Abstract
Microbiome engineering is increasingly being employed as a solution to challenges in health, agriculture, and climate. Often manipulation involves inoculation of new microbes designed to improve function into a preexisting microbial community. Despite, increased efforts in microbiome engineering inoculants frequently fail to establish and/or confer long-lasting modifications on ecosystem function. We posit that one underlying cause of these shortfalls is the failure to consider barriers to organism establishment. This is a key challenge and focus of macroecology research, specifically invasion biology and restoration ecology. We adopt a framework from invasion biology that summarizes establishment barriers in three categories: (1) propagule pressure, (2) environmental filtering, and (3) biotic interactions factors. We suggest that biotic interactions is the most neglected factor in microbiome engineering research, and we recommend a number of actions to accelerate engineering solutions.
Subject terms: Community ecology, Microbial ecology
Microbiome engineering is a rapidly evolving frontier for solutions to improve human health, agricultural productivity, and climate management. Microbiome engineering seeks to improve the function of an ecosystem by manipulating the composition of microbes. Two major challenges for successful microbiome engineering are (1) the design of a microbiome with improved function and (2) the establishment of an improved microbiome in a recipient system of interest. While multiple articles and reviews have addressed functional design [1–3], microbiome establishment has received less attention. Here, we propose a strategy to improve microbiome engineering by focusing on microbial establishment and leveraging insights from macrobial ecology.
Two general engineering strategies are to manipulate indigenous microbes [4] or to introduce new members [5]. The latter involves the design and delivery of inoculants (a.k.a., probiotics in medical and agricultural arenas) and is a rapidly growing biotechnology sector. In their most general form, both strategies have been practiced crudely for thousands of years in human health [6] and agriculture [7]. However, despite current technical advances, inoculants frequently still fail to establish or confer long-lasting (months to years) modifications to ecosystem function [8]. We argue that this repeated failure is in part driven by lack of emphasis on establishment of inoculants.
The problem of organism establishment in recipient ecosystems is not unique to microbiome engineering; it has roots in macrobiology, particularly invasion biology and restoration ecology. We propose that adopting a cross-disciplinary conceptual framework to identify barriers to organism establishment, and then prioritizing these barriers through targeted research will accelerate successful microbiome engineering. In addition, recognizing differences in terminology and experimental design within and across disciplines will facilitate research integration across diverse ecosystems and scales. The components of a more holistic strategy are discussed below.
Conceptual framework of barriers to organism establishment
Pinpointing and overcoming barriers to inocula establishment are important research priorities for successful microbiome engineering. Barriers to establishment have been studied extensively in both invasion biology and restoration ecology. Invasion biology aims to understand the mechanisms that promote or deter invasive species establishment [9]. Restoration ecology aims to restore beneficial plants, animals, insects, and/or microbial communities in ecosystems where they have previously been displaced or depleted [10]. Over time, these fields have produced numerous theoretical frameworks to explain successful species establishment [9, 11]. The overlapping foundational theory in these fields presents an opportunity for cross-disciplinary synergy with microbiome engineering. Although the goals of microbiome engineering and restoration/invasion ecology are often different (e.g., establishment of a new microorganism, consortium, and/or set of functional properties in microbiome engineering vs. prevention of establishment and/or removal of invading species and preservation of native biota in restoration/invasion ecology), the value in a cross-disciplinary approach to microbiome engineering that considers restoration/invasion ecology is in identifying emergent properties of the ecosystem that can facilitate or inhibit establishment. Furthermore, microbial ecology can advance foundational theory by overcoming common constraints on the scope and pace of macrobial ecology research, such as the relatively slow growth rates of macroscopic organisms and the difficulty of performing multi-species manipulations. Although microbial studies of factors influencing establishment are still relatively rare, foundational studies are emerging [12].
A useful theoretical framework for species establishment in microbiome engineering would identify the abiotic and biotic levers that engineers could use for successful community manipulation. Frameworks that vary in granularity have been developed within invasion biology. At one end of the spectrum, one synthesis outlined 33 mechanistic hypotheses for establishment success [13]. At the other end of the spectrum, a widely accepted framework posits that invasion success is mainly determined by three factors: (1) propagule pressure, (2) environmental filtering, and (3) biotic interactions [14]. Propagule pressure describes dispersal potential, which determines the spread of organisms to novel areas, either through natural- or human-mediated movement. Environmental filtering describes the compatibility of an organism with a new environment (e.g., suitable temperature or moisture range). Biotic interactions encompass a range of interchanges that can occur between introduced organisms and residents. Using this three-factor framework from invasion biology, in the next section we illustrate some complexity underlying these factors, illustrating the need for strategic prioritization as a first step to guide microbiome engineering (Table 1, Table S1).
Table 1.
Overview of factors impeding organism establishment, potential engineering solutions. A comprehensive source of examples of studies which illustrate barriers to establishment and solutions is provided in Table S1.
| Ecological principle | Factor impeding establishment | Potential engineering goal |
|---|---|---|
| Propagule pressure | ||
| • Dose |
a. Stochastic extinction b. Density-dependent competitiveness |
a. Add a higher dose of the inoculant b. Add a lower dose of the inoculant |
| • Frequency |
a. Succession makes niche ephemeral b. Biotic disturbance (i.e., inoculation) is needed to open a niche |
a. Add inoculant more frequently b. Add inoculant at certain timepoint(s) |
| • Delivery mode | a. Inoculants do not reach or do not stay in intended location |
a. Alter delivery mode b. Increase dose |
| Environmental filtering | ||
| • Disturbance | High-turnover of organisms (low residence time) |
a. Persistent delivery of inoculant is necessary b. Create a protected physical space c. Inoculant with characteristics resistant to disturbance |
| • Niche Breadth | Inoculated organism requires a specific resource that is absent |
a. Engineer inoculant that has a larger niche breadth b. Pre-adapt inoculant to available environment c. Add resource specific to inoculant (‘pre-biotic’) |
| Biotic interactions | ||
| • Antagonism via Competition | Direct competition exists between resident organisms and inoculants |
a. Remove/disturb resident microbes b. Increase “competitive” trait of inoculation (e.g., antibiotic production, biofilm formation) c. Both a & b d. Add resources to support the inoculant during establishment period or beyond e. Pre-adapt inoculant to available environment f. Create a protected space |
| • Antagonism via Antibiotics | Antibiotic-producing residents debilitate the inoculant |
a. Make inoculant resistant b. Disrupt resident(s) (reduce antibiotic(s) production) c. Create a protected space |
| • Antagonism via Predation | Predation by resident microbes |
a. Make the inoculant resistant b. Remove predators prior to inoculation c. Create a protected space |
| • Facilitation | Inoculant requires ‘services’ provided by another organism which is not present | a. Add in an additional organism serving as a ‘keystone’ species to modify interactions of the target inoculant and other organisms and/or modify the environment |
Prioritizing barriers to establishment
Potential barrier: propagule pressure
Dose and frequency
Propagule pressure (PP) is a measure of dispersal used to describe the magnitude (dose) and pattern of the arrival (frequency) of invasive individuals. PP is one of the most commonly tested factors in macroorganism invasion biology and is often linked to invasion success [15]. It also plays a role in restoration ecology [11]. For example, increased seeding rates can aid restoration of native plant communities (e.g., [16]), but is not always effective [17]. Increased PP of a single or a few microbial invaders may also increase establishment success [18]. However, independent modeling and experimental work suggest that PP in multi-species microbial invasions has restricted or minimal impacts on establishment and community functioning [19, 20].
There are a number of mechanisms by which PP in theory could influence establishment. Increasing PP can overcome the effects of ecological drift—random births and deaths that change the relative abundances of species in a community over time—also known as demographic stochasticity, which generally increases for smaller populations such as newly introduced species [21, 22]. Increasing PP can also mitigate the impacts of environmental stochasticity, unpredictable spatiotemporal fluctuations in environmental conditions [21]. A higher dose may increase the likelihood that sufficient inoculum reaches the desired establishment location [23] or may impact density-dependent competitiveness by affecting quorum sensing behaviors either positively or negatively [24]. The temporal frequency of dose events is an alternative route to enhance dosage while also addressing uncertain timing of niche access.
Delivery mode
Delivery mode may influence establishment by affecting dispersal range or dose viability. If the delivery mode is insufficient, inoculants may not reach the intended location of establishment or may be debilitated when they arrive. Parallels in plant invasion biology and restoration ecology include natural seed coats that enhance plant dispersal range by animal vectors [25] or artificial seed coatings that prevent desiccation or delay germination [26]. Delivery modes in microbiome engineering include direct addition of inoculants as free cells in liquids, lyophilized cells on solids (e.g., on animal feed or on seed surfaces), or protected cells (e.g., within seeds or gel beads) [27]. A delivery mode may be chosen to enhance the probability of microbial inoculant establishment based on environmental conditions and/or the traits of an inoculant [8].
Potential barrier: environmental filtering
Environmental filtering (EF) refers to the selection of organisms that are compatible with the existing environment or are able to rapidly modify local conditions to fit requirements. Incompatible immigrants become extinct. EF can limit invasive plant establishment (e.g., [28]) and often explains restoration failures (e.g., [11]). Invasion success is expected to increase if invaded environments match those of a species optimum range [29]. In restoration, environmental filters are commonly manipulated to facilitate establishment of target restoration species [30] and/or prevent invader establishment [31]. An example of this phenomenon among microbes is the role of host specificity in microbial invasion success [32]. In both macro and microbiology, the most broadly successful invaders tend to have larger habitat ranges than non-successful invaders [33], which typically corresponds to tolerance of diverse conditions. EF conventionally includes abiotic (physical, chemical) and biotic factors (e.g., interactions with resident species). However, for engineering purposes it is useful to separate biological interactions (BI) from other modes of EF when considering barriers to establishment and potential solutions, because BI are generally harder to control for than abiotic factors. When EF is a barrier to microbiome engineering, either the expected longevity of engineering or the environmental conditions must be adjusted. Below, we summarize two aspects of EF to consider for microbiome engineering: niche availability and disturbance.
Niche availability
A lack of niche space for an inoculant is an obvious barrier to establishment. Understanding the temporal and spatial distributions of niche availability may be key to tailoring inoculant establishment [34]. Identification of a target niche space for inoculants through in situ strain characterization and subsequent isolation has led to success in a number of bioaugmentation efforts [35] and more recently, high-throughput phenotyping assays have been used [36]. If the target niche for an inoculant exists but is already filled, displacing the resident competitors may be required (e.g., [37]; see Biotic Interactions).
Alternatively, a niche can be created. Niche availability can be manipulated in some cases by altering the abundance of a single or multiple resource(s), as is done with prebiotics [4]. In host-associated communities, the host can provide a substrate to recruit health-promoting taxa, as illustrated by oligosaccharides in human breast milk that foster the growth of Bifidobacterium infantis strains in infant guts [38]. Similarly, there is increasing evidence that plant roots secrete metabolites to shape rhizosphere microbial composition [39]. Prebiotics can also be used to create novel niches for exotic inoculants, resulting in a combined pre- and probiotic known as a synbiotic. For example, porphyran—a marine polysaccharide—was used to establish an exogenous Bacteriodetes strain in mice guts [40] and xenobiotic compounds have been applied to support microbes of interest in industrial fermentations [41]. Continual delivery of an exotic resource can enable long-term stability of a “specialist” organism [42]. Conversely, short-term delivery of an exogenous resource may aid ‘generalist’ microbial inoculants that have a wider niche breadth but need a resource supplement during the transitional establishment period [43]. An alternative to manipulating niche availability is to exploit microbial inoculants that modify the environment, constructing their own niche [44] or a niche for other inoculants ([45], see Biotic Interactions).
Disturbance
Disturbance, here defined as perturbation of physical (e.g., structure, flow rates, temperature) or chemical (e.g., nutrients, pH, oxygen) properties of the environment, has the potential to increase species turnover. Consequently, disturbance is either a useful tool to displace unwanted residents [46]; see Biotic Interactions) or a barrier that impedes inoculant establishment. In highly disturbed environments, long-term establishment may be an unrealistic goal and persistent delivery of inoculants may be unavoidable. For example, in phyllospheres, soils, or rivers, ‘washouts’ of microbes may occur from heavy or frequent precipitation events or other physical disruptions [47]. Periodic chemical disturbances, such bile secretion in the gut [48] or application of pesticides in soils [49] can also impact inoculant establishment. Providing inoculants with an enduring physical haven that allows continuous dispersal into surrounding space may overcome disturbance barriers, as seen in some macrobial and microbial systems [16, 50]. This strategy is enhanced when microbial inoculants possess superior abilities for attachment [51], biofilm formation [52], or stress resistance [53].
Potential barrier: biotic interactions
The variety of BI is complex but can be broadly categorized as either antagonistic or facilitative [54]. Within these categories, interactions can be described as direct or indirect in relation to an organism of interest. For example, antagonistic direct interactions include mechanisms such as predation or competition, while antagonistic indirect interactions include environmental modifications like antibiotic production or pH changes may adversely affect competitors and non-competitors alike. In macrobial ecology, the impacts of direct effects have been more commonly considered e.g., [55], but research on indirect effects is increasing e.g., [56]. There is also growing recognition that the number or complexity of interactions may affect establishment in macrobial [57] and microbial systems [58]. In simple systems with cultured microbes, antagonistic interactions are abundant and are a driving force in community assembly and stability [59]. Antagonism also plays a role in functioning of more complex microbial systems [60] and may be used in microbiome engineering [61]. Like antagonism, facilitative interactions can also be direct or indirect. For example, the presence of a keystone species that provides a resource supporting an inoculant is direct facilitation, whereas a general modification of the environment (e.g., a change in soil pH) that benefits an inoculant is indirect facilitation. The distinction between direct and indirect BI is important for microbiome engineering because preoccupation with direct interactions can overlook important indirect barriers and solutions.
Antagonism via simple competition
The complexity of competition can be categorized by the mechanisms organisms use to capture growth limiting resources. In the simple case, competition involves differences in search capability (i.e., motility) and/or resource capture efficiency (e.g., nutrient uptake via transporters). Increasing the number of competitors for a growth-limiting substrate may reduce the ability of an inoculant to capture a sufficient quota of resource to survive [62]. A general trend observed in multiple ecosystems is that increased resident community diversity generally reduces invasibility because diverse communities leave less free niche space [63, 64]. For microbiome engineering solutions to overcome a competition barrier might include creating a protected physical space or adding resources to support the inoculant during an establishment period [58, 65].
Antagonism via antibiotics
Complex competition involves additional strategies to undermine competitors. Strategies include secretion of antibiotics, signaling compounds that adversely affect the metabolism of other species (e.g., quorum signals or volatile organic compounds), or siderophores that create new growth limitations for competitors. Antibiotic production is the best studied strategy. In pairwise interactions of cultured isolates, antagonistic antibiotic production occurred in approximately half of Bacillus [66] and Streptomyces [67] isolates. Antagonistic growth inhibition tends to increase with phylogenetic similarity of species [68], presumably because competition is greatest among close relatives with similar traits, a concept shared across microbial and macrobial biology. The impact of antibiotics in natural communities is illustrated by the production of andrimid by marine bacteria, which inhibits Vibrio cholerae growth [69]. The extent to which antibiotic production is a barrier to establishment of inoculants in microbiome engineering remains to be established, but has been shown in at least one instance [70] and might be routinely assayed by inhibition assays with filtrates from a resident community. Active antagonism may also be an engineering tool for establishment, illustrated by an artificial biocontrol strain used to remove unwanted bacteria in a community [71].
Antagonism via predation
Another type of antagonistic interaction that may influence the ability of inoculants to establish is predation. Predation by protists, bacteria, and fungi is likely to be density dependent, not taxon-specific, although cases of the latter have been documented [61]. Predation by host-specific viruses may also impact invader success. Phages vary in host range but generally display host specificity. While phage are known to impact the diversity and function of microbial communities [72], they are still rarely considered when assessing controls on microbiome succession [73]. Phages may stabilize the co-existence of competing bacteria by preventing dominance of a single species—an example of the “kill-the-winner” hypothesis [74]. In microbiome engineering, establishing inoculants with “naïve immunity” may fail owing to lysis by phages in the resident community [75]. Conversely, predation can be a tool for tailored removal of residents that impede establishment of desired strains, by building on concepts from work using phages for biocontrol of pathogens in humans, on foods, in aquaculture, and on plants (e.g., [76]).
Facilitation
Stable facilitative interactions between microorganisms are only expected in restricted cases [77]. For example, environmental conditions can mediate a tradeoff between facilitation and competition, where harsher environments foster microbial facilitation [78]. Another view posits that microbial communities are able to organize into metabolically cohesive units where consortia with positive feedback loops use resources in a stable manner and minimize competition through resource specialization and exclusion of resource generalists [79]. For microbiome engineering, facilitative interactions could be leveraged, for example by creating highly specialized microbial units where organisms are filling different niches.
Summary
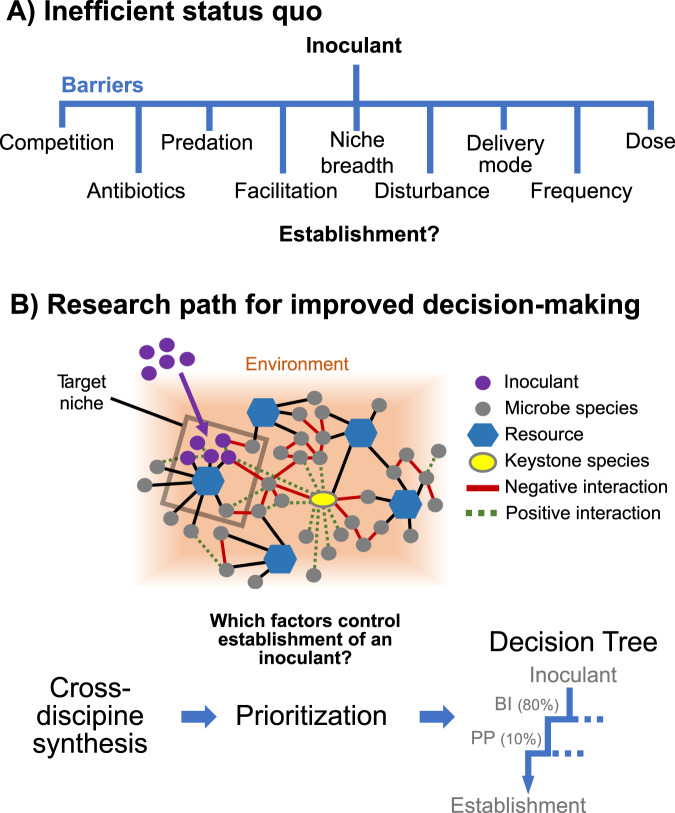
The framework of PP, EF, and BI is a simple way of categorizing a long list of ecological phenomena that can be barriers (or engineering tools) for establishment of inocula in resident communities (Table 1, Table S1). The overview above highlights a few of the many sub-factors that have been documented in macrobial ecology [13]. Given the large number and context-dependence of factors that may impede organism establishment, ranking barriers at the highest level (PP, EF, and BI) in a decision tree framework is a critical first step to accelerate microbiome engineering and avoid random testing of factors (Fig. 1).
Fig. 1. Barriers to organism establishment and a path forward for microbiome engineering.
A The current state of research focused on inoculum establishment. Without quantitative prioritization, potential barriers like the illustrated examples are investigated randomly. B Path forward to improve establishment success. Prioritization of barriers includes quantifying their relative impact in order to create decision trees that can simultaneously rank barriers and summarize the potential impact of overcoming each barrier in a multi-factor solution path. Values in parentheses on the decision tree illustrate an example of percentage impact of a barrier on inoculum establishment, guiding microbiome engineering investments.
Decision trees are increasingly being used in macrobial ecology to manage species invasions and ecological restoration. For example, some decision trees prioritize actions based on the potential severity of invasive species on ecosystem services [80]. In a restoration ecology examples, decision trees based on seed dormancy attributes [81] or optimal habitat characteristics for bird nesting [82] can guide restoration plans. Many decision trees available to managers in macrobial ecology include a decision-making step related to establishment (e.g., is there adequate time for a seedling to establish during its typical growing season?).
Building decision trees in microbiome engineering requires studies that test the relative importance of factors on establishment success. For example, a recent study found that the relative importance of PP and BI varied by organism domain, bacteria versus fungi [19]. Another study found that the impacts of PP depended on the competitive interactions of residents and inoculants [18]. Findings from many studies like these must be averaged to elucidate general rules for decision trees, but such a pursuit is inefficient without planning for quantitative synthesis.
Breaking barriers: synthesis and recommendations
Overall, we posit that increasing focus on BI will lead to increased success in inoculant establishment and improve outcomes in microbiome engineering (Fig. 1B). While BI are commonly considered in invasion biology and restoration ecology, manipulating BI is an under-explored mechanism in microbiome engineering, likely due to the abundance and complexity of organism interactions in any given microbiome. Greater success in manipulating early succession ecosystems (e.g., infant compared to adult guts) [38, 65] may be due in part to their lower complexity, which would point to the importance of BI. Assessing the number and type of BI relevant for engineering complex microbiomes is challenging because of the lack of direct measurement capabilities. Network reconstruction techniques based on correlated species abundances are often used to infer interactions (e.g., [83]), but further development is needed to move beyond speculative inferences [84]. Manipulation of microbial keystone species deserves attention as a potential solution in microbiome engineering as this concept has proven to be a powerful strategy in restoration ecology [85]. Although keystone microbial taxa have been predicted in co-occurrence networks [86] and linked to compositional shifts [87], few studies have confirmed the physiological role of keystone microbes in a community [88].
In order to confirm the role of BI in microbiome engineering outcomes and refine a decision tree of barriers to organism establishment we recommend a number of actions. First, greater attention is needed in microbial research to measure establishment (persistence and/or proliferation) of inocula over long timescales (e.g., months to years) with whole-community measurement techniques that offer broader insight and more standardized experimental design and reporting to facilitate meta-analyses. Second, a careful cross-discipline synthesis—i.e., meta-analyses of microbial studies that measure the dependence of inocula establishment on aspects of BI, as well as PP and EF, that would illuminate knowledge gaps is needed. We expand on these concepts in the sections below.
Recommendations for future research
To accelerate microbiome engineering across different applications, there is a need to target knowledge gaps that are contingent on experimental design. Gaps range from complexity of the inoculum to multifactorial manipulation strategies. We highlight four gaps. (1) To date, most microbial establishment studies have one or a few inoculant species. In contrast, transplant of entire microbiomes (i.e., fecal transplants and activated sludge transplants) is an increasing practice for some medical and wastewater treatment applications, respectively, yet knowledge of how these complex invasions impact community composition and functioning is limited. (2) Often microbiome engineering studies measure functional changes, not inoculant establishment. Monitoring inoculants is needed to determine if failure to achieve desired functional changes is due to lack of establishment or instead, to attenuation of desired functions in established inoculants. (3) More insight into the temporal dynamics of functional changes is needed. It is common practice to assess function at only one or two timepoints. Furthermore, the time interval between manipulation and functional assessment varies from hours to weeks. In many studies functional measurements occur immediately following probiotic intervention and longer-term impacts are not assessed, limiting insights into barriers to establishment. (4) It is likely that multiple factors simultaneously contribute to the failure or success of species establishment (e.g., see [18]). Thus, multifactorial experiments that test the interactive effects of establishment barriers (or engineering solutions) are needed and may be especially helpful in building decision trees.
Translating among fields
Prioritizing establishment barriers through cross-discipline synthesis depends on the capacity to find related knowledge that is obscured by discipline-specific jargon. For example, medical research on microbiome engineering typically uses the term “probiotics” [89], while agricultural studies use the terms “microbial inoculants”, “plant growth promoting microbes” (PGPM’s), “biocontrol agents”, plant “biostimulants” or “probiotics” [90]. To facilitate cross-discipline synthesis, we compiled some key concepts, terms, and definitions (Table S2)—a step towards making related knowledge findable, accessible, interoperable, and reusable (FAIR), as recommended by the National Microbiome Data Collaborative (NMDC; [91]).
Experimental design
Some standardization of experimental design features (e.g., independent and dependent variables, replication, testing scale, and temporal sampling) can also facilitate synthesis. At present, wide variation in experimental design [92] impedes this goal. For example, functional changes in the gut microbiome might be assessed in some studies qualitatively from statements of symptom relief among patients [93], whereas other studies may quantify changes in the concentration of a specific analyte [94]. Microbial community composition is often reported in a qualitative way; whereas variance partitioning would facilitate quantification of probiotic impacts among studies. Furthermore, use of positive controls (mock communities) and/or internal standards in amplicon sequencing for taxonomic profiling can improve the quantitative insights of this common and relatively low-cost measurement technique [95]. Including a link in publications to an easily accessible data table summarizing experimental design can also facilitate data syntheses within and across disciplines. A database of searchable studies that include the experimental design and results with permanent identification records and rich contextual metadata (somewhat analogous to clinicalstudies.info.nih.gov/) would greatly accelerate progress. Publicly accessible genomic repositories are a prime example of how data aggregation and standardized formatting across disparate studies is leading to groundbreaking discoveries [96]. Data attribution guidelines that give researchers credit for datasets used in subsequent work [96, 97] is another useful step.
Conclusions
Microbiome engineering is a rapidly expanding field. There are notable cases of success in microbiome engineering for human health, in particular in the infant gut [38, 65], bioremediation [98], wastewater engineering [99], and agriculture [100]. However, inoculants often fail to establish or to modify ecosystem functioning over significant time periods [8]. Dispersal, environmental, and biotic barriers to organism establishment likely contribute to failures. Given the complex suite of possible barriers, developing a decision tree to prioritize barriers is a top priority to guide engineering. This priority may be aided by cross-disciplinary synthesis because disparate fields are tackling similar challenges. However, to leverage research and unite findings across fields there is a need to recognize differences in terminology and to standardize reporting of tested factors and magnitude of effects. Lastly, increased attention to the types of experiments performed and extended time-course measurements of inoculant establishment will provide insights that accelerate successful microbiome engineering across a range of applications.
Supplementary information
Acknowledgements
This work was supported by a Los Alamos National Laboratory Postdoc Fellowship 20180746PRD3 to MBNA., by SFA grant 2018LANLF255 from the U.S. Department of Energy Office of Biological and Environmental Research to JD. SL was supported by a startup grant from the University of Oregon. JBE was supported by USDA National Institute of Food and Agriculture (NIFA) grant number 2021-67013-34815-0 and USDA NIFA Hatch project number CA-D-PPA-2464-H. SJH was supported by a National Science Foundation Rapid Response Research (RAPID) grant, grant number 1929843. We thank reviewers for their helpful comments and suggestions on earlier versions of the manuscript.
Author contributions
MBNA and JD wrote the first draft of the manuscript. SL, DEW, KLF, SJH, KLW, JBE, JD, and MBNA contributed to ideas and manuscript editing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01088-5.
References
- 1.Inda ME, Broset E, Lu TK, de la Fuente-Nunez C. Emerging frontiers in microbiome engineering. Trends Immunol. 2019;40:952–73. doi: 10.1016/j.it.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, Loffler FE, O’Malley MA, et al. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol. 2019;17:725–41. doi: 10.1038/s41579-019-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu ZG, Egidi E, Liu HW, Kaur S, Singh BK. New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol Adv. 2019;37:107371. doi: 10.1016/j.biotechadv.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Enam F, Mansell TJ. Prebiotics: tools to manipulate the gut microbiome and metabolome. J Ind Microbiol Biotechnol. 2019;46:1445–59. doi: 10.1007/s10295-019-02203-4. [DOI] [PubMed] [Google Scholar]
- 5.Ke J, Wang B, Yoshikuni Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021;39:244–61. doi: 10.1016/j.tibtech.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9:1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel OM, Castrillo G, Paredes SH, Gonzalez IS, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–63. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaminsky LM, Trexler RV, Malik RJ, Hockett KL, Bell TH. The inherent conflicts in developing soil microbial inoculants. Trends Biotechnol. 2019;37:140–51. doi: 10.1016/j.tibtech.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Kolar CS, Lodge DM. Progress in invasion biology: predicting invaders. Trends Ecol Evol. 2001;16:199–204. doi: 10.1016/s0169-5347(01)02101-2. [DOI] [PubMed] [Google Scholar]
- 10.Cairns J, Heckman JR. Restoration ecology: the state of an emerging field. Annu Rev Environ Resour. 1996;21:167–89. [Google Scholar]
- 11.Wainwright CE, Staples TL, Charles LS, Flanagan TC, Lai HR, Loy X, et al. Links between community ecology theory and ecological restoration are on the rise. J Appl Ecol. 2018;55:570–81. [Google Scholar]
- 12.Mallon CA, Le Roux X, van Doorn GS, Dini-Andreote F, Poly F, Salles JF. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 2018;12:728–41. doi: 10.1038/s41396-017-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enders M, Hutt MT, Jeschke JM. Drawing a map of invasion biology based on a network of hypotheses. Ecosphere. 2018;9:e02146. [Google Scholar]
- 14.Catford JA, Jansson R, Nilsson C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers Distrib. 2009;15:22–40. [Google Scholar]
- 15.Wittmann MJ, Metzler D, Gabriel W, Jeschke JM. Decomposing propagule pressure: the effects of propagule size and propagule frequency on invasion success. Oikos. 2014;123:441–50. [Google Scholar]
- 16.Hulvey KB, Leger EA, Porensky LM, Roche LM, Veblen KE, Fund A, et al. Restoration islands: a tool for efficiently restoring dryland ecosystems? Restor Ecol. 2017;25:S124–S34. [Google Scholar]
- 17.Funk JL, Hoffacker MK, Matzek V. Summer irrigation, grazing and seed addition differentially influence community composition in an invaded serpentine grassland. Restor Ecol. 2015;23:122–30. [Google Scholar]
- 18.Jones ML, Ramoneda J, Rivett DW, Bell T. Biotic resistance shapes the influence of propagule pressure on invasion success in bacterial communities. Ecology. 2017;98:1743–9. doi: 10.1002/ecy.1852. [DOI] [PubMed] [Google Scholar]
- 19.Albright MBN, Sevanto S, Gallegos Graves LV, Dunbar J. Biotic interactions are more important than propagule pressure in microbial community invasions. Mbio. 2020;11:e02089–20. doi: 10.1128/mBio.02089-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila JCC, Jones ML, Patel M, Bell T, Rosindell J. Uncovering the rules of microbial community invasions. Nat Ecol Evol. 2019;3:1162–71. doi: 10.1038/s41559-019-0952-9. [DOI] [PubMed] [Google Scholar]
- 21.Simberloff D. The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst. 2009;40:81–102. [Google Scholar]
- 22.Zhou JZ, Ning DL. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev. 2017;81:e00002–17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comeau Y, Greer CW, Samson R. Role of inoculum preparation and density on the bioremediation of 2,4-D-contaminated soil by bioaugmentation. Appl Microbiol Biotechnol. 1993;38:681–7. [Google Scholar]
- 24.Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86:1267–79. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 25.Kreitschitz A, Haase E, Gorb SN. The role of mucilage envelope in the endozoochory of selected plant taxa. Sci Nat-Heidelb. 2021;108:2. doi: 10.1007/s00114-020-01709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gornish E, Arnold H, Fehmi J. Review of seed pelletizing strategies for arid land restoration. Restor Ecol. 2019;27:1206–11. [Google Scholar]
- 27.Ali M, Oshiki M, Rathnayake L, Ishii S, Satoh H, Okabe S. Rapid and successful start-up of anammox process by immobilizing the minimal quantity of biomass in PVA-SA gel beads. Water Res. 2015;79:147–57. doi: 10.1016/j.watres.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Gallien L, Mazel F, Lavergne S, Renaud J, Douzet R, Thuiller W. Contrasting the effects of environment, dispersal and biotic interactions to explain the distribution of invasive plants in alpine communities. Biol Invasions. 2015;17:1407–23. doi: 10.1007/s10530-014-0803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cadotte MW, Campbell SE, Li SP, Sodhi DS, Mandrak NE. Preadaptation and naturalization of nonnative species: Darwin’s two fundamental insights into species invasion. Annu Rev Plant Biol. 2018;69:661–84. doi: 10.1146/annurev-arplant-042817-040339. [DOI] [PubMed] [Google Scholar]
- 30.Fick SE, Day N, Duniway MC, Hoy-Skubik S, Barger NN. Microsite enhancements for soil stabilization and rapid biocrust colonization in degraded drylands. Restor Ecol. 2020;28:S139–S49. [Google Scholar]
- 31.Vasquez E, Sheley R, Svejcar T. Creating invasion resistant soils via nitrogen management. Invas Plant Sci Man. 2008;1:304–14. [Google Scholar]
- 32.Zhao X, Wang W, Blaine A, Kane ST, Zijlstra RT, Ganzle MG. Impact of probiotic Lactobacillus sp. on autochthonous lactobacilli in weaned piglets. J Appl Microbiol. 2019;126:242–54. doi: 10.1111/jam.14119. [DOI] [PubMed] [Google Scholar]
- 33.Muthukrishnan R, Hansel-Welch N, Larkin DJ. Environmental filtering and competitive exclusion drive biodiversity-invasibility relationships in shallow lake plant communities. J Ecol. 2018;106:2058–70. [Google Scholar]
- 34.Pereira FC, Berry D. Microbial nutrient niches in the gut. Environ Microbiol. 2017;19:1366–78. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson IP, van der Gast CJ, Ciric L, Singer AC. Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol. 2005;7:909–15. doi: 10.1111/j.1462-2920.2005.00804.x. [DOI] [PubMed] [Google Scholar]
- 36.Bell TH, Bell T. Many roads to bacterial generalism. Fems Microbiol Ecol. 2021;97:fiaa240. [DOI] [PubMed]
- 37.Campieri M, Rizzello F, Venturi A, Poggioli G, Ugolini F, Helwig U, et al. Combination of antibiotic and probiotic treatment is efficacious in prophylaxis of post-operative recurrence of Crohn’s disease: a randomized controlled study vs mesalamine. Gastroenterology. 2000;118:A781–A. [Google Scholar]
- 38.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. Msphere. 2017;2:e00501–17. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018;23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–8. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw AJ, Lam FH, Hamilton M, Consiglio A, MacEwen K, Brevnova EE, et al. Metabolic engineering of microbial competitive advantage for industrial fermentation processes. Science. 2016;353:583–6. doi: 10.1126/science.aaf6159. [DOI] [PubMed] [Google Scholar]
- 42.Umu OCO, Rudi K, Diep DB. Modulation of the gut microbiota by prebiotic fibres and bacteriocins. Micro Ecol Health Dis. 2017;28:1348886. doi: 10.1080/16512235.2017.1348886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sriswasdi S, Yang CC, Iwasaki W. Generalist species drive microbial dispersion and evolution. Nat Commun. 2017;8:1162. doi: 10.1038/s41467-017-01265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNally L, Brown SP. Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos Trans R Soc B. 2015;370:20140298. doi: 10.1098/rstb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shahab RL, Brethauer S, Luterbacher JS, Studer MH. Engineering of ecological niches to create stable artificial consortia for complex biotransformations. Curr Opin Biotechnol. 2020;62:129–36. doi: 10.1016/j.copbio.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Shade A, Peter H, Allison SD, Baho DL, Berga M, Burgmann H, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Upton RN, Bach EM, Hofmockel KS. Spatio-temporal microbial community dynamics within soil aggregates. Soil Biol Biochem. 2019;132:58–68. [Google Scholar]
- 48.Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. Am J Clin Nutr. 2001;73:399s–405s. doi: 10.1093/ajcn/73.2.399s. [DOI] [PubMed] [Google Scholar]
- 49.Tripathi S, Srivastava P, Devi R, Bhadouria R. Influence of synthetic fertilizers and pesticides on soil health and soil microbiology. In: Prasad MNV (ed). Agrochemicals detection, treatment and remediation. (Butterworth-Heinemann, 2020) pp 25-54.
- 50.Dykhuizen DE, Hartl DL. Selection in chemostats. Microbiol Rev. 1983;47:150–68. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao D, Wu SG, Feng WW, Jakovlic I, Tran NT, Xiong F. Adhesion and colonization properties of potentially probiotic Bacillus paralicheniformis strain FA6 isolated from grass carp intestine. Fish Sci. 2020;86:153–61. [Google Scholar]
- 52.Wang XY, Cao ZP, Zhang MM, Meng L, Ming ZZ, Liu JY. Bioinspired oral delivery of gut microbiota by self-coating with biofilms. Sci Adv. 2020;6:eabb1952. doi: 10.1126/sciadv.abb1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali SA, Singh P, Tomar SK, Mohanty AK, Behare P. Proteomics fingerprints of systemic mechanisms of adaptation to bile in Lactobacillus fermentum. J Proteom. 2020;213:103600. doi: 10.1016/j.jprot.2019.103600. [DOI] [PubMed] [Google Scholar]
- 54.Wisz MS, Pottier J, Kissling WD, Pellissier L, Lenoir J, Damgaard CF, et al. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biol Rev. 2013;88:15–30. doi: 10.1111/j.1469-185X.2012.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Funk JL, Cleland EE, Suding KN, Zavaleta ES. Restoration through reassembly: plant traits and invasion resistance. Trends Ecol Evol. 2008;23:695–703. doi: 10.1016/j.tree.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Northfield TD, Laurance SGW, Mayfield MM, Paini DR, Snyder WE, Stouffer DB, et al. Native turncoats and indirect facilitation of species invasions. Proc Biol Sci. 2018;285:20171936. doi: 10.1098/rspb.2017.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gagnon K, Rinde E, Bengil EGT, Carugati L, Christianen MJA, Danovaro R, et al. Facilitating foundation species: the potential for plant-bivalve interactions to improve habitat restoration success. J Appl Ecol. 2020;57:1161–79. [Google Scholar]
- 58.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–23. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Bayona L, Comstock LE. Bacterial antagonism in host-associated microbial communities. Science. 2018;361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 60.Maynard DS, Crowther TW, Bradford MA. Competitive network determines the direction of the diversity-function relationship. Proc Natl Acad Sci USA. 2017;114:11464–9. doi: 10.1073/pnas.1712211114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feichtmayer J, Deng L, Griebler C. Antagonistic microbial interactions: contributions and potential applications for controlling pathogens in the aquatic systems. Front Microbiol. 2017;8:2192. doi: 10.3389/fmicb.2017.02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchslin HP, Schneider C, Egli T. In glucose-limited continuous culture the minimum substrate concentration for growth, s(min), is crucial in the competition between the enterobacterium Escherichia coli and Chelatobacter heintzii, an environmentally abundant bacterium. ISME J. 2012;6:777–89. doi: 10.1038/ismej.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beaury EM, Finn JT, Corbin JD, Barr V, Bradley BA. Biotic resistance to invasion is ubiquitous across ecosystems of the United States. Ecol Lett. 2020;23:476–82. doi: 10.1111/ele.13446. [DOI] [PubMed] [Google Scholar]
- 64.Eisenhauer N, Schulz W, Scheu S, Jousset A. Niche dimensionality links biodiversity and invasibility of microbial communities. Funct Ecol. 2013;27:282–8. [Google Scholar]
- 65.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–12. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 66.Perez-Gutierrez RA, Lopez-Ramirez V, Islas A, Alcaraz LD, Hernandez-Gonzalez I, Olivera BCL, et al. Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 2013;7:487–97. doi: 10.1038/ismej.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Safferman RS, Morris ME. Evaluation of natural products for algicidal properties. Appl Microbiol. 1962;10:289–92. doi: 10.1128/am.10.4.289-292.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russel J, Roder HL, Madsen JS, Burmolle M, Sorensen SJ. Antagonism correlates with metabolic similarity in diverse bacteria. Proc Natl Acad Sci USA. 2017;114:10684–8. doi: 10.1073/pnas.1706016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Long RA, Rowley DC, Zamora E, Liu JY, Bartlett DH, Azam F. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl Environ Microbiol. 2005;71:8531–6. doi: 10.1128/AEM.71.12.8531-8536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Wardenburg JB. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–91. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez-Igual R, Bernal-Bayard J, Rodriguez-Paton A, Ghigo JM, Mazel D. Engineered toxin-intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat Biotechnol. 2019;37:755–60. doi: 10.1038/s41587-019-0105-3. [DOI] [PubMed] [Google Scholar]
- 72.Koskella B. New approaches to characterizing bacteria-phage interactions in microbial communities and microbiomes. Environ Microbiol Rep. 2019;11:15–6. doi: 10.1111/1758-2229.12706. [DOI] [PubMed] [Google Scholar]
- 73.Soundararajan M, von Bunau R, Oelschlaeger TA. K5 Capsule and lipopolysaccharide are important in resistance to T4 phage attack in probiotic E. coli strain nissle 1917. Front Microbiol. 2019;10:2783. doi: 10.3389/fmicb.2019.02783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. [Google Scholar]
- 75.Marsh P, Wellington EMH. Phage-host interactions in soil. FEMS Microbiol Ecol. 1994;15:99–107. [Google Scholar]
- 76.Balogh B, Jones JB, Iriarte FB, Momol MT. Phage therapy for plant disease control. Curr Pharm Biotechnol. 2010;11:48–57. doi: 10.2174/138920110790725302. [DOI] [PubMed] [Google Scholar]
- 77.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–50. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Piccardi P, Vessman B, Mitri S. Toxicity drives facilitation between 4 bacterial species. Proc Natl Acad Sci USA. 2019;116:15979–84. doi: 10.1073/pnas.1906172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pascual-Garcia A, Bonhoeffer S, Bell T. Metabolically cohesive microbial consortia and ecosystem functioning. Philos Trans R Soc B. 2020;375:20190245. doi: 10.1098/rstb.2019.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martinez-Harms MJ, Bryan BA, Balvanera P, Law EA, Rhodes JR, Possingham HP, et al. Making decisions for managing ecosystem services. Biol Conserv. 2015;184:229–38. [Google Scholar]
- 81.Kildisheva OA, Dixon KW, Silveira FAO, Chapman T, Di Sacco A, Mondoni A, et al. Dormancy and germination: making every seed count in restoration. Restor Ecol. 2020;28:S256–S65. [Google Scholar]
- 82.Maslo B, Handel SN, Pover T. Restoring beaches for Atlantic coast piping plovers (Charadrius melodus): a classification and regression tree analysis of nest-site selection. Restor Ecol. 2011;19:194–203. [Google Scholar]
- 83.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–50. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 84.Carr A, Diener C, Baliga NS, Gibbons SM. Use and abuse of correlation analyses in microbial ecology. ISME J. 2019;13:2647–55. doi: 10.1038/s41396-019-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ, et al. Trophic downgrading of planet Earth. Science. 2011;333:301–6. doi: 10.1126/science.1205106. [DOI] [PubMed] [Google Scholar]
- 86.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herren CM, McMahon KD. Keystone taxa predict compositional change in microbial communities. Environ Microbiol. 2018;20:2207–17. doi: 10.1111/1462-2920.14257. [DOI] [PubMed] [Google Scholar]
- 88.Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract-identification of keystone and foundation taxa. Microbiome. 2015;3:44. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kopp-Hoolihan L. Prophylactic and therapeutic uses of probiotics: a review. J Am Diet Assoc. 2001;101:229–41. doi: 10.1016/S0002-8223(01)00060-8. [DOI] [PubMed] [Google Scholar]
- 90.Woo SL, Pepe O. Microbial consortia: promising probiotics as plant biostimulants for sustainable agriculture. Front Plant Sci. 2018;9:1801. doi: 10.3389/fpls.2018.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wood-Charlson EM, Anubhav, Auberry D, Blanco H, Borkum MI, Corilo YE, et al. The National Microbiome Data Collaborative: enabling microbiome science. Nat Rev Microbiol. 2020;18:313–4. doi: 10.1038/s41579-020-0377-0. [DOI] [PubMed] [Google Scholar]
- 92.Brussow H. Probiotics and prebiotics in clinical tests: an update. F1000Res. 2019;8:1157. [DOI] [PMC free article] [PubMed]
- 93.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 94.Weingarden AR, Chen C, Bobr A, Yao D, Lu YW, Nelson VM, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–G9. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hutchinson MI, Bell TAS, Gallegos-Graves L, Dunbar J, Albright M. Merging fungal and bacterial community profiles via an internal control. Microb Ecol. 2021; e-pub ahead of print 2021; 10.1007/s00248-020-01638-y. [DOI] [PubMed]
- 96.Nayfach S, Roux S, Seshadri R. A genomic catalog of Earth’s micobiomes. Nat Biotechnol. 2021;39:499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Azubuike CC, Chikere CB, Okpokwasili GC. Bioremediation techniques-classification based on site of application: principles, advantages, limitations and prospects. World J Microbiol Biotechnol. 2016;32:180. doi: 10.1007/s11274-016-2137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Henze M, Gujer W, Mino T, Van Loosdrecht MCM. Activated sludge models ASM1, ASM2, ASM2d and ASM, Vol 121. 2000. IWA Scientific and Technical Report 9, IWA publishing, London.
- 100.Orozco-Mosqueda MD, Rocha-Granados MD, Glick BR, Santoyo G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res. 2018;208:25–31. doi: 10.1016/j.micres.2018.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.