Abstract
Overexpression of antiapoptotic BCL2 family proteins occurs in various hematologic malignancies and contributes to tumorigenesis by inhibiting the apoptotic machinery of the cells. Antagonizing BH3 mimetics provide an option for medication, with venetoclax as the first drug applied for chronic lymphocytic leukemia and for acute myeloid leukemia. To find additional hematologic entities with ectopic expression of BCL2 family members, we performed expression screening of cell lines applying the LL-100 panel. Anaplastic large cell lymphoma (ALCL) and primary effusion lymphoma (PEL), 2/22 entities covered by this panel, stood out by high expression of MCL1 and low expression of BCL2. The MCL1 inhibitor AZD-5991 induced apoptosis in cell lines from both malignancies, suggesting that this BH3 mimetic might be efficient as drug for these diseases. The ALCL cell lines also expressed BCLXL and BCL2A1, both contributing to survival of the cells. The combination of specific BH3 mimetics yielded synergistic effects, pointing to a novel strategy for the treatment of ALCL. The PI3K/mTOR inhibitor BEZ-235 could also efficiently be applied in combination with AZD-5991, offering an alternative to avoid thrombocytopenia which is associated with the use of BCLXL inhibitors.
Subject terms: Cancer, Oncology, Pathogenesis
Introduction
Apoptosis or programmed cell death is a process which eliminates old cells or cells that are otherwise no longer needed or detrimental. Decisive for the fate of the individual cell is the interaction between pro- and antiapoptotic members of the BCL2 family1. Many forms of cancer have developed mechanisms to prevent mitochondrial apoptosis by altering the equilibrium between pro- and antiapoptotic proteins2. Expression levels and functional importance of individual antiapoptotic proteins are considered to be different in the different types of tumors1.
The aberrant expression of BCL2 and of other members of the BCL2 family is well-described in follicular lymphoma (FL), chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), and in acute myeloid leukemia (AML)3,4. Mimicking proapoptotic BH3-only proteins, BH3 mimetics antagonize the function of antiapoptotic BCL2 family proteins, providing a new opportunity for treatment of these diseases3,4. Antagonizing BCL2, venetoclax was the first BH3 mimetic approved for CLL and then—in combination with hypomethylating drugs—also for AML1,5. Various new drugs targeting BCL2 family members are currently evaluated in clinical trials1.
Aim of our study was to identify hematologic entities not yet in the focus of broad interest, but with a conspicuous expression pattern of antiapoptotic BCL2 family members, indicating a potential for the application of BH3 mimetics. To delineate disease-specific expression patterns of antiapoptotic genes in hematologic tumors, we screened cell lines from the leukemia-lymphoma 100 (LL-100) panel. In 2019, we had developed this panel of cell lines covering 22 entities of leukemia and lymphoma malignancies as novel platform for the development of targeted therapies6.
RNA-seq analysis of the LL-100 panel showed that cell lines derived from two rare lymphomas, anaplastic large cell lymphoma (ALCL) and from primary effusion lymphoma (PEL) stood out by high expression of MCL1. MCL1 had already been described as essential mediator for the survival of multiple myeloma (MM) and of AML7–9. BH3 mimetics targeting MCL1 are promising options for the therapy of both diseases5,8. We describe the effects of the MCL1 inhibitor AZD-5991 on growth and apoptosis in ALCL and PEL cell lines. Furthermore, we discuss under which circumstances the combination of AZD-5991 with other BCL2 family inhibitors might be of clinical benefit. Inhibitors against the antiapoptotic AKT pathway are an alternative for combination treatment with AZD-5991.
Results and discussion
Expression of MCL1 and BCL2 in cell lines of the LL-100 panel
Overexpression of antiapoptotic BCL2 family genes promotes survival of cells in hematological malignancies10,11. Deregulation of BCL2 in hematopoietic tumors could be traced back to the translocation t(14;18), amplification or hypomethylation of the gene2.
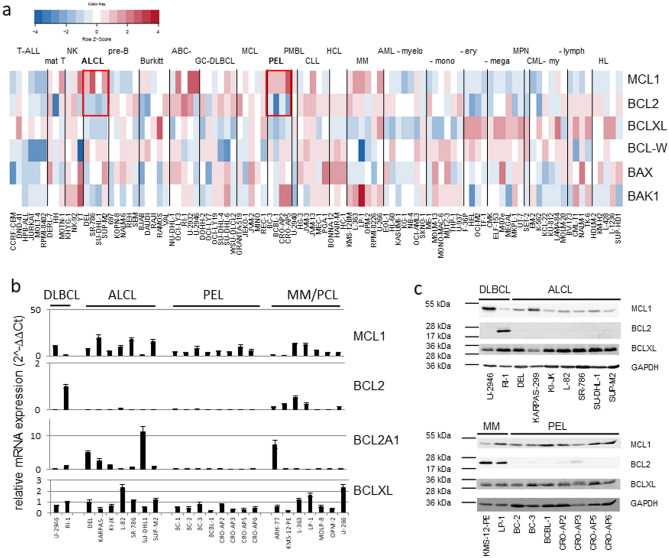
MCL1 is one of the eminent BCL2 family members which, when overexpressed, contributes significantly to the survival of malignant cells7,12. To find potential targets of BH3 mimetics, we examined the expression of BCL2 family genes in the LL-100 cell lines panel. Remarkably, all ALCL and PEL cell lines showed an MCL1high/BCL2low expression pattern (Fig. 1a). Quantitative reverse transcriptase (qRT) PCR analysis with an extended number of cell lines confirmed expression of MCL1 and absence of BCL2 in ALCL and PEL cell lines (Fig. 1b). Western blot analysis verified this finding at the protein level (Fig. 1c).
Figure 1.
Expression of BCL2 family genes in the LL-100 panel. (a) Heat map of RNA-seq gene expression data of BCL2 family members. Framed in red are expression levels of MCL1 and BCL2 in ALCL and PEL. (b) Results of qRT-PCR showing expression of BCL2 family members. (c) Western blot analysis showing the protein expression of MCL1, BCL2 and BCLXL in ALCL and PEL cell lines. DLBCL and MM cell lines were included as controls. Note that ALCL and PEL cell lines are consistently MCL1pos/BCL2neg.
MCL1 amplification in ALCL and PEL cell lines
MCL1 is the dominant antiapoptotic BCL2 family member in MM, more critical than BCL2 and BCLXL7,8. MCL1 is also essential for survival of AML cells5,9,13,14 and of Philadelphia chromosome positive acute lymphocytic leukemia (ALL) cells15.
The MCL1 gene is located on chromosome 1 (q21.3), in a region which is often amplified in human cancer16. According to our quantitative genomic PCR, 6/7 ALCL cell lines and 6/8 PEL cell lines carry gains of MCL1 (Suppl. Table 1). In most cell lines, the gains result from global tri- or tetraploidies or from aneuploidies of chromosome 1 (Suppl. Table 1). But the ALCL cell line with highest MCL1 protein expression levels (KARPAS-299) carries indeed a strong amplification of the MCL1 locus (9n), suggesting that focal gain can play a role in the aberrant overexpression of MCL1 (Fig. 1c, Suppl. Table 1).
Providing an alternative explanation for MCL1 expression in ALCL, the cytokine-induced activation of the TYK2/STAT1 pathway triggers MCL1 in this disease17.
Effect of BH3 mimetics on PEL cell lines
MCL1 and BCL2 are antiapoptotic BCL2 family members. MCL1 is overexpressed in ALCL and PEL tumors and protects cells from apoptosis18–20. Figure 1c shows that cell lines from both hematologic entities are MCL1pos/BCL2neg, suggesting that they might represent disease-specific models to study the efficiency of MCL1 inhibitors.
Inhibitors of antiapoptotic BCL2 family proteins (BH3 mimetics) are emerging agents for the treatment of BCL2 dependent tumors. Different compounds are in clinical trials, with the BCL2 inhibitor Venetoclax (ABT-199) being the first BH3 mimetic approved by the FDA for CLL1.
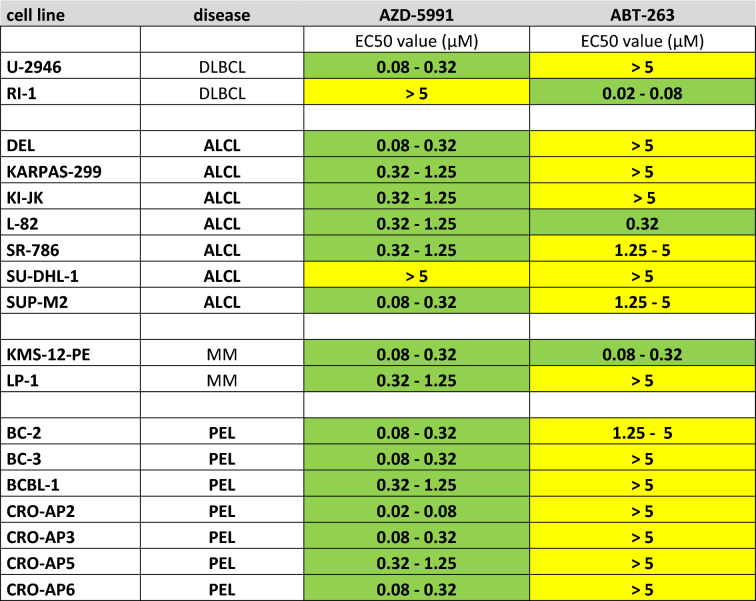
Before testing ALCL and PEL cell lines, we examined the growth inhibitory effects of three MCL1 inhibitors (AT-101, AZD-5991, UMI-77) and of one BCL2/BCLXL inhibitor (ABT-263) on the diffuse large B cell lymphoma (DLBCL) cell lines U-2946 (MCL1pos/BCL2neg) and RI-1 (MCL1low/ BCL2pos) (Fig. 1c). These cell lines were chosen as controls, as we had assessed their sensitivity for BH3 mimetics in an earlier study21. ABT-263 inhibited growth of the BCL2pos cell line (Ri-1), AZD-5991 efficiently inhibited growth of the MCL1pos cell line (U-2946) (Table 1, Suppl. Fig. 1a–c). The activity of caspases 3/7 was induced accordingly, confirming that the effects of the drugs on cell counts resulted from apoptosis (Suppl. Fig. 1b,c). Two MCL-1 inhibitors (AT-101 and UMI-77) were active in the µM range (data not shown) and hence no longer used in this study.
Table 1.
Effects of AZD-5991 and ABT-263 on growth of lymphoma cell lines.
EC50 values were determined after 48 h by life cell imaging. Green indicates inhibitor sensitivity of cell line (EC50 < 1.25 µM); yellow indicates resistance (EC50 ≥ 1.25 µM). EC50 values for BI-97C1 targeting various BCL2 family members including BCL2A1 were identical between the ALCL cell lines L-82 and SU-DHL-1 (1.25–5 µM).
Western blot analysis showed that PEL cell lines are MCL1pos/BCL2neg (Fig. 1c). Therefore, it was not surprising that all PEL cell lines tested (7/7) were sensitive to the MCL1 inhibitor AZD-5991 (EC50 < 1.25 µM) and resistant to the BCL2 inhibitor ABT-263 (EC50 ≥ 1.25 µM) (Table 1).
PEL is a rare and aggressive non-Hodgkin lymphoma, with poor prognosis22,23. PEL cell lines exhibit a characteristic gene expression profile including high levels of CD138, PRDM1 and SLAMF7, the latter being a potential candidate for targeted antibody therapy6. We show here that the MCL1 inhibitor AZD-5991 induces apoptosis in PEL cell lines. These results confirmed the importance of MCL1 for survival of PEL20.
Thus, PEL patients might benefit from targeted therapy with MCL1 antagonists. Nevertheless, our cell lines data need to be confirmed in primary tumor samples.
Effect of BH3 mimetics on ALCL cell lines
Like PEL cell lines, also ALCL cell lines are MCL1pos/BCL2neg (Fig. 1c). However, only five out of seven ALCL cell lines were sensitive to the MCL1 inhibitor and resistant to the BCL2 inhibitor, begging the question why cell line SU-DHL-1 was resistant for both inhibitors and why cell line L-82 responded to both inhibitors (Table 1).
ABT-263 inhibits not only BCL2, but also BCLXL and BCL-W. Therefore, expression of the latter two genes might explain responsiveness of a BCL2neg cell line to this drug. Western blot analysis allowed detection of BCLXL in ALCL cell lines, but showed only faint signals for BCL-W (Suppl. Fig. 2). In accordance, CT values of BCL-W, as determined by qRT-PCR, were decisively higher than those of MCL1 and BCLXL (> 5 CT value difference; data not shown). Therefore, we assessed BCLXL as candidate antiapoptotic mediator in ALCL.
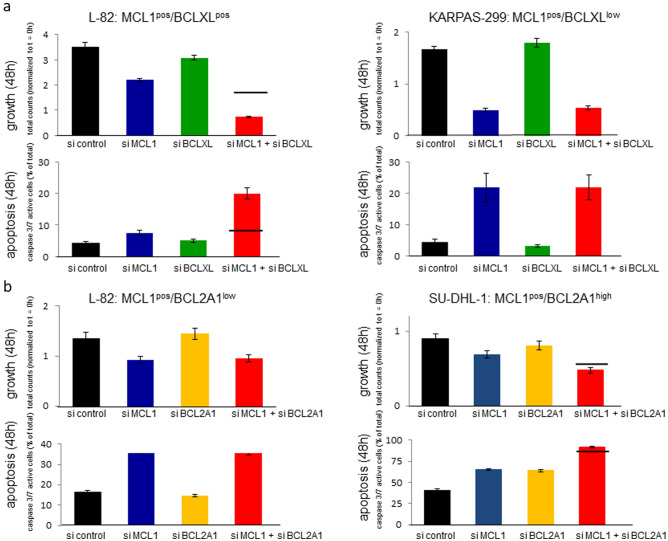
Knock-down experiments were performed to elucidate whether BCLXL was crucial for survival of the ALCL cell lines. The efficiency of knockdown with short interfering (si) RNA oligonucleotides was verified by qRT-PCR (Suppl. Fig. 3). MCL1 and BCLXL knockdown led to the reduction of growth in the MCL1pos/BCLXLpos cell line L-82 (Fig. 2a). Co-knockdown of both genes yielded more than additive effects with respect to growth arrest and induction of apoptosis (Fig. 2a). The MCL1pos/BCLXLlow cell line KARPAS-299 responded to knockdown of MCL1, but not to knockdown of BCLXL (Fig. 2a).
Figure 2.
SiRNA-mediated knockdown of BCL2 family members in ALCL cell lines. Life cell imaging shows effects of knockdown of BCL2 family members on growth (cell counts) and apoptosis (caspase 3/7 activity) 48 h after onset of treatment with siRNA oligos. (a) Knockdown of MCL1 and BCLXL; (b) knockdown of MCL1 and BCL2A1. Note synergistic effects of MCL1 and BCLXL knockdown in cell line L-82 and additive effects of MCL1 and BCL2A1 knockdown in cell line SU-DHL-1. The horizontal line shows the expected additive effects of the combination of siRNA oligos.
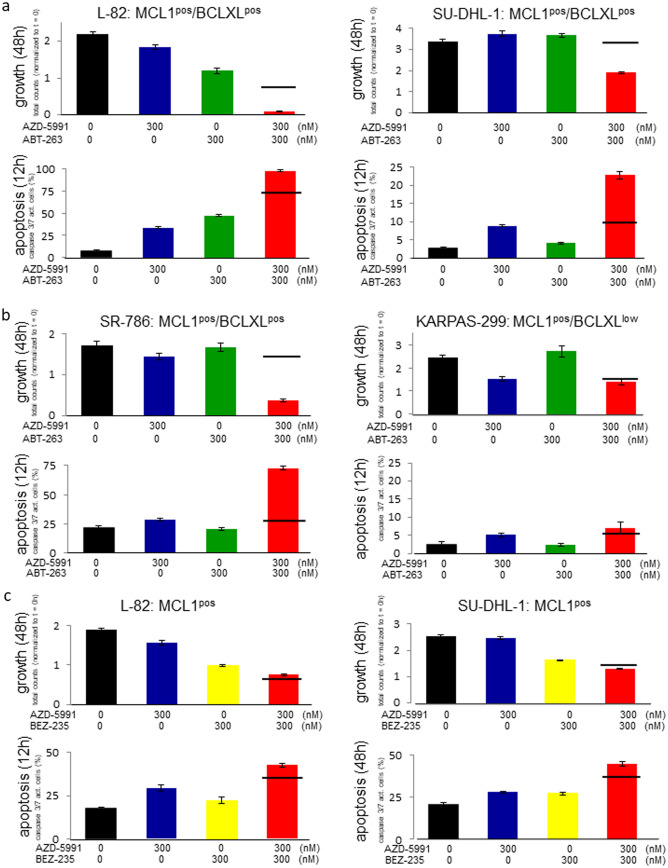
Accordingly, co-stimulation of L-82 cells with suboptimal concentrations of AZD-5991 (300 nM) and ABT-263 (300 nM) yielded synergistic antiproliferative and proapoptotic effects when compared to the effects of each inhibitor alone (Fig. 3a).
Figure 3.
Effect of AZD-5991 and ABT-263 on growth and apoptosis of ALCL cell lines. Life cell imaging data showing the effect of the MCL1 inhibitor AZD-5991 (300 nM) and the BCL2/BCLXL inhibitor ABT-263 (300 nM) on growth (48 h) and apoptosis (12 h) of (a) L-82, SU-DHL-1 and (b) SR-786, KARPAS-299. (c) shows effects of AZD-5991 and BEZ-235 on growth and apoptosis of cell lines L-82 and SU-DHL-1. Apoptosis was assessed by caspase 3/7 activity. The horizontal line shows the expected additive effects of the combination of both inhibitors. Note that the only cell line that does not respond to ABT-263 is the BCLXLlow cell line KARPAS-299.
Thus, knockdown and inhibitor experiments showed that BCLXL acts as antiapoptotic mediator in cell line L-82, cooperating with MCL1.
At first sight the notion that BCLXL acts as a major antiapoptotic mediator in ALCL seems to conflict with the observation that 5/6 ABT-263 resistant ALCL cell lines (EC50 ≥ 1.25 µM) expressed BCLXL (Fig. 1c, Table 1). These findings provoke the question why BCLXL exhibited an overt antiapoptotic function in L-82, but not in other BCLXLpos cell lines (Fig. 1c, Table 1). Interestingly, also the MCL1pos/BCL2pos MM cell line LP-1 tested AZD-5991 sensitive / ABT-263 resistant (Fig. 1c, Table 1), suggesting that MCL1 is the dominant molecule and that inhibitors targeting other antiapoptotic proteins are effective only if MCL1 is also inhibited.
To find out whether indeed ABT-263 resistance of ALCL cell lines resulted from the strong and dominant effect of MCL1, we costimulated all ALCL cell lines with suboptimal concentrations of AZD-5991 and ABT-263.
Regarding growth arrest, the combination of ABT-263 and AZD-5991 achieved more than additive effects in 4/6 BCLXLpos ALCL cell lines (L-82, SR-786, SU-DHL-1, KI-JK) (Fig. 3a,b, Suppl Fig. 4a). Synergistic activation of caspases was observed in even 5/6 (L-82, SR-786, SU-DHL-1, KI-JK, DEL) cell lines (Fig. 3a,b, Suppl. Fig. 4b). Costimulation with both drugs led to growth arrest also in SU-DHL-1 which was found to be resistant to either inhibitor alone (Table 1; Fig. 3a). Cell lines with weak expression levels of BCLXL (ALCL cell line KARPAS-299 and PEL cell line BC-3) did not respond significantly to ABT-263, neither alone nor in combination with the MCL-1 inhibitor (Fig. 3b, Suppl. Fig. 4a,b).
These results provide convincing evidence that besides MCL1 BCLXL acts as second antiapoptotic mediator in ALCL, with dominant effects of MCL1 in most cases.
BCLXL and BCL2A1: antiapoptotic mediators in ALCL
MCL1 had been described as the main antiapoptotic mediator in ALCL19. We show here that BCLXL acts as second antiapoptotic protein in this disease. However, the resistance of cell line SU-DHL-1 to MCL1 and BCLXL inhibitors when applied individually, suggests that still another protective protein might be active in this cell line. qRT-PCR analysis showed that cell line SU-DHL-1 stood out by high levels of BCL2A1 (Fig. 1b). Like BCLXL, BCL2A1 is regulated by the ALCL associated fusion protein NPM-ALK that sustains survival of the malignant cells24,25. All ALCL cell lines tested in this study carry the t(2;5)(p23;q35) encoding the NPM-ALK fusion gene.
BCL2A1 knockdown with siRNA oligonucleotides arrested growth and induced apoptosis in SU-DHL-1 (BCL2A1high), but not in L-82 (BCL2A1low) (Fig. 2b). Supporting the notion that BCL2A1 acts as antiapoptotic mediator in SU-DHL-1, this cell line also responded to the broad BCL2 family inhibitor BI-97C1 (legend to Table 1). BI-97C1 (Sabutoclax) is a drug which among other BCL2 family members also targets BCL2A126.
In conclusion, knockdown and inhibitor experiments show that MCL1 is not the only antiapoptotic BCL2 family member in ALCL. BCLXL and BCL2A1 protect from apoptosis as well. If validated in primary cells, our data suggest that treatment with a combination of inhibitors against MCL1, BCLXL and BCL2A1 might be an alternative option for patients.
Inhibition of PI3K/mTOR
The combination of specific BH3 mimetics or the use of inhibitors targeting multiple BCL2 family members might be a strategy for the treatment of ALCL. However, targeted therapies with BCLXL inhibitors have been associated with thrombocytopenia27. Therefore, we set out to elucidate whether the combination of the MCL1 inhibitor AZD-5991 and BEZ-235, the latter a dual inhibitor of PI3K and mTOR, might be an alternative to induce apoptosis in cells that are resistant to the MCL1 inhibitor alone.
The rationale for these experiments lies in earlier studies reporting that apoptosis in ALCL and DLBCL models was induced by downregulation of MCL1 following inhibition of the AKT/mTOR pathway28,29. Thus, we applied a dual PI3K/mTOR inhibitor (BEZ-235) to test whether this drug would repress MCL1 in our cell lines. We speculated that a double hit against MCL1, reducing expression (BEZ-235) and inhibiting its function (AZD-5991), might create cooperative effects.
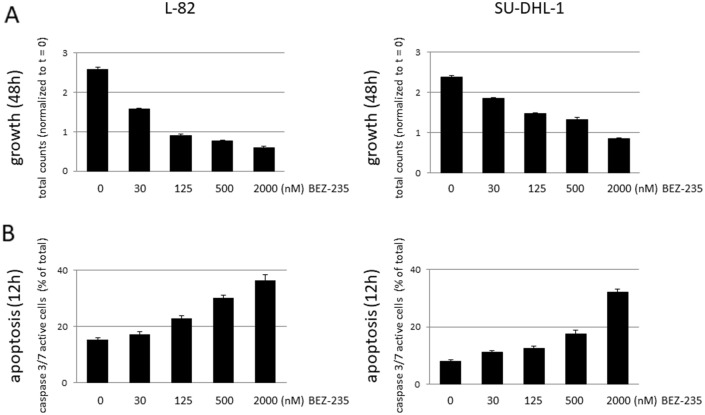
Indeed, BEZ-235 inhibited proliferation (48 h) and induced apoptosis (12 h) in a dose-dependent manner (Fig. 4). Low concentrations of BEZ-235 (30 nM, 24 h) induced dephosphorylation of AKT, the AKT target GSK3B and of the mTOR target S6 (Fig. 5a). Induction of apoptosis as assessed by caspase 3/7 activation paralleled dephosphorylation of AKT and its targets (Fig. 5b,c). However, apoptosis was not preceded by downregulation of MCL1 which we had expected based on the previously reported data28,29 (Fig. 5a,b). The MCL1 protein has a very short half-life30,31. Consequently, stimulation of cells with the global transcriptional inhibitor THZ1 (200 nM, 7.5 h) resulted in a dramatic drop of MCL1 (Suppl. Fig. 5). Even at high concentrations (500 nM, 24 h), BEZ-235 did not have such an effect on the expression of MCL1 (Fig. 5a). Also BCLXL and BCL2A1 were not influenced by BEZ-235 (Fig. 5a,b). Therefore, we conclude that the BEZ-235 induced apoptosis in ALCL cell lines was not caused by downregulation of MCL1, and also not by downregulation of BCLXL and BCL2A1.
Figure 4.
Effect of BEZ-235 on growth and apoptosis of ALCL cell lines. Life cell imaging data showing the effect of the PI3K/mTOR inhibitor BEZ-235 on (a) growth (48 h) and (b) apoptosis (12 h) of ALCL cell lines L-82 and SU-DHL-1. Apoptosis was assessed by caspase 3/7 activity.
Figure 5.
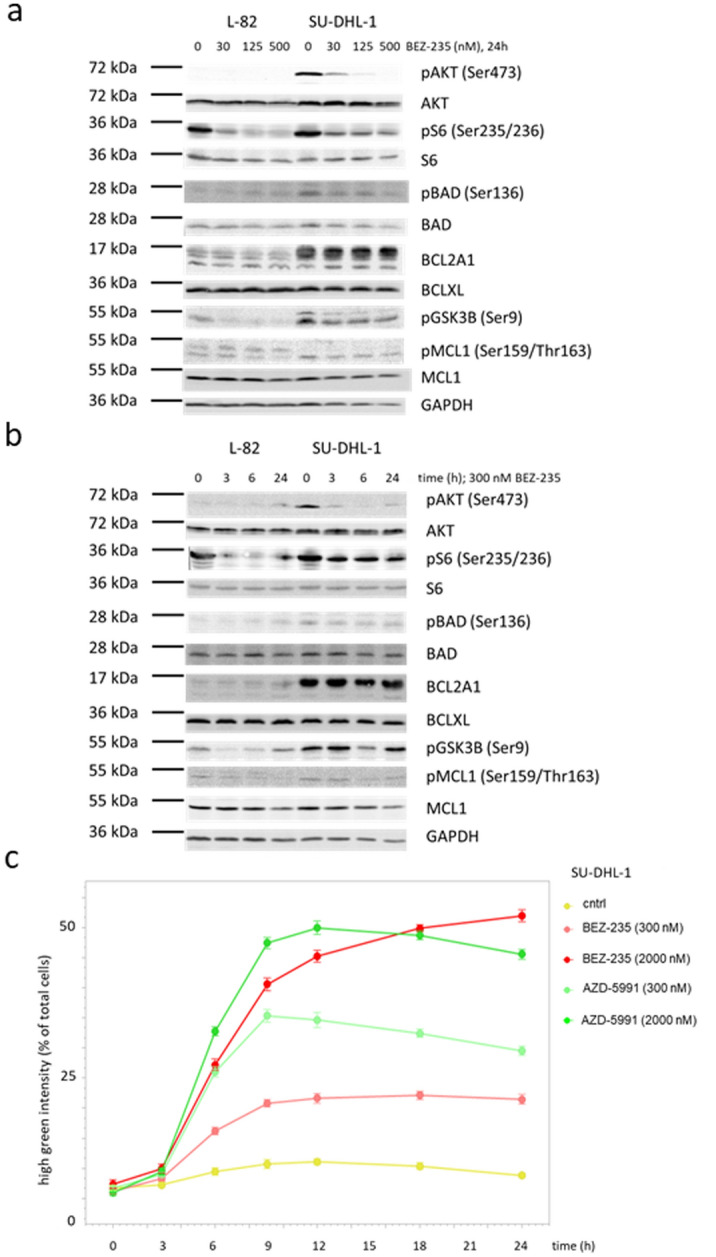
Effect of BEZ-235 on PI3K/AKT activity and on expression of BCL2 family members. Western blots (12% PAA gel) showing (a) dose- and (b) time-dependent effects of BEZ-235 on the expression of BCL2 family members and on phosphorylation of AKT, the mTOR target S6 and the BCL2 family member BAD. Dose-dependent effects were assessed after 24 h, time-dependent effects with a concentration of 300 nM BEZ-235. (c) Kinetics of apoptosis induced by BEZ-235 and AZD-5991 as determined by caspase 3/7 activity. Note that the kinetics of caspase 3/7 activity stimulated by BEZ-235 and AZD-5991 do not differ.
BEZ-235 (300 nM, 6 h) induced expression of several dozen genes in cell lines L-82 and SU-DHL-1 (Suppl. Table 2A,B). However, the upregulated genes are not downstream targets of FOXO1 (FKHR) and FOXO3 (FKHRL1), transcription factors which are inactivated by AKT-mediated phosphorylation32,33. Especially, we could not observe upregulation of the proapoptotic FOXO target BIM (Suppl. Fig. 6).
BAD is another target of AKT. Dephosphorylated BAD binds to the antiapoptotic BCL2 and BCLXL molecules, replacing proapoptotic BAX which then triggers apoptosis34–37. However, effects of BEZ-235 on phosphorylation of BAD could not be observed (Fig. 5a,b).
Thus, we could show that BEZ-235 induced dephosphorylation of AKT and of the mTOR target S6 in ALCL cell lines, but the downstream effectors mediating apoptosis still remain elusive. Important in the context of this study is that BEZ-235 and AZD-5991 showed additive effects on ALCL cell lines regarding arrest of cell growth and induction of apoptosis (Fig. 3c).
In sum, our data suggest that MCL1 inhibitors might be useful for the treatment of PEL. Furthermore, the combination of PI3K/AKT/mTOR pathway inhibitors and of MCL1 inhibitors might be an alternative for treatment of patients with ALCL.
Methods
Cell lines
Cell lines were taken from the stock of the cell lines bank (Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures). Cell lines were authenticated by DNA profiling and cytogenetics. Detailed references and cultivation protocols have been described previously38.
RNA-sequencing analysis
Total RNA was extracted via miRNeasy Mini Kit (Qiagen, Hilden, Germany) including DNase digestion. Library preparation and sequencing were commissioned to GATC Biotech (Cologne, Germany). RNA expression analysis was performed as described previously6. RNA-seq data were deposited at ArrayExpress E-MTAB-7721. For the heatmap visualization 0.1 was added to normalized expression levels prior to log2 transformation.
Growth and apoptosis
About 2 × 104 cells (in 50 µl) were seeded in triplicate in 96-well flat bottom microtiter plates. Effectors were added as 2 × concentrated solutions in a 50 µl volume. Growth and apoptosis were determined with the IncuCyte S3 Live-Cell analysis System (Sartorius, Essen Bioscience, Welwyn Garden City, UK). We used the Caspase 3/7 reagent (Essen Bioscience #4704) to detect apoptotic cells.
Knock-down experiments
Transfection with siRNA oligonucleotides was performed as described previously39. Gene-specific siRNA oligonucleotides and AllStars negative control siRNA were obtained from Qiagen. The siRNA (80 pmol) was transfected into 1 × 106 cells by electroporation using an EPI-2500 impulse generator (Fischer, Heidelberg, Germany) at 350 V for 10 ms. Treated cells were harvested after 24 h or 48 h.
Quantitative real-time PCR analysis
RNA was prepared using the RNeasy Mini kit (Qiagen). For mRNA quantification, reverse transcription was performed using the SuperScript II reverse transcriptase kit (Invitrogen, Karlsruhe, Germany). PCR was performed on a 7500 Applied Biosystems (Darmstadt, Germany). TaqMan probes (Applied Biosystems) were used to quantify human BAX (Hs00180269_m1), BCL2 (Hs00153350_m1), BCL2A1 (Hs00187845_m1), BCLW (Hs00187848_m1), BCLXL (Hs00236329_m1), CDKN1A (Hs00355783_m1), MCL1 (Hs03043899_m1), MDM2 (Hs01066930_m1) and NOXA1 (Hs00611456_g1) using TBP as endogenous control. DNA was isolated with the High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). Quantitative genomic PCR was performed with SYBR-green reagents (Applied Biosystems). Primers and PCR conditions for genomic PCR are shown in Table S3. Relative expression levels were calculated using the ΔΔCt method.
Western blot analysis
Samples were prepared as described previously40. Anti AKT (#4691), pAKT (Ser473) (#4060), BAD (#9239), pBAD (Ser136) (#4366), BCL2 (#4223), BCL2A1 (#14093), BCL-W (#2724), BCLXL (#2764), pGSK3B (Ser9) (#9323), MCL1 (#39224), pMCL1 (Ser159/Thr163) (#4579), S6 (#2317) and pS6 (Ser235/236) (#2211) antibodies were purchased from Cell Signaling (New England Biolabs, Frankfurt, Germany). Anti GAPDH (ab8245) antibody was from Abcam (Cambridge, UK). Specific bands on nitrocellulose membranes were visualized with the biotin/streptavidin–horseradish peroxidase system (GE Healthcare, Little Chalfont, UK) in combination with the “Renaissance Western Blot Chemoluminescence Reagent” protocol (Perkin Elmer, Waltham, MA, USA).
Expression array
Cell lines L-82 and SU-DHL-1 were stimulated with BEZ-235 (300 nM; 0 h, 3 h, 6 h). RNA was isolated using the RNeasy Mini kit (Qiagen). Quality and integrity of total RNA was controlled on Agilent Technologies 2100 Bioanalyzer (Agilent Technologies; Waldbronn, Germany). For biotin labelling 2–10 ng of total RNA were used according to GeneChip Pico Kit (Affymetrix). Then, 5.5 µg of biotinylated cDNA were fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. Samples were hybridized to an identical lot of Affymetrix Clariom S (400 Format) for 17 h at 45 °C. Hybridization was done for 16 h at conditions recommended by the manufacturer. Clariom S chips were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned using the Affymetrix GCS 3000. Image analysis was done by Affymetrix GeneChip Command Console Software (AGCC) and Affymetrix Expression Console Software. Clariom S arrays were analyzed via R/Bioconducor using the package oligo including multi-array average (RMA) background correction, quantile normalization and log2 transformation, whereas the annotation file was retrieved from Thermofisher (version na36, hg38, transcripts). Genes were filtered to at least log2FC < -1 in control versus 6 h after BEZ-235 treatment. The data were deposited with ArrayExpress under E-TAB-10903.
Supplementary Information
Author contributions
H.Q., study conception and design, manuscript writing; R.G., expression array analysis; V.H., acquisition of data; S.N., knock-down experiments; C.P., data query and filtering; C.C.U., analysis and presentation of data; M.Z., acquisition of data; H.G.D., LL-100 panel and provision of cell lines. All authors read and approved this manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-04916-6.
References
- 1.D’Aaguanno S, Del Bufalo D. Inhibition of anti-apoptotic Bcl-2 proteins in preclinical and clinical studies: Current overview in cancer. Cells. 2020;9:1287. doi: 10.3390/cells9051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klanova M, Klener P. BCL-2 proteins in pathogenesis and therapy of B-cell non-Hodgkin lymphomas. Cancers. 2020;12:938. doi: 10.3390/cancers12040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin VS, Xu ZF, Huang DCS, Thijssen R. BH3 mimetics for the treatment of B-cell malignancies—Insights and lessons from the clinic. Cancers. 2020;12:3353. doi: 10.3390/cancers12113353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parry N, Wheadon H, Copland M. The application of BH3 mimetics in myeloid malignancies. Cell Death Dis. 2021;12:222. doi: 10.1038/s41419-021-03500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewald L, Dittmann J, Vogler M, Fulda S. Side-by-side comparison of BH3 mimetics identifies MCL-1 as a key therapeutic target in AML. Cell Death Dis. 2019;10:917. doi: 10.1038/s41419-019-2156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quentmeier H, et al. The LL-100 panel: 100 cell lines for blood cancer studies. Sci. Rep. 2019;9:8218. doi: 10.1038/s41598-019-44491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derenne S, et al. Antisense strategy shows that Mcl-1 rather than Bcl-2 or Bcl-XL is an essential survival protein of human myeloma cells. Blood. 2002;100:194–199. doi: 10.1182/blood.V100.1.194. [DOI] [PubMed] [Google Scholar]
- 8.Gong JN, et al. Hierarchy for targeting prosurvival BCL2 family proteins in multiple myeloma: Pivotal role of MCL1. Blood. 2016;128:1834–1844. doi: 10.1182/blood-2016-03-704908. [DOI] [PubMed] [Google Scholar]
- 9.Glaser SP, et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26:120–125. doi: 10.1101/gad.182980.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hata AN, Engelman JA, Faber AC. The BCL2 family: Key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slomp A, Peperzak V. Role and regulation of pro-survival BCL-2 proteins in multiple myeloma. Front. Oncol. 2018;8:533. doi: 10.3389/fonc.2018.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Zebeeby A, et al. Targeting intermediary metabolism enhances the efficacy of BH3 mimetic therapy in hematologic malignancies. Haematologica. 2019;104:1016–1025. doi: 10.3324/haematol.2018.204701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tron AE, et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat. Commun. 2018;9:5341. doi: 10.1038/s41467-018-07551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei AH, et al. Targeting MCL-1 in hematologic malignancies: Rationale and progress. Blood Rev. 2020;44:100672. doi: 10.1016/j.blre.2020.100672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koss B, et al. Requirement for antiapoptotic MCL-1 in the survival of BCR-ABL B-lineage acute lymphoblastic leukemia. Blood. 2013;122:1587–1598. doi: 10.1182/blood-2012-06-440230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1143. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prutsch N, et al. Dependency on the TYK2/STAT1/MCL1 axis in anaplastic large cell lymphoma. Leukemia. 2019;33:696–709. doi: 10.1038/s41375-018-0239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rassidakis GZ, et al. Overexpression of Mcl-1 in anaplastic large cell lymphoma cell lines and tumors. Am. J. Pathol. 2002;160:2309–2310. doi: 10.1016/S0002-9440(10)61178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rust R, et al. High expression of Mcl-1 in ALK positive and negative anaplastic large cell lymphoma. J. Clin. Pathol. 2005;58:520–524. doi: 10.1136/jcp.2004.022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manzano M, et al. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat. Commun. 2018;9:3263. doi: 10.1038/s41467-018-05506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quentmeier H, et al. Diffuse large B cell lymphoma cell line U-2946: Model for MCL1 inhibitor testing. PLoS ONE. 2016;11:e0167599. doi: 10.1371/journal.pone.0167599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesarman E, Knowles DM. The role of Kaposi´s sarcoma-associated herpesvirus (KSHV HHV-8) in lymphoproliferative diseases. Semin. Cancer Biol. 1999;9:165–174. doi: 10.1006/scbi.1998.0118. [DOI] [PubMed] [Google Scholar]
- 23.Chen YB, Rahemtullah A, Hochberg E. Primary effusion lymphoma. Oncologist. 2007;12:569–576. doi: 10.1634/theoncologist.12-5-569. [DOI] [PubMed] [Google Scholar]
- 24.Coluccia AML, et al. BCL-XL down-regulation suppresses the tumorigenic potential of NPM-ALK in vitro and in vivo. Blood. 2004;103:2787–2794. doi: 10.1182/blood-2003-09-3144. [DOI] [PubMed] [Google Scholar]
- 25.Piva R, et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Invest. 2006;116:3171–3182. doi: 10.1172/JCI29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, et al. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018;423:47–59. doi: 10.1016/j.canlet.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 27.Roberts AW, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: Results of a phase I study of navitoclax in patients with relapsed or refractory disease. J. Clin. Oncol. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vega F, et al. Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase-positive anaplastic large cell lymphoma. Cancer Res. 2006;66:6589–6597. doi: 10.1158/0008-5472.CAN-05-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhary GS, et al. MCL-1 and BCL-XL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell. Death Dis. 2015;6:e1593. doi: 10.1038/cddis.2014.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang-Yen HF. Mcl-1: A highly regulated cell death and survival controller. J. Biomed. Sci. 2006;13:201–204. doi: 10.1007/s11373-005-9064-4. [DOI] [PubMed] [Google Scholar]
- 31.Huang T, et al. CDK7 inhibitor THZ1 inhibits MCL1 synthesis and drives cholangiocarcinoma apoptosis in combination with BCL2/BCL-XL inhibitor AB T-263. Cell Death Dis. 2019;10:206. doi: 10.1038/s41419-019-1424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuang CH, et al. Discovery of Akt kinase inhibitors through structure-based virtual screening and their evaluation as potential anticancer agents. Int. J. Mol. Sci. 2015;16:3202–3212. doi: 10.3390/ijms16023202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 34.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/S0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 35.Del Peso L, et al. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 36.Yang E, et al. Bad, a heterodimeric partner for BCL-XL and BCL-2 displaces BAX and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 37.Zha J, et al. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell. 1996;87:619–628. doi: 10.1016/S0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 38.Drexler, H.G. Guide to Leukemia-Lymphoma Cell Lines. 2nd Edition Braunschweig, eBook on CD (2010).
- 39.Nagel S, et al. Transcriptional activation of prostate specific homeobox gene NKX3–1 in subsets of T-cell lymphoblastic leukemia (T-ALL) PLoS ONE. 2012;7:e40747. doi: 10.1371/journal.pone.0040747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quentmeier H, et al. SET-NUP214 fusion in acute myeloid leukemia- and T-cell acute lymphoblastic leukemia-derived cell lines. J. Hematol. Oncol. 2009;2:3. doi: 10.1186/1756-8722-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.