Abstract
The inhibitor of nuclear factor-kappa B (NF-κB) kinase (IKK) is the core regulator of the NF-κB pathway against pathogenic invasion in vertebrates or invertebrates. IKKβ, -ε and -γ have pivotal roles in the Toll and immune deficiency (IMD) pathways. In this study, a homolog of IKKε (TmIKKε) was identified from Tenebrio molitor RNA sequence database and functionally characterized for its role in regulating immune signaling pathways in insects. The TmIKKε gene is characterized by two exons and one intron comprising an open reading frame (ORF) of 2,196 bp that putatively encodes a polypeptide of 731 amino acid residues. TmIKKε contains a serine/threonine protein kinases catalytic domain. Phylogenetic analysis established the close homology of TmIKKε to Tribolium castaneum IKKε (TcIKKε) and its proximity with other IKK-related kinases. The expression of TmIKKε mRNA was elevated in the gut, integument, and hemocytes of the last-instar larva and the fat body, Malpighian tubules, and testis of 5-day-old adults. TmIKKε expression was significantly induced by Escherichia coli, Staphylococcus aureus, and Candida albicans challenge in whole larvae and tissues, such as hemocytes, gut, and fat body. The knockdown of the TmIKKε messenger RNA (mRNA) expression significantly reduced the survival of the larvae against microbial challenges. Further, we investigated the induction patterns of 14 T. molitor antimicrobial peptides (AMPs) genes in TmIKKε gene-silencing model after microbial challenges. While in hemocytes, the transcriptional regulation of most AMPs was negatively regulated in the gut and fat body tissue of T. molitor, AMPs, such as TmTenecin 1, TmTenecin 4, TmDefensin, TmColeoptericin A, TmColeoptericin B, TmAttacin 1a, and TmAttacin 2, were positively regulated in TmIKKε-silenced individuals after microbial challenge. Collectively, the results implicate TmIKKε as an important factor in antimicrobial innate immune responses in T. molitor.
Keywords: Tenebrio molitor, IMD pathway, IKKε, antimicrobial peptides, RNAi
Introduction
The innate immune response represents the first line of defense in vertebrates and it is the only defense arsenal in invertebrates against microbial infections (Hoffmann et al., 1999). This is because of a lack of adaptive immune strategy in invertebrates that necessitates the over-reliance on the innate immune cascades for defense against microbial infection (Hoffmann et al., 1999; Li and Xiang, 2013). Antimicrobial peptides (AMPs) production represents one of the crucial effector mechanisms of innate immunity in insects. AMPs attribute insects most resistant to bacterial infections and shed applications as novel microbicides (Wu et al., 2018). Since the discovery of the first insect AMP called cecropin from the giant silk moth Hyalophora cecropia (Hultmark et al., 1980), over 150 AMPs have been identified, purified, and characterized from insects. Drosomycin, an antifungal peptide, and diptericin, an antibacterial peptide, have been identified and characterized in Drosophila melanogaster (Lemaitre et al., 1996; Nicolas et al., 1996). Attacin B, which is one of two attacin genes identified from Hyphantria cunea, is strongly induced by gram-positive and gram-negative bacteria (Kwon et al., 2008). Another attacin gene identified from Spodoptera exigua has antimicrobial activity against Escherichia coli DH5α strain, Pseudomonas cichorii, Bacillus subtilis, and Listeria monocytogenes (Bang et al., 2012). In the Tenebrio model, in silico analysis and induction patterns of AMP genes that include tenecin-1 (defensin family), -2 (coleoptericin family), -3 (thaumatin-like protein family), and -4 (attacin family), thaumatin-like protein (TLP)-1, and -2, Attacin-1a, -1b, and -2, Defensin and Defensin-like, Coleoptericin-A, -B, and -C, and Cecropin-2 have been studied (Kim et al., 1998, 2017; Roh et al., 2009; Chae et al., 2012; Johnston et al., 2013; Zhu et al., 2014; Noh and Jo, 2016; Jo et al., 2018; Maistrou et al., 2018; Jang et al., 2020a,b; Ali Mohammadie Kojour et al., 2021).
In insects, AMPs are induced through the activation of two key signaling cascade mechanisms – the Toll and immune deficiency (IMD) pathways. In Drosophila, the IκB kinase (DmIKK) complex, which is the major component of the IMD pathway, stimulates the activation of the nuclear factor-kappa B (NF-κB) protein Relish by phosphorylation (Georgel et al., 2001; Vidal et al., 2001; Chen et al., 2002; Choe et al., 2002, 2005; Gottar et al., 2002; Ramet et al., 2002; Cha et al., 2003; Leulier et al., 2003; Silverman et al., 2003; Kaneko et al., 2004). In addition, lipopolysaccharide (LPS) and peptidoglycan (PGN) from gram-negative bacteria stimulate the IMD pathway in Drosophila (Leulier et al., 2003; Kaneko et al., 2004). Further, IKKε in D. melanogaster not only plays a principal role in IMD regulation but also phosphorylates DIAP1 and controls janus kinase (JNK) activation and apoptosis (downstream of IMD) (Gan et al., 2021). This study demonstrated that the activation of the IMD pathway against sindbis virus (SINV) infection is highly dependent on the microbiota present in the gut (GT) of Aedes aegypti (Barletta et al., 2017). In another insect model, Bombyx mori, the expression of CecropinA1 is regulated by Relish in response to gram-negative bacteria (Hua et al., 2016). In addition, the direct functions of B. mori peptidoglycan recognition protein L1 (BmPGRP-L1) and BmIMD in the IMD pathway are suspected but not clearly identified (Zhan et al., 2018). In Plutella xylostella, IMD RNA interference (RNAi) affected the expression of the downstream genes of the IMD pathway (Lin et al., 2018). In fact, several studies have characterized the Toll and IMD pathway responses of diverse insects including Plautia stali stink bugs (Nishide et al., 2019), aphids (International Aphid Genomics Consortium, 2010), kissing bugs (Mesquita et al., 2015; Salcedo-Porras and Lowenberger, 2019), or other arthropods such as Tetranychus mites (Palmer and Jiggins, 2015; Santos-Matos et al., 2017) and shrimp (Li et al., 2019). Additionally, the crosstalk between Toll and IMD pathways in stink bugs by the RNAi experiments of Toll and IMD pathways-related genes, including PsImd, PsMyD88, PsDorsal, PsPGRP-L1a, PsPGRP-L1b, PsPGRP-L2, PsLysM, PsGNBP1 have been suggested (Nishide et al., 2019).
In the beetle Tribolium castaneum, the PGRP-LA may be a pivotal sensor of the IMD pathway for both gram-negative and gram-positive bacteria, and both PGRP-LC and -LE acts as IMD pathway-associated sensors, mainly for gram-negative bacteria (Koyama et al., 2015). Based on comparative genomic analysis, around 300 candidate defense proteins were identified and clustered depending on the immune pathway such as Toll, IMD, and JAK-STAT pathways (Zou et al., 2007). Inducible immune-related genes, including Toll, PGRP, and AMP genes such as ferritin, defensin, and others against crude LPS were identified using the suppression subtractive hybridization (SSH) method, and antifungal activity of recombinant TLP was assayed in T. castaneum (Altincicek et al., 2008).
In contrast, a comprehensive study of the IMD pathway in innate immune responses against infections by various pathogens has been partially performed in T. molitor with the functional characterization of TmPGRP-LE, TmIMD, TmIKKγ, and TmRelish (Tindwa et al., 2013; Jo et al., 2019; Ali Mohammadie Kojour et al., 2020; Keshavarz et al., 2020b,c,d; Ko et al., 2020). In this study, we identified the IκB kinase ε (IKKε) gene, one of the important components for the IMD pathway, from T. molitor RNA and DNA sequence database. We investigated the mRNA expression patterns of TmIKKε depending on different developmental stages, tissues, and microbial challenges to the host. Moreover, we investigated the effects of TmIKKε-specific knockdown on larval mortality, AMP production, and expression of NF-κB genes against various pathogens. The findings that IKKε knockdown beetles are especially susceptible to E. coli but not a gram-positive bacterium or fungus parallels a recent study wherein fruit flies lacking their major AMP genes were specifically susceptible to gram-negative bacteria, but not so much gram-positive bacteria or fungi (Hanson et al., 2019). Collectively, our data provide a better understanding of the IMD pathway in the Tenebrio innate immune response.
Materials and Methods
Insect Rearing
Tenebrio molitor larvae were reared in the dark at 27°C ± 1°C and 60% ± 5% relative humidity in an environmental chamber established in the laboratory. The reared larvae were fed an artificial diet (170 g wheat flour, 20 g roasted soy flour, 10 g protein, 100 g wheat bran, 0.5 g sorbic acid, 0.5 mL propionic acid, and 0.5 g chloramphenicol in 200 mL of distilled water, sterilized by autoclaving at 121°C for 15 min). Healthy 10th to 12th instar larvae (1.2–1.5 cm length) were used for experiments.
Preparation of Microorganisms
Three microorganisms—the gram-negative bacterium E. coli strain K12, gram-positive bacterium S. aureus strain RN4220, and the fungus C. albicans—were used for immune challenge experiments. E. coli and S. aureus were cultivated in Luria-Bertani (LB) broth (MB cell, South Korea). C. albicans suspension was prepared by culturing the fungi in Sabouraud dextrose broth (MB cell) at 37°C overnight. The microorganisms were harvested and washed twice in 1× phosphate-buffered saline (PBS) (8.0 g NaCl, 0.2 g KCl, 1.42 g Na2HPO4, 0.24 g of KH2PO4 in 1 L of distilled water, pH 7.0) and centrifuged at 1,700 × g for 10 min. The washed microorganisms were resuspended in 1× PBS and the optical density at 600 nm (OD600) of the suspension was measured using a spectrophotometer (Eppendorf, Germany). The concentration of microbial cells was adjusted to 1 × 106 cells/μL of E. coli and S. aureus and 5 × 104 cells/μL of C. albicans for the immune challenge studies. The relevant optimization of microbial concentration has been adjusted based on the previous studies (Chae et al., 2012; Jo et al., 2017; Park et al., 2019).
Identification and in silico Analysis of TmIKKε
The TmIKKε sequence was retrieved from T. molitor RNA sequencing (RNA-seq) (unpublished) and expressed sequence tag (EST) databases. Local-tblastn analysis was performed using T. castaneum IKKε amino acid sequence (EEZ99267.2) as a query. The full-length cDNA and deduced amino acid sequences of TmIKKε were determined using the blastx and blastp algorithm, respectively, on the National Center for Biotechnology Information (NCBI) website. Complementary DNA (cDNA) translation and predictions of the deduced protein were analyzed using BioPHP mini tools software (http://www.biophp.org). FGENESH eukaryotic gene prediction was used to predict the TmIKKε open reading frame (ORF) region. The domain architecture of the protein sequences was retrieved using the InterProScan domain analysis program. Representative IKKε protein sequences from other insects were obtained from GenBank and were used for multiple sequence alignments, and percentage identity analysis using Clustal X2.1 (Larkin et al., 2007). A phylogenetic tree was constructed based on the amino acid sequence alignments using the Maximum likelihood method (bootstrap trial set to 1000) with IKKε/TBK1 proteins from representative insects (Supplementary Table 1). The phylogram was analyzed using Tree Explorer view with the Molecular Evolutionary Genetics Analysis (MEGA) version 7.0 program (Kumar et al., 2016) (https://megasoftware.net).
Sample Collection and Microorganism Challenge
The TmIKKε mRNA expression was investigated in different developmental stages of T. molitor, eggs (EG), young instar larvae (YL; 10th–12th instar larvae), late-instar larvae (LL; 19th–20th instar larvae), prepupae (PP), 1- to 7-day-old pupae (P1–P7), and 1- to 5-day-old adults (A1–A5). TmIKKε mRNA expression was also measured in the different tissues that included integument (IT), hemocytes (HC), GT, fat body (FB), and Malpighian tubules (MT) dissected under a stereoscopic microscope (SMZ645, Nikon, Japan). The tissues were dissected from both LL and adults. Ovary (OV) and testis (TE) were additionally dissected from 5-day-old adults.
To investigate induction patterns of TmIKKε mRNA against microbial challenge, 1 × 106 cells/larva of E. coli and S. aureus, or 5 × 104 cells/larva of C. albicans were injected into 12th–15th instar larvae using microinjector with microcapillary. Samples (whole body, HC, GT, and FB) were collected at 3, 6, 9, 12, and 24 h following injection of microorganisms.
Total RNA Extraction and cDNA Synthesis
The total RNA was isolated from the developmental stages, tissues, and time-course samples using a Clear-S™ Total RNA extraction kit (Invirustech Co., Gwangju, South Korea) according to the manufacturer’s instructions. The total RNA (2 μg) was used as the template to synthesize cDNA using the Oligo(dT)12–18 primer on MyGenie96 Thermal Block (Bioneer, South Korea) and AccuPower® RT PreMix (Bioneer) according to the manufacturer’s instructions. The cDNA was stored at −20°C until required.
Expression and Induction Analysis of the TmIKKε mRNA
The relative expression level of TmIKKε mRNA was investigated by performing quantitative real-time polymerase chain reaction (qRT-PCR) using an AccuPower® 2× Greenstar™ qPCR Master Mix (Bioneer, Daejeon, Korea) and synthesized cDNAs, and TmIKKε gene-specific primers were designed using the Primer 3 plus program (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi) (Supplementary Table 2). The PCR conditions included an initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 30 s. The qRT-PCR assays were performed on an AriaMx Real-Time PCR System (Agilent Technologies, United States). The results were analyzed using AriaMx Real-Time PCR software. The 2–ΔΔCt method (Livak and Schmittgen, 2001) was employed to analyze the TmIKKε mRNA expression levels. The mRNA expression levels were normalized to those of T. molitor ribosomal protein L27a (TmL27a), which acted as an internal control. The results represent mean ± standard error (SE) of three biological replicates (3 pools of 20 T. molitor larvae).
TmIKKε Gene-Silencing
For the RNA interference (RNAi) experiments of TmIKKε, a double-strand RNA (dsRNA) fragment of TmIKKε gene was synthesized. Briefly, dsDNA fragment of TmIKKε was amplified using PCR with gene-specific primers conjugated with T7 promoter sequences (Supplementary Table 2). The primers were designed using Snapdragon software (https://www.flyrnai.org) to prevent any cross-silencing effects. The primary PCR for the TmIKKε gene was carried out using an AccuPower Pfu PCR PreMix (Bioneer) with cDNA and specific primers for the TmIKKε gene (Supplementary Table 2). The second PCR was conducted with primers tailed with T7 promoter sequences and 100× dilution of the second PCR products.
Polymerase chain reaction was conducted using an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 1 min, and extension at 72°C for 30 s on a MyGenie96 Thermal Block (Bioneer). The PCR products purified using the AccuPrep® PCR Purification Kit (Bioneer) were used to synthesize the dsRNA using the EZ™ T7 High Yield in vitro transcription kit (Enzynomics, South Korea). The dsRNA for enhanced green fluorescent protein (dsEGFP) synthesized from pEGFP-C1 plasmid DNA as described above acted as a negative control. The dsRNA products were purified using the phenol:chloroform:isoamyl alcohol (PCI) method, precipitated with 5 M ammonium acetate, and washed with 70 and 90% ethanol by centrifugation at 10,000 × g for 15 min at 4°C. The dried pellet was resuspended in DNase and RNase-free water. The synthesized dsRNA was stored at −20°C until required. For the knockdown of TmIKKε mRNA, 1 μg of dsRNA for EGFP and TmIKKε were injected into T. molitor 10th to 12th instar larvae.
Mortality Assay
To measure mortality, microorganisms (1 × 106 cells/μl of E. coli or S. aureus, and 5 × 104 cells/μl of C. albicans) were injected into TmIKKε gene-silenced T. molitor larvae. Dead larvae were counted each day for up to 10 days following the injection of the microorganisms. Ten insect larvae were used for each group in the mortality assay. Each assay was performed in triplicate. Kaplan–Meier method was used to plot cumulative survival curves of larvae after inoculation, the log-rank chi-squared test was used to assess differences in survival between treatments (Goel et al., 2010).
Effects of TmIKKε RNAi on the Expression of Antimicrobial Peptide and NF-κB Genes
To further characterize the function of the TmIKKε gene in the humoral innate immune response, the TmIKKε silenced individuals were challenged with microorganisms and the expression levels of 14 AMP genes as well as three NF-κB genes were investigated. After treatment of TmIKKε dsRNA into T. molitor 10th to 12th instar larvae, E. coli (1 × 106 cells/larva), S. aureus (1 × 106 cells/larva), or C. albicans (5 × 104 cells/larva) were injected into T. molitor larvae. Twenty-four hours post-injection, over 20 larvae (as a group) were dissected and the samples (HC, GT, and FB) were collected. 1× PBS was used as an injection control. The expression levels of the following 14 AMP genes were measured by qRT-PCR with 14 AMP gene-specific primers (Supplementary Table 2): TmTenecin 1, 2, 3, and 4 (TmTene1, 2, 3, and 4), TmDefensin and TmDefensin-like (TmDef and TmDef-like), TmColeoptericin A and B (TmColeA and B), TmAttacin 1a, 1b, and 2 (TmAtt1a, 1b, and 2), TmCecropin 2 (TmCec2), and TmThaumatin-like protein 1 and 2 (TmTLP1 and 2). In addition, the expression profiles of NF-κB genes such as TmDorsal isoform X1 and X2 (TmDorX1 and X2), and TmRelish, were investigated by qRT-PCR. A relative quantitative PCR was performed as mentioned above using the AMP and NF-κB gene specific primers.
Statistical Analyses
All experiments were carried out in triplicate and the data were subjected to one-way ANOVA. Tukey’s multiple range tests were used to evaluate the difference between groups (p < 0.05).
Results
Gene Organization, Open Reading Frame and in silico Analyses of TmIKKε
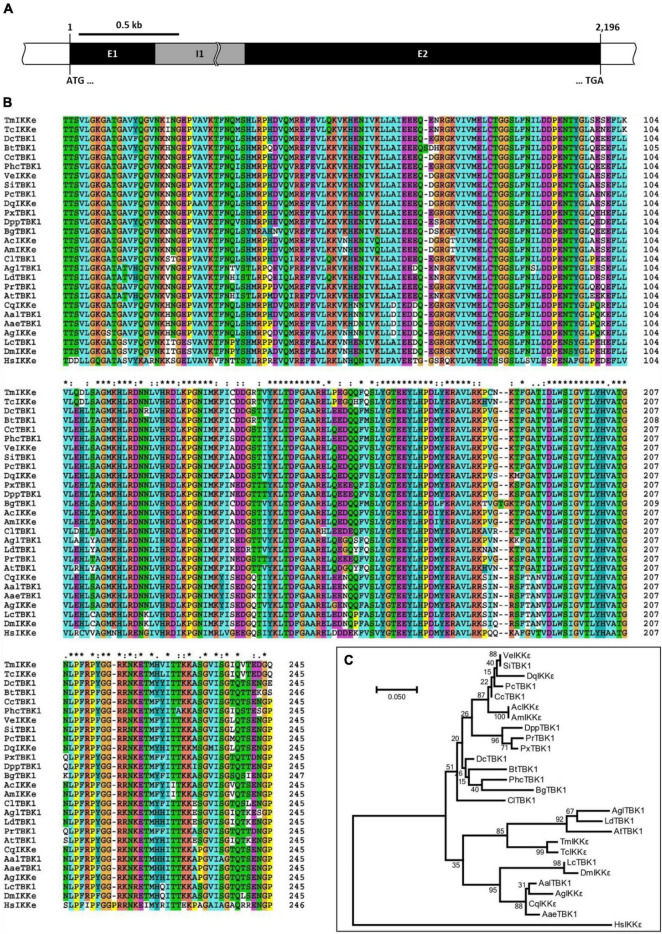
The organization of the TmIKKε gene was deciphered from the T. molitor nucleotide database using the TcIKKε amino acid sequence as a query in a tBLASTn analysis. TmIKKε includes two exons interrupted by a single intron (Figure 1A and Supplementary Figure 1). Exon 1 and exon 2 of TmIKKε were 423 bp and 1,773 bp, respectively. The ORF sequence of 2,196 bp starts with the “ATG” initiation codon and ends with the “TGA” stop codon. The ORF sequence of TmIKKε encoded a protein of 731 amino acid residues (Supplementary Figure 2). Domain analysis predicted a serine/threonine-protein kinase catalytic domain (residues 12–258), a ubiquitin-like domain (residues 310–387), and a TANK-binding kinase 1 coiled-coil domain 1 (residues 412–653) in TmIKKε protein. A 5′-untranslated region (UTR) sequence of 581 bp and 3′-UTR of 930 bp was also predicted for TmIKKε. The 3′-UTR sequence contains a consensus polyadenylation signal (AATAAA) located in 173–178 bp following termination codon (TGA). The TmIKKε cDNA sequence and deduced protein sequence have been submitted to GenBank (GenBank ID: MZ708789).
FIGURE 1.
Genomic organization and phylogenetics of the Tenebrio molitor IκB kinase ε (IKKε) (TmIKKε) gene. (A) Schematic representation of the TmIKKε gene. Two exons (E1 and E2) are separated by a single intron (I1). E1 starts with the translation start codon “ATG,” and E2 ends with the translation stop codon “TGA”. (B) Clustal X2-based multiple sequence alignment of IKKε/TBK1 proteins. “*”-Highly conserved residues; “:”-Conserved residues; “.”-Less conserved residues. The residues are colored automatically and represents physico-chemical properties of amino acid residues. (C) Maximum likelihood analysis based on the alignment of IKKε/TBK1 amino acid sequences. Bootstrap values (1,000 replicates) are indicated at the nodes.
The alignment of the predicted amino acid sequence (restricted to the serine/threonine protein kinase catalytic domain) of TmIKKε with IKKε and serine/threonine protein kinase TBK1 from other known insect IKKs revealed a high degree of conservation (Figure 1B). A phylogenetic tree was constructed using the Clustal X2 alignment file and MEGA 7.0 program to assess the evolutionary position of TmIKKε among the orthologs (Figure 1C). Homo sapiens IKKε (HsIKKε) was used as the outgroup in the phylogenetic analysis. Two clear clade divisions were observed. TmIKKε was placed with TcIKKε and TBK1 from other beetles (AglTBK1, LdTBK1, and AtTBK1). The same clade also placed the IKKε isoforms from the order Diptera in a separate cluster. The high bootstrap values supported the tree topology in this clade. While the mosquito IKKε/TBK1 formed one sub-cluster, the Drosophila and blowfly IKKε/TBK1 formed the other sub-cluster. In the second clade, the orthologs of other insect IKKε proteins were placed.
Expression of TmIKKε During Development and in Different Tissues
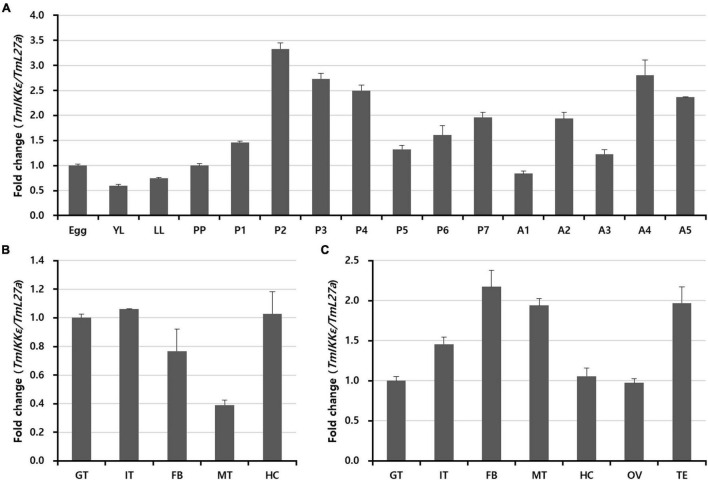
In order to examine the TmIKKε mRNA expression during development and in different tissues of T. molitor, we performed qRT-PCR using SYBR Green dye-binding assay. Developmental expression patterns indicate that TmIKKε mRNA was highly expressed in the pupal stage-2, -3, and -4 followed by variable expression in the adult stages (Figure 2A). Further, the expression of TmIKKε mRNA was measured in tissues of T. molitor late-instar larvae and 5-day old adults using qRT-PCR. In the late-instar larval tissues, the TmIKKε mRNA was expressed in a tissue-dependent manner (Figure 2B). TmIKKε mRNA was found to be higher (especially in FB, MT, and TE) in the 5-day old adult T. molitor (Figure 2C).
FIGURE 2.
Expression of TmIKKε during the developmental stages and in different tissues of T. molitor. (A) qRT-PCR was used to measure the expression of TmIKKε mRNA in the different developmental stages of the insect. YL, early instar larvae; LL, late-instar larvae; PP, pre-pupa; P1–P7, 1- to 7-day-old pupa; A1–A5, 1- to 5-day-old adult. (B) Expression of TmIKKε mRNA in tissues of T. molitor LL as analyzed using real-time polymerase chain reaction (qRT-PCR). (C) Expression of TmIKKε messenger RNA (mRNA) in the tissues of T. molitor adults as analyzed using qRT-PCR. GT, Gut; IT, integument; FB, fat body; MT, Malpighian tubules; HC, hemocytes. T. molitor ribosomal protein 27a (TmL27a) was used as an internal control to normalize the concentration of templates between samples. Vertical bars represent mean ± SE (n = 20).
Temporal Expression of TmIKKε After Microbial Infection
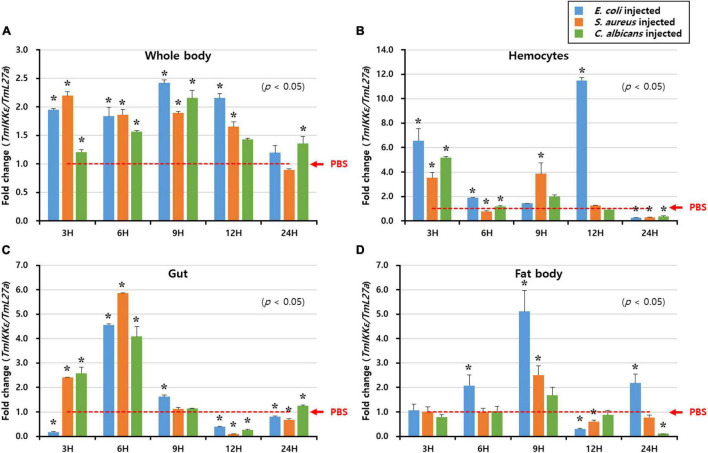
To understand the biological function of TmIKKε in innate immunity of T. molitor, the mRNA expression levels were monitored at different time-points (3, 6, 9, 12, and 24 h) in whole-larvae, HC, GT, and FB after exposure to PBS (as injection controls), and microorganisms (E. coli, S. aureus, and C. albicans) challenge (Figure 3). The expression level of TmIKKε at various time points was analyzed relative to PBS control. In the whole body, TmIKKε mRNA was induced early at 3 h and declined at 6, 9, and 12 h post-injection of microorganisms towards the level of PBS-injected control. At 24 h, the expression of TmIKKε mRNA was found to be lower compared to the expression at 3 h (Figure 3A). In hemocytes, TmIKKε mRNA was induced at 3 and 12 h post-injection by E. coli. S. aureus induced TmIKKε mRNA early at 3 and 9 h post-challenge (Figure 3B). But in the gut tissue, expression of TmIKKε mRNA was drastically high at 6 h-post-injection of S. aureus, E. coli, and C. albicans (Figure 3C). Expression of TmIKKε mRNA in the FB was higher at 9 h post-injections with E. coli and is decreased at 12 hpi relative even to 3 hpi (Figure 3D).
FIGURE 3.
Expression of TmIKKε in response to microbial challenge. Escherichia coli, Staphylococcus aureus, or Candida albicans were injected into 10th to 12th instar larvae, and samples were collected at specific time points (3, 6, 9, 12, and 24 h). TmIKKε mRNA expression in the whole body (A), hemocytes (B), gut (C), and fat body (D) of T. molitor larvae. The TmIKKε mRNA expression was normalized to that in the phosphate-buffered saline (PBS) group. TmL27a was used as an internal control. Data represent the mean ± SE. Significance is set at p < 0.05. *Denotes significant difference over the control.
Effects of TmIKKε RNAi on Larval Survivability
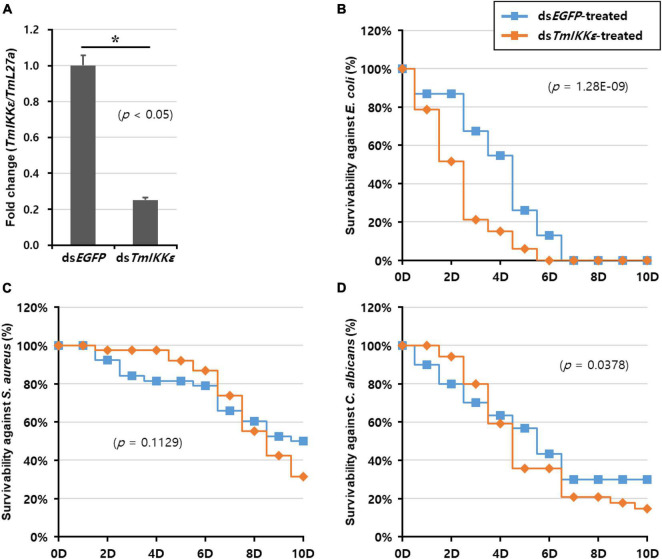
To further substantiate the function of TmIKKε in the host immunity against pathogens, we silenced TmIKKε mRNA by synthesizing dsRNA and injected it into the T. molitor larvae. The silencing of TmIKKε mRNA was compared relative to the injection of dsEGFP (as negative control) to a separate set of larvae. The RNAi efficiency of TmIKKε was found to be approximately 75% relative to the dsEGFP control in the whole body of the larvae (Figure 4A). In addition, the tissue-specific knockdown efficiency by injecting TmIKKε dsRNA was investigated, which showed the results that the expression of TmIKKε was significantly down-regulated by dsTmIKKε-treatment in all tissues including FB (86%), GT (68%), HC (89%), and integuments (84%) (Supplementary Figure 3).
FIGURE 4.
TmIKKε gene-silencing and larval survival against pathogenic challenge. (A) RNA interference (RNAi) efficiency of TmIKKε mRNA after dsTmIKKε injection into T. molitor measured using qRT-PCR. The EGFP dsRNA-treated larvae served as a negative control. Larval survival was measured until 10 days after injection with E. coli (B), S. aureus (C), and C. albicans (D). The Kaplan–Meier survival plots (log-rank chi-squared test) were used to analyze the larval mortality. *Denotes significant difference over control.
The survival of the larvae was recorded for 10 days after injection of E. coli, S. aureus, and C. albicans to TmIKKε-silenced larvae. After E. coli challenge to TmIKKε-silenced larvae, the mortality observed was 100% at 6-day post-challenge but was not significant compared to dsEGFP-treated larvae (log-rank chi-squared test; p = 1.28E-09) (Figure 4B). The mortality of TmIKKε-silenced larvae was 70% at 10-day post-challenge with S. aureus (log-rank chi-squared test; p = 0.1129) (Figure 4C). After infection with the fungus, C. albicans, the mortality was close to 90% in TmIKKε-silenced larvae (log-rank chi-squared test; p = 0.0378) (Figure 4D). The results indicate that depletion of TmIKKε caused increased larval mortality against E. coli, not S. aureus and C. albicans.
Antimicrobial Peptide Expression Levels in TmIKKε Knockdown T. molitor Larvae
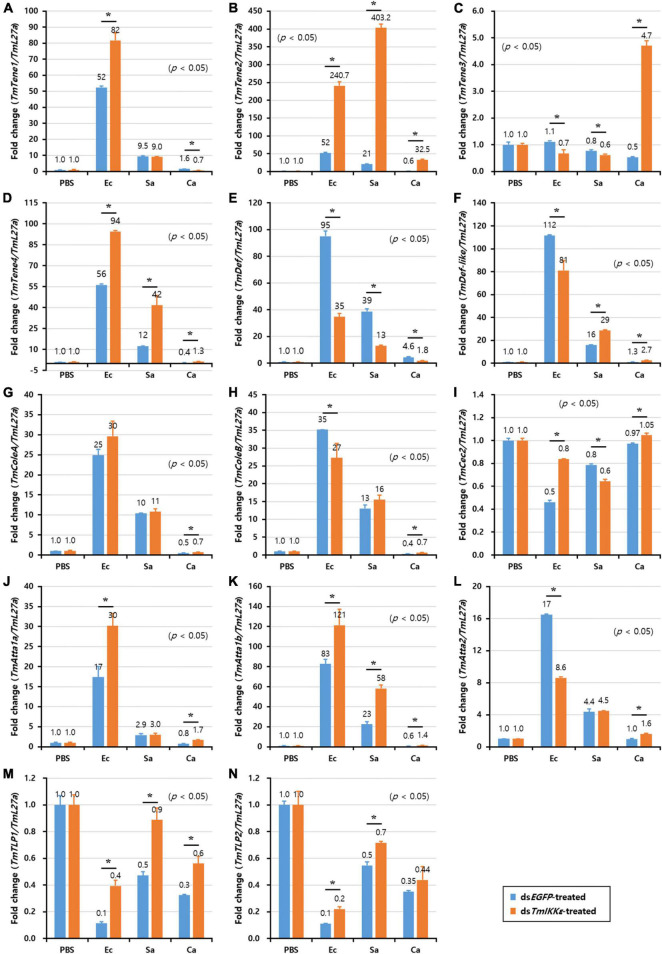
In order to investigate the requirement of TmIKKε gene in the regulation of AMP production in immune organs (hemocytes, gut, and fat body) of T. molitor larvae, we injected gram-negative bacteria E. coli, gram-positive bacteria S. aureus or fungus C. albicans into TmIKKε knockdown T. molitor larvae. The transcriptional expression levels of fourteen T. molitor AMP genes were measured in TmIKKε knockdown individuals in comparison with dsEGFP-treated group. In the hemocytes of T. molitor larvae, the expression of TmTene2 was significantly upregulated in TmIKKε knockdown individuals post-inoculation with E. coli, S. aureus, and C. albicans (Figure 5B). TmTene1 (Figure 5A) and TmTene3 (Figure 5C) expression were also upregulated post-inoculation of E. coli and C. albicans, respectively. Further, the expression of TmAtta1a was upregulated post-inoculation of E. coli (Figure 5J) and TmAtta1b upregulated post-inoculation of E. coli and S. aureus (Figure 5K) in TmIKKε knockdown individuals. The upregulation of AMPs in response to TmIKKε gene knockdown suggests negative regulation during the pathogenic challenge.
FIGURE 5.
The antimicrobial peptides (AMPs) expression levels in TmIKKε-knockdown T. molitor larval hemocytes upon microorganism challenge. E. coli (Ec), S. aureus (Sa), or C. albicans (Ca) were injected into TmIKKε-silenced T. molitor larvae. The transcriptional expression levels of TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmColeA (G), TmColeB (H), TmCec2 (I), TmAtta1a (J), TmAtta1b (K), TmAtta2 (L), TmTLP1 (M), and TmTLP2 (N) were measured using qRT-PCR. EGFP dsRNA was used as a silencing control, and TmL27a was used as an internal control. Data represent the mean ± SE of three independent biological replicates. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).
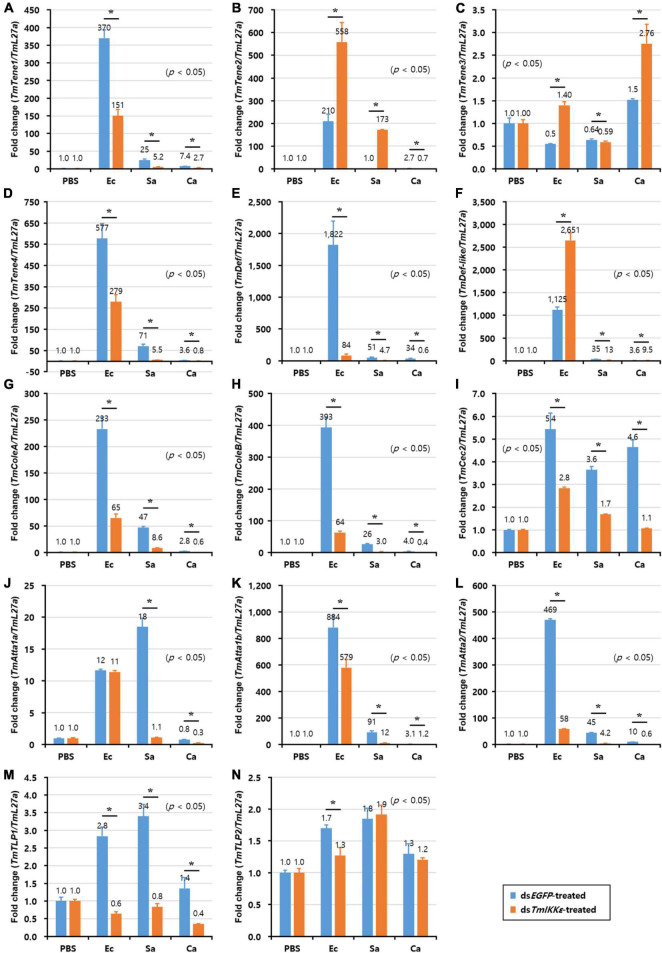
Alternatively, in the gut, the expression of eight AMP genes including TmTene1 (Figure 6A), TmTene4 (Figure 6D), TmDef (Figure 6E), TmColeA (Figure 6G), TmColeB (Figure 6H), TmAtta1a (Figure 6J), TmAtta1b (Figure 6K), and TmAtta2 (Figure 6L) out of the fourteen AMP genes were significantly decreased in TmIKKε knockdown individuals. On the other hand, the expression of only two AMP genes including TmTene2 (Figure 6B) and TmDef-like (Figure 6F) was significantly upregulated by TmIKKε RNAi. Overall, in the gut of T. molitor larvae, TmIKKε RNAi leads to decreased transcriptional regulation of most AMPs and might be putatively involved in the survival of the larvae against pathogenic stress. We also noticed the downregulation of TmIKKε transcripts in the gut tissue following systemic injection of dsTmIKKε. Moreover, in the silenced individuals, eight AMPs were downregulated suggesting putative role in gut immunity.
FIGURE 6.
AMP expression levels in TmIKKε-knockdown T. molitor larval gut upon microorganism challenge. E. coli (Ec), S. aureus (Sa), or C. albicans (Ca) were injected into TmIKKε-silenced T. molitor larvae. The transcriptional expression levels of TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmColeA (G), TmColeB (H), TmCec2 (I), TmAtta1a (J), TmAtta1b (K), TmAtta2 (L), TmTLP1 (M), and TmTLP2 (N) were measured using qRT-PCR. EGFP dsRNA was used as a silencing control, and TmL27a was used as an internal control. Data represent the mean ± SE of three independent biological replicates. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).
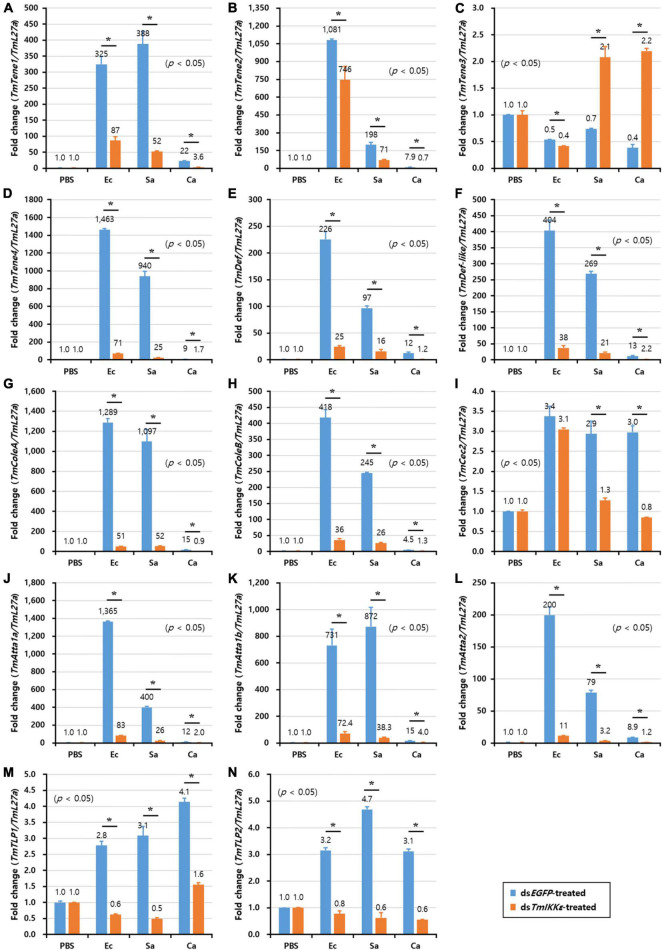
In T. molitor FB, the expression of twelve AMP genes including TmTene1 (Figure 7A), -2 (Figure 7B) and -4 (Figure 7D), TmDef (Figure 7E) and -2 (Figure 7F), TmColeA (Figure 7G) and -2 (Figure 7H), TmAtta1a (Figure 7J), -1b (Figure 7K), and -2 (Figure 7L), TmTLP1 (Figure 7M), and -2 (Figure 7N), out of fourteen AMP genes were significantly decreased by TmIKKε RNAi. Downregulation of AMPs in TmIKKε knockdown individuals after challenge with microorganisms ascertains the role of TmIKKε in the innate immunity of the insect.
FIGURE 7.
AMP expression levels in TmIKKε-knockdown T. molitor larval fat body upon microorganism challenge. E. coli (Ec), S. aureus (Sa), or C. albicans (Ca) were injected into TmIKKε-silenced T. molitor larvae. The transcriptional expression levels of TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmColeA (G), TmColeB (H), TmCec2 (I), TmAtta1a (J), TmAtta1b (K), TmAtta2 (L), TmTLP1 (M), and TmTLP2 (N) were measured using qRT-PCR. EGFP dsRNA was used as a silencing control, and TmL27a was used as an internal control. Data represent the mean ± SE of three independent biological replicates. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).
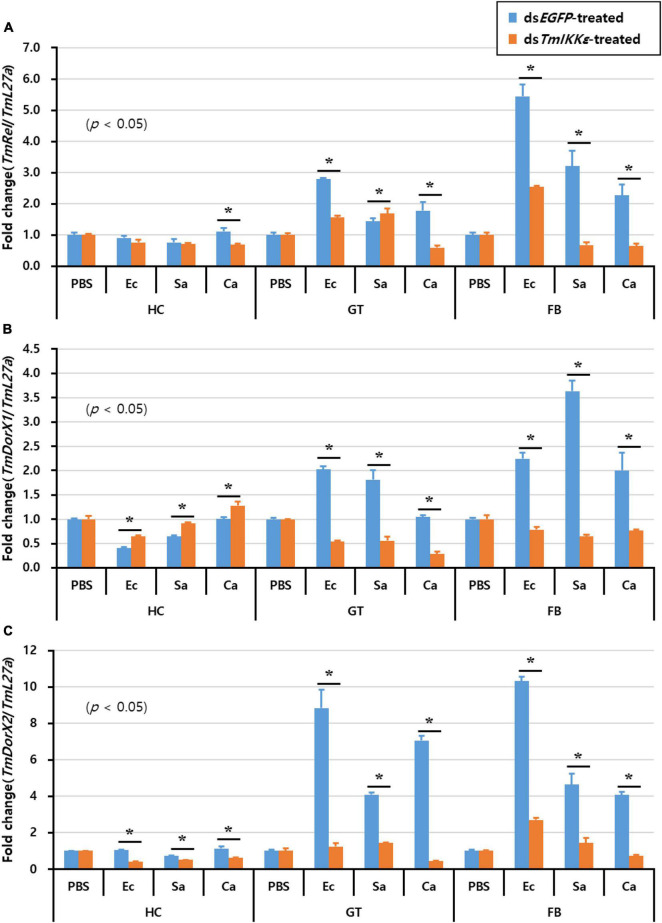
Effects of TmIKKε RNAi on the Expression of Tenebrio NF-κB Genes
Furthermore, to understand the effect of TmIKKε RNAi on expression of Tenebrio NF-κB genes, TmIKKε-silenced T. molitor larvae were challenged with microorganisms and the expression patterns of Tenebrio NF-κB genes such as TmRelish, TmDorX1, and TmDorX2 were investigated at 24 h by qPCR analysis. The results showed that the mRNA level of three NF-κB genes was dramatically decreased by TmIKKε RNAi in the FB of T. molitor larva. In addition, the expression of TmDorX1 and TmDorX2 transcripts were significantly decreased by TmIKKε RNAi in the GT (Figure 8).
FIGURE 8.
Tissue-specific induction patterns of Tenebrio NF-κB genes in TmIKKε dsRNA-treated T. molitor larvae against various pathogens. (A) Induction of TmRelish mRNA in the hemocytes, gut, and fat body of T. molitor larvae. (B) Induction of TmDorX1 mRNA in the hemocytes, gut, and fat body of TmIKKε-silenced T. molitor larvae. (C) Induction of TmDorX2 mRNA in the hemocytes, gut, and fat body of TmIKKε-silenced T. molitor larvae. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).
Discussion
The Toll and IMD pathways constitute an important defense arsenal to protect insects from non-self-discriminating pathogens. Our research group has been successful in elucidating key genes of the Toll and IMD intracellular pathways, which are relevant in the context of humoral immunity in the coleopteran pest T. molitor (Patnaik et al., 2013, 2014; Tindwa et al., 2013). Our focus has been on the transcriptional activation of AMPs elicited by diverse groups of microorganisms and mediated through the Toll/IMD signaling cascade (Jo et al., 2017, 2019; Keshavarz et al., 2019, 2020a,b, 2020d; Park et al., 2019; Ali Mohammadie Kojour et al., 2020; Edosa et al., 2020a,b; Ko et al., 2020). The IKK family of proteins act upstream of the NF-κB factor Relish in the IMD pathway by phosphorylating Relish and regulating the transcriptional activation of AMP genes. The involvement of the IKK isoforms IKKβ (ird5 in Drosophila) and IKKγ (Kenny in Drosophila) in phosphorylation of Relish has been described earlier (Erturk-Hasdemir et al., 2009; Kleino and Silverman, 2014). The IKKε isoform of the IKK family of proteins encodes a serine-threonine kinase that has been implicated in NF-κB activation. The isoform also forms an essential component of the interferon regulatory factor 3 (IRF3) signaling pathway (Fitzgerald et al., 2003; Seccareccia et al., 2014; Dubois et al., 2018). IKKε in the black carp (Mylopharyngodon piceus) was functionally characterized to participate in activating the expression of interferons in zebrafish and epithelioma papulosum cyprini (EPC) cells (Qu et al., 2015). TBK1, which is structurally identical to other IKK proteins, may also have a putative role as an important immunoregulator for IRF3 and IFNγ induction in chickens (Wang et al., 2017). However, the functional characterization of the IKKε homolog in insects is less described. In this study, we characterized the IKKε isoform in the coleopteran pest T. molitor by identifying its sequence, tissue distribution, and possible role in humoral immunity by studying the transcriptional regulation of 14 T. molitor AMP genes after microorganism challenge. We have also examined the expression of downstream NF-κB factors, including TmDorX1 and TmDorX2, which are involved in the Toll signaling pathway, and Relish, which is involved in the IMD signaling pathway, under TmIKKε-silenced conditions. Our results have improved understanding of the putative regulatory pathway involving TmIKKε.
The induction pattern of TmIKKε against injection of representative gram-negative bacteria, gram-positive bacteria, and fungi indicated that TmIKKε mRNA expression was mainly induced by E. coli challenge in the hemocytes. Furthermore, the knockdown of TmIKKε transcripts resulted in larval death upon E. coli challenge. The findings suggest that TmIKKε may be putatively involved in the defense against gram-negative bacteria. Interestingly, the TmIKKε transcript was significantly induced by S. aureus in hemocytes. This result may indicate the involvement of TmIKKε in the canonical IMD pathway. Further, the role of S. aureus in the activation of the Toll signaling cascade is well established in insects, including T. molitor (Patnaik et al., 2014). However, in the gut defense system of D. melanogaster. IMD pathway is required for clearance of S. aureus, possibly independently from AMP expression and via Duox system that produces reactive oxygen species (ROS) (Hori et al., 2018).
The transcriptional regulation of fourteen AMP genes in T. molitor in TmIKKε-silenced condition was investigated in hemocytes, gut, and fat body tissues post-injection with gram-negative bacteria E. coli, gram-positive bacteria S. aureus, and the fungus C. albicans. Interestingly, most AMP genes were not positively affected by TmIKKε RNAi in hemocytes, excepting to some extent TmDef. Other critical AMP genes were mostly negatively regulated after microorganisms challenge in TmIKKε-silenced individuals. Further, the inconsequential role of TmIKKε knockdown on the activation of the NF-κB genes including TmRelish, TmDorX1, and TmDorX2 suggest that TmIKKε is not required for AMP production in hemocytes. Contrastingly, in the gut, seven AMP genes were significantly downregulated by TmIKKε dsRNA-treatment post-injection of E. coli. We also find that the TmDorX1 and TmDorX2 mRNA (NF-κB regulator of Toll signaling pathway) were critically downregulated in the gut in TmIKKε-silenced individuals. This possibly suggests that TmIKKε regulates the transcriptional activation of seven AMP genes in the host gut in response to the systemic infection of microorganisms. Studies in mosquitoes highlight the promiscuous intervention of hemocytes in the activation of the anti-plasmodial gut immune system (Ramirez et al., 2014; Castillo et al., 2017). These pieces of evidence suggest the relationship between systemic infections and gut innate immune system.
In the fat body tissue of T. molitor larvae, the expression of ten AMP genes was downregulated and all the three NF-κB genes were significantly affected by TmIKKε RNAi in response to microbial challenge. These results indicate that TmIKKε is a key regulator for Toll and IMD pathways in the fat body of T. molitor larvae. Cross-talk between Toll and IMD pathways is proposed in D. melanogaster (Tanji et al., 2007), T. castaneum (Yokoi et al., 2012a,b), P. stali (Nishide et al., 2019), and T. molitor (Ko et al., 2020). It was also suggested that several AMP genes were co-regulated by those pathways in D. melanogaster (De Gregorio et al., 2002). Further, DmIKKε phosphorylates DIAP1 leading to DIAP1 degradation and apoptosis in development downstream of IMD. In Drosophila, the function of DmIKKε is not especially Toll or IMD related, particularly for its role in IMD activation (Kuranaga et al., 2006; Oshima et al., 2006). In addition, the activation of Toll and IMD pathways is totally dependent on invading microbes. For instance, the gram-positive bacteria including Micrococcus luteus, Bacillus subtilis, Bacillus megaterium, Enterobacter cloacae, and fungi such as Beauveria bassiana, Saccharomyces cerevisiae, Metarhizium anisopliae, and Geotrichum candidum activate the IMD pathway (Hedengren-Olcott et al., 2004). Drosophila Cecropin A was induced by the gram-positive bacteria M. luteus and S. aureus independent of the NF-κB factor Relish. The transcriptional activation of AMPs such as Cecropin A1 and Cecropin A2 in response to M. luteus infection required Relish and Dif, respectively. Even, it is well known that the Tenecin 3 protects T. molitor against infection by the fungus Beauveria bassiana exemplified by increased larval survivability (Maistrou et al., 2018). Contrastingly, our results showed that the TmIKKε RNAi does not mainly affect the expression of Tenecin 3 gene and larval mortality against fungal infection.
Taken together, TmIKKε plays a critical function in the production of nine AMPs in the fat body by regulating both Toll (TmDorX1 and -X2) and IMD (TmRelish) pathways. Interestingly, seven AMP genes were positively regulated by TmIKKε RNAi in the gut, carefully suggesting that the systemic infection may positively regulate AMP production in the gut through the Toll (TmDorX1 and -X2) pathway.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YH and YJ: conceptualization, methodology, supervision, and project administration. YH: software, validation, resources, and funding acquisition. HK and YJ: formal analysis and visualization. HK, KP, CK, and HJ: investigation. HK, KP, and CK: data curation. HK, YH, YJ, and BP: writing – original draft preparation. BP, SB, and YL: writing – review and editing. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (Grant No. 2018R1A2A2A05023367) and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Export Promotion Technology Development Program (Grant No. 617077-5), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.758862/full#supplementary-material
References
- Ali Mohammadie Kojour M., Han Y. S., Jo Y. H. (2020). An overview of insect innate immunity. Entomol. Res. 50 282–291. 10.1111/1748-5967.12437 [DOI] [Google Scholar]
- Ali Mohammadie Kojour M., Jang H. A., Edosa T. T., Keshavarz M., Kim B. B., Bae Y. M., et al. (2021). Identification, in silico characterization, and expression analysis of Tenebrio molitor Cecropin-2. Entomol. Res. 51 74–82. 10.1111/1748-5967.12476 [DOI] [Google Scholar]
- Altincicek B., Knorr E., Vilcinskas A. (2008). Beetle immunity: identification of immune-inducible genes from the model insect Tribolium castaneum. Dev. Comp. Immunol. 32 585–595. 10.1016/j.dci.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Bang K., Park S., Yoo J. Y., Cho S. (2012). Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, Spodoptera exigua (Hubner) (Insecta: Lepidoptera: Noctuidae). Mol. Biol. Rep. 39 5151–5159. 10.1007/s11033-011-1311-3 [DOI] [PubMed] [Google Scholar]
- Barletta A. B., Nascimento-Silva M. C., Talyuli O. A., Oliveira J. H., Pereira L. O., Oliveira P. L., et al. (2017). Microbiota activates IMD pathway and limits Sindbis infection in Aedes aegypti. Parasit. Vectors 10:103. 10.1186/s13071-017-2040-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J. C., Ferreira A. B. B., Trisnadi N., Barillas-Mury C. (2017). Activation of mosquito complement antiplasmodial response requires cellular immunity. Sci. Immunol. 2:eaal1505. 10.1126/sciimmunol.aal1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha G. H., Cho K. S., Lee J. H., Kim M., Kim E., Park J., et al. (2003). Discrete functions of TRAF1 and TRAF2 in Drosophila melanogaster mediated by c-Jun N-terminal kinase and NF-kappaB-dependent signaling pathways. Mol. Cell. Biol. 23 7982–7991. 10.1128/MCB.23.22.7982-7991.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J. H., Kurokawa K., So Y. I., Hwang H. O., Kim M. S., Park J. W., et al. (2012). Purification and characterization of tenecin 4, a new anti-Gram-negative bacterial peptide, from the beetle Tenebrio molitor. Dev. Comp. Immunol. 36 540–546. 10.1016/j.dci.2011.09.010 [DOI] [PubMed] [Google Scholar]
- Chen W., White M. A., Cobb M. H. (2002). Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J. Biol. Chem. 277 49105–49110. 10.1074/jbc.M204934200 [DOI] [PubMed] [Google Scholar]
- Choe K. M., Lee H., Anderson K. V. (2005). Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. U.S.A. 102 1122–1126. 10.1073/pnas.0404952102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. M., Werner T., Stoven S., Hultmark D., Anderson K. V. (2002). Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296 359–362. 10.1126/science.1070216 [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P. T., Tzou P., Rubin G. M., Lemaitre B. (2002). The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21 2568–2579. 10.1093/emboj/21.11.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois N., Berendsen S., Henry A., Nguyen M., Bours V., Robe P. (2018). I-Kappa-B kinase-epsilon activates nuclear factor-kappa B and STAT5B and supports glioblastoma growth but amlexanox shows little therapeutic potential in these tumors. Cancer Transl. Med. 4 1–8. 10.4103/ctm.ctm_3_18 [DOI] [Google Scholar]
- Edosa T. T., Jo Y. H., Keshavarz M., Bae Y. M., Kim D. H., Lee Y. S., et al. (2020a). TmSpz4 plays an important role in regulating the production of antimicrobial peptides in response to Escherichia coli and Candida albicans infections. Int. J. Mol. Sci. 21:1878. 10.3390/ijms21051878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edosa T. T., Jo Y. H., Keshavarz M., Bae Y. M., Kim D. H., Lee Y. S., et al. (2020b). TmSpz6 is essential for regulating the immune response to Escherichia Coli and Staphylococcus Aureus infection in Tenebrio molitor. Insects 11:105. 10.3390/insects11020105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk-Hasdemir D., Broemer M., Leulier F., Lane W. S., Paquette N., Hwang D., et al. (2009). Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. U.S.A. 106 9779–9784. 10.1073/pnas.0812022106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., et al. (2003). IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4 491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Gan T., Fan L., Zhao L., Misra M., Liu M., Zhang M., et al. (2021). JNK Signaling in Drosophila aging and longevity. Int. J. Mol. Sci. 22:9649. 10.3390/ijms22179649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P., Naitza S., Kappler C., Ferrandon D., Zachary D., Swimmer C., et al. (2001). Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1 503–514. 10.1016/S1534-5807(01)00059-4 [DOI] [PubMed] [Google Scholar]
- Goel M. K., Khanna P., Kishore J. (2010). Understanding survival analysis: Kaplan-Meier estimate. Int. J. Ayurveda Res. 1 274–278. 10.4103/0974-7788.76794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M., Gobert V., Michel T., Belvin M., Duyk G., Hoffmann J. A., et al. (2002). The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416 640–644. 10.1038/nature734 [DOI] [PubMed] [Google Scholar]
- Hanson M. A., Dostalova A., Ceroni C., Poidevin M., Kondo S., Lemaitre B. (2019). Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife 8:e44341. 10.7554/eLife.44341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedengren-Olcott M., Olcott M. C., Mooney D. T., Ekengren S., Geller B. L., Taylor B. J. (2004). Differential activation of the NF-kappa B-like factors relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J. Biol. Chem. 279 21121–21127. 10.1074/.M313856200jbc [DOI] [PubMed] [Google Scholar]
- Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999). Phylogenetic perspectives in innate immunity. Science 284 1313–1318. 10.1126/science.284.5418.1313 [DOI] [PubMed] [Google Scholar]
- Hori A., Kurata S., Kuraishi T. (2018). Unexpected role of the IMD pathway in Drosophila gut defense against Staphylococcus aureus. Biochem. Biophys. Res. Commun. 495 395–400. 10.1016/j.bbrc.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Hua X. T., Ma X. J., Xue R. J., Cheng T. C., Wang F., Xia Q. Y. (2016). Characterization of the Bombyx mori Cecropin A1 promoter regulated by IMD pathway. Insect Sci. 23 297–304. 10.1111/1744-7917.12210 [DOI] [PubMed] [Google Scholar]
- Hultmark D., Steiner H., Rasmuson T., Boman H. G. (1980). Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 106 7–16. 10.1111/j.1432-1033.1980.tb05991.x [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H. A., Park K. B., Kim B. B., Ali Mohammadie Kojour M., Bae Y. M., Baliarsingh S., et al. (2020a). Bacterial but not fungal challenge up-regulates the transcription of Coleoptericin genes in Tenebrio molitor. Entomol. Res. 50 440–449. 10.1111/1748-5967.12465 [DOI] [Google Scholar]
- Jang H. A., Park K. B., Kim B. B., Ali Mohammadie Kojour M., Bae Y. M., Baliarsingh S., et al. (2020b). In silico identification and expression analyses of Defensin genes in the mealworm beetle Tenebrio molitor. Entomol. Res. 50 575–585. 10.1111/1748-5967.12468 [DOI] [Google Scholar]
- Jo Y. H., Kim Y. J., Park K. B., Seong J. H., Kim S. G., Park S., et al. (2017). TmCactin plays an important role in Gram-negative and -positive bacterial infection by regulating expression of 7 AMP genes in Tenebrio molitor. Sci. Rep. 7:46459. 10.1038/srep46459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y. H., Park S., Park K. B., Noh M. Y., Cho J. H., Ko H. J., et al. (2018). In silico identification, characterization and expression analysis of attacin gene family in response to bacterial and fungal pathogens in Tenebrio molitor. Entomol. Res. 48 45–54. 10.1111/1748-5967.12287 [DOI] [Google Scholar]
- Jo Y. H., Patnaik B. B., Hwang J., Park K. B., Ko H. J., Kim C. E., et al. (2019). Regulation of the expression of nine antimicrobial peptide genes by TmIMD confers resistance against Gram-negative bacteria. Sci. Rep. 9:10138. 10.1038/s41598-019-46222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. R., Makarova O., Rolff J. (2013). Inducible defenses stay up late: temporal patterns of immune gene expression in Tenebrio molitor. G3 (Bethesda) 4 947–955. 10.1534/g3.113.008516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Goldman W. E., Mellroth P., Steiner H., Fukase K., Kusumoto S., et al. (2004). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20 637–649. 10.1016/s1074-7613(04)00104-9 [DOI] [PubMed] [Google Scholar]
- Keshavarz M., Jo Y. H., Edosa T. T., Bae Y. M., Han Y. S. (2020a). TmPGRP-SA regulates antimicrobial response to bacteria and fungi in the fat body and gut of Tenebrio molitor. Int. J. Mol. Sci. 21:2113. 10.3390/ijms21062113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Jo Y. H., Edosa T. T., Han Y. S. (2020b). Tenebrio molitor PGRP-LE plays a critical role in gut antimicrobial peptide production in response to Escherichia coli. Front. Physiol. 11:320. 10.3389/fphys.2020.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Jo Y. H., Edosa T. T., Han Y. S. (2020c). Two roles for the Tenebrio molitor relish in the regulation of antimicrobial peptides and autophagy-related genes in response to Listeria monocytogenes. Insects 11:188. 10.3390/insects11030188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Jo Y. H., Patnaik B. B., Park K. B., Ko H. J., Kim C. E., et al. (2020d). TmRelish is required for regulating the antimicrobial responses to Escherichia coli and Staphylococcus aureus in Tenebrio molitor. Sci. Rep. 10:4258. 10.1038/s41598-020-61157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Jo Y. H., Park K. B., Ko H. J., Edosa T. T., Lee Y. S., et al. (2019). TmDorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 9:16878. 10.1038/s41598-019-53497-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Lee Y. T., Lee Y. J., Chung J. H., Lee B. L., Choi B. S., et al. (1998). Bacterial expression of tenecin 3, an insect antifungal protein isolated from Tenebrio molitor, and its efficient purification. Mol. Cells 8 786–789. [PubMed] [Google Scholar]
- Kim D. H., Noh M. Y., Park K. B., Jo Y. H. (2017). Expression profiles of two thaumatin-like protein (TmTLP) genes in responses to various micro-organisms from Tenebrio molitor. Entomol. Res. 47 35–40. 10.1111/1748-5967.12197 [DOI] [Google Scholar]
- Kleino A., Silverman N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42 25–35. 10.1016/j.dci.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H. J., Jo Y. H., Patnaik B. B., Park K. B., Kim C. E., Keshavarz M., et al. (2020). IKKγ/NEMO is required to confer antimicrobial innate immune responses in the yellow mealworm, Tenebrio Molitor. Int. J. Mol. Sci. 21:6734. 10.3390/ijms21186734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H., Kato D., Minakuchi C., Tanaka T., Yokoi K., Miura K. (2015). Peptidoglycan recognition protein genes and their roles in the innate immune pathways of the red flour beetle, Tribolium castaneum. J. Invertebr. Pathol. 132 86–100. 10.1016/j.jip.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E., Kanuka H., Tonoki A., Takemoto K., Tomioka T., Kobayashi M., et al. (2006). Drosophila IKK-related kinase regulates non-apoptotic function of caspases via degradation of IAPs. Cell 126, 583–596. 10.1016/j.cell.2006.05.048 [DOI] [PubMed] [Google Scholar]
- Kwon Y. M., Kim H. J., Kim Y. I., Kang Y. J., Lee I. H., Jin B. R., et al. (2008). Comparative analysis of two attacin genes from Hyphantria cunea. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151 213–220. 10.1016/j.cbpb.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. (1996). The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86 973–983. 10.1016/s0092-8674(00)80172-5 [DOI] [PubMed] [Google Scholar]
- Leulier F., Parquet C., Pili-Floury S., Ryu J. H., Caroff M., Lee W. J., et al. (2003). The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4 478–484. 10.1038/ni922 [DOI] [PubMed] [Google Scholar]
- Li C., Wang S., He J. (2019). The two NF-kappaB pathways regulating bacterial and WSSV infection of shrimp. Front. Immunol. 10:1785. 10.3389/fimmu.2019.01785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Xiang J. (2013). Signaling pathways regulating innate immune responses in shrimp. Fish Shellfish Immunol. 34 973–980. 10.1016/j.fsi.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Lin J., Xia X., Yu X. Q., Shen J., Li Y., Lin H., et al. (2018). Gene expression profiling provides insights into the immune mechanism of Plutella xylostella midgut to microbial infection. Gene 647 21–30. 10.1016/j.gene.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maistrou S., Paris V., Jensen A. B., Rolff J., Meyling N. V., Zanchi C. (2018). A constitutively expressed antifungal peptide protects Tenebrio molitor during a natural infection by the entomopathogenic fungus Beauveria bassiana. Dev. Comp. Immunol. 86 26–33. 10.1016/j.dci.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Mesquita R. D., Vionette-Amaral R. J., Lowenberger C., Rivera-Pomar R., Monteiro F. A., Minx P., et al. (2015). Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. U.S.A. 112 14936–14941. 10.1073/pnas.1506226112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E., Nappi A. J., Lemaitre B. (1996). Expression of antimicrobial peptide genes after infection by parasitoid wasps in Drosophila. Dev. Comp. Immunol. 20 175–181. [DOI] [PubMed] [Google Scholar]
- Nishide Y., Kageyama D., Yokoi K., Jouraku A., Tanaka H., Futahashi R., et al. (2019). Functional crosstalk across IMD and toll pathways: insight into the evolution of incomplete immune cascades. Proc. R. Soc. B Biol. Sci. 286:20182207. 10.1098/rspb.2018.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh M. Y., Jo Y. H. (2016). Identification and sequence analysis of two thaumatin-like protein (TmTLP) genes from Tenebrio molitor. Entomol. Res. 46 354–359. 10.1111/1748-5967.12198 [DOI] [Google Scholar]
- Oshima K., Takeda M., Kuranaga E., Ueda R., Aigaki T., Miura M., et al. (2006). IKKε regulates F-actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr. Biol. 16, 1531–1537. 10.1016/j.cub.2006.06.032 [DOI] [PubMed] [Google Scholar]
- Palmer W. J., Jiggins F. M. (2015). Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol. Biol. Evol. 32 2111–2129. 10.1093/molbev/msv093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Jo Y. H., Park K. B., Ko H. J., Kim C. E., Bae Y. M., et al. (2019). TmToll-7 plays a crucial role in innate immune responses against gram-negative bacteria by regulating 5 AMP genes in Tenebrio molitor. Front. Immunol. 10:310. 10.3389/fimmu.2019.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik B. B., Kang S. M., Seo G. W., Lee H. J., Patnaik H. H., Jo Y. H., et al. (2013). Molecular cloning, sequence characterization and expression analysis of a CD63 homologue from the coleopteran beetle, Tenebrio molitor. Int. J. Mol. Sci. 14 20744–20767. 10.3390/ijms141020744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik B. B., Patnaik H. H., Seo G. W., Jo Y. H., Lee Y. S., Lee B. L., et al. (2014). Gene structure, cDNA characterization and RNAi-based functional analysis of a myeloid differentiation factor 88 homolog in Tenebrio molitor larvae exposed to Staphylococcus aureus infection. Dev. Comp. Immunol. 46 208–221. 10.1016/j.dci.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Qu Y., Zhou M., Peng L., Li J., Yan J., Yang P., et al. (2015). Molecular cloning and characterization of IKKepsilon gene from black carp Mylopharyngodon piceus. Fish Shellfish Immunol. 47 122–129. 10.1016/j.fsi.2015.08.033 [DOI] [PubMed] [Google Scholar]
- Ramet M., Manfruelli P., Pearson A., Mathey-Prevot B., Ezekowitz R. A. B. (2002). Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E-coli. Nature 416 644–648. 10.1038/nature735 [DOI] [PubMed] [Google Scholar]
- Ramirez J. L., Garver L. S., Brayner F. A., Alves L. C., Rodrigues J., Molina-Cruz A., et al. (2014). The role of hemocytes in Anopheles gambiae antiplasmodial immunity. J. Innate Immun. 6 119–128. 10.1159/000353765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh K. B., Kim C. H., Lee H., Kwon H. M., Park J. W., Ryu J. H., et al. (2009). Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J. Biol. Chem. 284 19474–19481. 10.1074/jbc.M109.007419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Porras N., Lowenberger C. (2019). The innate immune system of kissing bugs, vectors of chagas disease. Dev. Comp. Immunol. 98 119–128. 10.1016/j.dci.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Santos-Matos G., Wybouw N., Martins N. E., Zele F., Riga M., Leitao A. B., et al. (2017). Tetranychus urticae mites do not mount an induced immune response against bacteria. Proc. Biol. Sci. 284:20170401. 10.1098/rspb.2017.0401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seccareccia E., Pinard M., Wang N., Li S., Burnier J., Dankort D., et al. (2014). The inhibitor of kappa B kinase-epsilon regulates MMP-3 expression levels and can promote lung metastasis. Oncogenesis 3:e116. 10.1038/oncsis.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N., Zhou R., Erlich R. L., Hunter M., Bernstein E., Schneider D., et al. (2003). Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 278 48928–48934. 10.1074/jbc.M304802200 [DOI] [PubMed] [Google Scholar]
- Tanji T., Hu X., Weber A. N., Ip Y. T. (2007). Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 27 4578–4588. 10.1128/MCB.01814-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindwa H., Patnaik B. B., Kim D. H., Mun S., Jo Y. H., Lee B. L., et al. (2013). Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection. Int. J. Mol. Sci. 14 22462–22482. 10.3390/ijms141122462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S., Khush R. S., Leulier F., Tzou P., Nakamura M., Lemaitre B. (2001). Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 15 1900–1912. 10.1101/gad.203301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu Q., Xu X., Hu Y., Hou Q., Zhu Y., et al. (2017). Ctenopharyngodon idella IKKbeta interacts with PKR and IkappaBalpha. Acta Biochim. Biophys. Sin. (Shanghai) 49 729–736. 10.1093/abbs/gmx065 [DOI] [PubMed] [Google Scholar]
- Wu Q., Patocka J., Kuca K. (2018). Insect Antimicrobial peptides, a mini review. Toxins (Basel) 10:461. 10.3390/toxins10110461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi K., Koyama H., Ito W., Minakuchi C., Tanaka T., Miura K. (2012a). Involvement of NF-kappaB transcription factors in antimicrobial peptide gene induction in the red flour beetle, Tribolium castaneum. Dev. Comp. Immunol. 38 342–351. 10.1016/j.dci.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Yokoi K., Koyama H., Minakuchi C., Tanaka T., Miura K. (2012b). Antimicrobial peptide gene induction, involvement of Toll and IMD pathways and defense against bacteria in the red flour beetle, Tribolium castaneum. Results Immunol. 2 72–82. 10.1016/j.rinim.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan M. Y., Yang P. J., Rao X. J. (2018). Molecular cloning and analysis of PGRP-L1 and IMD from silkworm Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 215 19–30. 10.1016/j.cbpb.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Wu G. X., Zhang Z. (2014). Upregulation of coleoptericin transcription in Tenebrio molitor parasitized by Scleroderma guani. J. Asia Pac. Entomol. 17 339–342. 10.1016/j.aspen.2014.03.001 [DOI] [Google Scholar]
- Zou Z., Evans J. D., Lu Z., Zhao P., Williams M., Sumathipala N., et al. (2007). Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8:R177. 10.1186/gb-2007-8-8-r177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.