF, CKAP5, GTSE1, PKMYT1, TUBB | −18.57 | −15.61 |
AURKA, aurora kinase A; CDC25C, cell division cycle 25C; CENPF, centromere protein F; CKAP5, cytoskeleton associated protein 5; E2F8, E2F transcription factor 8; ESPL1, extra spindle pole bodies like 1, separase; FEN1, flap structure-specific endonuclease 1; GTSE1, G2 and S-phase expressed 1; KIF11, kinesin family member 11; KIF20A, kinesin family member 20A; PKMYT1, protein kinase, membrane associated tyrosine/threonine 1; RANBP1, RAN binding protein 1; TACC3, transforming acidic coiled-coil containing protein 3; TRIP13, thyroid hormone receptor interactor 13; TUBB, tubulin β class I; UBE2C, ubiquitin conjugating enzyme E2 C; UCP2, uncoupling protein 2.
Clinical significance of the genes modulated in IFN-γ- or ETO-treated and PDIA1-silenced MDA-MB-231 cells
Similar analysis to that described for the MCF-7 cells was carried out for the upregulated or downregulated genes in the IFN-γ- or ETO-treated MDA-MB-231 cells with PDIA1 silenced compared with IFN-γ- or ETO-treated cells transfected with scramble siRNA that exhibited statistically significant (P<0.05) KM results are shown in Table II, and the KM curves plotted for these genes in ERα-negative patients are presented in Fig. 6. KM survival curves were plotted for the upregulated or downregulated genes in MDA-MB-231 cells under various oxidative stress conditions in patients with ERα-negative breast cancer (Fig. 6A and B). High expression of the coiled-coil-helix -coiled-coil-helix domain containing 3 (CHCHD3), heat shock protein 90 β family member 1 (HSP90B1), lysosomal-associated transmembrane protein 5 (LAPTM5) (Fig. 6A), protein tyrosine phosphatase receptor type J (PTPRJ), ring finger protein 213 (RNF213), spectrin repeat containing nuclear envelope protein 1 (SYNE1), vacuolar ATPase assembly factor VMA21 (VMA21) and X-linked inhibitor of apoptosis (XIAP) (Fig. 6B) genes was associated with statistically significant (P<0.05) longer OS in the patients with ERα-negative breast cancer. On the other hand, high gene expression of insulin-like growth factor-binding protein 3 (IGFBP3) was associated with lower OS probability in these patients (Fig. 6A).
Figure 6.
(A and B) Kaplan-Meir survival curves in patients with ERα-negative breast cancer for genes upregulated or downregulated in PDIA1-silenced and etoposide (ETO)- or interferon γ (IFN-γ)-treated MDA-MB-231 breast cancer cells. PDIA1, protein disulfide isomerase A1; ER, estrogen receptor.
The Pearson correlation coefficients between PDIA1 gene expression and the upregulate or downregulated genes in the PDIA1-silenced MDA-MB-231 cells under oxidative stress conditions (Table II) were investigated using the cBioPortal platform. To carry out this analysis, only samples originating from patients with ERα-negative breast cancer were included, but no statistical significance or data were available to manually calculate the correlation between PDIA1 gene expression and these genes in the patients with ERα-negative breast cancer. Thus, it was hypothesized that the Pearson correlation coefficients between PDIA1 gene expression with that of the genes shown in Table II were positive, meaning that in the presence of PDIA1, HSP90B1, IGFBP3, LAPTM5, RNF213 and SYNE1 genes would be downregulated, while CHCHD3, PTPRJ, VMA21 and XIAP would be upregulated, in patients with ERα-negative breast cancer.
Direct associations between PDIA1 and CHCHD3, PTPRJ, VMA21 and XIAP indicated that these genes can induce tumor-suppressive effects in patients with ERα-negative breast cancer since the KM plots in these patients showed that the high expression of these genes was associated with longer OS (Fig. 6A and B). PDIA1 would mediate tumor-suppressive effects in patients with ERα-negative breast cancer patients via IGFBP3 as well since this gene in the presence of PDIA1 is downregulated (Table II) and the KM plot in these patients showed that low expression of IGFBP3 was associated with longer OS (Fig. 6A). In addition, direct association between PDIA1 and HSP90B1, LAPTM5, RNF213 and SYNE1 gene expression suggested that these genes confer oncogenic effects to patients with ERα-negative breast cancer since the KM plots for these patients indicated that low expression was associated with lower OS probability (Fig. 6A and B).
The Metascape bioinformatics tool was used to identify the pathways affected by the genes modulated by PDIA1 to confer anti- or pro-carcinogenic effects in patients with ERα-negative breast cancer. This analysis revealed that PDIA1 may exert tumor-suppressive effects in patients with ERα-negative breast cancer by interfering with the NF-kB signaling pathway (XIAP), glycolysis (IGFBP3), mitochondrial biogenesis (CHCHD3) and negative regulation of cell migration (IGFBP3, P4HB, PTPRJ) (Table V). Metascape analysis also showed that PDIA1 may trigger pro-oncogenic effects by forming signaling networks with calmodulin-3 (CALM3), HSP90B1, LAPTM5 and RNF213 genes, which could affect pathways such as the adaptive immune system and cytokine signalling in the immune system, as well as activating signal transduction in the immune response (Table VI).
Table V.
Gene Ontology pathways and gene networks through which PDIA1 confers anti-oncogenic effects to ERα-negative breast cancer patients.
| Term | Description | Hits | LogP | Log (q-value) | |
|---|---|---|---|---|---|
| 1 | hsa04064 | NF-κB signaling pathway | XIAP | −16.64 | −3.35 |
| 2 | GO:0006006 | Glucose metabolic process | IGFBP3 | −8.6 | −8.6 |
| 3 | R-HSA-1592230 | Mitochondrial biogenesis | CHCHD3 | −2.71 | −2.71 |
| 4 | GO:0030336 | Negative regulation of cell migration | IGFBP3, PTPRJ | −1.90 | −0.96 |
| 5 | GO:0030155 | Regulation of cell adhesion | P4HB, PTPRJ | −8.96 | −1.03 |
CHCHD3, coiled-coil-helix-coiled-coil-helix domain containing 3; IGFBP3, insulin like growth factor binding protein 3; P4HB, prolyl 4-hydroxylase subunit β; PTPRJ, protein tyrosine phosphatase receptor type J; XIAP, X-linked inhibitor of apoptosis.
Table VI.
Gene Ontology pathways and gene networks through which PDIA1 confers pro-oncogenic effects to ERα-negative breast cancer patients.
| Term | Description | Hits | Log P | Log (q-value) | |
|---|---|---|---|---|---|
| 1 | R-HSA-1280218 | Adaptive immune system | CALM3, RNF213 | −4.22 | −0.94 |
| 2 | R-HSA-1280215 | Cytokine signaling in immune system | HSP90B1 | −2.91 | −0.16 |
| 3 | GO:0006874 | Cellular calcium ion homeostasis | CALM3, HSP90B1 | −3.52 | −0.50 |
| 4 | GO:0002757 | Immune response-activating signal transduction | HSP90B1, LAPTM5 | −2.86 | −0.25 |
CALM3, calmodulin 3; RNF213, ring finger protein 213; HSP90B1, heat shock protein 90 β family member 1; LAPTM5, lysosomal protein transmembrane 5.
Drugs targeting genes involved in pathways conferring oncogenic activity in breast cancer patients
To investigate whether approved or experimental drugs targeting the genes involved in the pathways conferring oncogenic activity in the patients with ERα-positive (Table VII) or ERα-negative (Table VIII) breast cancer, the genes associated with PDIA1 and its oncogenic effects were investigated in each subgroup of patients with breast cancer using the DRUGSURV (52) bioinformatics tool. This investigation revealed that in the patients with ERα-positive breast cancer, drugs targeting the function of the AURKA, FEN1, KIF11, PKMYT1 and TUBB genes, which are involved in the regulation of cell cycle progression, would be selectively effective for this group of patients. On the other hand, drugs targeting the function of the HSP90B1 gene, which is involved in the regulation of toll-like receptor translocation through the endoplasmic reticulum to the plasma membrane (53), would be preferential for the patients with ERα-negative breast cancer.
Table VII.
Approved drugs directly targeting genes associated with PDIA1 that confer pro-oncogenic effects to ERα-positive breast cancer patients.
| Gene symbol | Drugs targeting genes associated with PDIA1 with potential anti-carcinogenic activity in ERα-positive breast cancer patients. |
|---|---|
| AURKA | Phosphonothreonine, alisertib, cenisertib, enzastaurin, fostamatinib |
| FEN1 | Idarubicin, quinacrine, masoprocol, mitoxantrone |
| KIF11 | Monastrol |
| PKMYT1 | Dasatinib |
| TUBB | Colchicine, vinblastine, albendazole, podofilox, vinorelbine, vincristine |
AURKA, aurora kinase A; FEN1, flap structure-specific endonuclease 1; KIF11, kinesin family member 11; PKMYT1, protein kinase, membrane associated tyrosine/threonine 1; TUBB, tubulin β class I.
Table VIII.
Approved drugs targeting directly or indirectly genes associated with PDIA1 that confer pro-oncogenic effects to ERα-negative breast cancer patients.
| Gene symbol | Drugs targeting genes associated with PDIA1 with potential anti-carcinogenic activity in ERα-negative patients |
|---|---|
| HSP90B1 | Rifabutin |
HSP90B1, heat shock protein 90 β family member 1.
Discussion
In our previous study, we described the effect of protein disulfide isomerase A1 (PDIA1) silencing on reactive oxygen species (ROS) generation, glutathione (GSH) levels, mitochondrial membrane potential, adenosine triphosphate (ATP) production and human leukocyte antigen G (HLA-G) surface levels in MCF-7 and MDA-MB-231 cells treated with etoposide or interferon (IFN)-γ in the presence or absence of PDIA1 (4). We and others reported that PDIA1 exerts differential effects in estrogen receptor (ER)α-positive vs. triple-negative breast cancer (TNBC) cells (4,10). These effects in the ERα-positive cells are mediated possibly through the physical interaction between PDIA1 and the ERα and the regulation of the transcriptional activity of the receptor (26) whereas the impact of the PDIA1 in the ERα-negative cells is mediated through estrogen-independent pathways. To solidify our previous observations indicating the differential PDIA1 effects in a type and genetic background tumor-dependent mode (4) and taking into account the fact that PDIA1 exerts these differential effects functioning as a transcriptional co-modulator (27,33,54), we silenced PDIA1 gene expression in ERα-positive MCF-7 and TNBC MDA-MB-231 cells and submitted total mRNA isolated from these cells to RNA-seq analysis. The RNAseq experiments were performed in PDIA1 siRNA or scramble transfected MCF-7 and MDA-MB-231 cells treated with either IFN-γ or etoposide. A limitation of this study is that the PDIA1 mRNA expression levels in untreated MCF-7 and MDA-MB-231 transfected with scramble or PDIA1 siRNA were not analyzed. The results indicated that the number of genes that were upregulated or downregulated in the PDIA1-silenced MCF-7 cells was higher compared to that identified in the MDA-MB-231 cells suggesting that the main effects of PDIA1 on gene expression were mediated by the activity of the ERα transcription factor whereas in the TNBC cells through other transcription factors associated with PDIA1 such as hypoxia-inducible factor 1α (HIF-1α) (18), nuclear factor (erythroid-derived 2)-like 2 (NRF2) (32) and nuclear factor (NF)-κB activity (27).
To overcome the limitation of the lack of patient recruitment in the present study, the clinical significance of the modified genes was investigated by plotting KM survival curves to identify whether high or low expression of each one of these genes affected the overall survival (OS) probability in ERα-positive or ERα-negative patients. Furthermore, the pathways that the modulated genes were involved in to confer anti-carcinogenic or pro-oncogenic effects were identified using bioinformatic tools and grouped into four categories. The first group included the genes that were downregulated in the PDIA1-silenced MCF-7 breast cancer cells and therefore upregulated in the presence of PDIA1 in the ERα-positive breast cancer patients and their high expression was found to be associated with short survival probability; or downregulated in the PDIA1-silenced MCF-7 breast cancer cells and therefore upregulated in the presence of PDIA1 in breast cancer patients and their high expression was found to be associated with long OS probability. The genes that were downregulated in the ERα-positive breast cancer patients (upregulated in the PDIA1-silenced MCF-7 cells) and their low expression was associated with reduced OS; or downregulated in the ERα-positive breast cancer patients (upregulated in the PDIA1-silenced MCF-7 cells) and their low expression was associated with long OS probability formed the second category. The same categories forming the third and fourth groups were created for the genes exerting negative or positive effects on the OS of ERα-negative patients. Although several genes were found to be upregulated or downregulated in both the MCF-7 and MDA-MB-231 cells, the genes whose high expression resulted in statistically significant probability of short or long OS in the ERα-positive or ERα-negative patients were different.
To investigate the possibility that different gene networks in ERα-positive vs. ERα-negative breast cancer patients are associated with tumor-suppressive or oncogenic effects, we employed the Metascape bioinformatic tool (51). The results of this analysis indicated that the pathways induced to confer tumor-suppressive effects in the ERα-positive patients are mainly cell-substrate adhesion (SMAD6) (55-57) regulation of growth (PTPRS, TKT) (58,59) and the response to reactive oxygen species (PYCR1, PRDX2, UCP2) (60). Tumor-suppressive effects in the ERα-negative breast cancer patients were found to be mediated by pathways related to positive regulation of cell death (IGFBP3) (61), NF-κB signaling pathway (XIAP) (62), mitochondrial biogenesis (CHCHD3) (63) and negative regulation of cell migration (PTPRJ) (64).
The gene networks with which PDIA1 was associated to induce tumorigenesis in the ERα-positive breast cancer patients regulate pathways controlling cell cycle progression (65). Aurora kinase-A (AURKA) gene amplification or mutations are common aberrations in breast cancer, especially in ERα-positive breast carcinomas (66). Cyclin A2 (CCNA2) is overexpressed in various types of cancer including breast cancer reducing the OS of ERα-positive breast cancer patients and induces resistance to tamoxifen treatment (67). The CDC25C phosphatase participates in the regulation of the cell cycle progression from the G2 to M phase by dephosphorylating and activating the cyclin B1/CDK1 complex. Overexpression of Cdc25A in breast carcinoma patients is associated with poor survival (68). The KIF11 gene is involved in the control of cell cycle progression by promoting centrosome separation and its high expression is associated with poor prognosis of breast cancer patients (69). Tubulin α1a (TUBA1A) is one of three alpha-tubulin genes which are the major components of microtubules. Analysis of the expression profile of breast cancer tumors has indicated that TUBA1A is upregulated in tumor tissues compared to tumor-adjacent normal breast tissues (70).
The oncogenic activities of PDIA1 in ERα-negative patients are exerted through its association with genes involved in the regulation of the immune response (HSP90B1, LAPTM5 RNF213) (71-73). Aberrant regulation of gene expression of genes involved in modulating the immune response in breast tumors could lead to a low level of tumor infiltrating lymphocytes (TILs) and hence immune response evasion and breast cancer aggressiveness (74,75). It should be noted that analysis of the correlation between PDIA1 gene expression and that of the genes upregulated or downregulated in the PDIA1-silenced MDA-MB-231 cells was not carried out because data for these genes were not available in the ERα-negative patients or the Pearson correlation coefficient for those that data were available was not statistically significant.
To further investigate the possibility of differential targeting of the oncogenic pathways in ERα-positive vs. ERα-negative breast cancer patients, the existence of drugs targeting the genes implicated in oncogenic pathways were investigated using the DRUGSURV tool (52). The drugs targeting oncogenic pathways in the ERα-positive patients include those that target cell cycle progression such as cenisertib, enzastaurin, and rostamatinib, whereas drugs targeting genes involved in immune response pathways such as rifabutin could be used for the treatment of ERα-negative patients.
In conclusion, PDIA1 functions as a transcriptional cofactor regulating the transcriptional activity of the ER in ERα-positive breast cancers and HIF-1α, NRF2 and NF-κB activity in ERα-negative breast cancers. The consequence of this is that diverse oncogenic pathways are induced by PDIA1 in the ERα-positive vs. the ERα-negative breast cancers allowing the hypothesis that these two types of breast cancer can be treated differentially by drugs targeting cell cycle progression (ERα-positive breast cancers) or evasion of immunosurveillance (ERα-negative breast cancers).
Acknowledgments
We thank Dr T. Geladopoulos for reading the manuscript and providing constructive suggestions.
Funding Statement
No funding was received.
Availability of data and materials
RNA-seq data are available and can be accessed at the link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188914.
Authors' contributions
Conceptualization of the study concept was carried out by MKD, CD and RA. Data curation was conducted by EYB, CD, and RA. Formal analysis and confirmation of the data were conducted by EYB and RA. Funding acquisition was achieved by CD and RA. Investigations, including bioinformatics database search, data collection and data analysis, were the responsibility of EYB and RA. Research methodology was the responsibility of EYB, CD, and RA. Project administration was conducted by CD and RA. Resources were the responsibility of CD and RA. Supervision was undertaken by MF, MKD, and CD. Visualization was conducted by EYB, RA, MKD and CD. Writing of the original draft was carried out by EYB, MKD, CD, and RA. Writing, review and editing were carried out by EYB, MF, MKD, CD, and RA. All authors read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
- 1.Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2009;106:14831–14836. doi: 10.1073/pnas.0900742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretti AI, Laurindo FR. Protein disulfide isomerases: Redox connections in and out of the endoplasmic reticulum. Arch Biochem Biophys. 2017;617:106–119. doi: 10.1016/j.abb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Hudson DA, Gannon SA, Thorpe C. Oxidative protein folding: From thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic Biol Med. 2015;80:171–182. doi: 10.1016/j.freeradbiomed.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alhammad R, Khunchai S, Tongmuang N, Limjindaporn T, Yenchitsomanus PT, Mutti L, Krstic-Demonacos M, Demonacos C. Protein disulfide isomerase A1 regulates breast cancer cell immunorecognition in a manner dependent on redox state. Oncol Rep. 2020;44:2406–2418. doi: 10.3892/or.2020.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grek C, Townsend DM. Protein disulfide isomerase superfamily in disease and the regulation of apoptosis. Endoplasmic Reticulum Stress Dis. 2014;1:4–17. doi: 10.2478/ersc-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang I, Pottekat A, Poothong J, Yong J, Lagunas-Acosta J, Charbono A, Chen Z, Scheuner DL, Liu M, Itkin-Ansari P, et al. PDIA1/P4HB is required for efficient proinsulin maturation and ß cell health in response to diet induced obesity. ELife. 2019;8:e44528. doi: 10.7554/eLife.44528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kukita K, Tamura Y, Tanaka T, Kajiwara T, Kutomi G, Saito K, Okuya K, Takaya A, Kanaseki T, Tsukahara T, et al. Cancer-associated oxidase ERO1-α regulates the expression of MHC class I molecule via oxidative folding. J Immunol. 2015;194:4988–4996. doi: 10.4049/jimmunol.1303228. [DOI] [PubMed] [Google Scholar]
- 8.Park B, Lee S, Kim E, Cho K, Riddell SR, Cho S, Ahn K. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell. 2006;127:369–382. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Kutomi G, Kajiwara T, Kukita K, Kochin V, Kanaseki T, Tsukahara T, Hirohashi Y, Torigoe T, Okamoto Y, et al. Cancer-associated oxidoreductase ERO1-α promotes immune escape through up-regulation of PD-L1 in human breast cancer. Oncotarget. 2017;8:24706–24718. doi: 10.18632/oncotarget.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee E, Lee DH. Emerging roles of protein disulfide isomerase in cancer. BMB Rep. 2017;50:401–410. doi: 10.5483/BMBRep.2017.50.8.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Sankar S, Neamati N. Protein disulfide isomerase: A promising target for cancer therapy. Drug Discov Today. 2014;19:222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Wise R, Duhachek-Muggy S, Qi Y, Zolkiewski M, Zolkiewska A. Protein disulfide isomerases in the endoplasmic reticulum promote anchorage-independent growth of breast cancer cells. Breast Cancer Res Treat. 2016;157:241–252. doi: 10.1007/s10549-016-3820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thongwatchara P, Promwikorn W, Srisomsap C, Chokchaichamnankit D, Boonyaphiphat P, Thongsuksai P. Differential protein expression in primary breast cancer and matched axillary node metastasis. Oncol Rep. 2011;26:185–191. doi: 10.3892/or.2011.1266. [DOI] [PubMed] [Google Scholar]
- 14.Zou H, Wen C, Peng Z, Shao YY, Hu L, Li S, Li C, Zhou HH. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol Rep. 2018;39:501–510. doi: 10.3892/or.2017.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X, Wang J, Zhuang J, Ma X, Zheng N, Song Y, Xia W. P4HB modulates epithelial-mesenchymal transition and the β-catenin/snail pathway influencing chemoresistance in liver cancer cells. Oncol Lett. 2020;20:257–265. doi: 10.3892/ol.2020.11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia W, Zhuang J, Wang G, Ni J, Wang J, Ye Y. P4HB promotes HCC tumorigenesis through downregulation of GRP78 and subsequent upregulation of epithelial-to-mesenchymal transition. Oncotarget. 2017;8:8512–8521. doi: 10.18632/oncotarget.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Li T, Zhang L, Shangguan F, Shi G, Wu X, Cui Y, Wang X, Wang X, Liu Y, et al. Targeting the functional interplay between endoplasmic reticulum oxidoreductin-1α and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine. 2019;41:408–419. doi: 10.1016/j.ebiom.2019.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Wu Y, Lin YH, Guo S, Ning PF, Zheng ZC, Wang Y, Zhao Y. Prognostic value of hypoxia-inducible factor-1 alpha and prolyl 4-hydroxylase beta polypeptide overexpression in gastric cancer. World J Gastroenterol. 2018;24:2381–2391. doi: 10.3748/wjg.v24.i22.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yousef EM, Tahir MR, St-Pierre Y, Gaboury LA. MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer. 2014;14:609. doi: 10.1186/1471-2407-14-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartels AK, Göttert S, Desel C, Schäfer M, Krossa S, Scheidig AJ, Grötzinger J, Lorenzen I. KDEL receptor 1 contributes to cell surface association of protein disulfide isomerases. Cell Physiol Biochem. 2019;52:850–868. doi: 10.33594/000000059. [DOI] [PubMed] [Google Scholar]
- 21.Khan HA, Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2014;2:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiersma VR, Michalak M, Abdullah TM, Bremer E, Eggleton P. Mechanisms of translocation of ER chaperones to the cell surface and immunomodulatory roles in cancer and autoimmunity. Front Oncol. 2015;5:7. doi: 10.3389/fonc.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brigelius-Flohé R, Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal. 2011;15:2335–2381. doi: 10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K. Endoplasmic reticulum stress response and transcriptional reprogramming. Front Genet. 2015;5:460. doi: 10.3389/fgene.2014.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu X, Wang P, Zhu BT. Protein disulfide isomerase is a multi-functional regulator of estrogenic status in target cells. J Steroid Biochem Mol Biol. 2008;112:127–137. doi: 10.1016/j.jsbmb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Schultz-Norton JR, McDonald WH, Yates JR, Nardulli AM. Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor α structure and function. Mol Endocrinol. 2006;20:1982–1995. doi: 10.1210/me.2006-0006. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi T, Watanabe Y, Waga I. Protein disulfide isomerase suppresses the transcriptional activity of NF-κB. Biochem Biophys Res Commun. 2004;318:46–52. doi: 10.1016/j.bbrc.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y, Li C, Gu M, Wang H, Chen W, Luo G, Yang G, Zhang Z, Zhang Y, Xian G, et al. Protein disulfide isomerase silence inhibits inflammatory functions of macrophages by suppressing reactive oxygen species and NF-κB pathway. Inflammation. 2018;41:614–625. doi: 10.1007/s10753-017-0717-z. [DOI] [PubMed] [Google Scholar]
- 29.Kranz P, Neumann F, Wolf A, Classen F, Pompsch M, Ocklenburg T, Baumann J, Janke K, Baumann M, Goepelt K, et al. PDI is an essential redox-sensitive activator of PERK during the unfolded protein response (UPR) Cell Death Dis. 2017;8:e2986–e2986. doi: 10.1038/cddis.2017.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Yang J, Zhang Q, Xu Q, Lu L, Wang J, Xia W. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol Med Rep. 2019;19:231–237. doi: 10.3892/mmr.2018.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frasor J, Weaver A, Pradhan M, Dai Y, Miller LD, Lin CY, Stanculescu A. Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer. Cancer Res. 2009;69:8918–8925. doi: 10.1158/0008-5472.CAN-09-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee LC, Weng YT, Wu YR, Soong BW, Tseng YC, Chen CM, Lee-Chen GJ. Downregulation of proteins involved in the endoplasmic reticulum stress response and Nrf2-ARE signaling in lymphoblastoid cells of spinocerebellar ataxia type 17. J Neural Transm (Vienna) 2014;121:601–610. doi: 10.1007/s00702-013-1157-z. [DOI] [PubMed] [Google Scholar]
- 33.Campos JLO, Doratioto TR, Videira NB, Filho HVR, Batista FA, Fattori J, de C Indolfo N, Nakahira M, Bajgelman MC, Cvoro A, et al. Protein disulfide isomerase modulates the activation of thyroid hormone receptors. Front Endocrinol (Lausanne) 2019;9:784. doi: 10.3389/fendo.2018.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto S, Imaoka S. Protein-disulfide isomerase regulates the thyroid hormone receptor-mediated gene expression via redox factor-1 through thiol reduction-oxidation. J Biol Chem. 2013;288:1706–1716. doi: 10.1074/jbc.M112.365239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura T, Horibe T, Sakamoto C, Shitara Y, Fujiwara F, Komiya T, Yamamoto A, Hayano T, Takahashi N, Kikuch M. Evidence for mitochondrial localization of P5, a member of the protein disulphide isomerase family. J Biochem. 2008;144:187–196. doi: 10.1093/jb/mvn057. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y, Simmen T. Mechanistic connections between endoplasmic reticulum (ER) redox control and mitochondrial metabolism. Cells. 2019;8:1071. doi: 10.3390/cells8091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman RJ, Malhotra JD. Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1843:2233–2239. doi: 10.1016/j.bbamcr.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer NG, Schilling T, Miralles F, Eder C. Mechanisms underlying interferon-γ-induced priming of microglial reactive oxygen species production. PLoS One. 2016;11:e0162497–e0162497. doi: 10.1371/journal.pone.0162497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin HJ, Kwon HK, Lee JH, Anwar MA, Choi S. Etoposide induced cytotoxicity mediated by ROS and ERK in human kidney proximal tubule cells. Sci Rep. 2016;6:34064. doi: 10.1038/srep34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief Bioinform. 2012;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stel VS, Dekker FW, Tripepi G, Zoccali C, Jager KJ. Survival analysis I: The kaplan-meier method. Nephron Clin Pract. 2011;119:c83–c88. doi: 10.1159/000324758. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.31–31.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV. DRUGSURV: A resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis. 2014;5:e1051. doi: 10.1038/cddis.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebersole JL, Novak MJ, Orraca L, Martinez-Gonzalez J, Kirakodu S, Chen KC, Stromberg A, Gonzalez OA. Hypoxia-inducible transcription factors, HIF1A and HIF2A, increase in aging mucosal tissues. Immunology. 2018;154:452–464. doi: 10.1111/imm.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiefel H, Bondong S, Hazin J, Ridinger J, Schirmer U, Riedle S, Altevogt P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adh Migr. 2012;6:374–384. doi: 10.4161/cam.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blom M, Reis K, Heldin J, Kreuger J, Aspenström P. The atypical Rho GTPase RhoD is a regulator of actin cytoskeleton dynamics and directed cell migration. Exp Cell Res. 2017;352:255–264. doi: 10.1016/j.yexcr.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Filippou PS, Karagiannis GS, Constantinidou A. Midkine (MDK) growth factor: A key player in cancer progression and a promising therapeutic target. Oncogene. 2020;39:2040–2054. doi: 10.1038/s41388-019-1124-8. [DOI] [PubMed] [Google Scholar]
- 58.Wang ZC, Gao Q, Shi JY, Guo WJ, Yang LX, Liu XY, Liu LZ, Ma LJ, Duan M, Zhao YJ, et al. Protein tyrosine phosphatase receptor S acts as a metastatic suppressor in hepatocellular carcinoma by control of epithermal growth factor receptor-induced epithelialmesenchymal transition. Hepatology. 2015;62:1201–1214. doi: 10.1002/hep.27911. [DOI] [PubMed] [Google Scholar]
- 59.Tseng CW, Kuo WH, Chan SH, Chan HL, Chang KJ, Wang LH. Transketolase regulates the metabolic switch to control breast cancer cell metastasis via the α-ketoglutarate signaling pathway. Cancer Res. 2018;78:2799–2812. doi: 10.1158/0008-5472.CAN-17-2906. [DOI] [PubMed] [Google Scholar]
- 60.Yi JW, Park JY, Sung JY, Kwak SH, Yu J, Chang JH, Kim JH, Ha SY, Paik EK, Lee WS, et al. Genomic evidence of reactive oxygen species elevation in papillary thyroid carcinoma with Hashimoto thyroiditis. Endocr J. 2015;62:857–877. doi: 10.1507/endocrj.EJ15-0234. [DOI] [PubMed] [Google Scholar]
- 61.Wang SH, Chen YL, Hsiao JR, Tsai FY, Jiang SS, Lee AYL, Tsai HJ, Chen YW. Insulin-like growth factor binding protein 3 promotes radiosensitivity of oral squamous cell carcinoma cells via positive feedback on NF-κB/IL-6/ROS signaling. J Exp Clin Cancer Res. 2021;40:95. doi: 10.1186/s13046-021-01898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofer-Warbinek R, Schmid JA, Stehlik C, Binder BR, Lipp J, de Martin R. Activation of NF-kappa B by XIAP, the X chromosome-linked inhibitor of apoptosis, in endothelial cells involves TAK1. J Biol Chem. 2000;275:22064–22068. doi: 10.1074/jbc.M910346199. [DOI] [PubMed] [Google Scholar]
- 63.Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneble N, Müller J, Kliche S, Bauer R, Wetzker R, Böhmer FD, Wang ZQ, Müller JP. The protein-tyrosine phosphatase DEP-1 promotes migration and phagocytic activity of microglial cells in part through negative regulation of fyn tyrosine kinase. Glia. 2017;65:416–428. doi: 10.1002/glia.23100. [DOI] [PubMed] [Google Scholar]
- 65.Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, Mantovani R, Domany E, Rotter V, Pilpel Y. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol Syst Biol. 2005;1:2005.0022. doi: 10.1038/msb4100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 67.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One. 2014;9:e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cangi MG, Cukor B, Soung P, Signoretti S, Moreira G, Jr, Ranashinge M, Cady B, Pagano M, Loda M. Role of the Cdc25A phosphatase in human breast cancer. J Clin Invest. 2000;106:753–761. doi: 10.1172/JCI9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rapley J, Nicolàs M, Groen A, Regué L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nami B, Wang Z. Genetics and expression profile of the tubulin gene superfamily in breast cancer subtypes and its relation to taxane resistance. Cancers (Basel) 2018;10:274. doi: 10.3390/cancers10080274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin T, Qiu Y, Peng W, Peng L. Heat shock protein 90 family isoforms as prognostic biomarkers and their correlations with immune infiltration in breast cancer. Biomed Res Int. 2020;2020:2148253. doi: 10.1155/2020/2148253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glowacka WK, Alberts P, Ouchida R, Wang JY, Rotin D. LAPTM5 protein is a positive regulator of proinflammatory signaling pathways in macrophages. J Biol Chem. 2012;287:27691–27702. doi: 10.1074/jbc.M112.355917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma P, Barlow WE, Godwin AK, Parkes EE, Knight LA, Walker SM, Kennedy RD, Harkin DP, Logan GE, Steele CJ, et al. Validation of the DNA damage immune response signature in patients with triple-negative breast cancer from the SWOG 9313c trial. J Clin Oncol. 2019;37:3484–3492. doi: 10.1200/JCO.19.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leung KL, Verma D, Azam YJ, Bakker E. The use of multi-omics data and approaches in breast cancer immunotherapy: A review. Future Oncol. 2020;16:2101–2119. doi: 10.2217/fon-2020-0143. [DOI] [PubMed] [Google Scholar]
- 75.Sabbatino F, Liguori L, Polcaro G, Salvato I, Caramori G, Salzano FA, Casolaro V, Stellato C, Col JD, Pepe S. Role of human leukocyte antigen system as a predictive biomarker for checkpoint-based immunotherapy in cancer patients. Int J Mol Sci. 2020;21:7295. doi: 10.3390/ijms21197295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data are available and can be accessed at the link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188914.