Abstract
Limiting postprandial glycemic response (PPGR) is an important intervention in reducing the risk of chronic metabolic diseases and has been shown to impart significant health benefits in people with elevated levels of blood sugar. In this study, we collected gut microbiome activity data by assessing the metatranscriptome, and we measured the glycemic responses of 550 adults who consumed more than 30,000 meals, collectively, from omnivore or vegetarian/gluten-free diets. We demonstrate that gut microbiome activity, anthropometric factors, and food macronutrients modulate individual variation in glycemic response. We employ two predictive models, including a mixed-effects linear regression model (R = 0.77) and a gradient boosting machine model (Rtrain = 0.80/R2train = 0.64; Rtest = 0.64/R2test = 0.40), which demonstrate variation in PPGR between individuals when ingesting the same foods. All features in the final mixed-effects linear regression model were significant (p < 0.05) except for two features which were retained as suggestive: glutamine production pathways (p = 0.08) and the interaction between tyrosine metabolizers and carbs (p = 0.06). We introduce molecular functions as features in these two models, aggregated from microbial activity data, and show their statistically significant contributions to glycemic control. In summary, we demonstrate for the first time that metatranscriptomic activity of the gut microbiome is correlated with PPGR among adults.
Keywords: Postprandial blood glucose, Gut microbiome, Metatranscriptome, Glycemic response, Machine learning, Artificial intelligence
Plain Language Summary
Blood sugar dysregulation is caused by various underlying conditions, including type 2 diabetes, and this may lead to extended periods of hypoglycemia or hyperglycemia, which can be harmful or deadly. Clinically, glycemic control is a primary therapeutic target for dysglycemia, and food and nutrition are frequent interventions used to reduce postprandial blood glucose excursions. Primary determinants of postprandial glycemic response (PPGR) include dietary carbohydrates, individual phenotypes, and individual molecular characteristics which include the gut microbiome. Typical investigations of gut microbiomes depend on analysis methods which have poor taxonomic resolution, cannot identify certain microorganisms, and are prone to errors. In this study, each RNA molecule was identified and counted, allowing quantitative strain-level taxonomic classification and molecular pathway analysis. The primary goal of the study was to assess the impact of microbial functional activity on PPGR. The study was conducted in the USA and involved a multiethnic population of healthy adults with HbA1c levels below 6.5. All participants received 14-day omnivore diets or vegetarian/gluten-free diets, depending on nutritional requirements (omnivore diets include meat while vegetarian/gluten-free diets exclude both gluten and meat). Over this timeframe, blood glucose levels were measured in 15-min intervals, 24 h per day, capturing postprandial responses for more than 27,000 meals, including more than 18,000 provided meals which spanned a wide range of foods and macronutrient characteristics. Computational modeling demonstrated the statistical significance of all features and identified new features which may be relevant to glycemic control. These results show, for the first time, that a person’s glycemic response depends on individual traits, including both their anthropometrics and their gut metatranscriptome, representing the activity of gut microbiomes.
Key Summary Points
| Why carry out this study? |
| Controlling postprandial glycemic response (PPGR) is a crucial part of mitigating the current epidemic of metabolic diseases including obesity, type 2 diabetes, hypertension, as well as cardiovascular and liver diseases. |
| Tools facilitating the mass adoption of dietary choices to maintain normal glycemic levels would be an important step towards halting the hyperglycemia epidemic. Previous studies have considered metagenomic data, and ours is the first metatranscriptomic study. |
| The primary goal of this study was to determine the impact of microbial gene expression (at the functional level) on PPGR. |
| What was learned from the study? |
| We demonstrate for the first time that metatranscriptomic activity of the gut microbiome is correlated with PPGR among adults. |
| Our study identifies new microbial features, linking molecular pathways to glycemic control, including the fucose metabolism pathway and the indoleacetate production pathways. |
Introduction
From a public health perspective, preventing elevated levels of blood glucose is a crucial part of mitigating the current epidemic of metabolic diseases including obesity, type 2 diabetes, hypertension, cardiovascular and liver diseases. Across the US population, 9.4% of people are diabetic and 26% are prediabetic, and this points to a large disease burden with associated healthcare costs [1]. Daily food choices play the largest role in determining overall blood glucose levels, and thus risk for various diseases [2–5]. Other factors affecting food choices, and thus risk for various diseases like diabetes, include socioeconomic background and ethnicity [6, 7]. Affordable and accessible tools facilitating the mass adoption of dietary choices to maintain normal glycemic levels would be an important step towards halting the hyperglycemia epidemic.
Typical dietary recommendations and nutritional information largely focus on food characteristics alone, such as calorie and carbohydrate content. However, there is increasing evidence that glycemic response to the same foods varies significantly between individuals. Recent studies [8, 9] have shown that postprandial glycemic response (PPGR) is driven not only by the glycemic index of food but also by individual phenotypes and molecular characteristics, including the gut microbiome, which may have a role in energy metabolism and insulin regulation [10]. These studies aligned with existing research and demonstrated an ability to predict PPGR through associations with standardized meal PPGRs (specifically, KEGG pathways [11] which represented: chemotaxis and flagellar assembly; ABC transporters; and type II and type III secretion systems). They also reported that diets high in positively charged amino acids were associated with high PPGR whereas diets high in negatively charged amino acids were associated with low PPGR. These studies evaluated PPGR in the context of specific populations (Israeli and US Midwestern), they implemented a small number of standardized meals, and their conclusions depended on 16S or metagenomic data from the gut microbiome. Often, gut microbiome analyses like these depend on 16S rRNA gene sequencing, but this method provides poor taxonomic resolution of microbiomes which typically contain many strains with very diverse gene content [12]. In addition, metagenomic methods are unable to identify some microorganisms (e.g., RNA viruses) and can only predict gene expression based on the gene content, which can be highly erroneous [13]. Metatranscriptomic methods offer a comprehensive lens of the gut microbiome, with a specific focus on genes that are actively transcribed. However, metatranscriptomics has not been widely used in clinical studies because of challenges posed by RNA instability, the need to remove diverse ribosomal RNAs in stool samples, and complex bioinformatic analyses.
In this study, we demonstrate that glycemic response depends on individual differences, including gut microbiome activity (i.e., metatranscriptomics of the gut microbiome) as well as anthropometrics. We cultivate a multiethnic study cohort from multiple study sites across the USA. Our diet design includes 104 unique pre-designed meals from across two distinct diet types—omnivore (which includes meat) and vegetarian/gluten-free (which excludes both gluten and meat). For our primary goal, we sought to determine the impact of microbial gene expression on PPGR, which we report in terms of functional microbiome features, including KEGG gene functional orthologs (or “KOs”) [11] and microbial taxonomy. We employ metatranscriptomics [14], which directly measures RNA molecules and provides the primary sequence and read counts for each transcript [15–17]. The metatranscriptomics approach we employ measures the complete set of gene transcripts (RNA) from a sample, and in microbiome samples, this technique facilitates quantitative strain-level classification and functional pathway analysis of microbes, which we accomplish through sequence alignments to publicly available reference databases. We aggregate collections of these microbial pathway features into curated scores, capturing functional characteristics as described in the literature [18]. Gut microbiome activity is measured by metatranscriptomics and includes all curated microbiome features and scores. With the goal of being directly interpretable, we provide a concise statistical explanation that links PPGR to nutrients, phenotypes, and gut microbiome activity, through a mixed-effects linear regression model. We also present a gradient boosting machine model, optimized for predictive accuracy. Finally, we identify significant functional microbiome features related to prediction of PPGR, indicating that microbiome activity affects the processing of carbohydrates and contributes to variation in baseline blood sugar. To our knowledge, this is the first study to demonstrate the application of gut metatranscriptomics in a population-scale dietary study.
Methods
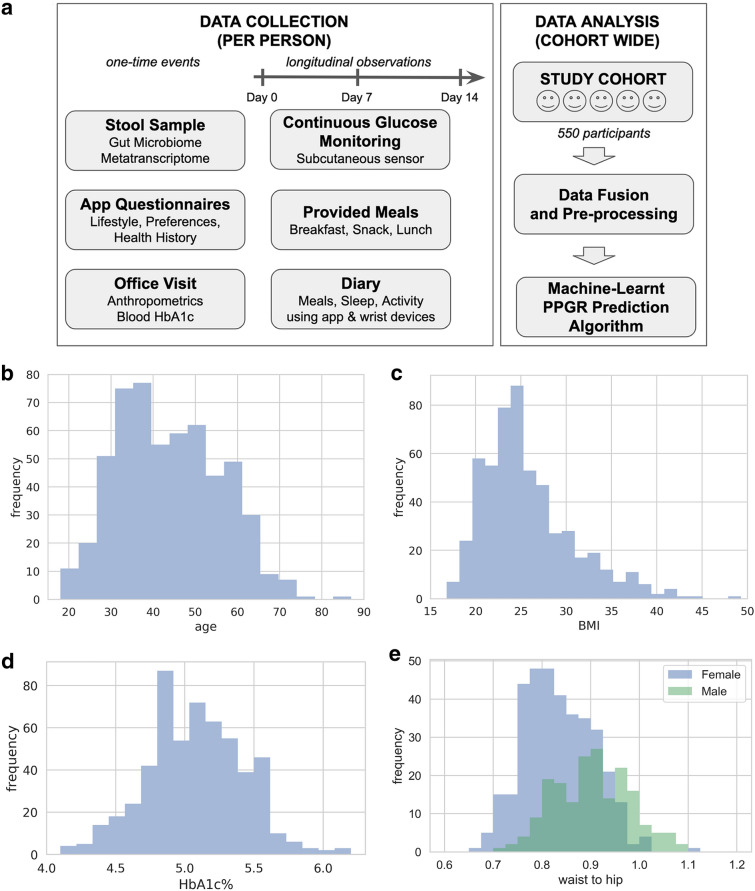
As shown in Fig. 1, we recruited 550 adults (66% female) from the general public through multiple study sites across the USA, and we tracked their food intake, sleep, activity, and glycemic response for up to 2 weeks. Four hundred participants were Caucasian, and of the remaining 150 participants, 37% were Asian, 33% were Hispanic, and 30% were Black or Other. The study was designed and carried out in accordance with the ethical principles of the Declaration of Helsinki and its later amendments, and was approved by a federally accredited institutional review board (IRB; 00011543). All samples and metadata were obtained from human subjects at least 18 years old and residing in the USA at the time of participation. All study participants consented to participating in the study. From each participant, we obtained a stool sample at participation enrollment, as well as a comprehensive questionnaire describing their lifestyle, preferences, and health history. We collected blood glucose measurements over 14 days, every 15 min for 24 h per day, using a continuous glucose monitor (CGM) sensor that measures glucose levels within the range of 40–500 mg/dL [19].
Fig. 1.
Study design and population characteristics. a The study cohort had 550 adult participants (66% female). Each study participant provided a stool sample, filled out questionnaires, and made an office visit. Then over 14 days, participants consumed pre-designed meals that were provided, they monitored their blood glucose response, and they kept a diary of their meals, sleep, and activity. At the end of the study, all the data streams were fused, pre-processed, and analyzed as described in this paper. The following exclusion criteria were used: age < 18; dietary restrictions that would prevent adherence to any of the study diets; antibiotic use 1 month prior to or during study; skin disease (e.g., contact dermatitis) that precludes proper attachment of the CGM; pregnancy; active neoplastic disease; active neuropsychiatric disorder; myocardial infarction or cerebrovascular accident in the 6 months prior; pre-diagnosed type I or type II diabetes mellitus; HbA1c ≥ 6.5; or unwilling/incapable of following instructions. b Age distribution with mean of 43.8 years (SD 12.115). c 28% of the study population had BMI > 25 and 18% had BMI > 30. d 4% of the study population were pre-diabetic with HbA1c% > 5.7. e Waist-to-hip ratio distribution with mean of 0.901 (SD 0.076) for men and 0.832 (SD 0.071) for women
Study Definitions
Participants were able to select from an omnivore diet (which includes meat) and a vegetarian/gluten-free diet (which excludes both gluten and meat). There were multiple phases of the study, which allowed us to design multiple meal plans for each of the two diets. For breakfasts, morning snacks, and lunches, all participants followed one of these two diets, and these pre-designed meals are referred to as “provided meals.” All other food (consumed more than 2.5 h before or after meals) is referred to as “free meals,” and this includes any other snacks, beverages, meals, and desserts which were not part of the provided meals. For each diet, a 14-day meal plan was designed to broadly cover diverse macronutrient ratios and randomize their consumption across the timeline (Tables 1 and 2). Each meal was typified with “High” (H) and “Low” (L) categorical bins, and these were assigned by thresholding the macronutrient values, based on the proportion of each macronutrient and the daily recommended allowance of that macronutrient. The characters in each four-letter meal composition “type” respectively refer to carbs, fiber, protein, and fat; for example, a meal with “high carbs, low fiber, high protein, and low fat” was categorized as “high, low, high, low” (HLHL). In all, there were 13 of these meal composition types.
Table 1.
Example meals
| Day | Description | |
|---|---|---|
| Type8 (HLLL) | Day 3 | Glucose drink (40 g of glucose + water) |
| Day 6 | Banana (1 banana, peeled) | |
| Day 11 |
White rice (1/2 cup, cooked) Black pepper (optional) |
|
| Type5 (HHLL) | Day 2 |
Orange (1 orange, peeled) Banana (1 banana, peeled) |
| Day 5 | Apple (2 apples) | |
| Day 7 |
Oatmeal (1 pkg, warm or cold) Blueberries (1/2 cup) Apple (1 apple) Orange (1 orange, peeled) |
|
| Type11 (HHHL) | Day 3 |
White rice (2/3 cup, cooked) Black beans (3/4 cup, warm or cold) Garlic (1 clove) Chicken sausage (4 links, cooked) |
| Day 10 |
White rice (1/2 cup, cooked) Chickpeas (1 + 1/2 cups, raw or steamed) |
|
| Day 12 |
Oatmeal (1 pkg, warm or cold) Sprouted grain bread (3 slices) Egg (1 egg, poached or hard boiled) Orange (1 orange, peeled) |
|
| Type4 (HHHH) | Day 4 |
Chickpeas (1 + 1/4 cups, raw or steamed) Chicken sausage (4 links, cooked) Apple (1 apple) Tortilla strips (5 strips) |
| Day 7 |
White rice (3/4 cup, cooked) Turkey (1/4 cup, measure before cooking) Black beans (1/2 cup, warm or cold) Sunflower seeds (1/4 cup) |
|
| Day 10 |
Apple (1 apple) Black beans (1/3 cup, warm or cold) Potato (1 potato, baked—no skin) Beef (1/2 cup, measure before cooking) |
In this omnivore diet, example meals are shown for several of the provided macronutrient groups (carbs–fiber–protein–fat)
Table 2.
Daily breakdown for meal plan
| Breakfast | Morning snack | Lunch | ||||
|---|---|---|---|---|---|---|
| Day 1 | Type1 | (HLHL) | Type2 | (LLHL) | Type3 | (HHLH) |
| Day 2 | Type7 | (LLLH) | Type5 | (HHLL) | Type9 | (LLHH) |
| Day 3 | Type8 | (HLLL) | Type11 | (HHHL) | Type3 | (HHLH) |
| Day 4 | Type4 | (HHHH) | Type13 | (LHHL) | Type9 | (LLHH) |
| Day 5 | Type10 | (HLLH) | Type5 | (HHLL) | Type6 | (LHLH) |
| Day 6 | Type7 | (LLLH) | Type8 | (HLLL) | Type9 | (LLHH) |
| Day 7 | Type5 | (HHLL) | Type13 | (LHHL) | Type4 | (HHHH) |
| Day 8 | Type1 | (HLHL) | Type2 | (LLHL) | Type3 | (HHLH) |
| Day 9 | Type10 | (HLLH) | Type13 | (LHHL) | Type12 | (HLHH) |
| Day 10 | Type6 | (LHLH) | Type11 | (HHHL) | Type4 | (HHHH) |
| Day 11 | Type7 | (LLLH) | Type8 | (HLLL) | Type12 | (HLHH) |
| Day 12 | Type11 | (HHHL) | Type10 | (HLLH) | Type6 | (LHLH) |
| Day 13 | Type1 | (HLHL) | Type2 | (LLHL) | Type12 | (HLHH) |
Each participant consumed 39 provided meals, which included 1 glucose drink, 6 repeat provided meals, and 32 unique provided meals. Meals with similar composition types were distributed across the meal plan, with days 1 and 8 representing repeated meal days
Study Meals
As described in Tables 1 and 2, participants were provided pre-designed breakfasts, snacks, and lunches (provided meals) over 14 days (day 0 to day 13). After lunch, participants were allowed to eat whatever they wanted (free meals) without further guidance on the composition, and day 0 consisted of only free meals. Provided meals accounted for 66% of all meals and free meals accounted for 34% of all meals.
Both provided and free meals were recorded by all participants during the entire study period, using a smartphone app (Bitesnap [20]). Macronutrient and micronutrient information were provided by the smartphone app platform for further analysis. Provided meals were pre-loaded into the app for convenience, while free meals were loaded by users through selection of custom dishes, ingredients, and quantities.
Over the course of their 14-day meal plans, we asked all participants to avoid the following activities and interventions: (1) intense exercise within 2.5 h before or 2.5 h after meals; (2) introduction of new probiotics or prebiotics; (3) all vitamins and supplements (with a specific detail to avoid interfering substances as defined in [19]); (4) over-the-counter medication. We also asked all participants to inform a study coordinator if they were prescribed antibiotics during the study.
In order to test our methodology across a range of diets and to support a range of participant preferences, we provided two diet types—omnivore and vegetarian/gluten-free—in different phases of the study. In all, we provided 104 unique, pre-designed meals: 71 meals were part of the omnivore diet and 33 meals were part of the vegetarian/gluten-free diet. A total of 140 participants signed up for the omnivore diet and 410 signed up for the vegetarian/gluten-free diet.
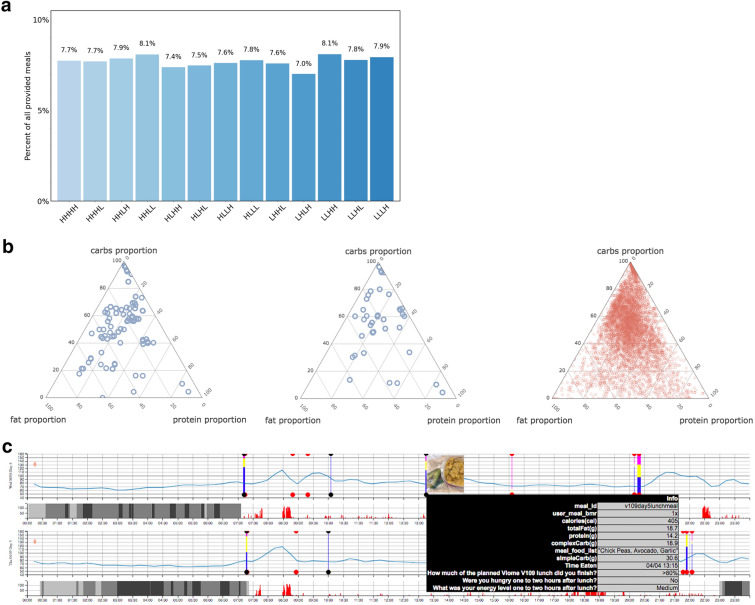
In an effort to maximize coverage and diversity of macronutrient ratios across meals, we designed the provided meals using a high (H)/low (L) determination of each of the four macronutrients—carbs, fiber, protein, and fat—resulting in 13 compositional types as shown in Fig. 2a. In this paper, we use the terms carbs and carbohydrates interchangeably to mean net carbohydrates (i.e., excluding fiber). “High” and “Low” categorical bins were assigned from thresholding of meal composition, based on the proportion of each macronutrient and its daily recommended allowance [21]. For example, day 1 breakfast was given a composition type of “high, low, high, low” (HLHL), respectively, in carbs, fiber, protein, and fat. All 13 of these meal composition types were included in every meal plan.
Fig. 2.
Study meal design and content. a Percentage of each meal category within all 18,000+ provided meals. Categories were defined using proportions of carbs (H/L), fiber (H/L), proteins (H/L), and fats (H/L). b Macronutrient proportions of all 71 unique pre-designed omnivore meals (top, blue), all 33 unique pre-designed vegetarian/gluten-free meals (middle, blue), and all free meals (bottom, red). Provided meals account for 66% of all meals, and free meals 34%. c Data collected for a single participant over 2 days (out of 14). Each row is a single day. The blue curve is the CGM reading collected every 15 min. Vertical bars are meal events, showing carbs proportion (blue), fat proportion (yellow), and protein proportion (pink). The user interface also visualizes a picture of the meal and the nutrient details. Gray bars represent light and deep sleep. The red histogram next to sleep bars is the tracked physical activity
An example meal plan is shown in Table 1 along with each composition type; over the 14 days, each participant was provided with a total of 39 meals, including one glucose drink (day 0 consisted of only free meals). Additional design strategy went into the meal plans. Days 1 and 8 of each meal plan were designed as replicate days (Table 2), and meals on these 2 days were completely identical (on day 8, each participant consumed the same breakfast, snack, and lunch meals they were assigned for day 1). Beyond these six repeat meals and the one glucose drink, each participant also consumed 32 unique provided meals which stratified across the 13 meal composition types (Table 2).
Significant effort was made to incorporate as many meal composition types and food staples as possible. The distribution of macronutrients (Fig. 2b) shows the coverage of the provided meals across the space of diverse macronutrient ratios. Implementing two diets (omnivore and vegetarian/gluten-free) in multiple study phases also allowed us to further diversify those meals repeated on days 1 and 8, such that a total of nine different types of replicate meals were assigned (Table 3), and each included high levels of just one or two macronutrients.
Table 3.
Macronutrient breakdown for repeat meals
| Description | Net Carb (g) | Fiber (g) | Protein (g) | Fat (g) |
|---|---|---|---|---|
| Bagel, eggs, mango | 71 | 7.9 | 21.3 | 10 |
| Rice, avocado, apple | 62.4 | 18.9 | 8.5 | 29.8 |
| Apple sauce, grapes, banana | 52.8 | 5.8 | 1.8 | 0.5 |
| Pea protein, mango, banana | 46.8 | 5.1 | 26.3 | 2.6 |
| Quinoa and veggies, lentils | 36 | 16.5 | 17 | 2 |
| Tortilla, almond butter | 25 | 5 | 9 | 20.5 |
| Grain bread, chicken | 24 | 6 | 24 | 8 |
| Celery, hummus | 5.4 | 2.6 | 2.7 | 2.7 |
| Pea protein | 3 | 0 | 24 | 2.3 |
Macronutrient information for the 9 repeat meals which were consumed twice by study participants. Actual servings were adjusted on the basis of the participant’s basal metabolic rate (BMR). Net carbs represent total meal carbohydrates, excluding fiber. Values for carbs, fiber, protein, and fat represent the respective macronutrient weights (grams) of each meal, computed from the manufacturer nutrition fact labels
Figure 2c is an example that shows all study data for a single participant over a 2-day period, fused into a single visualization. This visualization helped the study administrators to visually inspect the data and ensure that data was properly captured and lined up. On the basis of this visual inspection, we observed that some of the CGMs malfunctioned with consistent lack of signal; or in some cases, the events captured within the smartphone app were out of sync with the glucose curves, which indicated that the participant did not capture the data as instructed (at the point of meal consumption). In either of these cases, we discarded the respective meal data.
Meal data was pre-processed as follows. After discarding meals that were clearly from malfunctioning CGMs or from erroneous data capture, we ended up with 27,630 total meals, with 18,208 provided meals and 9422 free meals. All provided meals were at least 2.5 h apart (participants were instructed). Free meals that were within 30 min were merged, and those within 90 min of each other discarded. After all of this pre-processing, the final dataset included approximately 50 meals from each participant.
Postprandial Glycemic Response
We used incremental area under the curve (iAUC) to quantify glycemic response, which is a standard assessment of PPGR [22, 23]. We define this as the integrated area under the CGM curve over 120 min, relative to the baseline CGM measurement at one timepoint before the meal (i.e., the area above baseline and below the glycemic response curve). This measurement can be negative as a result of a decline in blood sugar level below the baseline, especially after activity, and due to noise from the CGM device.
Microbiome Features
Participant stool samples were processed using our metatranscriptomic method [14] to yield raw microbiome data features including KOs [11] and microbial taxonomy. Our cloud-based bioinformatic pipeline performs both read QC/trimming and host read filtering, ahead of alignments and classifications. Following this, classification occurs at three taxonomic ranks (strain, species, and genus) through sequence alignment to a custom database of more than 110,000 genomes. Functional assignments (KOs) are obtained through alignment to the IGC [24] and KEGG databases. For the samples provided by the 550 participants in this study, 6587 unique microbial KOs were detected, with a mean of 2941.9 per sample (SD 541.9); and 1047 unique species were detected, with a mean of 122.7 (SD 40.5).
Collections of these raw microbiome molecular pathway features were aggregated into custom microbiome scores, designed to capture the functional characteristics as described in the literature [18]. For example, the microbiome balance score is an aggregate assessment of overall ratios for both active beneficial and active harmful microbes, as well as select diversity metrics. This score is binary with a value of “Low” or “Normal”. All microbiome scores are generated by taking metatranscriptomic data as input, and applying an expert-designed scoring algorithm developed at Viome [18] to derive an overall activity level.
Metabolic and signaling pathway activities are scored using expression levels of genes, encoding specific microbial molecular pathway functions (primarily KEGG ortholog mappings), compared with a reference cohort of samples supplied by Viome customers. Scores measure the quantity and expression levels of specific KEGG KOs, selected owing to their specific directional enzymatic roles, pathway topology, or significance in the functional literature. The score values included in this study are increased when a greater number of key genes are expressed, and when their expression increases.
Statistical Analyses
Linear Multilevel Mixed-Effects Model Development
Fixed-effect predictors included nutrient characteristics of meals, anthropometric predictors, contextual factors, and microbiome features. Nutrient characteristics were reported once per meal in terms of absolute grams of net carbohydrates, fiber, protein, and fat. Anthropometric factors included BMI and age, as well as the interactions between each of these and net carbohydrates of meals. Contextual factors included sleep and activity levels, as well as the interactions between each of these and net carbohydrates of meals. Microbiome features included expert-designed scores, such as abundance of tyrosine- or fructose-metabolizing taxa, as well as activity for metabolic activity related to fucose, indoleacetate, or glutamine. Microbiome features also included interactions between net carbohydrates of meals and each of tyrosine metabolizers, fucose metabolism, and indoleacetate production.
The predictive model was incrementally developed using the lme4 package in R [25]. In several passes, the model was layered with (1) carbohydrates and anthropometric predictors; (2) carbohydrate interactions, additional nutrients, and contextual factors; and (3) expert-designed microbiome features. After each round, likelihood-ratio tests were used to assess whether contributions of features were significant. Candidate microbiome features were introduced to the model based on t test (binary) or regression (continuous) analyses, depending on the nature of each feature.
Gradient Boosting Machine Development
The final model was further advanced by improving feature selection and feature importance using the python package XGBoost [26]. As part of this process, additional meals, nutrients, and other features were incorporated into the model. Additions included (1) slope of CGM readings in 90 min leading up to a meal; (2) proportions for each macronutrient; (3) enhanced granularity of carbohydrates (i.e., starch and sugar abundance); (4) enhanced granularity of fats (i.e., monounsaturated fat and polyunsaturated fat abundance); (5) vitamin C and calorie abundance; and (6) heavy physical activity and sleep status up to 6 h before a meal and over the duration of the PPGR; (7) KOs selected with univariate linear regression with p value of the coefficient less than 0.01. Model improvement was judged by comparing R and R2 values after averaging across five random train/test splits of the data with nested cross validation.
Results
Meal Effects
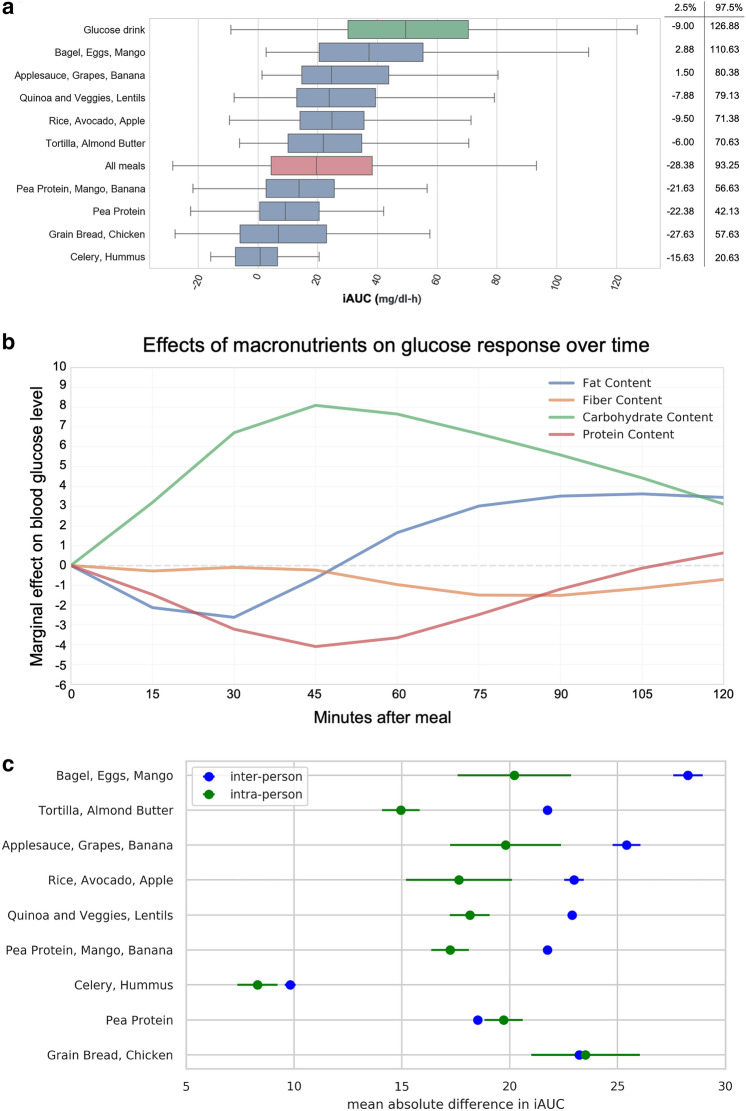
The iAUC values for all meals, the repeated meals from Table 3, and the glucose drink can be found in Fig. 3a. Figure 3b was constructed after modeling iAUC with standardized macronutrient values (i.e., z scores). Linear regression of iAUC on all four of the standardized macronutrients was performed at each timepoint after the meal. The plot shows the learned weights for each standardized macronutrient at each timepoint, and this reveals the magnitude and time-course of the macronutrient effects. Meals with more carbohydrates led to increased postprandial glucose (PPG), peaking 45 min after the meal, while meals with more fiber led to a diminished and delayed PPG. Protein and fat suppressed and delayed the response.
Fig. 3.
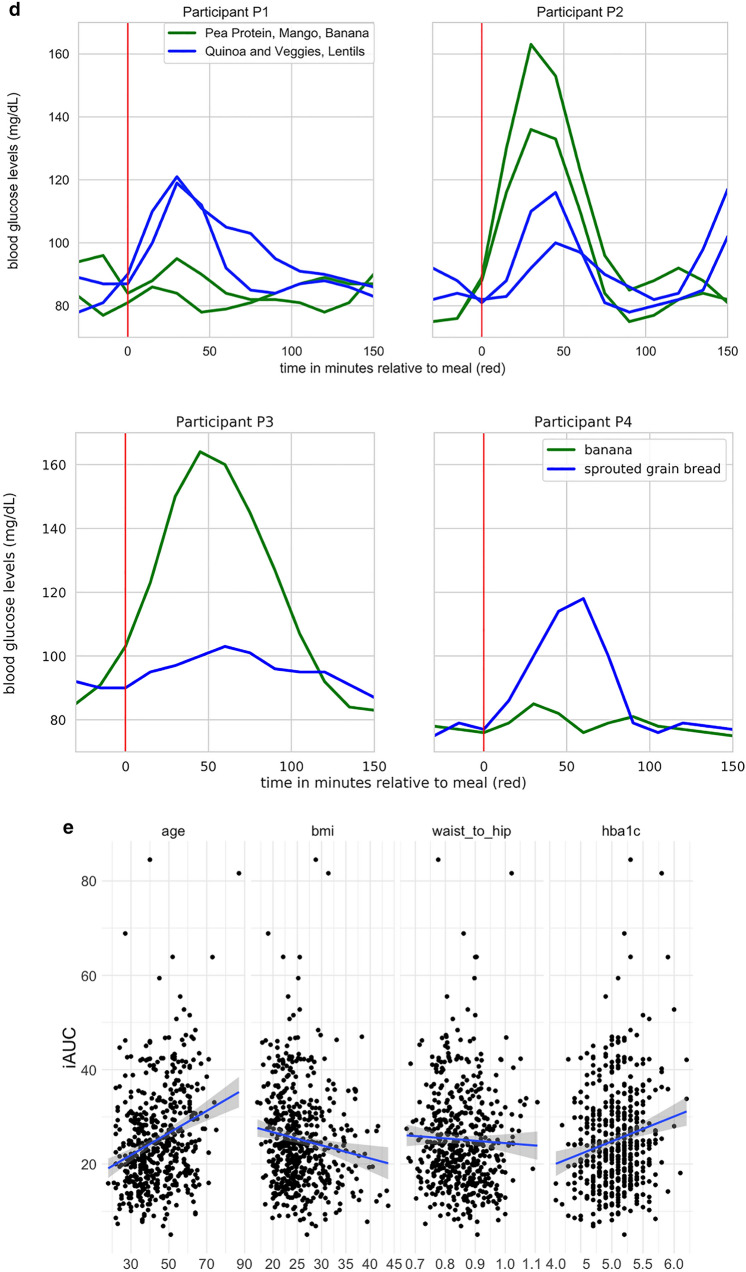
Overview of postprandial glycemic response data. a iAUC values (in mg/dl-h) for all meals, provided and free (red bar); the 9 repeated meals from Table 3, (blue bars); and the glucose drink (green bar). Boxplots show the interquartile range; whiskers cover the middle 95th percentile. b Marginal effect of macronutrients on glucose response over time, across all meals, across all participants. Each timepoint is a linear regression of iAUC on all four standardized macronutrients. c Inter- and intra-person variability for 9 repeated provided meals. Inter-person variability represents differences across multiple people. Intra-person variability represents differences for a single person between the two times each meal was consumed. X axis is the mean absolute difference in iAUC. Points indicate the mean absolute difference in response between two consumptions of the meal by one person (green), and mean absolute difference in response between all pairs of different people (blue). Bars indicate standard error. Y axis is in descending order of difference between inter-person and intra-person variability. d Examples of individual variation in glycemic response. Participants P1 and P2: Ingestion of two of the repeated meals (blue and green lines) result in opposite blood glucose response in two participants (top). Participants P3 & P4: Ingestion of two free meals results in opposite blood glucose response in two participants (bottom). e Relationships between anthropometric characteristics and per-participant average iAUC across all meals (provided free)
Figure 3c illustrates the intra-person variability in responses for meals that were repeated within the provided diet (Table 3). Intra-person variability is the difference in a participant’s response to a single meal when eaten on two occasions.
Predictive Model Development and Evaluation
In this section we first present a linear multilevel mixed-effects or hierarchical model [27] of PPGRs based on the data described above. The linear model allows us to provide a concise description of the relationships between nutrients, anthropometrics, microbiome activity, and PPGR. Additionally, it allows us to derive significance statistics, testing whether each predictor is relevant to the determination of the PPGR.
Importantly, the inclusion of random effects captures individual variation in PPGR due to unobserved factors that may affect the outcome (and these include unknown properties of the individual, provided meals, free meals, or measurement devices). Inclusion of random effects is essential for conservative hypothesis testing of both the relevance and magnitude of our fixed effects, especially in a repeated-measures design where each person and each meal provides many data points. Our experimental design calls for a multilevel model because of this repetition; each PPGR observation is at a lower level, nested within one person and within one meal (each is a higher level), in a crossed or fully factorial experimental design (see Fig. 4a). For example, without the multilevel model, we could not conclusively test the importance of any variable that is constant across all data taken from one person, such as microbiome features.
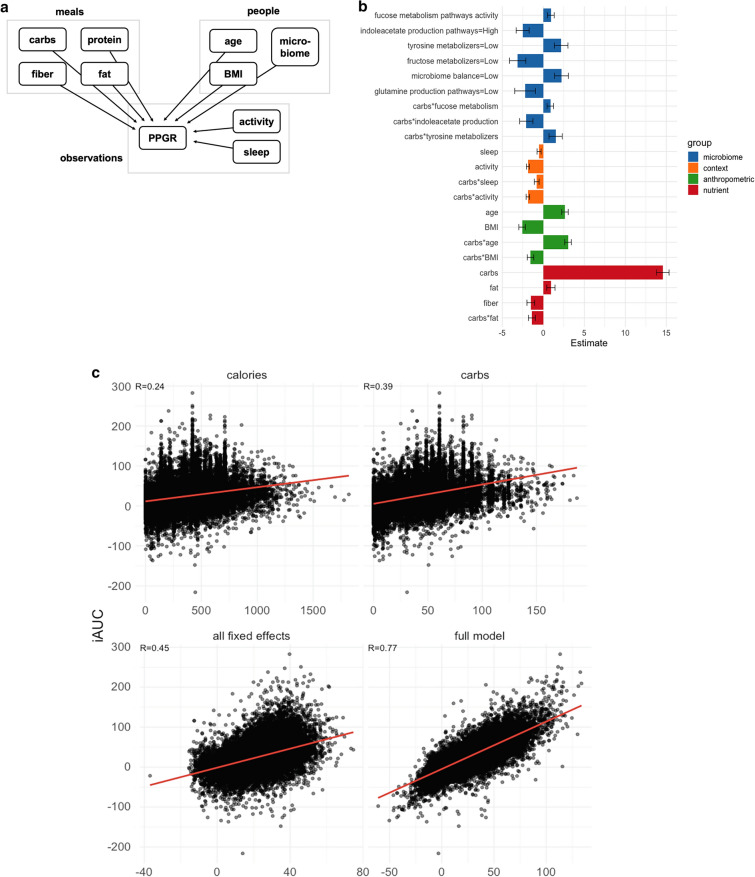
Fig. 4.
Models for predicting PPGR. a Nesting of predictors in our repeated-measures crossed experimental design. Plates (boxes) indicate repetition: e.g., there is one measurement of carbs, protein, fiber, fat for each meal. Arrows indicate the possibility of dependence: here, PPGR is estimated as a function of all other variables. b Fixed-effect estimates for all predictors included in the final model. Continuous predictors are standardized (mean is zero, units are standard deviations), meaning the expected response changes by the value of the coefficient when the predictor changes by one standard deviation. All microbiome scores except fucose are binary scores, meaning the coefficient is the difference in expected response between the two levels of the score. Error bars show standard error of the estimate. c Actual glycemic response (iAUC) against (a) calories, (b) carbs, (c) predictions from the model including all fixed effects (standard linear regression), and (d) the fit of the full mixed-effects model. d (left) Model predictions as a function of carbs and age, holding all other predictors at the baseline. (right) Model predictions as a function of carbs for 10 randomly sampled people, taking into account all person-level fixed and random effects and holding all other nutrient and context variables at the baseline. Two users are highlighted (red and blue) to draw attention to the flip in their predicted response to the two example meals A and B annotated in yellow. e Actual vs predicted iAUC for a gradient boosting machine (GBM) model. Performance shown is on the best test fold across 5 random splits, with data from 82% of the users used to train and 18% held out for evaluation
Model development was performed using the lme4 package in R [25]. The model was built incrementally. In the first pass, we determined appropriate transformations for the nutritional and anthropometric variables by visualizing the relationship between each predictor and the iAUC. The effect of carbohydrates appeared to be well described by the square root transformation, while other predictors were left untransformed. We then fitted a model of these nutritional and anthropometric predictors as fixed effects, with random intercepts and slopes, allowing the nutritional effects to vary between people and the anthropometric effects to vary between meals. Likelihood ratio tests showed no evidence for any random slopes of anthropometrics by meal (i.e., responses to all meals are similarly influenced by the participant’s age, BMI, etc.).
The largest effects were associated with carbs. Thus, in the second pass, we introduced interactions between carbs and all other nutrients as fixed effects, with random slopes by participant. On the basis of visual inspection of their relationship with iAUC, we also introduced two additional fixed effects for response-level “context” measurements: minutes of activity during the 2 h following the meal and minutes of sleep during the 3 h before the meal. We removed effects that were not significant, as determined by likelihood ratio test at p < 0.05.
In the third pass, we introduced additional fixed effects for our microbiome features, which included a number of expert-designed scores measuring the activity of crucial metabolic pathways and functions (see “Discussion” section). A candidate set of scores to introduce to the model were identified by predicting participants’ average iAUC from each score using t test (binary) or regression (continuous). There were 15 scores with a significant association which were tested in the full model. After controlling for all other predictors, six scores remained significant or marginally significant.
Fixed-effect coefficients from the final model are displayed in Fig. 4b. Positive coefficients indicate a greater predicted PPGR. All predictors were significant by a likelihood-ratio test at p < 0.05, except for glutamine production pathways (p = 0.08) and the interaction between tyrosine metabolizers and carbs (p = 0.06) which are kept in this model as suggestive, and these do not substantially affect the estimation of other coefficients.
Several of the significant predictors were microbiome scores. One of the scores, named microbiome balance, is an aggregate assessment of the microbiome, quantifying overall beneficial and harmful activities (based on the literature), as well as select diversity metrics. When it was “Low,” this binary score showed a negative association with PPGR. Two other scores (fucose metabolism and indoleacetate production) were quantified from overall activity levels of a given set of microbial pathways.
The fucose metabolism score considers the expression levels of all enzymes (encoded by transcripts) that contribute to processing and catabolic conversions of fucose—a glycan that microbes may obtain from the host’s gut lining or from food components. This quantitative score showed a positive association with PPGR.
The indoleacetate production score considers the expression levels of all the enzymes (encoded by transcripts) that contribute to production of indole-acetate (or indole acetic acid, IAA). When it was “High,” this binary score showed a negative association with PPGR.
The glutamine score was derived with the same approach as the indoleacetate score, and it considers the expression levels of enzymes (encoded by transcripts) that contribute to the production of glutamate (or glutamic acid). The glutamine production score was marginally significant, and when it was “Low,” this binary score implied a higher PPGR.
Microbial scores for tyrosine and fructose metabolizers are based on categorical functional bins of active microbes known to metabolize tyrosine and fructose, respectively. When “Low,” the tyrosine metabolizer score was directly related to elevated PPGR. In contrast, the fructose metabolizer score showed an inverse relationship, such that a “Low” score indicated elevated PPGR.
Figure 4c compares different approaches to predicting iAUC. The first two plots show single predictor models (calories or carbs). We also present predictions from the fixed-effect part of our model after zeroing out certain components. Using all nutrient predictors achieves a similar outcome as using the square root of carbs alone (both R = 0.41, not pictured). Including the microbiome features yields a small, but significant, improvement in fit (R = 0.42, not pictured), and adding the other fixed-effect predictors (age, BMI, sleep, activity, microbiome) improves the fit further (R = 0.45, Fig. 4c bottom-left). Finally, the full model with random effects (best linear unbiased predictions) fits the data very well (R = 0.77, Fig. 4c bottom-right).
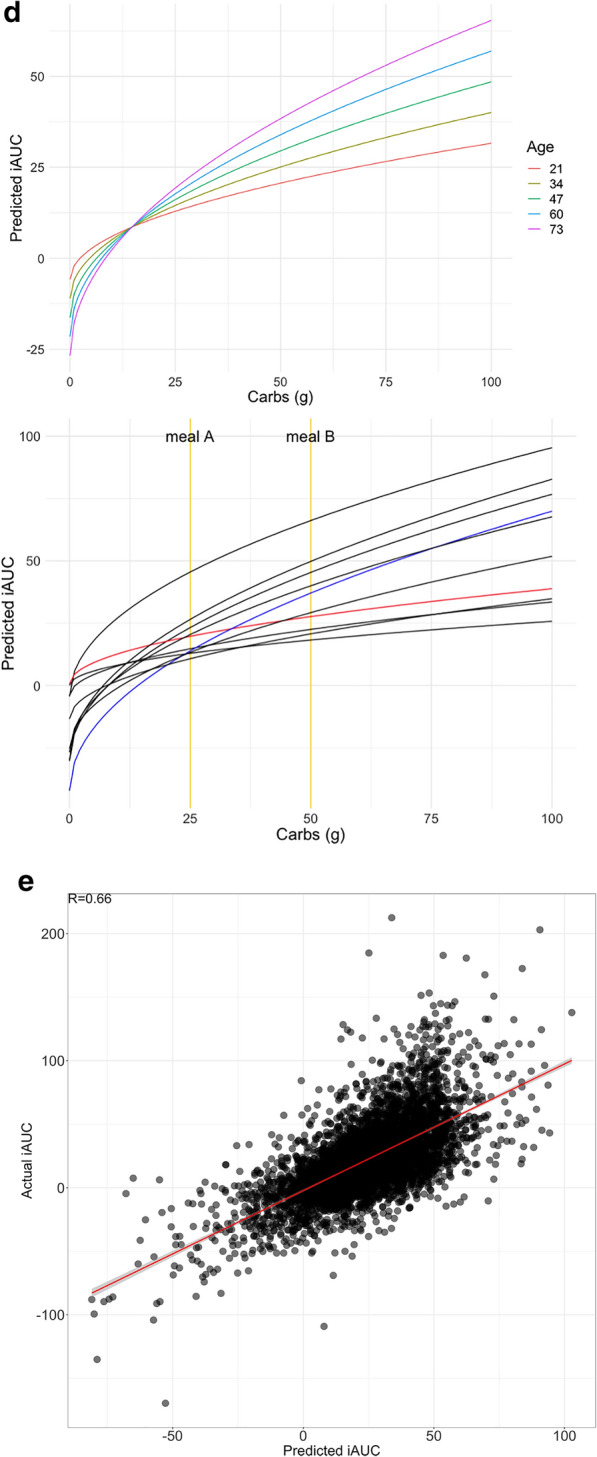
Figure 4d (top) shows the influence of age on the relationship between carbs and glycemic response. Figure 4d (bottom) illustrates the extent of individual differences in this relationship, taking into account all predictors. Red and blue lines highlight two hypothetical meals (A and B; similar to repeat meals from Table 3 with carbs approximately 25 g and 50 g, respectively). These illustrate that two users can have the opposite glycemic responses (respectively, iAUC of 14 and 20 vs iAUC of 37 and 27) due to the crossover of their iAUC response lines between the two meals—this effect can be seen in Fig. 3d using raw data for two sets of meals.
A Model Optimized for Prediction Accuracy Over Explanation
As described above, mixed-effects linear models are valuable, because they are easily interpreted and allow statements about the statistical significance of predictors. In this section, we present another model for the same data which does not offer these benefits, but it achieves greater predictive accuracy due to its richer modeling framework. This is a gradient boosting machine [26], and it is built on the same predictors already discussed, along with a number of additional features. These features encompass the microbiome (individual organisms and gene transcript levels), nutrients (weight of meal; subtypes of carbs and fats; micronutrients; specific compounds like caffeine and alcohol), and context (more detailed representations of sleep and activity). Following Zeevi et al. [8], we also add two further predictors which encode prior blood glucose levels: the CGM reading immediately before the meal, and the linear slope of CGM readings over the previous 90 min. After removing predictors that exhibited low variance, were highly correlated with each other, or were not correlated with the outcome, a total of 1446 were included in the model.
Training data was from 82% of users, while test data was from the remaining 18% of users who were held-out for evaluation. Hyperparameters which controlled learning rate, number of trees, and tree depth were estimated using cross-validation on the training set. Averaging across five such random train/test splits of the data, the model achieves R = 0.80 (R2 = 0.64) with training data, and R = 0.64 (R2 = 0.40) on held-out test data. Performance on training and test data is shown in Fig. 4e.
Discussion
We set out to study PPGR variation based on individual differences, focusing on differences in gut microbiome activity, and we were the first to accomplish this using the metatranscriptome. We made a few key design choices for the study, including (a) 14 days of monitoring, (b) multiple diet types—omnivore and vegetarian/gluten-free, and (c) a large proportion (66%) of provided (pre-designed) meals.
The number of provided meals (104) is considerably larger in our study as compared to previous studies (e.g., [8, 9] had only four standardized meals). This diet design allowed us to collect more precise readings of consumed meals rather than entirely depending on the smartphone diet tracking app. This choice also allowed us to quantify individual PPGR differences between people in response to the same food, reducing the risk that observed differences reflect nutritional differences between meals. Secondly, as shown in Fig. 2a, b, our diet design also provided broad coverage across the space of diverse macronutrient proportions (carbs, fiber, protein, fat), with the intention of teasing out the independent and interacting effects of the macronutrients. And finally, our diet design allowed us to retain greater control of the meals, since we also wanted to study the effect of multiple diet types: omnivore and vegetarian/gluten-free (this is ongoing work, not reported here).
Our data shows that accounting for individual differences is crucial in providing a full description of PPGR. This is evident, for example, in the data presented for our nine repeat meals (Table 3); each of these was a combination of food staples consumed at least twice by all participants. While intra-person variability for a given meal is substantial (Fig. 3c, green), this variability is small relative to the inter-person variability for the same meals (blue). We can conclude that while many factors affect PPGR, some of these factors are individual differences which must be accounted for by differences between people and their lifestyles, not properties of the meal alone. The two meals where intra-person variability is close to inter-person variability are meals that contain little to no carbohydrates.
Relationships Between iAUC and Phenotype and Food Features
We see the expected relationship between age and iAUC in Fig. 3e (first panel). The relationship we report between iAUC and BMI/waist-to-hip ratio (Fig. 3e second and third panels) is the opposite of previous findings [8]. We hypothesize that this may reflect the self-reported good health and high exercise rate within this study population, as determined from the questionnaire completed at participation enrollment. We see the expected increase in average iAUC with HbA1c in our study population (Fig. 3e, fourth panel). However, in our analysis there is no significant effect of HbA1c on iAUC after controlling for other predictors. This may be because the study population was selected for HbA1c in the normal range (< 6.5).
As expected, the bulk of variation in the PPGR is explained by the amount of carbs ingested, and by interactions with fat content in food and other factors that modulate the effect of carbs. Increased fiber resulted in overall lower PPGR, and while increased fat had little marginal effect by itself, it interacted with carbs to suppress the effect of ingested carbs on the PPGR. The time-course of this effect (Fig. 3b) suggests this may happen because fat and protein flatten and delay the digestion of carbs, pushing some of the PPGR out of the 2-h window considered here [28, 29]. Protein has a numerically negative effect on iAUC, but this was not significant after controlling for other predictors, so it was removed from the final model. Older people generally exhibited a higher PPGR as well as a higher PPGR per unit of carbs ingested (Fig. 4d). Activity after meal consumption, as well as sleep immediately before eating, both resulted in lower PPGR to carbs, which is consistent with known metabolic effects of circadian rhythms [30].
The fact that PPGR is better predicted by the square root of carbs than untransformed carbs is reminiscent of a standard model of gastric emptying in which the volume of food passing from the stomach per unit of time is linear in the square root of its volume [31].
Relationships Between iAUC and Microbiome Features
Microbiome features that were significantly related to prediction of PPGR include microbiome balance, fucose metabolism pathways, fructose metabolizers, tyrosine metabolizers (marginal), indoleacetate production pathways, and glutamine production pathways (marginal). Of these, fucose, indoleacetate, and tyrosine (marginal) scores interact with carbs, indicating the microbiome or correlated properties that affect the processing of carbs, and they also lead to an overall difference in baseline blood sugar.
The microbiome balance score was one of the significant features in predicting PPGR. Microbiome balance scores that were “Low” usually resulted from either an imbalance of relative activities of beneficial vs. harmful microbes or from lower quantity and diversity of microbial organisms. The relationship between a suboptimal gut microbiome and higher PPGR is in line with the current literature [32–34] implicating the role of gut health in glycemic regulation.
The fucose metabolism pathway score showed a direct relationship with PPGR. Fucose is a sugar molecule that various microbial organisms can use as an energy source [35]. When other carbohydrate sources are not available, gut microbiota can switch to using fucose that can be obtained from the host’s gut mucosal lining. This process is often carried out by microbes known as mucin degraders, such as certain species of Ruminococcus [36]. We therefore hypothesize that higher fucose consumption activity, as reflected by the fucose metabolism pathway score, may be associated with microbiomes of individuals who are either more likely to fast or whose body and internal ecosystem resemble the conditions of fasting or calorie deprivation. This may explain its association with higher PPGR. More research is needed to establish relationships between microbial metabolism of gut sugars and the host’s tendency to show higher glucose spikes in the blood after meals.
The indoleacetate production pathways score incorporates the role and significance of expressed genes in the context of microbial indoleacetate production. The algorithm considers all pathway contributions to the production of indole acetic acids (IAAs) and scores them using gene expression as input data. A “High” indoleacetate score correlates to lower PPGR. This is consistent with known anti-inflammatory properties of IAA [37, 38]. In addition, both inflammatory activities in the gut and systemic low-grade inflammation are implicated in the development of type 2 diabetes and other metabolic disorders [39, 40]. Moreover, IAAs have been directly implicated in PPGR, and some findings suggest a hypoglycemic action of indole-3-acetic acid in diabetes mellitus [41].
Beneficial products of protein fermentation include indoles and indoleacetate, though especially products from tryptophan metabolism [42]. In one study, intraperitoneal administration of indole-3-propionic acid, indole-3-butyric acid, and indole-3-acetic acid was associated with hypogIycemia in normal and alloxandiabetic mice, while l-tryptophan and kynurenic acid had no effect [43].
The interpretation we offer here is dependent on the individual’s microbiome function. If a given person’s microbiota mainly exhibits conversion of tryptophan to beneficial indoles and indole-acetate molecules (those capable of reducing inflammation and glycemic effects), then it may benefit such people to consume tryptophan (in the form of food or supplement). On the other hand, if the microbiota mainly exhibits conversion of tryptophan to more of the pro-inflammatory triggers, then consuming tryptophan may not be suitable to mitigate glycemic effects or inflammation in general.
Tyrosine metabolizer and fructose metabolizer scores categorically bin active microbes into groups, according to functional characteristics. We have observed that these groups tend to reflect the host’s habitual diet. Hence, “Low” tyrosine metabolizers may suggest a diet that is low on protein sources of tyrosine. We also hypothesize that the inverse relationship between the fructose metabolizer score and PPGR may be due to a diet that is low in fructose or carbohydrates that serve as fructose precursors. It is not yet clear how a diet that is rich in fructose or deficient in tyrosine may influence PPGR, and more studies are needed.
When it was “Low,” the glutamine production pathways score showed a direct relationship with higher PPGR. Microbial glutamine production has not been directly linked to PPGR in humans. However, glutamine is considered an important nutrient for gut health and has been included in various supplements used by clinical healthcare practitioners to prevent or heal “leaky gut” [44, 45]. More research is needed to understand the molecular mechanisms that may be responsible for higher PPGR in individuals with low microbial glutamine production activity in the gut.
The microbiome features revealed by our glycemic response model may influence PPGR directly or indirectly. Although it is challenging to delineate causal mechanisms, there may be functional patterns that connect the significant scores with gut health, intestinal barrier integrity, and inflammation. Inflammation and stress response may be implicated in elevation of blood glucose (either due to cortisol pathway or other mechanisms). Knowing which foods elicit higher PPGR can offer valuable guidance in diet selection. However, to affect the root cause of PPGR, the specific mechanisms connecting nutrients to the gut microbiome as well as to inflammation and PPGR will need to be taken into account. We seek to confirm and validate these mechanisms. An understanding of which microbiome features are significant will pave the path to precise personalization of food and supplement recommendations.
Modeling Methods and Model Evolution
The mixed-effects linear model presented first in the previous section was deliberately chosen to better understand the incremental effects of the significant features, especially the functional gut microbiome activity features. We are not aware of any prior literature that demonstrates the statistical significance of the microbiome in the context of a predictive model for PPGR. Prior studies [8] using only ensemble methods, like gradient boosting machine models, represent the state of the art in accurate prediction of PPGR (which we also show in Fig. 4e). These models suffer from difficulty of interpretation, including determination of which features significantly contribute to the predicted outcome.
We are in the process of increasing the generalizability of our findings by collecting further data from underrepresented subpopulations such as people with pre-diabetes, people reporting poor overall health, and older participants. With the goal of continuous improvement, we will rebuild and revalidate our model on the basis of this new expanded data. The current paper provides a first snapshot of the collected data, and we will use the additional data to consolidate the current model, as well as potentially surface new relevant predictors. Future steps include validating the model by employing an unseen cohort. Finally, we will also perform a blinded, randomized-controlled dietary intervention based on this predictive model, in order to look for improvements in PPGR as well as alterations to the gut microbiota.
Conclusions
Most significantly, this paper makes the following contributions:
Demonstrates for the first time that metatranscriptomic activity of the gut microbiome contributes to individual variation in glycemic response among adults.
Suggests new microbial features that may help uncover molecular mechanisms of glycemic control.
All features included in our mixed-effects linear regression model are significant to individual glycemic response, where fixed effects represent the measured properties and random effects are included to account for further variation.
Our predictive accuracy of individual glycemic response was improved with a gradient boosting machine, which allowed us to incorporate additional raw data and maximize our accuracy.
Demonstrates that PPGR is driven by the properties of an individual in addition to the food’s macronutrient content, as measured with 104 unique pre-designed meals within omnivore and vegetarian/gluten-free diet types and within a multiethnic population.
These data will be used in the future to identify additional specific microbial pathways that affect the glycemic response which could be targeted with therapeutics. Depending on the observed effects, the therapeutics could include small molecule inhibitors, small molecule supplements, phages, vaccines, or probiotics, for example.
Acknowledgements
We thank all participants in the study. We are grateful to the Viome study team—(alphabetically by first name) Ashleigh Winter, Barry Zhao, Braidon Waggoner, Brendan Zapp, Calita Quesada, Erica Castillo, Gabby Hands, James Horne, Kelly Nebgen, Marcela Walker, Miranda Intrator, Pete Lichtman, Ryan Toma, Sabria Hankins, Tristen Hill, and YehShen McShan—for handling the considerable logistics during the data collection and annotation phases. We thank Matteo Rinaldi and Sha Yu Han for initial data exploration and analyses; Subha Krishnan for curating the microbiome pathway literature and features; and Debra Heald and Samika Savanur for their clinical input.
Funding
This research and Rapid Service Fee were funded by Viome Inc.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published. The authors did not receive any medical writing or editorial assistance during the writing of this article.
Authorship Contributions
G.B., H.M., and M.V. designed the study. A.S. managed the study logistics including sample and clinical data collection. M.V. performed the lab analysis. H.L. developed the meal plans. E.P. organized and analyzed the nutrient data, and contributed to both the clinical data collection pipeline and the writing. Y.C. validated the statistical outcomes and contributed to writing. N.K. developed the bioinformatics pipeline. H.T., S.G., M.G., V.G., and I.S. performed the analyses, developed the machine learning models, generated visualizations, and contributed to writing. A.P. and H.M. interpreted the features and contributed to writing. H.T. and G.B. guided the analysis and led the manuscript writing.
Prior Presentation
This manuscript has been presented earlier as a preprint at the following link: https://www.biorxiv.org/content/10.1101/641019v4.
Disclosures
While contributing to this study, all authors were employees of Viome Inc, a commercial for-profit company. Hal Tily, Eric Patridge, Ying Cai, Vishakh Gopu, Stephanie Gline, Matvey Genkin, Haely Lindau, Alisson Sjue, Iordan Slavov, Ally Perlina, Niels Klitgord, Helen Messier, Momchilo Vuyisich, Guruduth Banavar were stockholders of Viome. Currently: Hal Tily is with Ōura Health; S.G. is with The New York Times; M.G. is attending The City University of New York; Haely Lindau is with Haely Lindau Nutrition; Iordan Slavov is with Cogitativo; Ally Perlina is with CureMatch; Niels Klitgord is with Monoceros Biosystems; Helen Messier is with Altum Medical.
Compliance with Ethics Guidelines
The study was designed and carried out in accordance with the ethical principles of the Declaration of Helsinki and its later amendments, and was approved by a federally accredited institutional review board (IRB; 00011543). All samples and metadata were obtained from human subjects at least 18 years old and residing in the USA at the time of participation. All study participants consented to participating in the study.
Data Availability
The datasets generated during and/or analyzed during the current study are not publicly available due to the fact that all participants in this study are Viome customers, and Viome is bound by the privacy policy with customers, available at https://www.viome.com/privacy-policy. According to this policy, Viome cannot automatically share individual-level sample data or individual-level meta data of any customer with third parties including publishers of scientific manuscripts. However, if the data is specifically requested via https://www.viome.com/vri/data-access, we may be able to share a summary and portions of the data.
References
- 1.Centers for Disease Control and Prevention. National diabetes statistics report, 2020. Atlanta, GA: Centers for Disease Control and Prevention, US Department of Health and Human Services. 2020; p 12–5. https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf. Accesed 1 Sept 2021.
- 2.Korsmo-Haugen HK, Brurberg KG, Mann J, Aas AM. Carbohydrate quantity in the dietary management of type 2 diabetes: a systematic review and meta-analysis. Diabetes Obes Metab. 2019;21(1):15–27. doi: 10.1111/dom.13499. [DOI] [PubMed] [Google Scholar]
- 3.Livesey G, Taylor R, Hulshof T, Howlett J. Glycemic response and health—a systematic review and meta-analysis: relations between dietary glycemic properties and health outcomes. Am J Clin Nutr. 2008;87(1):258S–S268. doi: 10.1093/ajcn/87.1.258S. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig DS, Hu FB, Tappy L, Brand-Miller J. Dietary carbohydrates: role of quality and quantity in chronic disease. BMJ. 2018;13:361. doi: 10.1136/bmj.k2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi X, Al-Ghazzewi FH, Tester RF. Dietary fiber, gastric emptying, and carbohydrate digestion: a mini-review. Starch. 2018;70(9–10):1700346. doi: 10.1002/star.201700346. [DOI] [Google Scholar]
- 6.Kris-Etherton PM, Petersen KS, Velarde G, et al. Barriers, opportunities, and challenges in addressing disparities in diet-related cardiovascular disease in the United States. J Am Heart Assoc. 2020;9(7):e014433. doi: 10.1161/JAHA.119.014433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker RJ, Garacci E, Ozieh M, Egede LE. Food insecurity and glycemic control in individuals with diagnosed and undiagnosed diabetes in the United States. Prim Care Diabetes. 2021;15(5):813–18. [DOI] [PMC free article] [PubMed]
- 8.Zeevi D, Korem T, Zmora N, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Mendes-Soares H, Raveh-Sadka T, Azulay S, et al. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Netw Open. 2019;2(2):e1881102. doi: 10.1001/jamanetworkopen.2018.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suez J, Shapiro H, Elinav E. Role of the microbiome in the normal and aberrant glycemic response. Clin Nutr Exp. 2016;1(6):59–73. doi: 10.1016/j.yclnex.2016.01.001. [DOI] [Google Scholar]
- 11.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. Available from http://www.kegg.jp/ or http://www.genome.jp/kegg/. Accessed 1 Sept 2021. [DOI] [PMC free article] [PubMed]
- 12.Knight R, Vrbanac A, Taylor BC, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 13.Bervoets I, Charlier D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev. 2019;43(3):304–339. doi: 10.1093/femsre/fuz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatch A, Horne J, Toma R, et al. A robust metatranscriptomic technology for population-scale studies of diet, gut microbiome, and human health. Int J Genom. 2019;1:2019. doi: 10.31219/osf.io/8vd6x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosalbes MJ, Durbán A, Pignatelli M, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 2011;6(3):e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashiardes S, Zilberman-Schapira G, Elinav E. Use of metatranscriptomics in microbiome research. Bioinform Biol Insights. 2016;10:BBI-S34610. doi: 10.4137/BBI.S34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, Wurtzel O, Singh K, et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods. 2010;7(10):807–812. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- 18.Connell J, Toma R, Ho C, et al. Evidence-based precision nutrition improves clinical outcomes by analyzing human and microbial molecular data with artificial intelligence. bioRxiv. 2021 doi: 10.1101/2021.04.24.441290. [DOI] [Google Scholar]
- 19.Abbott. Freestyle Libre. 2016. https://www.accessdata.fda.gov/cdrh_docs/pdf15/p150021c.pdf. Accessed 1 Sept 2021.
- 20.Karlsson M. Bitesnap - photo food journal, June 2020. https://getbitesnap.com/. Accessed 1 Sept 2021.
- 21.Lupton JR, Brooks JA, Butte NF, Caballero B, Flatt JP, Fried SK. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein, and amino acids. Washington, DC: National Academy Press; 2002. pp. 589–768. [Google Scholar]
- 22.Cheng JT, Li Y, Cheng JT. Merit of incremental area under the curve (iAUC) in nutrition is varied in pharmacological assay—a review. Clin J Dia Care Control. 2018;1(2):180008. [Google Scholar]
- 23.Wolever TM, Jenkins DJ. The use of the glycemic index in predicting the blood glucose response to mixed meals. Am J Clin Nutr. 1986;43(1):167–172. doi: 10.1093/ajcn/43.1.167. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 25.Bates D, Maechler M, Bolker B, et al. Package ‘lme4’. Convergence. 2015;12(1):437. [Google Scholar]
- 26.Chen T, Guestrin C. XGBoost: a Scalable Tree Boosting System. In: KDD’16: Proceedings of the 22nd ACM SIGKDD International Conference on knowledge discovery and data mining, August 2016, pp 785–794.
- 27.Gelman A, Hill J. Data analysis using regression and hierarchical/multilevel models. New York: Cambridge; 2007. [Google Scholar]
- 28.Franz MJ. Protein: metabolism and effect on blood glucose levels. Diabetes Educ. 1997;23(6):643–651. doi: 10.1177/014572179702300603. [DOI] [PubMed] [Google Scholar]
- 29.Wakhloo AK, Beyer J, Diederich C, Schulz G. Effect of dietary fat on blood sugar levels and insulin consumption after intake of various carbohydrate carriers in type I diabetics on the artificial pancreas. Dtsch Med Wochenschr. 1984;109(42):1589–94. doi: 10.1055/s-2008-1069418. [DOI] [PubMed] [Google Scholar]
- 30.Van Cauter E, Polonsky KS, Scheen AJ. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev. 1997;18(5):716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins A. The pattern of gastric emptying: a new view of old results. J Physiol. 1966;182(1):144–149. doi: 10.1113/jphysiol.1966.sp007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 33.Larsen N, Vogensen FK, Van Den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 35.Chen YM, Zhu Y, Lin EC. NAD-linked aldehyde dehydrogenase for aerobic utilization of l-fucose and l-rhamnose by Escherichia coli. J Bacteriol. 1987;169(7):3289–3294. doi: 10.1128/jb.169.7.3289-3294.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crost EH, Tailford LE, Monestier M, et al. The mucin-degradation strategy of Ruminococcus gnavus: the importance of intramolecular trans-sialidases. Gut microbes. 2016;7(4):302–312. doi: 10.1080/19490976.2016.1186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan S, Ding Y, Saedi N, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018;23(4):1099–1111. doi: 10.1016/j.celrep.2018.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitfield-Cargile CM, Cohen ND, Chapkin RS, et al. The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes. 2016;7(3):246–261. doi: 10.1080/19490976.2016.1156827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez LL, Garrie K, Turner MD. Type 2 diabetes–an autoinflammatory disease driven by metabolic stress. Biochim Biophys Acta Mol Basis Dis. 2018;1864(11):3805–23. doi: 10.1016/j.bbadis.2018.08.034. [DOI] [PubMed] [Google Scholar]
- 40.Tuomainen M, Lindström J, Lehtonen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes. 2018;8(1):1–5. doi: 10.1038/s41387-018-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirsky IA, Diengott D. Hypoglycemic action of indole-3-acetic acid by mouth in patients with diabetes mellitus. Proc Soc Exp Biol Med. 1956;93(1):109–110. doi: 10.3181/00379727-93-22678. [DOI] [PubMed] [Google Scholar]
- 42.Russell WR, Duncan SH, Scobbie L, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57(3):523–535. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 43.Silverstein MN, Wakim KG, Bahn RC, Decker RH. Role of tryptophan metabolites in the hypoglycemia associated with neoplasia. Cancer. 1966;19(1):127–133. doi: 10.1002/1097-0142(196601)19:1<127::AID-CNCR2820190114>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Kim MH, Kim H. The roles of glutamine in the intestine and its implication in intestinal diseases. Int J Mol Sci. 2017;18(5):1051. doi: 10.3390/ijms18051051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao R, Samak G. Role of glutamine in protection of intestinal epithelial tight junctions. J Epithel Biol Pharmacol. 2012;5(Suppl 1-M7):47. doi: 10.2174/1875044301205010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available due to the fact that all participants in this study are Viome customers, and Viome is bound by the privacy policy with customers, available at https://www.viome.com/privacy-policy. According to this policy, Viome cannot automatically share individual-level sample data or individual-level meta data of any customer with third parties including publishers of scientific manuscripts. However, if the data is specifically requested via https://www.viome.com/vri/data-access, we may be able to share a summary and portions of the data.