Abstract
The Covid-19 pandemic is the defining global health crisis of our time. Chest X-Rays (CXR) have been an important imaging modality for assisting in the diagnosis and management of hospitalised Covid-19 patients. However, their interpretation is time intensive for radiologists. Accurate computer aided systems can facilitate early diagnosis of Covid-19 and effective triaging. In this paper, we propose a fuzzy logic based deep learning (DL) approach to differentiate between CXR images of patients with Covid-19 pneumonia and with interstitial pneumonias not related to Covid-19. The developed model here, referred to as CovNNet, is used to extract some relevant features from CXR images, combined with fuzzy images generated by a fuzzy edge detection algorithm. Experimental results show that using a combination of CXR and fuzzy features, within a deep learning approach by developing a deep network inputed to a Multilayer Perceptron (MLP), results in a higher classification performance (accuracy rate up to 81%), compared to benchmark deep learning approaches. The approach has been validated through additional datasets which are continously generated due to the spread of the virus and would help triage patients in acute settings. A permutation analysis is carried out, and a simple occlusion methodology for explaining decisions is also proposed. The proposed pipeline can be easily embedded into present clinical decision support systems.
Keywords: Chest X-ray, Convolutional Neural Network, Covid-19, explainable Artificial Intelligence, Fuzzy logic, Portable systems
1. Introduction
Coronavirus disease (Covid-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was first identified in Wuhan, Hubei Province, China in December, 2019 and its rapid worldwide spread has resulted in a global pandemic [1]. Globally there have been nearly 240 million confirmed infections and 4.8 million deaths attributed to the disease according to the COVID-19 Dashboard from John Hopkins University as of 15 October, 2021 [2]. Primary symptoms include fever, fatigue, shortness of breath and loss of taste and/or smell [3]. In some cases, the infection can lead to severe respiratory complications, primarily pneumonia, requiring hospitalisation and often admission to intensive care units (ICU). Unfortunately, the capacity of ICU wards is limited; as a consequence many healthcare systems are being overwhelmed [4]. Many countries around the world have enacted strict physical distancing policies, including national lockdowns. The primary diagnostic tool for Covid-19 is a real-time reverse transcription polymerase chain reaction (RT-PCR). In clinical practice, chest X-ray (CXR) and computed tomography (CT) scans are often employed as screening tools and for detection of Covid-19 pneumonia. CXRs have been employed for many decades as portable tools as they are cost-effective and allowing for a quick bed-side evaluation of patients’ lungs [5]. The obtained images are mostly manually analysed by radiologists, which is both labour and time intensive. Hence, there is much interest around the development of computer-aided diagnosis tools, in particular to assist early detection of Covid-19 related pneumonia from medical images. Lung damages due to Covid-19 can be observed in both CXR and CT images and typically appear as bilateral ground-glass opacification associated with possible consolidation. As such identifying multifocal air-space disease in CXR images could be an indicator of a Covid-19 infection. Recent studies have also shown that features involving the lung hilum, thickening of the lung texture, and pulmonary fibrosis can support diagnosis from CXR images [6].
In recent months, there is growing interest in developing approaches based on Artificial Intelligence (AI) to automate detection of Covid-19 pneumonia from CXR images. In particular, Deep Learning (DL) based techniques have been extensively studied, and proposed as efficient data-driven image processing-based solutions [7], including medical areas [8], [9], [10], [11], [12].
Relevant literature studies that have employed these techniques in CXR images are reviewed and summarised in Table 1 . Wang et al. [13] developed COVID-Net to perform a 3-way classification of patients with Covid-19, non-Covid-19 infection (e.g., viral, bacterial) and no infection, achieving 93.3% accuracy. Heidari et al. [14], employed a 3-way transfer learning VGG16-based convolutional neural network (CNN) to classify preprocessed CXR images of subjects with: pneumonia due to Covid-19, non-Covid-19 pneumonia and healthy controls, reporting accuracy rate up to 94.5%. Similarly, Apostolopoulos et al. [15] exploited pre-trained CNNs (VGG19, MobileNet v2) and used transfer learning to perform a 3-way classification; while, Ismael et al. [16] used pre-trained deep CNN models (ResNet18, ResNet50, ResNet101, VGG16, and VGG19) to classify CXR images of patients affected by Covid-19 and healthy subjects. Zabirul Islam et. [17] combined a CNN (used as feature extraction module) and a long short-term memory (LSTM, used as classifier) to perform a 3-way classification to detect Covid-19 from CXR images. Karthik et al. [18] proposed a channel shuffled dual-branched residual CNN to identify specific pneumonia classes using Covid-19 pneumonia, viral pneumonia, bacterial pneumonia cases and normal CXRs, reporting 99.8% accuracy. Khan et al. [19] proposed CoroNet, a deep CNN based on the pre-trained Xception model to also identify Covid-19 from CXRs. They performed two multi-classifications of: (1) Covid-19 pneumonia vs. bacterial pneumonia vs. viral pneumonia vs. normal and, (2) Covid-19 pneumonia vs. non-Covid-19 pneumonia vs. normal, reporting 89.6% and 95.0% accuracy, respectively. Oh et al. [20] developed a patch-based CNN (i.e., ResNet-18) approach with a few trainable parameters to diagnose Covid-19 from CXR. They performed a 4-way classification, normal vs. bacterial pneumonia vs. tuberculosis vs. viral pneumonia. Ozturl et al. [21] implemented a CNN based DarkNet model and performed binary classification (Covid-19 pneumonia vs. normal) and multi-class classification (Covid-19 pneumonia vs. normal vs. pneumonia), achieving an accuracy of 98.1% and 87.0%, respectively. Minaeea et al. [22] used transfer learning to train ResNet18, ResNet50, SqueezeNet, and DenseNet-121 to classify Covid-19 pneumonia from non-Covid-19 cases (these included normal cases and other diseases as well) from CXR images. Similarly, Boudrioua [23] used deep transfer learning for classifying Covid-19, non-Covid-19 pneumonia and normal cases. Ezzat et al. [24] proposed the gravitational search optimization-DenseNet121-Covid-19 to perform 2-way classification between Covid-19 pneumonia and non-Covid-19 cases (included normal, SARS, ARDS, Pneumocystis, Streptococcus, pneumonia), achieving high performance (93.4% accuracy). Marques et al. [25] developed a CNN by using the EfficientNet architecture to perform both binary (Covid-19 vs. normal) and multi-classification (Covid-19 vs. pneumonia vs. normal) reporting accuracies of 99.6% and 96.7%, respectively. Das et al. [26] proposed a CNN model, termed Truncated Inception Net, which achieved 99.9% accuracy in detecting Covid-19 from combined pneumonia and normal XRs; and, an accuracy of 99.9% in classifying Covid-19 from combined pneumonia, tuberculosis, and normal XRs. Babukarthik et al. [27] used the genetic deep CNN to differentiate between Covid-19 and healthy lungs using CXR images, reporting a very high accuracy (98.8%). Mporas and Naronglerdrit [28] evaluated several pre-trained deep CNNs for discriminating Covid-19 pneumonia from non-Covid-19 cases (including normal lungs, viral and bacterial pneumonia) from CXRs, reporting accuracies up to 99.9%. Recently, Hussain et al. [29] proposed CoroDet, a 22-layer CNN for binary classification (Covid-19 vs. normal) and multi-classifications (Covid-19 vs. normal vs. non-Covid-19 pneumonia and Covid-19 vs. normal vs. non-Covid-19 pneumonia vs. non-Covid-19 bacterial pneumonia) achieving an accuracy rate up to 94.2%. Similarly, Umer et al. [30] developed COVINet, a CNN for performing the 2-way (Covid-19 vs. normal), 3-way (Covid-19 vs. normal vs. virus pneumonia) and 4-way (Covid-19 vs. normal vs. virus pneumonia vs. bacterial pneumonia) classifications. Chakraborty et al. [31] proposed Corona-Nidaan, a deep CNN to detect Covid-19, pneumonia and normal cases, achieving 95% accuracy. Mukherjee et al. [32] designed a shallow CNN to detect Covid-19-positive cases from a non-Covid-19 class, consisting of normal, viral and bacterial pneumonia cases, with an accuracy of 99.69%; whereas, Keles et al. [33] focused on the problem of 3-way classification (Covid-19, viral pneumonia, and normal), reporting a high accuracy of 97.6% with the proposed COV19-ResNet model.
Table 1.
Deep Learning approaches to Covid-19 diagnosis from Chest X-Rays.
| Reference | Method | Classification | Accuracy |
|---|---|---|---|
| Wang et al. [13] | COVID-Net | 3-way (Covid-19, non-Covid-19 pneumonia (e.g., viral, bacterial, etc.), normal) | 93.3% |
| Heidari et al. [14] | Transfer learning VGG16-based CNN | 3-way (Covid-19, non-COVID-19 pneumonia, normal) | 94.5% |
| Apostolopoulos et al. [15] | Pretrained CNNs (i.e., VGG19, MobileNet v2) and transfer learning | 3-way (Covid-19, non-COVID-19 pneumonia, normal) | 96.7% |
| Ismael et al. [16] | Pretrained deep CNNs (ResNet18, ResNet50, ResNet101, VGG16, and VGG19) | 2-way (Covid-19, normal) | 92.6% |
| Islam et. [17] | CNN + LSTM | 3-way (Covid-19, non-COVID-19 pneumonia, normal) | 99.4% |
| Karthik et al. [18] | Dual-branched residual CNN | 4-way (Covid-19, viral pneumonia, bacterial pneumonia, normal) | 97.9% |
| Khan et al. [19] | CoroNet | 4-way (Covid-19 pneumonia, bacterial pneumonia, viral pneumonia, normal) 3-way (Covid-19 pneumonia, non-Covid-19 pneumonia, normal) | 89.6%, 95.0% |
| Oh et al. [20] | ResNet-18 | 4-way (viral pneumonia + Covid-19, bacterial pneumonia, tuberculosis, normal) | 91.9 % |
| Ozturl et al. [21] | DarkNet | 2-way (Covid-19, normal) 3-way (Covid-19, non-Covid-19 pneunomia, normal) | 98.1%, 87.0% |
| Minaeea et al. [22] | Transfer learning based ResNet18, ResNet50, SqueezeNet, and DenseNet-121 | 2-way (Covid-19,non-Covid-19 cases (these included normal cases and other diseases)) | 90.0% |
| Boudrioua [23] | Deep transfer learning based CNNs | 3-way (Covid-19, non-COVID-19 pneumonia, normal) | 99.5% (sensitivity) |
| Ezzat et al. [24] | Gravitational search optimization (GSA) -DenseNet121-Covid-19 | 2-way (Covid-19, non-COVID-19 pneumonia) | 93.4% |
| Marques et al. [25] | CNN + EfficientNet | 2-way (Covid-19, normal) 3-way (Covid-19, non-Covid-19 pneunomia, normal) | 99.6%, 96.7% |
| Das et al. [26] | Truncated Inception Net | 2-way (Covid-19, combined non-Covid-19 pneunomia, normal) 2-way (Covid-19, combined non-Covid-19 pneunomia, Tuberculosis, normal) | 99.9%, 99.9% |
| Babukarthik et al. [27] | Genetic deep CNN | 2-way (Covid-19, normal) | 98.8% |
| Mporas and Naronglerdrit [28] | pre-trained deep CNNs | 2-way (Covid-19, non-COVID-19 pneumonia i.e. normal, viral and bacterial pneumonia) | 99.9% |
| Hussain et al. [29] | CoroDet | 2-way (Covid-19, normal) 3-way (Covid-19, normal, non-Covid-19 pneumonia) 4-way (Covid-19, normal, non-Covid-19 pneumonia, non-Covid-19 bacterial pneumonia) | 99.1%, 94.2%, 91.2% |
| Umer et al. [30] | COVINet | 2-way (Covid-19, normal) 3-way (Covid-19, normal, virus pneumonia) 4-way (Covid-19, normal, virus pneumonia, bacterial pneumonia) | 97%, 90%, 85% |
| Chakraborty [31] | Corona-Nidaan | 3-way (Covid-19, normal, pneumonia,) | 95% |
| Mukherjee [32] | Shallow CNN | 2-way (Covid-19, non-COVID-19 pneumonia i.e. normal, viral and bacterial pneumonia) | 99.7% |
| Keles [33] | COV19-ResNet | 3-way (Covid-19, normal, viral pneumonia) | 97.6% |
Most of the above reported studies carried out multi-classification or binary classification, included healthy individuals and used pre-trained CNNs. Although limited by lower resolution, portable CXRs are increasingly being used for Covid-19 patients, especially in countries with limited access to reliable Covid-19 testing and structural lack of hospitalization [34]. They also avoid the need for decontamination of the radiology room/ward, which is necessary when using for standard X-ray equipment. However, research into detection of disease from portable CXRs is limited and is more challenging due to the low resolution and increased variability in orientation [34]. Furthermore, early disease detection is important for regulating triaging [34]. The aim of this paper is to address the challenge of automated detection of Covid-19 pneumonia from portable CXRs, without taking normal subjects into consideration in the learning procedure. It is indeed evident that the differences between healthy lungs and pneumonia are easier to detect in images than differentiating diverse kinds of disease showing similar effects. Guidelines on the diagnosis of pneumonia due to Covid-19 from portable CXRs are under development; however, according to a recent review, some typical words used by experts when reporting such patients include: patchy, hazy, irregular, widespread ground glass opacities and reticular opacities [34]. Hazy pulmonary opacities in CXR can sometimes be blurred which makes detection more difficult. This is particularly relevant in the case of portable equipment. In order to use DL algorithms to handle such hazy regions and irregularities of portable CXR images, there is a need for formal methods. In addition, although several DL-based approaches have been successfully applied for Covid-19 detection by analyzing CXR images, it is worth mentioning that, from a diagnostic point of view, it is essential to evaluate the extent of the lung area affected by Covid-19 in order to determine the optimal drug administration dose. However, these extensions are not always perfectly defined and often have areas characterized by shades that make the edges uncertain and/or imprecise. In this context, Fuzzy Logic (FL)-based image processors are universally recognized as reliable tools to handle such problems. FL is a computational paradigm based on a multi-valued approach that is designed to manage problems that are impacted by uncertainty, irregularities and/or inaccuracy [35]. Thus, in order to handle vagueness, ambiguities and uncertainties that are particularly significant in portable CXR images, a fuzzy edge detection procedure is introduced. Indeed, preprocessing CXR images using fuzzy logic improves the edge detection step, where the boundaries are not well defined or are characterized by locally variable shades [36]. To this end, here, a hybrid approach based on Fuzzy Logic and a Deep Learning is proposed.
In particular, a customized deep CNN, herein termed CovNNet, has been designed to perform binary classification between pneumonia due to Covid-19 and Interstitial Pneumonias not due to Covid-19 (referred to as “No-Covid-19”). A hybrid architecture is used, where a block features is extracted from CovNNet trained on raw CXR images, and a second block of features is extracted from images pre-processed by fuzzy algorithms. The two blocks are then combined in a unique vector to train a multilayer feedforward neural network.
The major contributions of the paper can be summarized as follows:
-
•
acquisition and processing of CXRs images acquired by means of portable devices and recorded from patients diagnosed with Covid-19 pneumonia and non-Covid-19 interstitial pneumonias.
-
•
development of a fuzzy-edge detection procedure to overcome uncertainties and ambiguities in CXR raw images.
-
•
development of an innovative method based on the use of DL and fuzzy approaches to design an effective classification system for early detection of Covid-19 from portable CXRs.
-
•
development of CovNNet, i.e. a neural network which is able to extract the most relevant features from raw CXRs (i.e., CXR-features) and pre-processed fuzzy images (i.e., fuzzy-features). These are used as input to the classification network to discriminate between CXR images of patients affected by Covid-19 and CXR images of patients affected by non-Covid-19 interstitial pneumonias.
-
•
interpretation of the achieved results by means of an explainable Artificial Intelligence approach (xAI) to support clinicians.
-
•
development of a fuzzy-assisted DL system with potential future applications in clinical practice.
The rest of the paper is organized as follows. Section 2 describes the database used. Section 3 introduces the methodology, including the image pre-processing step, the fuzzy-based approaches and CovNNet. It also includes a permutation analysis. Section 4 reports the achieved results. Section 5, 6 consist of the discussion and conclusion, respectively.
2. Experimental database
For this study, a slightly unbalanced database of portable CXR images comprising a total of 121 images (one per subject), with 64 images belonging to patients diagnosed with Covid-19 pneumonia (aged between 40 and 90 years old) and 57 belonging to patients diagnosed with non-Covid-19 interstitial pneumonias (aged between 30 and 90 years old), were collected at the Advanced Diagnostic and Therapeutic Technology Department of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria, Italy, from January 2018 to May 2020. Furthermore, an additional database of 34 Covid-19 images was made available for validating purposes. The GOM Ethics Committee approved our study in January 2021. Patients with confirmed Covid-19 infection via RT-PCR and patients with confirmed pneumonia not due to Covid-19 were identified and enrolled in the study. All patients underwent CXR and the images were retrieved from the picture archiving and communication system, and were standardly anonymized. The main CXR findings in Covid-19 pneumonia included multifocal bilateral ground glass opacities with patchy consolidations, prominent peripheral sub-pleural distribution, and predilection of the posterior part of the lower lobe. The images of No-Covid-19 pneumonia cases were variable and some included peripheral bilateral ground glass opacities.
3. Methodology
3.1. Chest X-ray pre-processing
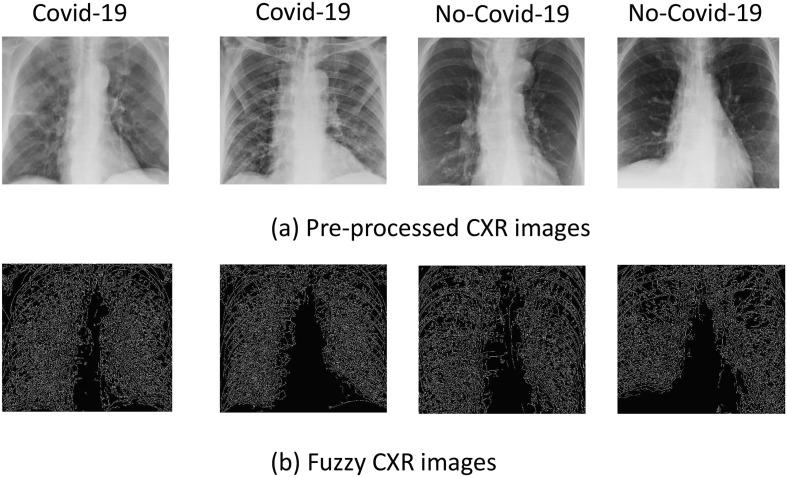
Fig. 1 shows examples of CXR images of some patients with Covid-19 pneumonia and some patients with interstitial pneumonias (No-Covid-19). Retrieved CXRs included text and empty black areas around the image which may negatively impact classification. As such each CXR image was manually processed by an expert operator to only include the entire lung region and to remove any text or black edges to produce an image of a size 800 × 900 pixels. It is important to note that some of the patients had external devices (e. g., pacemaker) which if in the lung region were not removed.
Fig. 1.
Examples of original (a) and pre-processed (b) CXR images.
3.2. A fuzzy-enhanced deep learning approach
3.2.1. Fractal dimension
CXR images, especially those from portable equipment, invariably show a blurred texture that may hinder interpretation by clinicians. In this sub-section, the fuzzy logic paradigm is introduced and takes advantage of introducing a nonlinear model which is able to handle uncertainties and ambiguities in raw images. From a mathematical viewpoint, the strongly irregular edges of the objects within the image can be characterized as fractals. To facilitate the description of structural patterns of the CXR image, a fractal analysis is then carried out. The underlying fractality of the image can be expressed numerically by its fractal dimension (FD): specifically, high FD values correspond to higher complexity degrees of the pattern being taken into account. Here, FD is measured by using the box-counting technique [37], defined as where 1D2 and M is the least number of distinct boxes (with edge 0) able to cover the CXR image completely. The FD is the slope in the diagram.
3.2.2. Fuzzy edge detection
In the previous sub-section, we argued that fractality can represent a formal method to characterize CXR images, particularly from portable devices. Actually, a computational approach based on fuzzy logic can also be used to handle ambiguity and/or vagueness in the images by introducing suitable adaptive membership functions, to characterize the fuzziness of the analyzed image. Here, a formal algorithm to extract fuzzy features from the images is presented. The aim is to obtain a set of features that will be used as additional inputs to the machine learning processor. These fuzzy features will presumably add information to the features extracted by a fully data-driven approach, particularly considering the different textures of the two classes and the roughness of the boundaries. Specifically, a fuzzy edge detection procedure based on a fuzzy entropy formulation is introduced. The procedure consists of the following steps.
Step 1. A 2D grayscale image I, comprising an array of pixels aij, each represented by a level of brightness ranging from [0–255], is normalized so that /255[0,1]. Once the normalized images (here denoted as ) are fuzzified by a fuzzy membership function , a fuzzy template (FT) procedure over is applied by placing the centre of the fuzzy template at each pixel over .
Step 2. The fuzzy divergence value D, between each element of (i.e., an image window) and the fuzzy template is computed. The minimum value is then selected as:
| (1) |
where
| (2) |
and
| (3) |
Step 3. Step 2 is repeated for all of the fuzzy templates (there are 16 templates in order to take into account each possible direction of the edge).
Step 4. The maximum value among all 16 minimum divergences values is selected.
Step 5. The fuzzy template corresponding to the maximum value is centered over the image taken into account.
Step 6. The procedure is repeated from Step 2 to Step 5, for all the pixels positions, so that a fuzzy divergence matrix is constructed.
Step 7. The thresholding of the fuzzy divergence matrix is performed by exploiting both fuzzy entropy and fuzzy divergence. Then, a fuzzy thresholding procedure based on the minimization of fuzzy entropy and fuzzy divergence is developed. In particular, once the fuzzy divergence matrix is obtained, for each threshold , a square matrix (-sized), , located on is set. By considering another square matrix (of the same size), , located on another pixel , their distance, , is computed by fuzzy divergence, . Finally, the average value of all fuzzy divergences is determined by moving in all possible positions is computed. Furthermore, the average value achieved by moving in all possible positions (indicated by ) is also estimated. This procedure is repeated for ()-square windows obtaining . Hence, the fuzzy entropy (FE) is computable as the negative natural logarithm () of the conditional probability that two similar -dimensional patterns remain similar for (). Thus:
| (4) |
from which
| (5) |
Taking into account (5), is achieved by
| (6) |
Specifically, inspired by [38] where a two-dimensional entropy was exploited for assessing image texture through irregularity, the new procedure here exploits the fuzzy divergence concept as a distance between two fuzzified images, taking into account the distance between the first image and the second one (and vice versa). Finally, for fuzzifying each inspected image, an S-shaped membership function has been devised by exploiting the concept of fuzzy entropy minimization coupled to noise reduction [36], [39], [40]. In order to fuzzify , an S-shaped membership function is used. Note that it depends on three shape parameters , and (with ), which are set adaptively. For each pixel of can be defined as:
| (7) |
Fig. 2 shows the effect of the fuzzification and the related fuzzy edge detection procedure on sample CXR images.
Fig. 2.
Examples of pre-processed CXR images (a) and fuzzy CXR images (b), obtained by applying the proposed fuzzy edge detection procedure.
3.2.3. A convolutional neural network for Covid-19 detection (CovNNet)
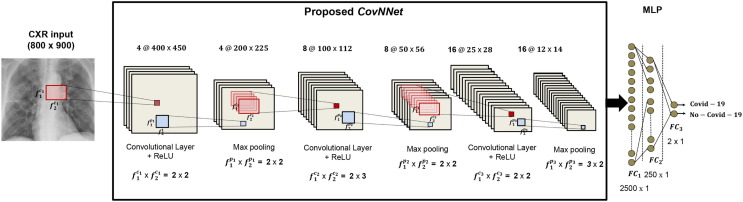
CXR images, pre-processed according to the algorithm described in Section 3.1, and sized 800 × 900, are used as input to a custom CNN, referred to as CovNNet. It consists of three convolutional layers each followed by a rectified linear unit (ReLU) activation function layer, three max pooling layers (subsampling level), two fully connected layers and a final softmax layer for performing binary classification Covid-19 vs. No-Covid-19. The architecture is illustrated in Fig. 3 . The first convolutional layer has four filters sized 2 × 2, stride 2, padding parameter equal to 0 and generates four features maps sized 400 × 450; the first max pooling layer has a filter of size 2 × 2, stride 2 and reduces the input features space to 200 × 225. The second convolutional layer has eight filters sized 2 × 3, stride 2, padding parameter equal to zero and results in eight feature maps of dimension 100 × 112; whereas, the subsequent max pooling stage is composed of a filter size 2 × 2, stride of 2, producing eight features maps sized 50 × 56. Finally, the third convolutional layer, contains sixteen filters sized 2 × 2 that moves with a step size 2, thus generating sixteen feature maps sized 25 × 28. The last max pooling layer down-samples the input maps from 25 × 28 to 12 × 14 by using a filter sized 3 × 2 and a stride 2. The extracted features (denoted to as CXR-features) are then vectorized (12 × 14 × 16 = 2688) and used as input to MLP with two hidden fully connected layers consisting of 2500 and 250 units, respectively, and a softmax output layer. The proposed CovNNet was designed and implemented in MATLAB R2019a. The training procedure was performed by using the Adaptive Momentum (Adam) estimation algorithm with default hyper-parameters (in this case, first moment exponential decay = 0.9, second moment exponential decay = 0.999, learning rate = 0.001, = 10−8) on a workstation with Intel Core i7-8700 K CPU 3.70 GHz and 64 GB RAM using two NVIDIA GeForce RTX 2080 Ti graphics processing units (GPUs). CovNNet was trained for about 100 iterations, until the convergence of the loss function. It is worth noting that the topology of the proposed CNN as well the learning parameters were set up after several experimental tests with a trial and error strategy. Different CNN configurations were developed and tested by iteratively modifying the number of convolutional, ReLU and max-pooling layers; the number and size of filters; and the number of hidden layers and neurons in the MLP. More specifically, it was observed that a small number of processing layers with a high number of filters were not able to extract discriminating features from input CXR images, causing a reduction in performance. Hence, we decided to make the CNN deeper, keeping a limited number of filters of small dimension. The best classification score was achieved with the configuration reported in Fig. 3, comprising three modules of convolutional, ReLU and max pooling layers, followed by a 2-hidden MLP with a softmax output layer for binary classification purposes. Two approaches were used:
-
•
Approach 1 for concatenation of CXR and fuzzy images
Given a subject, the CXR and the corresponding fuzzy image (produced by the fuzzy edge detection methodology), are arranged in a volume sized × × , where = 800 and = 900 represent the height and width of the CXR/fuzzy image, respectively; while, = 2 denotes the number of images stratified (i.e., the CXR and the related fuzzy image). Hence, the volumetric data (sized 800 × 900 × 2), are used to train and test the proposed CovNNet (described in Section 3.2.3) as shown in Fig. 4 .
-
•
Approach 2 for concatenation of CXR and fuzzy features
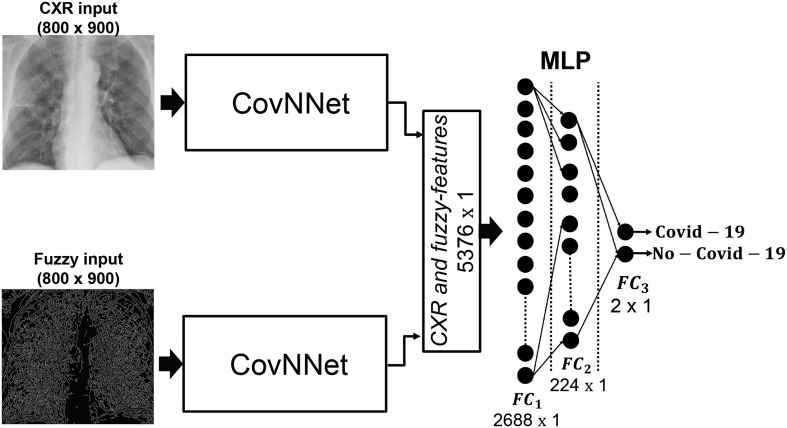
Given a subject, the CXR and the corresponding fuzzy image are used as input to the proposed CovNNet in order to extract the most relevant features directly from CXR and fuzzy images. The extracted features denoted to as CXR-features sized 1 × 2688 and the fuzzy-features sized 1 × 2688, respectively, are concatenated into a single vector sized 1 × 5376 (i.e., CXR and fuzzy features vector) and further classified by means of a standard MLP, that includes two hidden layers of 2688 and 224 neurons, respectively, and an output softmax layer, as shown in Fig. 5 .
Fig. 3.
Lay-out of the Covid-19 vs. No-Covid-19 pneumonia classification system based on a CNN approach (CovNNet)..
Fig. 4.
Fuzzy-enhanced CNN classification system (approach 1): CXR and fuzzy images of the same patient are stratified in a volume of data × × = 800 × 900 × 2 and used as input to the proposed CovNNet followed by a standard MLP for performing the 2-way classification task: Covid-19 vs. No-Covid-19.
Fig. 5.
Fuzzy-enhanced CNN classification system (approach 2): CXR and fuzzy images of the same patient are used as input to two CovNNet here employed to automatically extracts the most relevant CXR-features from CXR images and fuzzy-features from fuzzy images. Such features are concatenated and used as input to a standard MLP for performing the 2-way classification task: Covid-19 vs. No-Covid-19.
3.3. Analysis of fuzzy CNN models for Covid-19 detection via explainable Artificial Intelligence (xAI)
Machine learning models are generally opaque and suffer from lack of transparency and explainability. Such opaqueness, commonly referred to as black-box behaviour, is the subject of the growing domain of explainable Artificial Intelligence (xAI). The term explainability denotes any technical development of transparency in AI methodologies that is capable of highlighting which part contributed to achieve a specific performance and possibly why [41]. In medical field, the notion of causability defined by Holzinger et al. [42], also needs to be introduced. Causability measures the quality of explanations, that is, the level of causal understanding acquired by a user from an explanation, achieved by xAI. Hence, it is worth highlighting that explainability is a characteristic of a model whilst causability is a characteristic of a person. Mapping such concepts entails a new conceptualization of human-AI interface [41]. However, in the medical field in particular, there is another challenge required to be dealt with, namely, the integration of heterogeneous data and features in a multi-modal (MM) fashion. A MM strategy is also known to contribute to clinicians’ decision making. In this sense, here, we propose a hybrid representation space based on the extraction and concatenation of different features (i.e., CXR and fuzzy features) in an attempt to enhance the Covid-19 classification performance. Furthermore, since a technical explanation of the system is particularly important in clinical settings, a sensitivity analysis is carried out to infer information about the relevance of the images’ regions, allowing for suitable interpretability and more informed decision-making by clinicians.
In particular, an occlusion sensitivity analysis (OSA) was proposed to justify the behaviour of the proposed CNN [43]. This procedure consists of studying the deterioration of the classification performance, when systematically occluding parts of the input image through a moving mask. Given an input image and the corresponding class predicted by the previously trained CNN, for every successive location of the occluding mask, the classification is performed again by feeding the occluded image into the trained CNN and measuring the inducted modifications in the classification score. These changes can be used to plot a heat or saliency map. Every pixel in the saliency map is generated starting from the corresponding pixel in the original input image. The coloration of the pixels in the saliency map range from blue (low relevance) to red (high relevance). When a given area in the input image is occluded by the patch and the ability of the network to correctly classify the image gets worse, such an area is recognised as relevant for classification and hence associated with a high saliency, and vice versa.
3.4. Performance evaluation metrics
In this study, the available experimental database was used to train and test the proposed CNN enhanced through the fuzzy based approaches. The performance of the whole systems were reported using standard metrics. Specifically, sensitivity, specificity, positive predicted value (PPV), negative predicted value (NPV) and accuracy were measured as:
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
where TP, TN, FP and FN stand for true positive, true negative, false positive and false negative, respectively. In this work, TP refers to the images recorded from patients affected by Covid-19; TN refers to the images recorded from patients affected by No-Covid-19 interstitial pneumonias; FP are the No-Covid-19 samples misclassified as Covid-19; whereas, FN are Covid-19 samples erroneously classified as No-Covid-19. In order to avoid overfitting, the k-fold cross validation technique was used. The database was randomly partitioned into 8 groups (i.e., k = 8). Each test set included 30% of the data, and the remaining 70% was used for training.
3.5. Permutation analysis
A permutation-based p-value statistical analysis is performed for evaluating the reliability of results. The permutation test estimates the p-value under a specific null hypothesis for assessing the probability that the performance metric would be achieved by chance, due to the assumption of randomness of the k-fold cross-validation technique. The null hypothesis supposes the independence between labels and features. To this end, permutation distributions are generated by destroying the association between target and input, i.e., by shuffling the labels and estimating the performance metric. The p-value is estimated as the proportion of times that the performance metric, achieved by using shuffled label data (pm), performs equally or better than the performance metric calculated with the original un-shuffled labels data ():
| (13) |
where is the indicator function and is the number of permutations. The null hypothesis is rejected when p-value (typically = 0.5). It is important to note that all possible permutation distributions should be generated to compute the exact p-value. However, here, due to computational constraints, the permutation analysis is performed on = 100 permutations.
4. Results
4.1. Classification performance
Table 2 shows the classification results achieved by the developed CovNNet and the proposed fuzzy enhanced DL approaches. Good discrimination performance were observed by the proposed CovNNet when CXRs were used as input, achieving sensitivity of 76.021.4%, specificity of 75.015.6%, PPV of 83.58.7%, NPV of 72.215.0% and accuracy rate up to 75.69.9%. In the fuzzy-enhanced DL approaches, a reduction of performance was reported when concatenating the CXR and fuzzy images in a single volume of data (Approach 1). This achieved sensitivity of 68.030.6%, specificity of 66.134.2%, PPV of 81.515.3%, NPV of 62.414.6% and accuracy rate up to 67.212.5%. In contrast, the combination of the extracted CXR and fuzzy features from CovNNet when CXR and the related fuzzy images were used as input (Approach 2), reported the highest classification performance, with regard to sensitivity (82.511.9%), specificity (78.66.9%), PPV (85.24.5%), NPV (77.110.4%) and accuracy (80.96.2%).
Table 2.
Classification performance in terms of sensitivity, specificity, PPV, NPV and accuracy of the proposed models. Best results are reported in boldface.
| Method | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|
| CovNNet | 76.021.4% | 75.015.6% | 83.58.7% | 72.215.0% | 75.69.9% |
| Fuzzy-CovNNet (Approach 1) | 68.030.6% | 66.134.2% | 81.515.3% | 62.414.6% | 67.212.5% |
| Fuzzy-CovNNet (Approach 2) | 82.511.9% | 78.66.9% | 85.24.5% | 77.110.41% | 80.96.2% |
It is important to note that the reported results were achieved by applying the 8-folds cross validation method and were expressed as mean values standard deviation. The Approach 2 also clearly reduces the variances of the estimated performance metrics. Table 3 summarizes the computing cost required to train each model. It is worth noting that, although the proposed fuzzy based DL system (Approach 2) is the most time-consuming in terms of training-time when compared with other benchmark approaches, it took approximately only 50 s.
Table 3.
Computing cost of each algorithm.
| Method | Time Cost |
|---|---|
| CovNNet | 10 s |
| Fuzzy-CovNNet (Approach 1) | 18 s |
| Fuzzy-CovNNet (Approach 2) | 50 s |
Nevertheless, in order to demonstrate that the proposed fuzzy enhanced DL-based framework was statistically significant the p-value permutation test was computed. As reported in Section 3.5, = 100 permutations were generated and for each iteration the accuracy performance metric was estimated. p-value was calculated according to Eq. (13) by counting the number of accuracies equal or greater than 80.9%. Simulation results demonstrated that p-value 0.05. Hence, the null hypothesis was rejected leading to the conclusion that the proposed system was statistically significant. Furthermore, in order to assess the validity of the proposed fuzzy-enhanced DL system (Approach 2, Fig. 5) a new set of 34 Covid-19 CXR images was used as input. It is to be noted that such set of images were not included neither in the train set nor in the test set. Results showed that the proposed fuzzy-enhanced DL network was able to successfully classify the new images reporting only 3 false negatives.
4.2. Explainable predictions of Covid-19
The input images (sized 800 × 900) are systematically occluded by means of a 160 × 180 pixel grey mask sliding over the input image with a vertical and horizontal stride of 80 pixels (values corresponding to 20% and 10% of the image size, respectively). For each position of the mask, the 2-way discrimination task (i.e. Covid-19 vs No-Covid-19 image) was performed through the proposed pre-trained CNN (Section 3.2.3). Fig. 6 shows examples of saliency maps obtained for the test images (run with the best accuracy in the k-fold cross-validation process). Each saliency map was superimposed on the grey scale CXR image. Generally, the lung area in No-Covid-19 images is associated with low saliency (blue coloration), whereas the same area is generally associated with a relatively high saliency in Covid-19 images.
Fig. 6.
Saliency maps obtained for sample images that were correctly classified by the CNN. Each saliency map is overlapped to the corresponding RX image. The coloration of the pixels in the saliency map ranges from blue (low relevance) to red (high relevance). The true class (Covid-19/No-Covid-19) can be read in the title displayed on top of every image.
5. Discussion
This study explored the potential of deep learning and fuzzy techniques to enable clinicians differentiate between Covid-19 and infectious pneumonias (No-Covid-19) based on portable CXRs. The dataset used consisted of CXR images of 64 patients diagnosed with Covid-19 and 57 patients with interstitial pneumonias, provided by the Advanced Diagnostic and Therapeutic Technology Department of the Grande Ospedale Metropolitano (GOM) of Reggio Calabria, Italy. Despite some state-of-the-art works also considering the “normal” category, discriminating healthy lungs from pneumonia would reasonably be an easy task for any classification system but would not be necessary for doctors in clinical practice. Furthermore, CXR images analysed here, were recorded using portable devices with the possibility of making recordings directly at home. It is worth mentioning that portable CXRs are not usually required or necessary in clinical triage, however, in this difficult situation of the pandemic emergency, portable CXRs are planned and reserved only for those subjects that show symptoms of Covid-19. In addition, in real-world scenario, healthy individuals undergo X-ray (XR) examinations in Radiology Department with dedicated XR unit in a standing position. Hence, the quality of XR images may result different than those acquired by bedside XR examinations of hospitalized patients. Further, bedside CXR can show clinical signs of stasis (bronchial texture accentuation) and non-real size of cardiac shadow. Therefore, collecting normal CXRs from portable diagnostics for performing a 3-way classification study is very difficult. Furthermore, from a strict DL perspective, as the automatic extraction carries out features discriminating the classes, the presence of strong differences from normal to pneumonia CXRs will reduce the efficiency in differentiating the two diseases as the system will favor the difference with normal cases. In other words, the learning procedure would be biased
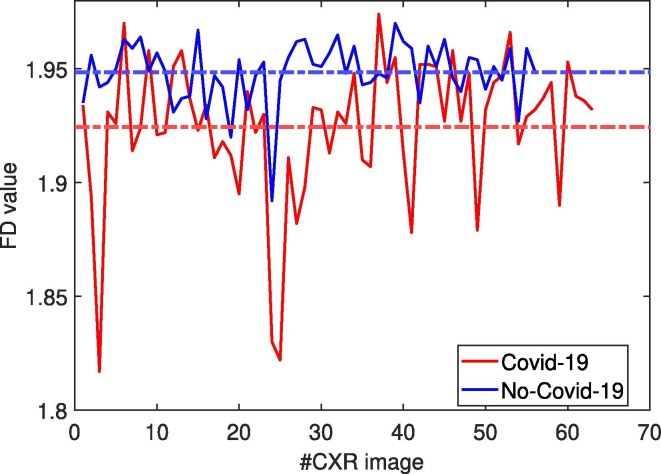
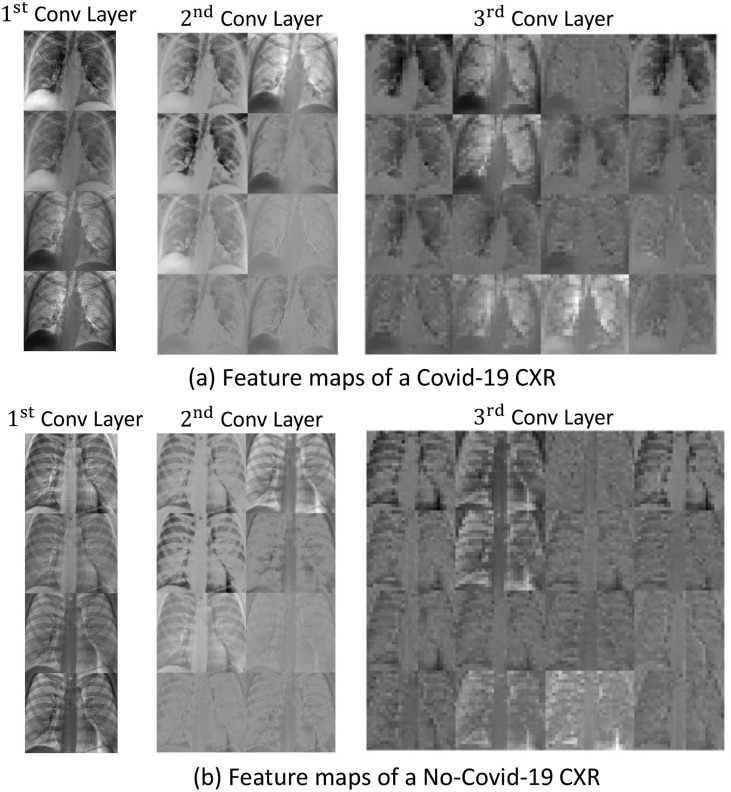
It was noted that the various regions within the images show blurred and irregular contours, which may complicate the diagnosis. For coping with this aspect, the concept of fractality was introduced to formally characterize both the boundaries of the regions and the texture of the whole image. Fig. 7 show the distributions of the FD values for the Covid-19 and No-Covid-19 images, respectively. It is observed that it is not possible to easily separate the two classes by means of a simple threshold, although the mean values of the two distributions are quite different. Hence, an enhanced fuzzy based deep learning strategy was employed. Specifically, a custom CNN, referred as CovNNet, was developed to receive as input Covid-19 and No-Covid-19 CXR images and learn the most relevant Covid-19 and No-Covid-19 features from CXR images. Fig. 8 shows examples of the features learned by CovNNet on a Covid-19 and No-Covid-19 CXR image extracted from the first, second and third convolutional layer. Encouraged by the promising results achieved, a hybrid fuzzy and deep learning framework was introduced. In particular, two strategies were developed: one used CXR and fuzzy images stratified in a volume of data as input to the customized CovNNet; a second one used CXR and fuzzy images to train the proposed CovNNet in order to extract the most significant features denoted as CXR-features and fuzzy-features, respectively. Experimental results reported that the concatenation of such CXR and fuzzy features used as input to a standard MLP, achieved the highest performance in classifying Covid-19 vs. No-Covid-19 (accuracy rate up to 81%). Moreover, 34 additional Covid-19 CXR images were used to validate the proposed fuzzy-enhanced DL approach. 31 images were indeed correctly classified as belonging to Covid-19 group and only 3 images were missclassified. To the best of our knowledge, this is the first work that attempts to develop a fuzzy-enhanced DL framework for detecting pneumonia due to Covid-19, and differentiating it from interstitial pneumonias using portable CXR images. Additionally, in an attempt to provide an explanation of the achieved results a simple xAI was also used. Specifically, OSA was carried out to highlight the area of CXR image mostly involved in discriminating between Covid-19 and No-Covid-19 classes. As can be seen in Fig. 6 most of the correctly classified Covid-19 CXR images showed high saliency (red coloration) in the lung area and vice versa.
Fig. 7.
Distribution of the FD values for Covid-19 (red) and No-Covid-19 (blue) CXR images. Dashed lines represent the average FD of Covid-19 and No-Covid-19 class.
Fig. 8.
Feature maps learned by the three convolutional layers of CovNNet on a Covid-19 (a) and No-Covid-19 CXR image (b). Note that the convolutional layers generate four, eight and sixteen feature maps sized 400 × 450, 100 × 112 and 25 × 28, respectively. The learning procedure seems to assign a highest resolution to the feature maps generated from Covid-19 images. Some feature maps are evidently devoted to detect the zones where lung hilum, thickening of the lung texture, pulmonary fibrosis are present, some others simply highlight diffuse opacities bilaterally.
Despite the encouraging results, this study has a number of limitations. The primary limitation was the small size of the dataset (121 CXR images) used to evaluate our approach. As expected, the quality of portable CXRs was variable due to rotation and patient position. In addition, the No-Covid-19 class only included acute interstitial pneumonias. In future, learning from a larger linked dataset will help improve classification performance and allow for benchmarking.
6. Conclusion and future work
In this study, a fuzzy enhanced deep learning-based framework was proposed to differentiate between CXRs of Covid-19 pneumonia and interstitial pneumonias not due to Covid-19. CXR and fuzzy images generated by using a formal fuzzy edge detection method were used as an input to the developed CovNNet model and the most significant features were automatically extracted. Experimental results showed that by mixing CXR and fuzzy-features, higher classification performance was achieved, with an accuracy rate of up to 81%. It is worth mentioning that recent performance measures proposed in [44], that are particularly suitable for imbalanced datasets (such as those used in this work), will be explored in order to further improve the understanding and interpretation of our developed model. Moreover, further works include: integration of recent face mask detection method for Covid-19 prevention and control in public [45]; validation of the approach by means of larger and linked datasets to develop systems for triaging patients in acute settings and integration with other state-of-the-art hybrid AI approaches, supervised and unsupervised models (e.g. [46], [47], [48], [49], [50], [51]). Finally, since medical datasets are often uncertain and incomplete [52], the validity of achieved results using automatic machine learning algorithms may be questionable. Hence, hybrid interactions between human intelligence and machine intelligence can be of great importance to enhance the knowledge discovery process. To this end, interactive machine learning and in particular human-in-the-learning-loop based strategies [53], [54] will be investigated. In addition, since the European Commission in its the recent White Paper on AI points out the “European Approach” to AI with the intention of EU legislation beging to preserve human dignity and privacy protection especially in the medical field, an in-depth analysis of legal issues in the medical AI will be also taken into account [55].
CRediT authorship contribution statement
Cosimo Ieracitano: Conceptualization, Methodology, Investigation, Implementation, Software, Validation, Visualization, Formal analysis, Writing – original draft, Writing-review-editing. Nadia Mammone: Conceptualization, Methodology, Investigation, Formal-analysis, Writing – original draft, Writing – review & editing. Mario Versaci: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Giuseppe Varone: Data curation, Writing – original draft, Writing – review & editing. Abder-Rahman Ali: Formal-analysis, Writing – original draft, Writing – review & editing. Antonio Armentano: Data curation, Resources, Writing – review & editing. Grazia Calabrese: Data curation, Resources, Writing – review & editing. Anna Ferrarelli: Data curation, Resources, Writing – review & editing. Lorena Turano: Data curation, Resources, Writing – review & editing. Carmela Tebala: Data curation, Resources, Writing – review & editing. Zain Hussain: Visualization, Writing – review & editing. Zakariya Sheikh: Visualization, Writing – review & editing. Aziz Sheikh: Visualization, Writing – review & editing. Giuseppe Sceni: Investigation, Data curation, Resources, Writing – review & editing. Amir Hussain: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. Francesco Carlo Morabito: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing, Project administration, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to thank the anonymous reviewers for their insightful comments and suggestions. This work was supported by POR Calabria FESR FSE 2014–2020 – Grant C39B18000080002, PON 2014–2020, COGITO project – Grant Ref. ARS0100836. Hussain would like to acknowledge the support of the UK Engineering and Physical Sciences Research Council (EPSRC) – Grants Ref. EP/M026981/1, EP/T021063/1, EP/T024917/1.
Biographies

Cosimo Ieracitano received the M.Eng. (summa cum laude) and Ph.D. (with additional label of Doctor Europaeus) degrees from the University Mediterranea of Reggio Calabria (UNIRC), Italy, in 2013 and 2019, respectively. He is currently working as an Assistant Professor at the DICEAM Department of the same University (UNIRC). Dr. Ieracitano is also Honorary Senior Postdoctoral Research Fellow at the Edinburgh Napier University (UK) and at the Nottingham Trent University (UK), since 2019. Formerly, he was a Research Fellow at the University Mediterranea of Reggio Calabria, a Visiting Ph.D. Fellow at the University of Stirling (UK) and a Visiting Master Student at ETH Zurich (Switzerland). He has co-authored one international patent and has publications in peer reviewed international journals/conference proceedings in several fields of engineering: artificial intelligence, biomedical signal processing, brain computer interface, information theory and material informatics. He is actively involved in organizing international conferences, serving as General Chair for the 2022 International Conference of Applied Intelligence and Informatics (AII2022), Local Arrangements Chair for the IEEE WCCI2020 (the world’s largest IEEE CIS technical event in computational intelligence), Publicity Chair for the AII2021, IEEE EdgeDL2021 and IEEE DASH2020. He is Lead Guest Editor for Cognitive Computation and Sensors MDPI. Dr. Ieracitano is also Associate Editor for the International Journal of Electronics and Communications and Academic Editor for the Journal of Healthcare Engineering.

Nadia Mammone is currently an Assistant Professor at the Mediterranea University of Reggio Calabria (Italy), lecturer in Electrical Engineering and Neural Engineering. From 2014 to 2018 she was the Principal Investigator of a research project on advanced EEG processing, funded by the Italian Ministry of Health, at IRCCS Centro Neurolesi Bonino-Pulejo in Messina (Italy). Formerly, she was a Post-Doc Fellow in Biomedical and Electrical Engineering at the Mediterranea University of Reggio Calabria. She received her PhD in Informatics, Biomedical and Telecommunications Engineering in 2007 from the same University, with a dissertation that was awarded the Caianiello Prize from the Italian Neural Networks Society (SIREN). She was a visiting PhD Fellow at the Computational NeuroEngineering Laboratory (CNEL, University of Florida, USA) in 2005 and 2008 and a visiting Post-Doc Fellow at the Communication and Signal Processing Research Group (Department of Electrical and Electronic Engineering, Imperial College, London, UK) in 2015. Between 2008 and 2013, she was Adjunct Professor of “Neural Networks and Fuzzy Systems” at the University “Mediterranea” of Reggio Calabria, of “Electronics and Instrumentation for laboratories of Neurophysio-pathology” and of “Biomedical Signal Processing” at the Faculty of Medicine (University of Catanzaro, Italy). Her research interests include brain computer interfaces, neural and adaptive systems, biomedical signal processing, information and complex network theory.

Mario Versaci (Senior Member, IEEE) received the degree in civil engineering from the Mediterranea University of Reggio Calabria, Italy, in 1994, the Ph.D. degree in electronic engineering from the Mediterranea University of Reggio Calabria, in 1999, and the Laurea degree in mathematics from the University of Messina, Italy, in 2013. He currently serves as an Associate Professor of electrical engineering with the Mediterranea University of Reggio Calabria, where he serves as the Scientific Head of the NDT/NDE Laboratory. His research interests include soft computing techniques for NDT/NDE and image processing. He is a member of the Italian Society for Industrial and Applied Mathematics.

Giuseppe Varone received the Ph.D. degree in New Magnetic Resonance Imaging and Bioinformatics Techniques Applied to Neurosciences from the Magna Graecia University of Catanzaro, Italy, 2021. He got a master’s degree (summa cum laude) in biomedical engineering in 2016, a master’s in biomedical electronics in 2014, and a bachelor’s degree in computer science in 2012. He is a researcher fellow at the University of Chieti-Pescara, Italy. Recently, he was at Aalto University and the Eberhard-Karls University of Tübingen as a Visiting Postdoctoral Fellow in the framework of the Connect to Brain European Project for TMS-EEG experiments. His current research interests include Data acquisition, transcranial magnetic stimulation (TMS), machine learning, biomedical signal processing, and the development of hardware/software tools for real-time TMS data analysis.

Abder-Rahman Ali received the B.Sc. degree in computer science from the University of Jordan, Jordan, the M.Sc. degree in software engineering from DePaul University, USA. He is currently pursuing a Ph.D. in Computing Science from the University of Stirling, U.K., where he worked on developing machine learning and image processing-based approaches for the early detection of melanoma. His research interest is in medical image analysis started during his master’s degree, where he conducted research at the Intelligent Multimedia Processing Laboratory, DePaul University, related to content-based image retrieval for lung nodules. His research interest is also in melanoma started after doing an Internship at RWTH Aachen University, Germany. He is very passionate about the idea of developing solutions that would aid in the early detection of cancer.

Antonio Armentano is the Head of the Department of Diagnostic and Therapeutic Techniques and Director of the Department of Neuroradiology at the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). His field of interest lies especially in the diagnosis and therapy of neurological diseases. He uses MRI with advanced technologies of Spectroscopy, Perfusion and Diffusion and CT with advanced software dedicated to the study of neurovascular diseases. He has undertaken, in the Department that he directs, a new line of study and deepening of radiomics and radiogenomics methods on lung and brain tumors.

Grazia Calabrese is a radiologist at the Radiology Complex Operating Unit of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). Her work includes predominantly abdominal and thoracic emergencies, as well as elective activities relating to the oncology, breast and musculoskeletal fields, through the integrated use of available technologies (X-Ray, US, CT and MR).In the last few years her work has also extended to the study of Covid-19 patients using chest X-Ray and CT, in collaboration with other specialists.

Anna Ferrarelli is a radiologist at the Radiology Complex Operating Unit of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). Her work includes predominantly abdominal and thoracic emergencies, as well as elective activities relating to the oncology, breast and musculoskeletal fields, through the integrated use of available technologies (X-Ray, US, CT and MR).In the last few years her work has also extended to the study of Covid-19 patients using chest X-Ray and CT, in collaboration with other specialists.

Lorena Turano is a radiologist at the Radiology Complex Operating Unit of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). Her work includes predominantly abdominal and thoracic emergencies, as well as elective activities relating to the oncology, breast and musculoskeletal fields, through the integrated use of available technologies (X-Ray, US, CT and MR).In the last few years her work has also extended to the study of Covid-19 patients using chest X-Ray and CT, in collaboration with other specialists.

Carmela Tebala is a radiologist at the Radiology Complex Operating Unit of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). Her work includes predominantly abdominal and thoracic emergencies, as well as elective activities relating to the oncology, breast and musculoskeletal fields, through the integrated use of available technologies (X-Ray, US, CT and MR).In the last few years her work has also extended to the study of Covid-19 patients using chest X-Ray and CT, in collaboration with other specialists.

Zain Hussain obtained his BSc (Hons) in Medicine from the University of St Andrews, UK and MBChB Medicine from the University of Edinburgh, UK, in 2017 and 2020 respectively. He is a Foundation Doctor (Year 2) in Ninewells Hospital, and a Foundation Fellow for the UK Royal College of Psychiatrists. He has been an Associate Faculty Member of the Clinical Educator Programme since 2020, and undertakes a range of teaching and research activities in Edinburgh Medical School. He sits on the steering group of the Undergraduate Certificate in Medical Education (UCME), is the founding Co-ordinator of the FY Clinical Skills Tutor Scheme (CEP & EUMES), and works on a range of medical education research projects. He is also an SSC1 Tutor and Project Lead on the SSC5b “How to Teach” PAL. He has previously held appointments as a Visiting Researcher at the Boston University School of Public Health, USA (2020) and the School of Computer Science and Engineering, Nanyang Technological University, Singapore (2018), and as a Visiting Student Researcher at the Nuffield Department of Surgical Sciences, University of Oxford, UK (2015-17).

Zakariya Sheikh is a third year medical student at the University of Edinburgh currently intercalating in public health. His research interests include epidemiology and medical informatics.

Aziz Sheikh is Professor of Primary Care Research & Development and Director of the Usher Institute at The University of Edinburgh. He is Honorary Consultant in Paediatric Allergy at the Royal Hospital for Sick Children, NHS Lothian, Director of the 16-university Asthma UK Centre for Applied Research, Director of the NIHR Global Respiratory Health Unit (RESPIRE), Co-Director of the NHS Digital Academy, and Director of BREATHE – The Health Data Research Hub for Respiratory Health. Aziz has substantive academic and health policy interests in leveraging the potential of health information technology and data science to transform the delivery of health care and improve population health, which have led him to work with numerous governments, the World Health Organization (WHO) and the World Bank. He serves on the Steering Group and as Chair of the Evaluation Group of the WHO’s 3rd Global Safety Challenge on Medication Safety and is an adviser to the WHO’s Eastern Mediterranean Regional Office. He is an editorial board member of PLOS Medicine, Medical Care, Health Informatics Journal and BMC Medicine, and is Editor-in-Chief of Nature Partnership Journal: Primary Care Respiratory Medicine. He was Guest Editor of PLOS Medicine’s theme issue on Machine Learning in Healthcare and Biomedicine (2018) and BMC Medicine’s theme issue on Big Data in Healthcare (2019), and serves on The Lancet’s Commission on the Future of the NHS. He chaired the World Innovation Summit for Health (WISH) Forum on Data Science and Artificial Intelligence in Healthcare. Aziz has held research grants in excess of 150m, has over 1,000 peer-reviewed publications, and has a H-index of 100. He is a ‘Highly Cited Researcher’. He has been honoured with fellowships from 8 learned societies and was made an Officer of the Order of the British Empire in 2014.

Giuseppe Sceni is the Director of the Medical Physics Department of the Grande Ospedale Metropolitano (GOM) Bianchi-Melacrino-Morelli of Reggio Calabria (Italy). His work interests concern the dosimetric techniques in Radiotherapy, with particular reference to the dosimetry of small fields in conditions of non-electronic equilibrium (used in radiotherapy/radiosurgery stereotactic techniques), dosimetry in radiodiagnostics and Nuclear Medicine. Among the interests in the field of Physics in Radiotherapy he deals with the optimization of the planning of radiotherapy treatments through the use of GPU-Based treatment plan systems that use advanced algorithms (Montecarlo methods) and multimodal imaging. In the field of advanced dosimetric techniques, he has dealt with two-dimensional dosimetry techniques – based on ionization chamber detector arrays – aimed to the pre-treatment dosimetric verification of VMAT, SRS, IMRT plans. advanced has dealt with two-dimensional dosimetry techniques – based on ionisation chamber detector arrays – aimed at the pretreatment dosimetric verification of VMAT, SRS, IMRT plans.

Amir Hussain is Professor and founding Director of the Centre of AI and Data Science at Edinburgh Napier University, UK. His research interests are cross-disciplinary and industry-led, aimed at developing cognitive data science and AI technologies to engineer smart healthcare and industrial systems of tomorrow. He has (co)authored three international patents and over 500 publications, including around 200 journal papers, and over 20 Books/monographs. He has led major national, EU and internationally funded projects, and supervised over 35 PhD students to-date. He is founding Chief Editor of Cognitive Computation journal (Springer Nature), and serves on the editorial board a various other leading journals, including Elsevier’s Information Fusion and IEEE Transactions on: AI; Emerging Topics in Computational Intelligence: and Systems, Man and Cybernetics (Systems). Amongst other distinguished roles, he is an elected Executive Committee member of the UK Computing Research Committee (national expert panel of the IET and the BCS for UK computing research), General Chair of IEEE WCCI 2020 (the world’s largest IEEE CIS technical event in computational intelligence, comprising IJCNN, IEEE CEC and FUZZ-IEEE), and the IEEE UK and Ireland Chapter Chair of the IEEE Industry Applications Society.

Francesco Carlo Morabito (M’89 – SM’00) was the Dean with the Faculty of Engineering and Deputy Rector with the University Mediterranea of Reggio Calabria, Reggio Calabria, Italy, where he is currently a Full Professor of Electrical Engineering. He is also serving as the Vice-Rector for International and Institutional Relations. He has authored or co-authored over 400 papers in international journals/conference proceedings in various fields of engineering (radar data processing, nuclear fusion, biomedical signal processing, nondestructive testing and evaluation, machine learning, and computational intelligence). He has co-authored 15 books and holds three international patents. Prof. Morabito is a Foreign Member of the Royal Academy of Doctors, Spain, in 2004, and a member of the Institute of Spain, Barcelona Economic Network, in 2017. He served as the Governor of the International Neural Network Society for 12 years and as the President of the Italian Network Network Society from 2008 to 2014. He is a member on the editorial boards of various international journals, including the International Journal of Neural Systems, Neural Networks, International Journal of Information Acquisition, and Renewable Energy.
Communicated by Zidong Wang
References
- 1.Cucinotta D., Vanelli M. Who declares COVID-19 a pandemic. Acta Bio Medica: Atenei Parmensis. 2020;91(1):157. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Covid-19 dashboard by the center for systems science and engineering (csse) at johns hopkins university (jhu). https://coronavirus.jhu.edu/map.html, accessed: 2021-10-15.
- 3.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Internal Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. Phua, L. Weng, L. Ling, M. Egi, C.-M. Lim, J.V. Divatia, B.R. Shrestha, Y.M. Arabi, J. Ng, C.D. Gomersall, et al., Intensive care management of coronavirus disease 2019 (covid-19): challenges and recommendations, The Lancet Respiratory Med. [DOI] [PMC free article] [PubMed]
- 5.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C.S.Y., Lui M.M.-S., Lee J.C.Y., Chiu K.W.-H., Chung T., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;201160 doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varela-Santos S., Melin P. A new approach for classifying coronavirus covid-19 based on its manifestation on chest x-rays using texture features and neural networks. Inf. Sci. 2020;545:403–414. doi: 10.1016/j.ins.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 8.Zeng N., Li H., Peng Y. A new deep belief network-based multi-task learning for diagnosis of alzheimer’s disease. Neural Comput. Appl. 2021:1–12. [Google Scholar]
- 9.Zeng N., Li H., Wang Z., Liu W., Liu S., Alsaadi F.E., Liu X. Deep-reinforcement-learning-based images segmentation for quantitative analysis of gold immunochromatographic strip. Neurocomputing. 2021;425:173–180. [Google Scholar]
- 10.Zeng N., Wang Z., Zhang H., Kim K.-E., Li Y., Liu X. An improved particle filter with a novel hybrid proposal distribution for quantitative analysis of gold immunochromatographic strips. IEEE Trans. Nanotechnol. 2019;18:819–829. [Google Scholar]
- 11.Bi X., Zhao X., Huang H., Chen D., Ma Y. Functional brain network classification for alzheimer’s disease detection with deep features and extreme learning machine. Cogn. Comput. 2020;12(3):513–527. [Google Scholar]
- 12.Shen L., Shi J., Dong Y., Ying S., Peng Y., Chen L., Zhang Q., An H., Zhang Y. An improved deep polynomial network algorithm for transcranial sonography–based diagnosis of parkinson’s disease. Cogn. Comput. 2020;12(3):553–562. [Google Scholar]
- 13.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Scientific Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest x-ray images with preprocessing algorithms. Int. J. Med. Inf. 2020;104284 doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest x-ray images. Expert Syst. Appl. 2020;114054 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (covid-19) using x-ray images. Inf. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karthik R., Menaka R., Hariharan M. Learning distinctive filters for COVID-19 detection from chest x-ray using shuffled residual CNN. Appl. Soft Comput. 2020;106744 doi: 10.1016/j.asoc.2020.106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;105581 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Oh, S. Park, J.C. Ye, Deep learning COVID-19 features on cxr using limited training data sets, IEEE Trans. Med. Imag. [DOI] [PubMed]
- 21.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;103792 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Minaee, R. Kafieh, M. Sonka, S. Yazdani, G.J. Soufi, Deep-covid: Predicting COVID-19 from chest x-ray images using deep transfer learning, arXiv preprint arXiv:2004.09363. [DOI] [PMC free article] [PubMed]
- 23.M.S. Boudrioua, COVID-19 detection from chest X-Rays images using CNNs models: Further evidence from deep transfer learning, Available at SSRN 3630150.
- 24.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of covid-19 disease based on gravitational search optimization. Appl. Soft Comput. 2020;106742 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through efficientnet convolutional neural network. Appl. Soft Comput. 2020 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das D., Santosh K., Pal U. Truncated inception net: COVID-19 outbreak screening using chest x-rays. Phys. Eng. Sci. Med. 2020:1–11. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. Babukarthik, V.A.K. Adiga, G. Sambasivam, D. Chandramohan, J. Amudhavel, Prediction of COVID-19 using genetic deep learning convolutional neural network (GDCNN), IEEE Access. [DOI] [PMC free article] [PubMed]
- 28.I. Mporas, P. Naronglerdrit, COVID-19 identification from chest X-Rays, in: 2020 International Conference on Biomedical Innovations and Applications (BIA), IEEE, 2020, pp. 69–72.
- 29.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. Corodet: A deep learning based classification for covid-19 detection using chest x-ray images. Chaos Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umer M., Ashraf I., Ullah S., Mehmood A., Choi G.S. Covinet: a convolutional neural network approach for predicting covid-19 from chest x-ray images. J. Ambient Intell. Humanized Comput. 2021:1–13. doi: 10.1007/s12652-021-02917-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty M., Dhavale S.V., Ingole J. Corona-nidaan: lightweight deep convolutional neural network for chest x-ray based covid-19 infection detection. Appl. Intell. 2021:1–18. doi: 10.1007/s10489-020-01978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee H., Ghosh S., Dhar A., Obaidullah S.M., Santosh K., Roy K. Shallow convolutional neural network for covid-19 outbreak screening using chest x-rays. Cogn. Comput. 2021:1–14. doi: 10.1007/s12559-020-09775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keles A., Keles M.B., Keles A. COV19-CNNet and COV19-ResNet: Diagnostic inference engines for early detection of covid-19. Cogn. Comput. 2021:1–11. doi: 10.1007/s12559-020-09795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Jacobi, M. Chung, A. Bernheim, C. Eber, Portable chest x-ray in coronavirus disease-19 (covid-19): A pictorial review, Clin. Imaging. [DOI] [PMC free article] [PubMed]
- 35.Zadeh L.A. Fuzzy logic. Computer. 1988;21(4):83–93. [Google Scholar]
- 36.Versaci M., Morabito F.C., Angiulli G. Adaptive image contrast enhancement by computing distances into a 4-dimensional fuzzy unit hypercube. IEEE Access. 2017;5:26922–26931. [Google Scholar]
- 37.J. Feder, Fractals, Springer Science & Business Media, 2013.
- 38.Silva L., Senra Filho A., Fazan V., Felipe J., Junior L.M. Two-dimensional sample entropy: Assessing image texture through irregularity. Biomed. Phys. Eng. Express. 2016;2(4) [Google Scholar]
- 39.Versaci M., Morabito F.C. Image edge detection: A new approach based on fuzzy entropy and fuzzy divergence. Int. J. Fuzzy Syst. 2021:1–19. [Google Scholar]
- 40.Versaci M., Calcagno S., Morabito F. International Conference on Computer Analysis of Images and Patterns, Springer. 2015. Image contrast enhancement by distances among points in fuzzy hyper-cubes; pp. 494–505. [Google Scholar]
- 41.Holzinger A., Malle B., Saranti A., Pfeifer B. Towards multi-modal causability with graph neural networks enabling information fusion for explainable ai. Inf. Fusion. 2021;71:28–37. [Google Scholar]
- 42.Holzinger A., Langs G., Denk H., Zatloukal K., Müller H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev.: Data Min. Knowl. Discov. 2019;9(4) doi: 10.1002/widm.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M.D. Zeiler, R. Fergus, Visualizing and understanding convolutional networks, in: European conference on computer vision, Springer, 2014, pp. 818–833.
- 44.A.M. Carrington, D.G. Manuel, P.W. Fieguth, T. Ramsay, V. Osmani, B. Wernly, C. Bennett, S. Hawken, M. McInnes, O. Magwood, et al., Deep roc analysis and auc as balanced average accuracy to improve model selection, understanding and interpretation, arXiv preprint arXiv:2103.11357.
- 45.Wu P., Li H., Zeng N., Li F. Fmd-yolo: An efficient face mask detection method for covid-19 prevention and control in public. Image Vis. Comput. 2021;104341 doi: 10.1016/j.imavis.2021.104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammone N., Ieracitano C., Morabito F.C. A deep CNN approach to decode motor preparation of upper limbs from time–frequency maps of EEG signals at source level. Neural Networks. 2020;124:357–372. doi: 10.1016/j.neunet.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 47.Ieracitano C., Morabito F.C., Hussain A., Mammone N. A hybrid-domain deep learning-based BCI for discriminating hand motion planning from EEG sources. Int. J. Neural Syst. 2021;31(09):2150038. doi: 10.1142/S0129065721500386. [DOI] [PubMed] [Google Scholar]
- 48.Ramirez-Quintana J.A., Madrid-Herrera L., Chacon-Murguia M.I., Corral-Martinez L.F. Brain-computer interface system based on p300 processing with convolutional neural network, novel speller, and low number of electrodes. Cogn. Comput. 2020:1–17. [Google Scholar]
- 49.Ieracitano C., Mammone N., Hussain A., Morabito F.C. A novel multi-modal machine learning based approach for automatic classification of EEG recordings in dementia. Neural Networks. 2020;123:176–190. doi: 10.1016/j.neunet.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 50.Mammone N., De Salvo S., Bonanno L., Ieracitano C., Marino S., Marra A., Bramanti A., Morabito F.C. Brain network analysis of compressive sensed high-density EEG signals in ad and mci subjects. IEEE Trans. Industr. Inf. 2018;15(1):527–536. [Google Scholar]
- 51.Mahmud M., Kaiser M.S., Hussain A., Vassanelli S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans. Neural Networks Learn. Syst. 2018;29(6):2063–2079. doi: 10.1109/TNNLS.2018.2790388. [DOI] [PubMed] [Google Scholar]
- 52.Holzinger A. Springer; 2014. Biomedical informatics: discovering knowledge in big data. [Google Scholar]
- 53.Holzinger A. Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Informatics. 2016;3(2):119–131. doi: 10.1007/s40708-016-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holzinger A., Plass M., Kickmeier-Rust M., Holzinger K., Crisan G.C., Pintea C.-M., Palade V. Interactive machine learning: experimental evidence for the human in the algorithmic loop. Appl. Intell. 2019;49(7):2401–2414. [Google Scholar]
- 55.D. Schneeberger, K. Stöger, A. Holzinger, The european legal framework for medical AI, in: International Cross-Domain Conference for Machine Learning and Knowledge Extraction, Springer, 2020, pp. 209–226.