Abstract
Brain metastases comprise 40% of all metastatic tumours and breast tumours are among the tumours that most commonly metastasise to the brain, the role that epigenetic gene dysregulation plays in this process is not well understood. We carried out 450 K methylation array analysis to investigate epigenetically dysregulated genes in breast to brain metastases (BBM) compared to normal breast tissues (BN) and primary breast tumours (BP). For this, we referenced 450 K methylation data for BBM tumours prepared in our laboratory with BN and BP from The Cancer Genome Atlas. Experimental validation on our initially identified genes, in an independent cohort of BP and in BBM and their originating primary breast tumours using Combined Bisulphite and Restriction Analysis (CoBRA) and Methylation Specific PCR identified three genes (RP11-713P17.4, MIR124-2, NUS1P3) that are hypermethylated and three genes (MIR3193, CTD-2023M8.1 and MTND6P4) that are hypomethylated in breast to brain metastases. In addition, methylation differences in candidate genes between BBM tumours and originating primary tumours shows dysregulation of DNA methylation occurs either at an early stage of tumour evolution (in the primary tumour) or at a later evolutionary stage (where the epigenetic change is only observed in the brain metastasis). Epigentic changes identified could also be found when analysing tumour free circulating DNA (tfcDNA) in patient’s serum taken during BBM biopsies. Epigenetic dysregulation of RP11-713P17.4, MIR3193, MTND6P4 are early events suggesting a potential use for these genes as prognostic markers.
Subject terms: Cancer, Genetics, Molecular biology, Biomarkers, Molecular medicine, Oncology
Introduction
More than 90% of cancer related deaths are attributed to metastases and about 80% of breast cancer deaths occur from metastases1. 15–25% of breast tumours metastasise to the brain during the course of disease2 and despite aggressive treatment strategies with surgical resection, stereotactic radiosurgery and whole brain radiotherapy (WBRT), the prognosis of breast to brain metastasis (BBM) patients remains poor3. Due to the challenges associated with the treatment of brain tumours, it is crucial to find novel prognostic markers that will inform the clinical management of breast cancer patients.
The seed and soil theory of metastases indicates that metastasing tumour cells have affinity for specific organs4,5. Metastases arise from metastatic initiating cells, which intravasate from the primary sites and remain as undetectable disseminated tumour cells before evolving into clinically visible lesions in distant organs6. These dormant cells are often found as micrometastases in bone marrow and lymph nodes many years after primary tumour treatment7–9.
Primary breast tumour hormone receptor status (Estrogen receptor (ER), Progesterone Receptor (PR) and Human EGFR Receptor 2 (HER2)) can be used as prognostic markers for brain metastasis risk10,11 and BBM occurs more frequenty in patients with ER–, HER2 + or triple negative breast tumours (ER-/PR-/HER2-)12. Of the different tumour subtypes, triple negative breast tumours have the worst prognosis13, and have faster rates of metastases compared to HER2 + patients14,15. However, following primary tumour treatment, and a period of apparent dormancy (in some cases lasting > 10 years), ER + tumours frequently metastasise to the brain16–18. Therefore, identifying genomic alterations that occur uniquely in primary tumours that eventually metastasise to specific organs could help clinical management of the disease18.
We have previously used a candidate-gene approach to identify epigenetically dysregulated genes in BBM19. Following from this study, we wished to carry out a non-selective, genome-wide analysis to identify novel epigenetic changes that are common in breast tumours that metastasis to the brain. To identify candidate genes that are differentuialy methylated when comparing primary breast tumours and BBM we have carried out 450 K-methylation array analysis of BBM tumours and compared the genome-wide methylation status of these tumours to primary breast tumours from the Cancer Genome Atlas (TCGA). We then determined the methylation status of candidate genes in BBM and their originating primary breast tumour. This analysis has led to the identification of DNA methylation alterations that commonly occur in primary breast tumours that eventualy metastasis to the brain. We have also identified epigenetic changes that occur only after the tumor cells have disseminated from the primary breast tumours. In addition, as a first step towards the development of a non-invasive prognostic tool, we have validated the methylation status of the genes identified in patients’ serum. The identified epigenetic alterations may be used as potential non-invasive markers and new therapeutic targets for BBM patients.
Materials and methods
Patients and samples
Thirty fresh-frozen metastatic brain tumours (BBM) that originated from primary breast tumours (BP) were provided by The Walton Research Tissue Bank (WRTB), Liverpool and Brain Tumour North West (BTNW) Tissue Bank, Preston; BBM tumours were labelled BBM1 to BBM30. Formalin fixed paraffin embedded (FFPE) originating primary tumours (BP) from individual patients corresponding to their brain metastases (matched-pairs) were available for 11 of these tumours (BBM1, BBM2, BBM5, BBM7, BBM8, BBM10, BBM11, BBM12, BBM13, BBM14 and BBM15). These primary and BBM pairs were labelled as individual patients such as patients 1, patient 2, patients 5 etc. Patients’ serum was available for BBM1, BBM2, BBM5, BBM6, BBM7, BBM8, BBM10, BBM11, BBM12, BBM13. Serum was collected at the time of BBM Surgery. Receptor status information is available for 9 of the 11 primary tumour pairs, six of these are ER + ve, one is triple negative, one is ER/PR-ve, and one is HER2 + ve (with ER/PR unknown status). Additional clinical information is available through our previous publication19. The time between primary tumour surgery and removal of the brain metastasis ranges from 2 to 10 years.
An independent cohort of 40 primary breast tumours (BP) analysed during this study were ductal carcinomas; their clinical characteristics were described previously19. Molecular characterization was available for 20 of these tumours, 15 of these are ER + ve and three are triple negative. No brain metastases were observed in any of these patients. Seventeen of these patients had been screened for metastasis for ≥ 5 years from the time of primary tumour surgery and nine had been screened for ≥ 10 years.
The research ethics committee (North Wales REC: 11/WNo03/2) approved tissues from the research banks and informed consent was obtained from each patient. The project was carried out following local ethical approval (University of Wolverhampton Life Sciences Ethics Committee: LSEC/201,011/43). This study was conducted according to the principles expressed in the Declaration of Helsinki.
Genomic DNA/RNA extraction
Genomic DNA was extracted from fresh-frozen BBM tumours using The DNA isolation kit from cells and tissues (Roche, Germany) as previously described19. Briefly, 25 mg of tissue was homogenised using lysis buffer and incubated at 37 °C for 30 min followed by the addition of Proteinase K and RNase solution. The samples were then centrifuged and processed according to manufacturer’s instructions. For FFPE samples, a FFPE DNA extraction kit (Qiagen, USA) was used as previously described19. Briefly, a small block of samples embedded with paraffin was cut into thin sections and mixed with xylene followed by 100% ethanol. The samples were then processed according to manufacturer’s instructions. The tumour-free circulating DNA from the patients’ serum was extracted using ZR serum DNA kit (Zymo research, USA). Briefly, 2 ml plasma from each patient was transferred to a conical shaped 50 ml universal tube. 8 ml of genomic lysis buffer and 10 μl of zymoBeads were added to each sample and placed in a shaker for two hours at room temperature. The samples were then processed further according to manufacturer’s instructions. Similarly, the total RNA was extracted using EZ-RNA extraction kit (Biological Industries, Israel). Briefly, fresh-frozen tumours were homogenized using lysis buffer followed by addition of extraction solution. The samples were then centrifuged and processed according to manufacturer’s instructions. The concentration of DNA and RNA was measured using nanodrop2000 (Thermo Scientific, USA).
Illumina BeadChip 450 K HumanMethylation array
Of 30 samples received, twenty-three BBM samples were used to assess the genome-wide methylation profilings of > 485 K individual CpG loci using Illumina BeadChip 450 K HumanMethylation Array (450 K array). The 450 K array profiling was carried out at Cambridge Genomic Services (CGS), UK. Chip processing was carried out based on 450 K array design according to manufacturer’s instructions. Signal intensities generated by Illumina GenomeStudio were converted to β-values and BeadStudio software was used to remove biases between the Infinium I and II probes. In order to remove the technical biases between the 450 K array downloaded from the TCGA (14 normal breast tissues and 14 primary breast tumours) and our array on 23 BBM, further normalisation was carried out using R statistical package20. Processing of array data was performed with the R packaged RnBeads (version 0.99.10) and normalized using the SWAN normalization option. The hg19 genome annotations were used during the analysis and sex chromosome data was excluded21,22. Data in figures and tables are derived from gene–gene comparisons using standard analysis options in the package. The tumour barcode for the tumour data downloaded from TCGA is given in Supplementary Fig. 1.
Initial screening of candidate genes signatures hypermethylated or hypomethylated in brain metastases compared to primary breast tumours and normal breast tissues
In order to generate an initial candidate list of genes that are either hypomethylated or hypermethylated in BBM, we compared CpG methylation between normal breast tissues (BN), primary breast tumour tissues (BP) and breast to brain metastases (BBM) samples. For this, we downloaded 450 K methylation array data from The Cancer Genome Atlas (TCGA) for 14 normal breast tissues and 14 primary breast tumours (Supplementary Fig. 1A, B. Our analysis aimed to identify individual CpGs that gained methylation (hypermethylated) in BBM compared to primary breast tumour and normal breast tissue samples (see Supplementary Fig. 1C for an example). We also looked for individual CpGs that have lost methylation (hypomethylated) in BBM compared to primary breast tumors and normal breast tissues. (See supplementary Fig. 1D for an example). In order to generate an initial list of CpGs, that have either gained or lost methylation in BBM, we retrieved individual CpGs (array probes) with a β-value of ≥ 0.4 in 50% of the BBM tumours and < 0.4 in 50% of the primary tumours and normal tissue; previous studies have shown that β-values of ≥ 0.4 are associated with silencing of genes or significant loss of expression23–26. From this list of methylated CpGs we retained only those CpGs that had a diference in β-value of ≥ 0.15 between the metastatic and primary tumour sets. Similarly, In order to identify candidate hypomethylated loci in BBM, we selected CpG probes that had β-values < 0.4 in BBM and β-values of ≥ 0.4 in normal breast tissues and primary tumour tissues in at least 50% of each sample sets and then retained the CpG loci that have an average β-value differences of ≥ 0.15 between BBM and, normal breast tissues and primary breast tumours. The CpG loci that met these criteria and were significnaly differentialy methylated (P ≤ 0.05) across the different tissue types (Supplementary Table 1A) were carried forward for further analysis.
Bisulfite conversion of DNA
Bisulfite conversion of genomic DNA from tumours was carried out using the EZ DNA methylation kit (Zymo Research Corp., USA). Briefly, 300 ng of genomic DNA was mixed with bisulphite conversion reagent and was incubated for 16 h at 50 °C. The samples were then processed according to manufacturer’s instructions. Bisulphite conversion of tumour-free circulating DNA (tfcDNA) from patients’ serum was carried out using Epitect Bisulfite kit (Qiagen, USA). Briefly, 200 ng DNA from the plasma was added to 200 μl sterile PCR tube and was mixed up with 85 μl bisulphite conversion reagent (bisulphite mix), 85 μl DNA protect buffer and RNase free water making up total volume of 140 μl. The samples were then processed according to manufacture’s instructions. Fully methylated, positive controls were generated by incubating genomic DNA with DNA methyltransferase, in the presence of S-Adenosyl methionine (SAM) (New England bio lab, USA) for 2 h at 37 °C prior to bisulfite conversion.
Experimental validation of methylation status of individual genes
The methylation status of each gene corresponding to differentially methylated probes was determined by Combined Bisulphite and Restriction Analyses (CoBRA). Semi-nested PCR was carried out and DNA methylation status of candidate genes was determined by digesting CoBRA PCR products with BstUI or TaqI restriction enzymes (Fermentas, UK).
CoBRA primers were designed based on the standard primary designing criteria used in analyzing bisulphite converted DNA27–29. The CoBRA primers were designed in such a way that the region analysed included the specific CpG identified by the 450 K array and additional local CpGs to enable the reliable determination methylation status by restriction digestion (Supplementary Table 2A). The methylation status of genes in patients’ serum was determined using Methylation Specific PCR (Supplementary Table 2B).
Experimental validation of expression of selected genes
Expression analysis of candidate genes was carried out using quantitative-reverse transcription PCR (qRT PCR). Due to the limited size of the tumor biopsies available, RNA was available only for BBM13, BBM15, BBM16, BBM20 and BBM30. Prior to the PCR, cDNA was prepared using QuanTect reverse transcription kit (QIAGEN, USA). β-actin gene was used as internal control for mRNA expression. The Ct value obtained were converted to the relative quantity of targets genes normalized with respect to internal control, and relative to a control sample. Due to the lack of normal tissue control samples (associated normal brest tissue was avalble only as FFPE), a BBM samples with median level of of expression for each qRTPCR among the samples set, was used as a control sample to calculate fold enrichment of other samples.
Statistical analysis
Initial statistical tests to generate candidate CpG loci was carried out using the statistical package available in R21. During the experimental validation, only the samples where methylation differences were statistically significant between primary breast tumours and BBM were taken further. Fisher’s exact test was used to determine the statistical significance of methylation between BP versus BBM samples. P ≤ 0.05 was considered statistically significant.
Patient survival analyses
Survival analyses of breast cancer patients were carried out to investigate the correlation of expression of candidate genes with clinical prognosis using online KM-plotter tools30. KM-plotter is an online tool capable of measuring the prognostic indication of 22,277 genes based on gene expression data and survival information of 6234 breast cancer patients (Affymetrix HGU133A and HGU133 microarrays datasets) downloaded from Gene Expression Omnibus (GEO)31. This tool segregates patients on high or low expressing groups based on median or a lower/upper quartile as a cut off to measure a statistical significance of gene expression with patient survival. Logrank P < 0.05 was considered statistically significant.
Results
Identification of differentially methylated genes in BBM
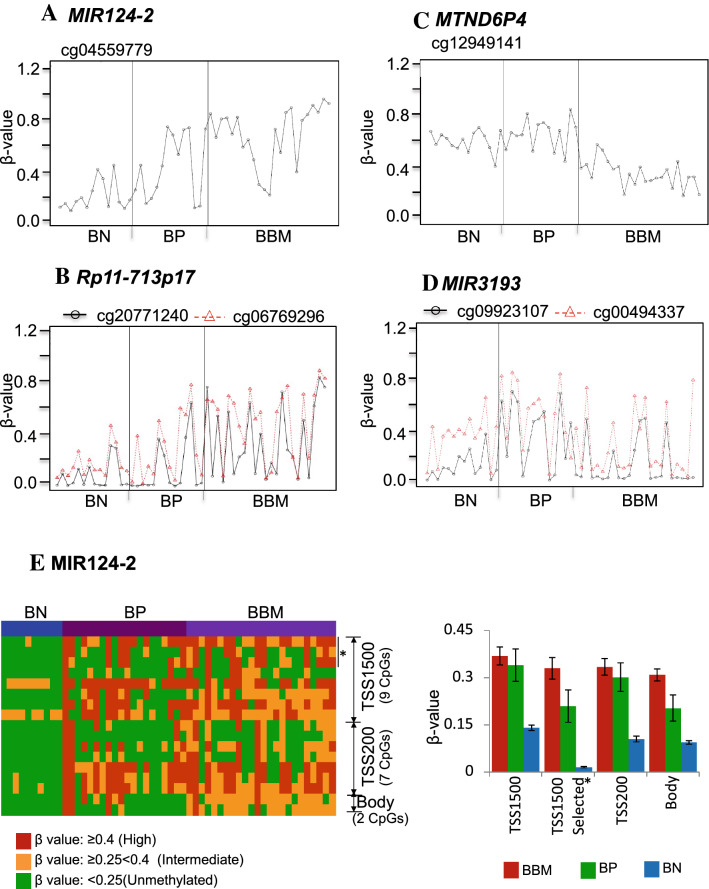
We identified differentialy methylated CpGs between breast-to-brain metastases (BBM) and non-metastatic primary breast tumours (BP) and normal breast tissue (BN) by comparing Illumina BeadChip 450 K methylation data that we generated for 23 BBM to data for normal breast tissues and primary tumours from TCGA. First, an initial screening of genes based on β-value (see method section for details) was carried out to identify CpG loci that were either hypermethylated (Fig. 1A,B) or hypomethylatled in BBM (Fig. 1C,D) compared to normal breast tissues and primary breast tumours. This identified 26 candidate BBM hypermethylayted loci (14 genes) and 58 BBM hypomethyaled loci (40 genes) (p ≤ 0.05; Supplementary Table 1A). In order to concentrate on loci consisting of the most frequently occurring and largest β-value differences between the two tumour types, we further generated a candidate list that contained loci that are either hypermethylated in BBM (β-value > 0.4) in ≥ 50% of tumours analysed or hypomethylated in BBM ((β-value < 0.4) in ≥ 50% of tumours analysed compared to BP (See Fig. 1A–D and Supplementary Fig. 2 for representative data of genes that matched these criteria). This shortlisting identified 6 hypermethylated genes that are frequently methylated in BBM samples compared to BN and BP and 9 hypomethylated genes that are infrequently methylateld in BBM compapred to BN and BP samples (Supplementary table 1B).
Figure 1.
Genome-wide DNA methylation analyses by 450 K array for BBM, primary breast tumours (BP) and normal breast tissues (BN) identified CpG loci which have either gained methylation (hypermethylated) or have lost methylatioin (hypomethylated) in BBM compared to BP and BN tumours. Representative examples of gene-associated CpG loci that have gained methylation (hypermethylated) in BBM (A,B) or have lost methylation (hypomethylated) in BBM (C,D) compared to BP and BN (as measured by β-value). (E) Regional methylation pattern of MIR124-2 showing the the location of all CpG loci present on the 450 K array relative to the transcription start of the gene and the differing methylation levels for each loci in BBM, BP and BN. * denotes the location of the differentially methylated CpG that was identified by the bioinformatic analysis of 450 K-array data from BN, BP and BBM.
In addition, we examined the methylation level of all CpG loci available based on the 450 K array design that are associated with our candidate genes which we refer to as the regional methylation level of each gene. The 450 K array design provides annotations for each CpG includin genomic location of the CpGs in the array. We retrieved all CpGs corresponding to our candidate genes and outlined where each of the CpG sites are located in relation to the gene structure: TSS1500, (1500 nucleotides upstreams of transcription start site), TSS200 (200 nucleotides upstream of transcription start site), 1st Exon, or body regions (downstream of the TSS or the first Exon) individually for all samples in relation to the gene structure (Fig. 1E, supplementary Fig. 3, and supplementary Fig. 9 Left panel). We determined an average methylation level of each CpGs in each sample type i.e. BN, BP and BBM in individual structural regions of each gene (Fig. 1E, supplementary Fig. 3, supplementary Fig. 9; right panel). Regional methylation maps have been constructed only for those candidate genes where there is sufficient 450 K probe density to generate informative figures.
RP11-713P17.4, MIR124-2, NUS1P3 are hypermethylated in brain metastases compared to primary breast tumours
All candidate genes were validated for their methylation status using Combined Bisulpite and Restriction Analysis (CoBRA), this is a robust and reliable method that is not prone to false positive results32. Primers were designed to amplify the probe regions identified in the 450 K arrays using standard criteria published previously19,23.
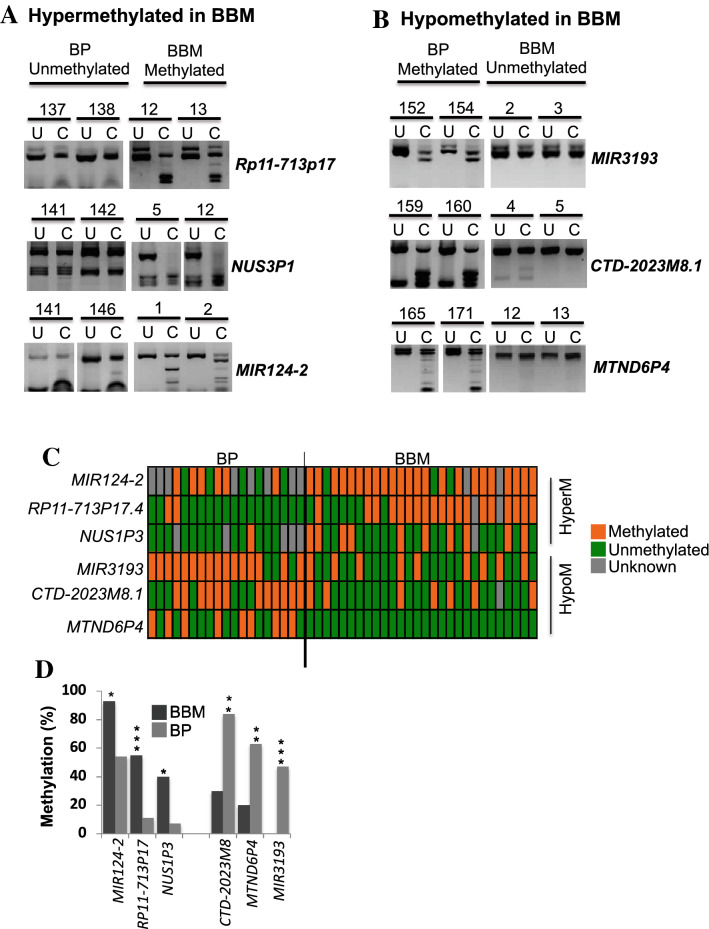
First, we validated the six shortlisted candidate hypermethylated genes (see supplementary table 1B), by CoBRA in 30 BBM, that included the 23 BBM samples used in the methylation array. Then, using the CoBRA method, the methylation level of these genes was examined in an independent cohort of 20 primary breast tumours (BP) with no evidence of metastatic progression. Comparison of the results from these two tumour groups allowed us to determine if the methylation of these genes is more commonly found in the brain metastases than in BP. Of the 6 candidate genes, 2 were infrequently methylated in BP tumours; RP11-713P17.4 and NUS1P3 (Fig. 2A,D and Supplementary Fig. 4B, S4C, S10A-C). These genes were found to be methylated in 10% and 7% of primary tumours but 73% and 55% of BBM respectively. While MIR124-2 was methylated in 55% of the primary tumours and 88% of BBM (Fig. 2A–D, Supplementary Fig. 4A). This suggests that RP11-713P17.4 and NUS1P3 are frequently methylated in BBM but not in BP, and MIR124-2 methylation frequency is enriched BBM compared to primary breast tumours (Fig. 2A–D and Supplementary Fig. 4A–C, Supplementary Table 1C). The methylation status of genes validated by CoBRA that were not differentially methylated are presenterd in Supplementary Fig. 6.
Figure 2.
Representative examples of experimental validation of methylation status of candidate genes hypermethylated in breast to brain metastases (BBM) in a cohort of non-metastatic primary breast tumours (BP) (n = 20) and BBM (n = 30) using Combined Bisulphite and Restriction Analyses (CoBRA). (A) The two genes RP11-713P17.4 and NUS1P3 are infrequently methylated in BP and frequently methylated in BBM (In addition, MIR124-2 methylation is enriched in BBM compared to primary breast tumours). In contrast, (B) MIR3193, CTD-2023M8, and MTND6P are frequently methylated in BP and infrequently methylated (hypomethylated) in BBM. (C) An overview of the methylation status of genes that are hypermethylated or hypomethylated in BBM in all BBM and BP samples analysed by CoBRA (each box represents a tumour). (D) Summary of methylation status of genes in primary and metastatic brain tumours, *: Indicates statistical significant difference between primary and BBM tumours. BBM: Breast to Brain Metastases, BP: Primary breast tumours, U: Uncut/control sample, C: cut by restriction enzyme, , HyperM: hypermethylated in BBM, HypoM: Hypomethylated in BBM.
MIR3193, CTD-2023M8.1 and MTND6P4 are hypomethylated in brain metastases compared to primary breast tumours
Our methylation array analysis identified a panel of nine candidate genes that were hypometylated in BBM compared to BP and BN (see supplementary table 1B). As above, these genes were also validated using CoBRA in 30 BBM that included 23 BBM samples used in the methylation array and in an independent cohort of 20 primary breast tumours. This CoBRA analysis identifed three genes that were frequently methylated in BP and infrequently methylated in BBM tumours; CTD-2023M8.1, MIR3193 and MTND6P4 were methylated in 26%, 29% and 0% of BBM samples respectively (n = 30) (Fig. 2B–D, and Supplementary Fig. 4D–F, S11A-C) and were methylated in 63%, 67% and 47% non-metastatic BP respectively (n = 20) (Fig. 2B–D Supplementary Fig. 4D-F).
In summary, experimental validation of gene methylation status in BBM and primary breast tumours has identified six genes that are differentially methylated in BBM compared to BP (Fig. 2C,D). Of these, the promoter regions of three genes (MIR124-2, RP11-713P17.4, and NUS1P3) were hypermethylated in BBM and three genes (MIR3193, CTD-2023M8.1 and MTND6P4) were hypomethylated in BBM relative to the primary tumours (Fig. 2C,D). As was seen following the initial 450 K array analysis, the differences in the methylation status between BBM and BP for these genes analysed by CoBRA were statistically significant i.e. MIR124-2 (p = 0.03), RP11-713P17.4 (p = 0.0001), NUS1P3 (p = 0.004), MIR3193 (p = 0.0003), CTD-2023M8.1 (p = 0.01) and MTND6P4 (P = 0.0001) (Fig. 2D, Supplementary Table 1C).
Methylation status of candidate genes in BBM samples and their originating primary breast tumours
To determine if the common BBM-associated methylation events are also detectable in the primary tumours that the metastases are deriverd from, the methylation status of these genes was determined in BP and associated secondary BBM tumours from 11 individuals.
Primary tumour material was available in the form of FFPE sections. We could bisulphite convert, amplify and analyse the promoter regions for RP11-713P14.4, MIR3193, MTND6P4 and CTD-2023M8.1 in these corresponding primary tumours. However, we were unable to amplify some regions in all 11 samples and MIR124-2 and NUS1P3 promoter regions were refracrtive to amplification in all FFPE primary samples.
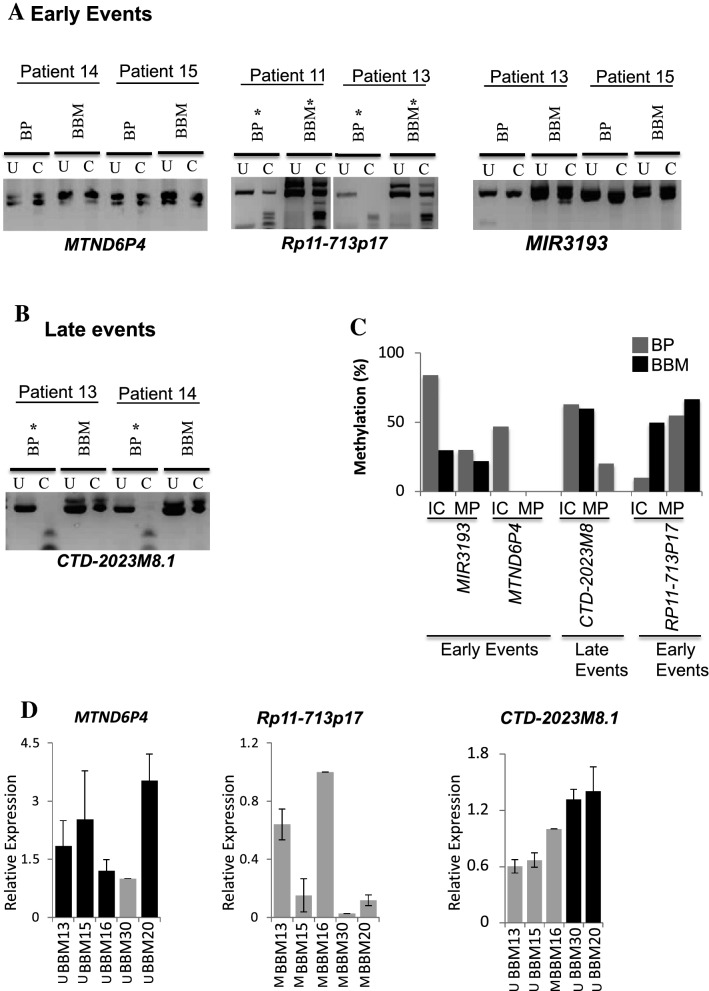
MTND6P4 and MIR3193 are frequrently methylated in primary breast tumours tumurs (9/19, 16/19; see Fig. 2). However, we found that these regions are commonly unmethylated in metastasis-originating primary tumours (0/5, 3/9) and their corresponding BBM tumours (Fig. 3A; left panel, 3C and Supplementary Fig. 5A,C). Similarly, RP11-713P17.4 is infrequently methylated in primary breast tumours (non-metasatic; 2/19 see, Fig. 2). However, in the primary tumours that proceed to metastasise to the brain this region is found to be methylated (3/6) (Fig. 3A; right panel, 3C, Supplementary Fig. 5B). These results suggest that the differential methylayion we observed between unrelated primary tumours an BBM may be a result of differences that occur early during the developmet of the tumours that metastasize as they are common to the originating primary tumour and the associated metastatic tumour. From hearin we will refere to these as early events (in metastatic tumour evolution).
Figure 3.
Methylation status of BBM-Hypermethylated and BBM-Hypomethylated genes in BBM and their originating BP tumours from individual patients. (A) Early events; Methylation of MTND6P4 (left panel), RP11-713P14.4 (middle panel) and MIR3193 (right panel) in originating BP is the same as found in their corresponding BBM in individual patients (B) Late events; Methylation status of CTD-2023M8.1 in orginating BP is different to their corresponding BBM tumours (loss of methylation at this region occurs in the metastasis). (C) Summary of methylation status of genes denoting early events and late events in individual patients/matched pairs (MP) and an independent cohort (IC) of primary breast tumours. (D) Expression analyses of genes by quantative reverse transcriptase PCR (qRT-PCR) in representative BBM tumours showing methylation-expression correlation, tumours with genes that are methylated (M) have a low level of expression. BP: Breast Primary tumour, BBM: Brain Metastases, U: Uncut/Control sample, C: cut by methylation specific restriction enzyme, *: Methylated samples, IC: independent cohort, MP: matched pairs from individual patients.
CTD-2023M8.1 is frequently methylated in primary tumours with no history of distant metastasis (12/19) but infrequently methylated in BBM (7/27) (see Fig. 2). CTD-2023M8.1 was also found to be methylated in metastasis-originating primary tumours However, CTD-2023M8.1 was not methylatyed in the corresponding BBM. This Suggests that this genomic change (the loss of methylation) was selected for after the metastasising tumour cells had left the primary tumour (Fig. 3B,C, Supplementary Fig. 5D). From hearin we will refere to this as a late event (in metastatic tumour evolution).
In addition, we carried out quantitative reverse transcription PCR (qRT PCR) to determine the expression status of these genes in the same cohort of BBM tumours that was used for 450 K methylation and experimental validation of methylation status (Fig. 3D, Supplementary Fig. 7). The expression was normalized against β-actin. As there was no RNA available from the primary tumors the fold change was determined relative to the expression level of a median ΔCT value for each transcript analysed. We have found that the genes that have promoter methylation have relatively low RNA levels (relative expression < 1) in those tumours. Similarly, the genes which are unmethylated, are expressed in those tumours (relative expression > 1). We have found that some of the samples/ genes that are not methylation have also low level expression which could be attributed to genomic changes other than the DNA methylation (Fig. 3D, Supplementary Fig. 7).
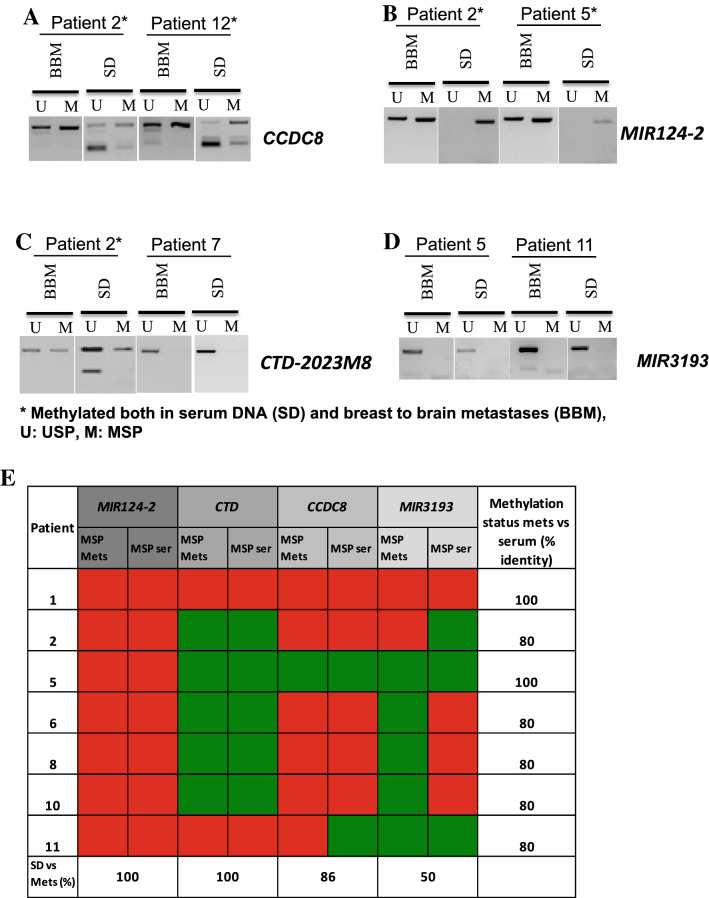
Examination of methylation status of candidate genes in tumour free circulating (tfc) DNA
In order to investigate if the methylation status of candidate genes in BBM is similar to their methylation status in circulating DNA isolated from patients’ serum (at the time of metastasis surgery), we determined the methylation status of MIR124-2, CTD-2023M8 and MIR3193 and CCDC8 in patients’ serum (CCDC8 had previously been identified as methylated in BBM in our earlier study19) by methylation-specific PCR (MSP). Serum was available from those patients whose originating primary tumours and BBM were also available. The methylation status of tfc DNA isolated from serum at MIR1242, CTD-2028M8, CCDC8 and MIR3193 was the same as that seen in BBM in 100%, 100%, 83% and 50% of the samples respectively (Fig. 4A–E, Supplementary Figs. 8, 12). It is important to note that while we belive this analysis a useful in showing that these epigenetic marks can be identified in tfc DNA, this analysis is limited as matched blood samples to the retrospectively collected primary breast tumours were not available.
Figure 4.
Methylation status of candidate genes in BBM, and tumour free circulationg DNA (tfcDNA) in patients’ serum determined by Methylation Specific PCR (MSP). (A,B) Methylation status of candidate genes denoting early events (CCDC8 and MIR124-2) and (C,D) late events (CTD-2023M8 and MIR3193) in BBM and their corresponding tumour free circulating DNA in individual patients (presence of a PCR product in the MSP lane is indicative of methylation in the region analysed). (E) Summary of methylation status of BBM and serum DNA in individual patients; Red suqares indicate methylation identified, Green squares indicate no methylation identified. BBM: Brain Metastases, SD: Serum DNA, U: Unmethylation Specific PCR; USP, M: Methylation Specific PCR; MSP, *: Methylated samples.
Expression status of MIR124-2 gene correlates to the clinical prognosis of patients
We wished to carry out survival analyses of breast cancer patients to investigate the correlation of expression of candidate genes with clinical prognosis using KM-plotter tools using data from the Gene Expression Omnibus (GEO)31. However, prognosis data was not available for mir3193 and NUS1P3 or the log-non-coding RNAs that we have identified. Data was avalalber for MIR124-2 and, in addition, we also performed KM analysis for three genese we have previously found to be frequently methylated in BBM (BNC1, CCDC8 and GALNT9)19.
Kaplan–Meier analyses showed that low expression of MIR124-2 (that we have shown here to be frequently hypermethyated in BBM) correlates to poorer clinical outcomes (Fig. 5). We have found that the high expression of MIR124-2 correlates to better relapse-free survival (RFS) of breast cancer patients (p = 0.00004 (Fig. 5A). Furthermore, we carried out combined prognosis of the four candidate genes (BNC1, CCDC8, GALNT9, MIR124-2), three of which (BNC1, CCDC8, GALNT9) were reported to have metastatic suppressive functions in our previous study19. This combined analysis shows that higher combined expression of these 4 genes together correlated to better RFS (P = 0.0046) (Fig. 5B). In addition, we carried out survival analyses of ER + and ER- patients separately to investigate if expression of our candidate genes is associated with ER receptor expression in breast cancer patients; the data indicates that the expression of MIR124 is independent of estrogen receptor (ER + or ER-) status in breast cancer patients (Fig. 5C).
Figure 5.
Loss of expression of BBM-hypermethylated genes (candidate metastasis suppressor genes) correlates with survival in breast cancer patients. (A) Kaplan–Meier analysis shows that low expression of MIR124-2 correlates to poor relapse free survival (RFS) of breast cancer patients (p = 0.00004). (B) Kaplan–Meier analysis using a combined four-metastasis suppressor gene signature (BNC1, CCDC8, GALNT9 and MIR124-2) shows that loss of expression of these genes in combination correlates to poor RFS of breast cancer patients (p = 0.0046). (C) Low expression of MIR124-2 is associated with poor prognosis of ER + and ER- breast cancer patients (p = 0.007 and 0.017 respectively).
Discussion
The aim of this study was to identify genes that are frequently epigeneticly dysregulated in breast to brain metastases (BBM) using a genome-wide approache. Our study identified 6 genes (RP11-713P17.4, MIR124-2, NUS1P3, MIR3193, MTND6P4 and CTD-2023M8.1) of which, RP11-713P17.4, MIR124-2 and NUS1P3 were frequently hypermethylated in BBM whereas MIR3193, MTND6P4 and CTD-2023M8.1 were frequently hypomethylated BBM compared to BP and BN. Notably, all of these genes identified were non-protein coding genes; two microRNAs (MIR3193 and MIR124-2), two long intergenic non-coding RNA (lincRNA) genes (RP11-713P17.4 and CTD-2023M8.1) and two pseudogenes (MTND6P4 and NUS1P3).
Recently dysregulated lncRNAs have been shown to be associated with various cancer types33,34, EMTs and metastases35. There is growing evidence that non-coding RNA could be a class of novel biomarkers or therapeutic targets in multiple cancers36,37 and prevous studies have reported that non-coding RNAs are epigenetically dysregulated in cancer38,39. This study has identified non-coding RNAs that are dysregulated by DNA methylation that, potentially, could be used as a DNA methylation prognostic markes in BBM.
The methylation level of non-protein coding genes MIR124-2, NUS1P3 and RP11-713P17.4 was enriched in BBM tumours compared to primary breast tumours. Previous studies have reported that MIR124 is associated with inhibition of invasion and metastases of breast and lung cancers40, oral squamous cell carcinoma41 and pancreatic adenocarcinoma42. MIR124 is epigenetically dysregulated in hepatocellular carcinoma (HCC) cell lines43 and its silencing is also associated with poor clinical prognosis of colorectal carcinoma44. Recently, the methylation status of MIR124-2 has been proposed as a prognostic marker associated with cervical cancer in human papoloma Virus (HPV) positive women45,46. MIR124-2 is one of three independent precursors genes; MIR124-1, MIR124-2, and MIR124-3, which are processed to form MIR124. Interestingly, the other precursors MIR124-1 and MIR124-3 are also methylated in BBM compared to BP and BN (Supplementary Fig. 3) suggesting that MIR124 is dysregulated in BBM. Recent studies have reported that MIR124-2 suppresses proliferation, aggressiveness, apoptosis, and invasion of osteosarcoma47,48. MIR124 is abundantly expressed in the nervous system where it contributes to regulation of alternative splicing and plays a crucial role in the differentiation of progenitor neuronal cells49,50. MIR124 also contributes to glial cells quiescence and is involved in repression of migration and invasion of various cancers through its targets49.
NUS1P3 is a processed pseudogene of its parental gene NUS1(Dehydrodolichyl Diphosphate Synthase Subunit). NUS1, also known as NOGO-B Receptor is expressed in most tissues44,51,52. NUS1 down regulates epithelial markers such as E-cadherin and increases mesenchymal markers contributing to EMT in cervical cancer promoting invasion and metastasis53. In addition, NUS1 dysregulation is associated with various cancer types54 including ER/PR/HER2 positive breast tumours55. There is growing evidence that pseudogenes are dysregulated in cancer and this dysregulation may modulate thire intereaction with either parental genes or other gene loci regulating transcriptional, and post transcriptional activities56,57. In our study, NUS1P3 is unmethylated in primary tumours suggesting its expression, which may lead to increased expression of NUS1. Furthermore, increased expression of NUS1 could promote Epithelial to Mesenchymal Transition (EMT) in breast cancer contributing to invasion and metastases to the brain. It is possible that NUS1P3 acts as a competitive endogenous RNA (ceRN)A for NUS1 and the silencing of NUS1P3 in metastases found in the brain lead to down-regulation of NUS1 concorant with Mesenchymal to Epithelial Transition (MET). These findings are consistent with our finding that silencing of MIR124-2 and NUS1P3 through promoter methylation in BBM samples may provide a selective advantage for metastasised tumours to survive and to proliferate in the brain microenvironment.
RP11-713P17.4 is a long intergenic non-coding RNA (lincRNA) gene. LincRNAs are gene-associated transcripts that are associated with open chromatin marks such as histone modification sites and epigenetic regulation of transcription, RNA stability, and recruitment of protein complexes58–60. LincRNA are associated with crucial biological functions such as cellular growth and differentiation, development, and apoptosis59,60. There is growing evidence of epigenetic reglations of lincRNAs in cancers61,62, and further mechanistic studies are required to investigate the molecular mechanistic role of RP11-713P17.4 in BBM and cancer metastases.
Three non-protein coding genes MIR3193, MTND6P4 and CTD-2023M8.1 are hypomethylated in BBM compared to primary tumours and normal breast tissues. The microRNA, MIR3193 was one of 209 novel micro RNAs identified by deep sequencing melanomas63. Furthermore, MIR3193 is frequently upregulated in glioma compared to normal brain tissue64.
MTND6P4 is a processed pseudogene of its parental gene MTND6 (ND6) that codes for the protein Mitochondrially Encoded NADH Dehydrogenase 6 (MTND6). MTND6 provides a quinone binding sites and is one of the six subunits (ND1-ND6) of the complex I in electron transport chain (ETS) in mitochondria. Mutations in MTND6 are associated with an increase in metastatic potential that was associated with low NADH and high reactive oxygen species (ROS) in lung and breast cancer cell lines65,66 and heptatocellular carcinoma67. Mutation in one of the subunits of ETS leads to low oxidative phosphorylation and increased glycolytic activity of mitochondria contributing to aggressiveness of childhood Acute Lymphoblastic Leukemia68. It is possible that epigenetic dysregulation of MTND6P4 may contribute to an energy shift towards glycolysis leading to acidosis with microenvionment changes that provide powerful growth advantages and invasive potential to the tumour cell69. A principle emerging role of pseudogenes is to act as competitive endogenous RNAs (ceRNA), a sponge for molecules that interact with mRNA (such as miRNA) thus positively influencing the expression of their parental gene70. MTND6P4 may regulate its parental gene MTND6 and hypomethylation and subsequent overexpression of MTND6P4 may lead to changes in oxidative phosphorylation activies, glycolysis and brain microenvironment contributing to growth advantages to tumours via upregulation of MTND6.
In our study, MIR3193, MTND6P4 and CTD-2023M8.1 were frequently methylated in a cohort of non-metastatic primary breast tumours and infrequently methylated in BBM. The two genes MIR3193 and MTND6P4 were commonly unmethylated in BBM and their originating primary breast tumours in individual patients suggesting that the demethylation/hypomethylation of MIR3193 and MTND6P4 is an early event during tumour evolution. It further suggests that MIR3193 and MNTND6P4 have metastatic promoter function, which are silenced in normal breast tissues and primary breast tumours due to methylation. Similarly, CTD-2023M8.1 is frequently methylated in non-metastatic primary breast tumours. It is also frequently methylated in metastasis-originating primary tumours. However, it is frequently unmethylated in BBM. This suggests that the hypomethylation of CTD-2023M8.1 in BBM is a late event that may occur only after the tumour cells have left the primary site.
RP11-713P17.4 is infrequently methylated in primary breast tumours and normal breast tissues but frequently methylated in BBM and their originating primary tumours in individual patients suggesting that the promoter hypermethylation of RP11-713P17.4 is an early event during BBM. The novel non-protein coding genes identified in this study may regulate invasion and metastasis directly or by regulating other protein coding genes. However, their regulatory functions and their target genes have not been reported before. Functional studies are required to determine the role of these genes in BBM.
In addition, our study shows that patients’ serum could be useful to detect the methylation status of tumour associated circulating DNA in BBM suggesting a potential method of prognostic analysis. For this, a panel of genes could possibly be developed as prognostic markers for BBM. However in this study, patients’ serum taken at the time of primary tumour diagniosis was not available to determine if the methylation status of these genes could be used as non-invasive prognostic markers.
Taken together, our study has identified a panel of six novel non-protein coding genes (miRNAs, pseudogenes and long intergenic/non-coding RNAs) of which RP11-713P17.4, NUS1P3, MIR3193, MTND6P4 and CTD-2023M8.1 have been reported for the first time as epigenetically dysregulated genes in cancer and in metastases. MIR124-2 has previously been reported as an epigenetically dysregulated gene in cancer43–46,71–74, however, its role in breast cancer metasetases has not been reported previously. The non-coding RNA genes75 are a part of broad epigenetic network that are emerging as critical regulators in human diseases and cancers76,77. The genes that we have identified may be of use in the development of prognostic biomarkers and as therapeutic targets. Larger-scale studies will be required to determine the usefulness of these findings to translate to the clinic.
Supplementary Information
Author contributions
R.P.P. designed and carried out the experiments, analyzed data and prepared manuscript, I.O. and V.B. carried out experiments, D.H. and L.Y. analysed data, T.P.D., K.M.A., C.D., A.R.B., M.D.J., I.B., F.L. collaborated the studies, provided experimental and study inputs, prepared clinical specimens, J.L.D. and T.J.W. collaborated the studies, provided experimental and study inputs, M.R.M. conceived and directed the studies, designed expeiments, and prepared manuscripts, all authors contributed in reviewing the manuscripts.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05050-z.
References
- 1.Chen W, et al. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018;2(1):4. doi: 10.1038/s41698-018-0047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler AF, et al. The biology of brain metastases-translation to new therapies. Nat. Rev. Clin. Oncol. 2011;8(6):344–356. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leone JP, Leone BA. Breast cancer brain metastases: the last frontier. Exp. Hematol. Oncol. 2015;4:33. doi: 10.1186/s40164-015-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 5.Paget S. The distribution of secondary growths in cancer of the breast: 1889. Cancer Metastasis. Rev. 1989;8(2):98–101. [PubMed] [Google Scholar]
- 6.Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlimok G, et al. Micrometastatic cancer cells in bone marrow: in vitro detection with anti-cytokeratin and in vivo labeling with anti-17-1A monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 1987;84(23):8672–8676. doi: 10.1073/pnas.84.23.8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer. 2014;14(9):611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leong SP, Tseng WW. Micrometastatic cancer cells in lymph nodes, bone marrow, and blood: Clinical significance and biologic implications. CA Cancer J. Clin. 2014;64(3):195–206. doi: 10.3322/caac.21217. [DOI] [PubMed] [Google Scholar]
- 10.Gamez-Pozo A, et al. Functional proteomics outlines the complexity of breast cancer molecular subtypes. Sci. Rep. 2017;7(1):10100. doi: 10.1038/s41598-017-10493-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark SE, et al. Molecular subtyping of DCIS: heterogeneity of breast cancer reflected in pre-invasive disease. Br. J. Cancer. 2011;104(1):120–127. doi: 10.1038/sj.bjc.6606021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartorius CA, et al. Estrogen promotes the brain metastatic colonization of triple negative breast cancer cells via an astrocyte-mediated paracrine mechanism. Oncogene. 2016;35(22):2881–2892. doi: 10.1038/onc.2015.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palma G, et al. Triple negative breast cancer: looking for the missing link between biology and treatments. Oncotarget. 2015;6(29):26560–26574. doi: 10.18632/oncotarget.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae SY, et al. Poor prognosis of single hormone receptor- positive breast cancer: similar outcome as triple-negative breast cancer. BMC Cancer. 2015;15:138. doi: 10.1186/s12885-015-1121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaedcke J, et al. Predominance of the basal type and HER-2/neu type in brain metastasis from breast cancer. Mod. Pathol. 2007;20(8):864–870. doi: 10.1038/modpathol.3800830. [DOI] [PubMed] [Google Scholar]
- 16.Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat. Rev. Cancer. 2011;11(5):352–363. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XH, et al. Metastasis dormancy in estrogen receptor-positive breast cancer. Clin. Cancer Res. 2013;19(23):6389–6397. doi: 10.1158/1078-0432.CCR-13-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigelt, B., J.L. Peterse, and L.J. van 't Veer, Breast cancer metastasis: markers and models. Nat Rev Cancer, 2005. 5(8): 591–602. [DOI] [PubMed]
- 19.Pangeni RP, et al. The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain. Clin. Epigenet. 2015;7:57. doi: 10.1186/s13148-015-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R core team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2018.
- 21.Assenov Y, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014;11(11):1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pangeni RP, et al. Genome-wide methylomic and transcriptomic analyses identify subtype-specific epigenetic signatures commonly dysregulated in glioma stem cells and glioblastoma. Epigenetics. 2018;13(4):432–448. doi: 10.1080/15592294.2018.1469892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris MR, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29(14):2104–2117. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricketts CJ, et al. Genome-wide CpG island methylation analysis implicates novel genes in the pathogenesis of renal cell carcinoma. Epigenetics. 2012;7(3):278–290. doi: 10.4161/epi.7.3.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pangeni RP, et al. G9a regulates tumorigenicity and stemness through genome-wide DNA methylation reprogramming in non-small cell lung cancer. Clin. Epigenet. 2020;12(1):88. doi: 10.1186/s13148-020-00879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark SJ, et al. High sensitivity mapping of methylated cytosines. Nucl. Acids Res. 1994;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang T, et al. MST4 Phosphorylation of ATG4B Regulates Autophagic Activity, Tumorigenicity, and Radioresistance in Glioblastoma. Cancer Cell. 2017;32(6):840–855. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sastry NG, et al. LY6K promotes glioblastoma tumorigenicity via CAV-1-mediated ERK1/2 signaling enhancement. Neuro Oncol. 2020;22(9):1315–1326. doi: 10.1093/neuonc/noaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucl. Acids Res. 2007;35:760. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong Z, Laird PW. COBRA: A sensitive and quantitative DNA methylation assay. Nucl. Acids Res. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan X, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S.J., et al., Long noncoding RNAs in cancer metastasis. Nat Rev Cancer, 2021. [DOI] [PMC free article] [PubMed]
- 36.Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-Derived Small RNAs in Cancer: New Potential Biomarkers and Target for Therapy. Adv Cancer Res. 2017;135:173–187. doi: 10.1016/bs.acr.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179(5):1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, et al. Non-coding RNAs as mediators of epigenetic changes in malignancies. Cancers (Basel) 2020;12(12):1. doi: 10.3390/cancers12123657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, et al. The landscape of DNA methylation-mediated regulation of long non-coding RNAs in breast cancer. Oncotarget. 2017;8(31):51134–51150. doi: 10.18632/oncotarget.17705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han ZB, et al. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151. Cell Physiol. Biochem. 2013;31(6):823–832. doi: 10.1159/000350100. [DOI] [PubMed] [Google Scholar]
- 41.Hunt S, et al. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 2011;585(1):187–192. doi: 10.1016/j.febslet.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 43.Furuta M, et al. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, et al. miR-124 and miR-506 inhibit colorectal cancer progression by targeting DNMT3B and DNMT1. Oncotarget. 2015;6(35):38139–38150. doi: 10.18632/oncotarget.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Strooper LMA, et al. Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol. Oncol. 2016;141(2):341–347. doi: 10.1016/j.ygyno.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zummeren MV, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124–2 genes in cervical cancer and precursor lesions. Mod. Pathol. 2018;1:1. doi: 10.1038/s41379-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 47.Yu B, Jiang K, Zhang J. MicroRNA-124 suppresses growth and aggressiveness of osteosarcoma and inhibits TGF-beta-mediated AKT/GSK-3beta/SNAIL-1 signaling. Mol. Med. Rep. 2018;17(5):6736–6744. doi: 10.3892/mmr.2018.8637. [DOI] [PubMed] [Google Scholar]
- 48.Meng Q, et al. The effects of TRAF6 on proliferation, apoptosis and invasion in osteosarcoma are regulated by miR-124. Int. J. Mol. Med. 2018;41(5):2968–2976. doi: 10.3892/ijmm.2018.3458. [DOI] [PubMed] [Google Scholar]
- 49.Makeyev EV, et al. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deo M, et al. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006;235(9):2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 51.Schwab ME. Functions of Nogo proteins and their receptors in the nervous system. Nat. Rev. Neurosci. 2010;11(12):799–811. doi: 10.1038/nrn2936. [DOI] [PubMed] [Google Scholar]
- 52.Huber ABW, Brosamle C, Oertle T, Schwab ME. Expression Pattern of Nogo. J. Neurosci. 2002;22(9):3553–3567. doi: 10.1523/JNEUROSCI.22-09-03553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao W, et al. Nogo-B promotes the epithelial-mesenchymal transition in HeLa cervical cancer cells via Fibulin-5. Oncol. Rep. 2013;29(1):109–116. doi: 10.3892/or.2012.2069. [DOI] [PubMed] [Google Scholar]
- 54.Oertle T, Merkler D, Schwab ME. Do cancer cells die because of Nogo-B? Oncogene. 2003;22(9):1390–1399. doi: 10.1038/sj.onc.1206278. [DOI] [PubMed] [Google Scholar]
- 55.Wang B, et al. Expression of NgBR is highly associated with estrogen receptor alpha and survivin in breast cancer. PLoS ONE. 2013;8(11):e78083. doi: 10.1371/journal.pone.0078083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao-Jie L, et al. Pseudogene in cancer: real functions and promising signature. J. Med. Genet. 2015;52(1):17–24. doi: 10.1136/jmedgenet-2014-102785. [DOI] [PubMed] [Google Scholar]
- 58.O'Leary, V.B., et al., PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. (2015). [DOI] [PubMed]
- 59.Marchese FP, Huarte M. Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics. 2014;9(1):21–26. doi: 10.4161/epi.27472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics. 2014;9(1):13–20. doi: 10.4161/epi.26700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 62.Ali MM, et al. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat. Commun. 2018;9(1):883. doi: 10.1038/s41467-018-03265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark MS, et al. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS ONE. 2010;5(3):9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu DM, et al. MircoRNA-1275 promotes proliferation, invasion and migration of glioma cells via SERPINE1. J. Cell Mol. Med. 2018;1:1. doi: 10.1111/jcmm.13760. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320(661):1. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 66.Yuan Y, et al. Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer. 2015;15:346. doi: 10.1186/s12885-015-1349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, et al. Heteroplasmy and Copy Number Variations of Mitochondria in 88 Hepatocellular Carcinoma Individuals. J. Cancer. 2017;8(19):4011–4017. doi: 10.7150/jca.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jarviaho T, et al. Novel non-neutral mitochondrial DNA mutations found in childhood acute lymphoblastic leukemia. Clin. Genet. 2018;93(2):275–285. doi: 10.1111/cge.13100. [DOI] [PubMed] [Google Scholar]
- 69.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 70.An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell Mol. Med. 2017;21(1):185–192. doi: 10.1111/jcmm.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia X, et al. MicroRNA-124: An emerging therapeutic target in cancer. Cancer Med. 2019;8(12):5638–5650. doi: 10.1002/cam4.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panagopoulou M, et al. Deciphering the methylation landscape in breast cancer: Diagnostic and prognostic biosignatures through automated machine learning. Cancers (Basel) 2021;13(7):1. doi: 10.3390/cancers13071677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oltra SS, et al. Methylation deregulation of miRNA promoters identifies miR124-2 as a survival biomarker in Breast Cancer in very young women. Sci. Rep. 2018;8(1):14373. doi: 10.1038/s41598-018-32393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, et al. Molecular and epigenetic profiles of BRCA1-like hormone-receptor-positive breast tumors identified with development and application of a copy-number-based classifier. Breast Cancer Res. 2019;21(1):14. doi: 10.1186/s13058-018-1090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 76.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 77.Diaz-Lagares A, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA. 2016;113(47):E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.