Abstract
The load of Epstein-Barr virus (EBV) in peripheral blood mononuclear cells of transplant recipients represents a predictive parameter for posttransplant lymphoproliferative disorders (PTLD). The aim of our work was to develop a rapid and reliable PCR protocol for the quantification of cell-associated EBV DNA in transplant recipients. In contrast to previous studies, a protocol that facilitated quantification independent of photometric nucleic acid analysis was established. We took advantage of the real-time PCR technology which allows for single-tube coamplification of EBV and genomic C-reactive protein (CRP) DNA. EBV copy numbers were normalized by division by the amount of CRP DNA, with the quotient representing the actual amount of amplifiable genomic DNA per reaction. Coamplification of CRP DNA did not result in a diminished detection limit for EBV. By using the protocol without normalization, EBV copy numbers in 4 out of 10 PTLD patients were within the normal range determined with data for 114 transplant recipients that served as controls. After normalization, however, all of the PTLD patients had a higher viral load than the control population, indicating an increased sensitivity of the assay. Moreover, EBV copy numbers obtained for one patient by conventional quantification and suggestive of relapsing PTLD were within normal range after normalization. We conclude that normalization of PCR signals to coamplified genomic DNA allows a more accurate quantification of cell-bound EBV.
Epstein-Barr virus (EBV)-induced posttransplant lymphoproliferative disorders (PTLD) are a rare but often fatal complication of immunosuppression after bone marrow and organ transplantation. Clinically, PTLD ranges from an infectious mononucleosis-like syndrome—which might respond to reduction of immunosuppression—to a primary extranodal presentation of the disease with a poor prognosis (17). Early diagnosis of PTLD is still a prerequisite for successful treatment, despite advances in therapy (9, 17). Recent studies have demonstrated a direct relationship between the extent of EBV load in peripheral blood mononuclear cells (PBMC) and the risk of developing PTLD (1, 2, 11, 20–22, 24). Although PTLD patients usually exhibit uncommonly high levels of EBV DNA, new evidence that an increased viral load alone might not be related to PTLD development has emerged (18). Thus, an accepted level of EBV load predictive of PTLD development has not been established as yet.
Conventional PCR-based quantification of viral DNA only allows semiquantitative results, and time-consuming hybridization and blotting steps are necessary after amplification (18, 22, 26, 27). These and other disadvantages of conventional quantification have now been overcome by the real-time TaqMan PCR technology using the ABI Prism 7700 sequence detection system (SDS) (8). This technology has already been applied to the detection and quantification of EBV in plasma and cellular DNA (13, 16, 19, 28). Quantification of cell-associated viruses, however, has remained imprecise, since the amount of viral genomes has to be related to the amount of total DNA, which is usually determined by photometry. Photometric nucleic acid analysis is subject to many disturbance factors, and its use thus decreases the validity of total DNA quantification.
The aim of our work was to overcome the inaccurate quantification of cell-associated EBV copies due to erroneous photometric DNA quantification. We have developed a coamplification protocol which allows exact quantification of EBV genomes and total genomic DNA within a single tube. The coamplification protocol was applied to the quantification of EBV DNA in healthy blood donors, otherwise healthy organ recipients, and PTLD patients. By comparison to EBV quantification without normalization to genomic DNA, the diagnostic value of the two protocols with regard to PTLD development was analyzed.
MATERIALS AND METHODS
Cell lines and plasmids.
The EBV-positive Burkitt's lymphoma cell line Namalwa (CRL-1432; American Type Culture Collection) was used as the coamplification standard for EBV and genomic DNA. The pCMVEBNA plasmid (Invitrogen, Carlsbad, Calif.), which carries the complete EBV nuclear antigen 1 (EBNA1) gene, was used for calibration of the Namalwa DNA standard. pCMVEBNA is 5,452 bp long, which corresponds to a molecular weight of 3,598.3. Thus, 1 μg of plasmid represents approximately 1.67 × 1011 copies. As controls, we used the EBV-positive cell lines Daudi and Raji as well as the replication permissive cell lines B95-8, Akata, and P3HR-1/13. Furthermore, the EBV-negative Burkitt's lymphoma cell line BJAB, the endothelial cell line ECV, the chorion carcinoma cell lines BeWo, JAR, and JEG, and cytomegalovirus (CMV) Amplicheck DNA (TEBU, Frankfurt, Germany) as well as herpes simplex virus type 1 (HSV-1) and HSV-2 DNA were tested for specific amplification.
Patients' samples and serology.
Whole blood, PBMC, and B-lymphocyte DNA were prepared simultaneously from 1 EBV-seronegative and 11 EBV-seropositive healthy blood donors and were tested for the presence of EBV DNA. For 2 additional individuals, whole blood and PBMC DNA were examined. Another 3 blood donors provided material for B-cell isolation. PBMC were isolated from buffy coats by standard-density centrifugation and two wash steps in phosphate-buffered saline pH 7.2. B cells were enriched from the PBMC fraction by magnetic cell sorting using a B-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Fluorescence-activated cell sorter analysis revealed a median purity of 98.1% ± 7.0% for the B-cell preparation (data not shown). B-cell-derived DNA of 17 healthy EBV-seronegative donors from a former study (10) was also investigated for EBV-specific DNA. PBMC from 114 heart transplant (HTx) patients (65 female and 49 male; median age, 55 years) were isolated from 9 ml of EDTA-anticoagulated blood. At the time of blood sampling, none of the patient had signs of infection or rejection crisis. All of the HTx patients were EBV seropositive, and 34 (30%) showed serological signs of EBV reactivation (see below).
The PTLD patients from whom samples were collected were children (one heart, two kidney, and six liver transplant recipients; six female and three male; median age, 6 years). PTLD diagnosis was based on clinical and histological criteria as described previously (5). Data for each patient originate from samples obtained at the time of histological diagnosis of PTLD. One liver transplant patient presented with a relapse of PTLD after a clinically stable period of 4 months. Samples of this patient were regarded as two cases of PTLD. The viral load in one patient with primary EBV infection and subsequent PTLD was monitored over a period of 40 weeks.
Serological investigation of patients and healthy controls consisted of anti-EBNA1 immunoglobulin G (IgG) and anti-early antigen (EA) IgG, IgA, and IgM testing using enzyme-linked immunosorbent assays with recombinant antigens (Biotest AG, Dreieich, Germany). A positive result for anti-EBNA1 IgG was defined as EBV seropositivity. Reactivation of latent EBV infection in anti-EBNA1 IgG-positive individuals was defined as either a positive reaction with anti-EA IgM (optical density > 0.5) or a positive reaction with anti-EA IgM regardless of the optical density value, as long as this result was accompanied by additional positive results with anti-EA IgA and anti-EA IgG.
DNA extraction and quantification.
PBMC from patients were isolated within 24 h after blood sampling, and pellets were stored at −20°C until further processing. DNA from all material used was extracted with the QIAamp blood Mini kit (Qiagen, Hilden, Germany). Two hundred microliters of whole blood, or up to 5 × 106 cells, were loaded onto each isolation column according to the manufacturer's recommendations. The DNA was eluted in 200 μl of Aqua Dest. However, 50 μl of distilled water was used for elution of samples with a low PBMC yield. Photometric DNA quantification was performed in duplicate for every sample by means of a dynamic spectral photometer (Hitachi, Tokyo, Japan).
Real-time quantitative PCR.
A coamplification system was developed for EBV DNA quantification by amplifying part of the single BamHI-K rightward (BKRF1) open reading frame (encoding EBNA1) together with human C-reactive protein (CRP) DNA. Coamplification of EBNA1 and CRP DNA was distinguished by means of specific probes labeled with two different reporter dyes. The EBNA1 probe and the CRP probe were respectively labeled with VIC and FAM, two fluorescent dyes whose spectra emitted after laser excitation at 488 nm are easily distinguished by the ABI Prism 7700 SDS (Perkin-Elmer [PE] Applied Biosystems, Weiterstadt, Germany). Primer-probe combinations were designed using Primer Express software (PE Applied Biosystems). The sequence data were obtained from the GenBank sequence database (accession number V01555 for EBV strain B95-8 and accession number M11880 for human CRP). For the EBNA1 gene, several 3′-end sequence variations derived from biopsy material of various malignant EBV-positive tumors have been reported so far (3, 4, 6, 25, 30). Using the OMIGA software program, version 1.1.3 (Oxford Molecular, Oxford, United Kingdom), all sequences published in the GenBank sequence database (n = 25) were compared for mismatches, and the primer-probe combinations for EBNA1 were situated in the most conserved regions. Altogether, a guanidine-to-adenine substitution in one sequence at position 18 of the forward primer and a cytosine-to-guanidine substitution in nine sequences at position 3 of the probe were accepted for the primer-probe design. The primer-probes sequences have already been published elsewhere (28). With regard to the greatest sensitivity, the optimal annealing-extension temperature (from 58 to 64°C) was determined. In addition, chessboard titration of the magnesium chloride concentration ranging from 3.0 to 7.0 mM and of the EBNA1 probe concentration ranging from 50 to 300 nM as well as EBNA1 primer titration (from 100 to 300 nM) were also performed.
Fluorogenic PCR reactions were set up in a reaction volume of 50 μl. Individual master mix concentrations and thermal cycler conditions were as described previously (28). Fluorescent probes were custom-synthesized by PE Applied Biosystems and by TIB MOLBIOL, Berlin, Germany. PCR primers were purchased from TIB MOLBIOL. Ten microliters of cell-derived DNA was analyzed at a photometrically determined concentration of 50 μg/ml. The amplifications were carried out in an ABI Prism 7700 SDS. Each sample was analyzed in duplicate unless otherwise mentioned. At least three negative water blanks (no-template controls) were included in every analysis. Threshold values were calculated as the upper 10-fold standard deviation (SD) of the background fluorescence signal measured over all cycles defined by the baseline. The baseline was set manually from cycle 3 to the cycle before the exponential increase of the first PCR kinetics was to be observed. The threshold cycle (Ct), which is proportional to the starting copy numbers (8) and is defined as the PCR cycle at which the fluorescence signal of the PCR kinetics exceeds the threshold value of the respective analysis, was used for quantification.
Calibration curves were run in parallel and in duplicate with each analysis, using plasmid dilution series or DNA extracted from the Namalwa cell line. Each diploid Namalwa cell harbors two integrated EBV copies/cell, providing equivalent amounts of genomic and viral single-copy genes (14, 29). By coamplification of semilogarithmic dilutions of Namalwa DNA, usually ranging from 101.5 to 105 pg of DNA per analysis, two separate calibration curves for EBNA1 and CRP were calculated. Accordingly, the amount of EBNA1 and CRP DNA determined by coamplification was expressed in picograms of Namalwa DNA. Concentrations of cell-derived EBV DNA were expressed in numbers of EBV copies per microgram of DNA, where the number of micrograms of DNA was either the amount of total DNA determined photometrically or the amount of genomic DNA calculated by CRP DNA coamplification. The number of EBV copies using the coamplification system was calculated using the following equation: C = [AEBV/(ACRP × 3.3)] × 106, in which C represents the target concentration (in number of EBV copies per microgram of PBMC DNA), AEBV represents the amount of EBNA1-specific DNA (in picograms of Namalwa DNA), ACRP represents the amount of CRP-specific DNA (in picograms of Namalwa DNA), and 3.3 is the conversion factor for single-copy genes (in picograms of DNA per copy) (23). Spectral compensation for real-time measurement was activated in every coamplification experiment as recommended by the manufacturer.
Statistical analysis.
Detection limits of the EBNA1 protocol with and without coamplification of genomic CRP DNA were determined by means of Probit analysis. This statistical calculation is based on repetitive determinations (at least eight) of six different target DNA concentrations near the expected detection limit. Detection limits of the target were calculated by plotting the observed detection frequencies against the respective target DNA concentrations. Plasmid concentrations ranging from 100 to 102.5 copies per reaction were used for EBNA1. Plasmid copies were diluted in semilogarithmic intervals. Plasmid dilutions always contained 0.5 μg of EBV-negative DNA derived from an EBV-seronegative donor (10). The software program SPSS for Windows 8.0 (SPSS Inc., Richmond, Calif.) was used for statistical analysis.
RESULTS
Development and statistical analysis of quantitative coamplification protocol for EBNA1 and genomic DNA.
The specificities of the selected primer-probe combinations were determined by examining DNA extracted from several EBV-positive and -negative cell lines. As expected, positive signals were observed for the B-cell lines Namalwa, Akata, P3HR-1/13, Raji, B95-8, and Daudi, with the order reflecting increasing numbers of EBV genomes per microgram of DNA. No amplification was observed using DNA from the cell line BJAB, BeWo, JAR, JEG, or ECV or when CMV or HSV-1 or -2 DNA was used. The single-tube coamplification protocol was then established while taking into account the following considerations. (i) The detection limit of the target sequence, i.e. EBV, should not be diminished by amplification of the internal reference (genomic CRP DNA). To this end, CRP primer concentrations were limited to terminate CRP amplification after a few cycles, avoiding inhibition of EBNA1 amplification. In Table 1, the 50, 95, and 99% detection limits for EBNA1 with and without coamplification are shown. Probit analysis demonstrates that the detection limits of EBNA1 in the presence of high amounts of EBV-negative DNA are not significantly altered by simultaneous coamplification of genomic CRP DNA. (ii) CRP DNA detection should be independent of the coamplified amount of EBV DNA. Therefore, 0.5 μg of DNA derived from an EBV-negative donor (10) was spiked with increasing amounts of the plasmid carrying the gene encoding EBNA1. Three replicates for each plasmid concentration were used. The Ct values for CRP remained stable over a range of from 101 to 106 EBNA1 plasmid copies, thus excluding a significant influence of EBNA1 amplification on the detection of CRP DNA (data not shown).
TABLE 1.
Amplification of the EBNA1 gene with and without coamplification of human genomic CRP DNA
| Detection limit (%)a | No. of EBNA1 plasmid copies per reaction
|
|
|---|---|---|
| EBNA1-CRP coamplification | EBNA1 amplification | |
| 50 | 3.2 | 2.0 |
| 95 | 5.5 | 5.5 |
| 99 | 8.6 | 7.2 |
Probit analysis was used for calculation of the detection limits as described in the text.
Using all of the established parameters, we determined the linear range of the single-tube coamplification system. Typical coamplification experiments showed valid detection of EBNA1 and CRP within a range from 100.5 × 105 to 5 × 105 pg of Namalwa DNA.
Finally, the coefficients of variation (CV) for the coamplification protocol were determined and compared to EBV quantification without normalization to genomic DNA. For CV determination, four experiments, each analyzing five replicates of 1 ng of Namalwa DNA were performed. Calibration curves employing Namalwa DNA as well as the pCMVEBNA plasmid were run in parallel for each experiment and were used for separate quantification. Table 2 shows the respective CV of EBNA1 quantification with and without normalization to genomic CRP DNA. Taken together, intra- and interassay CV of EBNA1 quantification were not significantly reduced by normalization to genomic DNA and were within the range of results obtained with the use of other protocols. In addition, the four experiments revealed a mean value of 303.0 EBV copies per nanogram of Namalwa DNA when calibrated against the EBNA1 plasmid pCMVEBNA.
TABLE 2.
Quantification of 1 ng of Namalwa DNAa
| Gene(s) amplified | DNA or plasmid used for calibration | CV (%)
|
|
|---|---|---|---|
| Intraassay | Interassay | ||
| EBNA1 and CRP | Namalwa DNA | 16.6 | 10.8 |
| EBNA1 | Namalwa DNA | 13.7 | 4.4 |
| EBNA1 | EBNA1 plasmid | 13.1 | 19.1 |
Simultaneously calibrated against three different calibrators: Namalwa DNA either normalized to coamplified genomic CRP DNA or not and the plasmid carrying the gene for EBNA1, pCMVEBNA.
Quantification of EBV genomes in healthy blood donors, immunosuppressed HTx transplant recipients, and patients with PTLD.
First, we investigated which peripheral cell fraction was a reliable source of EBV DNA detection by using the coamplification technique. To this end, whole blood, PBMC, and B lymphocytes were simultaneously collected from healthy individuals and prepared. EBV detection frequencies are summarized in Table 3. Only B-cell-derived DNA was sufficient for reliable EBV detection in nonimmunocompromised EBV-seropositive individuals. EBV DNA negativity in the peripheral blood was unequivocally related to EBV seronegativity. None of the seronegative adults were positive for EBV DNA, and 100% of the healthy EBV-seropositive adults gave positive PCR signals when their B-cell-derived DNA was analyzed. Considerably different quantitative results for the healthy adult population were obtained with different calibrators. EBV copy numbers calibrated against the EBNA1 plasmid ranged from 0 to 37 copies/μg of PBMC DNA (median, 7 copies/μg) and from 0 to 53 copies/μg of PBMC DNA (median, 12 copies/μg) using Namalwa DNA, but revealed, on average, two- to threefold-higher levels when normalized to amplifiable genomic CRP DNA, with values ranging from 0 to 264 copies/μg of PBMC DNA (median, 27 copies/μg).
TABLE 3.
Frequencies of detection of EBV DNA with the coamplification systema
| DNA source | No. of samples positive/no. tested for healthy blood donors that were:
|
|
|---|---|---|
| EBV seronegative | EBV seropositive | |
| Whole blood | 0/1 (0) | 6/13 (46.2) |
| PBMC | 0/1 (0) | 11/13 (84.6) |
| B cells | 0/18 (0) | 14/14 (100) |
Results shown are for DNA extracted from the three sources listed. Detection frequencies (values are percentages) are shown in parentheses.
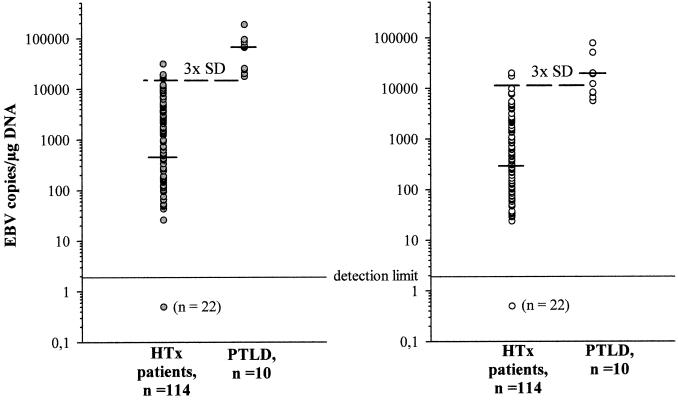
After examination of the healthy individuals, consecutively collected samples from 114 HTx recipients were analyzed for EBV load. PBMC-derived DNA was used for all further studies, since B-cell isolation from peripheral blood was impractical for routine analysis. Quantification was performed by using the coamplification system, so that the results were either normalized to the amount of coamplified CRP DNA or not. As shown in Fig. 1, EBV copy numbers substantially varied in immunosuppressed HTx recipients, ranging from 0 to 31,549 copies/μg of DNA (median ± SD, 454 ± 4,932 copies/μg) when normalized to amplifiable genomic DNA and from 0 to 19,946 copies/μg of DNA (median ± SD, 290 ± 2,974 copies/μg) without normalization. PTLD patients exhibited significantly higher levels of EBV DNA, with values ranging from 17,674 to 187,478 copies/μg of DNA (median ± SD, 67,445 ± 51,470 copies/μg) when normalized to amplifiable genomic DNA and from 5,595 to 78,090 copies/μg of DNA (median ± SD, 15,665 ± 25,007 copies/μg) without normalization.
FIG. 1.
EBV copy numbers in healthy immunosuppressed individuals and in patients with PTLD. The viral load in the PBMC of 114 otherwise healthy HTx recipients and of 10 patients with PTLD was determined. EBV copy numbers were calibrated against Namalwa DNA, and the results were either normalized to the amount of coamplified genomic DNA (dark gray circles) or not (white circles). Shown are individual values, with the short dashes indicating median values of the respective patient groups and the broken lines indicating the threefold SD level of the HTx control group. 3x, threefold.
The main goal of this study was to investigate whether normalization of EBV copy numbers to the actual amount of amplifiable genomic DNA improves the diagnostic value of EBV load determination with regard to PTLD development. To this end, the threefold SD levels of the HTx control group were calculated for both quantitative protocols—i.e., 14,796 copies/μg of DNA for quantification with normalization and 8,922 copies/μg of DNA without normalization—and were considered to be the normal range for immunosuppressed but otherwise healthy individuals. As shown in Fig. 1, EBV copy numbers in 4 out of 10 PTLD patients were within the normal range of the control group by quantification without normalization. After normalization, however, all PTLD patients were beyond the normal range. Statistical analysis of the threefold SD level for diagnosis of PTLD can be summarized with the following values: for normalizing numbers of EBV copies to coamplified genomic CRP DNA levels, sensitivity, 100%; specificity, 96%; positive predictive value, 67%; negative predictive value, 100%; and for conventional quantification, sensitivity, 60%; specificity, 97%; positive predictive value, 67%; negative predictive value, 97%.
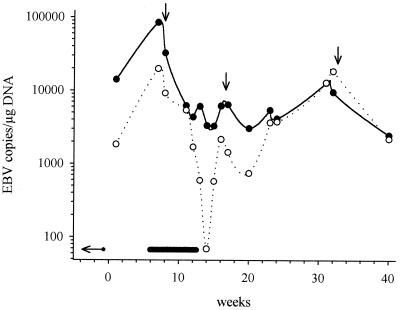
Finally, the time-course of EB viral load was investigated for one kidney transplant patient who developed PTLD after primary EBV infection. For both quantitative protocols, Fig. 2 demonstrates that PTLD diagnosis 7 weeks after the primary infection was accompanied by the highest EBV copy numbers observed during a 40-week follow-up. Tapering of immunosuppression led to a significant decrease of the EBV load, indicating regression of lymphoproliferation. By means of the normalization protocol, after PTLD regression, copy numbers reached a stable level of approximately 4,000 copies/μg of DNA, which suggests there was no further disease activity. In contrast, EBV copies upon conventional quantification varied considerably during the follow-up and even reached initial PTLD levels after 31 to 32 weeks. At that time, EBV load was found to be beyond the normal range of the HTx control group. The clinical course of the patient, however, was stable the entire time; no evidence of a PTLD relapse was detected by clinical examination or imaging.
FIG. 2.
EBV copy numbers in one patient with primary EBV infection and PTLD after kidney transplantation: comparison of quantification with and without normalization to genomic DNA. The viral load of one kidney transplant recipient who developed PTLD after primary EBV infection was monitored over a period of 40 weeks. Shown are results either normalized to the amount of coamplified genomic DNA (black circles) or not (white circles). The time points of primary EBV infection (horizontal arrow) PTLD duration (bold line) and tapering of immunosuppression (vertical arrows) are indicated.
DISCUSSION
Through the presence of two or more fluorescent dye-labeled probes, the TaqMan PCR technology allows the simultaneous detection of different sequences within a single tube (8). Thus, the amplification signal of an internal reference may be directly used for normalization of the amount of target. This approach has been integrated into our single-tube coamplification protocol. It is the first one published that overcomes all the disadvantages of previous quantitative EBV PCR protocols. It combines a control for DNA amplifiability with an internal calibrator for relative quantification. Furthermore, it is independent of varying DNA and RNA isolation efficiencies and exact photometric nucleic acid analysis, both of which significantly reduce the quantification precision. The coamplification protocol was based on the detection of the single open reading frame BKRF1 encoding the gene for EBNA1. Polymorphisms of EBNA1 have been the subject of interest in the context of certain malignancies (3, 4, 6, 25, 30). The primer-probe combination for EBNA1 was situated in the most conserved region of BKRF1, providing a very high detection sensitivity for different EBV strains. For amplification of genomic DNA, the CRP single-copy gene was selected, since it is well conserved and no deletions have been reported to date (15).
Until now, relative quantification of DNA viruses was dependent on externally added competitors and time-consuming post-PCR handling (1, 2, 22, 26). Although the TaqMan PCR technology has had great impact on the ability to quantify relative levels of mRNA (7, 8), it has rarely been used for relative quantification of DNA sequences, especially DNA viruses. At present, there is only one study which normalized EBV amplification signals to the quantitative results of an internal reference; however, this control was derived from a separate reaction (16). Therefore, in contrast to our protocol, it did not exclude pipetting errors and tube-to-tube variations as a cause of erroneous quantification.
DNA from the Burkitt's lymphoma cell line Namalwa was used for quantification by our coamplification system. For determination of the amount of Namalwa DNA, we had to rely on photometric nucleic acid analysis. However, calibration of the Namalwa standard using the pCMVEBNA plasmid revealed a mean value of 303.0 EBV copies per ng of Namalwa DNA. Given a DNA content of 6.6 pg per diploid cell (23), this corresponds exactly to 2 EBV copies per cell. Therefore, the number of EBV DNA copies determined by normalization to genomic DNA may be interpreted in absolute numbers.
By using our real-time PCR protocol, we found significantly higher EBV copy numbers in the PBMC of transplant recipients than would be suggested by semiquantitative PCR (1, 2, 22, 26). Rowe et al., for example, found a viral load of between <8 and 600 copies/μg of DNA by conventional PCR in pediatric transplant patients (22), whereas another group reported a median EBV DNA value of 20 copies/μg of DNA (range, <10 to 3,000 copies/μg) in an adult transplant population (2). The increase in EBV load observed in the present study (median, 290 copies/μg of DNA; range, 0 to 19,946 copies/μg [without normalization]) is readily explained by the use of the more sensitive real-time PCR technique. However, these results are in contrast to those of Kimura et al., who reported a mean level of only 20 copies/μg of DNA by real-time PCR (13). Interestingly, normalization of EBV copy numbers resulted in a further increase of the EBV load in our healthy transplant population (median, 454 copies/μg of DNA; range, 0 to 31,549 copies/μg of DNA). It must be noted that this increase was not observed in general, which argues against biased results (Fig. 2). On the contrary, a systematic influence of coamplification on the detection limit of EBV as well as on the amount of genomic DNA was ruled out. Furthermore, the CV of the coamplification protocol were within the range of other quantitative real-time PCR assays (Table 1). Nevertheless, it should be emphasized that quantification of EBV DNA normalized to genomic DNA is still a relative determination and that the unit of measurement, i.e., number of copies per microgram of DNA, does not correlate with the number of micrograms of DNA determined by photometry.
Finally, we addressed the question of whether normalized quantification improves the predictive value of EBV load determination for the diagnosis of PTLD. The specificity and the positive and negative predictive values did not differ significantly for the two protocols. However, sensitivity was considerably increased by coamplification. It is of importance that the threefold SD level resulted from a homogenous population of adult HTx patients but was applied to a heterogenous group of pediatric PTLD patients. However, the aim of our study was to compare two quantitative procedures, not to establish a normal and pathological range of EBV load in immunosuppressed individuals. Nevertheless, our results confirm those from other studies which had already indicated a strong relationship between the extent of EBV load and the onset of PTLD (1, 2, 11, 18, 20–22, 24). Our data underlines the importance of viral load monitoring for patients at high risk for developing PTLD.
A remarkable difference was observed comparing the viral load in follow-up samples of one patient with PTLD. Upon normalization, the EBV copy numbers remained rather stable after recovery from the disease but fluctuated substantially and even reached PTLD levels without normalization. This was in striking contrast to the clinical course of the patient who did not show any signs of a disease relapse. The fluctuating results achieved by conventional quantification are most likely explained by erroneous photometry due to contaminated DNA preparations (preparations contaminated with, e.g., ethanol, hemoglobin, or other proteins). In the context of this patient, it should be noted that the EBV load in the periphery seems tightly controlled and remains stable without underlying pathology for a period of years (12). Therefore, the longitudinal course of the EBV load determined by normalization seems to reflect the in vivo situation of this patient more appropriately. Further studies are required to analyze the longitudinal course of the EBV load by normalized quantification in a large population of healthy individuals and transplant recipients in relation to disease activity and reactivation of EBV.
In conclusion, single-tube coamplification of an internal reference and normalization to the amount of genomic DNA significantly improves quantification of cell-associated EBV genomes. This approach offers many advantages over conventional quantification systems and might also improve quantification of other cell-bound viruses, such as CMV.
ACKNOWLEDGMENTS
Part of this work was supported by the “Deutsche Krebshilfe” grant 70-2297-Wa I.
We are indebted to G. Offner, Department of Pediatrics, Hannover School of Medicine, Hannover, Germany; P. Emrich, Department of Pediatrics, Technical University of Munich, Munich, Germany; and T. Voigt, Department of Pediatrics, University Hospital Essen, Essen, Germany for providing DNA samples from PTLD patients. Furthermore, we thank Una Doherty, Glasgow, United Kingdom, and K. Chang, Shreveport, La., for critically reviewing the manuscript.
REFERENCES
- 1.Bai X, Hosler G, Rogers B B, Dawson D B, Scheuermann R H. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem. 1997;43:1843–1849. [PubMed] [Google Scholar]
- 2.Baldanti F, Grossi P, Furione M, Simoncini L, Sarasini A, Comoli P, Maccario R, Fiocchi R, Gerna G. High levels of Epstein-Barr virus DNA in blood of solid-organ transplant recipients and their value in predicting post-transplant lymphoproliferative disorders. J Clin Microbiol. 2000;38:613–619. doi: 10.1128/jcm.38.2.613-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatia K, Raj A, Gutiérrez M I, Judde J G, Spangler G, Venkatesh H, Magrath I T. Variation in the sequence of Epstein Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt's lymphomas. Oncogene. 1996;13:177–181. [PubMed] [Google Scholar]
- 4.Chang K L, Chen Y Y, Chen W G, Hayashi K, Bacchi C, Bacchi M, Weiss L M. EBNA-1 gene sequences in Brazilian and American patients with Hodgkin's disease. Blood. 1999;94:244–250. [PubMed] [Google Scholar]
- 5.Craig F E, Gulley M L, Banks P M. Posttransplantation lymphoproliferative disorders. Am J Clin Pathol. 1993;99:265–271. doi: 10.1093/ajcp/99.3.265. [DOI] [PubMed] [Google Scholar]
- 6.Gutiérrez M I, Raj A, Spangler G, Sharma A, Hussain A, Judde J G, Tsao S W, Yuen P W, Joab I, Magrath I T, Bhatia K. Sequence variations in EBNA-1 may dictate restriction of tissue distribution of Epstein-Barr virus in normal and tumor cells. J Gen Virol. 1997;78:1663–1670. doi: 10.1099/0022-1317-78-7-1663. [DOI] [PubMed] [Google Scholar]
- 7.Härtel C, Bein G, Kirchner H, Klüter H. A human whole-blood assay for analysis of T-cell function by quantification of cytokine mRNA. Scand J Immunol. 1999;49:649–654. doi: 10.1046/j.1365-3083.1999.00549.x. [DOI] [PubMed] [Google Scholar]
- 8.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 9.Heslop H E, Rooney C M. Adoptive cellular immunotherapy for EBV lymphoproliferative diseases. Immunol Rev. 1997;157:217–222. doi: 10.1111/j.1600-065x.1997.tb00984.x. [DOI] [PubMed] [Google Scholar]
- 10.Jabs W J, Paulsen M, Wagner H J, Kirchner H, Klüter H. Analysis of Epstein-Barr virus (EBV) receptor CD21 on peripheral B lymphocytes of long-term EBV− adults. Clin Exp Immunol. 1999;116:468–473. doi: 10.1046/j.1365-2249.1999.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenagy D N, Schlesinger Y, Weck K, Ritter J H, Gaudreault-Keener M M, Storch G A. Epstein-Barr virus DNA in peripheral blood leukocytes of patients with posttransplant lymphoproliferative disease. Transplantation. 1995;60:547–554. doi: 10.1097/00007890-199509270-00005. [DOI] [PubMed] [Google Scholar]
- 12.Khan G, Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Is EBV persistence in vivo a model for B cell homeostasis? Immunity. 1996;5:173–179. doi: 10.1016/s1074-7613(00)80493-8. [DOI] [PubMed] [Google Scholar]
- 13.Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Matsuyama T, Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence J B, Villnave C A, Singer R H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 15.Lei K J, Liu T, Zon G, Soravia E, Liu T Y, Goldman N D. Genomic DNA sequence for human C-reactive protein. J Biol Chem. 1985;260:13377–13383. [PubMed] [Google Scholar]
- 16.Lo Y M D, Chan L Y S, Lo K W, Leung S F, Zhang J, Chan A T C, Lee J C K, Hjelm N M, Johnson P J, Huang D P. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 17.Lucas K G, Pollock K E, Emanuel D J. Post-transplant EBV-induced lymphoproliferative disorders. Leukemia Lymphoma. 1998;25:1–8. doi: 10.3109/10428199709042491. [DOI] [PubMed] [Google Scholar]
- 18.Lucas K G, Burton R L, Zimmerman S E, Wang J, Cornetta K G, Robertson K A, Lee C H, Emanuel D J. Semiquantitative Epstein-Barr virus (EBV) polymerase chain reaction for the determination of patients at risk for EBV-induced lymphoproliferative disease after stem cell transplantation. Blood. 1998;91:3654–3661. [PubMed] [Google Scholar]
- 19.Niesters H G M, van Esser J, Fries E, Wolthers K C, Cornelissen J, Osterhaus A D M E. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol. 2000;38:712–715. doi: 10.1128/jcm.38.2.712-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preiksaitis J K, Diaz-Mitoma F, Mirzayans F, Roberts S, Tyrrell D L J. Quantitative oropharyngeal Epstein-Barr virus shedding in renal and cardiac transplant recipients: relationship to immunosuppressive therapy, serologic responses, and the risk of posttransplant lymphoproliferative disorder. J Infect Dis. 1992;166:986–994. doi: 10.1093/infdis/166.5.986. [DOI] [PubMed] [Google Scholar]
- 21.Riddler S A, Breinig M C, McKnight J L C. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood. 1994;84:972–984. [PubMed] [Google Scholar]
- 22.Rowe D T, Qu L, Reyes J, Jabbour N, Yunis E, Putnam P, Todo S, Green M. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J Clin Microbiol. 1997;35:1612–1615. doi: 10.1128/jcm.35.6.1612-1615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 24.Savoie A, Perpete C, Carpentier L, Joncas J, Alfieri C. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood. 1994;83:2715–2722. [PubMed] [Google Scholar]
- 25.Snudden D K, Smith P R, Lai D, Ng M H, Griffin B E. Alterations in the structure of the EBV nuclear antigen, EBNA1, in epithelial cell tumors. Oncogene. 1995;10:1545–1552. [PubMed] [Google Scholar]
- 26.Stevens S J, Vervoort M B, van den Brule A J, Meenhorst P L, Meijer C J, Middeldorp J M. Monitoring of Epstein-Barr virus DNA load in peripheral blood by quantitative competitive PCR. J Clin Microbiol. 1999;37:2852–2857. doi: 10.1128/jcm.37.9.2852-2857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner H J, Bein G, Bitsch A, Kirchner H. Detection and quantification of latently infected B lymphocytes in Epstein-Barr virus-seropositive, healthy individuals by polymerase chain reaction. J Clin Microbiol. 1992;30:2826–2829. doi: 10.1128/jcm.30.11.2826-2829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wandinger K P, Jabs W, Siekhaus A, Bubel S, Trillenberg P, Wagner H J, Wessel K, Kirchner H, Hennig H. Association between clinical disease activity and Epstein-Barr virus reactivation in MS. Neurology. 2000;55:178–184. doi: 10.1212/wnl.55.2.178. [DOI] [PubMed] [Google Scholar]
- 29.Whitaker A M. The chromosomes of the Namalwa cell line. J Biol Stand. 1985;13:173–175. doi: 10.1016/s0092-1157(85)80024-x. [DOI] [PubMed] [Google Scholar]
- 30.Wrightham M N, Stewart J P, Janjua N J, Pepper S D V, Sample C, Rooney C M, Arrand J R. Antigenic and sequence variation in the C-terminal unique domain of the Epstein-Barr virus nuclear antigen EBNA-1. Virology. 1995;208:521–530. doi: 10.1006/viro.1995.1183. [DOI] [PubMed] [Google Scholar]