Abstract
Actinobacillus actinomycetemcomitans is considered to be one of the major oral putative pathogens, especially in cases of juvenile periodontitis. This microorganism requires nutritionally complex media for growth, and therefore the media for its primary isolation usually include blood agar or serum in their base. In this study we present a new medium, Dentaid-1, which improves the detection of A. actinomycetemcomitans in periodontal samples. In its composition, blood and serum have been omitted, hence reducing its cost and making it a more restrictive medium against the growth of other microorganisms with high nutritional requirements. The growth yields of pure cultures of the bacteria on Dentaid-1 were comparable to those on nonselective blood agar. Moreover, clinical efficacy was evaluated in subgingival samples from 77 subjects with adult periodontitis. Dentaid-1 detected A. actinomycetemcomitans in 24 subjects, while a previously described tryptic soy-serum-bacitracin-vancomycin agar detected the microorganism in only 19 subjects (79.1%). Dentaid-1 is a low-cost, noninhibitory formula for the improved diagnosis and monitoring of patients subgingivally infected by this important oral putative pathogen.
In the last few years, substantial evidence has emerged that Actinobacillus actinomycetemcomitans may be, along with Porphyromonas gingivalis, a major oral putative pathogen, as judged by this organism's rare occurrence in periodontally healthy individuals (25). A. actinomycetemcomitans has been isolated from adult periodontitis lesions, but less frequently and in lower numbers than from lesions in juvenile periodontitis subjects (21, 26). Treatment failures have been associated with the failure to reduce the amount of the microorganism in treated sites (14, 17). Furthermore, data on the transmission of A. actinomycetemcomitans from host to host (5, 6, 32) and new evidence of its role as an infectious agent involved in disease development at extraoral sites (35) are providing one of the strongest associations between this oral pathogen and periodontal and systemic diseases.
Existing data on the presence of A. actinomycetemcomitans in clinical infections have been obtained by using both selective and nonselective media for their isolation. Since A. actinomycetemcomitans is found in small proportions and because its growth can be inhibited in vitro by common oral streptococcal species (23), selective culture media are useful tools in the detection and enumeration of this bacterium. Although new molecular techniques are extremely sensitive in the detection of target bacteria (32, 33, 34), culture techniques are still the methods of reference for studying viable cells and a prerequisite for determining the antimicrobial susceptibility of a given pathogen.
Among the selective culture media described for the isolation and enumeration of A. actinomycetemcomitans (11, 16, 23), tryptic soy-serum-bacitracin-vancomycin (TSBV) medium (23) has been the most widely used in the analysis of any kind of oral samples (4, 19) in studies throughout the world (2, 10, 13, 26). TSBV medium has been defined as a low-inhibitory medium compared with a nonselective blood agar medium (23). Moreover, it allows for the direct detection of catalase activity on the primary isolation plate, facilitating the presumptive identification of A. actinomycetemcomitans (23). Catalase activity is a key assay in distinguishing between A. actinomycetemcomitans and the morphologically similar Haemophilus aprophilus (23). The genus Actinobacillus is included in the Pasteurellaceae family. These microorganisms require nutritionally complex media for primary isolation (15). Usually blood agar or serum has been the base for the design of selective media (11, 16, 23).
The aim of the present study was to evaluate a new selective culture medium for A. actinomycetemcomitans that lacks both blood and serum. It was designed to confirm the following expectations: optimal growth of A. actinomycetemcomitans and suppression of oral flora that should be equal to or better than what is observed when using TSBV medium as the reference medium.
MATERIALS AND METHODS
Bacterial strains.
The A. actinomycetemcomitans strains used were ATCC 33383 and NCTC 10981, obtained from the American Culture Collection, Rockville, Md., and from the National Collection of Type Cultures, London, England, respectively. Additionally, 20 clinical isolates from our laboratory were included. The 22 strains were maintained at −85°C and subcultured three times at 48-h intervals on brain heart infusion agar (BHIA) (Difco Laboratories, Detroit, Mich.) before testing. Plates were incubated in a CO2 incubator (5% CO2) (Sanyo Electric Co., Ltd.).
Selective media.
Dentaid-1 was prepared using BHIA to which the following compounds were added: 5 g of yeast extract, 1.5 g of sodium fumarate (Sigma Chemical Co., St. Louis, Mo.), and 1 g of sodium formate (Sigma) per liter. The medium was autoclaved for 15 min at 121°C. The final pH was 7.2 ± 0.2. Once the medium was cooled to 50°C, vancomycin (Sigma) was added to a final concentration of 9 μg/ml. TSBV medium was prepared according to the original description of the medium (23).
Recovery efficiency of pure A. actinomycetemcomitans cultures.
BHIA subcultures of the 22 previously described strains were inoculated into freshly prepared tubes containing brain heart infusion broth (Difco), and the cell density was adjusted to 108 to 109 cells per ml. Suspensions were dispersed by mixing with a vortex mixer for 30 s and then serial 10-fold diluted in phosphate-buffered saline (PBS). Adequate dilutions were spread in duplicate by using a spiral plater (Countermat; IUL Instruments) and incubated for 72 h in a 5% CO2 incubator. The media used were; TSBV, Dentaid-1, and an anaerobic blood agar (BHIA, 5% horse blood, 5 mg of hemin per liter, 10 mg of menadione per liter). Bacterial counts were numbered as CFU/milliliter. For each strain, the yield of growth on the two selective media was compared with that found on the blood agar and is expressed as a relative-growth-supporting-ability (RGSA) value, which was determined as the logarithm of the ratio of the number of colonies on the blood agar to the number of colonies on the selective agar (11, 12).
Clinical specimens.
Subgingival plaque samples from 77 patients with untreated (before mechanical treatment of scaling and root planning) or treated (1 to 3 months after treatment) adult periodontitis were received for microbiological diagnosis at our laboratory from several private dental clinics. One pooled sample per patient was obtained by sampling the four deepest periodontal pockets (one in each quadrant) and using paper points (two paper points per site) as previously described (9). The eight paper points per patient were transferred to 2 ml of reduced transport fluid (28), which was transported and processed within 24 h. In the laboratory, samples were dispersed with a vortex mixer for 30 s and serially diluted in 10-fold steps in prereduced PBS. Appropriate dilutions were plated in parallel on TSBV and Dentaid-1. After incubation for 72 h at 37°C in a 5% CO2 incubator, the plates were examined for the presence and enumeration of A. actinomycetemcomitans. The contaminant flora (rest of the flora) was quantified in positive samples. Counts on clinical samples were also numbered as CFU/milliliter.
An A. actinomycetemcomitans presumptive identification was first made on the basis of colonial morphology. Presumptive identification continued with determining the catalase activity at 72 h of incubation on discrete colonies on the primary isolation plate. Catalase-positive and catalase-negative colonies resembling A. actinomycetemcomitans were subcultured on BHIA (Difco), and after 24 to 48 h of incubation the catalase activity was confirmed upon subculture. Strains were also tested for lactose fermentation (ONPG [o-nitrophenyl-β-d-galactopyranoside]) (27), and ONPG-positive strains were identified by Gram stain, aerotolerance, and rapid enzymatic methods (Innovative Diagnostic Systems, Inc., Norcross, Ga.). Subsequently, lactose-negative colonies resembling A. actinomycetemcomitans were confirmed by using a species-specific PCR (8, 20).
PCR.
For PCR analysis of presumptive A. actinomycetemcomitans, a colony was resuspended in 100 μl of sterile distilled water. Then, 3.2 μl of this suspension was used in each PCR reaction. The primers used for PCR were designed to identify A. actinomycetemcomitans targeted 16S rRNA (3) were as follows: the forward primer sequence was 5′-AAACCCATCTCTGAGTTCTTCTTC-3′, and the reverse primer sequence was 5′-ATGCCAACTTGACGTTAAAT-3′. These primers gave an expected amplification product of 557 bp. PCR amplification was carried out in a reaction volume of 25 μl consisting of 3.2 μl of the initial sample in water for a final volume of 20.1 μl and 4.9 μl of the reaction mixture containing 1× PCR buffer [67 mM Tris-HCl, pH 8.8; 16 mM (NH4)2SO4, 0.01% Tween 20; 1.5 mM MgCl2], 0.6 U of EcoTaq DNA polymerase (ECOGEN), 0.25 mM concentrations of the deoxynucleoside triphosphates, and 80 pmol of each primer. PCR cycling was carried out in a GeneAmp PCR System 2400 (Perkin-Elmer, Barcelona, Spain). After an initial denaturation step of 94°C for 5 min, 35 amplification cycles of denaturation at 94°C for 30 s, annealing of primers at 55°C for 30 s, and primer extension at 72°C for 30 s were carried out, followed by a final primer extension step at 72°C for 7 min. Reaction products were either stored at −20°C or analyzed immediately.
Negative control samples consisting of the standard mixture with 3.2 μl of sterile distilled water were included in each batch of samples analyzed by PCR.
RESULTS
The 20 clinical isolates used for recovery efficiency purposes were all gram negative, capnophilic short rods, and non-lactose fermenting and rendered the band of a 557-bp amplification product by PCR assay (3). Catalase activity was positive for all but one.
The mean RGSA values for A. actinomycetemcomitans on TSBV and Dentaid-1 were 0.81 (Standard deviation [SD] = 1.61) and 0.06 (SD = 0.11), respectively. The difference was statistically significant (P = 0.03, Student paired t test). The RGSA value for Dentaid-1 suggests no inhibition compared to blood agar.
For the 77 specimens collected from periodontal pockets, a good relationship was found in the detection of A. actinomycetemcomitans in TSBV and Dentaid-1. From 24 positive subgingival samples, 19 were detected in parallel by TSBV and Dentaid-1, and five additional positive samples were found on Dentaid-1. In summary, A. actinomycetemcomitans was found in 24.7% (n = 19) of the samples when they were assayed with TSBV and in 31.2% (n = 24) when Dentaid-1 was used.
Colonies on TSBV after 72 h of incubation are rough, circular, and convex with slightly irregular edges; they appear as small colonies with dark borders and a common star-shaped inner structure (23). In some strains, the morphology at 72 h on Dentaid-1 can differ from this appearance, showing smooth, circular, spherical colonies with an enhanced dark border and an incipient star inner structure. A total of 46 subcultures (21 from TSBV and 25 from Dentaid-1) from clinical specimens were performed for presumptive identification from selected colonies resembling A. actinomycetemcomitans on TSBV and Dentaid-1. From these, 43 subcultures (19 from TSBV and 24 from Dentaid-1) were ONPG negative and were confirmed by PCR as being A. Actinomycetemcomitans (3).
From the 43 subcultures, 41 subcultures on BHIA after 48 h of incubation in 5% CO2 showed strong catalase activity, and 2 were catalase negative, corresponding to strains from TSBV and Dentaid-1 from the same sample. Three subcultures were ONPG positive: two strains isolated from TSBV were identified as H. aphrophilus and Haemophilus segnis, whereas one strain isolated from Dentaid-1 was identified as H. segnis.
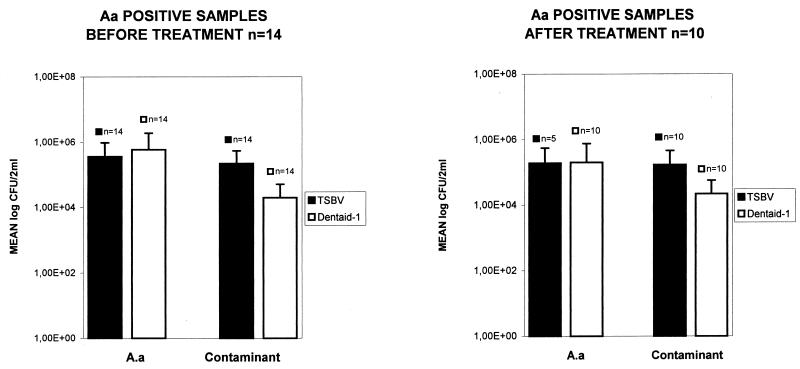
When A. actinomycetemcomitans-positive samples were grouped according to patients before or after mechanical treatment, differences between the two media appeared only in the prevalence of the bacteria in the latter group, as shown in Table 1. Dentaid-1 detected twice as many A. actinomycetemcomitans-positive samples as did TSBV in these particular samples. As shown in Table 1, no differences in the mean recovery of viable CFU of A. actinomycetemcomitans per milliliter were observed (P = 0.5, Student paired t test). However, higher mean percentages of incidence of the bacteria were calculated from Dentaid-1 in both groups, which represents statistically significant differences (P = 0.005, Student paired t test). This suggests a lower recovery of contaminant flora by Dentaid-1 in comparison to TSBV. This is clearly shown in Fig. 1, where the mean log CFU of both A. actinomycetemcomitans and contaminant flora per milliliter are represented. The differences between contaminant flora on Dentaid-1 with respect to contaminant flora on TSBV are statistically significant (P = 0.007, Student paired t test).
TABLE 1.
Recovery and percentage of total cultivable A. actinomycetemcomitans microrganisms in positive subgingival samples from patients with adult periodontitis
| Treatment group and medium | No. of positive samples (n = 24) | Mean CFU/ml ± SD (105) | Mean % ± SD |
|---|---|---|---|
| Before mechanical treatment | |||
| TSBV | 14 | 1.6 ± 2.8 | 43.6 ± 38. |
| Dentaid-1 | 14 | 3.3 ± 7.0 | 81.7 ± 31. |
| After mechanical treatment | |||
| TSBV | 5 | 0.9 ± 1.8 | 37.2 ± 38.0 |
| Dentaid-1 | 10 | 1.0 ± 2.8 | 71.3 ± 40.6 |
FIG. 1.
Recovery of A. actinomycetemcomitans and contaminant flora (rest of flora) on Dentaid-1 in comparison to TSBV in samples before or after mechanical treatment.
DISCUSSION
Development of a selective medium for the specific recovery of a given pathogen is primarily based on the selection of an adequate nutritive base and a proper inhibition system for reducing contaminant flora. Potential drawbacks with selective media are overselectivity or insufficient selectivity.
In the literature, Trypticase soy agar supplemented with either blood (16) or serum (11, 23) has been the nutritive base applied to selective media designed for the isolation of A. actinomycetemcomitans. Preliminary work carried out in our laboratory indicated that BHIA by itself is an excellent nutritive base that allows for good growth of pure cultures of the bacteria. When 5-g/liter doses of yeast extract are added, colonial development is comparable to that observed in TSBV (23). This fact allowed us to omit blood and serum from the nutritive base of the selective medium Dentaid-1. Moreover, if blood and serum are omitted from the formula, contaminant flora with high nutritional requirements are controlled and the cost per plate greatly decreased. The TSBV medium described by Slots (23) which omits the blood content of the previous MGB (malachite green-bacitracin) selective medium described by Mandell and Socransky (16), results in the inhibition of hemin-requiring Haemophilus species (23). Dentaid-1 suppresses the growth of H. aphrophilus and Haemophilus paraphrophilus by 3 log orders with respect to both anaerobic blood agar and TSBV (data not shown). Vancomycin plus formate and/or fumarate, as described below, and the use of a BHIA nutritive base lacking both blood and serum were found to be responsible for such suppression. Interestingly, H. aphrophilus is described as being morphologically similar to A. actinomycetemcomitans (23).
Bacitracin and vancomycin are antibiotics commonly used in the formulation of microbiological media (7) and particularly for the isolation of A. actinomycetemcomitans due to their expected role against a wide range of mainly gram-positive flora (19). Holm et al. (11) improved inhibition, including the inhibition of more gram-negative bacteria, by complementing bacitracin and vancomycin activity with carbenicillin, fusidic acid, and spiramycin in the same formula. This medium, called A-medium, is described as a modification of Slots' TSBV medium (23) and inhibits the growth of Capnocytophaga spp. and Neisseria spp. These species are found in large quantities as contaminant microorganisms in samples taken from periodontal pockets (22, 23, 31). In spite of the various antibiotics included, A-medium resulted only in slightly higher inhibition of the growth of A. actinomycetemcomitans compared with TSBV, as expressed by RGSA values of 1.12 and 0.76, respectively. These RGSA values for TSBV are similar to the value obtained in the present study. In consequence, our results also agree with the previous observations of Holm et al. (11), which indicated The suppression of A. actinomycetemcomitans grown on TSBV compared with blood agar. In our study, five pure cultures of A. actinomycetemcomitans (22.7%) experienced high suppression on TSBV. Between 2 and 5 logs of inhibition were observed, contributing to the high RGSA value obtained for TSBV. Possible explanations may be either methodological differences, as suggested by Holm et al. (11), or TSBV's overselectivity for some strains, as indicated by our results.
The vancomycin content of Dentaid-1 was chosen for its efficacy in eliminating streptococcal species (16, 18, 23) and the high resistance of A. actinomycetemcomitans to this antibiotic, with a MIC90 value of ≥64 μg/ml (16).
Vancomycin as a sole inhibitory agent has been previously used in Hammond's selective medium for the oral putative pathogen Campylobacter rectus (B. F. Hammond and D. Mallonee, Abstract, J. Dent. Res. 67:327, abstr. 1712). Our first observations of A. actinomycetemcomitans pure culture grown on Hammond's medium (Hammond and Mallonee, abstract) formulated on a BHIA base indicated an unexpected slow colonial development, which seems to be due to an ingredient in the formula other than vancomycin. Hammond's medium incorporates vancomycin (9 μg/ml) as a selective agent and an SH2 indicator system (ferrous sulfate, 0.2 g/liter, and sodium thiosulfate, 0.3 g/liter) as a differential marker with sodium formate (2 g/liter) and sodium fumarate (3 g/liter) as energy sources. Since our preliminary studies showed that formate and/or fumarate delays colonial development of A. actinomycetemcomitans from 24 to 48 h, the dosage of formate and fumarate was studied in a liquid medium (data not shown) to allow for good growth of A. actinomycetemcomitans. Furthermore, some strains belonging to other gram-negative species, mainly H. aphrophilus and H. paraphrophilus, were suppressed. Formate and fumarate sodium salts are usually included in cultivation media as an energy source for formate- and/or fumarate-requiring species (29; Hammond and Mallonee, abstract). Since vancomycin or formate-fumarate by themselves do not have such inhibitory qualities, we postulate that their combination can have a synergistic effect upon strains of certain gram-negative species in subgingival samples. No references have been found in the literature regarding this possible synergistic effect.
Adult periodontitis patients were chosen in order to challenge the efficacy of Dentaid-1 under the worst possible conditions. In the 24 positive clinical samples studied, Dentaid-1 suppressed contaminant flora by 10-fold compared to TSBV. Before mechanical treatment (scaling and root planning), A. actinomycetemcomitans was detected by Dentaid-1 and TSBV in the same patients and showed similar recovery rates. Surprisingly, after mechanical treatment, The prevalence of the bacteria was 100% higher in Dentaid-1 than in TSBV. Dentaid-1's greater suppression of contaminant flora, and TSBV's suppression of some strains of A. actinomycetemcomitans may be an explanation for these results. Furthermore, after mechanical treatment, subgingival pockets should be recolonized by species that are more inhibited by the new proposed medium. Streptococcal species are the most commonly encountered subgingival species, and they are known to be inhibitory for A. actinomycetemcomitans growth in vitro (23). Experimental work on comparing the species suppressed by Dentaid-1 and those suppressed by TSBV was beyond the scope of the present study and will be the subject of further specific study.
The presumptive identification performed here took into account colonial morphology, catalase activity, and lactose fermentation as previously described (1, 24) and is considered to help in the rapid and accurate screening of A. actinomycetemcomitans from either TSBV or Dentaid-1 selective media. We found two catalase-negative strains growing on both Dentaid-1 and on TSBV from clinical isolates. Although catalase-negative strains have been reported as relatively rare (30), they must be considered in order to achieve a correct microbiological diagnosis.
Finally, and in order to complement the presumptive identification of A. actinomycetemcomitans colonies isolated on Dentaid-1, we performed specific PCR which confirms our results. According to other authors (8, 20), the extreme specificity of PCR has been found to be particularly useful for the identification of suspected pathogens, supplying inconclusive or unexpected biochemical patterns as previously reported (20).
In conclusion, the new proposed medium, Dentaid-1, improves the detection of A. actinomycetemcomitans inexpensively, with a noninhibitory formula, and can be of considerable aid in microbiological diagnosis and in monitoring patients subgingivally infected with this bacterium.
ACKNOWLEDGMENT
We thank Ann Bangle for her contribution in correcting the manuscript.
REFERENCES
- 1.Alcoforado G A P, McKay T L, Slots J. Rapid method for detection of lactose fermenting oral microorganisms. Oral Microbiol Immunol. 1987;2:35–38. doi: 10.1111/j.1399-302x.1987.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 2.Ali R W, Johannessen A C, Dahlén G G, Socransky S S, Skaug N. Comparison of the subgingival microbiota of periodontally healthy and diseased adults in Northern Cameroon. J Clin Periodontol. 1997;24:830–835. doi: 10.1111/j.1600-051x.1997.tb01197.x. [DOI] [PubMed] [Google Scholar]
- 3.Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Asikainen S, Chen C, Alaluusua S, Slots J. Can one acquire periodontal bacteria and periodontits from a family member? JADA. 1997;128:1263–1271. doi: 10.14219/jada.archive.1997.0403. [DOI] [PubMed] [Google Scholar]
- 5.Asikainen S, Alaluusua S, Saxén L. Recovery of Actinobacillus actinomycetemcomitans from teeth, tongue and saliva. J Periodontol. 1991;62:203–206. doi: 10.1902/jop.1991.62.3.203. [DOI] [PubMed] [Google Scholar]
- 6.Asikainen S, Chen C. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol 2000. 1999;20:65–81. doi: 10.1111/j.1600-0757.1999.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 7.Atlas R M. Selective components. In: Parks L C, editor. Handbook of microbiological media. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 7–10. [Google Scholar]
- 8.Bogen G, Slots J. Black-pigmented anaerobic rods in closed periapical lesions. Int Endodontol J. 1999;32:204–210. doi: 10.1046/j.1365-2591.1999.00216.x. [DOI] [PubMed] [Google Scholar]
- 9.Haffajee A D, Socransky S S. Effect of sampling strategy on the false-negative rate for detection of selected subgingival species. Oral Microbiol Immunol. 1992;7:57–59. doi: 10.1111/j.1399-302x.1992.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 10.Hagiwara S, Takamatsu N, Tominaga Y, Umeda M. Subgingival distribution of periodontopathic bacteria in adult periodontitis and their susceptibility to minocycline-HCl. J Periodontol. 1998;69:92–99. doi: 10.1902/jop.1998.69.1.92. [DOI] [PubMed] [Google Scholar]
- 11.Holm A, Rabe P, Kalfas S, Edwarsson S. Improved selective culture media for Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J Clin Microbiol. 1987;25:1985–1988. doi: 10.1128/jcm.25.10.1985-1988.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalfas S, Edwardsson S. Agar media that indicate acid production from sorbitol by oral microorganisms. J Clin Microbiol. 1985;22:959–961. doi: 10.1128/jcm.22.6.959-961.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamma J J, Nakou M, Manti F A. Microbiota of rapidly progressive periodontitis lesions in association with clinical parameters. J Periodontol. 1994;65:1073–1078. doi: 10.1902/jop.1994.65.11.1073. [DOI] [PubMed] [Google Scholar]
- 14.Kornman K S, Robertson P B. Clinical and microbiological evaluation of therapy for juvenile periodontitis. J Periodontol. 1985;56:443–446. doi: 10.1902/jop.1985.56.8.443. [DOI] [PubMed] [Google Scholar]
- 15.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: The Williams & Wilkins Co.; 1984. [Google Scholar]
- 16.Mandell R L, Socransky S S. A selective medium for Actinobacillus actinomycetemcomitans and the incidence of the organism in juvenile periodontitis. J Periodontol. 1981;52:593–598. doi: 10.1902/jop.1981.52.10.593. [DOI] [PubMed] [Google Scholar]
- 17.Mandell R L, Tripodi L S, Savitt E, Goodson J M, Socransky S S. The effect of treatment on Actinobacillus actinomycetemcomitans in localized juvenile periodontitis. J Periodontol. 1986;57:94–99. doi: 10.1902/jop.1986.57.2.94. [DOI] [PubMed] [Google Scholar]
- 18.Mensa J, Gatell J M, Jiménez de Anta M T, Prats G. Caracteristicas de los antimicrobianos. In: Masso S A, editor. Guía de terapéutica antimicrobiana. 7th ed. Barcelona, Spain: Editorial Masson; 1997. p. 44. [Google Scholar]
- 19.Müller H P, Lange D E, Müller R F. Actinobacillus actinomycetemcomitans recovery from extracrevicular locations of the mouth. Oral Microbiol Immunol. 1993;8:344–348. doi: 10.1111/j.1399-302x.1993.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 20.Riggio M P, MacFarlane T W, Mackenzier D, Lennon A, Smith A J, Kinane D. Comparison of polymerase chain reaction and culture methods for detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in subgingival plaque samples. J Periodontol Res. 1996;31:496–501. doi: 10.1111/j.1600-0765.1996.tb01415.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodenburg J P, van Winkelhoff A J, Winkel E G, Goene R J, Abbas E, de Graaff J. Occurrence of Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in severe periodontitis in relation to age and treatment history. J Clin Periodontol. 1990;17:392–399. doi: 10.1111/j.1600-051x.1990.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 22.Slots J. Subgingival microflora and periodontal disease. J Periodontol. 1979;6:351–382. doi: 10.1111/j.1600-051x.1979.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 23.Slots J. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1982;15:606–609. doi: 10.1128/jcm.15.4.606-609.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slots J. Rapid identification of important periodontal microorganisms by cultivation. Oral Microbiol Immunol. 1986;1:48–55. doi: 10.1111/j.1399-302x.1986.tb00318.x. [DOI] [PubMed] [Google Scholar]
- 25.Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontal disease: introduction. Periodontol 2000. 1999;20:7–13. doi: 10.1111/j.1600-0757.1999.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 26.Slots J, Feik D, Rams T E. Actinobacillus actinomycetemcomitans and Bacteroides intermedius in human periodontitis: age relationship and mutual association. J Clin Microbiol. 1990;17:659–662. [PubMed] [Google Scholar]
- 27.Summanen P, Baron E J, Citron D M, Strong C, Wexler H M, Finegold S M. Wadsworth anaerobic bacteriology manual. 5th ed. Belmont, Calif: Star Publishing Company; 1993. Biochemical test procedures; p. 152. [Google Scholar]
- 28.Syed S A, Loesche W J. Survival of human dental plaque flora in various transport media. Appl Microbiol. 1972;24:638–644. doi: 10.1128/am.24.4.638-644.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner A C R, Visconti R A, Socransky S S, Holt S C. Classification and identification of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus by cluster analysis and deoxyribonucleic acid hybridizations. J Periodontol Res. 1982;17:585–596. doi: 10.1111/j.1600-0765.1982.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 30.Tanner A. Media for cultivation of Eikenella corrodens formate and fumarate requiring species of one bacteria. Oral Microbiol Immunol. 1987;2:187–189. doi: 10.1111/j.1399-302x.1987.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 31.Tempro P J, Slots J. Selective medium for the isolation of Haemophilus aphrofilus from the human periodontium and other oral sites and the low proportion of the organism in the oral flora. J Clin Microbiol. 1986;23:777–782. doi: 10.1128/jcm.23.4.777-782.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiroco E M B, Sivakumar M, Preus H R. The distribution and transmission of Actinobacillus actinomycetemcomitans in families with localized juvenile periodontitis. J Clin Periodontol. 1998;25:99–105. doi: 10.1111/j.1600-051x.1998.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 33.Umeda M, Contreras A, Chen C, Bakker I, Slots J. The utility of whole saliva to detect the oral presence of periodontopathic bacteria. J Periodontol. 1998;69:828–833. doi: 10.1902/jop.1998.69.7.828. [DOI] [PubMed] [Google Scholar]
- 34.Umeda M, Chen C, Bakker I, Contreras A, Morrison J L, Slots J. Risk indicators for harboring periodontal pathogens. J Periodontol. 1998;69:1111–1118. doi: 10.1902/jop.1998.69.10.1111. [DOI] [PubMed] [Google Scholar]
- 35.Van Winkenhoff A J, Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in non-oral infections. Periodontol 2000. 1999;20:122–135. doi: 10.1111/j.1600-0757.1999.tb00160.x. [DOI] [PubMed] [Google Scholar]