Abstract
Ammonia oxidizers are key players in the global nitrogen cycle, yet little is known about their ecological performances and adaptation strategies for growth in saline terrestrial ecosystems. This study combined 13C-DNA stable-isotope probing (SIP) microcosms with amplicon and shotgun sequencing to reveal the composition and genomic adaptations of active ammonia oxidizers in a saline-sodic (solonetz) soil with high salinity and pH (20.9 cmolc exchangeable Na+ kg−1 soil and pH 9.64). Both ammonia-oxidizing archaea (AOA) and bacteria (AOB) exhibited strong nitrification activities, although AOB performed most of the ammonia oxidation observed in the solonetz soil and in the farmland soil converted from solonetz soil. Members of the Nitrosococcus, which are more often associated with aquatic habitats, were identified as the dominant ammonia oxidizers in the solonetz soil with the first direct labeling evidence, while members of the Nitrosospira were the dominant ammonia oxidizers in the farmland soil, which had much lower salinity and pH. Metagenomic analysis of “Candidatus Nitrosococcus sp. Sol14”, a new species within the Nitrosococcus lineage, revealed multiple genomic adaptations predicted to facilitate osmotic and pH homeostasis in this extreme habitat, including direct Na+ extrusion/H+ import and the ability to increase intracellular osmotic pressure by accumulating compatible solutes. Comparative genomic analysis revealed that variation in salt-tolerance mechanisms was the primary driver for the niche differentiation of ammonia oxidizers in saline-sodic soils. These results demonstrate how ammonia oxidizers can adapt to saline-sodic soil with excessive Na+ content and provide new insights on the nitrogen cycle in extreme terrestrial ecosystems.
Subject terms: Soil microbiology, Biogeochemistry, Microbial ecology
Introduction
Chemolithoautotrophic ammonia oxidizers are key players in ammonia oxidation, the first and often rate-limiting step of nitrification, which is a central process in the global nitrogen cycle [1, 2]. Three major groups of ammonia oxidizers, ammonia-oxidizing archaea (AOA), canonical ammonia-oxidizing bacteria (AOB), and complete ammonia-oxidizing Nitrospira (comammox), have been characterized as active players in autotrophic ammonia oxidation, with the activities of AOA and/or AOB dominating ammonia oxidation in soils depending on environmental conditions. Generally, one of the most important factors determining the distribution and activity of AOA and AOB in soil is pH [3, 4], and several terrestrial ammonia oxidizer clades have adapted to extreme conditions. For example, strains of the AOA lineage Nitrosotalea and a gammaproteobacterial AOB have been isolated from acidic soils and possess adaptations for growth at low pH [5–7], while ecological investigations have confirmed the important contributions of related clades to nitrification in acidic soils [8]. Ecological investigations have also revealed the presence of diversified alkaliphilic ammonia oxidizers, within both the AOA and AOB, in soils with pH values of up to 9 [9, 10], but their activity and ecological importance in such environments remain unclear.
Saline ecosystems represent globally distributed habitats and are often highly productive in terms of microbial diversity and related biogeochemical processes [11–14]. Notably, the area of soils affected by high salinity and sodicity is close to 1 billion hectares, accounting for nearly 7% of the Earth’s land surface [15]. However, there are limited reports on the ecological performance and niche specialization of functional microbial guilds, including ammonia oxidizers, in such ecosystems [14]. Previous information on ammonia-oxidizing microbes in saline ecosystems is mainly from aquatic environments, while knowledge of ammonia-oxidizing microbes in terrestrial saline systems remains lacking [16]. For instance, AOA from marine ecosystems, including the genera Nitrosopumilus [17, 18] and Ca. Nitrosopelagicus [19], appear to be salt-tolerant based on cultivation studies, but no strains of soil AOA have been reported to grow under high saline conditions [20]. Betaproteobacterial Nitrosomonas strains enriched from desert soil have also demonstrated salt tolerance [21]. In addition, Nitrosococcus species within the Gammaproteobacteria are predominantly found in marine systems and adapted to high salt concentrations [22, 23]. Nitrosococcus-related AOB have also been found to have a high salt tolerance in saline wastewater at low pH [24] and detected in a paddock soil with carbonate accumulations [25] and saline alkaline soils of a former lake [26]. However, several recent studies have also shown that some Nitrosococcus-related and other gammaproteobacterial AOB are present in non-saline systems, such as a farm biofilter [27], tea field soil [7], and grassland soil [28]. Furthermore, how AOA and AOB respond to salt stress remains unclear. Although some studies have shown that AOA are more adaptive to hypersaline conditions than AOB [20, 29], other researchers have observed the opposite [30]. These results indicate that ammonia oxidizers may differ in salt tolerance, although the mechanisms leading to the difference remain unresolved.
The saline-sodic area in the western Songnen Plain in China is one of the three major regions with saline-sodic soils worldwide, with more than three million hectares of salt-affected soils [31]. Soils in this area typically have excessive Na2CO3 and NaHCO3 contents, leading to an extreme environment of both Na+ toxicity and high pH stress [32]. In the present study, we investigated the ammonia oxidizers present in a natural hypersaline, alkaline field from this area, and compared them to those in a converted agricultural land in the same region with significantly reduced pH and salt content. Using 13C-tracing microcosms and sequence analysis, we identified the composition of the active ammonia-oxidizing communities in both soils and obtained insights into the genetic capacity of salt-tolerant soil ammonia oxidizers.
Materials and methods
Site description and soil characteristics
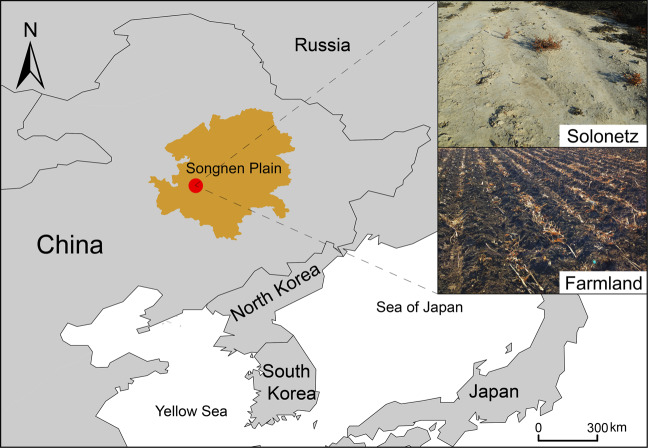
The sampling site is located at the Da’an Sodic Land Experiment Station of the Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Da’an County, Jilin Province, China (45°35′ N, 123°51′ E, Fig. 1). Samples were collected in December 2017 from two systems: (i) solonetz soil (natural saline-sodic soil) with sparse native vegetation and (ii) farmland soil that was converted from solonetz soil and has been cultivated with a maize-watermelon rotation system for >40 years. Triplicate plots (1 m × 1 m) were selected from the solonetz and farmland fields, and composite samples of five soil cores (0–20 cm) were taken at random from each plot before homogenizing through a 2.0 mm sieve and stored at 4 °C until further use. Methods for determining the soil properties are described in the Supplementary Information. All soil properties are listed in Table 1.
Fig. 1. Schematic diagram showing the sampling sites of agricultural soil that was reclaimed from typical saline-sodic soils in Northeastern China.
The area in brown yellow represents the Songnen Plain, one of the three major regions with saline-sodic soil in the world. The solid circle in red within the Songnen Plain refers to the long-term reclamation experiment field of the Da’an Sodic Land Ecological Experiment Station of Northeast, Chinese Academy of Sciences. Soil samples were collected from the Solonetz saline-sodic field (upright panel) that contains no vegetation cover for >40 years as a control, and farmland (downright panel) that has been maintained for maize-watermelon rotation system more than 40 years.
Table 1.
Physiochemical and biological properties of soils.
| Soil | Solonetz | Farmland |
|---|---|---|
| Exchangeable sodium percentage (ESP, %) | 26.9*** | 0.511 |
| Exchangeable Na+ (ENa, cmolc·kg−1) | 20.8*** | 0.376 |
| Soil pH | 9.64*** | 7.76 |
| Soil bulk density (SBD, g·cm−3) | 1.38** | 1.24 |
| HCO3− (mmol·L−1) | 3.94** | 1.36 |
| CO32− (mmol·L−1) | 6.79*** | 0.00 |
| Microbial biomass carbon (MBC, mg·kg−1) | 62.8 | 194*** |
| Microbial biomass nitrogen (MBN, mg·kg−1) | 19.4 | 29.1** |
| MBC/MBN | 3.26 | 6.70*** |
| Cation exchange capacity (CEC, cmolc·kg−1) | 77.2** | 73.4 |
| Total carbon (TC, mg·g−1) | 13.6 | 13.5 |
| Total nitrogen (TN, mg·g−1) | 0.393 | 0.872*** |
| TC/TN | 34.7*** | 15.5 |
| Available phosphorus (AP, g·kg−1) | 6.57 | 25.5*** |
| Available potassium (AK, mg·kg−1) | 171** | 155 |
| Archaeal amoA gene abundance (106 copies g−1) | 3.35 | 495*** |
| Bacterial amoA gene abundance (107 copies g−1) | 17.0 | 330*** |
| Nitrification potential (NP, μg N g−1 day−1) | 23.5* | 17.1 |
Significance levels are *p < 0.05, **p < 0.01, and ***p < 0.001.
Soil microcosms for stable-isotope probing of active ammonia oxidizers
Triplicate microcosms were constructed for both the solonetz and farmland soils as previously described [33]. Briefly, fresh soil equivalent to 5.0 g dry weight soil was incubated at a 60% maximum water-holding capacity in a 120-ml serum bottle tightly capped with a butyl stopper at 28 °C in the dark. The soil microcosms were incubated with a 5% (v/v) isotopically-enriched labeled 13C–CO2 (Sigma-Aldrich, St. Louis, MO, USA) or 12C–CO2 headspace in the absence or presence of 100 pa acetylene (C2H2), an inhibitor of microorganisms that use ammonia monooxygenase. The bottles were flushed with pressurized synthetic air (20% O2, 80% N2) for 1 min to maintain oxic conditions and resupplied with 13C– or 12C–CO2 at 7-day intervals. In addition, the soil microcosms were amended with 100 μg urea-N per gram of dry soil on a weekly basis, starting from the first day of incubation. For the 13CO2 amended microcosms, the supplemented CO2 and urea (Shanghai Research Institute of Chemical Industry, Shanghai, China) were >99-atom% 13C-labeled. Isopycnic density gradient centrifugation with 2.0 μg of extracted DNA was performed as previously described [33]. Additional details are provided in the Supplementary Information.
Quantification and sequencing of 16S rRNA and amoA genes
The abundance of prokaryote 16S rRNA, archaeal amoA, and bacterial amoA genes were determined using quantitative PCR with the primer sets 515F/907R [33], Arch-amoAF/Arch-amoAR [34], and A189F/A682R [35], respectively (Table S1). The amplification efficiencies for all genes were 91.5–100%, with R2 values of 0.991–1.000. The 16S rRNA gene was amplified for sequencing using the primer pair 515F/907R [33] with a barcoded (12 bp) forward primer. Archaeal and bacterial amoA genes were amplified for clone library construction with the same primer sets used in quantitative PCR (Table S1). Detailed descriptions of the reaction conditions, sequencing protocols, and sequence analyses are provided in the Supplementary Information.
Metagenome sequencing and analysis
The 13C-DNA of the heavy fractions (5–7 for both AOA and AOB in the solonetz soil, and 5–7 and 8–10 for AOB and AOA in the farmland soil, respectively) from the DNA-SIP soil microcosms were used to construct libraries for metagenomic sequencing using a VAHTS Universal Plus DNA Library Prep Kit for Illumina (Vazyme Biotech, Nanjing, Jiangsu, China) following the manufacturer’s instructions. The size of each metagenome was 32.05±1.59 Gb, resulting in a total data output of 288.45 Gb. The metagenome sequencing and analysis are described in detail in the Supplementary Information.
Statistical analysis
One-way analysis of variance (ANOVA) with Duncan’s post hoc test was performed for multiple comparisons using the statistical package SPSS version 23.0, and p < 0.05 was considered to indicate a significant difference. Distance-based linear modeling (DISTLM) was performed to identify the potential abiotic driver(s) for different community compositions of ammonia oxidizers.
Results
Changes in saline-sodic soil properties under agricultural reclamation
Agricultural reclamation resulted in significant alleviation of soil salinity and sodicity (Table 1). The mean concentration of exchangeable sodium (ENa) decreased significantly by 55.3-fold from 20.8 in the solonetz soil to 0.376 in the farmland soil. Moreover, the exchangeable sodium percentage (ESP), carbonate and bicarbonate contents also showed consistently significant declines. In addition, significant decreases in soil bulk density (SBD) and pH were observed (Table 1). Both microbial biomass carbon (MBC) and nitrogen (MBN) of the solonetz soils were much lower than those of the farmland soils. Furthermore, the mean abundance of AOB amoA genes was 19.4-fold lower in the solonetz soil than in the farmland soil, while AOA amoA gene abundance was 147.8-fold lower (Table 1). Intriguingly, nitrification potential was significantly higher (1.4-fold) in the solonetz soil than in the farmland soil (Table 1).
13C-labeling of active ammonia oxidizers
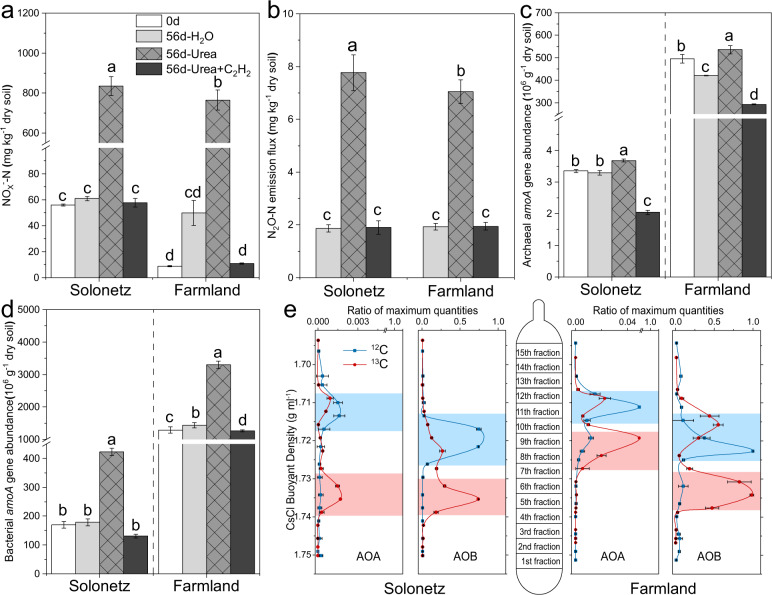
Soil nitrification was assessed as the net change in NOx−-N content (Fig. 2a). Urea fertilization led to a remarkable accumulation of NOx−-N at approximately 800 μg g−1 dry soil in both solonetz and farmland soils, whereas no NOx−-N accumulation was detected in the water-amended (control) soils (Fig. 2a). The presence of C2H2 completely inhibited NOx--N production, and the stoichiometric relationship between NH4+-N accumulation in C2H2-inhibited soils (Fig. S1) and NOx--N production (Fig. 2a) indicated that autotrophic nitrification predominated in both soils. Similar results were also observed for N2O emissions (Fig. 2b), and the weekly emission flux was generally higher in solonetz soils than in farmland soils (Fig. S2). The abundance of amoA genes of both AOA (Fig. 2c) and AOB (Fig. 2d) showed an increasing trend only in the urea-amended soils, particularly for AOB during the 56-day incubation period.
Fig. 2. Stable-isotope probing (SIP) of active ammonia oxidizers in solonetz and farmland soil.
Changes in the concentration of soil nitrite- plus nitrate-N (NOx−-N) (a), N2O emissions (b), and amoA gene abundances (c, d) of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in SIP microcosms over an incubation period of 56 days. The 13C-DNA of AOA and AOB was revealed by quantitative analysis of archaeal and bacterial amoA gene abundances across the entire buoyant density gradient of the fractionated DNA from SIP microcosms at day 56 (e). SIP microcosms were incubated with either 13C (CO2 and urea) or 12C (CO2 and urea), and an equal volume of H2O instead of urea solution was amended as a control to monitor nitrification activity due to ammonia released from soil mineralization. The designations “0 d” and “56 d” denote days 0 and 56, respectively. “56 d-H2O” and “56 d-urea” indicate samples from microcosms that received water or urea every seven days, respectively. The designation “56d-Urea+C2H2” represents the sample at day 56 from SIP microcosms incubated with both urea and 100 Pa C2H2. Different letters above the columns indicate significant differences (p < 0.05) (a, b). Different letters above the columns in each soil indicate significant differences (p < 0.05) (c, d). The data are normalized units (e) using the ratio of the amoA gene copy number in each DNA gradient fraction to the maximum quantity of two soils, and the “13C-DNA” (“heavy DNA”) and “12C-DNA” (“light DNA”) fractions are indicated by the shaded rectangles in red and blue, respectively.
Following ultracentrifugation of the total DNA extracted from 13C-labeled and 12C-control microcosms, quantification of amoA gene distribution as a function of the buoyant density of the DNA demonstrated labeling of growing ammonia oxidizers in both urea-amended soils (Fig. 2e). For the solonetz soil, high peaks of AOA and AOB amoA genes occurred in the 13C-labeled ‘heavy’ DNA (fractions 5–7) from the 13C-microcosms when compared to those from 12C-microcosms. The distribution of amoA genes in farmland soil DNA also showed distinct labeling patterns, as the abundance of 13C-AOA amoA genes appeared in DNA fractions 8–10, whereas 13C-AOB amoA genes remained in DNA fractions 5–7 (Fig. 2e). Notably, no labeling of comammox amoA genes was detected despite the use of different primers and PCR conditions [36, 37] (Fig. S3).
Population dynamics of active ammonia oxidizers
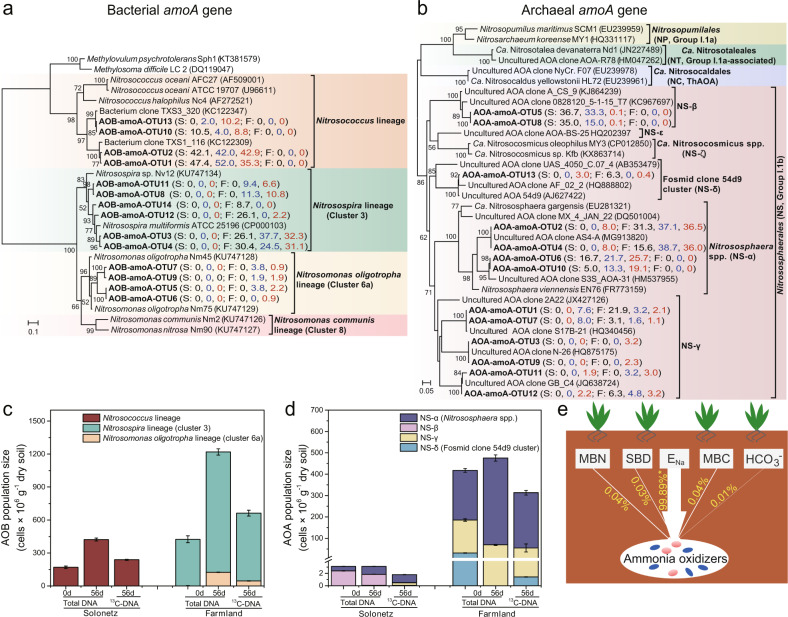
Phylogenetic analysis of amoA genes in total DNA (Day 0 and Day 56) and 13C-DNA at Day 56 revealed distinct changes in the community structure of active ammonia oxidizers in solonetz soil upon agricultural reclamation (Fig. 3). The population size of distinct phylotypes was further determined based on their relative proportion (Fig. 3a, b) and the total abundance of ammonia oxidizers as inferred from amoA gene abundance (Table S2). In the solonetz soil, the populations of AOB were exclusively within the Nitrosococcus-related Gammaproteobacteria, while betaproteobacterial AOB were not detected under any conditions by sequence analysis of either 16S rRNA (Fig. S4a) or amoA (Fig. 3a) genes. Urea amendment stimulated significantly the autotrophic growth of Nitrosococcus-related AOB in this solonetz soil (Fig. 3c), whereas Nitrosospira-related Betaproteobacteria predominated the AOB populations in the farmland soil (Fig. 3c).
Fig. 3. Population dynamics of active ammonia oxidizers in solonetz and farmland soil.
Total DNA (day 0 and 56) and 13C-DNA (day 56) were sequenced for phylogenetic identification of ammonia oxidizers in soils based on amoA genes of AOB (a) and AOA (b). Numbers in black, blue, and red represent the relative abundance of each operational taxonomic unit (OTU) sequences to the total amoA gene sequences in the total DNA at day 0, total DNA at day 56, and 13C-DNA, respectively. “S” and “F” in parentheses refer to solonetz and farmland soil, respectively. For instance, the designation of AOB-amoA-OTU-1 (S: 47.4, 52.0, 35.3; F:0, 0, 0) indicates that OTU-1 of AOB accounts for 47.4, 52.0, and 35.3% of the total AOB amoA gene sequences in the total DNA at day 0, total DNA at day 56 and 13C-DNA from solonetz soils, respectively. The population size of AOB (c) and AOB (d) was determined by multiplying the relative abundance of different lineages/clusters by the total AOA or AOB abundance (Table S2). OTUs were clustered at 93% identity. The phylogeny of AOA and AOB was generated using IQtree 1.6.12 with the best fit SYM+I+G4 and TPM2u+F+G4 model selected using the BIC. Bootstraps are based on 1000 replicated trees. Methylovulum psychrotolerans Sph1 and Methylosoma difficile LC 2 were included as an outgroup within the class Gammaproteobacteria. The results of distance-based linear modeling (DISTLM) analysis of the ammonia oxidizer compositions using the physicochemical properties of soils as predictor variables (e), where the explanatory proportion of each variable is shown beside the arrow line. The significance level is *p < 0.05. MBN microbial biomass nitrogen, SBD soil bulk density, ENa exchangeable Na+ content, MBC microbial carbon, HCO3− HCO3− content.
Analysis of amoA (Fig. 3b) and 16S rRNA genes (Fig. S4b) indicated that all AOA fell within the order Nitrososphaerales [38]. The AOA in the solonetz soil consisted of clades NS-α (Nitrososphaera spp.), NS-β, and NS-γ, but only clades NS-α and NS-γ exhibited growth during microcosm incubation (Fig. 3d). The AOA in the farmland soils were in clades NS-α, NS-γ, and NS-δ (Fosmid clone 54d9 cluster), and showed autotrophic growth after urea fertilization (Fig. 3d). In both soils, the activity of AOA was dominated by clade NS-α, as suggested by the 13C-DNA-SIP results (Fig. 3d). The changes in these active populations were further supported by analysis of AOB and AOA in the 16S rRNA gene amplicon analysis of total DNA and 13C-DNA (Fig. S5, Table S3, Supplementary results).
Moreover, the DISTLM analysis indicated that the concentration of exchangeable Na+ (ENa) alone could explain 99.89% of the variation in active communities (Fig. 3e), suggesting that salt-tolerance was a major factor in shaping the niche separation of ammonia-oxidizing prokaryotes.
Metagenome assembly of active ammonia oxidizers
Metagenomic analysis of 13C-DNA further demonstrated a significant shift in active ammonia oxidizers in the two soils. Taxonomic classification of the scaffold sequences (1340–1752 nt on average, Table S4) showed that the dominant clades of AOB were affiliated with Nitrosococcus and Nitrosospira in the solonetz and farmland soils, respectively, while the dominant clades of AOA were closely associated with NS-α in both soils (Table S5), and largely consistent with the amplicon sequencing results (Fig. 3, Fig. S5).
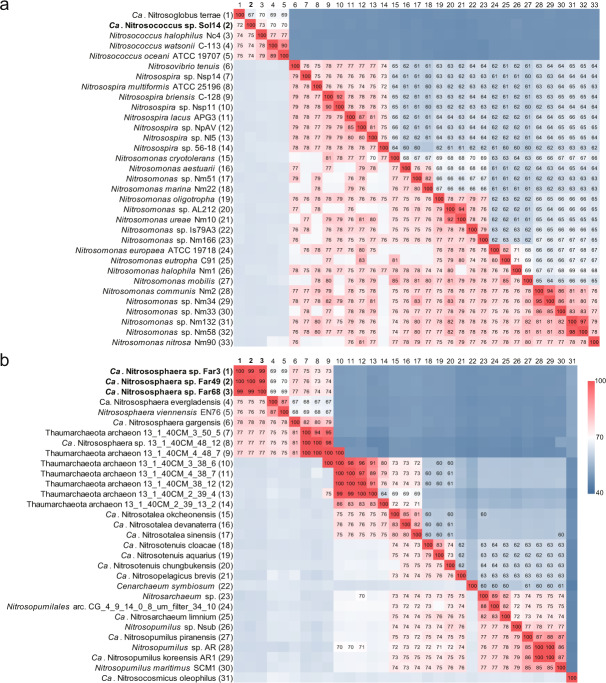
A total of eight genome bins of ammonia oxidizers were obtained (Table 2). Three Nitrosococcus and two NS-α MAGs were recovered from the solonetz soil, but only one Nitrosococcus genome (bin14) was of high quality (91.7% completeness and 0.5% contamination) (Table 2). The genome-wide average nucleotide identities (gANI) between pairwise sets of three Nitrosococcus MAGs (Fig. S6) were higher than the species threshold of 95% [39], indicating that the three MAGs should represent closely related strains within the same species. Similarly, gANI and genome-wide average amino acid identities (gAAI) between bin14 and all known cultured Nitrosococcus representatives (Table S6) were 74–75% [39] and 70–73% (higher than the genus cutoff of approximately 65%) [40, 41], respectively (Fig. 4a), suggesting the Nitrosococcus MAGs represented a new species. This finding was further supported by the phylogenetic analysis of a concatenation of 120 conserved bacterial marker proteins (Fig. S7a) [42]. This Nitrosococcus MAG was thus designated “Candidatus Nitrosococcus sp. Sol14”.
Table 2.
Characteristics of ammonia oxidizer genome bins from 13C-labeled metagenomes of solonetz and farmland soils.
| Soil | Taxonomic identity | Completeness (%) | Contamination (%) | GC (%) | Size (Mbp) | Genes | |
|---|---|---|---|---|---|---|---|
| Solonetz | AOB | Ca. Nitrosococcus sp. Sol14 | 91.7 | 0.5 | 51.4 | 2.71 | 2521 |
| Nitrosococcus bin19 | 62.2 | 0.1 | 52.3 | 1.94 | 1783 | ||
| Nitrosococcus bin74 | 65.1 | 0.2 | 52.3 | 1.99 | 1843 | ||
| AOA | Nitrososphaera bin12 | 58.1 | 0 | 47.3 | 1.13 | 1327 | |
| Nitrososphaera bin62 | 60.2 | 0 | 47.4 | 1.13 | 1323 | ||
| Farmland | AOA | Ca. Nitrososphaera sp. Far3 | 97.3 | 1.9 | 51.3 | 1.59 | 2092 |
| Ca. Nitrososphaera sp. Far49 | 96.1 | 1.0 | 51.4 | 1.54 | 2048 | ||
| Ca. Nitrososphaera sp. Far68 | 98.5 | 1.9 | 51.1 | 1.65 | 2168 | ||
Only genome bins that were above 50% completion with less than 10% contamination are included.
Fig. 4. Genome-wide, pairwise comparisons of the average nucleotide identity (gANI) and average amino acid identity (gAAI) values between MAGs (highlighted in bold) and known genomes of ammonia oxidizers.
a Symmetrical matrix of pairwise gANI and gAAI between AOA MAGs (Ca. Nitrososphaera sp. Far3, Ca. Nitrososphaera sp. Far49 and Ca. Nitrososphaera sp. Far68) and known AOA genomes (Table S6). The gANI is presented in the lower left triangle and values ≥70. The gAAI is presented in the upper right triangle and values ≥60 are provided. b Symmetrical matrix of pairwise gANI and gAAI between AOB MAG (Ca. Nitrosococcus sp. Sol14) and known AOB genomes (Table S6). The gANI is presented in the lower left triangle and values ≥70. The gAAI is presented in the upper right triangle and values ≥60 are provided.
Three high-quality NS-α-related MAGs were obtained from the farmland soil (Table 2) and shared a gANI of 99–100%, indicating that they represented closely related strains of the same species. These MAGs likely represented a novel species according to the pairwise comparison of gANI (75–78%) and gAAI (69–77%) with known NS-α representatives (Fig. 4b, Table S6), which was verified by phylogenetic analysis (Fig. S7b) of a concatenation of 122 conserved archaeal marker proteins [42]. We propose “Candidatus Nitrososphaera sp. Far49”, “Candidatus Nitrososphaera sp. Far3” and “Candidatus Nitrososphaera sp. Far68” as names for these three AOA MAGs, which we collectively refer to as “Candidatus Nitrososphaera sp. FarX” hereafter. In addition, the two NS-α bins of the solonetz soil and Ca. Nitrososphaera sp. FarX were not the same species according to a pairwise comparison of gANI and gAAI (Fig. S6).
Salt-tolerance mechanisms in active ammonia oxidizers
Comparative metagenomic analysis of active ammonia oxidizers identified gene repertoires that could potentially enable increased salt-tolerance of Nitrosococcus strains compared to Nitrosospira and NS-α (Table S7). As there was no MAG available for AOB in the farmland soils, we parsed all the genes from the Nitrosospira-affiliated scaffolds in the metagenome dataset for comparison (Table S7). Metabolic reconstruction revealed the key role of three modes of saline adaption in driving niche differentiation of ammonia oxidizers in saline-sodic soil (Fig. 5).
Fig. 5. Metabolic reconstruction of active ammonia-oxidizing bacteria and archaea in response to agricultural reclamation of Solonetz saline-sodic soil.
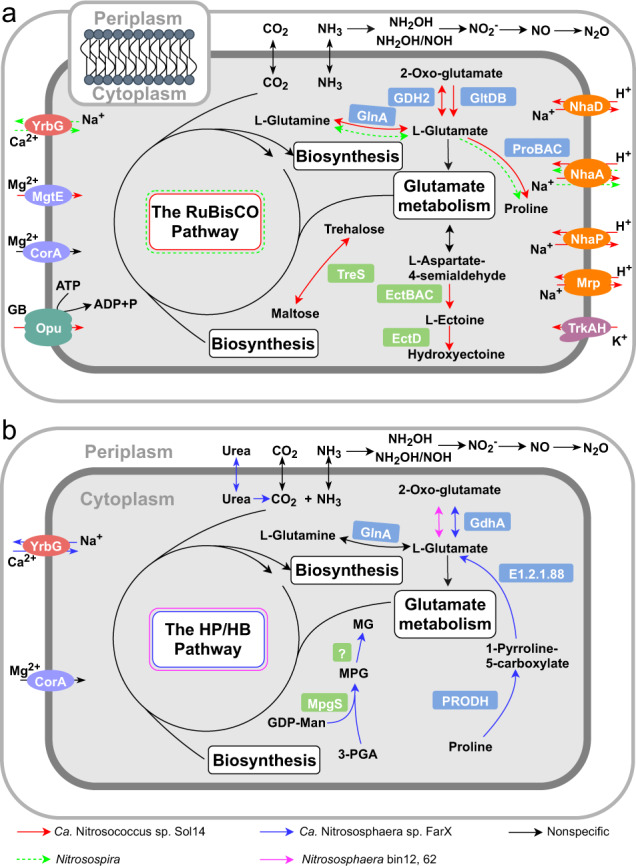
Cell metabolism diagrams of AOB (a) and AOA (b) were constructed from the genome annotation of Ca. Nitrosococcus sp. Sol14, Ca. Nitrososphaera sp. FarX and Nitrososphaera bin12, 62 and the scaffold annotation of the genus Nitrosospira. Putative adaptations to high salinity and selected core metabolic pathways of ammonia oxidizers are shown. NhaA NhaA Na+/H+ antiporter, NhaD NhaD Na+/H+ antiporter, NhaP NhaP Na+/H+ antiporter, Mrp Mrp Na+/H+ antiporter, Trk Trk K+ uptake system, MgtE/CorA magnesium uptake mediated by facilitated diffusion, Opu glycine betaine uptake transporter, TreS trehalose synthase, EctA diaminobutyrate acetyl transferase, EctB diaminobutyrate transaminase, EctC ectoine synthase, EctD ectoine hydroxylase, GDH2 and GdhA glutamate dehydrogenase, GlnA glutamine synthetase, GltDB glutamate synthase, ProA glutamate-5-semialdehyde dehydrogenase, ProB glutamate-5-kinase, ProC pyrroline-5-carboxylate reductase, YrbG Ca2+/Na+ antiporters, PRODH proline dehydrogenase, E1.2.1.88 1-pyrroline-5-carboxylate dehydrogenase, MpgS mannosyl-3-phosphoglycerate synthase, question mark (?) uncharacterized phosphatase, GB glycine betaine. See Table S7 for detailed gene presence/absence.
Na+ extrusion
The Ca. Nitrosococcus sp. Sol14 genome encodes proteins involved in four Na+ extrusion mechanisms, i.e., NhaA, NhaD, NhaP, and Mrp Na+/H+ antiporters [43–45] (Fig. 5). However, Nitrosospira from the farmland soil appeared to possess only NhaA according to the annotation of the scaffold genes (Fig. 5). None of Na+ extrusion mechanisms in Ca. Nitrosococcus sp. Sol14 was detected in Ca. Nitrososphaera sp. FarX and NS-α bins of the solonetz soil (Fig. 5).
Inorganic compatible solute uptake
The Trk transporter is a major transport system for K+ accumulation in cells [46], and the genes (trkAH) encoding these proteins were identified in Ca. Nitrosococcus sp. Sol14 but were absent in all the MAGs of AOA and Nitrosospira scaffolds (Fig. 5). Genes encoding MgtE and CorA proteins for Mg2+ uptake [47] were identified in the Ca. Nitrosococcus sp. Sol14 genome, while the Nitrosospira and AOA MAGs contained genes encoding only CorA (Fig. 5). In addition, Ca. Nitrososphaera sp. FarX and Nitrosospira (Fig. 5) might import Ca2+ and export Na+ by YrbG [47].
Organic compatible solute transport and biosynthesis
The identification of opuCA genes only in Ca. Nitrosococcus sp. Sol14 suggests the potential to import glycine betaine [47]. The genes (treS and ectABCD) encode proteins involved in the synthesis of the compatible solutes trehalose, ectoine and hydroxyectoine [44, 48, 49] and were identified only in Ca. Nitrosococcus sp. Sol14 (Fig. 5). Glutamate is also a compatible solute [50], which may be synthesized through the reversible reactions of glutamate dehydrogenase (GDH2) and glutamine synthetase (GlnA) [50] and the reaction catalyzed by glutamate synthase (GltDB) in Ca. Nitrosococcus sp. Sol14 (Fig. 5a). However, Ca. Nitrososphaera sp. FarX might encode another glutamate dehydrogenase (GdhA) and GlnA (Fig. 5b). GdhA and GlnA were also identified in NS-α bins from the solonetz soil and Nitrosospira (Fig. 5), respectively. In addition, Ca. Nitrosococcus sp. Sol14 might be able to convert glutamate into proline as another compatible substance, catalyzed by glutamate 5-kinase (ProB), glutamate-5-semialdehyde dehydrogenase (ProA), and pyrroline-5-carboxylate reductase (ProC) (Fig. 5a). Nitrosospira may also perform this conversion because the key enzyme (ProA) was detected (Table S7). In contrast, only Ca. Nitrososphaera sp. FarX could convert proline into glutamate through 1-pyrroline-5-carboxylate dehydrogenase (E1.2.1.88, PCD) and proline dehydrogenase (PRODH) (Fig. 5b). Moreover, Ca. Nitrososphaera sp. FarX may synthesize the compatible solute mannosylglycerate (MG), as the gene encoding the key enzyme mannosyl-3-phosphoglycerate synthase (MpgS) was detected (Fig. 5b) [51].
Genomes of previously characterized ammonia oxidizers in the NCBI database were also examined for the presence of genes encoding the above-described proteins (Fig. 6). These salt-tolerant mechanisms were unevenly distributed among ammonia-oxidizing microorganisms, especially Na+ extrusion mechanisms, which are relatively scarce (Fig. 6).
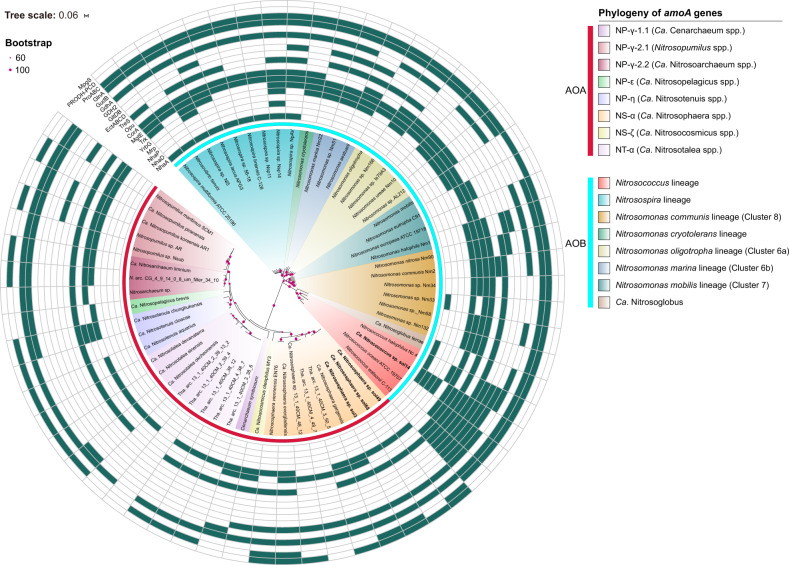
Fig. 6. Meta-analysis of salt resistance genes in phylogenetically distinct ammonia-oxidizing bacteria and archaea.
Phylogenetic tree of amoA genes from typical AOA and AOB species, with known genomes (see Table S6), including those from the MAGs in this study, highlighted in bold. Representative amoA sequences were phylogenetically analyzed with MEGA version 7.0 using the neighbor-joining method and the maximum composite likelihood model with 1000 replicates to generate bootstrap values. Outside the tree, the phylogenetic grouping of AOA and AOB is shown in the first internal ring with the colored strip in red and blue, respectively. The salt tolerance proteins identified in active ammonia oxidizers in this study (Fig. 5) are shown from the internal 2nd ring to the 20th ring including those encoding proteins of NhaA; NhaD; NhaP; Mrp; YrbG; Trk; MgtE; CorA; Opu; TreS; EctABCD; GltDB; GDH2; GdhA; GudB; GlnA; ProABC; PRODH-PCD; and MpgS. The presence and absence of these proteins in ammonia oxidizers are indicated in green and white, respectively. GudB, glutamate dehydrogenase (K00260); PCD, E1.2.1.88/1-pyrroline-5-carboxylate dehydrogenase.
Discussion
Salinity-based niche differentiation of ammonia oxidizers
Our results showed a significant shift in the active ammonia-oxidizing communities in solonetz soil under reclamation. Stable-isotope probing revealed the growth and activity of both AOB and AOA in saline-sodic soil. In particular, Nitrosococcus- and Nitrosospira-affiliated clades dominated the active AOB in the solonetz and farmland soils, respectively, and were most likely the primary contributors to ammonia oxidation, considering the higher population size of AOB than AOA in these two soils (Fig. 3c, d) and the higher specific cell activities of AOB than AOA usually observed in pure cultures [8, 52, 53]. Furthermore, the increased ammonia availability caused by alkaline pH in both soils is likely to favor AOB over both AOA and comammox [54, 55]. Nitrosospira is frequently identified as the major contributor to ammonia oxidation in soils, especially those under agricultural management [33, 56, 57]. The estimated cell apparent activity of AOA far exceeded the activities of pure cultures, further suggesting the dominant activity of Nitrosococcus in solonetz soils (Table S2). Gammaproteobacterial AOB are predominantly found in marine environments from which all previously cultured Nitrosococcus species were isolated [22, 23]. Intriguingly, Nitrosococcus-related and other gammaproteobacterial AOB have been demonstrated using molecular surveys and physiological studies to be present in a diverse range of habitats, including wastewaters [24], saline alkaline soils [26], a biofilter [27], a tea field [7] and rangelands [25, 28], also indicating that gammaproteobacterial AOB may have a higher diversity and more widespread distribution than previously appreciated (Fig. S8). Nonetheless, the ecophysiologies of these gammaproteobacterial AOB remain poorly understood in complex natural environments. Our labeling results provide the first direct evidence of Nitrosococcus dominating autotrophic ammonia-oxidizing activity in upland soil under salt stress. The NS-α clade was the dominant group of AOA in both soils investigated in our study, which is consistent with previous findings in alkaline farmland soil [33].
ENa is potentially the most critical factor driving the niche specialization of different ammonia-oxidizer clades in these two soils (Fig. 3e). The solonetz soil exhibited a 40-fold higher ENa than the converted farmland soil, which may have selected microbes with different tolerances to elevated salinity. Similarly, high salinity has been observed in wastewater reactors [24], marine ecosystems [22, 58], and salt lakes [23], which also contain dominant or highly abundant Nitrosococcus organisms similar to those in the solonetz soil. The adaptation of AOA to high salinity has also been previously demonstrated in aquatic systems based on the isolation of several strains of marine AOA [17, 50, 59–61], and ecological studies have shown high abundances of Nitrosopumilales-related AOA in marine systems, saline wastewater and floodplains [62–64], NS-α dominated groups in mangrove sediment [65] and the dominant activity of a Nitrosocosmicus-related clade (NS-ζ) following salinization of activated sludge [20]. Interestingly, the NS-α and NS-γ clades were present consistently in both soils, suggesting that these clades of AOA could adapt to a wide range of environmental conditions and may be important in maintaining the diversity and activity of AOA in soils subjected to severe environmental disturbances, e.g., during land conversion. In addition, growth of NS-δ was detected in the farmland soil, while NS-β was present but did not grow during incubations of the solonetz soil (Fig. 3c, d). Therefore, NS-β is likely specialized for highly saline environments and replaced by NS-γ in soils with much less salinity, leading to the reassembly of communities of AOA following agricultural conversion. The average cell apparent activity of total active ammonia oxidizers in the solonetz soil was 4.5-fold higher than that in the farmland soil (Table S2), which suggested that active ammonia oxidizer communities dominated by Nitrosococcus-related AOB in the solonetz soil possessed a much higher average specific-cell-energy-yield efficiency and appeared to have more energy to overcome the limitation of the low energy-yield of nitrification and tolerate salt stress [66].
Genomic adaptations to salt stress
High salinity leads to high osmotic pressure that can severely constrain the survival of ammonia oxidizers in such environments [50, 66]. In our study, comparative genomics analysis indicated that a range of different mechanisms are adopted by ammonia oxidizers to cope with excessive Na+ and high osmotic stress. NhaA, NhaD, NhaP and Mrp, identified in Ca. Nitrosococcus sp. Sol14, represents a group of membrane transporter proteins, with secondary Na+/H+ antiporter activity energized by the proton motive force for exporting Na+ and importing H+ (Fig. 5) [67]. NhaA, NhaD, and NhaP are encoded by single genes and are widely distributed in a wide variety of microorganisms involved in the maintenance of ion homeostasis under high sodium stress [43, 45], while Mrp is typically encoded by six or seven genes and has been shown to play an important role in Na+ resistance [67]. As these antiporters are involved in H+ uptake in exchange for cytoplasmic Na+ (Fig. 5, Fig. S9, Supplementary results), they could also contribute to maintaining a lower intracellular pH compared with the exceedingly high alkaline saline-sodic soils [44, 68, 69]. The variety of Na+ transport mechanisms identified in the Nitrosococcus MAG may allow it to survive under severe salt stress even in oligotrophic conditions because the energetic cost of establishing ionic gradients for salt tolerance is less than that of biosynthesizing compatible solutes [70]. The multiple Na+/H+ antiporters, which were very scarce or absent in Nitrosospira and NS-α in our soils and other known ammonia oxidizers (Fig. 6), may contribute to protecting Nitrosococcus against Na+ toxicity and high pH stress. In addition, adjusting the pH and osmotic homeostasis with the same transporter is obviously an effective strategy for efficient energy utilization, particularly considering the low energy yield of ammonia oxidation [50, 70].
Inorganic compatible solutes can be absorbed by cells to maintain osmotic equilibrium when the external salt concentration is high, whereas inorganic ions are maintained mostly outside the cells [46, 71, 72]. For instance, cytoplasmic K+ is less toxic to enzyme activity and metabolic function than Na+ [73], and many bacteria actively accumulate K+ to achieve osmotic equilibrium at elevated saline concentrations [74, 75]. Similarly, the Trk system harbored by Ca. Nitrosococcus sp. Sol14 is widely distributed in haloarchaea [47] and has been demonstrated to play a vital role in Na+ resistance in the halophilic bacterium Halomonas elongate of the family Halomonadaceae [46] that was also detected in the solonetz soil (Table S8), suggesting that K+ uptake likely alleviated the salt stress of Ca. Nitrosococcus sp. Sol14. Moreover, Mg2+ uptake transporters possessed by the ammonia oxidizers in our soils may also alleviate Na+ stress and appear to be important in stabilizing halophilic enzymes in many microbes [47]. However, Mg2+ transport does not seem to be a specific factor in the salt tolerance of ammonia oxidizers in the solonetz soil, because almost all ammonia-oxidizing microorganisms contain Mg2+ uptake transporters (Fig. 6). In addition, the Ca. Nitrososphaera sp. FarX and Nitrosospira (Fig. 5) in our soil possess YrbG antiporters to extrude Na+ by Ca2+ uptake, but this process might function only under high Ca2+ content conditions [47], which are not available in saline-sodic soil. YrbG proteins were consistently detected in 41 ammonia oxidizers (Fig. 6) but not in the genome of Ca. Nitrosococcus sp. Sol14, possibly indicating that YrbG was not an effective antiporter for salt tolerance in our soil.
Glycine betaine is another compatible solute that many prokaryotes and eukaryotes are known to accumulate under salt stress to regulate their osmotic pressure. For example, in the presence of a high concentration of sodium chloride, the addition of glycine betaine could improve the salt-tolerance of Rhizobium meliloti Be 151 [76]. Opu was identified in Ca. Nitrosococcus sp. Sol14 and two other Nitrosococcus strains from marine ecosystems [22, 77] as a transport system for glycine betaine (Fig. 6), and may be a commonly adopted salt tolerance mechanism for Nitrosococcus. Among all active ammonia oxidizers in our study, only Ca. Nitrosococcus sp. Sol14 appeared to possess the enzyme for the biosynthesis of trehalose, which is an organic molecule produced in many microorganisms and is typically associated with halotolerance [48, 78]. Both ectoine and hydroxyectoine are osmolytes [49] that are synthesized by Nitrosopumilus maritimus in response to increased osmotic stress [50, 79]. The gene cluster ectABCD encodes proteins for the biosynthesis of ectoine and hydroxyectoine from the substrate aspartate semialdehyde [80, 81], and was detected in Ca. Nitrosococcus sp. Sol14 and all three known Nitrosococcus genomes (Fig. 6) but absent in the MAGs of AOA and Nitrosospira scaffolds in our soils (Fig. 5). TreS and EctABCD proteins might confer an advantage to Nitrosococcus activity and competition over other ammonia oxidizers in our solonetz soil. Moreover, the cytoplasmic glutamate concentrations in some bacteria increase after exposure to highly osmotic media [82]. Similarly, glutamate may be synthesized in Nitrosopumilus by reversible reactions of glutamate dehydrogenase and glutamine synthetase [50], which were also identified in Ca. Nitrosococcus sp. Sol14 and Ca. Nitrososphaera sp. FarX (Fig. 5). Analysis of the Nitrosospira scaffolds and AOA MAGs of the solonetz soil indicated that these organisms may be able to synthesize glutamate via glutamine synthetase and glutamate dehydrogenase, respectively (Fig. 5). However, Nitrosopumilus may also take up and synthesize glutamate through a glutamate/aspartate symporter and the ornithine-glutamate reaction, respectively [50], which were not found in Ca. Nitrosococcus sp. Sol14 or AOA MAGs or Nitrosospira (Fig. 5). It is noteworthy that only some AOB, including Ca. Nitrosococcus sp. Sol14, have the potential to synthesize glutamate through the reaction of glutamate synthase [83] (Fig. 6). These multiple reactions may indicate that glutamate plays an important role in the adaptation of Ca. Nitrosococcus sp. Sol14 to its highly saline habitat.
Proline is a vital osmoprotectant for many Gram-positive bacteria, such as the moderately halophilic Salinicoccus roseus and Salinicoccus hispanicus [84]. Many bacteria increase their proline concentrations under osmotic stress by synthesizing or taking up proline [82]. Proline synthesis proteins [85] were identified in Ca. Nitrosococcus sp. Sol14. Nitrosospira in our soils may also synthesize proline based on the identified key enzyme (ProA) (Table S7) and the wide distribution of ProABC in known Nitrosospira genomes (Fig. 6). These results potentially illustrate that proline synthesis is not the key mechanism underlying the salt resistance of Ca. Nitrosococcus sp. Sol14. Interestingly, as in Nitrosopumilus [50], proteins that convert proline to glutamate were detected in Ca. Nitrososphaera sp. FarX, suggesting the potential for osmolyte switching [50]. However, the significance of this reaction with regard to salt tolerance is unclear. Ca. Nitrososphaera sp. FarX may be capable of forming MG, which is absent in Ca. Nitrosococcus sp. Sol14 and Nitrosospira (Fig. 5), as a compatible solute for cellular osmotic adjustment and thermal protection; this inference is based on the identification of a key enzyme similar to that found in Nitrososphaera and Nitrosocaldus lineages [51, 86]. MG is distributed widely among thermophilic and hyperthermophilic organisms [87]. However, in a few Rhodothermus marinus strains, MG accumulates only at supraoptimal growth temperatures during salt stress, suggesting that the synthesis of MG may require a higher temperature [87]. As the average annual temperature of the sites used in this study is only 4.7 °C [31], it seems unlikely that AOA use MG for effective defense against salt stress.
In summary, diverse salt tolerance mechanisms are key to the competitive adaptation of Ca. Nitrosococcus sp. Sol14 to its highly saline environment. Ca. Nitrosococcus sp. Sol14 contains more Na+/H+ antiporters than other Nitrosococcus representatives, suggesting greater salt-tolerance. These results also indicate that the ecological significance of active Ca. Nitrosococcus sp. Sol14 could be largely represented by pure culture studies, wherein the maximum salt tolerance of Nitrosococcus representatives (80–180 cmol per liter) [77, 88] was shown to be much higher than that of Nitrosospira briensis (only 25 cmol per liter) [88], which has far fewer salt-resistance mechanisms.
Conclusions
The present study showed salt tolerance-based niche differentiation of soil ammonia oxidizers. The Nitrosococcus species, which are predominantly found in marine environments and salt lakes, was demonstrated in soil ecosystems with markedly high sodium salt content and high pH. The targeted reconstruction of metagenome-assembled 13C-labeled genomes revealed that Nitrosococcus in saline-sodic soil possesses a more sophisticated assembly of salt tolerance mechanisms including Na+/H+ antiporters, a K+ uptake system and the transport and biosynthesis of organic compatible solutes (glycine betaine, trehalose, ectoine, hydroxyectoine, and glutamate) than other ammonia oxidizers (Thaumarchaeota and Nitrosospira) detected in the soil. These findings extend our understanding of important salt-tolerant microbes contributing to the nitrogen cycles, and suggest that the ecological importance of gammaproteobacterial ammonia oxidizers need to be re-assessed in salt-affected environments.
Supplementary information
Acknowledgements
We thank Dr. Wei Gao for the help on soil sample collection. We are also grateful to Profs. Brett Baker and Yuanfeng Cai for discussion on metagenomics analysis. We thank Dr. Yong-Xin Liu and the WeChat subscription ID “meta-genome” for some metagenomic analysis methods. We also gratefully acknowledge the helpful comments and careful corrections from Prof. Graeme Nicol, and the technical support from Rong Huang, Zhiying Guo, Yufang Sun, Deling Sun and Ruhai Wang of the Analytical Center of the Institute of Soil Science. This study was supported by the National Natural Science Foundation of China (41530857), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA28020203) and the Key Deployment Project of the Chinese Academy of Sciences (KFZD-SW-112).
Author contributions
XS and ZJ designed the study. XS performed the experiments and analyzed the data. JZ, XZ, and WX helped data mining. QB constructed the metagenome-assembled genomes. BZ helped acquire soil data. ZJ and J-BZ supervised the project and approved the final version. XS wrote the manuscript with input from all authors.
Data availability
Raw 16S rRNA and amoA gene sequences were deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject ID PRJNA641227. Metagenomics and metagenome-assembled genomes (MAGs) data are available at MG-RAST under the study names DAAN_WGS and DAAN_MAGs, respectively.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01079-6.
References
- 1.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 2.Stein LY, Klotz MG. The nitrogen cycle. Curr Biol. 2016;26:R94–R98. doi: 10.1016/j.cub.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–69. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 4.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–78. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 5.Lehtovirta-Morley LE, Ge C, Ross J, Yao H, Nicol GW, Prosser JI. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol. 2014;89:542–52. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–7. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayatsu M, Tago K, Uchiyama I, Toyoda A, Wang Y, Shimomura Y, et al. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 2017;11:1130–41. doi: 10.1038/ismej.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–31. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, et al. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:21206–11. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aigle A, Prosser JI, Gubry-Rangin C. The application of high-throughput sequencing technology to analysis of amoA phylogeny and environmental niche specialisation of terrestrial bacterial ammonia-oxidisers. Environ Microbiome. 2019;14:3. doi: 10.1186/s40793-019-0342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antony CP, Kumaresan D, Hunger S, Drake HL, Murrell JC, Shouche YS. Microbiology of Lonar Lake and other soda lakes. ISME J. 2013;7:468–76. doi: 10.1038/ismej.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montanarella L, Chude V, Yagi K, Krasilnikov P, Panah SKA, Mendonca-Santos MDL, et al. Status of the World’s Soil Resources (SWSR) - Main Report. 2015.
- 13.Vera-Gargallo B, Chowdhury TR, Brown J, Fansler SJ, Durán-Viseras A, Sánchez-Porro C, et al. Spatial distribution of prokaryotic communities in hypersaline soils. Sci Rep. 2019;9:1769. doi: 10.1038/s41598-018-38339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 2010;4:829–938. doi: 10.1038/ismej.2010.3. [DOI] [PubMed] [Google Scholar]
- 15.Metternicht GI, Zinck JA. Remote sensing of soil salinity: potentials and constraints. Remote Sens Environ. 2003;85:1–20. [Google Scholar]
- 16.Shi YL, Liu XR, Zhang QW. Effects of combined biochar and organic fertilizer on nitrous oxide fluxes and the related nitrifier and denitrifier communities in a saline-alkali soil. Sci Total Environ. 2019;686:199–211. doi: 10.1016/j.scitotenv.2019.05.394. [DOI] [PubMed] [Google Scholar]
- 17.Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 18.Bayer B, Vojvoda J, Offre P, Alves RJE, Elisabeth NH, Garcia JAL, et al. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J. 2016;10:1051–63. doi: 10.1038/ismej.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR, et al. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: An ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci USA. 2015;112:1173–8. doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan KL, Gao JF, Li DC, Fan XY. The dominance of non-halophilic archaea in autotrophic ammonia oxidation of activated sludge under salt stress: a DNA-based stable isotope probing study. Bioresour Technol. 2019;291:8. doi: 10.1016/j.biortech.2019.121914. [DOI] [PubMed] [Google Scholar]
- 21.Nejidat A. Nitrification and occurrence of salt-tolerant nitrifying bacteria in the Negev desert soils. FEMS Microbiol Ecol. 2005;52:21–29. doi: 10.1016/j.femsec.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Ward BB, O’Mullan GD. Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing gamma-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl Environ Micro. 2002;68:4153–7. doi: 10.1128/AEM.68.8.4153-4157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koops HP, Böttcher B, Möller UC, Pommerening-Röser A, Stehr G. Description of a new species of Nitrosococcus. Arch Microbiol. 1990;154:244–8. [Google Scholar]
- 24.Fumasoli A, Bürgmann H, Weissbrodt DG, Wells GF, Beck K, Mohn J, et al. Growth of Nitrosococcus-related ammonia oxidizing bacteria coincides with extremely low pH values in wastewater with high ammonia content. Environ Sci Technol. 2017;51:6857–66. doi: 10.1021/acs.est.7b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivera NL, Prieto L, Bertiller MB, Ferrero MA. Sheep grazing and soil bacterial diversity in shrublands of the Patagonian Monte, Argentina. J Arid Environ. 2016;125:16–20. [Google Scholar]
- 26.Pérez-Hernandez V, Hernandez-Guzman M, Serrano-Silva N, Luna-Guido M, Navarro-Noya YE, Montes-Molina JA, et al. Diversity of amoA and pmoA genes in extremely saline alkaline soils of the former lake Texcoco. Geomicrobiol J. 2020;37:785–97. [Google Scholar]
- 27.Picone N, Pol A, Mesman R, van Kessel MAHJ, Cremers G, van Gelder AH, et al. Ammonia oxidation at pH 2.5 by a new gammaproteobacterial ammonia-oxidizing bacterium. ISME J. 2020;15:1150–64. doi: 10.1038/s41396-020-00840-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan H, Liu HY, Liu YW, Zhang QC, Luo Y, Liu XM, et al. Understanding the relationships between grazing intensity and the distribution of nitrifying communities in grassland soils. Sci Total Environ. 2018;634:1157–64. doi: 10.1016/j.scitotenv.2018.04.117. [DOI] [PubMed] [Google Scholar]
- 29.Santos JP, Mendes D, Monteiro M, Ribeiro H, Baptista MS, Borges MT, et al. Salinity impact on ammonia oxidizers activity and amoA expression in estuarine sediments. Estuar Coast Shelf Sci. 2018;211:177–87. [Google Scholar]
- 30.Ye L, Zhang T. Ammonia-oxidizing bacteria dominates over ammonia-oxidizing archaea in a saline nitrification reactor under low DO and high nitrogen loading. Biotechnol Bioeng. 2011;108:2544–52. doi: 10.1002/bit.23211. [DOI] [PubMed] [Google Scholar]
- 31.Luo S, Wang S, Tian L, Shi S, Xu S, Yang F, et al. Aggregate-related changes in soil microbial communities under different ameliorant applications in saline-sodic soils. Geoderma. 2018;329:108–17. [Google Scholar]
- 32.Wang WJ, He HS, Zu YG, Guan Y, Liu ZG, Zhang ZH, et al. Addition of HPMA affects seed germination, plant growth and properties of heavy saline-alkali soil in northeastern China: comparison with other agents and determination of the mechanism. Plant Soil. 2011;339:177–91. [Google Scholar]
- 33.Xia W, Zhang C, Zeng X, Feng Y, Jia Z. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 2011;5:1226–36. doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–8. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–8. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 36.Fowler SJ, Palomo A, Dechesne A, Mines PD, Smets BF. Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environ Microbiol. 2018;20:1002–15. doi: 10.1111/1462-2920.14033. [DOI] [PubMed] [Google Scholar]
- 37.Zhao ZR, Huang GH, He SS, Zhou N, Wang MY, Dang CY, et al. Abundance and community composition of comammox bacteria in different ecosystems by a universal primer set. Sci Total Environ. 2019;691:145–55. doi: 10.1016/j.scitotenv.2019.07.131. [DOI] [PubMed] [Google Scholar]
- 38.Alves RJE, Minh BQ, Urich T, von Haeseler A, Schleper C. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat Commun. 2018;9:1517. doi: 10.1038/s41467-018-03861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–31. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo C, Rodriguez-R LM, Konstantinidis KT. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014;42:e73. doi: 10.1093/nar/gku169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroda T, Mizushima T, Tsuchiya T. Physiological roles of three Na+/H+ antiporters in the halophilic bacterium Vibrio parahaemolyticus. Microbiol Immunol. 2005;49:711–9. doi: 10.1111/j.1348-0421.2005.tb03662.x. [DOI] [PubMed] [Google Scholar]
- 44.Daebeler A, Kitzinger K, Koch H, Herbold CW, Steinfeder M, Schwarz J, et al. Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant. Nitrospira ISME J. 2020;14:2967–79. doi: 10.1038/s41396-020-0724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padan E, Venturi M, Gerchman Y, Dover N. Na+/H+ antiporters. Biochim Biophys Acta. 2001;1505:144–57. doi: 10.1016/s0005-2728(00)00284-x. [DOI] [PubMed] [Google Scholar]
- 46.Kraegeloh A, Amendt B, Kunte HJ. Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T. J Bacteriol. 2005;187:1036–43. doi: 10.1128/JB.187.3.1036-1043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, et al. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PloS Genet. 2014;10:e1004784. doi: 10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardoso FS, Castro RF, Borges N, Santos H. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology. 2007;153:270–80. doi: 10.1099/mic.0.29262-0. [DOI] [PubMed] [Google Scholar]
- 49.Sadeghi A, Soltani BM, Nekouei MK, Jouzani GS, Mirzaei HH, Sadeghizadeh M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol Res. 2014;169:699–708. doi: 10.1016/j.micres.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Ngugi DK, Blom J, Alam I, Rashid M, Ba-Alawi W, Zhang G, et al. Comparative genomics reveals adaptations of a halotolerant thaumarchaeon in the interfaces of brine pools in the Red Sea. ISME J. 2015;9:396–411. doi: 10.1038/ismej.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–45. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 52.Glover HE. The relationship between inorganic nitrogen oxidation and organic carbon production in batch and chemostat cultures of marine nitrifying bacteria. Arch Microbiol. 1985;142:45–50. [Google Scholar]
- 53.Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C, et al. Isolation of ‘Candidatus Nitrosocosmicus franklandus’, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol. 2016;92:fiw057. doi: 10.1093/femsec/fiw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hink L, Gubry-Rangin C, Nicol GW, Prosser JI. The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J. 2018;12:1084–93. doi: 10.1038/s41396-017-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol. 2008;10:1601–11. doi: 10.1111/j.1462-2920.2008.01578.x. [DOI] [PubMed] [Google Scholar]
- 57.Jia Z, Conrad R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–71. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 58.Millero FJ, Feistel R, Wright DG, McDougall TJ. The composition of Standard Seawater and the definition of the Reference-Composition Salinity Scale. Deep-Sea Res Part I-Oceanogr Res Pap. 2008;55:50–72. [Google Scholar]
- 59.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco bay estuary. J Bacteriol. 2012;194:2121–2. doi: 10.1128/JB.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsutani N, Nakagawa T, Nakamura K, Takahashi R, Yoshihara K, Tokuyama T. Enrichment of a novel marine ammonia-oxidizing archaeon obtained from sand of an eelgrass zone. Microbes Environ. 2011;26:23–29. doi: 10.1264/jsme2.me10156. [DOI] [PubMed] [Google Scholar]
- 61.Park BJ, Park SJ, Yoon DN, Schouten S, Damste JSS, Rhee SK. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Micro. 2010;76:7575–87. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parada AE, Fuhrman JA. Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor. ISME J. 2017;11:2510–25. doi: 10.1038/ismej.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu YJ, Whang LM, Fukushima T, Chang SH. Responses of ammonia-oxidizing archaeal and betaproteobacterial populations to wastewater salinity in a full-scale municipal wastewater treatment plant. J Biosci Bioeng. 2013;115:424–32. doi: 10.1016/j.jbiosc.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Cardarelli EL, Bargar JR, Francis CA. Diverse Thaumarchaeota dominate subsurface ammonia-oxidizing communities in semi-arid floodplains in the western United States. Micro Ecol. 2020;80:778–92. doi: 10.1007/s00248-020-01534-5. [DOI] [PubMed] [Google Scholar]
- 65.Wang HT, Gilbert JA, Zhu YG, Yang XR. Salinity is a key factor driving the nitrogen cycling in the mangrove sediment. Sci Total Environ. 2018;631-2:1342–9. doi: 10.1016/j.scitotenv.2018.03.102. [DOI] [PubMed] [Google Scholar]
- 66.Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol. 2011;13:1908–23. doi: 10.1111/j.1462-2920.2010.02365.x. [DOI] [PubMed] [Google Scholar]
- 67.Ito M, Guffanti AA, Oudega B, Krulwich TA. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol. 1999;181:2394–402. doi: 10.1128/jb.181.8.2394-2402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–43. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005;9:345–54. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- 70.Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol R. 1999;63:334–48. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mackay MA, Norton RS, Borowitzka LJ. Organic osmoregulatory solutes in Cyanobacteria. J Gen Microbiol. 1984;130:2177–91. [Google Scholar]
- 72.Sadler M, McAninch M, Alico R, Hochstein LI. The intracellular Na+ and K+ composition of the moderately halophilic bacterium, Paracoccus halodenitrificans. Can J Microbiol. 1980;26:496–502. doi: 10.1139/m80-083. [DOI] [PubMed] [Google Scholar]
- 73.Brown AD. Compatible solutes and extreme water stress in eukaryotic micro-organisms. Adv Micro Physiol. 1978;17:181–243. doi: 10.1016/s0065-2911(08)60058-2. [DOI] [PubMed] [Google Scholar]
- 74.Reed RH, Warr SRC, Richardson DL, Moore DJ, Stewart WDP. Multiphasic osmotic adjustment in a euryhaline cyanobacterium. FEMS Microbiol Lett. 1985;28:225–9. [Google Scholar]
- 75.Welsh DT, Herbert RA. Osmoadaptation of Thiocapsa roseopersicina OP-1 in batch and continuous culture: Accumulation of K+ and sucrose in response to osmotic stress. FEMS Microbiol Ecol. 1993;13:151–7. [Google Scholar]
- 76.Sauvage D, Hamelin J, Larher F. Glycine betaine and other structurally related compounds improve the salt tolerance of Rhizobium meliloti. Plant Sci Lett. 1983;31:291–302. [Google Scholar]
- 77.Campbell MA, Chain PSG, Dang H, Sheikh EI, Norton AF, Ward JM, et al. MG. Nitrosococcus watsonii sp. nov., a new species of marine obligate ammonia-oxidizing bacteria that is not omnipresent in the world’s oceans: calls tovalidate the names’Nitrosococcus halophilus’ and ‘Nitrosomonas mobilis’. FEMS Microbiol Ecol. 2011;76:39–48. doi: 10.1111/j.1574-6941.2010.01027.x. [DOI] [PubMed] [Google Scholar]
- 78.Arguelles JC. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol. 2000;174:217–24. doi: 10.1007/s002030000192. [DOI] [PubMed] [Google Scholar]
- 79.Widderich N, Czech L, Elling FJ, Könneke M, Stöveken N, Pittelkow M, et al. Strangers in the archaeal world: osmostress-responsive biosynthesis of ectoine and hydroxyectoine by the marine thaumarchaeon Nitrosopumilus maritimus. Environ Microbiol. 2016;18:1227–48. doi: 10.1111/1462-2920.13156. [DOI] [PubMed] [Google Scholar]
- 80.Bursy J, Pierik AJ, Pica N, Bremer E. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem. 2007;282:31147–55. doi: 10.1074/jbc.M704023200. [DOI] [PubMed] [Google Scholar]
- 81.Kol S, Merlo ME, Scheltema RA, de Vries M, Vonk RJ, Kikkert NA, et al. Metabolomic characterization of the salt stress response in Streptomyces coelicolor. Appl Environ Micro. 2010;76:2574–81. doi: 10.1128/AEM.01992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–47. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saum SH, Sydow JF, Palm P, Pfeiffer F, Oesterhelt D, Muller V. Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately halophilic bacterium Halobacillus halophilus. J Bacteriol. 2006;188:6808–15. doi: 10.1128/JB.00781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol R. 1998;62:504–44. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahan MJ, Csonka LN. Genetic analysis of the proBA genes of Salmonella typhimurium: physical and genetic analysis of the cloned proB+A+ genes of Escherichia coli and of a mutant allele that confers proline overproduction and enhanced osmotolerance. J Bacteriol. 1983;156:1249–62. doi: 10.1128/jb.156.3.1249-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Empadinhas N, Pereira PJB, Albuquerque L, Costa J, Sa-Moura B, Marques AT, et al. Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity. Mol Microbiol. 2011;79:76–93. doi: 10.1111/j.1365-2958.2010.07432.x. [DOI] [PubMed] [Google Scholar]
- 87.Santos H, da Costa MS. Compatible solutes of organisms that live in hot saline environments. Environ Microbiol. 2002;4:501–9. doi: 10.1046/j.1462-2920.2002.00335.x. [DOI] [PubMed] [Google Scholar]
- 88.Koops HP, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. The Lithoautotrophic Ammonia-Oxidizing Bacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds). The Prokaryotes: a Handbook on the Biology of Bacteria, 3rd edn. New York, USA: Springer Science+Business Media; 2006, pp 778–811.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA and amoA gene sequences were deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject ID PRJNA641227. Metagenomics and metagenome-assembled genomes (MAGs) data are available at MG-RAST under the study names DAAN_WGS and DAAN_MAGs, respectively.