Abstract
Cyclic nucleotide-gated (CNG) channels transduce light-induced chemical signals into electrical signals in retinal cone and rod photoreceptors. Structures of native CNG channels, which are heterotetramers formed by CNGA and CNGB subunits, have not been obtained. Here we report a high-resolution cryo-EM structure of the human cone CNG channel in the apo closed state. The channel contains three CNGA3 and one CNGB3 subunits. R403 in the pore helix of CNGB3 projects into an asymmetric selectivity filter and forms hydrogen bonds with two pore-lining backbone carbonyl oxygens. R442 in S6 of CNGB3 protrudes into and occludes the pore below the hydrophobic cavity gate previously observed in homotetrameric CNGA channels. Interestingly, R403Q is a disease mutation, and R442 is replaced by glutamine in some animal species with dichromatic or monochromatic vision. These and other unique structural features and the disease link conferred by CNGB3 indicate a critical role of CNGB3 in shaping cone photoresponses.
INTRODUCTION
Vertebrate retinal cone and rod photoreceptors mediate vision in bright and dim light, respectively, and cones are responsible for color vision 1. In retinal photoreceptors, light activation of photopigments decreases the intracellular cGMP concentration and closes cyclic nucleotide-gated (CNG) channels 2,3. This phototransduction converts light stimulation into electrical signals that are transmitted to the brain 1,4,5. CNG channels belong to the voltage-gated ion channel superfamily but they are largely insensitive to membrane voltage and are activated by intracellular cGMP or cAMP 4,6–9. CNG channels are nonselective for monovalent cations and are permeable to Ca2+, which paradoxically also blocks monovalent cation currents 2,4,6–15. Cone and rod CNG channels have similarities and differences in their molecular and biophysical properties, which contribute to the different responses of cones and rods to light 1,4,7,8,16,17. The cone CNG channel is composed of CNGA3 (A3) and CNGB3 (B3) subunits, and the rod channel of CNGA1 (A1) and CNGB1 (B1) subunits 4,8,17–21. A1 and A3 subunits form functional homomeric channels when expressed heterologously and have been studied extensively 4,8,17,18. B1 and B3 subunits do not form functional homomeric channels on their own and are therefore regarded as regulatory subunits, but they profoundly affect the biophysical properties of their respective heteromeric channels, including ion selectivity, Ca2+ permeation and block, ligand discrimination and affinity, and spontaneous open probability 19–23. CNGB subunits also affect regulation of the heteromeric channels by Ca2+/calmodulin, phosphoinositides, and matrix metalloproteinases 23–28. Numerous inherited mutations in retinal CNG channel genes have been associated with degenerative visual disorders such as retinitis pigmentosa and achromatopsia (ACHM) 4,8,18,29, and disease-associated mutations (DAMs) have been found not only in CNGA but also in CNGB subunits 4,18,29–38. Despite their functional importance, it is largely unclear how CNGB subunits affect native CNG channel properties.
Recently, cryo-EM structures of eukaryotic CNG channels in the apo closed state and cGMP-bound open state have been solved, including TAX-4, a CNGA subunit in Caenorhabditis elegans 39,40, and human A1 channels 41. These structures provide significant insights into the molecular mechanisms of CNG channel properties, including ligand activation and ion selectivity and permeation. However, these structures are of homomeric CNGA subunits and therefore do not faithfully and fully represent heteromeric native cone and rod CNG channels. Up to date, structures of heteromeric native CNG channels have not been obtained, most likely owing to the presence of CNGB subunits and the inherent asymmetry. Clearly, structures of native CNG channels are needed for a more accurate and comprehensive understanding of their biophysical properties and biological functions. In this study, we report a 2.93 Å-resolution cryo-EM structure of the full-length human cone A3/B3 CNG channel in the apo closed state. This structure reveals some novel features that may shed light on the role of B3 in shaping cone CNG channel properties and responses, and provides a new framework for further mechanistic investigation of cone CNG channel physiology and channelopathy.
RESULTS and DISCUSSION
Overall structure of apo A3/B3
Full-length human A3 and B3 were coexpressed in and purified from HEK 293 cells. To ensure that only A3/B3 heteromeric complexes were purified, A3 was connected to an N-terminal MBP tag and B3 to an N-terminal 2× Strep tag, and the A3/B3 heteromeric protein was purified by a two-step affinity purification followed by gel filtration. Details of protein expression and purification, cryo-EM data collection and processing, and atomic model refinement and validation can be found in Table 1, Extended Data Figs. 1 and 2, and Methods.
Table 1.
Cryo-EM data collection, refinement and validation statistics.
| CNGA3/CNGB3 apo (EMD-24468, PDB 7RHS) | |
|---|---|
|
| |
| Data collection and processing | |
| Magnification | 105,000 × |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 57.63 |
| Defocus range (μm) | −0.6 to −2.2 |
| Pixel size (Å) | 0.8247 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 3,982,214 |
| Final particle images (no.) | 205,492 |
| Map resolution (Å) | 2.93 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 2.7 to 3.1 |
| Refinement | |
| Initial model used | De novo structure |
| Model resolution (Å) | 3.06 |
| FSC threshold | 0.143 |
| Map sharpening B factor (Å2) | −103 |
| Model composition | |
| Non-hydrogen atoms | 14,767 |
| Protein residues | 1,804 |
| Sugar moieties | 3 |
| Ion (Na+) | 1 |
| B factors (Å2) | |
| Protein | 85.39 |
| Sugar moieties | 89.59 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.945 |
| Validation | |
| MolProbity score | 1.28 |
| Clashscore | 3.47 |
| Poor rotamers (%) | 0.06 |
| Ramachandran plot | |
| Favored (%) | 97.22 |
| Allowed (%) | 2.78 |
| Disallowed (%) | 0 |
When reconstituted into liposomes, the purified A3/B3 proteins produced cGMP-induced currents, which were inhibited by 100 μM L-cis-diltiazem (DTZ), a blocker of native cone CNG channels and heterologously expressed A3/B3 channels (Extended Data Fig. 3a, b). The average single-channel conductance is 41 pS (Extended Data Fig. 3c, d), similar to the values (41–42 pS) reported previously of A3/B3 channels expressed in HEK 293 cells or oocytes 21,22. The reconstituted channels preferred cGMP over cAMP, showing strong activation by 10 μM cGMP but little activation by 10 μM cAMP, with a single-channel open probability (Po) of 0.28 and 0.004 with 10 μM cGMP and 10 μM cAMP, respectively (Extended Data Fig. 3e, f). These properties are qualitatively or even quantitatively similar to those of native cone CNG channels or heteromeric A3/B3 channels expressed in oocytes and HEK 293 cells 4,8,21,23,42. However, because of markedly different experimental conditions and because the purified channel proteins underwent harsh treatments (such as dehydration and rehydration) during liposome reconstitution, quantitative comparisons of our liposome recordings and previous recordings of native cone CNG channels or heterologously expressed A3/B3 channels are inadequate. Nevertheless, our liposome recordings demonstrate that the purified proteins form functional, A3/B3 heteromeric channels.
A cryo-EM density map of apo A3/B3 was obtained at an overall resolution of 2.93 Å with imposed C1 symmetry (Fig. 1a). The quality of the densities is high in the center (Extended Data Fig. 1h) and in most functional regions (Extended Data Fig. 2). We were able to model amino acids A158-L611, I159-L611 and I159-E614 of the three A3 subunits and L206-L646 of the B3 subunit in the final structure. The remaining residues in the N- and C-termini were not modeled because of weak or missing densities. The unmodeled N- and C-terminal regions contain some functionally important domains, such as the calmodulin-binding sites and carboxy-terminal leucine-zipper (CLZ) domain 4,8.
Fig. 1 |. Cryo-EM structure of apo human A3/B3.
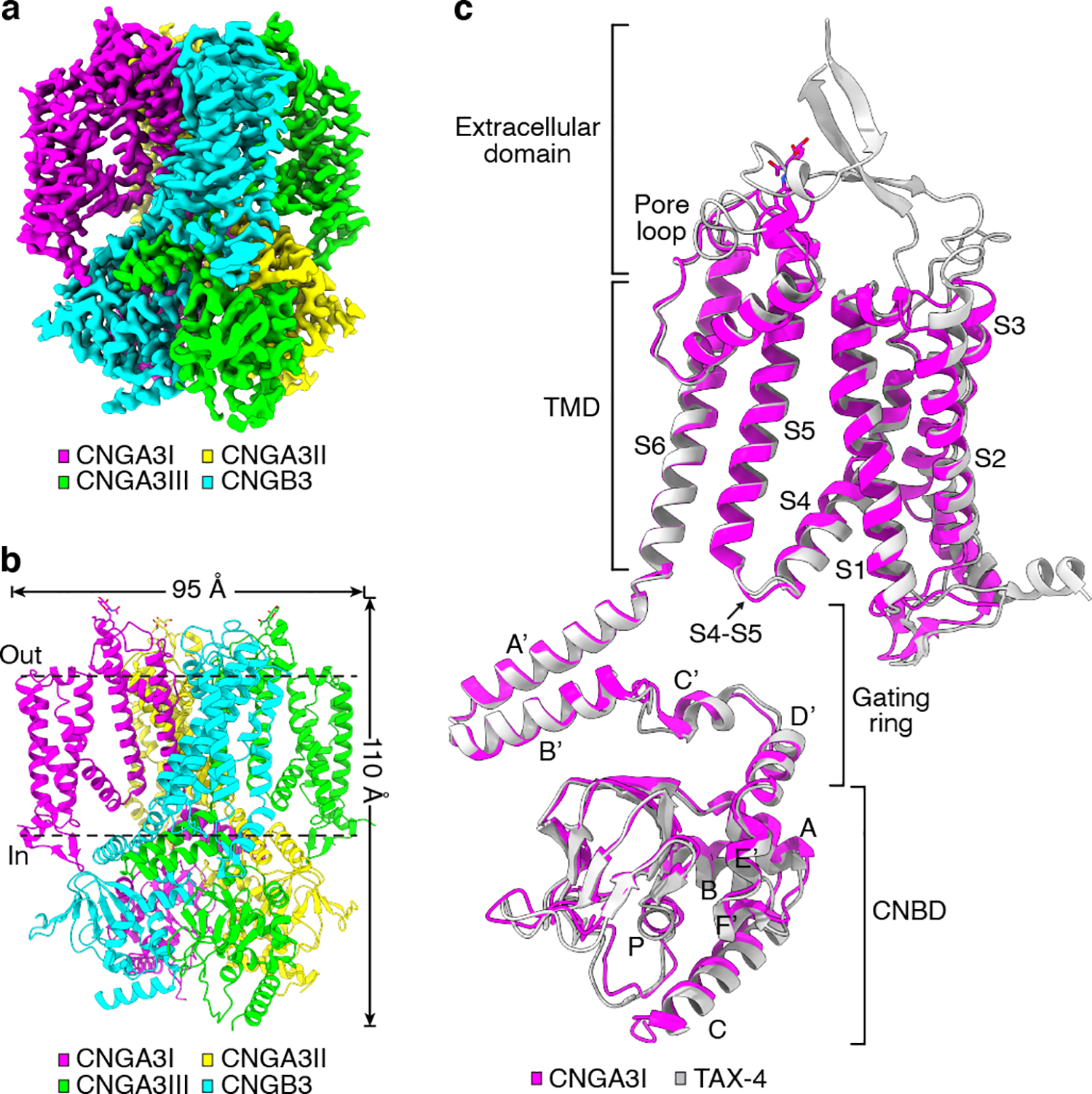
a, Cryo-EM density map of apo A3/B3. b, Apo structure, viewed parallel to the membrane. c, Superposition of apo A3I and TAX-4 (PDB ID: 6WEJ) protomers.
The channel is a tetramer comprising of three A3 subunits and one B3 subunit (Fig. 1a, b). As in homomeric TAX-4 and A1 channels, the subunits are organized in a non-domain swapped configuration, i.e., the voltage sensing-like domain interacts with the pore domain of the same subunit. Although there is a general agreement and acceptance that the rod CNG channel has a stoichiometry of three A1 and one B1 43–46, there has been a disagreement on the subunit stoichiometry of the cone channel, with competing results and proposals of a 2 A3:2 B3 stoichiometry 8,17,47 and a 3 A3:1 B3 stoichiometry 18,48. Our structure unambiguously shows a 3 A3:1 B3 stoichiometry. To differentiate the three A3 subunits, they are designated as A3I, A3II and A3III, respectively, starting clockwise from the B3 subunit when viewed from the extracellular side (Fig. 1a, b).
The overall structure of the heteromeric A3/B3 channel is similar to that of homomeric TAX-4 39,40 and A1 channels 41. This is evident when comparing the protomer structures of A3I and TAX-4 (Fig. 1c). As with TAX-4, A3 and B3 subunits can be arbitrarily divided into four structural layers from the outside to the inside: the extracellular domain, the transmembrane domain (TMD), the gating ring, and the cyclic nucleotide-binding domain (CNBD) (Fig. 1c). The gating ring is formed by helices A’-D’ of the C-linker, which encompasses helices A’-F’ (Fig. 1c).
A3 and B3 can be unambiguously distinguished
The structures of A3I, A3II and A3III are highly similar (Fig. 2a), but local differences exist, as will be described later. The overall structure of B3 is very similar to that of A3 (Fig. 2b); however, numerous differences in local structures are clearly noticeable (Fig. 2b–e). For example, different structures are observed in several loops in the CNBD (Fig. 2b), in helices A’B’ of the gating ring (Fig. 2b), in the cytoplasmic loop between S2 and S3 (Fig. 2b), in S1, S2 and the S1-S2 linker (Fig. 2c), and in S3 and S4 (Fig. 2d). Also, A3 is glycosylated at N339, which is located in the extracellular linker between S5 and the pore helix, but B3 is not glycosylated at this position (Fig. 2e). These and other structural differences collectively allowed for an unequivocal 3D reconstruction of the density map and subsequent atomic modeling. It has been reported that glycosylation of N339 protects A3 homomeric channels from proteolysis by matrix metalloproteinases 49.
Fig. 2 |. Comparison of A3 and B3 structures.
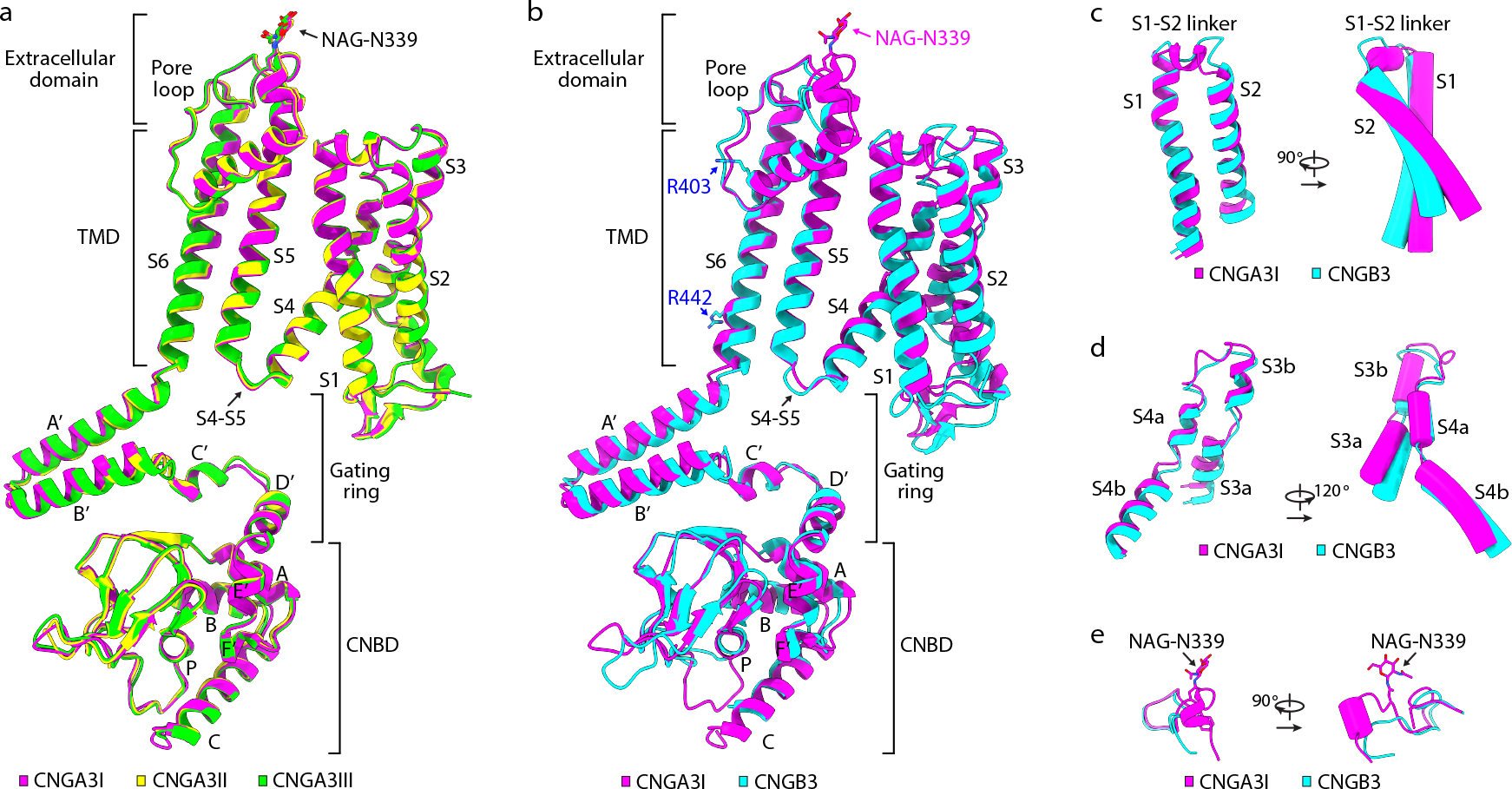
a, Superposition of A3I, A3II and A3III protomers. b, Superposition of A3I and B3 protomers. c-e, Differences between A3 and B3 in S1-S2 (c), S3-S4 (d), and the extracellular linker between S5 and the pore helix (e). NAG: N-acetyl-beta-D-glucosamine.
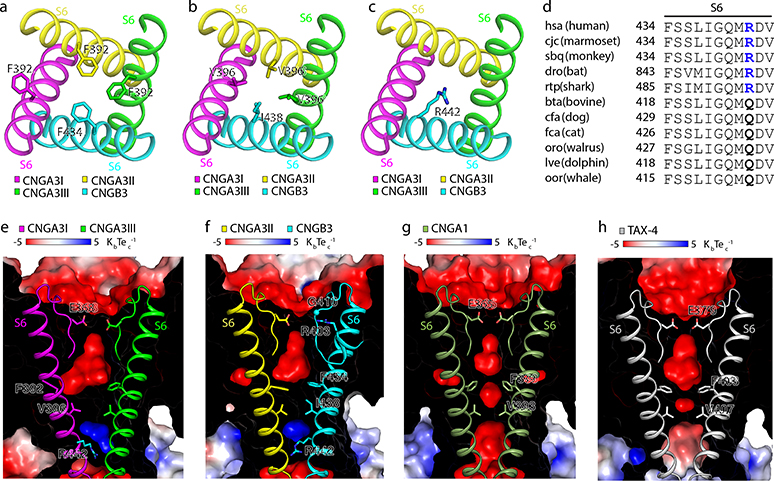
R442 in S6 of CNGB3 occludes the pore below the cavity gate
The ion conduction pathway of the A3/B3 channel is asymmetric along the pore axis due to the presence of B3 (Fig. 3a, b). The dimensions of the selectivity filter (SF) and central cavity of A3/B3 are generally similar to those of TAX-4 (Fig. 3c–e), but the distances between B3 and its diagonally opposed A3II are narrower than those between two diagonally opposed TAX-4 subunits (Fig. 3e). In TAX-4 and A1 homomeric channels, two hydrophobic S6 residues (F403 and V407 in TAX-4, and F389 and V393 in human A1) form a double-barrier hydrophobic gate in the central cavity 39,41. This cavity gate is also present in the unliganded A3/B3 channel, where it is formed collectively by F392 and V396 in A3 and F434 and I438 in B3, and this gate is fully closed (Figs. 3 and 4a, b). Curiously, the density map suggests that the side-chain of F392 of A3I can adopt one of two orientations (Extended Data Fig. 4). These alternate projections are probably caused by the presence of B3. The major orientation is a sideway projection and is used in the relevant illustrations. The minor orientation is a pore projection. Regardless the projection of F392 of A3I, the pore is tightly sealed at this position by the phenylalanines in the other three subunits (Figs. 3 and 4a). I438 of B3 has just one orientation and projects to the pore, as does V396 of A3; together, they form another tight constriction in the pore (Figs. 3 and 4b).
Fig. 3 |. The ion conduction pathway.
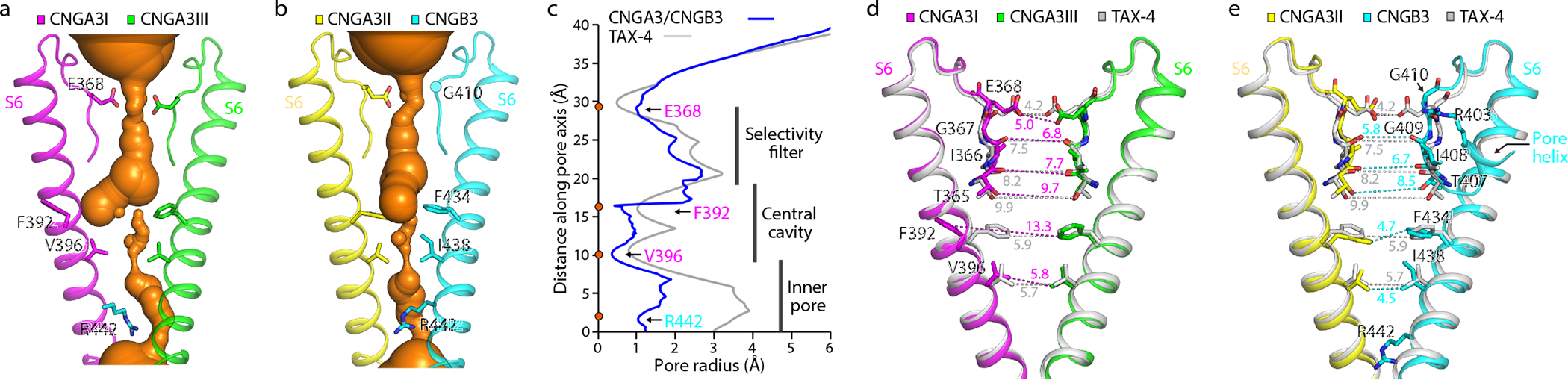
a, b, The solvent-accessible pathway generated with the HOLE program, shown in the center. c, Pore-size profiles of the solvent-accessible pathway in apo A3/B3 and TAX-4. The origin of the pore axis is set at the cytoplasmic end of S6. d, e, Comparison of the SF, central cavity and inner pore in apo A3/B3 and TAX-4. Distances (in Å) between the atoms are measured as the center-to-center distance of two diagonally opposed atoms.
Fig. 4 |. The cavity gate and inner gate.
a, b, Cavity gate in S6 (viewed from the extracellular side). c, Inner gate in S6 (viewed from the extracellular side). d, Amino acid sequence alignment of the gate-forming region of S6 of B3 from different species. Sequences and species abbreviations were obtained from the KEGG database (http://www.genome.ad.jp/kegg/). e-h, Electrostatic potential (calculated with APBS plug-in in PyMol) of the interior surface of the ion conduction pathway in apo A3/B3 (e, f), CNGA1 (g, PDB ID: 7LFT), and TAX-4 (h). Front halves of the channels were cut away and rear halves were overlayed with the SF and S6.
Unexpectedly, R442 of B3, located one α helical turn below I438, projects directly into the ion conduction pathway and blocks it, creating a possible additional positively charged gate below the hydrophobic cavity gate (Figs. 3 and 4c). The equivalent residue is also an arginine in B1 (Supplementary Fig. 1), suggesting that this putative positively charged gate is also present in the rod A1/B1 channel. This putative gate is missing in homomeric CNGA channels, as the analogous position is a glycine in A1, A2, A3 and TAX-4 and a serine in A4 (Supplementary Fig. 1). However, definitive proof of R442 being a gate in A3/B3 would require the determination of the cGMP-bound open state and further functional characterization.
Interestingly, R442 is replaced by a glutamine in some animal species and all these animals are dichromatic or monochromatic (Fig. 4d). A search of available data suggests that there is a strict correlation between the presence of glutamine at position 442 of B3 and dichromatic or monochromatic vision. At this point, this correlation is merely an observation and it is unclear whether this correlation indicates a causal relationship or a coincidence. On the other hand, a dichromatic or monochromatic vision does not always predict a glutamine at the analogous position of R442, as an arginine is present in some dichromatic or monochromatic animals such as monochromatic whale shark (Fig. 4d). It should also be noted that the main cause of dichromatic or monochromatic vision is the lack of a particular type of cone photoreceptor or mutations in a particular pigment in cone photoreceptors50,51.
The presence of R442 in the inner pore of A3/B3 creates a positive electrostatic potential that would strongly repel monovalent and divalent cations, in contrast to the electronegative or slightly electronegative inner pore of A1 and TAX-4 (Fig. 4e–h). That the cone CNG channel has a hydrophobic cavity gate and a possible electropositive inner gate signifies a stringent control of ion conduction in cones. Why the cone CNG channel has two gates and how these gates open in response to cGMP remain to be elucidated. We speculate that having two gates may be related to the fidelity, dynamics or adaptation of cone’s response to light.
The existence of a unique combination of a hydrophobic gate and a positively charged gate in A3/B3 may offer useful clues for developing therapeutic strategies for the treatment of degenerative visual disorders. Several DAMs in A3 and B3 have been shown to be gain-of-function mutations that alter A3/B3 channel activities or promote cone degeneration 22,52–55. We have found that an ACHM-associated mutation, R410W in A3 31,33, results in constitutive A3/B3 activity in the absence of cGMP and causes death of an immobilized cone cell line (unpublished observations; Xiangdong Zheng, Zhengshan Hu, Huan Li and Jian Yang). There may be more DAMs like R410W. Specific blockers or gating modulators targeting one or both gates in the A3/B3 channel may help retard cone degeneration caused by spontaneously active mutant channels.
A3/B3 has a unique selectivity filter
In TAX-4 and A1 channels, the SF is formed by four conserved amino acids, TIGE (Supplementary Fig. 1). The SF of A3/B3 is formed by T365, I366, G367 and E368 of A3 and T407, I408, G409 and G410 of B3 (Fig. 3d, e). Thus, instead of a symmetric ring of four glutamates, A3/B3 has an asymmetric ring of three glutamates (Fig. 5a, b). R403, located in the pore helix of B3, projects its side-chain toward the SF vestibule, occupying a position ~5 Å away from E368 of A3I and at a horizontal plane similar to that of E368 (Fig. 5a, b). Thus, the external entrance of the SF of A3/B3 is asymmetric and may be less electronegative than that of homomeric CNGA channels. The equivalent position of R403 is a leucine in CNGA subunits and a lysine in human B1, K841 (Supplementary Fig. 1). It will be interesting to examine whether K841projects to the SF vestibule as R403 does.
Fig. 5 |. The selectivity filter.
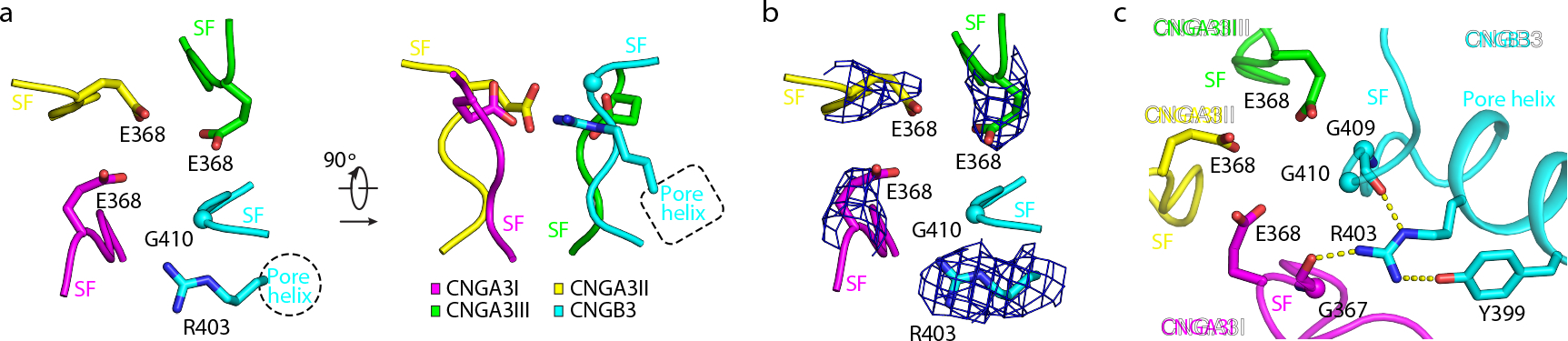
a, The SF viewed from the extracellular side (left) and parallel to the membrane (right). Only the side-chains of the four charged amino acids are shown. b, Densities of the side chains of the four charged residues, viewed from the extracellular side. Densities are shown as blue mesh and are contoured to 3σ. c, Interaction network of R403 of B3. Hydrogen bonds are shown as yellow dash lines.
The electron density of R403 is strong and definitive (Fig. 5b and Extended Data Fig. 2d) and reveals that R403 forms hydrogen bonds with the backbone carbonyl oxygens of G409 of B3 and G367 of A3I (Fig. 5c), both of which line the SF (Fig. 3d, e). R403 also forms a hydrogen bond as well as a π-cation interaction with Y399 in the pore helix of B3 (Fig. 5c). These interactions and the strategic location of R403 suggest that R403 plays a role in maintaining the SF structure and regulating ion selectivity and permeation. Notably, mutating R403 to glutamine produces various disease phenotypes such as progressive macular degeneration, macular dystrophy, cone dystrophy, or ACHM 37,56–61, depending on whether the mutation is monoallelic or biallelic and whether it is combined with an additional monoallelic mutation in CNGA3 37,61. A previous study shows that the A3_R403Q/B3 mutant channel has a 1.33-fold increase in cGMP affinity and a more pronounced outward rectification 52.
A3/B3 has asymmetric gating ring/TMD interactions
The structures of the C-linker and CNBD of A3 and B3 are generally similar to each other and to those of TAX-4 (Fig. 6a), A1 41, and a hyperpolarization-activated, cyclic nucleotide modulated channel 62. There are some differences in the loops connecting some of the β strands in the CNBD (Fig. 6a). As in TAX-4 and A1, the A’B’ helices of the C-linker of one subunit associate with the C’D’ helices of the C-linker of the clockwise (viewing from top down) neighboring subunit, forming the gating ring (Fig. 6b). These intersubunit interactions, first dubbed as the ‘elbow-shoulder’ interactions 62, are mainly hydrophobic interactions but also include salt bridges (Extended Data Fig. 5). These interactions are similar between the A3 intersubunit interfaces (Extended Data Fig. 5a, b) but are distinct between the A3/B3 interfaces (Extended Data Fig. 5c, d).
Fig. 6 |. The C-linker and CNBD.
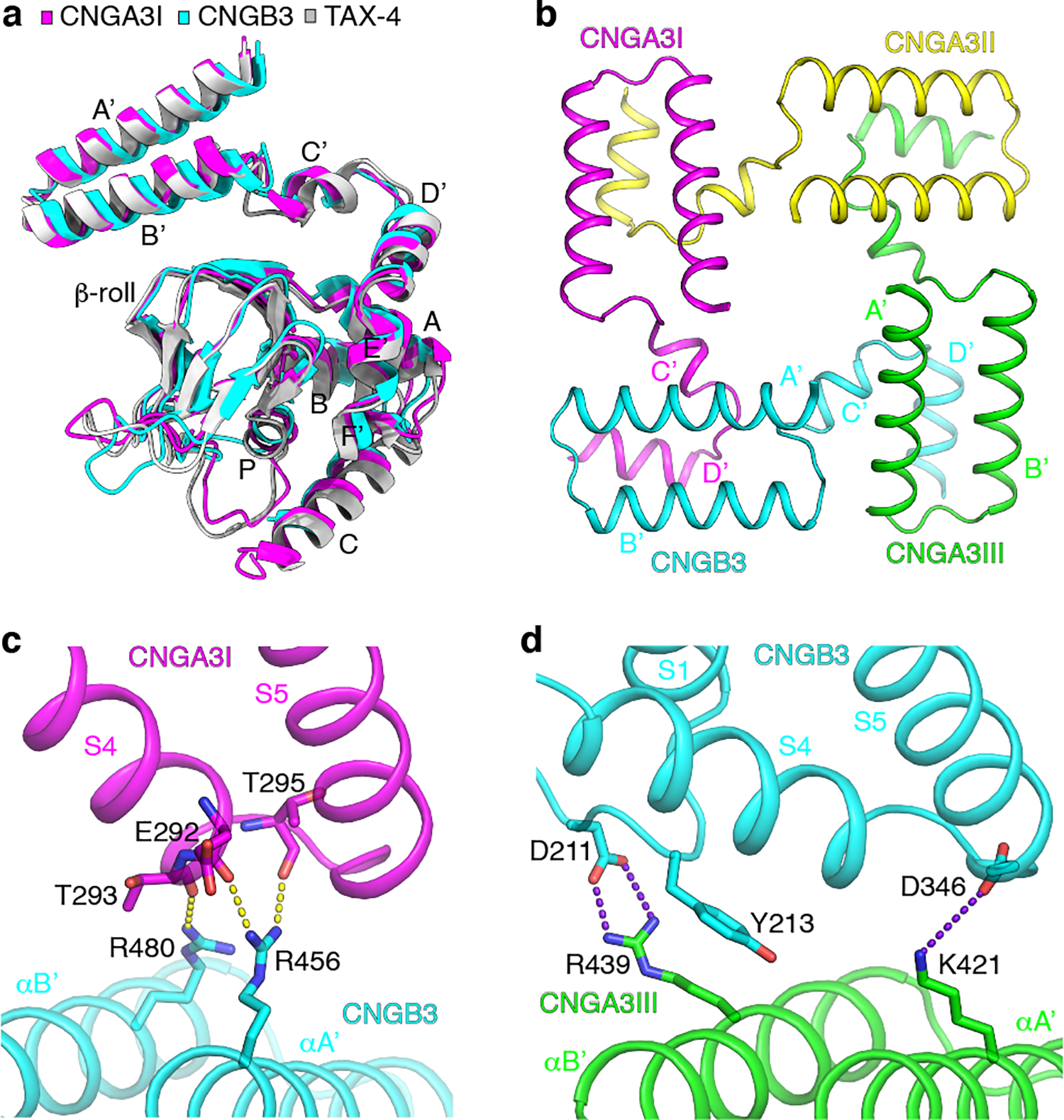
a, Comparison of the structures of the C-linker and CNBD in apo A3I, B3 and TAX-4. b, The gating ring viewed from the extracellular side. c, Interactions between helices A’B’ of the C-linker of B3 and S4-S5 of A3I. Hydrogen bonds are shown as yellow dash lines. d, Interactions between helices A’B’ of the C-linker of A3III and the TMD of B3. Salt bridges are shown as purple dash lines.
The A′ B′ helices of the gating ring interact with the TMD of the same clockwise neighboring subunit, but the interactions vary among different pairs of subunits (Fig. 6c, d). The strongest interactions occur between A3I and B3, where R456 and R480 in helices A’B’ of B3 form hydrogen bonds with the backbone carbonyl groups of E292, T293 and T295 in S4 and S5 of A3I (Fig. 6c). Strong interactions also occur between A3III and B3, where K421 in helix A’ of A3III forms a salt bridge with D346 in the S4-S5 linker of B3, and R439 in helix B’ of A3III forms a salt bridge and a π-cation interaction with D211 and Y213 in a loop immediately preceding S1 of B3 (Fig. 6d). No specific interactions are observed between the gating ring and TMDs of A3I and A3II and between A3II and A3III except weak Van der Waals contacts involving R410 in helix A’. As mentioned above, R410W is a DAM that renders the A3/B3 channel spontaneously active, suggesting that this mutation destabilizes the closed state and stabilizes the open state.
The gating ring plays a critical role in ligand activation of CNG channels 63–68. In TAX-4 and A1 channels, the gating ring undergoes large conformational changes upon the binding of cGMP to the CNBD, transmits its conformational changes to the TMD, and ultimately opens the cavity gate 39,41. The asymmetric and varied interactions between the gating ring and TMDs of different subunits raise the possibility of asymmetric gating of different subunits and may partly underpin some cone CNG channel properties, such as ligand efficacy and gating kinetics.
Concluding remarks
The structure of apo A3/B3 is a structure of a native vertebrate CNG channel and reveals some striking features that shed light on the question of why the native cone CNG channel has a B3 subunit. The pore projection of R403 and R442 of B3 endows the A3/B3 channel with an added positive charge at the external vestibule of an asymmetric SF and a putative positively charged gate below the hydrophobic cavity gate, in comparison to homomeric CNGA channels. Both of these features may play a role in regulating ion permeation, Ca2+ block and ligand gating. Structures of cGMP-bound A3/B3 channel and structure-guided functional studies will shed more light on the role of B3 in shaping cone CNG channel properties and cone photoresponses.
Methods
Molecular cloning
cDNAs encoding human A3 (NCBI Reference Sequence: NP_001289.1) and B3 (NCBI Reference Sequence: NP_061971.3) were amplified by RT-PCR from HEK 293T cells (ATCC) using SuperScript™ III First-Strand Synthesis System (ThermoFisher). For protein expression and purification, full-length A3 was cloned into the pEZT-BM vector 69 with an N-terminal MBP tag followed by a P3C cleavage site and a C-terminal FLAG tag; full-length B3 was cloned into the same vector with an N-terminal 2× Strep tag.
Protein expression and purification
Human A3/B3 were expressed in HEK 293S GnTi− cells (ATCC) using the BacMam system. Recombinant baculoviruses of full-length A3 and B3 were generated separately with Sf9 insect cells (Expression Systems) following the standard protocol. Harvested baculoviruses were amplified twice in Sf9 cells to obtain sufficient viruses for large-scale infection. HEK 293S GnTi− cells were grown in suspension at 37 °C. When cells reached a density of 2–2.5 × 106 cells mL−1, a baculovirus mixture of A3 and B3 at a ratio of 1:1.5 was added to the culture (10%, v/v). After incubation for 12–24 h, the culture was supplemented with 10 mM sodium butyrate to boost the expression, and was further incubated at 30 °C for 72 h before collection.
Purification of human A3/B3 was carried out at 4 °C. Cell pellet from 8 L culture was resuspended and lysed by stirring for 30 min in 200 mL hypotonic buffer (10 mM HEPES-Na, pH 8.58, 0.1 mM TCEP, 2 mM EGTA) supplemented with a protease inhibitor cocktail (Millipore Sigma). Membrane fraction was collected by centrifugation at 29,448 g for 40 min, and then homogenized with a Dounce homogenizer in 200 mL extraction buffer (50 mM HEPES-Na, pH 8.58, 150 mM NaCl, 2% DDM, 0.2% CHS, 0.1 mM TCEP, 2 mM EGTA). After incubation for 2 h, the solubilized membrane was clarified by ultracentrifugation at 111,338 g for 50 min. The supernatant was incubated with amylose resin (New England Biolabs) for 2 h with gentle agitation. The resin was collected by low-speed spin at 800 g, transferred into a gravity column, and washed with 20 CV wash buffer A (50 mM HEPES-Na, pH 8.58, 150 mM NaCl, 0.1% DDM, 0.01% CHS, 0.1 mM TCEP, 2 mM EGTA), followed by 30 CV wash buffer B (50 mM HEPES-Na, pH 8.58, 150 mM NaCl, 0.02% GDN, 0.1 mM TCEP, 2 mM EGTA). The protein was eluted from amylose resin with 30 mL wash buffer B containing 20 mM maltose, and then loaded onto a 1 mL StrepTrap HP column (Cytiva). The column was washed with 20 CV of wash buffer B before eluted with 20 mL wash buffer B containing 5 mM d-Desthiobiotin. Eluted protein was incubated with P3C protease overnight to cleave the MBP tag and was further purified by a Superose 6 column (Cytiva) equilibrated with a gel-filtration buffer (50 mM HEPES-Na, pH 8.58, 150 mM NaCl, 0.02% GDN, 0.1 mM TCEP, 2 mM EGTA). Fractions corresponding to hetero-tetrameric A3/B3 were pooled and concentrated to 1.5 mg mL−1 for cryo-EM analysis.
Cryo-EM sample preparation and data acquisition
Cryo-EM grids were prepared by applying 3 μL of human A3/B3 sample to a glow-discharged UltrAuFoil R0.6/1 300 mesh gold grid (Quantifoil Micro Tools). After waiting for 15 s, the grid was blotted for 10 s (double-sided, blot force 3) under 100% humidity and 4 °C using FEI Vitrobot Mark IV (FEI) and immediately plunged into liquid ethane cooled by liquid nitrogen. Micrographs were acquired by a Titan Krios microscope (ThermoFisher) operated at 300 kV and equipped with a K3 direct electron detector (Gatan) working at counting mode. Leginon 70 was used for data collection. A nominal magnification of 105,000 × was used for imaging the samples, corresponding to a final pixel size of 0.8247 Å on image. The defocus ranged from −0.6 μm to −2.2 μm. A total of 6,684 movies were collected. Each micrograph was dose-fractionated to 40 movie frames under a dose rate of 19.6 counts per pixel per second, with a total exposure time of 2 s and a frame exposure time of 50 ms, resulting in a total dose of 57.63 e−/Å2.
Image processing
Drift correction, beam-induced motion correction and dose-weighting were performed with MotionCor2 71 implemented in RELION 3.0 72. Contrast-transfer function parameters of the micrographs were estimated using Gctf 73. Particles were automatically picked by RELION 3.0. Initially, 3,982,214 particles were picked, 4 × binned (3.2988 Å), and then classified by two rounds 2D classification. After 2D classification, 763,081 particles corresponding to intact particles with evident secondary structures were selected. The selected particles were re-extracted, 2 × binned (1.6494 Å), and then applied to one round 3D classification (K=6) with C1 symmetry. After 3D classification, 361,299 particles from classes exhibiting recognizable continuous secondary structures were selected. 3D refinement was then performed in RELION 3.0 with C1 symmetry using the selected particles to generate a consensus 3D map. The resulting consensus map shows distinct CNG channel features and one subunit showing a different extracellular domain from the other three subunits was considered to be the B3 subunit. A pseudo model of B3 subunit was predicted by SWISS-MODEL 74 using reference of chain A from the apo structure of TAX-4 (PDB ID: 6WEJ). A mask of B3 subunit was generated in RELION 3.0 using a 6 Å map converted from the pseudo model in UCSF Chimera 75. Focused 3D classification (K=4) was then performed by applying the generated mask on B3 subunit. After focused 3D classification, 205,492 particles in the dominant class showing the highest resolution were selected and re-extracted without binning (0.8247 Å). The final particles were applied to cryoSPARC v3.2.0 76 for 3D refinement. Finally, the density map of A3/B3 was refined at a resolution of 2.93 Å with C1 symmetry.
Model building, refinement, and validation
Two pseudo models for A3/B3 were individually predicted by SWISS-MODEL 74 using reference of the apo structure of TAX-4 (PDB ID: 6WEJ) and were then superimposed onto TAX-4 in PyMol 77. Three A3 protomers aligned to chain A, B, C of TAX-4 and one B3 protomer aligned to chain D were extracted and merged together resulting in an initial model for heterotetrameric A3/B3. The initial model was docked into the cryo-EM map of heterotetrameric A3/B3 using UCSF Chimera 75 and then refined against the cryo-EM map using phenix.real_space_refine 78 implemented in PHENIX 79 with the option simulated_annealing turned on. Afterwards, model building was carried out in Coot 80 using the tool of Real-Space Refinement Zone by manually adding missed residues, remodeling unmatched regions and adjusting geometry and rotamer outliers. Coordinates and individual B-factors were finally refined against the cryo-EM map of heterotetrameric A3/B3 using phenix.real_space_refine in PHENIX with the option secondary structure restraints turned on and simulated_annealing off. Overfitting of the atomic models was validated via previously described methods 81. In brief, atoms in the final model were randomly shifted by up to 0.2 Å, and the new model was then refined against one of the two half maps generated during the final 3D reconstruction. FSC values were calculated between the map generated from the resulting model and the two half maps, as well as the averaged map of the two half maps. The quality of the models was evaluated with MolProbity 82. All the figures were prepared in PyMol 77 or UCSF Chimera 75. Pore dimensions were analyzed using HOLE83 program. Cryo-EM data collection, analysis, and refinement statistics are shown in Table 1.
Electrophysiology
For inside-out patch-clamp recordings on giant unilamellar vesicles (GUV) (i.e., liposomes), purified A3/B3 channel proteins were reconstituted at a 1:100 (w/w) ratio into preformed Triton X-100 (0.11 %, w/v)-destabilized liposomes that were prepared from Brain Extract Polar (Avanti Polar Lipid) in 150 mM NaCl and 20 mM HEPES (pH 7.5 adjusted with NaOH). Triton X-100 was subsequently removed by Bio-Beads SM-2 (Bio-Rad). 3 μL of reconstituted proteoliposome was dehydrated on a clean cover slip overnight at 4 °C. The dried proteoliposome layer was then rehydrated at room temperature with a bath solution containing 270 mM NaCl, 30 mM KCl and 10 mM HEPES (pH 7.5 adjusted with NaOH) to form GUV. Single-channel recording pipettes were pulled from borosilicate glass (Corning Pyrex) using a micropipette puller (PC-10; Narishige) and were fire-polished to a resistance of 13−18 MΩ when filled with the bath solution. Giga-Ω seal was formed by gentle suction when the patch pipette made contact with the GUV. To obtain an inside-out single-layer of membrane patch, the pipette was pulled away from the GUV, and the tip was exposed to air for less than 1 s and put back into the bath solution. cGMP, cAMP, and DTZ were added using a home-made perfusion system to the inside of the membrane patch. Currents were amplified by Axopatch 200B and digitized by Digidata 1322A (Molecular Devices). Currents were low-pass filtered at 1 kHz and sampled at 10 kHz. pCLAMP 8.2 software (Molecular Devices) and Microsoft Excel was used for data acquisition and analysis. Single-channel current amplitude histograms were generated and fitted with the Gaussian distribution using Prism 7, and single-channel openings were determined using a 50% threshold of the full amplitude. Because most of the recordings lasted only a few minutes, the currents were mostly recorded with only one concentration of cGMP (10 μM) and at only one or a few voltages, typically −100 mV, −80 mV, +80 mV or +100 mV. Previous recordings of heterologously expressed CNGA3/CNGB3 channels show that their single-channels have a largely linear current-voltage relationship between −100 mV and +100 mV, with a slight inward or outward rectification between +70 and +100 mV 21,22; thus for analyzing single-channel conductance and open probability (Po), we pooled the results at different voltages together, with one voltage for each patch. Data were presented as mean ± SD.
Statistics and reproducibility
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. For all experiments presented as representative images in Extended Data Fig. 1b, c, 3–5 replicates were performed.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper. The cryo-EM density map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-24468. The coordinates of the atomic model have been deposited in the Protein Data Bank under the accession number 7RHS. Several structural coordinates in the PDB database were used in this study, which can be located by accession numbers 6WEJ and 7LFT. Source Data is available online.
Extended Data
Extended Data Fig. 1. Cryo-EM single-particle analysis of apo A3/B3.
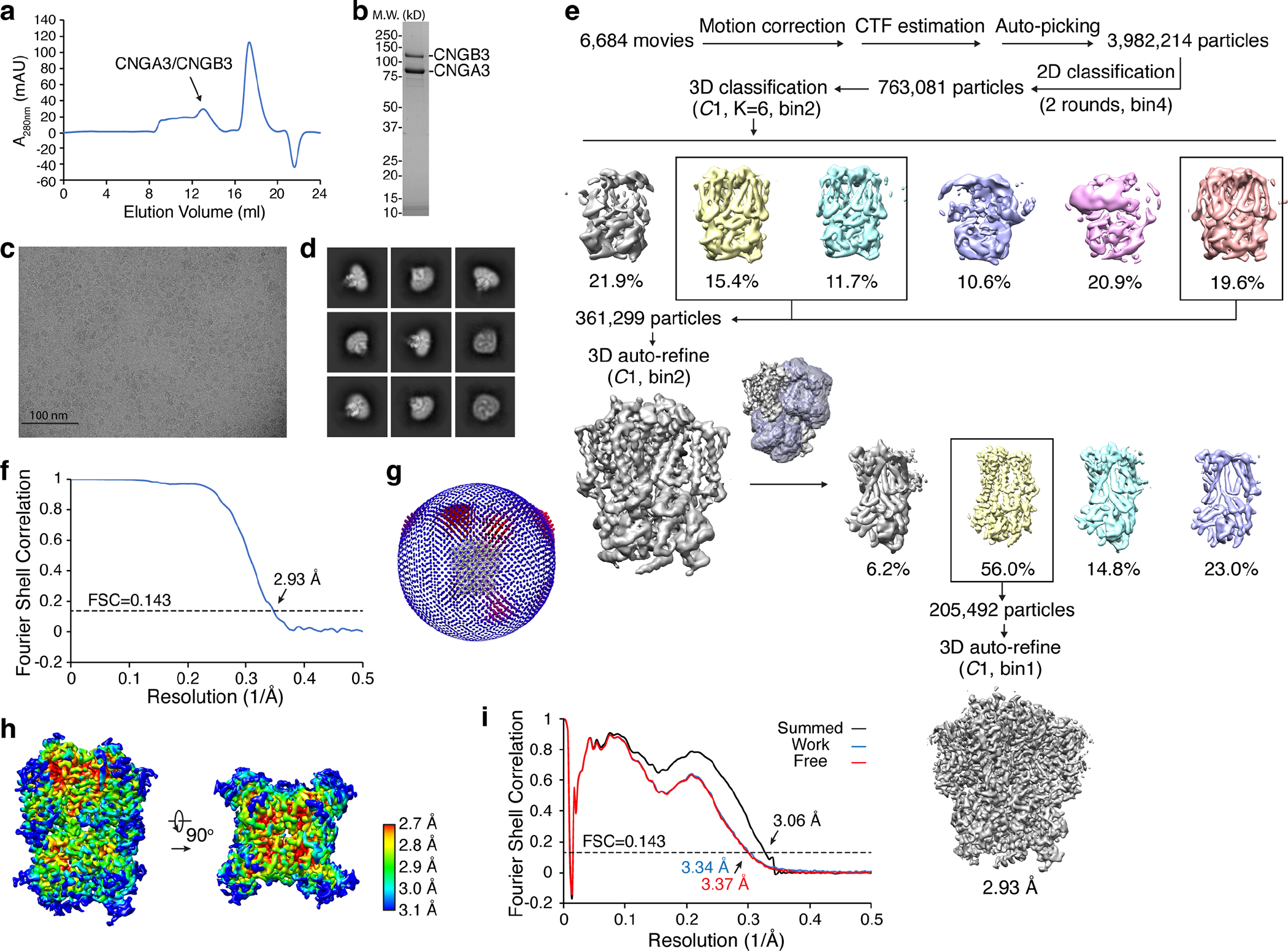
a, Gel filtration profile of heterotetrameric A3/B3 sample. b, SDS-PAGE of A3/B3 sample used for cryo-EM. c, A representative motion-corrected micrograph. Scale bar: 100 nm. d, Gallery of typical averages from 2D classification. e, Flow chart of cryo-EM image processing. f, FSC curve of the final 3D reconstructions. FSC threshold: 0.143. g, Euler angle distribution of particles used in the final 3D reconstruction. h, Local resolution of the final density map. i, FSC curves for cross-validation between maps and model. Black, model versus the summed map. Blue, model versus the half map that was used for model refinement (called ‘work’). Red, model versus another half map that was not used for model refinement (called ‘free’). FSC threshold: 0.143. Uncropped image for panel b is available as source data.
Extended Data Fig. 2. Cryo-EM density maps and fitting of atomic models of selected key regions in all four subunits of A3/B3.
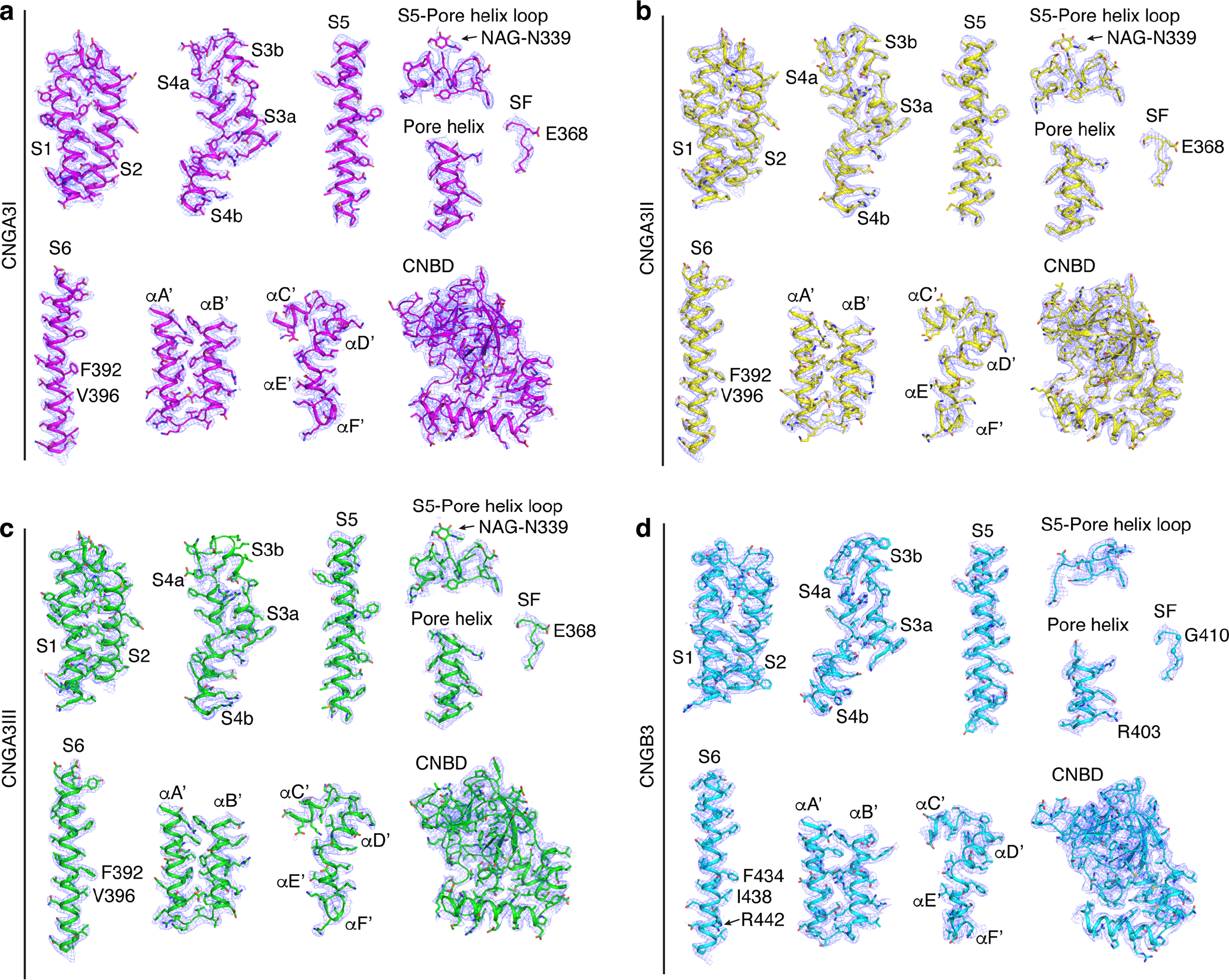
All maps were low-pass filtered to 2.93 Å, sharpened with a temperature factor of −103 Å2 and contoured at 4σ. NAG: N-acetyl-beta-D-glucosamine.
Extended Data Fig. 3. Liposome recordings of purified A3/B3 channel proteins.
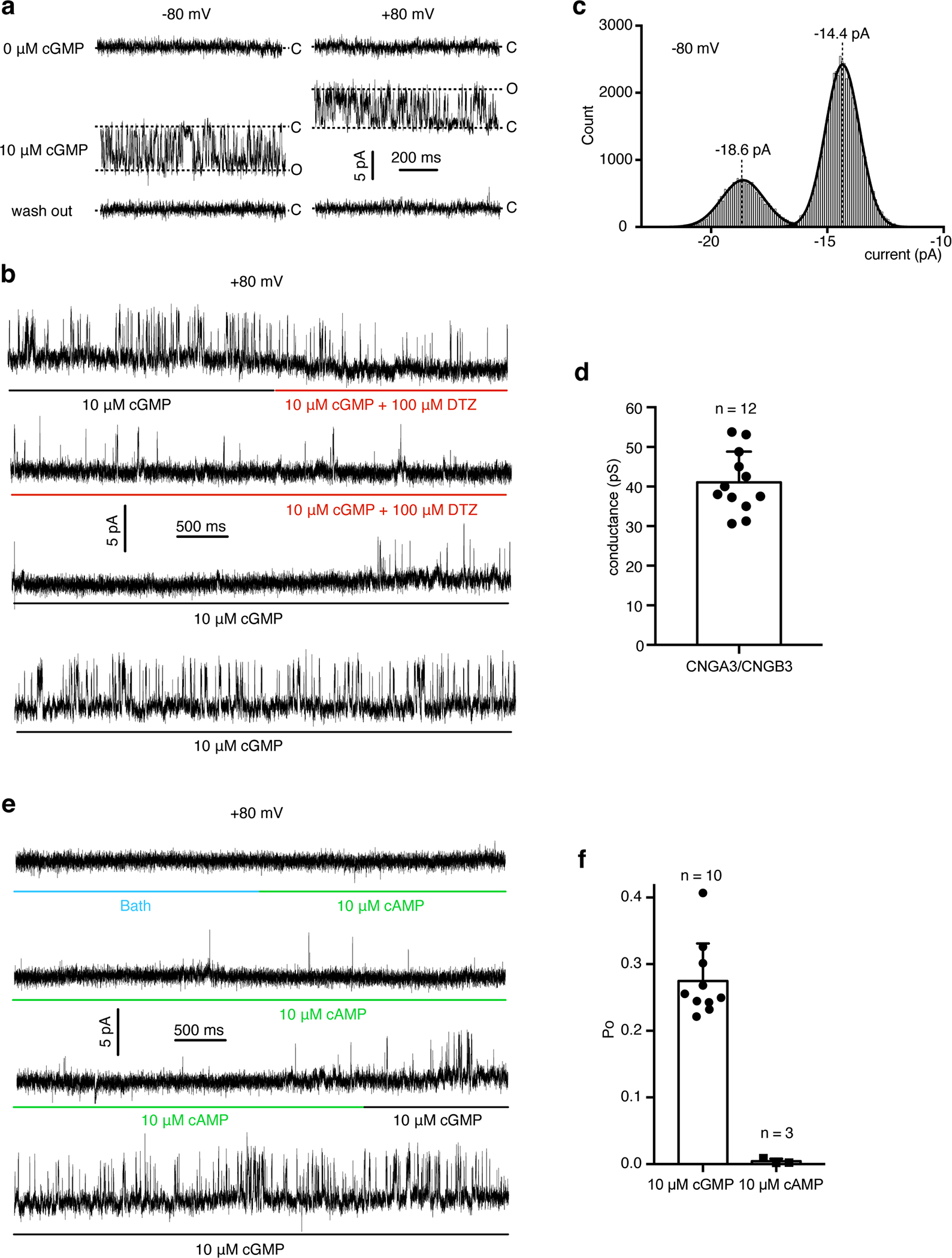
a, Single-channel currents of purified A3/B3 activated by 10 μM cGMP. A total of 29 patches were recorded. Of them, 17 showed no cGMP activity and 12 showed cGMP-activated activity. b, Continuous trace of A3/B3 single-channel current showing inhibition by L-cis-diltiazem (DTZ). c, Amplitude histogram of cGMP-induced currents at + 80 mV from one patch. d, Single-channel conductance of cGMP-induced currents. Data were presented as mean±SD. e, Continuous trace of the same patch as in (b) showing a preference of cGMP over cAMP. f, Single-channel open probability of currents evoked by 10 μM cGMP or 10 μM cAMP. Data were presented as mean±SD.
Extended Data Fig. 4. Alternative projections of F392 in S6 of A3I.
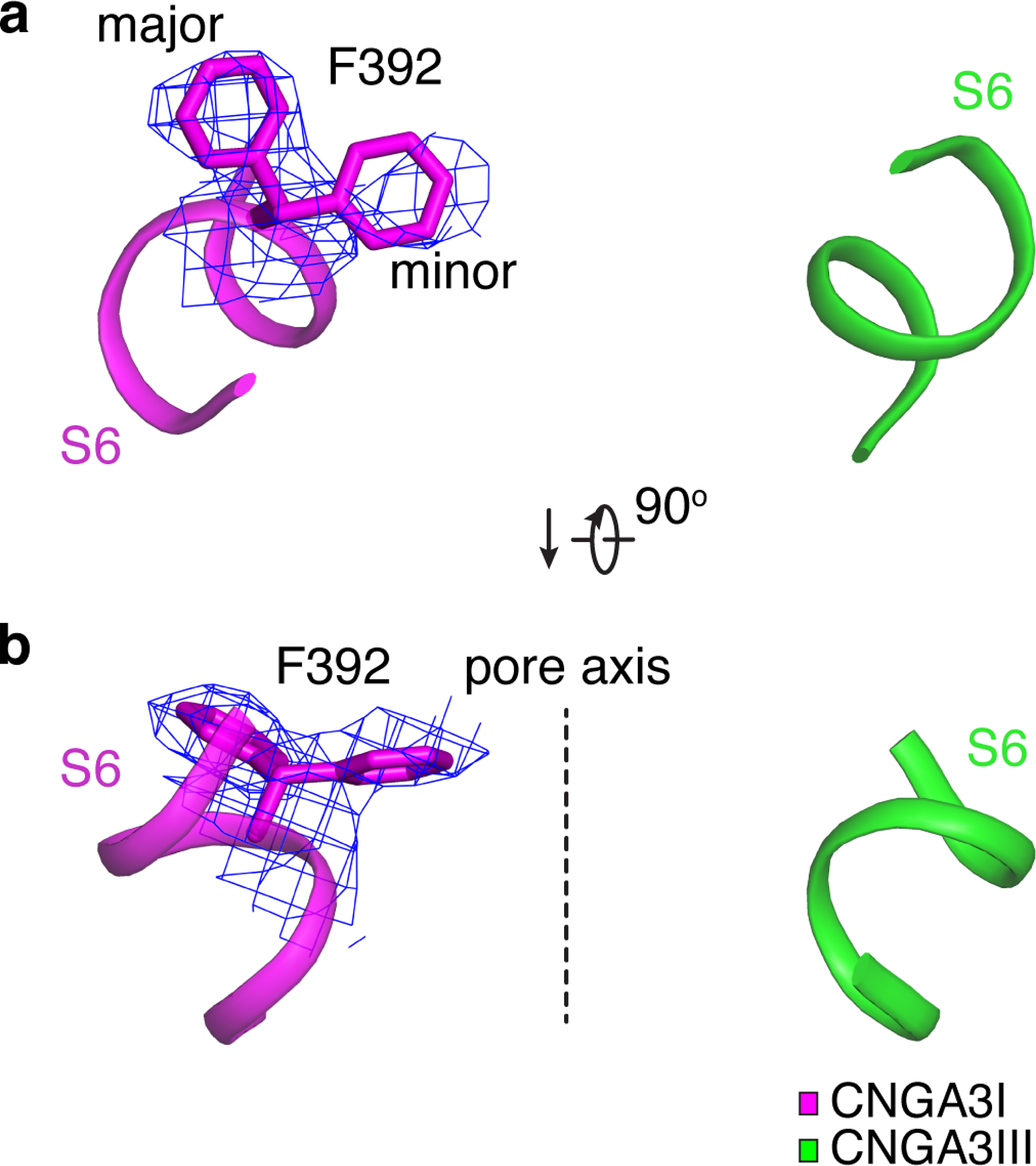
a, b, Close-up view of F392 in S6 of A3I from the extracellular side (a) and parallel to the membrane (b). Cryo-EM densities are shown as blue mesh and are contoured at 3σ.
The dash line marks the pore axis.
Extended Data Fig. 5. Interactions between A’B’ and C’D’ helices of the gating ring.
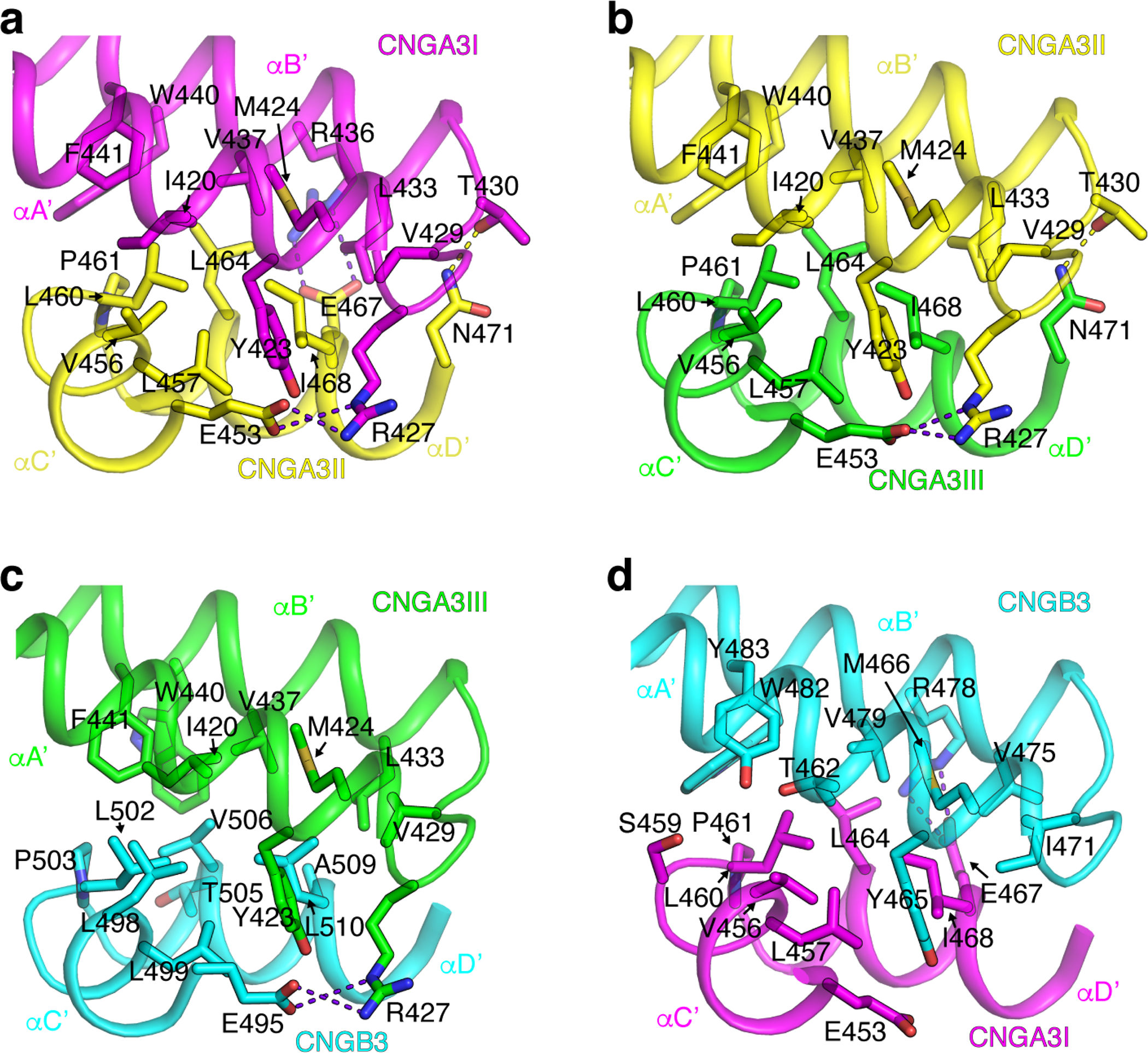
Side-chains of residues involved in A’B’/C’D’ interactions between A3I and A3II (a), A3II and A3III (b), A3III and B3 (c), and B3 and A3I (d) are shown as sticks. Hydrogen bonds and salt bridges are shown as yellow and purple dash lines, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health grants RO1EY027800 and RO1GM085234 (J.Y.). Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center (CUCEC) and at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at NYSBC, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310) with additional support from Agouron Institute (F00316) and NIH (OD019994). Some cryo-EM work was performed at the National Center for CryoEM Access and Training (NCCAT), supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539). We thank members of CUCEC, NYSBC and NCCAT for support and assistance in cryo-EM grid screening and data acquisition. We thank members of our laboratory for discussion during the course of this work.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
References
- 1.Yau KW & Hardie RC Phototransduction motifs and variations. Cell 139, 246–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fesenko EE, Kolesnikov SS & Lyubarsky AL Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature 313, 310–3 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Yau KW & Baylor DA Cyclic GMP-activated conductance of retinal photoreceptor cells. Annu Rev Neurosci 12, 289–327 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Kaupp UB & Seifert R Cyclic nucleotide-gated ion channels. Physiol Rev 82, 769–824 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Masland RH The Neuronal Organization of the Retina. Neuron 76, 266–280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaupp UB et al. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature 342, 762–6 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Zagotta WN & Siegelbaum SA Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci 19, 235–63 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Varnum MD & Dai G Cyclic nucleotide-gated channels. in The Hankbook of Ion Channels (eds. Zheng J & Trudeau MC) 361–382 (CRC Press, Boca Raton, 2015). [Google Scholar]
- 9.Dhallan RS, Yau KW, Schrader KA & Reed RR Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347, 184–7 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T & Gold GH A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature 325, 442–4 (1987). [DOI] [PubMed] [Google Scholar]
- 11.Goulding EH, Tibbs GR, Liu D & Siegelbaum SA Role of H5 domain in determining pore diameter and ion permeation through cyclic nucleotide-gated channels. Nature 364, 61–4 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Eismann E, Muller F, Heinemann SH & Kaupp UB A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc Natl Acad Sci U S A 91, 1109–13 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Root MJ & MacKinnon R Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron 11, 459–66 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Morrill JA & MacKinnon R Isolation of a single carboxyl-carboxylate proton binding site in the pore of a cyclic nucleotide-gated channel. J Gen Physiol 114, 71–83 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CS & MacKinnon R Divalent cation selectivity in a cyclic nucleotide-gated ion channel. Biochemistry 34, 13328–33 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Kaupp UB Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci 11, 188–200 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Biel M & Michalakis S Cyclic nucleotide-gated channels. Handb Exp Pharmacol, 111–36 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Michalakis S, Becirovic E & Biel M Retinal Cyclic Nucleotide-Gated Channels: From Pathophysiology to Therapy. Int J Mol Sci 19(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korschen HG et al. A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–36 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Chen TY et al. A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature 362, 764–7 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Gerstner A, Zong X, Hofmann F & Biel M Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J Neurosci 20, 1324–32 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng C, Rich ED & Varnum MD Achromatopsia-associated mutation in the human cone photoreceptor cyclic nucleotide-gated channel CNGB3 subunit alters the ligand sensitivity and pore properties of heteromeric channels. J Biol Chem 278, 34533–40 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Peng C, Rich ED, Thor CA & Varnum MD Functionally important calmodulin-binding sites in both NH2- and COOH-terminal regions of the cone photoreceptor cyclic nucleotide-gated channel CNGB3 subunit. J Biol Chem 278, 24617–23 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Chen TY et al. Subunit 2 (or Beta) of Retinal Rod Cgmp-Gated Cation Channel Is a Component of the 240-Kda Channel-Associated Protein and Mediates Ca2+-Calmodulin Modulation. Proceedings of the National Academy of Sciences of the United States of America 91, 11757–11761 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady JD et al. Interplay between PIP3 and calmodulin regulation of olfactory cyclic nucleotide-gated channels. Proc Natl Acad Sci U S A 103, 15635–40 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bright SR, Rich ED & Varnum MD Regulation of human cone cyclic nucleotide-gated channels by endogenous phospholipids and exogenously applied phosphatidylinositol 3,4,5-trisphosphate. Mol Pharmacol 71, 176–83 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Meighan PC, Meighan SE, Rich ED, Brown RL & Varnum MD Matrix metalloproteinase-9 and −2 enhance the ligand sensitivity of photoreceptor cyclic nucleotide-gated channels. Channels (Austin) 6, 181–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebrik TI, Botchkina I, Arshavsky VY, Craft CM & Korenbrot JI CNG-modulin: a novel Ca-dependent modulator of ligand sensitivity in cone photoreceptor cGMP-gated ion channels. J Neurosci 32, 3142–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohl S, Jagle H, Wissinger B & Zobor D Achromatopsia. in GeneReviews((R)) (eds. Adam MP et al.) (Seattle (WA: ), 2004). [Google Scholar]
- 30.Biel M & Michalakis S Function and dysfunction of CNG channels: insights from channelopathies and mouse models. Mol Neurobiol 35, 266–77 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kohl S et al. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet 19, 257–9 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Kohl S et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet 9, 2107–16 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Wissinger B et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet 69, 722–37 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson S et al. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J Med Genet 41, e20 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remmer MH, Rastogi N, Ranka MP & Ceisler EJ Achromatopsia: a review. Current Opinion in Ophthalmology 26, 333–340 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Kohl S et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet 13, 302–8 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Mayer AK et al. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum Mutat 38, 1579–1591 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Dryja TP et al. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A 92, 10177–81 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng XD et al. Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel. Nature Structural & Molecular Biology 27, 625–+ (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li MH et al. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue J, Han Y, Zeng W, Wang Y & Jiang Y Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 109, 1302–1313 e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes LW Block of the cyclic GMP-gated channel of vertebrate rod and cone photoreceptors by l-cis-diltiazem. J Gen Physiol 100, 783–801 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong H, Molday LL, Molday RS & Yau KW The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 420, 193–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weitz D, Ficek N, Kremmer E, Bauer PJ & Kaupp UB Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36, 881–9 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Zheng J, Trudeau MC & Zagotta WN Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 36, 891–6 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Shuart NG, Haitin Y, Camp SS, Black KD & Zagotta WN Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat Commun 2, 457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng C, Rich ED & Varnum MD Subunit configuration of heteromeric cone cyclic nucleotide-gated channels. Neuron 42, 401–10 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Ding XQ, Matveev A, Singh A, Komori N & Matsumoto H Biochemical characterization of cone cyclic nucleotide-gated (CNG) channel using the infrared fluorescence detection system. Adv Exp Med Biol 723, 769–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meighan SE, Meighan PC, Rich ED, Brown RL & Varnum MD Cyclic nucleotide-gated channel subunit glycosylation regulates matrix metalloproteinase-dependent changes in channel gating. Biochemistry 52, 8352–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoreson WB & Dacey DM Diverse Cell Types, Circuits, and Mechanisms for Color Vision in the Vertebrate Retina. Physiological Reviews 99, 1527–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nache V et al. Differential regulation by cyclic nucleotides of the CNGA4 and CNGB1b subunits in olfactory cyclic nucleotide-gated channels. Sci Signal 5, ra48 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Bright SR, Brown TE & Varnum MD Disease-associated mutations in CNGB3 produce gain of function alterations in cone cyclic nucleotide-gated channels. Mol Vis 11, 1141–50 (2005). [PubMed] [Google Scholar]
- 53.Meighan PC, Peng C & Varnum MD Inherited macular degeneration-associated mutations in CNGB3 increase the ligand sensitivity and spontaneous open probability of cone cyclic nucleotide-gated channels. Front Physiol 6, 177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Sherpa T & Varnum MD Disease-associated mutations in CNGB3 promote cytotoxicity in photoreceptor-derived cells. Mol Vis 19, 1268–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai G & Varnum MD CNGA3 achromatopsia-associated mutation potentiates the phosphoinositide sensitivity of cone photoreceptor CNG channels by altering intersubunit interactions. Am J Physiol Cell Physiol 305, C147–59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michaelides M et al. Progressive cone dystrophy associated with mutation in CNGB3. Invest Ophthalmol Vis Sci 45, 1975–82 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Nishiguchi KM, Sandberg MA, Gorji N, Berson EL & Dryja TP Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mutat 25, 248–58 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Khan NW, Wissinger B, Kohl S & Sieving PA CNGB3 achromatopsia with progressive loss of residual cone function and impaired rod-mediated function. Invest Ophthalmol Vis Sci 48, 3864–71 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Wiszniewski W, Lewis RA & Lupski JR Achromatopsia: the CNGB3 p.T383fsX mutation results from a founder effect and is responsible for the visual phenotype in the original report of uniparental disomy 14. Hum Genet 121, 433–9 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Thiadens AA et al. Comprehensive analysis of the achromatopsia genes CNGA3 and CNGB3 in progressive cone dystrophy. Ophthalmology 117, 825–30 e1 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Burkard M et al. Accessory heterozygous mutations in cone photoreceptor CNGA3 exacerbate CNG channel-associated retinopathy. Journal of Clinical Investigation 128, 5663–5675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zagotta WN et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–5 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Gordon SE & Zagotta WN A histidine residue associated with the gate of the cyclic nucleotide-activated channels in rod photoreceptors. Neuron 14, 177–83 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Gordon SE & Zagotta WN Localization of regions affecting an allosteric transition in cyclic nucleotide-activated channels. Neuron 14, 857–64 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Brown RL, Snow SD & Haley TL Movement of gating machinery during the activation of rod cyclic nucleotide-gated channels. Biophys J 75, 825–33 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zong X, Zucker H, Hofmann F & Biel M Three amino acids in the C-linker are major determinants of gating in cyclic nucleotide-gated channels. EMBO J 17, 353–62 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou L, Olivier NB, Yao H, Young EC & Siegelbaum SA A conserved tripeptide in CNG and HCN channels regulates ligand gating by controlling C-terminal oligomerization. Neuron 44, 823–34 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Paoletti P, Young EC & Siegelbaum SA C-Linker of cyclic nucleotide-gated channels controls coupling of ligand binding to channel gating. J Gen Physiol 113, 17–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morales-Perez CL, Noviello CM & Hibbs RE Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure 24, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 77.The PyMOL Molecular Graphics System v.2.3. (Schrödinger, LLC, 2019). [Google Scholar]
- 78.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallographica Section D-Structural Biology 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amunts A et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smart OS, Neduvelil JG, Wang X, Wallace BA & Sansom MS HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–60, 376 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper. The cryo-EM density map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-24468. The coordinates of the atomic model have been deposited in the Protein Data Bank under the accession number 7RHS. Several structural coordinates in the PDB database were used in this study, which can be located by accession numbers 6WEJ and 7LFT. Source Data is available online.