Abstract
We evaluated the ability of the AccuProbe (Gen-Probe, San Diego, Calif.) to detect Mycobacterium gordonae and Mycobacterium avium complex directly in liquid medium flagged positive by the MB/BacT (Organon Teknika Corp., Durham, N.C.). Seventy-one bottles from clinical specimens containing M. gordonae and 34 containing M. avium, confirmed by culture, were tested by direct AccuProbe assay for both organisms after additional incubation for ≥48 h and centrifugation at 4,500 × g for 15 min. Relative light unit (RLU) values were analyzed using the manufacturer's recommended cutoff of 30,000 RLU and a lower cutoff of 10,000 RLU. Using the 30,000 RLU cutoff, 55 of 71 (77.5%) specimens containing M. gordonae yielded positive results, whereas 28 of 34 (82.3%) M. avium complex specimens were correctly identified by direct probe. No specimens shown by culture to contain either M. gordonae or M. avium complex tested positive with the probe for the opposite organism (100% specificity). When the cutoff was lowered to 10,000 RLU, 67 of 71 M. gordonae (94.4%) and 32 of 34 M. avium complex (94.1%) specimens were correctly identified. This difference was significant for M. gordonae (P = 0.004) but not for M. avium complex (P = 0.26) compared to detection using the recommended RLU cutoff. Specificity was 100% for specimens containing M. gordonae that were tested with the M. avium complex probe using the 10,000 RLU cutoff, whereas specificity for specimens containing M. avium complex tested with the M. gordonae probe was 97%. Using a lower RLU cutoff for determining a positive result using the M. gordonae or M. avium complex probes when testing instrument-positive MB/BacT bottles directly will improve sensitivity without substantially compromising specificity.
The continuing global threat of tuberculosis has led to an urgent need for rapid and accurate diagnostic procedures for detection of mycobacterial growth (2, 3, 4). The MB/BacT (Organon Teknika Corp., Durham, N.C.) is a continuously monitored, nonradiometric colorimetric CO2 detection system with computerized database management that was approved for detection of mycobacterial growth in the United States in 1996 (3). An advantage of this system is the possibility of rapid identification of mycobacteria using nucleic acid hybridization to detect species-specific rRNA by chemiluminescence directly in culture bottles flagged positive by the instrument (3; K. Couchot and R. Talbot, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. U-7, p. 102). Nucleic acid probes (AccuProbe) sold commercially in the United States (Gen-Probe Inc., San Diego, Calif.) have been developed for the identification of the Mycobacterium tuberculosis complex, the Mycobacterium avium complex, Mycobacterium intracellulare, Mycobacterium gordonae, and Mycobacterium kansasii. Use of AccuProbe to identify mycobacteria directly from MB/BacT-positive bottles is not specifically endorsed by the manufacturer of either product, but it is potentially a major advantage that can reduce turnaround time for reporting a positive culture (2). Preliminary studies suggested that direct probing without further incubation or concentration of mycobacteria by centrifugation is adequate for identification of M. tuberculosis complex (2, 3), the organism for which greatest emphasis has been placed in development of these diagnostic products. However, the same cannot be said of M. gordonae and M. avium complex. Over a period of several months after we began using direct probes of positive MB/BacT bottles in 1998, we encountered 37 of 74 (50%) specimens containing M. gordonae and 8 of 33 (24.2%) containing M. avium complex that gave false-negative results using the manufacturer's cutoff of 30,000 relative light units (RLU) compared to probes of colonies eventually detected on solid medium.
Although M. gordonae is rarely of clinical significance (6), its ubiquity in the environment makes it one of the most common mycobacterial species isolated from clinical specimens, accounting for 57% of all positive mycobacterial cultures in our laboratory in 1999. Its morphology in acid-fast smears from positive MB/BacT bottles can sometimes be indistinguishable from that of M. tuberculosis complex, and it can therefore cause diagnostic confusion and the inconvenience of unnecessary patient isolation until it can be properly identified.
Due to concern about the ability of the AccuProbe to adequately identify both M. gordonae and M. avium complex, we undertook an evaluation to optimize the use of AccuProbe in MB/BacT-positive bottles using centrifugation, incubating for at least 48 h before probing, and modifying the cutoff for designation of a positive test.
MATERIALS AND METHODS
Study design.
Based on the poor analytical sensitivity of the M. gordonae and M. avium complex probes for accurate mycobacterial identification when using them to probe directly from instrument-positive bottles, we implemented a laboratory policy that mandated further incubation of MB/BacT bottles for at least 48 h after being flagged positive and centrifugation at 4,500 × g for 15 min prior to direct probing of the pellet. This was an attempt to improve the ability of AccuProbe to detect M. gordonae and M. avium complex by increasing the number of organisms in the tested sample. A total of 105 specimens encountered from June 1999 through March 2000 were probed for both M. gordonae and M. avium complex under these conditions. Direct probe results using this modified protocol were compared to species identifications using probes of mycobacterial colonies eventually grown from the same specimens.
Specimen processing and inoculation of cultures.
Specimens consisted primarily of respiratory secretions, including sputum, tracheal aspirates, bronchoalveolar lavage fluid, and lung tissue. All specimens were inoculated within 24 h of collection onto a Middlebrook 7H11/7H11 selective agar biplate (Remel, Lenexa, Kans.) and into MB/BacT bottles with a revised supplement kit as described previously by Benjamin et al. (3). Specimens positive by fluorescence with Auramine O fluorochrome (Remel) were confirmed with Kinyoun's modification of the Ziehl-Neelsen acid-fast stain.
Incubation and detection of positive specimens.
Inoculated MB/BacT bottles were placed in the MB/BacT incubator/cabinet for continuous monitoring for 6 weeks or until flagged positive, according to the manufacturer's instructions. Agar plates were incubated at 35°C in an atmosphere of 5 to 10% CO2 in air. Auramine O smears were prepared from all instrument-positive MB/BacT bottles. Positive Auramine O smears were confirmed with Kinyoun's stain. Positive bottles were subcultured to a Middlebrook agar plate (Remel) if there was no concomitant growth on the original biplate at the time that the bottle was designated positive by the MB/BacT. The 7H11 agar plates were examined twice weekly for a total of 8 weeks before being designated negative. All flagged bottles which grew nonmycobacterial organisms were stained weekly for the remainder of the 6 weeks with Auramine O to detect any mycobacteria growing in these bottles.
Mycobacterial species identification.
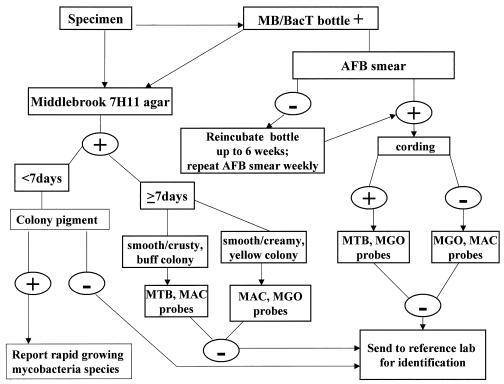
For purposes of this evaluation, M. gordonae and M. avium complex probes were both used to identify mycobacterial species present in each instrument-positive bottle. Specimens encountered during the study period that were found to contain other mycobacterial species either by use of the M. tuberculosis complex probe or by culture isolation of other nontuberculous mycobacteria were excluded from analysis. The M. tuberculosis complex probe has worked consistently for direct testing of positive MB/BacT bottles without false-negative results compared to probe of isolated colonies, so there was no need to evaluate it as part of this investigation. AccuProbe assays were performed in batches on a weekly basis according to the manufacturer's instructions and using all necessary controls. Smears of flagged bottles were evaluated to determine presumptive identification to guide AccuProbe selection for each isolate (1, 5). Bottles with cording were probed with M. tuberculosis complex and M. gordonae probes because we found that 10% of the M. gordonae isolates produced cords (data not shown). Bottles with no cording were probed with M. avium complex and M. gordonae probes. The identification scheme used in our laboratory is illustrated in Fig. 1.
FIG. 1.
Diagram illustrating the culture and identification scheme for mycobacterial cultures using the MB/BacT and AccuProbe with high-pressure liquid chromatography backup for identification. AFB, acid-fast bacillus; MGO, M. gordonae; MAC, C, M. avium complex.
Data analysis.
AccuProbe assay sensitivity and specificity for detection of M. gordonae and M. avium complex were calculated for two different diagnostic cutoffs in RLU for specimens that had been centrifuged following an additional 48 h or more of incubation after being flagged as positive, and results were compared by chi square.
RESULTS
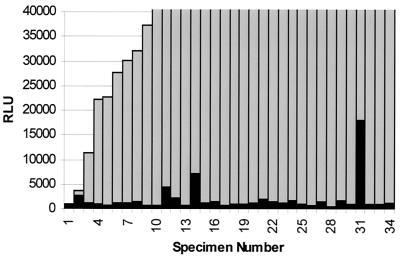
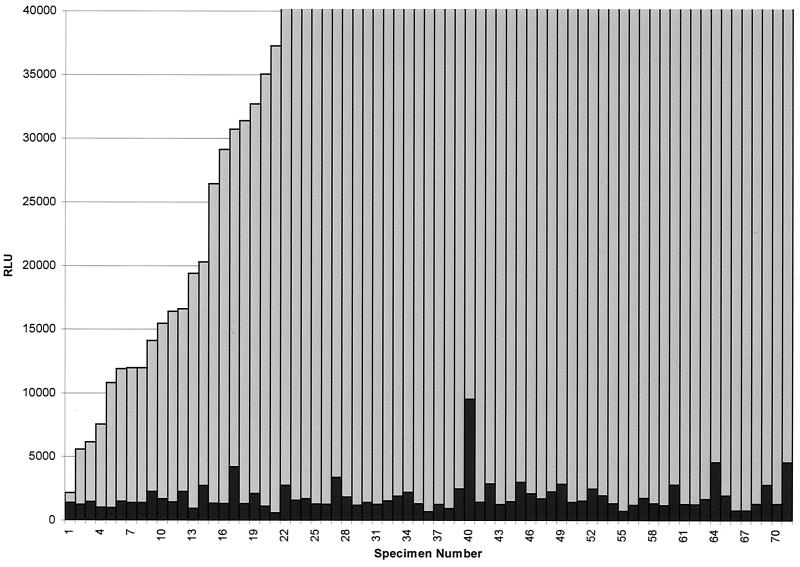
A total of 71 instrument-positive bottles confirmed to contain M. gordonae and 34 shown to contain M. avium complex by probe of colonies detected by culture were tested by direct AccuProbe assay for both organisms after additional incubation for ≥48 h and centrifugation at 4,500 × g for 15 min. Actual RLU values obtained by direct probe of positive bottles for both M. gordonae and M. avium complex are shown in Fig. 2 and 3. Results were analyzed using the manufacturer's recommended cutoff of 30,000 RLU for a positive test and a lower cutoff of 10,000 RLU to determine sensitivity and specificity for each probe. Using the manufacturer's recommended cutoff for a positive test, 71 specimens containing M. gordonae yielded 55 positive results when probed directly (77.5% sensitivity), whereas 28 of 34 M. avium complex specimens were correctly identified (82.3% sensitivity) by direct probe. No specimens were eventually shown by culture to contain either M. gordonae or M. avium complex that tested positive with the probe for the opposite organism (100% specificity). These sensitivity figures are somewhat improved over results shown previously for bottles probed directly without modifications to the procedure designed to increase the number of organisms but not sufficient for optimum diagnostic purposes.
FIG. 2.
AccuProbe assay results for M. avium complex and M. gordonae for 34 specimens culture positive for M. avium complex. Gray bars indicate RLU values obtained using the M. avium complex probe; black bars indicate RLU values obtained using the M. gordonae probe.
FIG. 3.
AccuProbe assay results for M. avium complex and M. gordonae for 71 specimens culture positive for M. gordonae. Gray bars indicate RLU values obtained using the M. gordonae probe; black bars indicate RLU values obtained using the M. avium complex probe.
When the cutoff level for a positive test was lowered to 10,000 RLU, 67 of 71 M. gordonae (94.4% sensitivity) and 32 of 34 M. avium complex (94.1% sensitivity) specimens were correctly identified. This difference was statistically significant for M. gordonae (P = 0.004) but not for M. avium complex (P = 0.26) compared to sensitivity for detection using the recommended RLU cutoff. Specificity was 100% for specimens containing M. gordonae that were tested with the M. avium complex probe using the 10,000 RLU cutoff. Seventy of 71 specimens that were positive for M. gordonae had RLU levels for the M. avium complex probe below 5,000 RLU. One specimen had a level of 9,539 RLU. Specificity for specimens containing M. avium complex that were tested with the M. gordonae probe using the 10,000 RLU cutoff was 97%. Among the 34 specimens positive for M. avium complex, 32 had levels below 5,000 RLU for the M. gordonae probe. One specimen was 7,158 RLU, and the other was 17,929 RLU (Fig. 2 and 3).
DISCUSSION
In this article, data are presented demonstrating the limitations of the M. gordonae and M. avium complex AccuProbes for identification of these common species of nontuberculous mycobacteria by probing directly from MB/BacT bottles on the day that they turned positive. Allowing bottles to incubate an additional 48 h or more and concentrating organisms by centrifugation improved performance somewhat, but some bottles apparently still did not have sufficient cell mass to allow accurate identification of isolates by AccuProbe. Thus, use of M. gordonae and M. avium complex probes for direct detection in positive MB/BacT bottles lacks the sensitivity necessary for routine diagnostic use when the manufacturer's recommended RLU cutoff to define a positive test is applied. Although not a focus of this evaluation, our experience with the M. tuberculosis complex probes used directly on positive MB/BacT bottles has demonstrated no problems compared to results obtained from testing colonies that eventually grow from primary inoculation or subculture, suggesting that M. tuberculosis complex is flagged at higher organism concentrations or that the probe for this species is more efficient. Since there are no other obvious ways to improve the quality of the specimen or the number of organisms, we sought to determine whether we could improve diagnostic sensitivity for M. gordonae and M. avium complex without compromising specificity by modifying the RLU cutoffs to designate a positive test. Reducing the RLU cutoff for the M. gordonae and M. avium complex probes from 30,000 to 10,000 RLU improved the sensitivity for each probe to 94% without substantially increasing the likelihood of a false-positive test with the probe not used. In the case of M. gordonae, this improvement in sensitivity was statistically significant. We believe that a cutoff level of 10,000 RLU is reasonable, since most of the false-negative direct probe results were between 10,000 and 30,000 RLU. We also observed that whenever a reading between 10,000 and 30,000 RLU was obtained, a second probe analyzed a few days later generally yielded positive results, but waiting longer would compromise the desired turnaround time for reporting results. Further lowering the cutoff to a figure such as 5,000 RLU for either M. gordonae or M. avium complex could make false-positive results more likely, based on our experience, since some M. gordonae isolates probed with M. avium complex and some M. avium complex isolates probed with M. gordonae yielded RLU figures that approached this value. In view of our findings in this evaluation, we have devised our own customized approach for use of AccuProbe for determining three of the most common mycobacterial species encountered in our laboratory, M. gordonae, M. avium complex, and M. tuberculosis complex, using the MB/BacT system. The system is shown graphically in Fig. 1.
As soon as the MB/BacT bottles are flagged positive, specimens are plated onto Middlebrook agar and a 7H11 selective biplate. Auramine O smears are examined by fluorescence microscopy and confirmed by Kinyoun stain if positive. If the smear is negative, bottles are reincubated for up to 6 weeks, and acid-fast smears are performed weekly. Plates are checked twice weekly for mycobacterial growth. If Gram stain indicates bacterial contamination at any point, we have not found it useful to attempt repeat decontamination, and the specimens are designated as contaminated and weekly Auramine O staining is performed to detect mycobacteria. If the acid-fast smear is positive, specimens can be probed directly from bottles after incubating for an additional 48 h or more and centrifuging to concentrate the mycobacteria. If colonies are visible on the agar plates on probing day, which is designated once weekly, probes of the colonies should be performed instead because of the greater organism density. Depending on which species is present, it may be necessary to use more than one probe to arrive at the correct identification.
The choice of the specific probe to be used initially in the MB/BacT bottle is made based on the morphology of the organism in the smear from the bottle, looking for the presence of cording as well as pigmentation of the pellet and colonies, if present. The prevalence of the different mycobacterial species in the region should also be considered. If cording is present in the smear from the bottle, the probability of M. tuberculosis complex is high, and it should be sought initially. However, M. gordonae cannot be excluded entirely, since it can show cording in about 10% of smears made from MB/BacT broths in our experience. The colony color in this case would help in choosing between these organisms. If the colonies or pellet are yellow, the probe for M. gordonae should be used initially, whereas if a buff color is noted, the probe for M. tuberculosis complex should be used initially. If no cording is present in the smear and yellow is present in the pellet or colonies, M. gordonae and M. avium complex probes should be used, as both can produce yellow colonies. In the relatively uncommon event of a specimen's containing more than one species of mycobacteria, examination of colonial morphology is important before finalizing a report, since use of a single probe directly with the liquid medium would not allow such differentiation. A lower diagnostic cutoff of 10,000 RLU is recommended when using the M. gordonae and M. avium complex but not the M. tuberculosis complex probe. Individual laboratories should perform internal studies using their own specimens to determine whether the lower cutoff for M. gordonae and M. avium complex probes is valid for their patient population. The risk of limiting the probes used is that an M. tuberculosis complex isolate may be missed. We have found that using a fairly liberal definition of cording and probing all specimens with cords for M. tuberculosis complex as well as reevaluating all cultures which are still unidentified after probing does not significantly delay identification of or misidentify M. tuberculosis complex-containing cultures.
ACKNOWLEDGMENTS
This work was supported in part by Gen-Probe Inc., San Diego, Calif., which supplied the reagents and nucleic acid probes that were used.
We thank Larry Gibbs, Jerry Kimbrell, Michelle Waller, Nancy Smith, Marilyn Horton, Sheila Johnson, Andrea Boozer, Eneida Brookings, and Shawn Banks for technical support.
REFERENCES
- 1.Badak F Z, Goksel S, Sertoz R, Guzelant A, Kizirgil A, Bilgic A. Cord formation in MB/BacT medium is a reliable criterion for presumptive identification of Mycobacterium tuberculosis complex in laboratories with high prevalence of M. tuberculosis. J Clin Microbiol. 1999;37:4189–4191. doi: 10.1128/jcm.37.12.4189-4191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badak F Z, Goksel S, Sertoz R, Nafile B, Ermertcan S, Cavusoglu C, Bilgic A. Use of nucleic acid probes for identification of Mycobacterium tuberculosis directly from MB/BacT bottles. J Clin Microbiol. 1999;37:1602–1605. doi: 10.1128/jcm.37.5.1602-1605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin W H, Jr, Waites K B, Beverly A, Gibbs L, Waller M, Nix S, Moser S A, Willert M. Comparison of the MB/BacT system with a revised antibiotic supplement kit to the BACTEC 460 system for detection of mycobacteria in clinical specimens. J Clin Microbiol. 1998;36:3234–3238. doi: 10.1128/jcm.36.11.3234-3238.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunello F, Favari F, Fontana R. Comparison of the MB/BacT and BACTEC 460 TB systems for recovery of mycobacteria from various clinical specimens. J Clin Microbiol. 1999;37:1206–1209. doi: 10.1128/jcm.37.4.1206-1209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaminski D A, Hardy D J. Selective utilization of DNA probes for identification of Mycobacterium species on the basis of cord formation in primary BACTEC 12B cultures. J Clin Microbiol. 1995;33:1548–1550. doi: 10.1128/jcm.33.6.1548-1550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne L G, Sramek H A. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev. 1992;5:1–25. doi: 10.1128/cmr.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]