Abstract
Pacheco's disease (PD) is a common, often fatal, disease of parrots. We cloned a virus isolate from a parrot that had characteristic lesions of PD. Three viral clones were partially sequenced, demonstrating that this virus was an alphaherpesvirus most closely related to the gallid herpesvirus 1. Five primer sets were developed from these sequences. The primer sets were used with PCR to screen tissues or tissue culture media suspected to contain viruses from 54 outbreaks of PD. The primer sets amplified DNA from all but one sample. Ten amplification patterns were detected, indicating that PD is caused by a genetically heterogeneous population of viruses. A single genetic variant (psittacid herpesvirus variant 1) amplified with all primer sets and was the most common virus variant (62.7%). A single primer set (23F) amplified DNA from all of the positive samples, suggesting that PCR could be used as a rapid postmortem assay for these viruses. PCR was found to be significantly more sensitive than tissue culture for the detection of psittacid herpesviruses.
Herpesviruses have been isolated from a range of wild and domestic birds, among which these viruses have been shown to cause considerable morbidity and mortality (11, 15, 19, 22, 32, 42). An avian herpesvirus of particular importance is the psittacid herpesvirus (PsHV), also known as the Pacheco's disease (PD) virus. PD was first recognized as an infectious disease of psittacine birds (parrots) in 1929 (29). Subsequently, it was proposed that the etiologic agent of PD was a herpesvirus (8, 36, 37). More recently, preliminary sequence data suggest that PsHV is an alphaherpesvirus (43). Although described in 1929, PD was not recognized again until the early 1970s in flocks of captive parrots in Florida (40, 41). Subsequently, this disease has been reported from multiple locations within the United States (5, 6, 15, 20, 26, 27, 30, 35, 39) and from around the world, in countries where Central and South American parrots have been imported (3, 7, 13, 14, 21, 23, 24, 25).
PD is almost exclusively a disease of psittacine birds. There is one report of a disease resembling PD in a toucan; however, the relationship between the herpesvirus found in this bird and PsHV is not known (4). Mortality in PD outbreaks varies from the loss of an individual bird to the loss of hundreds of parrots (3, 5–7, 13–15, 20, 21, 23–27, 30, 35, 39). Most diseased birds die suddenly without premonitory signs. When signs occur they are not specific and resemble other systemic infectious diseases. Parrots with signs of disease seldom survive. Although hematologic and serum biochemical changes have been suggestive of PD (12), the diagnosis of PD in live birds has only been made based on virus isolation from feces (10).
Gross necropsy findings of PD have been variable and nonspecific (3, 5, 7, 15, 24, 30, 35, 40, 41). Microscopically, PD has been characterized by moderate to marked, acute hepatic necrosis with minimal associated inflammation and the presence of intranuclear inclusion bodies. The abundance of inclusion bodies, however, has varied from parrot to parrot, and in some cases the inclusion bodies were either rare or absent entirely. Splenic necrosis, enteritis, pancreatitis, tracheitis, and air sacculitis have been variable features of PD (3, 5, 7, 15, 24, 30, 35, 40, 41). Additional assays that have been used to confirm the diagnosis of PD have included immunocytochemical staining of impression smears (15) and paraffin-embedded sections (33), electron microscopy of negatively stained supernatants of crushed tissues, and virus isolation (6, 15–18, 26, 30). In situ hybridization with PD-specific DNA probes has also been reported, but the sequence of these probes and how they were determined to be PD specific was not documented (34).
The epizootiology of PD is incompletely understood. Based on serologic data, it appeared that not all PsHV infections result in disease and that many birds that survived outbreaks were unapparently infected (10). It has been assumed that these birds shed virus either intermittently or continuously (10, 30). Several species of conures have been implicated as sources of outbreaks, but serologic data suggest that many species of parrots, including Amazon parrots and macaws, may also shed this virus (10).
In the last 10 years, it has become apparent that PD may be caused by more than one herpesvirus. Based on serologic cross-reactivity, five PsHV subtypes have been isolated from birds with PD-like lesions (16). Also, limited comparisons of herpesviruses isolated from parrots with PD by restriction enzyme digestion of their entire genome have demonstrated a significant degree of genetic polymorphism (1, 18). The observed variation in the distribution of lesions in birds with PD also suggests that there may be multiple variants of PsHV.
The use of a PCR that is capable of detecting PsHVs in tissues at necropsy and live birds would be of value in controlling the impact of these viruses. Rapid postmortem diagnosis would allow aviculturalists to immediately begin medicating exposed birds and to make management changes that would reduce mortality rates. PCR would also be a valuable adjunct to the pathologist when the histologic lesions were suggestive but not pathognomic for PD. In the live bird, if the PsHV could be found on mucous membranes or in the blood, then unapparently infected birds could be identified and isolated from susceptible birds. Detecting unapparently infected birds is particularly important, since reintroduction efforts of captive birds back into the wild are being contemplated. For these efforts to be successful, it will be essential to ensure that the released birds do not introduce PD to already-threatened native populations.
To date, comparative serology of PsHV isolates has only been done in Europe (16). The development of a PCR assay or assays that could distinguish between the various PsHVs would allow worldwide investigation of these viruses without the need for shipping virus isolates or serum from country to country. Currently, the PD vaccine (Psittimune PDV [Pacheco's Disease Vaccine]; Biomune Co., Lenexa, Kans.) available in North America is monovalent, and it has been suggested that vaccine prepared against serotype 1 did not protect against serotype 2 (25). A study of the diversity and prevalence of PsHV subtypes found in North America would provide valuable evidence as to whether a polyvalent vaccine is needed. Also, a PCR assay capable of amplifying DNA from all PsHVs would allow direct amplification of viral DNA from tissues for genetic studies, bypassing costly and time-consuming virus isolation and purification procedures.
We report here the partial sequence of a PsHV isolated from a parrot with histologic lesions characteristic of PD. These sequence data confirm that this PsHV is an alphaherpesvirus that has evolved substantially from the closest known virus, Gallid HV-1. From the sequence of this virus, five primer pairs were developed for use in PCR. Amplification patterns show that there is a predominant PsHV (variant 1) that causes PD, but there are at least nine less-common variants that are also etiologic agents of PD. A single primer set was found to amplify all 10 variants of the PsHV. PCR was also found to be a highly sensitive assay for the detection of PsHVs in tissues and culture fluids. In contrast, attempts to isolate these viruses in chicken embryo fibroblasts (CEFs) were only successful in 58% of the cases.
MATERIALS AND METHODS
Isolation of herpesviruses.
Virus isolation attempts were made from one or more tissues, including the liver, kidney, and spleen, from 31 parrots suspected to have PD. Each bird represented a different outbreak. When multiple birds were submitted from the same outbreak, only one of the birds was included in this study. CEFs derived from 11-day-old specific-pathogen-free embryos were grown until 75% confluent. The fibroblast monolayer was then incubated with a 10% (wt/vol) homogenate of liver, or combined liver and spleen, in cell culture medium for 1 h (30). The homogenate was aspirated from the cells, the cells were washed, and the monolayers were observed daily for 7 days. The presence of a herpesvirus in the CEF monolayers was confirmed by characteristic cytopathic effects and the presence of eosinophilic intranuclear inclusion bodies within the CEFs. Up to three blind passages were undertaken before it was concluded that the virus would not grow in this culture system.
Purification of reference virus.
The reference herpesvirus (PsHV-R) was isolated from the liver of an Amazon parrot (Amazona oratrix) with hepatic and splenic lesions characteristic of PD. Pancreatic, intestinal, and inguvial lesions were not seen. Herpesvirus infection was confirmed, in this bird, by visualization of herpesvirus virions by electron microscopy of supernatant from crushed tissues (Texas Veterinary Medical Diagnostic Laboratory [TVMDL], College Station, Tex.). The virus was isolated in CEF monolayers. Fluid and cellular debris from the second passage were frozen and thawed three times and cleared by centrifugation at 1,500 × g for 5 min, and the virus was pelleted by centrifugation at 40,000 × g for 4 h. The resulting pellet was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.4]), layered onto a 30% (wt/vol) sucrose-TE buffer cushion, and centrifuged at 40,000 × g for 1 h. The virus pellet was rinsed three times with TE buffer, resuspended in TE, and stored at −20°C. The virus presence in the pellet was confirmed by negative-contrast electron microscopy of an aliquot of the resuspended pellet.
DNA isolation.
The virus suspension was incubated overnight in 0.5% sodium dodecyl sulfate with 0.05 μl of protease K (Promega, Madison, Wis.) per ml at 65°C. The resulting mixture was extracted twice with 1:1 saturated phenol-chloroform. DNA was precipitated by the addition of a 0.1 volume of 10% (wt/vol) sodium acetate and 2.5 volumes 100% ethanol and resolubilized in TE (38).
Cloning reference virus restriction fragments.
PsHV-R DNA and puc18 plasmid (Invitrogen, Carlsbad, Calif.) were restricted with HindIII (Promega), ligated, and transformed into competent Escherichia coli (One Shot Competent E. coli; Invitrogen) using the manufacturer's suggested conditions. Selected recombinant colonies were cultured for 16 to 18 h in lactose broth supplemented with ampicillin (25 mg/ml). Plasmid DNA was isolated using the QIAQuick Gel Extraction Kit (Qiagen, Valencia, Calif.). To verify that the plasmids contained an insert, they were digested with HindIII. Digestion products were separated by electrophoresis in an agarose gel containing ethidium bromide and visualized with UV light (IS-500 Gel Documentation System; Alpha Innotech, San Leandro, Calif.).
Sequencing.
Three clones (9, 11, and 23) were selected for sequencing. Using forward and reverse universal puc primers and the ABI 377 DNA sequencer (Perkin-Elmer Cetus, Norwalk, Conn.), the sequences of both terminal portions of inserts 9 and 11 and the forward end of insert 23 were determined. The newly determined sequence was used to select new primers (MacVector 5.0; Oxford Molecular Group, Campbell, Calif.) that were used to sequence further into the some of the inserts. The sequences were compared to the known databases (GenBank [National Center for Biotechnology Information, Bethesda, Md.], Data Bank of Japan [Mishima, Shizuoka, Japan], and EMBL Nucleotide Sequence Submissions [Cambridge, United Kingdom]) using the Fasta3 (32) and BLASTX (2) programs. Sequence data were submitted to GenBank; the accession numbers are AF261752, AF261753, AF261754, AF261755, and AF261756.
PCR amplification of herpesvirus DNA.
PCR primer sets (Table 1) from the forward sequence of clones 9, 11, and 23 and the reverse sequence of clones 9 and 11 were selected (MacVector 5.0). Purified PsHV-R DNA and each respective clone were used as positive controls. DNA extracted from whole 4-day-old chicken embryos and DNA extracted from the blood of five incubator hatched parrots were used as negative controls. Primer set MCW45, a microsatellite marker for chicken embryonic myosin, was used to rule out the possible presence of chicken DNA in the whole-virus DNA preparation and as a positive control for chicken embryo DNA (M. X. Groenen, www.zod.wav.nl/vf/research/chicken).
TABLE 1.
PsHV derived PCR primers used in this study
| Primer set | Sequence (5′-3′) | Amplicon size (nt)a |
|---|---|---|
| Clone 9 (forward) | TGAGAGGATGTTCGCACGAGC | 489 |
| CGGCTCTTACTTCAACACGATAACC | ||
| Clone 9 (reverse) | GCACATGAGAGACTTCACAGGGC | 378 |
| TCTAACGCAGGGGAAGCTATACG | ||
| Clone 11 (forward) | CAAGTTCAAAACCGTCGC | 394 |
| TTGCCGCTTCTTCGTGCTC | ||
| Clone 11 (reverse) | AATAGGAGCTTTGGGGAGTTGC | 310 |
| ACAGCCTTTTCTGGGTGCG | ||
| Clone 23 (forward) | TTTGTCCCACACTTCGTC | 281 |
| ACTACTTTCGCTTTGGCG |
nt, nucleotides.
The primer sets were used to screen liver, spleen, and/or kidney from parrots diagnosed with PD. In one case, cloacal tissue was the only tissue screened. The diagnosis of PD was based on the presence of one or more of the following criteria: characteristic histologic lesions, detection of herpesvirus by electron microscopy, or detection of herpesvirus-infected cells in impression smears of the spleen or liver by use of a fluorescent-antibody conjugate prepared to a PD isolate. Tissues were collected from diagnostic submissions to Schubot Exotic Bird Center (Texas A&M University), to TVMDL, or directly to the authors. Tissues from 54 parrots, representing 54 independent outbreaks of PD, were collected for this study. The birds submitted represented 16 species; however, the species of 10 birds were not recorded. Genomic DNA was isolated with Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, Minn.) using manufacturer's protocols, rehydrated in H2O, and stored at −20°C until use. In three cases, unfixed tissues were not available. In one case, DNA was extracted directly from formalin-fixed liver; in another case, DNA was extracted from formalin-fixed paraffin-embedded liver (Puregene DNA Isolation Kit; Gentra Systems) using the manufacturer's protocols. The source of a single DNA sample was tissue culture medium from the first passage of the virus.
PCR amplifications were performed in a Programmable Thermal Block II (Lab-line Instruments, Inc., Melrose Park, Ill.) using the following protocol: an initial denaturation step of 94°C for 5 min, followed by 40 cycles of 60°C (58°C for primer set 11R) for 45 s, 72°C for 90 s, and 94°C for 30 s, followed by a final extension cycle of 72°C for 5 min, after which the reaction mix was immediately cooled to 4°C. Each 25-μl reaction mix contained 100 ng of genomic DNA, 25 pmol of each primer, 0.1 mM concentrations of each of the four deoxynucleotide triphosphates, 2.5 mM magnesium chloride, 0.75 U of Taq, and 1× buffer A (all reagents; Promega). Amplicons were separated by electrophoresis on a 1% agarose gel containing ethidium bromide, visualized with UV light, and then photographed. In all specimens for which amplification products were not seen for particular primers or for which the amplification product was of a different molecular mass than the control, the sample was reexamined two more times with the primer set in question.
Statistical analysis.
The chi-square test of independence was used to compare sensitivity of the PCR to that of culture (28). The percentages of variant 1 viruses and pooled variants 2 to 10 were compared between those of Amazon parrot origin and those of African gray parrot origin and between those of Amazon origin and viruses from all other parrot species using the chi-square test of independence. Finally, the percentages of culture-positive variant 1 viruses were compared to the percentages of culture-positive viruses of the pooled variants 2 to 10. Results were considered significant at P ≤ 0.05.
RESULTS
Virus purification.
In order to obtain pure virus for cloning, concentrated virus grown in CEFs was centrifuged through a sucrose cushion. Electron microscopy revealed that the pellet consisted entirely of herpesvirus virions. PCR of the DNA extracted from the virus using a set of primers specific for the chicken myoglobin gene failed to amplify a product, indicating that all cellular DNA had been eliminated.
PsHV sequence.
Virus DNA restricted with HindIII was cloned into a plasmid vector. Three cloned fragments of the PsHV-R, fragments 9 (ca. 4 kb), 11 (ca. 10 kb), and 23 (ca. 2 kb), were partially sequenced. The sequences were compared to the GenBank, Data Bank of Japan, and EMBL Nucleotide Sequence Submissions databases. The organization of PsHV-R was found to be that of an alphaherpesvirus (Table 2). The nucleotide sequences for five of six open reading frames most closely matched those of the Gallid HV-1. The sixth open reading frame coded for UL16, a host range protein. This open reading frame is not present in Gallid HV-1. This nucleotide sequence most closely matched the open reading frame for bovine herpesvirus 1 UL16. The sequence of the reverse end of clone 9 was speculated to fall within UL21. In the n and n+2 reading frames multiple stop codons were present. In the n+1 reading frame, stop codons were not found, suggesting that this sequence may be within an open reading frame. However, there was essentially no identity between this sequence any other sequences in the known data banks, so the precise location of this sequence in the PsHV is not known.
TABLE 2.
Open reading frames within the sequenced fragments of PsHV
| Clone | nta | Corresponding protein | Coding region | Portion of protein | Herpesvirus with closest homologyb (nt [%]) |
|---|---|---|---|---|---|
| 9 forward sequence | 851 | UL19; major capsid protein | 703–1 (R)c | N terminus | Gallid HV-1 (638 [61.8]) |
| 9 reverse sequence | 379 | UL21? | 1–379 | Unknown | None |
| 11 forward sequence | 882 | UL14; unknown function | 259–847 | Complete protein | Gallid HV-1 (361 [64.9]) |
| UL15A; DNA cleaving/packaging protein | 721–882 | N terminus | None | ||
| 11 reverse sequence | 427 | UL9; origin binding protein | 404–1 (R) | C terminus | Gallid HV-1 (303 [60.7]) |
| 23 forward sequence | 1,588 | UL17; DNA cleaving/packaging protein | 1–523 | C terminus | Gallid HV-1 (532 [56.4]) |
| UL16; host-range protein | 508–1531 | Complete protein | Bovine herpesvirus 1 (237 [54.9]) |
nt, nucleotides.
Number of nucleotides compared and percent homology.
R indicates that the open reading frame was found on the reverse strand.
This open reading frame is not present in Gallid HV-1.
PCR amplification of PsHV DNA.
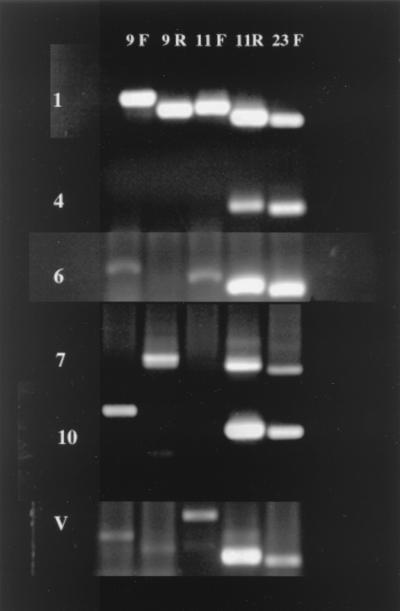
A set of PCR primers was selected from the DNA sequence of each forward and reverse sequence of clones 9 and 11 and in the forward sequence of clone 23 (Table 1). All primers amplified DNA from the reference virus (PsHV-R) and their respective clone. DNA isolated from a specific-pathogen-free chicken embryo and DNA isolated from the blood of five incubator-hatched parrot chicks did not amplify with these primer sets. These primers were then used to amplify DNA from tissues and cell culture media suspected to contain PsHVs. Of the 54 samples screened with the five PCR primer sets, amplification products were produced with one or more primer sets from 53 (98%) of the samples. PCR products were described as either positive, negative, or abnormal (different molecular mass than the control product). Ten patterns of amplification were observed (Table 3). We have called these PsHV variants 1 (PsHV v1) through PsHV variant 10 (PsHV v10). The four most common amplification patterns and two atypical patterns are shown (Fig. 1). Most viruses (61%) were detected with all five primer sets. PsHV v6 was the second most common variant (11%) and amplified with all but primer set 9R. The remaining variants were only represented by one (2%) to four (7.5%) of the viruses assayed. Primer set 23F detected all 53 viruses.
TABLE 3.
PCR amplification patterns of the 10 PsHV variants
| Variant | Amplification patterna of:
|
Total no. of samples per variant | ||||
|---|---|---|---|---|---|---|
| 9F | 9R | 11F | 11R | 23F | ||
| 1 | + | + | + | + | + | 33 |
| 2 | − | + | + | + | + | 4 |
| 3 | − | − | + | + | + | 1 |
| 4 | − | − | − | + | + | 3 |
| 5 | − | − | − | − | + | 1 |
| 6 | + | − | + | + | + | 6 |
| 7 | − | + | − | + | + | 1 |
| 8 | + | − | − | − | + | 1 |
| 9 | + | + | − | + | + | 2 |
| 10 | + | − | − | + | + | 1 |
+, Amplification product observed. −, amplification product not observed.
FIG. 1.
PCR amplification patterns from the three of most common PsHV variants (1, 4, and 6) and two variants represented by a single virus (7 and 10). The last row (V) demonstrates an atypical amplification pattern. Primer set 11F produced a larger-than-expected amplicon. Sequence analysis showed that the product was not herpesvirus DNA and, instead, it is suspected to be DNA of the host.
Atypical amplification products occurred in three birds, all of which were African gray parrots. In these birds primer set 11F did not produce an amplicon of the appropriate mass but did produce a larger one. This product was partially sequenced, and the sequence was not PsHV DNA, nor was it found to match any sequence found in the current DNA databases (data not shown). Therefore, these birds were considered to be negative with primer set 11F. One of these three African gray parrots also produced atypical amplification products with primer sets 9R and 11R. In this bird, amplicons of normal size were produced, as well as two amplicons of smaller molecular mass. The amplicons of appropriate molecular mass were partially sequenced and found to be identical to the PsHV-R sequence. The amplicons of smaller molecular mass were not sequenced. This virus was considered positive for 9R and 11R.
The majority of the viruses studied, 28, originated from Amazon parrots (Amazona spp.) (Table 4). The second most frequent source of virus, with seven submissions, was the African gray parrot (Psitticus erithacis). Seven other species of parrot were also represented. Three samples were labeled only as parrot or, in one case, as cockatoo species. The percentage of variant 1 viruses derived from Amazon parrots compared to the 10 other variants was statistically indistinguishable from the percentage of variant 1 viruses found in African gray parrots or those found in all other non-Amazon parrots combined.
TABLE 4.
Distribution of genera or species of bird as a function of PsHV variant
| PsHV variant | Distribution (no.) of genera or species
|
||||||
|---|---|---|---|---|---|---|---|
| Amazon species | African gray | Macaw species | Conure | Cockatoo species | Cockatiel | Other | |
| 1 | 16 | 2 | 5 | 3 | 2 | 2 | 3 |
| 2 | 1 | 3 | |||||
| 3 | 1 | ||||||
| 4 | 1 | 1 | 1 | ||||
| 5 | 1 | ||||||
| 6 | 5 | 1 | |||||
| 7 | 1 | 1 | |||||
| 8 | 1 | ||||||
| 9 | 2 | ||||||
| 10 | 1 | 1 | |||||
Microscopic lesions found in the tissues of the bird that was negative by PCR were not typical of PD. There was only scattered necrosis of hepatocytes and splenocytes, and intranuclear inclusion bodies were not seen. However, impression smears of the liver and spleen were positive using an immunofluorescent conjugate made against a PsHV. These impression smears were also positive for Chlamydia psittaci using a immunofluorescent conjugate. The sample used for PCR was not optimal since the original tissue collected from this bird was not available, so the frozen suspension used to inoculate the CEFs was used as the source of DNA.
Correlation of culture attempts and PCR amplification.
Isolation attempts were made from 31 fresh or frozen tissues. All but one of these samples was found to be positive by PCR. Of the positive PCR samples, virus was grown from 18 (58%). The only sample suspected to contain a PsHV that was not positive on PCR did not grow virus. PCR was found to be significantly more sensitive than culture in detecting PsHV in necropsy tissues (P ≤ 0.5). There was not a statistically significant difference between the number of variant 1 viruses that grew in cell culture and the number of other variants that grew in cell culture.
DISCUSSION
Sequence data reported here confirm the observations of VanDevanter et al. that the PsHV is an alphaherpesvirus (43). The sequence homology is greatest between PsHV-R and Gallid HV-1, indicating that they are derived from a single progenitor herpesviruses that branched from other known mammalian and avian herpesviruses. However, the presence of extended sequences within the PsHV-R that have virtually no homology to Gallid HV-1 and the presence of an open reading frame corresponding to UL16, which is absent in Gallid HV-1, suggests that these viruses evolved independently of each other for an extended period of time.
At least five serologically distinct PsHVs are believed to be the etiologic agent of PD (16). Restriction enzyme analysis of whole-virus DNA also suggests that there is more than one PsHV variant (1, 18). Our data document 10 PsHV variants. These variants are defined by the ability of the five PCR primer sets to detect their DNA. The failure of DNA amplification implies that the sequence of the primer sets was sufficiently different from that of the target virus that annealing did not occur. We have designated the variants PsHV v1 through v10, as a temporary measure, until the genetic data can be correlated with the serologic data. Not all of these variants are equally as likely to be found in birds with PD. PsHV v1 was present in 61% of the cases and was identified six times more frequently than the next most common virus variant. Although PsHV v1 is the most common variant, a significant number of PD cases reported here (39%) were caused by other variants. We are currently comparing the PsHV variants described here with the DNA amplification patterns found in previously reported, serologically distinct PsHVs.
Specific pathotypes with species specificity were not identified in this study. However, five of seven African gray isolates represented rare variants. These data are not statistically significant with the number of cases available for this study; however, we will continue to seek samples from African gray parrots with PD to determine if this trend is significant.
A single primer set, 23F, was able to detect virus in all but one of the samples (98%), suggesting that this single primer set could be used alone as a highly sensitive means of identifying PsHV in necropsy specimens and tissue culture fluid and possibly in the live bird. Rapid detection of PD outbreaks will result in early treatment of affected flocks, thereby minimizing further losses. We are currently investigating the possibility that these primer sets can be used to detect PsHVs in swabs of the mucous membranes of persistently infected birds. If this line of investigation proves fruitful, we will be able, for the first time, to detect and isolate potential sources of these devastating outbreaks.
Although primer set 23F was able to detect 98% of the samples examined in this study, we cannot rule out the possibility that there are other rare PsHVs causing PD that cannot be detected by primer set 23F. A single sample examined in this study was negative with 23F and the other primer sets. However, this sample was also culture negative and did not have the microscopic lesions consistent with PD. The only reason that it was suspected to contain a PsHV was that it was positive on immunofluorescence assay. The failure of this sample to be detected by our primers could be explained if it contained a divergent herpesvirus whose sequence was not recognized by the primers. We consider it more likely, however, that, because the sample was ground tissue that had been frozen and thawed several times, herpesvirus DNA, if present at all, was degraded past the point of detection.
An amplicon of increased molecular mass was produced by the 11F primer sets with DNA that originated from three African gray parrots (Fig. 1). This DNA was not herpesvirus DNA and is suspected to be African gray parrot DNA, although the sequence could not be matched with previously reported sequences. The origin of the amplicons of lesser molecular weight produced from a sample of a single African gray parrot by primer sets 9R and 11R were not sequenced, and their origin remains unknown.
In this study, herpesvirus DNA was readily detected in the liver and/or spleen and/or kidney. A previous study has shown that PsHVs can be isolated from multiple organ systems in parrots with PD and that many of these birds are viremic at the time of their death (17). Based on that report, it appears that, in addition to liver, spleen, and kidney, other tissues, including syrinx, lung, heart blood, and cerebellum are also excellent samples to examine by PCR. Previous investigations have shown that PsHV is shed in the feces of acutely and chronically ill birds, and the virus is suspected to be shed in the feces of persistently infected birds (10).
At least a portion of the UL17 gene appears to be highly conserved within the PsHV variants that we examined. This leaves open the possibility that the 23F primer set may also prove to be a useful tool for amplifying viral DNA from the as-yet-uncharacterized PsHVs (11, 19, 42) and possibly from the herpesviruses of other avian species.
Other authors have reported that the herpesviruses present in the tissues of parrots with PD are readily isolated in CEFs (6, 17, 20, 26, 30, 40). We were less successful, since only 61.3% of virus isolation attempts resulted in virus growth. The cause of this discrepancy is not known. Because of the relatively low success of virus isolation attempts, the sensitivity of PCR was superior to the sensitivity of virus isolation in this study. PCR also could be performed more quickly than virus isolation; PCR results could be obtained within a single day after the sample was provided.
In summary, the etiologic agents of PD are a genetically heterogeneous population of alphaherpesviruses. We have determined, however, that at least a portion of the UL17 open reading frame is highly conserved between these virus variants and that most or all of them can be detected with a single set of primers. These viruses are thus readily detected, using these primers and the PCR, in tissues from necropsy specimens and tissue culture media. Finally, the sequence data that we have generated now provides investigators and diagnosticians with the opportunity to develop other molecular diagnostic assays such as in situ hybridization.
ACKNOWLEDGMENTS
We acknowledge the following for their financial support of this research: the Department of Large Animal Medicine and Surgery and the Schubot Exotic Bird Health Center, Texas A&M University, the Association of Avian Veterinarians, the Midwestern Avian Research Exhibition, the Geraldine R. Dodge Foundation, the Central Indiana Cage-Bird Club, the Alaska Bird Club, the North County Aviculturalists, the Central Jersey Bird Club, the Long Island Bird Club, Semiconductor Equipment and Materials International, the Miami Valley Bird Club, Kathryne and Richard Thorpe, Barabra A. Brinker, Charles and Margaret Bloodworth, Paul T. and Jacqueline L. Frederickson, Nancy Miller, Martha Gravlee, Mary Lee Leinneweber, Mary Yerardi, Gail Padgett, Michael Ambrose, Rodica Stoicoiu, Joanie Doss, Elizabeth Wilson, Sally Spencer and Lyne Dicker, and Ronald and Linda Wilson.
REFERENCES
- 1.Aini L, Shih L M, Castro A E, Zee Y X. Comparison of herpesvirus isolates from falcons, pigeons, and psittacines by restriction endonuclease analysis. J Wildl Dis. 1993;29:196–202. doi: 10.7589/0090-3558-29.2.196. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canti M, Marchi R, Rampin T, Sironi G, Caniatti M, Cerutti F. Infezione da Herpesvirus in psittacidi riconducibile alla malattia de Pacheco. Zootec Int. 1992;3:88–93. [Google Scholar]
- 4.Charlton B R, Barr B C, Castro A E, David P L, Reynolds B J. Herpes viral hepatitis in a toucan. Avian Dis. 1990;34:787–790. [PubMed] [Google Scholar]
- 5.Chartwright M, Spraker T R, McCluggage D. Psittacine inclusion body hepatitis in an aviary. J Am Vet Med Assoc. 1985;187:1045–1046. [PubMed] [Google Scholar]
- 6.Cho B R, McDonald T L. Isolation and characterization of a herpesvirus of Pacheco's parrot disease. Avian Dis. 1980;24:268–277. [Google Scholar]
- 7.Dharma D N, Sudana I G. Hepatic intranuclear inclusion bodies in a cockatoo bird (Cacatua sulphurea) Avian Dis. 1983;27:301–303. [PubMed] [Google Scholar]
- 8.Findlay G M. Pacheco's parrot disease. Vet J. 1933;89:12. [Google Scholar]
- 9.Fuchs W, Mettenleiter T C. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL 10 gene product as a nonglycosylated and nonessential virion protein. J Gen Virol. 1999;80:2173–2182. doi: 10.1099/0022-1317-80-8-2173. [DOI] [PubMed] [Google Scholar]
- 10.Gaskin J, Raphael B, Major A, Hall G. Proceedings of the American Association of Zoo Veterinarians. Seattle, Wash: AAZV; 1981. Pacheco's disease: the search for the elusive carrier bird; pp. 24–28. [Google Scholar]
- 11.Gerlach H. Viral diseases. In: Ritchie B W, Harrison G J, Harrison L R, editors. Avian medicine: principles and application. Lake Worth, Fla: Wingers Publishing; 1994. pp. 862–948. [Google Scholar]
- 12.Godwin J S, Jacobson E R, Gaskin J M. Effects of Pacheco's parrot disease virus on hematologic and blood chemistry values of Quaker parrots (Myopsitta monachus) J. Zoo Wildl Anim Med. 1982;13:127–132. [Google Scholar]
- 13.Gómez-Villamandos J C, Mozos E, Sherra M A, Fernàndez A, Diaz F. Mortality in psittacine birds resembling Pacheco's disease in Spain. Avian Pathol. 1991;20:541–547. doi: 10.1080/03079459108418791. [DOI] [PubMed] [Google Scholar]
- 14.Gough R E, Alexander D J. Pacheco's disease in psittacine birds in Great Britain 1987 to 1991. Vet Rec. 1993;132:113–115. doi: 10.1136/vr.132.5.113. [DOI] [PubMed] [Google Scholar]
- 15.Graham D L. Acute avian herpesvirus infections. In: Kirk R W, editor. Current veterinary therapy VII. Philadelphia, Pa: The W. B. Saunders Co.; 1980. pp. 704–706. [Google Scholar]
- 16.Gravendyck M, Tritt S, Spenkoch-Piper H, Kaleta E F. Antigenic diversity of psittacine herpesviruses: cluster analysis of antigenic differences obtained from cross-neutralization tests. Avian Pathol. 1996;25:345–357. doi: 10.1080/03079459608419145. [DOI] [PubMed] [Google Scholar]
- 17.Gravendyck M, Balks E, Schröder-Gravendyck A-S, Eskens U, Frank H, Marchang R E, Kaleta E F. Quantification of the herpesvirus content in various tissues and organs, and associated post mortem lesions of psittacine birds which died during an epornithic of Pacheco's parrot disease (PPD) Avian Pathol. 1998;27:478–489. doi: 10.1080/03079459808419372. [DOI] [PubMed] [Google Scholar]
- 18.Günther B M F, Klupp B G, Gravendyck M, Lohr J E, Mettenleiter T C, Kaleta E F. Comparsion of the genomes of 15 avian herpesvirus isolates by restriction endonuclease analysis. Avian Pathol. 1997;26:305–316. doi: 10.1080/03079459708419213. [DOI] [PubMed] [Google Scholar]
- 19.Helfer D H, Schmitz J A, Seefeldt S L, Lowenstine L. A new viral respiratory infection in parakeets. Avian Dis. 1980;24:781–783. [PubMed] [Google Scholar]
- 20.Hiari K, Hitchner S B, Calnek B W. Characterization of paramyxo, herpes, and orbiviruses isolated from psittacine birds. Avian Dis. 1979;23:148–163. [PubMed] [Google Scholar]
- 21.Kaleta E F, Heffels U, Neumann U, Mikami T. Nachweis eines herpesvirus bie Amazonen (Amazona aestiva und Amazona ochrocephala) Zentbl Vet Med. 1980;27:405–411. [PubMed] [Google Scholar]
- 22.Kaleta E F. Herpesviruses of birds—a review. Avian Pathol. 1990;19:193–211. doi: 10.1080/03079459008418673. [DOI] [PubMed] [Google Scholar]
- 23.Kaleta E F, Brinkmann M B. An outbreak of Pacheco's parrot disease in a psittacine bird collection and an attempt to control it by vaccination. Avian Pathol. 1993;22:785–789. doi: 10.1080/03079459308418964. [DOI] [PubMed] [Google Scholar]
- 24.Krautwald E, Foerster S, Herbst W, Schildger B, Kaleta E F. Nachweis eines neuen Herpesvirus bei einem ungewöhnlichen Fall von Pachecoscher Krankheit bei Amazonen and Graupapageien. J Vet Med B. 1988;35:415–420. [PubMed] [Google Scholar]
- 25.Magnino S, Conzo G, Fiorietti A, Menna L F, Rampin T, Sironi G, Fabbi M, Kaleta E F. An outbreak of Pacheco's parrot disease in psittacine birds recently imported to Campania, Italy: isolation of psittacid herpesvirus 2. Zentbl Vet Reihe B. 1996;43:631–637. doi: 10.1111/j.1439-0450.1996.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 26.Martin H T, Early J L, Bridger J C. The isolation of herpesvirus from psittacine birds. Vet Rec. 1979;105:256–258. doi: 10.1136/vr.105.11.256-a. [DOI] [PubMed] [Google Scholar]
- 27.Miller T D, Miller D L, Naqi S A. Isolation of Pacheco's disease herpesvirus in Texas. Avian Dis. 1979;23:753–756. [PubMed] [Google Scholar]
- 28.Ott L. An introduction to statistical methods and data analysis. 3rd ed. Boston, Mass: PWS-Kent Publishing Co.; 1988. [Google Scholar]
- 29.Pacheco G, Bier O. Epizootie chex les perroquets du Brésil. Relations avec le psittacose. C R Soc Biol. 1930;105:109–111. [Google Scholar]
- 30.Panigrahy B, Grumbles L C. Pacheco's disease in psittacine birds. Avian Dis. 1984;28:808–812. [PubMed] [Google Scholar]
- 31.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plummer P J, Alefantis T, Kaplan S, O'Connell P, Shawky S, Schat K A. Detection of duck enteritis virus by PCR. Avian Dis. 1998;42:554–564. [PubMed] [Google Scholar]
- 33.Ramis A, Fondevila D, Tarres J, Ferrer L. Immunocytochemical diagnosis of Pacheco's disease. Avian Pathol. 1992;21:523–527. doi: 10.1080/03079459208418872. [DOI] [PubMed] [Google Scholar]
- 34.Ramis A, Latimer K S, Niagro F D, Campagnoli R P, Ritchie B W, Pesti D. Diagnosis of psittacine beak and feather disease (PBFD) viral infection, avian polyomavirus infection, adenovirus infection and herpesvirus infection in psittacine tissue using DNA in situ hybridization. Avian Pathol. 1994;23:643–657. doi: 10.1080/03079459408419034. [DOI] [PubMed] [Google Scholar]
- 35.Randall D J, Dagless M D, Jones H G R, MacDonald J W. Herpesvirus infection resembling Pacheco's disease in Amazon parrots. Avian Pathol. 1979;8:229–238. doi: 10.1080/03079457908418348. [DOI] [PubMed] [Google Scholar]
- 36.Rivers T M A. A recently described disease of parrots and parakeets differing from psittacosis. Proc Soc Exp Biol Med. 1931;29:155–156. [Google Scholar]
- 37.Rivers T M, Schwentker F F. A virus disease of parrots and parakeets differing from psittacosis. J Exp Med. 1932;55:911–924. doi: 10.1084/jem.55.6.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Senne D A, Pearson J E, Miller L D, Gustaeson G A. Virus isolation from pet birds submitted for importation into the United States. Avian Dis. 1983;27:731–744. [PubMed] [Google Scholar]
- 40.Simpson D F, Hanley J E, Gaskin J M. Psittacine herpesvirus infection resembling Pacheco's parrot disease. J Infect Dis. 1975;131:390–396. doi: 10.1093/infdis/131.4.390. [DOI] [PubMed] [Google Scholar]
- 41.Simpson D F, Hanley J E. Pacheco's parrot disease. Avian Dis. 1977;23:209–219. [PubMed] [Google Scholar]
- 42.Tsai S S, Park J H, Hirai K, Itakura C. Herpesvirus infections in psittacine birds in Japan. Avian Pathol. 1993;22:141–156. doi: 10.1080/03079459308418906. [DOI] [PubMed] [Google Scholar]
- 43.VanDevanter D R, Warrener P, Bennett L, Schultz E R, Coulter L, Garber R L, Rose T M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]