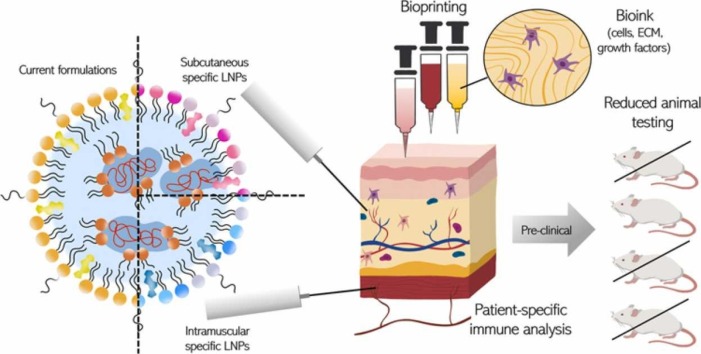
Graphical Abstract
Keywords: COVID-19, Vaccines, Lipid nanoparticles, Bioprinting, Administration route, mRNA
Abstract
BioNTech/Pfizer’s Comirnaty and Moderna’s SpikeVax vaccines consist in mRNA encapsulated in lipid nanoparticles (LNPs). The modularity of the delivery platform and the manufacturing possibilities provided by microfluidics let them look like an instant success, but they are the product of decades of intense research. There is a multitude of considerations to be made when designing an optimal mRNA-LNPs vaccine. Herein, we provide a brief overview of what is presently known and what still requires investigation to optimize mRNA LNPs vaccines. Lastly, we give our perspective on the engineering of 3D bioprinted validation systems that will allow faster, cheaper, and more predictive vaccine testing in the future compared with animal models.
COVID-19 mRNA lipid nanoparticle vaccines: where we are and challenges ahead
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19) pandemic started in 2019, has rapidly spread to more than 200 countries with destructive economic consequences and huge loss of lives [1]. SARS-CoV-2 has an incubation period that can last up to 33 days and a rapid transmission speed even from asymptomatic carriers. Approximately 15% of COVID-19 patients develop severe pneumonia and eventually ∼5% acute respiratory distress syndrome, septic shock, multiple organ failure, and even death (∼ 4 million confirmed deaths) [2]. Given these characteristics, scientists worked at unprecedented speed to create effective vaccines. Presently, there are over 230 COVID-19 vaccines under study worldwide. More than thirty years of research in RNA biology, genetic engineering, immunology, pharmacology, and nanoparticle technologies enabled the fast-tracked development of fully synthetic messenger RNA (mRNA)-based vaccines within just a few months. Besides mRNA-based vaccines, like Pfizer-BioNTech and Moderna, diverse strategies have been employed, including vaccines made of protein subunit with adjuvant, nonreplicating viral vectors (Astra Zeneca/Oxford ChadOx), virus-like-particles, DNA, inactivated- and live-attenuated virus [3]. Now in the spotlight as key components of the COVID-19 mRNA vaccines, lipid nanoparticles (LNPs) are the most advanced technological delivery platform enabling high levels of encapsulation, protection, and efficient in vivo transport. LNPs are nanoparticles composed of lipids that are proven to shield mRNA from enzymatic digestion and boost cellular uptake and protein expression by up to 1000-fold with respect to naked mRNA in animal models [4]. LNP-based vaccines are safe and easy to develop: their production involves handling genetic material only, not viral components. LNP-based vaccines can be produced quickly, which is a key benefit in the existing pandemic, where billions of doses are needed in a short time to immunize the world population.
Thus, compared to approaches based on viruses (e.g., Oxford/AstraZeneca), optimized LNP formulations could be adapted quickly to new SARS-CoV-2 mutants and simultaneously target different variants in a safer manner. Most importantly, mRNA-LNPs are considered drugs rather than biologics, and this feature is critical to expedite regulatory authorities’ approval.
Here we highlight key considerations necessary for designing vaccine delivery systems based on LNPs and explore areas for further optimization. A detailed description of mRNA design and optimization in COVID-19 vaccines has been provided in other reviews [5] and will not be discussed here. We will focus on LNPs optimization and then discuss how new 3D cell culture models for in vitro vaccine testing could replicate in vivo physiology and immune response in a way that could reduce the time to clinical trials and animal testing. Finally, the adoption of 3D models for the study of vaccines in pregnant women will be also discussed. We hope this report can serve as a practical resource for those interested in the development and testing of mRNA-LNP based vaccines.
The focus on delivery: considerations for designing an optimal lipid nanoparticle vaccine
The process of vaccine development is laborious and involves several stages [6]. However, when, on March 11th, 2020, COVID-19 was declared to be a global pandemic by the World Health Organization (WHO), it was recommended that some of the usual stages of vaccine development be skipped to accelerate development and regulatory assessment, approval, manufacturing and quality control. The race for COVID-19 vaccine development has generated a plethora of potential candidates. The two vaccines that have shown the most promising results in preventing COVID-19 infection are genetic vaccines consisting of mRNA encapsulated in LNPs: mRNA-1273/SpikeVax by Moderna and BNT162b2/Comirnaty by BioNTech/Pfizer. SpikeVax is composed of synthetic mRNA which codes for the full-length spike protein (S) of SARS-CoV-2, while Comirnaty codes for the SARS-CoV-2 receptor binding domain (RBD) that is the target of the virus neutralizing antibodies (nAb). Both the vaccines were granted ‘Emergency Use Authorization’ by the U.S. Food and Drug Administration (FDA) and, in December 2020, ‘conditional approval’ by the European Medicinal Agency (EMA).
These mRNA-loaded nanomedicines are formulated through microfluidic mixing and made of four main lipid components: a neutral lipid (e.g., DSPC), cholesterol, a PEG-lipid and an ionizable cationic lipid. Frontrunner drugs (e.g., Doxil®) showed that the neutral lipid and cholesterol provide LNPs with bilayer stability and fusogenic properties [7], [8]; while PEG lipids control particle size and prevent aggregation and fusion [9]. Ionizable lipids - the most important component of mRNA-LNPs [10] - are designed with a pKa below 7 to allow high encapsulation efficiencies of negatively charged mRNA during the mixing step (pH<6), while preserving a neutral charge at physiological pH, reducing toxicity associated with the cationic charge and extending the circulation lifetime of the LNP system.
At a cellular level, protonation occurring in acidic endosomal compartments favors interaction and mixing with anionic cellular lipids leading to intracellular release of mRNA.
In one of the fundamental steps in the process, libraries of different ionizable lipids are screened to identify the optimal lipid species to boost protein expression and induce better immune responses while maintaining low toxicity profiles [11], [12], [13]. In order to speed up the development process, this optimization step was skipped and Moderna and BioNTech/Pfizer turned to a lipid composition inspired by Patirisan (trade name Onpattro® by Alnylam Pharmaceuticals), an orphan drug with a short interfering RNA (siRNA)-LNP system, that was already approved by the FDA for the treatment of hereditary transthyretin (TTR) amyloidosis [14]. The main difference with respect to Onpattro® was the usage of new ionizable cationic lipids (ALC-0315 and SM-102 in BNT162b2/Comirnaty and mRNA-1273/SpikeVax respectively) [15]. However, a persistent theme in gene delivery has been the design of transport technologies according to biomedical demand. Design considerations are made about the payload to be delivered, the therapeutic target and the route of administration, i.e ., typically intramuscular (IM) or intravenous (IV).
It is well known that the encapsulated genetic payload affects size, shape and, most importantly, nanoscale arrangement of lipid systems. The exact arrangement of mRNA encapsulated in LNPs has been poorly addressed so far, while most information has been obtained with siRNA-LNP systems. Depending on the siRNA/lipid molar ratio (i.e., phosphate over ionizable cationic lipid molar ratios) and on the lipid ingredients, siRNA-LNPs may exhibit a multilamellar structure, a nanostructure core a homogeneous core shell [16], [17]. Some authors also reported the existence of cubic structures with Ia3d and Pm3n as crystallographic space groups where siRNA is confined within bicontinuous aqueous channels [18], [19]. According to previous findings [20] the superior efficient silencing of these structures was attributed to enhanced fusion with endosomal membranes promoted by the positive Gaussian modulus of the cubic phases KG> 0. However, whether and to what extent the lessons learned from the structure of siRNA-LNPs can apply to the mRNA-LNPs is under debate [17], [21]. mRNA is at least 100-fold larger than siRNA, and this may affect the inner structure of the LNP. Using cryo-TEM, small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS) Arteta et al. [22] suggested that mRNA is located inside water cylinders, which are coated by cationic lipids and proposed that this peculiar arrangement can determine instability under non-frozen storage conditions. While mRNA is certainly positioned within the particle interior, the nanoscale organization of lipids and mRNA remains ambiguous, with more research needed to confirm the morphology of mRNA-LNPs. Nanoscale arrangement is a critical determinant for the functional delivery of nucleic acids and it is a central dogma that distinct morphologies and nanostructures are associated with totally different transfection performances [23]. This shows how using the same standards (research results, rules, procedures, and protocols) of an approved drug (e.g., Onpattro®) can generate safe and well-functioning vaccines in a short time, but better understanding of particle attributes (e.g., structure-activity relationship) should be prioritized to optimize mRNA-LNPs and create effective immunization delivery systems.
Most of the time, the therapeutic target dictates the choice of the administration route as it influences organ distribution, expression kinetics and therapeutic outcomes [24]. The lipid composition of Onpattro® (a ionizable cationic lipid, a neutral lipid, cholesterol and a PEGylated lipid in proper proportions) was specifically designed for hepatocyte targeting following IV administration. A major challenge often linked with IV administration of LNPs is their short circulation time. Ionizable amino-lipids help maintain a more biocompatible neutral charge, while PEG-lipids also give particles a ‘stealth’ effect. This shields the LNP surface and limits the adsorption of serum proteins, ensuring protection against mononuclear phagocyte systemic uptake and thus contributing to increased circulation time [25]. However, both lipid components have their own drawbacks. For instance, ionizable cationic lipids are susceptible to temperature- and pH-dependent hydrolysis, which has a detrimental effect on particle stability, while high concentrations of PEG lipids prevent the delivery of RNA into cells (the “PEG dilemma”) [26]. Immune responses to PEG molecules have also been reported to result in the accelerated blood clearance (‘ABC’) phenomenon [27]. A major pitfall of IV administration in the context of immunomodulatory agents is the risk of systemic impact and cytokine storm [28], making this route less desirable for LNP-mRNA vaccine candidate development. Injection by IV also requires administration in a medical setting by a trained professional.
COVID-19 vaccine targets are primarily myocytes and keratinocytes but also antigen presenting cells (APC), such as dendritic cells (DCs) residing near the injection side. As such, for mRNA-LNP vaccines, typically IM administration is the most common route of delivery, allowing diffusion of LNPs into the lymph rather than the circulation. In the lymph mRNA-LNP vaccines have direct access to APCs and are not at risk of opsonization and systemic clearance by immune cells as it occurs in the bloodstream. These considerations explain why lipid formulations associated with minor protein expression after IV administration can exhibit the high levels of expression after IM administration [29]. This means that the use of ionizable and PEGylate lipids that are key components of LNPs given to patients by IV administration may not be necessary upon IM injection. Future research will be aimed at understanding whether cationic lipids and alternative materials to PEG-lipids such as zwitterionic materials [30] and artificial protein coatings [31] may even provide a superior efficacy of COVID-19 vaccine [32].
So far, the synthetic identity of LNP-based vaccines in terms of proportions of lipids or lipid/mRNA ratio has been developed through phenomenological approaches. Recent research has clarified the wide gap existing between benchtop discoveries and clinical success of LNP-based vaccines is partly due to partly a result of our poor understanding of the relationship between their synthetic identity and cellular activity. Decrypting this code is a priority to maximize the intracellular release of mRNA, the synthesis of the S protein and the stimulation of the immune system to produce nAb. In this regard, quantitative structure-activity relationship (QSAR) methods can be used to predict the relationships between the synthetic identity of mRNA-LNPs and their mechanism of action at a cellular level. mRNA-LNPs can be internalized by multiple mechanisms, including macropinocytosis, clathrin-mediated, caveolae-mediated endocytosis and temperature-independent fusion processes [33], [34]. Once inside the cell, the ability of mRNA-LNPs to escape from the autophagy-lysosomal pathway is still unclear. As endosomes mature and their pH drops to 6 or less ionizable lipids become protonated, changing from being neutral to positively charged. This protonation is likely accompanied by a change in the lipid molecular shape that, ultimately, promotes the formation of inverted highly-fusogenic structure [35], [36]. Recently, Maugeri et al. [37] also demonstrated that some internalized mRNA-LNPs are secreted into the extracellular matrix as extracellular vesicles (EVs). QSAR approaches assume that LNPs with comparable synthetic identities will interact similarly with cells thus eliciting similar physiological responses (e.g., protein expression and production of antibodies). We envision that QSAR investigations will enable in vitro prediction of bioactivity and accelerate the development of optimized mRNA-LNP vaccines against COVID-19 [38], [39].
Vaccine validation on 3D bioprinted tissues
Validation of delivery systems requires the establishment of proper in vitro models for formulation screening. Although 2D cell culture systems play a key role in advancing our understanding of molecular signaling, cellular morphology, and drug discovery, not all results are translatable to physiological in vivo systems [40]. Indeed, 2D cellular models lack the complex tissue structure consisting of neighboring cells, molecules gradients, and extracellular matrix (ECM). It has been demonstrated that nanoparticle uptake in 2D systems reaches a plateau in a few hours while continues to increase over time if the same cells are grown in 3D matrices [41]. These differences are likely the result of unique diffusive behavior, interaction with ECM components [42], [43], [44], and limited exposure of the cell surface to nanoparticle uptake in 2D cultures. It also seems that endocytic pathways are enhanced when cells are in a 3D arrangement [41].
3D cell cultures, as well as microfluidic and organ-on-chip systems, represent the current frontline in the recreation of in vivo cell arrangement, mechanical environment, and molecular turnover [45]. In vivo testing brings several disadvantages, including cost, lack of similarity with human response, for example for species-specific pathogens [46], and ethical concerns [47]. In 1959, the establishment of the 3 R principles (replacement, reduction, and refinement) set the basis for experimental research, with the replacement of animal models top of the priority list. Even if complete substitution of animal testing is still unimaginable, the creation of suitable 3D models would allow a reduction of the number of animals used in trials and speed up high throughput testing necessary in fast vaccine development. Several 3D models of lung tissue have been developed for the study of host response towards coronaviruses as well as multiorgan failure, as recently reviewed [48], [49].
3D cell culture is extremely technically and economically challenging. Recently, 3D additive manufacturing has been applied to biology, where it is referred to as bioprinting. It consists of a layer-by-layer positioning of cells and biochemicals with a high throughput workflow and customizable biological material, including patient-derived cells and body fluids [50]. Even if fabrication efficiency and resolution still do not match the same performance of robotics-based high-throughput screening platforms (10,000–100,000 tests per day), 3D bioprinting is continuously being improved [51]. Recently, Hwang et al. created can integrated 3D bioprinter based on microscale continuous optical printing capable of the large-scale production of biological samples production within multi-wells [51], [52].
Administration route dramatically impacts the performance of a vaccine formulation and consequently, 3D models should be chosen accordingly, e.g., for IM, subcutaneous (SC), or intradermal (ID) delivery. 3D bioprinting of muscular tissue has been recently reviewed [53], [54]. Ghobolova and colleagues demonstrated the feasibility of a human bio-artificial muscle model formed by mixing muscle cells, obtained from biopsies, in a fibrin hydrogel to observe the effect of micro-injections. Measured parameters included the release of the injected compounds and their metabolites over time [55]. This kind of model could be further improved by adding immune cells to study responses to the vaccine. Indeed, in only a few models have immune cells been inserted into muscular tissues [56]. Most interestingly, bone marrow-derived macrophages injected into adult-derived muscle 3D cultures enable near-complete tissue repair post-injury, limiting myofiber apoptosis and attenuating inflammatory environment [57]. It should also be noted that considerable progress has been made towards vascularization of 3D muscle tissues and that a proper blood supply could also help mimic cell recruitment during IM vaccination [58].
COVID-19 vaccines are usually IM administered but could be switched to SC or ID routes [59]. For example, the accidental SC injection of the BNT162b2/Comirnaty vaccine, resulted in high immunogenicity (98%) after the first dose in 790 patients [60]. In recent experiments in vivo, administration of fluorescently labeled HIV-1 envelope glycoprotein on liposomes was used to demonstrate that both SC and IM routes induce efficient immune cell activation, though antigen is delivered to different lymph nodes depending on the route [61], [62]. A few skin-targeted SARS-CoV-2 vaccines are being studied, based on DNA, mRNA, or viral subunits [59]. Indeed, the cutaneous microenvironment is an ideal target for vaccination since the skin contains a high density of APC and generates a high sustained innate immune response, even at distant organs. The use of 3D cell cultures also is advantageous to follow dynamic cell-cell interactions via multiple fluorescent labeling or other techniques designed to track interactions between immune cells [63], [64].
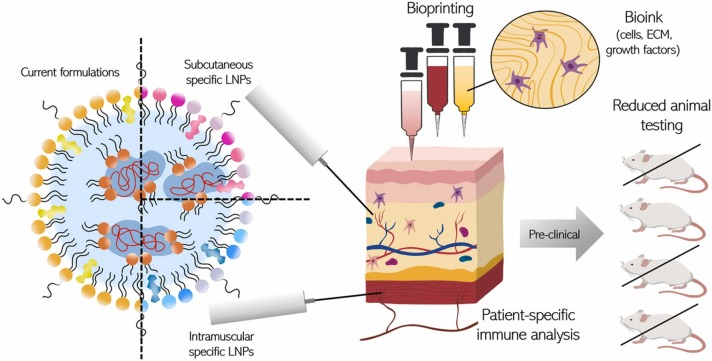
However, proper training is necessary for ID administration unless specific technologies like microneedle patches are adopted [65]. Certain adjuvants, which may be toxic with IM injection, could be feasible via SC or ID delivery [59]. The identification of route-specific adjuvants, as well as LNP specific formulations, will contribute significantly to next-generation vaccine development. 3D tissue models will enable a fundamental understanding of immune activation and overall efficiency to be obtained rapidly using different administration routes as is necessary in a pandemic situation ( Fig. 1).
Fig. 1.
LNP-specific delivery testing in 3D bioprinted tissue would reduce the animal testing and allow for analysis of patient-specific immune analysis.
Skin bioprinting, including the reconstruction of skin appendages, is a research field that has mainly focused on wound healing to replace autologous skin transplantation [66], [67]. Natural skin contains several types of cells, including Langerhans cells, dendritic cells, macrophages, T cells and pluripotent stem cells. Importantly, there are several differences between mouse and human skin immune systems in terms of the amount and position of T cells, and chemokines expressed. These differences may contribute to the failure of translating results obtained in animal models to humans [67]. Since a specialized medium is required to maintain different cell types and can influence marker expression, the combination of many cell types in one model is technically complicated. Concerning immune response, it has been demonstrated that skin differentiation cell media influence response to stimuli and migration of macrophages [68], [69]. Furthermore, spatial organization and the physical constraints provided by 3D culture influence gene expression and cell behavior therefore a controlled bioprinting technology is indispensable for skin reconstruction.
A full immunocompetent 3D skin, necessary to analyze COVID-19 vaccine response, should be composed of various immune cells and be correctly vascularized [70]. Indeed, blood and lymphatic vessels supply nutrients, oxygen and permit immune cell and cytokines uptake. Interestingly using a layer-by-layer technique and nanofilms of extracellular matrix, Matsusaki et al. assembled a multilayer model with human umbilical vein endothelial cells and dermal lymphatic microvascular endothelial cells [71]. In this model blood and lymph-like capillaries formed inside the dermis. Future developments should examine the correctly integration of immune cells together with vascularization with minimal interference between different elements. Concerning the validation of effective 3D models functionality and performance, Pourchet et al. demonstrated the capability of 3D printed formulation of gelatin, alginate and fibrinogen, to constitute a complete skin model that presents all the features of human skin, as demonstrated by immunostaining and molecular analysis [72]. Moreover, a 3D-printed functional human skin has been implanted in vivo in immunodeficient mouse and became indistinguishable from mouse skin after transplantation [73]. Whether these systems will effectively recapitulate human immunological response and substitute in vivo testing in the future is still an open question, however immunocompetent skin is currently available from companies such as Genoskin® for the testing of allergens, psoriasis and drug efficacy after subcutaneous injections, with the InflammaSkin® model able to establish the interplay between activated skin-resident T cells (Th17) and keratinocytes [74].
Finally, it should be mentioned that bioprinting technology can be also used to assess the effect of vaccination on newborns, in terms of safety and efficacy for protection by the placental passage of antibodies against SARS-CoV-2. Several case reports of 2021, reported antibodies in the cord blood after maternal vaccination [75], [76], [77]. In vivo study of blood-placenta barrier (BPB) passage is clearly impossible in human and difficult and time-consuming in animal models. Furthermore BPB physiology is complicated to reproduce with classical 2D systems [78]. In 2018, Arumugasaamy et al. fabricated a 3D model of BPB made of BeWo B30 cells and human umbilical vein endothelial cells in a gelatin methacrylate hydrogel support [48], [79]. This model was further improved by using primary placental fibroblasts within the biological membrane and primary human placental endothelial cells [80]. In the coming years, currently available 3D bioprinting techniques of tissues for vaccine testing will likely improve.
Concluding remarks and future perspectives
In summary, there is ample room to improve mRNA-based candidates for vaccines or therapeutics that are under clinical trials and at the preclinical stage [81]. We foresee that the knowledge gained from the development and clinical use of LNPs in SARS-CoV-2 vaccines gives new impetus to development of other LNP-based therapeutics. Of equal importance is the development of ever more accurate biomimetic 3D tissue models capable of simulating human immune response.
3D bioprinted models allow to optimization and choice of administration route. We are convinced that the enormous efforts that we are observing in the technological development of new methods such as 3D bioprinting will allow faster and more predictive vaccine testing than animal models in the future.
CRediT authorship contribution statement
Massimiliano Papi: Writing – review & editing. Daniela Pozzi: Writing – review & editing. Valentina Palmieri: Writing – original draft, Writing – review & editing. Giulio Caracciolo: Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Massimiliano Papi is an Associate Professor of Applied Physics at the Neuroscience Department at Università Cattolica del Sacro Cuore, Rome. Prof. Massimiliano Papi interests lie at the nexus of physics, biology and biotechnology. He received his degree in Physics (2003) and Ph.D. in Biophysics (2006) at the Sapienza University of Rome (Italy). During his post-doctoral formation, he carried out research in the area of biophysics, from single molecules to cells and tissues. Current main research interests are the development of delivery systems for macromolecular drugs, the cytotoxicity of nanomaterials and their interactions with biological systems and 3D printing. Prof. Papi is currently head of the 3D Printing Facility G-STEP of Policlinico Gemelli Foundation.

Daniela Pozzi is an Associate Professor of Applied Physics at the Molecular Medicine Department of the Sapienza University of Rome. She is co-founder and co-Principal investigator of the NanoDelivery Lab whose mission is to generate and disseminate knowledge in nanomedicine by combining physics, chemistry, biology, pharmacology and nanotechnology and their translation into innovative, clinically applicable therapeutics and diagnostics. She has been a recipient of grants from the Italian Ministry of Health and the Italian Foundation for Cancer Research (AIRC).

Dr. Valentina Palmieri is Researcher at Institute for Complex Systems (ISC) at National Research Council (Italy). VP is a medical biotechnologist and obtained a PhD in Oncobiology and Medical Oncology in 2014. She worked in collaboration with several national and international groups and obtained National awards and International Grants such as Fondazione Umberto Veronesi Post-Doctoral Grant (2018), European Society of Clinical Microbiology and Infectious Diseases Grant (2017) and Italian Ministry of Health. Valentina Palmieri is expert in nanomaterial synthesis and characterization and nanoparticle research, and her work is focused on graphene antimicrobial and diagnostics applications.

Giulio Caracciolo is an Associate Professor of Applied Physics at the Molecular Medicine Department of the Sapienza University of Rome. Over a period of 20 years, Dr. Caracciolo has investigated the structure-activity relationship of lipid-based systems for gene delivery applications. His current research is aimed at exploring the mechanisms governing the interaction of nanomaterials with living systems for prevention/treatment of life-threatening human conditions such as cancer and neurodegenerative diseases. He is a recipient of multiple grants from the Italian Foundation for Cancer Research (AIRC), other public agencies and private companies.
References
- 1.Dong Y., Dai T., Wang B., Zhang L., Zeng L., Huang J., Yan H., Zhang L., Zhou F. The way of SARS-CoV-2 vaccine development: success and challenges. Signal Transduct. Target. Ther. 2021;6:1–14. doi: 10.1038/s41392-021-00796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Y.-W., Xie Y., Tang L.-S., Pu D., Zhu Y.-J., Liu J.-Y., Ma X.-L. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Signal Transduct. Target. Ther. 2021;6:1–25. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald I., Murray S.M., Reynolds C.J., Altmann D.M., Boyton R.J. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. npj Vaccin. 2021;6:1–14. doi: 10.1038/s41541-021-00336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granados-Riveron J.T., Aquino-Jarquin G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaneri A.B., Johnson R.F., Wada J., Bollinger L., Jahrling P.B., Kuhn J.H. Middle east respiratory syndrome: obstacles and prospects for vaccine development. Expert Rev. Vaccin. 2015;14:949–962. doi: 10.1586/14760584.2015.1036033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson M., Omri A. The effect of different lipid components on the in vitro stability and release kinetics of liposome formulations. Drug Deliv. 2004;11:33–39. doi: 10.1080/10717540490265243. [DOI] [PubMed] [Google Scholar]
- 8.Briuglia M.-L., Rotella C., McFarlane A., Lamprou D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015;5:231–242. doi: 10.1007/s13346-015-0220-8. [DOI] [PubMed] [Google Scholar]
- 9.Gabizon A., Chemla M., Tzemach D., Horowitz A.T., Goren D. Liposome longevity and stability in circulation: effects on the in vivo delivery to tumors and therapeutic efficacy of encapsulated anthracyclines. J. Drug Target. 1996;3:391–398. doi: 10.3109/10611869608996830. [DOI] [PubMed] [Google Scholar]
- 10.Rietwyk S., Peer D. Next-generation lipids in RNA interference therapeutics. ACS Nano. 2017;11:7572–7586. doi: 10.1021/acsnano.7b04734. [DOI] [PubMed] [Google Scholar]
- 11.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramishetti S., Hazan‐Halevy I., Palakuri R., Chatterjee S., Naidu Gonna S., Dammes N., Freilich I., Kolik Shmuel L., Danino D., Peer D. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv. Mater. 2020;32 doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 13.Guimaraes P.P.G., Zhang R., Spektor R., Tan M., Chung A., Billingsley M.M., El-Mayta R., Riley R.S., Wang L., Wilson J.M. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Release. 2019;316:404–417. doi: 10.1016/j.jconrel.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 15.Thi T.T.H., Suys E.J.A., Lee J.S., Nguyen D.H., Park K.D., Truong N.P. Lipid-based nanoparticles in the clinic and clinical trials: from cancer nanomedicine to COVID-19 vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 17.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G.F.A., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez L., et al. Biocompatible nanovector of siRNA consisting of arginine-based cationic lipid for gene knockdown in cancer cells. ACS Appl. Mater. Interfaces. 2020 doi: 10.1021/acsami.0c06273. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Negro M., Kumar K., Barrán-Berdón A.L., Datta S., Kondaiah P., Junquera E., Bhattacharya S., Aicart E. Efficient cellular knockdown mediated by siRNA nanovectors of gemini cationic lipids having delocalizable headgroups and oligo-oxyethylene spacers. ACS Appl. Mater. Interfaces. 2016;8:22113–22126. doi: 10.1021/acsami.6b08823. [DOI] [PubMed] [Google Scholar]
- 20.Leal C., Bouxsein N.F., Ewert K.K., Safinya C.R. Highly efficient gene silencing activity of siRNA embedded in a nanostructured gyroid cubic lipid matrix. J. Am. Chem. Soc. 2010;132:16841–16847. doi: 10.1021/ja1059763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 22.Arteta M.Y., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., Hou S., Esposito A.A., Ketova T., Welsher K. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhang Z., Luo J., Han X., Wei Y., Wei X. mRNA vaccine: a potential therapeutic strategy. Mol. Cancer. 2021;20:1–23. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama H., Akita H., Harashima H. The polyethyleneglycol dilemma: advantage and disadvantage of PEGylation of liposomes for systemic genes and nucleic acids delivery to tumors. Biol. Pharm. Bull. 2013;36:892–899. doi: 10.1248/bpb.b13-00059. [DOI] [PubMed] [Google Scholar]
- 27.Lila A.S.A., Kiwada H., Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J. Control. Release. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., Grupp S.A., Mackall C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood, J. Am. Soc. Hematol. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke P.C., Lin S., Parak W.J., Davis T.P., Caruso F. A decade of the protein corona. ACS Nano. 2017;11:11773–11776. doi: 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- 31.Giulimondi F., Digiacomo L., Pozzi D., Palchetti S., Vulpis E., Capriotti A.L., Chiozzi R.Z., Laganà A., Amenitsch H., Masuelli L. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-11642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zukancic D., Suys E.J.A., Pilkington E.H., Algarni A., Al-Wassiti H., Truong N.P. The importance of poly (Ethylene glycol) and lipid structure in targeted gene delivery to lymph nodes by lipid nanoparticles. Pharmaceutics. 2020;12:1068. doi: 10.3390/pharmaceutics12111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pozzi D., Marchini C., Cardarelli F., Rossetta A., Colapicchioni V., Amici A., Montani M., Motta S., Brocca P., Cantù L. Mechanistic understanding of gene delivery mediated by highly efficient multicomponent envelope-type nanoparticle systems. Mol. Pharm. 2013;10:4654–4665. doi: 10.1021/mp400470p. [DOI] [PubMed] [Google Scholar]
- 34.Schlich M., Palomba R., Costabile G., Mizrahy S., Pannuzzo M., Peer D., Decuzzi P. Cytosolic delivery of nucleic acids: the case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 2021 doi: 10.1002/btm2.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jadhav S.G., Dowdy S.F. Overcoming delivery barriers with LNPs. Nat. Mater. 2021;20:575–577. doi: 10.1038/s41563-021-00988-3. [DOI] [PubMed] [Google Scholar]
- 36.Marchini C., Pozzi D., Montani M., Alfonsi C., Amici A., Amenitsch H., Candeloro De Sanctis S., Caracciolo G. Tailoring lipoplex composition to the lipid composition of plasma membrane: a Trojan horse for cell entry? Langmuir. 2010;26:13867–13873. doi: 10.1021/la1023899. [DOI] [PubMed] [Google Scholar]
- 37.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., Hölttä M., Skantze P., Johansson S., Sundqvist M. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caracciolo G., Farokhzad O.C., Mahmoudi M. Biological identity of nanoparticles in vivo: clinical implications of the protein corona. Trends Biotechnol. 2017;35:257–264. doi: 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Kamat S., Kumari M., Jayabaskaran C. Nano-engineered tools in the diagnosis, therapeutics, prevention, and mitigation of SARS-CoV-2. J. Control. Release. 2021;338:813–836. doi: 10.1016/j.jconrel.2021.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randall M.J., Jüngel A., Rimann M., Wuertz-Kozak K. Advances in the biofabrication of 3D skin in vitro: healthy and pathological models. Front. Bioeng. Biotechnol. 2018;6:154. doi: 10.3389/fbioe.2018.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belli V., Guarnieri D., Biondi M., Della Sala F., Netti P.A. Dynamics of nanoparticle diffusion and uptake in three-dimensional cell cultures. Colloids Surf. B Biointerfaces. 2017;149:7–15. doi: 10.1016/j.colsurfb.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W., Yang Y., Cui B. New perspectives on the roles of nanoscale surface topography in modulating intracellular signaling. Curr. Opin. Solid State Mater. Sci. 2021;25 doi: 10.1016/j.cossms.2020.100873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Goas M., Testard F., Taché O., Debou N., Cambien B., Carrot G., Renault J.-P. How do surface properties of nanoparticles influence their diffusion in the extracellular matrix? A model study in Matrigel using polymer-grafted nanoparticles. Langmuir. 2020;36:10460–10470. doi: 10.1021/acs.langmuir.0c01624. [DOI] [PubMed] [Google Scholar]
- 44.Tomasetti L., Breunig M. Preventing obstructions of nanosized drug delivery systems by the extracellular matrix. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201700739. [DOI] [PubMed] [Google Scholar]
- 45.Baker B.M., Chen C.S. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He B., Chen G., Zeng Y. Three-dimensional cell culture models for investigating human viruses. Virol. Sin. 2016;31:363–379. doi: 10.1007/s12250-016-3889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bédard P., Gauvin S., Ferland K., Caneparo C., Pellerin È., Chabaud S., Bolduc S. Innovative human three-dimensional tissue-engineered models as an alternative to animal testing. Bioengineering. 2020;7:115. doi: 10.3390/bioengineering7030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawko N., Plaskasovitis C., Stokes C., Abelseth L., Fraser I., Sharma R., Kirsch R., Hasan M., Abelseth E., Willerth S.M. 3D tissue models as an effective tool for studying viruses and vaccine development. Front. Mater. 2021;8:80. [Google Scholar]
- 49.Chakraborty J., Banerjee I., Vaishya R., Ghosh S. Bioengineered in vitro tissue models to study SARS-CoV-2 pathogenesis and therapeutic validation. ACS Biomater. Sci. Eng. 2020;6:6540–6555. doi: 10.1021/acsbiomaterials.0c01226. [DOI] [PubMed] [Google Scholar]
- 50.Bejoy A.M., Makkithaya K.N., Hunakunti B.B., Hegde A., Krishnamurthy K., Sarkar A., Lobo C.F., Keshav D.V.S., Dharshini G., Mascarenhas S. An insight on advances and applications of 3d bioprinting: a review. Bioprinting. 2021;24 [Google Scholar]
- 51.Gao G., Ahn M., Cho W.-W., Kim B.S., Cho D.-W. 3D printing of pharmaceutical application: drug screening and drug delivery. Pharmaceutics. 2021;13:1373. doi: 10.3390/pharmaceutics13091373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang H.H., You S., Ma X., Kwe L., Victorine G., Lawrence N., Wan X., Shen H., Zhu W., Chen S. High throughput direct 3D bioprinting in multiwell plates. Biofabrication. 2021;13:25007. doi: 10.1088/1758-5090/ab89ca. [DOI] [PubMed] [Google Scholar]
- 53.Palmieri V., Sciandra F., Bozzi M., De Spirito M., Papi M. 3D graphene scaffolds for skeletal muscle regeneration: future perspectives. Front. Bioeng. Biotechnol. 2020;8:383. doi: 10.3389/fbioe.2020.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang P., An J., Chua C.K., Tan L.P. Bioprinting of 3D in vitro skeletal muscle models: a review. Mater. Des. 2020;193 [Google Scholar]
- 55.Gholobova D., Gérard M., Decroix L., Desender L., Callewaert N., Annaert P., Thorrez L. Human tissue-engineered skeletal muscle: a novel 3D in vitro model for drug disposition and toxicity after intramuscular injection. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-30123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihaly E., Altamirano D.E., Tuffaha S., Grayson W. Semin. Cell Dev. Biol. Elsevier; 2021. Engineering skeletal muscle: building complexity to achieve functionality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juhas M., Abutaleb N., Wang J.T., Ye J., Shaikh Z., Sriworarat C., Qian Y., Bursac N. Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration. Nat. Biomed. Eng. 2018;2:942–954. doi: 10.1038/s41551-018-0290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gholobova D., Terrie L., Gerard M., Declercq H., Thorrez L. Vascularization of tissue-engineered skeletal muscle constructs. Biomaterials. 2020;235 doi: 10.1016/j.biomaterials.2019.119708. [DOI] [PubMed] [Google Scholar]
- 59.Korkmaz E., Balmert S.C., Sumpter T.L., Carey C.D., Erdos G., Falo L.D., Jr Microarray patches enable the development of skin-targeted vaccines against COVID-19. Adv. Drug Deliv. Rev. 2021;171:164–186. doi: 10.1016/j.addr.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedensohn L., Zur M., Timofeyev M., Burshtein S., Ben Michael Y., Fink N., Glassberg E. Sub-cutaneous Pfizer/BioNTech COVID-19 vaccine administration results in seroconversion among young adults. Vaccine. 2021;39:6210–6212. doi: 10.1016/j.vaccine.2021.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., Lin A., Eriksson B., Hedestam G.B.K., Wyatt R.T. Route of vaccine administration alters antigen trafficking but not innate or adaptive immunity. Cell Rep. 2020;30:3964–3971. doi: 10.1016/j.celrep.2020.02.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding Y., Li Z., Jaklenec A., Hu Q. Vaccine delivery systems toward lymph nodes. Adv. Drug Deliv. Rev. 2021;179 doi: 10.1016/j.addr.2021.113914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pasqual G., Chudnovskiy A., Tas J.M.J., Agudelo M., Schweitzer L.D., Cui A., Hacohen N., Victora G.D. Monitoring T cell–dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature. 2018;553:496–500. doi: 10.1038/nature25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Septiadi D., Bourquin J., Durantie E., Petri-Fink A., Rothen-Rutishauser B. A novel sample holder for 4D live cell imaging to study cellular dynamics in complex 3D tissue cultures. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-28206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caudill C., Perry J.L., Iliadis K., Tessema A.T., Lee B.J., Mecham B.S., Tian S., DeSimone J.M. Transdermal vaccination via 3D-printed microneedles induces potent humoral and cellular immunity. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2102595118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weng T., Zhang W., Xia Y., Wu P., Yang M., Jin R., Xia S., Wang J., You C., Han C. 3D bioprinting for skin tissue engineering: current status and perspectives. J. Tissue Eng. 2021;12 doi: 10.1177/20417314211028574. 20417314211028576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pupovac A., Senturk B., Griffoni C., Maniura‐Weber K., Rottmar M., McArthur S.L. Toward immunocompetent 3D skin models. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201701405. [DOI] [PubMed] [Google Scholar]
- 68.Moon S., Kim D.H., Shin J.U. In vitro models mimicking immune response in the skin. Yonsei Med. J. 2021;62:969–980. doi: 10.3349/ymj.2021.62.11.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffoni C., Neidhart B., Yang K., Groeber-Becker F., Maniura-Weber K., Dandekar T., Walles H., Rottmar M. In vitro skin culture media influence the viability and inflammatory response of primary macrophages. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-86486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akagi T., Nagura M., Hiura A., Kojima H., Akashi M. Construction of three-dimensional dermo–epidermal skin equivalents using cell coating technology and their utilization as alternative skin for permeation studies and skin irritation tests. Tissue Eng. Part A. 2017;23:481–490. doi: 10.1089/ten.TEA.2016.0529. [DOI] [PubMed] [Google Scholar]
- 71.Matsusaki M., Fujimoto K., Shirakata Y., Hirakawa S., Hashimoto K., Akashi M. Development of full‐thickness human skin equivalents with blood and lymph‐like capillary networks by cell coating technology. J. Biomed. Mater. Res. Part A. 2015;103:3386–3396. doi: 10.1002/jbm.a.35473. [DOI] [PubMed] [Google Scholar]
- 72.Pourchet L.J., Thepot A., Albouy M., Courtial E.J., Boher A., Blum L.J., Marquette C.A. Human skin 3D bioprinting using scaffold‐free approach. Adv. Healthc. Mater. 2017;6 doi: 10.1002/adhm.201601101. [DOI] [PubMed] [Google Scholar]
- 73.Cubo N., Garcia M., Del Cañizo J.F., Velasco D., Jorcano J.L. 3D bioprinting of functional human skin: production and in vivo analysis. Biofabrication. 2016;9:15006. doi: 10.1088/1758-5090/9/1/015006. [DOI] [PubMed] [Google Scholar]
- 74.Jardet C., David A., Braun E., Descargues P., Grolleau J., Hebsgaard J., Norsgaard H., Lovato P. Development and characterization of a human Th17–driven ex vivo skin inflammation model. Exp. Dermatol. 2020;29:993–1003. doi: 10.1111/exd.14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paul G., Chad R. Newborn antibodies to SARS-CoV-2 detected in cord blood after maternal vaccination–a case report. BMC Pedia. 2021;21:1–2. doi: 10.1186/s12887-021-02618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riviello C., Pontello V. Maternal and neonatal SARS‐CoV‐2 antibodies assessment after mRNA maternal vaccination in the third trimester of pregnancy. Int. J. Gynecol. Obstet. 2021;154:565–566. doi: 10.1002/ijgo.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soysal A., Bilazer C., Gönüllü E., Barın E., Çivilibal M. Cord blood antibody following maternal SARS-CoV-2 inactive vaccine (CoronaVac) administration during the pregnancy. Hum. Vaccin. Immunother. 2021;17:3484–3486. doi: 10.1080/21645515.2021.1947099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winter M., Jankovic-Karasoulos T., Roberts C.T., Bianco-Miotto T., Thierry B. Bioengineered microphysiological placental models: towards improving understanding of pregnancy health and disease. Trends Biotechnol. 2021;39:1221–1235. doi: 10.1016/j.tibtech.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 79.Arumugasaamy N., Ettehadieh L.E., Kuo C.-Y., Paquin-Proulx D., Kitchen S.M., Santoro M., Placone J.K., Silveira P.P., Aguiar R.S., Nixon D.F. Biomimetic placenta-fetus model demonstrating maternal–fetal transmission and fetal neural toxicity of zika virus. Ann. Biomed. Eng. 2018;46:1963–1974. doi: 10.1007/s10439-018-2090-y. [DOI] [PubMed] [Google Scholar]
- 80.Kreuder A.-E., Bolaños-Rosales A., Palmer C., Thomas A., Geiger M.-A., Lam T., Amler A.-K., Markert U.R., Lauster R., Kloke L. Inspired by the human placenta: a novel 3D bioprinted membrane system to create barrier models. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-72559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forni G., Mantovani A. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28:626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]