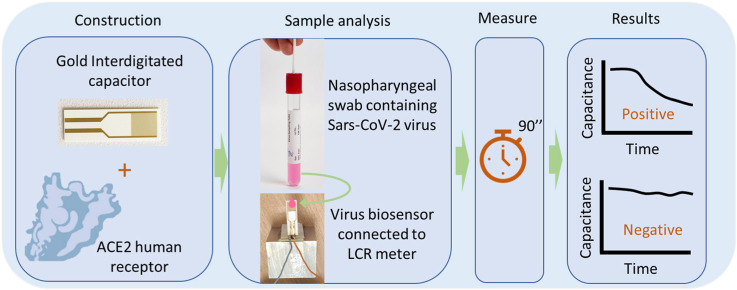
Abstract
The spread of the SARS-CoV-2 and its increasing threat to human health worldwide have necessitated the development of new technological tools to combat the virus. Particular emphasis is given to the development of diagnostic methods that monitor the spread of the virus rapidly and effectively. In this study, we report the development and testing of an antibody-free biosensor, based on the immobilization of ACE2 protein on the surface of gold interdigitated electrode. When the sensor was used in laboratory conditions for targeting the virus’ structural spike protein, it showed a limit of detection [LOD] of 750 pg/μL/mm2. Thereafter, the response of the sensor to swab and saliva samples from hospitalized patients was examined. The virus presence in the samples was confirmed by electrical effective capacitance measurements executed on the biosensor, and correlated with real-time PCR results. We verified that the biosensor can distinguish samples that are positive for the virus from those that are negative in a total of 7 positive and 16 negative samples. In addition, the biosensor can be used for semi-quantitative measurement, since its measurements are divided into 3 areas, the negative samples, the weakly positive and the positive samples. Reproducibility of the experiments was demonstrated with at least 3 replicates and stability was tested by keeping the sensor standby for 7 days at 4 °C before repeating the experiment. This work presents a biosensor that can be used as a fast-screening test at point of care detection of SARS-CoV-2 since it needs less than 2 min to provide results and is of simple operation.
Keywords: SARS-CoV-2 biosensor, Antibody free, Interdigitated electrodes, Detection
Graphical abstract
1. Introduction
Human susceptibility to certain viruses has been well-documented, and the effect of viruses on the human population varies from the relatively benign to deadly pandemics that may cause illness and loss of life. The 2020 pandemic caused by the spread of the SARS-CoV-2 has, to date, led to hundreds of thousands of reported deaths, and a major disruption of global commerce and economy, education and societal behavior in general. Transmission of the virus mainly occurs via airborne respiratory droplets or from contact with surfaces containing undamaged virus particles, also named virions. While viable tests for determining the presence of the virus in a human host have been developed, most of the methods used require either a significant amount of time to be completed or are implemented through complex and expensive methods.
The standard method that is used world-wide to detect SARS-CoV-2 is quantitative real-time PCR. This method is highly specific because it detects a portion of virus’ genome. To perform this analysis multiple steps are required along with several reagents, enzymes and machines able to amplify the cDNA and at the same time measure fluorescent light. From the time samples reach the analysis center, at least 5–6 h are required to obtain results. Despite the high accuracy and specificity, real-time PCR is an expensive and time-consuming method. The development of a quick and accurate detection assay is considered vital aiming to control the possible sources of infection in order to design effective measures to prevent further transmission.
For this reason, the need to develop easy-to-use devices for cheap, fast and efficient detection has been emphasized (Morales-Narváez and Dincer, 2020), and electrochemical biosensors are proposed to fulfill this purpose (Imran et al., 2021). Even though research aimed to find solutions for a rapid and reliable detection method has been boosted by the unprecedent effects of SARS-CoV-2 on global economy, up to this point few devices have been developed for the detection of the of native virus particles (Fathi-Hafshejani et al., 2021) (Mavrikou et al., 2021) (Mak et al., 2020) (Seo et al., 2020) (Rashed et al., 2021), including biosensors like lateral flow devices (LFID) and enzyme linked immunosorbent assays (ELISA). However what seems to be more exciting is the detection of certain biomarkers associated with SARS-CoV-2 (Garg et al., 2021).
It is well known that ACE2 protein is the membrane surface receptor used by SARS-CoV-2 to bind and infect human cells (Lan et al., 2020) (Devaux et al., 2020). The binding happens through the virus’ structural spike (S) protein, which has very high and specific affinity for ACE2 protein, as the virus is positively charged, whereas ACE2 is negatively charged (Xie et al., 2020). In particular, the extracellular domain of ACE2 interacts with its N-terminal region with the receptor binding domain (RBD) of S protein (Lan et al., 2020). Therefore the use of ACE2 as binding element for native SARS-CoV-2 particles represents a marvel opportunity to set up a biosensor for rapid and efficient virus detection.
Interdigitated electrodes (IDEs) are simple and very sensitive which makes them one of the most favored transducers in the field of biological and chemical sensors (Mazlan et al., 2017). Τhey are used as capacitance or impedance biosensors for various applications (Sontimuang et al., 2011) (Quershi et al., 2009) (Khan et al., 2016), including virus detection (Huy et al., 2011) (Wang et al., 2009) (Birnbaumer et al., 2009) (Yao and Fu, 2014) (Cheng et al., 2017) (Wang et al., 2017). IDEs have been also used for SARS-CoV-2 antibodies (Rashed et al., 2021) and nucleocapsid protein (Ramanathan et al., 2022) detection. Graphene IDE biosensors have also been developed, targeting S protein, based on its interaction with SARS-CoV-2 antibodies (Sharma et al., 2021).
Interdigitated electrodes serve as the transducer, when used as capacitive biosensors. A receptor is immobilized on the electrode surface and the interaction of an analyte with it brings about a change in the material's dielectric properties or in the thickness of the dielectric layer (Berggren et al., 2001). The basic equation according to which the IDE capacitance changes is given by Equation (1) (Tsouti et al., 2011):
| (1) |
where ε0 is the vacuum permittivity, εr is the relative permittivity of the medium between the plates, A is the electrodes' surface area and d is the distance between the electrodes. According to equation (1), when the distance between the electrodes increases, the total capacitance decreases.
In this study a label-free affinity-based capacitive IDE sensor is developed for detecting both recombinant SARS-CoV-2 S protein and the native virions themself with high sensitivity and selectivity in a total time of less than 2 min. ACE2 enzyme is immobilized on IDE surface and used as the bioreceptor for virus' S protein. ACE2 immobilized gold IDEs' surface serves as the transducer. When a SARS-CoV-2 particle or S protein molecule binds to ACE2 receptor a displacement of the counter ions around the capacitive electrode results in a decrease in its effective capacitance (Mattiasson and Hedström, 2016). The higher the amount of virus molecules bound to ACE2 is, the greater is the decrease in the transducer's capacitance (and therefore the change of the total impedance), detected as an electric signal. At first, preparative experiments were conducted using SARS-CoV-2 S protein. In the second phase, analytical experiments were conducted using real virion containing samples obtained from hospitalized patients. The results of the biosensor testing of the samples were compared to the results of real-time PCR on the same samples.
2. Materials and methods
2.1. Materials
The sensor includes 2 separate components connected via electrical wires. The first component is the biological part which is able to physically interact with virions particles; the second is the electronic part that detects the interaction between the biological component and virion particles via changes of capacitance and impedance.
For the biological part, the following components have been used: the human extracellular domain of ACE2 protein with tags; the amino acid L-Cysteine in powder form; and reagents named MES, EDC and PBS.
ACE2 protein (InvivoGen, San Diego, USA) was dissolved in distilled water at a concentration of 50 ng/μl. SARS-Cov-2 Spike RBD (RBD) (InvivoGen, San Diego, USA) was dissolved in distilled water at a concentration of 25 ng/μl. L-cysteine (Sigma-Aldrich, St. Louis, USA) was dissolved in distilled water at a concentration of 25 mM. EDC (N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide) (Sigma-Aldrich, St. Louis, USA) at a concentration of 0.877 g/ml was dissolved in water at a concentration of 0.1 M. MES (2-(N-Morpholino)ethanesulfonic acid and 4-Morpholineethanesulfonic acid) was purchased from Sigma-Aldrich (St. Louis, USA). Cell culture grade PBS (Phosphate Buffered saline) was purchased from Gibco (Carlsbad, USA). Bovine Serum Albumin (BSA) (Sigma-Aldrich, St. Louis, USA) was dissolved in distilled water at a concentration of 50 ng/μl. Gold interdigitated electrodes were purchased from DropSens (Asturias, Spain), cat. N.: PW-IDEAU50. Each IDE has a finger width and spacing of 50 μm, with a total number of 70 fingers, a total electrode length of 7 mm, and electrode surface area of 8.45 mm2.
2.2. Grafting ACE2 protein on interdigitated gold surface
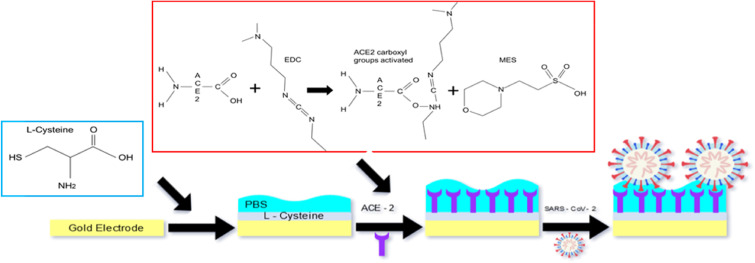
To clean the electrodes' surface, interdigitated capacitors were immersed in a solution of 100% isopropanol for 5 min and then rinsed in Milli-Q water. 30 μL (25 mM) of L-Cysteine were placed on the electrode surface and let to form bonds between gold and the amino acid's thiol groups. The reaction was stopped when all L-cysteine solution dried out and three washes were performed using PBS to remove unbound material. Subsequently, a solution containing 5 μL (50 ng/ml) of ACE2, 5 μL (0.877 g/ml) of EDC and 10 μL (0.1 M) of MES was placed on top of the L-Cysteine layer. Specifically, EDC and MES activate the carboxylic terminus (-COOH) of ACE2. Once active the carboxylic terminus of ACE2 reacts with the amine group (-NH2) of L-Cysteine forming a covalent peptide bond. Due to the cross reactivity between C- and N- termini of both ACE2 and L-Cysteine, unwanted covalent bonds were formed also among the C-terminal of L-Cysteine and the N-terminal of ACE2. After each step, the biosensor was rinsed 3 times in Milli-Q water, in order to remove unbound molecules from its surface. The IDE with immobilized ACE2 or L-Cysteine alone was covered with 20 μl of PBS and kept wet until its use (Fig. 1 ). Some of the biosensors were kept standby for 7 days at 4 °C in order to be used for the biosensor stability calibration.
Fig. 1.
Schematic IDE ACE2-based Sars-CoV-2 biosensor.
2.3. SARS-CoV-2 biological fluid and clinical samples collection
For specimen's collection it was used the Citoswab transport medium VTM 3 ml product code 2118-0019 and Citoswab collection swab product code 2122-0009 (WellKang, Dover UK). A total of 23 clinical specimens were collected from 22 subjects from Konstantopoulio General Hospital, a CV-19 Reference Hospital in Greece. All specimens had the suspected COVID-19 case definition set by the World Health Organization (World Health Organization, 2020). Sampling was accomplished to our premises with all the required precautions. Nasopharyngeal swabs were collected following the standard procedure to harvest nasopharyngeal fluids using sterile swab and transport solutions and they were subsequently placed in 2 ml of transport medium. Specimen processing was performed in a class II biological safety cabinet using biosafety level three (BSL3) work practices. All samples were collected according the safety standards by specialized personnel at the Konstantopoulio General Hospital (Athens, Greece).
2.4. Protocol used to measure spike protein solutions and clinical samples
From each swab sample or solution containing S protein (various concentrations in the range 100 pg/μL – 10 ng/μL), 20 μl of sample were deposited in the interdigitated capacitor surface and the effective capacitance was monitored in real-time. The measurements were performed at room temperature and were extended for 1–2 min. The tests were performed in a blind-blind mode: the swab samples were tested first with the biosensor and then undergone real-time PCR tests. In order to examine the reproducibility of the experiments, each sample was demonstrated with 3 replicates. The capacitance values illustrated in the results section correspond to the average value measured, whereas the error bars correspond to the standard deviation. Capacitance measurements were performed using an HP LCR Meter (Hewlett Packard, model 4284A Precision). Two golden needles were connected with the LCR machine and they were placed at the pads of the capacitor. As the relative dielectric permittivity of the interdigitated capacitor is dependent on the testing frequency (Mazlan et al., 2017), a standard 1 kHz frequency was selected with the amplitude of 50 mV. For better display of results capacitance was normalized. That is, the capacitance values presented in the results section are divided by the maximum capacitance value of each experiment. For example, if the maximum capacitance value measured in an experiment was 700 nF, then a 350 nF capacitance would be equal to 0.5 or 50%.
2.5. Biosensor selectivity
In order to verify the biosensor's selectivity, a second IDE biosensor was covered with 20 μl of BSA solution and effective capacitance was measured. Subsequently, on top of a third sensor 20 μl of a solution (1:1) containing both BSA and S protein was added and effective capacitance was measured. Solutions containing BSA, S protein and both of them were let on the sensor surface for 20 min in order to ensure a proper reaction with ACE2. After this time period, PBS was used to wash the three biosensors and 20 μl of PBS were left on in order to avoid measurement errors due to water evaporation. In addition, to assess the selectivity of the Biosensor assay, 5 clinical samples (adenovirus, influenza virus A/H1N1, parainfluenza virus, varicella-zoster virus and rubella virus) collected and diagnosed from the Hospital were used. The capacitance response of all the clinical samples was similar to the negative samples, indicating that the method was specific for SARS-CoV-2 (Supplemental Fig. S1).
2.6. RNA extraction and real-time PCR
Nucleic acids were isolated from clinical specimens using an automatic extractor (GXT NA Extraction Kit DNA/RNA 200 virus). The RNA was converted to cDNA and subsequently amplified using the YouSeq® SARS-CoV-2 COVID-19 RT-qPCR kit (YouSeq, Winchester, England). The oligonucleotide primers and probes for specific detection of SARS-CoV-2 are designed to detect regions of RNA-dependent RNA (RdRP) labelled with ROX and envelope protein (E) genes labelled with FAM of the SARS-CoV-2 genome. The kit also contains primers and probe (labelled with VIC/HEX) for detection of human RNA as an endogenous internal control for confirming specimen integrity, nucleic acid isolation, possible inhibitions reverse transcription, amplification and detection.
2.7. Standard curve and limit of detection (LOD)
The real-time PCR with standard curve was generated by serial 10-fold dilutions of synthetic positive controls with known copy numbers (106 to 10 copies/μL) (YouSeq, Winchester, England). These dilutions were tested using 10 replicates and they were used as quantification standards to construct the standard curve by plotting the copy number against the corresponding threshold cycle values (Ct). For the biosensor standard curve determination, the biosensor response for 5 PBS solutions of 20 μL was tested, each containing different concentrations of S protein (ng/mL).
2.8. Biomedical ethics issues
The collection of clinical data was correlated with the laboratory research results and was conducted in such a way as to fully guarantee the patients’ anonymity and personal data confidentiality.
3. Results and discussion
3.1. Interdigitated capacitance measurements and changes with spike RBD protein
When interdigitated capacitor was measured with only air on its surface, its capacitance ranges on the scale of few pF. When measurements were performed on IDE containing PBS and L-Cysteine on top of the gold surface, a capacitance increase of about 350 nF was observed. After ACE2 was immobilized on top of L–Cysteine monolayer, an additional capacitance increase of about 300 nF was detected, bringing the total capacitance up to 650 nF (Fig. 2 ). This increase in capacitance is justified by the fact that as a solution of proteins (ACE2) in PBS is added on the electrode surface, the dielectric constant of the material increases since chemical bonds are formed, resulting in a total capacitance increase (Qureshi et al., 2010) (Sontimuang et al., 2011) (Quershi et al., 2009). Apart from that, the ACE2 protein and the bound S protein result in an additional resistivity, parallel to the pure capacitance, thus allowing for the monitoring of an effective capacitance or impedance of the sensing element. However, the measurements in the employed experimental apparatus were realized in terms of capacitance measurements and corresponding monitoring.
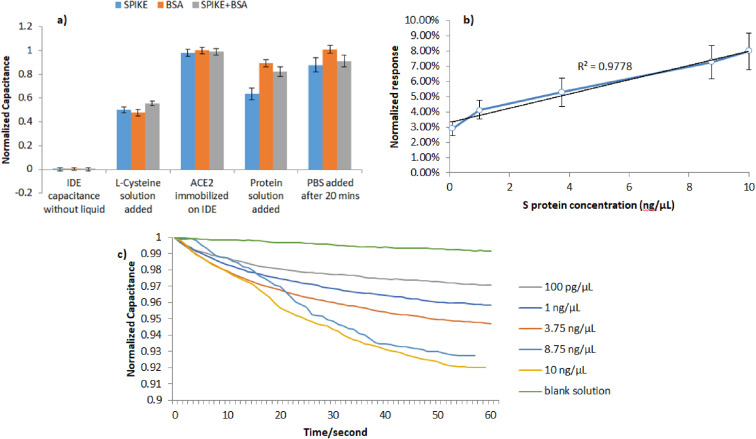
Fig. 2.
Capacitance changes with S protein. a) Selective response of the sensor towards BSA and S protein; b) Normalized response of the sensor for S protein; c) Real time detection of S protein.
Initially, experiments were conducted targeting the SARS-CoV-2 S protein to verify the selectivity and the sensitivity of the biosensor. The addition of S protein (10 ng/μL) and its binding to molecules present in the medium bind ACE2 molecules, results in an effective capacitance decrease since S proteins displace ions interaction with ACE2 molecules. L-Cysteine and ACE2 form stable chemical bonds (e.g. covalent bond) among them and with gold electrodes, whereas S and PBS interact with ACE2 with weak transient interactions (e.g. ionic or electric bonds).
As illustrated in Fig. 2a, the sensor seems to be selective to S protein since, when electrical measurement was performed after 20 min, its effective capacitance was reduced by at least 10% when a solution containing S protein was added on top of the sensor, compared to the absence of capacitance change when a solution containing BSA was added. Furthermore, the reaction seems to take place even when BSA is present in the S protein solution, confirming the selectivity of ACE2 protein for S protein. The calibration curve of the capacitance change due to different dilutions of S protein is illustrated in Fig. 2b. Strong linear correlations (r2 ≥0.9778) were obtained between the S protein concentration and the biosensor capacitance change. The analytical decrease of the capacitance over time is illustrated in Fig. 2c. The biosensor sensitivity to S protein is calculated at 750 pg/μL/mm2, since standard deviation of the blank solution is 0, 01 and the slope of the calibration curve is 0,0047 μL/ng. The sensors that were kept standby for 7 days at 4 °C showed a similar response to the sensors used directly.
3.2. SARS-Cov-2 virions detection
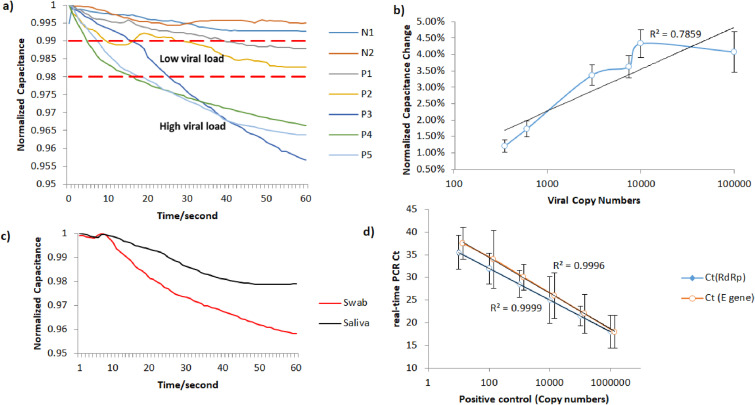
According to the real-time PCR method, 16 samples were negative and 7 of them were positive to SARS-CoV-2. It was observed that for negative swabs the effective capacitance was either increasing or slightly decreasing. In cases where the effective capacitance was decreasing, the range of such changes was very low, in the order of <1% respect to the initial value. The capacitance response over time of the positive to the virus samples and two negative samples with typical behavior is illustrated in Fig. 3 a, where the positive swab samples are divided into 2 groups according to the viral load. For the positive samples that had a viral load of at least 103 virus copy numbers/μL, it was observed that the effective capacitance was constantly and quickly decreasing. However, for the positive swab samples with a lower viral load, the effective capacitance was slowly decreasing and the total decrease was sometimes comparable to blank solution. Concerning negative samples, N1 response shows typical negative response, whereas N2 refers to a positive swab sample that was placed on a capacitor missing the ACE2 layer; therefore, it was observed that its behavior was similar to the ones of the negative samples. Fig. 3b illustrates the total capacitance change caused by the positive swab samples, depending on the viral copy numbers/μL in each sample, according to the real-time PCR measurements. To verify if the sensor response can be affected by the typology of sample collected from a positive patient, we used both a swab and a saliva sample collected from the same positive subject. These samples were tested both with the real-time PCR method and the biosensor. Fig. 3c illustrates the biosensor response for the 2 samples. Both samples were found by the real-time PCR to have a high viral load, leading to a reduction in capacitance, which was more evident in the swab samples with respect to the saliva one.
Fig. 3.
Sensor response towards SARS-CoV-2. a) Samples are distinguished in 3 regions; Negative to SARS-CoV-2, Positive with low viral load and positive with high viral load; b) Normalized response of the sensor for SARS-CoV-2; c) Capacitance change over time for swab and saliva sample of the same person; d) Calibration curve of Ct vs Virus Copy Numbers.
Capacitance reduction can be explained in two ways: the first is related to the displacement of the counter ions because of S protein binding on ACE2, as reported in the literature (Mattiasson and Hedström, 2016); the second, our hypothesis, is related to the decrease of ACE2 oscillation due to the applied electric voltage to interdigitated electrodes, caused by S protein or virus assembly on the ACE 2 layer.
3.3. Results for real-time PCR
The linearity and efficiency of the real-time PCR were determined by generating a standard curve in which serial 10-fold dilutions of positive control were tested, as is illustrated in Fig. 3d. The standard curve was generated by plotting the real-time PCR threshold cycle numbers (Ct) of each dilution against the known copy numbers of positive control. The resulting slope showed a linear relationship over 5 orders of magnitude, ranging from 106 to 10 copies/μL with a correlation coefficient R2 > 0.99. The detection rate was 100% for up to 10 copies/μL having 10/10 replicates positive for E and RdRp genes.
Strong linear correlations (r2 ≥ 0.99) were obtained between CT values and transcript quantity. Assay reproducibility and repeatability was tested by using replicate 10-fold serial dilutions of the RNA transcripts evaluated for each dilution point in triplicate on three different days. At the lower copy detection limit for SARS-CoV-2 and assay reproducibility exceeded 95%.
As illustrated in Table 1 , all positive RNA samples tested, were positive for the human gene which was included as internal control to evaluate the quality of clinical specimens (nasopharyngeal swabs and saliva) and nucleic acid extraction. The Ct Value of positive specimens ranged from 18 to 34 that corresponded to 106- 102 copy numbers. A 75% of the specimens tested comprised 106- 103 copy numbers of E and RdRp amplicons.
Table 1.
Results for real-time PCR.
| Sample | Ct (Internal Control) | Ct (RdRp) | Ct (E gene) |
|---|---|---|---|
| P1 | 29 ± 0.2 | 31 ± 0.2 | 32 ± 0.1 |
| P2 | 30 ± 0.2 | 30 ± 0.1 | 31 ± 0.2 |
| P3 | 26 ± 0.1 | 25 ± 0.2 | 26 ± 0.1 |
| P4 | 23 ± 0.2 | 28 ± 0.3 | 28 ± 0.1 |
| P5 | 26 ± 0.1 | 27 ± 0.1 | 27 ± 0.2 |
| Swab | 27 ± 0.2 | 18 ± 0.2 | 18 ± 0.3 |
| Saliva | 27 ± 0.2 | 25 ± 0.1 | 24 ± 0.2 |
Our biosensor benefits from using the natural receptor instead of antibodies (Sharma et al., 2021) (Seo et al., 2020), as in this way it can be used to detect all strains of the virus. When compared to other SARS-CoV-2 biosensors based on interdigitated electrodes (Wang et al., 2017) (Rashed et al., 2021), the novelty of our research is related to the ability to detect directly the superficial viral S protein and not viral DNA or the antibodies produced against viral proteins.
When compared to classical biochemical tests used on rapid antigen tests which are only qualitative (visual observation), our biosensor benefits from its significantly faster response, as well as from the possibility to provide an electronic measurement which can be used to make quantitative estimation, and can be shared and integrated with health-related databases.
However, when compared to antibody based sensors for the rapid detection of SARS-CoV-2, our biosensor exhibits a slightly reduced sensitivity for the case of samples with viral load of less than 103 virus copy numbers/μL.
4. Conclusion
The objective of this study was to develop an antibody-free capacitive biosensor for the rapid detection of SARS-CoV-2 native virions. Even though at the time this study was concluded, several biosensors based on antibodies against S protein have been developed, they may fail in the detection of SARS-CoV-2 variants. Indeed, the majority of genetic changes are detected on distinct S protein domains and pose serious concerns also for the vaccine efficacy (Li et al., 2021), which ultimately stimulates the production of Spike-targeting antibodies. Therefore, the biosensor was realized by immobilizing ACE2 protein on top of interdigitated electrodes, which binds with high specificity SARS-CoV-2 and with minor, but still consistent, other coronavirus like SARS-CoV-1 (Ortega et al., 2020) (Anand et al., 2020). We are aware that ACE2 may be the receptor used by other pathogens to infect the human body, but we never intended the use of such sensor as solely tool to detect a SARS-CoV-2 infection. Indeed, the standard method to certify the presence of SARS-CoV-2 remains the real-time PCR. Therefore, our device could be used for rapid screening in order to clearly differentiate negative subjects from potential SARS-CoV-2 positive subjects. It is worth noting to mention that the widely used rapid antigenic SARS-CoV-2 detection tests may also recognize other coronavirus species because most systems recognize the N protein (highly conserved among the beta-Coronavirus genus to which SARS-CoV-2 belongs) leading to false positive results (Bai et al., 2021) (Mahmoudinobar et al., 2021).
Using the real-time PCR method as a diagnostic standard, the results demonstrated that the biosensor was able to detect clearly the SARS-CoV-2 virion in about 1 min, in swab samples with a viral load as low as 103 virus copy numbers/μL, or saliva samples with a viral load as low as 104 virus copy numbers/μL. Furthermore, the biosensor demonstrated good selectivity towards SARS-CoV-2 S protein, as it was not affected by the presence in the reaction of BSA protein. Future research on the topic should focus on noise compensation by optimizing the electrical measurement, which can improve the sensor response for samples with viral load below 103 virus copy numbers/μL (swab) or less than 104 virus copy numbers/μL (saliva).
CRediT authorship contribution statement
A. Georgas: Conducting sensor development and experiments. E. Lampas: Collecting swab. D.P. Houhoula: Performing RT PCR. A. Skoufias: samples, Aiding in sensor experiments. S. Patsilinakos: Supervising experiments with patients samples. I. Tsafaridis: Providing materials for sensors. G.P. Patrinos: Commenting on qRT-PCR. N. Adamopoulos: Providing materials for sensors. A. Ferraro: Co-conceiving the sensor with, supervising the whole work. E. Hristoforou: binding ACE2 on gold surface, Co-conceiving the sensor with.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: This work has been supported by Katharsis Technologies Inc. and Galenica SA.
Acknowledgments
This work has been supported by Katharsis Technologies Inc. and Galenica SA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114021.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Anand S.P., Chen Y., Prévost J., Gasser R., Beaudoin-Bussières G., Abrams C.F., Pazgier M., Finzi A. Interaction of human ACE2 to membrane-bound SARS-CoV-1 and SARS-CoV-2 S glycoproteins. Viruses. 2020 doi: 10.3390/v12101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z., Cao Y., Liu W., Li J. The sars-cov-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13 doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren C., Bjarnason B., Johansson G. Capacitive biosensors. Electroanalysis. 2001;13:173–180. doi: 10.1002/1521-4109(200103)13:3%3c173::AID-ELAN173%3e3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Birnbaumer G.M., Lieberzeit P.A., Richter L., Schirhagl R., Milnera M., Dickert F.L., Bailey A., Ertl P. Detection of viruses with molecularly imprinted polymers integrated on a microfluidic biochip using contact-less dielectric microsensors. Lab Chip. 2009;9:3549–3556. doi: 10.1039/b914738a. [DOI] [PubMed] [Google Scholar]
- Cheng C., Cui H., Wu J., Eda S. A PCR-free point-of-care capacitive immunoassay for influenza A virus. Microchim. Acta. 2017;184:1649–1657. doi: 10.1007/s00604-017-2140-4. [DOI] [Google Scholar]
- Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi-Hafshejani P., Azam N., Wang L., Kuroda M.A., Hamilton M.C., Hasim S., Mahjouri-Samani M. Two-dimensional-material-based field-effect transistor biosensor for detecting COVID-19 virus (SARS-CoV-2) ACS Nano. 2021;15:11461–11469. doi: 10.1021/acsnano.1c01188. [DOI] [PubMed] [Google Scholar]
- Garg M., Sharma A.L., Singh S. Advancement in biosensors for inflammatory biomarkers of SARS-CoV-2 during 2019–2020. Biosens. Bioelectron. 2021;171:112703. doi: 10.1016/j.bios.2020.112703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huy T.Q., Hanh N.T.H., Thuy N.T., Chung P. Van, Nga P.T., Tuan M.A. A novel biosensor based on serum antibody immobilization for rapid detection of viral antigens. Talanta. 2011;86:271–277. doi: 10.1016/j.talanta.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran S., Ahmadi S., Kerman K. Electrochemical biosensors for the detection of sars-cov-2 and other viruses. Micromachines. 2021;12:1–23. doi: 10.3390/mi12020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.R.R., Khalilian A., Kang S.W. Fast, highly-sensitive, and wide-dynamic-range interdigitated capacitor glucose biosensor using solvatochromic dye-containing sensing membrane. Sensors. 2016;16:265. doi: 10.3390/s16020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li R., Liu J., Zhang H. The challenge of emerging SARS-CoV-2 mutants to vaccine development. J. Genet. Genomics. 2021 doi: 10.1016/j.jgg.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudinobar F., Britton D., Montclare J.K. Protein-based lateral flow assays for COVID-19 detection. Protein Eng. Des. Sel. 2021;34:1–10. doi: 10.1093/protein/gzab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiasson B., Hedström M. Capacitive biosensors for ultra-sensitive assays. TrAC Trends Anal. Chem. (Reference Ed.) 2016;79:233–238. doi: 10.1016/j.trac.2015.10.016. [DOI] [Google Scholar]
- Mavrikou S, et al. Clinical application of the Novel Cell-Based Biosensor for the Ultra-Rapid Detection of the SARS-CoV-2 S1 Spike Protein Antigen: a Practical Approach. Biosensors. 2021 doi: 10.3390/bios11070224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazlan N.S., Ramli M.M., Abdullah M.M.A.B., Halin D.S.C., Isa S.S.M., Talip L.F.A., Danial N.S., Murad S.A.Z. Interdigitated electrodes as impedance and capacitance biosensors: a review. AIP Conf. Proc. 1885. 2017 doi: 10.1063/1.5002470. [DOI] [Google Scholar]
- Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163 doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quershi A., Gurbuz Y., Kang W.P., Davidson J.L. A novel interdigitated capacitor based biosensor for detection of cardiovascular risk marker. Biosens. Bioelectron. 2009;25:877–882. doi: 10.1016/j.bios.2009.08.043. [DOI] [PubMed] [Google Scholar]
- Qureshi A., Niazi J.H., Kallempudi S., Gurbuz Y. Label-free capacitive biosensor for sensitive detection of multiple biomarkers using gold interdigitated capacitor arrays. Biosens. Bioelectron. 2010;25:2318–2323. doi: 10.1016/j.bios.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Ramanathan S., Gopinath S.C.B., Ismail Z.H., Md Arshad M.K., Poopalan P. Aptasensing nucleocapsid protein on nanodiamond assembled gold interdigitated electrodes for impedimetric SARS-CoV-2 infectious disease assessment. Biosens. Bioelectron. 2022;197:113735. doi: 10.1016/j.bios.2021.113735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S. Il. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sharma P.K., Kim E.-S., Mishra S., Ganbold E., Seong R.-S., Kaushik A.K., Kim N.-Y. Ultrasensitive and reusable graphene oxide-modified double-interdigitated capacitive (DIDC) sensing chip for detecting SARS-CoV-2. ACS Sens. 2021;6:3468–3476. doi: 10.1021/acssensors.1c01437. [DOI] [PubMed] [Google Scholar]
- Sontimuang C., Suedee R., Dickert F. Interdigitated capacitive biosensor based on molecularly imprinted polymer for rapid detection of Hev b1 latex allergen. Anal. Biochem. 2011;410:224–233. doi: 10.1016/j.ab.2010.11.043. [DOI] [PubMed] [Google Scholar]
- Tsouti V., Boutopoulos C., Zergioti I., Chatzandroulis S. Capacitive microsystems for biological sensing. Biosens. Bioelectron. 2011;27:1–11. doi: 10.1016/j.bios.2011.05.047. [DOI] [PubMed] [Google Scholar]
- Wang L., Veselinovic M., Yang L., Geiss B.J., Dandy D.S., Chen T. A sensitive DNA capacitive biosensor using interdigitated electrodes. Biosens. Bioelectron. 2017;87:646–653. doi: 10.1016/j.bios.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Wang Y., Lassiter K., Li Y., Hargis B., Tung S., Berghman L., Bottje W. Interdigitated array microelectrode based impedance immunosensor for detection of avian influenza virus H5N1. Talanta. 2009;79:159–164. doi: 10.1016/j.talanta.2009.03.017. [DOI] [PubMed] [Google Scholar]
- World Health Organization WHO COVID-19: case definitions: updated in public health surveillance for COVID-19, published 16 December 2020. World Health Organization. 2020 [Google Scholar]
- Xie Y., Karki C.B., Du D., Li H., Wang J., Sobitan A., Teng S., Tang Q., Li L. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front. Mol. Biosci. 2020;7:1–14. doi: 10.3389/fmolb.2020.591873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.Y., Fu W.L. Biosensors for hepatitis B virus detection. World J. Gastroenterol. 2014;20:12485–12492. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.