Abstract
β-defensins are antimicrobial peptides presenting in vertebrate animals. They participate in innate immunity, but little is known about them in reptiles, including snakes. Although several β-defensin genes were described in Brazilian snakes, their function is still unknown. The peptide sequence from these genes was deduced, and synthetic peptides (with approximately 40 amino acids and derived peptides) were tested against pathogenic bacteria and fungi using microbroth dilution assays. The linear peptides, derived from β-defensins, were designed applying the bioisosterism strategy. The linear β-defensins were more active against Escherichia coli, Micrococcus luteus, Citrobacter freundii, and Staphylococcus aureus. The derived peptides (7–14 mer) showed antibacterial activity against those bacteria and on Klebsiella pneumoniae. Nonetheless, they did not present activity against Candida albicans, Cryptococcus neoformans, Trychophyton rubrum, and Aspergillus fumigatus showing that the cysteine substitution to serine is deleterious to antifungal properties. Tryptophan residue showed to be necessary to improve antibacterial activity. Even though the studied snake β-defensins do not have high antimicrobial activity, they proved to be attractive as template molecules for the development of antibiotics.
Keywords: β-defensins, snakes, antimicrobial activity, bioisosterism, peptides
1. Introduction
With the frightening advent of the global increase of microbial resistance to conventional antibiotics, the search for alternatives has become of utmost importance, and the industry, as well as the regulatory authorities, are realizing the potential of antimicrobial peptides. Since the last decade, antimicrobial peptides have been trialed in clinical phases [1].
In drug development, peptides’ properties have been considered more convenient due to their high affinity for the target and selective biological activities [2]. Bacterial resistance is a global health problem due to the indiscriminate use of antibiotics in humans, as well as in animals and in agricultural production that needs multipronged solutions [3]. To contribute to this campaign, many antimicrobial peptides have been discovered and some are in the clinical development phase [4]. For instance, molecules from innate immunity, such as cathelicidins present in snake venoms, have been reported as potentially active against some bacterial strains [5,6,7,8,9,10], including antibiofilm activity [11,12].
Snakes have developed many active peptides for predation and in their venom composition, many classes of proteins as phospholipases A2 (PLA2) [13], L-amino-acid oxidase (LAAO) [14], metalloproteases [15], cathelicidins [8] and crotamine [16,17,18] are also described as showing antimicrobial activity. Many attempts have been made to shorten these proteins and discover the active site [19,20,21,22]. Crotamine is a small basic myotoxin present in the venom of the rattlesnake C. durissus terrificus and has a β-defensin structure [23,24] and antimicrobial activity [16,17,18]. Crotamine and small basic myotoxins from this family are unique and only present in rattlesnake venom [23].
β-defensins from various vertebrates have been studied, and little is known about these molecules and the innate immunity in snakes. β-defensin-like genes with unknown functions have also been described in Brazilian snakes using a PCR approach [25,26,27]. In the current study, we evaluated the antimicrobial activity of snake β-defensins, both from rattlesnake venom (crotamine) and non-venom β-defensins. Its primary sequences were deduced from genomic sequences and synthesized, except for crotamine purified from rattlesnake venom. Moreover, the amino acid sequences were used to design shorter derived peptides, which can be obtained using simpler and more economical procedures. The snake β-defensins and derived peptides were assayed against microorganisms relevant to the snake’s biology and human health because they can cause opportunistic infections.
2. Results
The sequences of mature peptides were deduced from gene codifying sequences and synthesized, except crotamine purified from venom. They were tested in linear form because the linearization could not affect the antibacterial activity, and the linear form facilitates the development of shorter peptides. The alkylation was done to avoid the dimerization and the intramolecular cyclization of peptides with free thiol groups. Besides, the linear form facilitates the development of shorter peptides. Alkylated peptides were purified by high-performance liquid chromatography (HPLC) and analyzed by MALDI-TOF-MS.
The β-defensins were tested using a microbroth dilution assay. It consists of incubating the bacteria (4.105 colony-forming unit—CFU/mL) and the peptide (from 1 to 512 µg/mL) in liquid broth. After the incubation at 37 °C during the night, the bacterial growth was detected by spectrophotometry at 600 nm. If the measure is similar to the broth, it indicates bacterial growth. The molarity was calculated only to defensins that showed antibacterial activity to compare with the derived peptides. The β-defensins were tested against B. jararaca oral flora because it would be of biological and medical interest. In addition, we tested on E. coli ATCC 25922 and M. luteus that are common Gram-negative and Gram-positive bacteria used in antibacterial tests.
Table 1 shows the antibacterial effect of the snake β-defensins from venom (crotamine) and tissues indicating the minimal inhibition concentration that is the lowest concentration of peptide that results in no visible bacterial growth. It was possible to observe that the linearization of crotamine did not modify its antibacterial activity significantly. These β-defensins did not inhibit the bacterial growth of Providencia rettgeri, Serratia marcescens, Morganella morganii, and Klebsiella pneumonia up to 512 µg/mL, so the results were not shown in the table. M. luteus was the most sensitive bacterium to these peptides. The most active defensins present the highest cationic net charge, DefBm02 (+11), DefbBm03 (+10), DefbLm02 (+8), and crotamine (+7). On the contrary, the β-defensins DefbBju01 (+7), DefbBn02 (+2), and crotasin (−1) did not show antibacterial activity against any strains used in this study. In addition, the substitution in 32nd position, Arg (defbBm02) for Gln (defbBm03), has caused the loss of activity against Staphylococcus aureus.
Table 1.
Antibacterial effect expressed by the minimal inhibitory concentration (MIC) of snake β-defensins on bacteria.
| MIC (µM) | ||||
|---|---|---|---|---|
| Samples | E. coli | M. luteus | C. freundii | S. aureus |
| Crotamine (+7) | 13.3 | 1.7 | 6.7 | >105 |
| Linear crotamine | 26.6 | 1.7 | 26.7 | >105 |
| DefbBm02 (+11) | 3.2 | 0.8 | 1.6 | 12.8 |
| DefbBm03 (+10) | 1.6 | 1.6 | 3.2 | >103 |
| DefbBd03 (+6) | >115 | 0.9 | >115 | >115 |
| DefbBj01 (+4) | >105 | 3.2 | >105 | >105 |
| DefbBju01 (+7) | >111 | >111 | >111 | >111 |
| DefbBn02 (+2) | >113 | >113 | >113 | >113 |
| DefbLm01 (+2) | >113 | 28.4 | >113 | >113 |
| DefbLm02 (+8) | 3.4 | 0.9 | 3.4 | 6.8 |
| DefbPm (+6) | >109 | 6.8 | >109 | >109 |
| DefbTs (+3) | >122 | 15.3 | >122 | >122 |
| hBd02 (+6) | 15.6 | 2 | >124 | >124 |
| Crotasin (−1) | >108 | >108 | >108 | >108 |
The MIC was determined to be the lowest concentration, resulting in no visible bacterial growth in microbroth dilution assay [28]. The assays were performed in three independent experiments. All the β-defensins tested did not inhibit the growth of Providencia rettgeri, Serratia marcescens, Morganella morganii, and Klebsiella pneumonia.
The snake β-defensins crotamine, defbBm02, defbBm03, and defbLm03 were chosen to design the derived peptides for the next step due to their performance in the antibacterial assay. The crotamine 3D model was used as a template for designing the derived peptides because the 3D structure deposited in PDB presented a better resolution (1.70 Å). The software used to build the peptides has a tool to mutate, grow or delete amino acid residues, based on the sequence to be constructed. Furthermore, many algorithms clean the geometry to get a minimum energy structure to start the properties’ calculations. These peptides are presented in Table 2. They derived from the C-terminal portion of chosen β-defensins (Table 1). The design considered the bioisosterism strategy [29], for instance, the substitution of SH group (Cys) to OH group (Ser). Linear peptide constructions of different sizes (7 to 14 aa) were considered to verify which minor sequence would retain the antimicrobial activity. The peptides were commercially purchased, and their purities were tested by HPLC and mass spectrometry.
Table 2.
Peptides derived from β-defensins applying the bioisosterism strategy.
| Peptide | Derived from | Sequence | Hidrofobicity (Kcal/mol) | Net Charge | MW (Da) |
|---|---|---|---|---|---|
| PS1 (13 aa) | Crotamine | SRWRWKSSKKGSG | +19.88 | +5 | 1549 |
| PS2 (14 aa) | defbBm02 | SQMGRMSSRRRFGK | +19.34 | +5 | 1683 |
| PS3 (12 aa) | defbLm02 | SGPGRRSSRRRK | +23.57 | +6 | 1399 |
| PS4 (14 aa) | defbBm03 | SQMGQMSSRRRFGK | +18.30 | +4 | 1655 |
| PS5 (13 aa) | PS3 | SGPGRRSSRRRWK | +21.48 | +6 | 1585 |
| PS6 (7 aa) | PS1 | SRWRWKS | +11.06 | +3 | 1005 |
The primary structure does not illustrate a structure of interaction. The molecular surfaces translate the molecular form since the calculations consider a water molecule running through the van der Waals radius of each atom of the molecule. The molecular form property, consequently, is dependent on the 3D structure and can be used to visualize those differences.
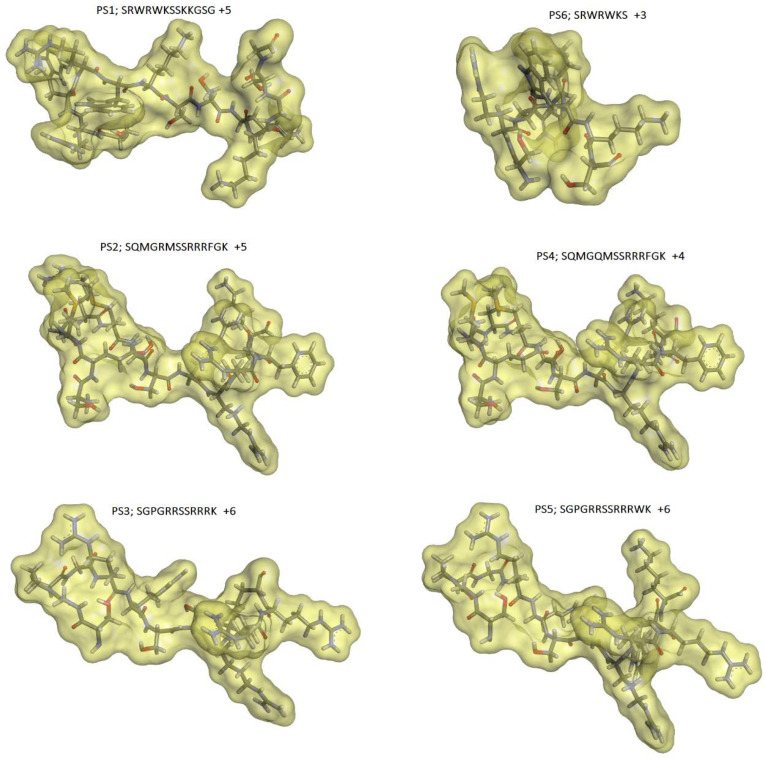
The coordinates of the polypeptide crotamine from the Brazilian rattlesnake C. durissus terrificus [24], retrieved from Protein Data Bank (PBD ID 4GV5; resolution at 1.70 Å) [30], were used as starting geometry to construct the three-dimensional (3D) molecular models of the defensins derived peptides. The crotamine C-terminal portion, containing the fragments Cys30-Gly42 (13 aa), was used to build up the peptides’ 3D molecular models (see Material and Methods section). For instance, the Cys residues (SH group) were mutated into Ser (OH group), generating the peptide PS1, and the PS6 peptide was obtained by extracting Ser37-Gly42 from PS1. PS6 (7 aa) corresponds to the minor sequence assayed in this study. The molecular model of each peptide and its respective calculated solvent-accessible molecular surface are shown in Figure 1.
Figure 1.
Solvent accessible molecular surfaces using a probe of 1.4 Å (water molecule radius). The peptides are presented in stick models, where hydrogen atoms are in white, oxygens in red, nitrogen atoms in blue, sulfur in orange, and carbon atoms in gray. The molecular surfaces are translucid and presented in yellow color (Discovery Studio Visualizer 4.0 program, Accelrys Software Inc., 2005–2013, San Diego, CA, USA).
The peptide PS1 (SRWRWKSSKKGSG) is a bioisoster of the crotamine terminal portion (substitution of SH(Cys) to OH(Ser) group), and PS6 (SRWRWKS) shares the same sequence of PS1 from the 1st to 7th positions. The calculated solvent accessible surface area and molecular volume values were 1295. 61 Å2 and 1551.53 Å3 for PS1; 838.26 Å2 and 1013.05 Å3 for PS6, respectively. As mentioned above, PS6 is the minor peptide of this set. The peptides PS2 (SQMGRMSSRRRFGK) and PS4 (SQMGQMSSRRRFGK) have 14 amino acid residues and differ from one another only in the fifth position. PS2 has an arginine (positively charged) in the fifth position, whereas PS4 has a glutamine (polar; non-charged residue). The solvent-accessible surface area and molecular volume values found for PS2 were 1385.25 Å2 and 1672.60 Å3; and for PS4 were 1356.84 Å2 and 1633.52 Å3, respectively. The peptides PS3 (SGPGRRSSRRRK) and PS5 (SGPGRRSSRRRWK) share the same amino acid sequence from the 1st to 11th positions. PS5 has a tryptophan residue before the last residue, lysine (K). The solvent-accessible surface area and molecular volume values found for PS3 were 1179.24 Å2 and 1387.56 Å3; and for PS5 were 1355.29 Å2 and 1562.72 Å3, respectively. Concerning the solvent-accessible molecular surface or surface area values, the peptides can be classified in the following crescent order: PS6 < PS1 < PS3 < PS5 < PS4 < PS2. This property is related to both molecular shape and solvation process, which are important in the ligand–target recognition process.
These β-defensin-derived peptides were tested against bacteria using the microbroth dilution assay from 2.75 to 700 µg/mL. The results showed that these peptides were not active against M. morganii and P. rettgeri but presented various antibacterial properties against Gram-positive and Gram-negative bacteria (Table 3).
Table 3.
Antibacterial effect expressed by the minimal inhibitory concentration (MIC) of short peptides derived from snake β-defensins on bacteria.
| MIC (µM) | |||||
|---|---|---|---|---|---|
| Peptides | E. coli | S. aureus | M. luteus | K. pneumoniae | C. freundii |
| PS1 | 56.5 | 56.5 | 28.4 | >452 | 113 |
| PS2 | 415 | 415 | 26.1 | >415 | >415 |
| PS3 | >500 | 125 | 31.5 | >500 | >500 |
| PS4 | >423 | >423 | 26.6 | >423 | >423 |
| PS5 | 55.2 | 27.7 | 13.9 | 27.7 | 110 |
| PS6 | 697 | 349 | 43.8 | >697 | >697 |
The MIC was determined as the lowest concentration that results in no visible bacterial growth in microbroth dilution assay [28]. The assays were performed in three independent experiments. All the short peptides tested did not inhibit the growth of Providencia rettgeri, Serratia marcenses, and Morganella morganii.
The derived peptides were also tested against fungi species using the classical resazurin microtiter assay plate method. It is based on the color change of the rezazurin dye that in blue indicates the absence of growth in at least 90%, and pink refers to microorganism growth. MIC is determined as the minimal concentration of the sample that can prevent the color change from blue to pink and refers to the inhibition of 90% of microorganisms. The samples did not inhibit the fungal growth and were considered ineffective until the highest assayed concentrations (250 µM) (Table 4).
Table 4.
Antifungal effect expressed by the minimal inhibitory concentration (MIC90) of short peptides derived from snake β-defensins on yeasts and filamentous fungi.
| MIC90 (µM) | ||||
|---|---|---|---|---|
| Peptides | Candida albicans | Cryptococcus neoformans | Trichophyton rubrum | Aspergillus fumigatus |
| PS1 | >250 | >250 | >250 | >250 |
| PS2 | >250 | >250 | >250 | >250 |
| PS3 | >250 | >250 | >250 | >250 |
| PS4 | >250 | >250 | >250 | >250 |
| PS5 | >250 | >250 | >250 | >250 |
| PS6 | >250 | >250 | >250 | >250 |
MIC The minimal inhibitory concentration (MIC90) was defined as the lowest concentration that prevents the rezazurin’s change in color from blue to pink due to the inhibition of at least 90% of the microorganism’s growth [31]. The assays were performed in three independent experiments.
Observing the molecular shapes (Figure 1) expressed by the obtained solvent surface area values, peptides PS2 and PS4 (14 aa) are the most related and showed similar activities. The replacement of amino acid residues at the fifth position, R5Q, to a polar non-charged residue showed to be deleterious to maintain the antibacterial activity against E. coli and S. aureus (Table 3). These results indicate the importance of the basic residue to retain the activity. On the other hand, the PS2 and PS4 inhibitory activity values on M. luteus were practically the same (Table 3), suggesting the R5Q substitution is not crucial for that interaction profile.
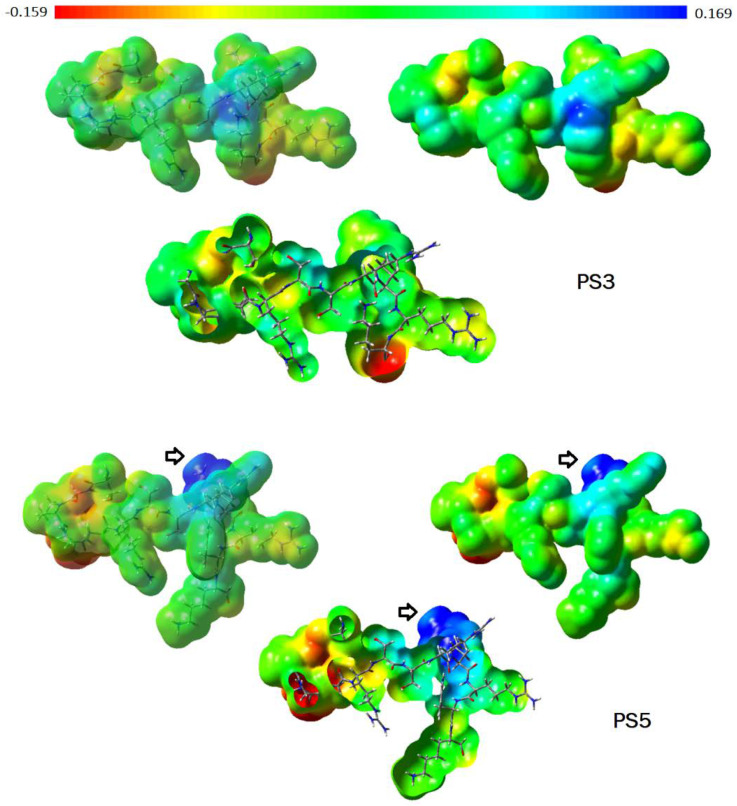
The PS3 (12 aa) and PS5 (13 aa) peptides differ from one another by the tryptophan (W) insertion at the 12th position, and the PS5 sequence modification can be visualized through the molecular shape (Figure 1). The sequence change has provided a significant improvement in the PS5 activity profile in comparison to PS3. PS5 has presented interesting MIC values against E. coli (55.2 µM), S. aureus (27.7 µM), M. luteus (13.9 µM), and K. pneumoniae (27.7 µM). Regarding C. freundi, the MIC value was higher (110 µM). The map of electrostatic potential (MEP) was calculated onto the molecular surface of both peptides, and the changes related to the electronic density distribution are presented in Figure 2. The insertion of tryptophan has contributed to exposing the positively charged region of PS5 (intense blue, lower electronic density distribution; arrow in Figure 2).
Figure 2.
Maps of electrostatic potential (MPEs) were calculated onto the molecular surface of PS3 and PS5 peptides. Regarding the color range, higher electronic density distribution regions are displayed as intense red color (−0.159), and lower electronic density distribution regions are shown in intense blue color (+0.169) (Gaussian 03W, Gaussian, Inc., Pittsburgh, PA, USA, 2003; GaussView 05, Gaussian, Inc., Pittsburgh, PA, USA, 2002–2008).
The PS1 peptide (13 aa) has presented interesting antibacterial activity against E. coli (56.5 µM), S. aureus (56.5 µM), and M. luteus (28.4 µM). As for PS5, regarding C. freundi, the MIC value was higher (113 µM). In comparison to PS1, the PS6 peptide (7 aa), having the sequence core SRWRWKS, did not show any activity against C. freundi. The minor sequence has retained indeed some antibacterial activity.
Unfortunately, none of the derived peptides have shown antifungal activity against the tested fungi, suggesting that the presence of Cys residues in the sequence is important to that kind of activity profile.
3. Discussion
The β-defensins were tested against B. jararaca oral flora because it would be of biological and medical interest. It is known that microorganism infection is one of the snakebite complications due to bacteria or fungi found in the snake’s oral cavity [32,33]. In Brazil, over 80% of snake envenomations are caused by Bothrops snakes, and around 10% of the snakebites evolve to infections [34]. Among bacteria isolated from abscesses, the most reported were M. morganii, E. coli, Providencia sp., Klebsiella sp. [34]. Interestingly, no β-defensins tested in this study showed antibacterial activity against M. morganii nor K. pneumoniae. Although bacteria were also isolated from the venom of C. durissus terrificus [35], envenomations by this species (C. durissus terrificus) do not usually cause infection or macroscopic necrosis in the bite site [34]. The toxins present in C. durissus terrificus venom: crotamine, PLA2, LAAO [36], and cathelicidins [22] could help the asepsis snakebite wound.
The linearization did not abolish the antibiotic activity of crotamine and the majority of snake β-defensins studied herein. The MIC against Gram-positive and -negative bacteria is shown in Table 1. The β-defensins presenting lower net charge values (crotasin, −1, defbBn02, +2) did not show antibacterial activity against any bacteria tested or showed a weak antibacterial effect with high MIC (defbLm01, +2). On the other side, the β-defensins defbBm02 (+11) and defbLm02 (+8) showed the best antibacterial activity against the bacteria E. coli, C. freundii, M. luteus, and S. aureus with MIC in the range of 0.8 to 12.8 µM. Interestingly, DefbBju, with a net charge of +7, did not inhibit any bacteria tested in this study, indicating that the 3D structure could be essential to its antibacterial activity. The Gram-positive bacteria M. luteus was the most sensitive to snake β-defensins while S. aureus was inhibited only by defbBm02 and defbLm02. The Gram-negative bacteria E. coli and C. freundii were inhibited by β-defensins with a net charge higher than +7. The basicity of AMPs is related to antibacterial activity, the higher the positive charge higher the inhibition of bacteria growth, but there are indications that the disulfide bridges modulate the antimicrobial activity [37] and the balance of hydrophilic and hydrophobic surfaces [38]. The net charge of β-defensins was thoroughly discussed by Huang et al. [39]. The activity of linear crotamine corroborates the antibacterial activity of reduced crotamine [17] as well as antifungal activity [18].
Although S. aureus is sensitive to several toxins from snake venoms such as PLA2 [13,40,41,42,43,44,45], LAAO [46,47,48], and cathelicidins [8]; crotamine did not inhibit the growth of this bacterium; however, the β-defensins from snakes defbLm02 and defbBm02 showed antibacterial activity with MIC of 32 and 64 µg/mL respectively. Although native crotamine did not show activity against S. aureus, fragments of this toxin inhibited the growth of this bacterium [18].
Although K. pneumoniae is inhibited by cathelicidins [5,49,50], PLA2 [40,41,42,43,45], and LAAO [51,52,53], no β-defensin tested in this work showed antibacterial activity against this species. The same was true for S. marcensens, sensitive to LAAO [53,54], but not to cathelicidins [5,50]. M. morganii, sensitive to the venom of Montivipera bornmuelleri [55], was also resistant to snake β-defensins and crotamine.
As most of the bacteria tested were isolated from the oral flora of B. jararaca, the lack of antibacterial activity or even weak activity of these molecules may be because these β-defensins only control the coexistence of animals with these microorganisms and the tested snake β-defensin could have other functions in the animal. Many biological functions are described to antimicrobial peptides such as modulation of inflammatory responses, wound healing, angiogenesis promotion [56], regeneration of a lizard tail [57,58], and sperm function in mammals [59], but these biological activities depend on the 3D structure of the β-defensins [60].
The goal of the design of short peptides based on our β-defensins is to get more accessible and cheaper manufacturing as the advantages of small molecules [61]. Peptides P1 to P6 (14 to 7 residues) were tested against bacteria and fungi.
The use of β-defensin fragments caused the decrease of antibacterial activity (see MIC values in Table 3), which was expected since the isolated fragments did not have the same behavior as a conformational organized protein structure. However, it also led to positive results: PS1 showed antibacterial activity against C. freundii and S. aureus differently from crotamine, as well PS5 that inhibited the growth of K. pneumoniae, unlike defbLm02. The decreased antibacterial activity presented by smaller analogs in comparison to the original was also reported to the β-defensin HBD3 [60,62]. A probable cause of the decrease may be the proteolysis that these peptides may be subject to since they were not structurally protected [63]. The peptide with the highest performance was PS5, its only difference to PS3 being an introduction of Trp between basic residues, which may have improved the charge net facilitating the interaction with the bacterial membrane, as happened with the decamer derived from HBD28 [64]. Wessolowski et al. [65] observed that introducing Trp residue in the sequence increases the antibacterial activity, and the cyclization increases the antibacterial activity and the selectivity of peptides. Interestingly, the substitution of Arg for Gln (defbBm02 to defbBm03) did not alter the activity against E. coli, M. luteus, and C. freundii, but it seemed essential to inhibit S. aureus by defbBm02. On the other hand, the same substitution was critical for PS4 (derived from defbBm03) to inhibit S. marcenses.
It is known that fungi are sensitive to toxins from snake venoms, and LAAO [48], PLA2 [43], cathelicidins [5,50], and crotamine [18] were already evaluated on C. albicans. Other sensitive fungi are Aspergillus niculans [5] and Cryptococcus neoformans [18]. In contrast, C. neoformans was resistant to cathelicidins [51]. Despite the crotamine fragment (residues 27–39) having inhibited the growth of C. neoformans [18], the substitution of Cys by Ser in crotamine fragments was deleterious for the antifungal activity [66] in the same way this substitution also abolished the activity against C. albicans [67]. Unfortunately, this was true to all the peptides designed in this work showing that this substitution is deleterious to antifungal activity.
We observed that the C-terminal segment of β-defensins is essential to antimicrobial activity, as also observed by Mandal et al. [68]. Additional molecular modifications seem to be necessary to improve the conformational arrangement and to aid the establishment of the structure–property/activity relationships. Based on that, novel promising peptides can be designed to have better antibacterial activity (higher potency). However, to improve the antibacterial activity of derived peptides, it is necessary to protect them from proteolysis by chemical alteration, C-terminal amidation [64], or cyclizing the peptide by disulfide bonds [63], as also the inclusion of Trp in the sequence to ameliorate the balance between hydrophobicity and hydrophilic or to stabilize the peptide structure favoring the membrane interaction. The substitution of Cys residue by Ala should be also considered, since it has been reported to increase the antibacterial activity of short peptides, as well as the substitution of Leu and Ile by Trp [69].
A feature of the β-defensin family is the 3D structure conserved by the disulfide bridges of the cysteine motif and a significant variation of amino acid sequences [70]. These molecules are good templates for development studies since biological targets have already been selected by nature [71].Furthermore, its mechanism of action, which causes the rupture of the bacterial membrane and can bind to different targets as DNA, makes it difficult for the bacteria to develop resistance [56,70].
In summary, the studied snake β-defensins do not have optimum antimicrobial activity, but they proved to be attractive as template molecules for the development of antibiotics. Our data indicate that the short peptides show specific activity on prokaryote cells but not on eukaryote cells. Based on that, the snake β-defensins C-terminal portion, if optimized, could be indeed used as a bioactive agent.
4. Material and Methods
Twelve peptides (GenBank Accession number is shown in Table 5) were synthesized by Biomatik (Wilmington, NC, USA) and coded as crotasin, DefbBd03, DefbBj01, DefbBju01, DefbBm02, DefbBm03, DefbBn02, DefbLm01, DefLm02, DefbPm, DefbTs and hBd02. The amino acid sequences were deduced from genes and are presented in Table 3. The synthetic peptides were treated with 45 mM DTT by 15 min at 50 °C [72] followed by alkylation using 100 mM IAA (iodoacetamide) 15 min, at room temperature. Alkylated peptides were purified by high performance liquid chromatography (HPLC) and analyzed by MALDI-TOF-MS. This step was performed at the Laboratory of Applied Toxinology—Instituto Butantan with the supervision of Dr. Pedro I. da Silva Jr.
Table 5.
β-defensins sequences deduced from genes and biochemical characteristics.
| Peptide/ Snake |
GenBank Accession Number | Amino Acid Sequence | Hidrofobicity Kcal/mol |
Net Charge at pH 7 | MW Da |
|---|---|---|---|---|---|
| Crotamine/ C. durissus |
YKQCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG | +49.14 | +8 | 4886 | |
| Crotasin/ C. durissus |
AF250212 | QPQCRWLDGFCHSSPCPSGTTSIGQQDCLWYESCCIPRYEK | +28.91 | −1 | 4708 |
| defbBd03/ B. diporus |
KC117160 | QPECLRQGGMCRPRLCPYVSLGQLDCQNGHVCCRKKPRK | +37.08 | +5 | 4456 |
| defbBj/ B. jararaca |
KC117163 | QEECLQQGGFCRLIRCPFGYDSLEQQDCRKGQRCCIRKPRK | +45.05 | +4 | 4874 |
| defbBju/ B. jararacussu |
KC117165 | QRRCHQKGGMCLPGPCPPGYDSLGQQDCRRGQKCCIKRFGK | +43.85 | +7 | 4591 |
| defbBm02/ B. mattogrossensis |
KC117167 | QRRCRQRRGICRPRPCPPENFSLGRLDCQMGRMCCRRRFGK | +39.97 | +11 | 4978 |
| defbBm03/ B. mattogrossensis |
KC117168 | QRRCRQRRGICRPRPCPPENFSLGRLDCQMGQMCCRRRFGK | +38.93 | +10 | 4950 |
| defbBn02/ B. neuwiedi |
KC117169 | QPECCQEGGICHSKQCPLGYSSLGRLDCQLGQRCCIRIFGK | +33.65 | +2 | 4513 |
| defbLm01/ L. muta |
KC117171 | QEWCRGLGGFCSFYQCRPGHDLGPQDCWPERRCCRWGK | +33.69 | +2 | 4515 |
| defbLm02/ L. muta |
KC117172 | QGQCHQQRGRCFLHQCPLSHYFLGRLDCGPGRRCCRRRK | +36.90 | +8 | 4655 |
| defbPm/ P. mertensis |
KX664436 | QRICLGGRGFCHSTPCPRSTIDYGKKDCWGSLRCCEPKRPGK | +42.13 | +6 | 4695 |
| defbTs/ T. strigatus |
KX664429 | QDLCHNLGGRCFRNRCSWSLRNHGGQDCPWGSVCCKP | +31.16 | +3 | 4188 |
| hBD02/ Human |
AF071216 | DPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKP | +31.07 | +6 | 4104 |
Brazilian pitvipers Crotalus (C.), Bothrops (B.), Lachesis (L.) and the colubrides Phalotris (P.), and Thamnophis (T.). The codifying sequences of genes were used to deduce the amino acid sequence and the Signal P software [74] used to determine the mature β-defensins. The hidrofobicity, net charge, and Molecular Weight were theoretical calculated using PepDraw software [http://pepdraw.com/ by Thomas C. Freeman, Jr. Accessed on 27 September 2021].
Crotamine was purified from the C. durissus terrificus venom (purchased from CEVAP, Botucatu, SP, Brazil) and purified as described [73]. Briefly, crotamine was purified from crude venom by size exclusion on a Sephacryl S200 column (GE Healthcare, Uppsala, Sweden), followed by cation exchange chromatography on a 1-mL Resource S on FPLC system (Akta Purifier System, GE Healthcare). The identity and purity of crotamine were confirmed by MS analysis. The reduction and alkylation were proceed as described above.
Crotamine, defbm02, defBm03, and defbLm02 were chosen to design the short peptides due to the best antibacterial activity among the β-defensins tested. Previous studies have indicated the C-terminal of the human β-defensin HBD3 and crotamine as the mandatory region of antimicrobial activity [18,22,60,66,67].
The short peptides, derived from β-defensins (Table 2), were designed applying the bioisosterism strategy [29]; for instance, the substitution of SH group (Cys) by OH group (Ser), regarding the C-terminal portion of the β-defensins (crotamine, defbBm02, defbLm02, defbBm03). Bioisoster groups or substituents share chemical or physical similarities, producing similar biological properties. It was considered linear peptide constructions having different sizes (7 to 14 aa) to verify which would be the minor sequence able to retain the defensins biological activity exploited. The designed peptides were synthesized by GenOne Biotechnologies. The amino acid sequences are presented in Table 1.
4.1. Molecular Modeling and Molecular Properties Calculation
The coordinates of the polypeptide crotamine from the Brazilian rattlesnake C. durissus terrificus [24] were retrieved from Protein Data Bank (PBD ID 4GV5; resolution at 1.70 Å) [30] and used as starting geometry to extract the fragment Cys30-Gly42 (13 aa), which were employed to build up the peptides’ 3D molecular models. The Cys residues were mutated into Ser, generating the peptide PS1. The PS6 peptide was obtained by extracting Ser37-Gly42 from PS1. PS6 (7 aa) corresponds to the minor sequence designed and assayed in this study. The other fragments were constructed using the tool build-mutate or build-grow available in the Discovery Studio Visualizer 4.0 software (Accelrys Software Inc., 2005–2013). The geometries were optimized, and partial atomic charges were assigned using the CHARMM force field [63], included in Discovery Studio (Accelrys Software Inc., 2005–2013).
Furthermore, the electrostatic potential (EP) property was calculated for the PS3 and PS5 peptides to visualize the changes in electronic density distribution concerning the amino acid substitution patterns (PS5 has one more residue, Trp, in comparison to PS3). The charges from electrostatic potential using a grid-based method (CHELPG [64]) were calculated employing the ab initio method Hartree-Fock/3-21G* basis set (Gaussian 03W software; Gaussian, Inc., Pittsburgh, PA, USA, 2003). The EP maps were calculated onto the peptides’ molecular surfaces using GaussView 05 software (Gaussian, Inc., Pittsburg, PA, USA, 2002–2008). The interpretation of EP maps is based on a color scheme, where regions having higher electronic density distribution are presented as an intense red color (negatively charged), whereas regions with lower electronic density distribution are shown as an intense blue color (positively charged). Since the EP property has been calculated onto the PS3 and PS5 molecular surfaces, their molecular shapes were also assessed. Moreover, the molecular volume (intrinsic molecular property) of each peptide considering the van der Waals radii was also calculated employing Discovery Studio Visualizer 4.0 software (Accelrys Software Inc., 2005–2013). Of note, the molecular shape and electronic properties are among the primary molecular properties in the ligand–receptor recognition process.
4.2. Antibacterial Activity
The antibacterial activity of the alkylated peptides was tested against Gram-negative (G−) bacteria (Klebsiella pneumonia, Serratia marcescens, Morganella morganii, Providencia rettgeri, Citrobacter freundii, Escherichia coli ATCC 25922 and Gram-positive (G+) bacteria (Micrococcus luteus A270 and Staphylococcus aureus) using microbroth assay [28]. In a 96-well plate, 90 µL of 10% tryptone soy broth (TSB) with 4 × 105 colony-forming unit (CFU/mL) were mixed with serial twofold dilutions of β-defensins duplicate in concentrations from 512 to 1 µg/mL. The derived peptides coded as PS1, PS2, PS3, PS5, and PS6 (Table 1) were tested against these bacteria in serial twofold dilutions with concentrations bellow 700 µg/mL. After overnight incubation at 37 °C, the turbidity of the bacterial culture was read at 600 nm (Epoch microplate reader, Biotek). The minimal inhibitory concentration (MIC) of each peptide was determined as the lowest concentration that results in no visible bacterial growth. The MIC resulted from three independent experiments. The bacteria K. pneumonia, S. marcescens, M. morganii, P. rettgeri, C. freundii, M. luteus A270 and S. aureus were isolated from Bothrops jararaca buccal flora and kindly provided by Dr. Márcia R. Franzolin (Laboratory of Bacteriology—Instituto Butantan). M. luteus was kindly provided by Dr. Pedro I. da Silva Jr. (Laboratory of Applied Toxinology—Instituto Butantan).
4.3. Antifungal Activity
The antifungal activity was evaluated by the microdilution assay as previously described [31] on clinical strains of Candida albicans (IOC 4525), Cryptococcus neoformans (IOC 4528), Trichophyton rubrum (IOC 4527) and Aspergillus fumigatus (IOC 4526). The fungi were cultivated in potato dextrose agar at 28 °C according to the Clinical and Laboratory Standards Institute recommendations [75]. Fungal suspensions were prepared in RPMI 1640 culture media; the CFU/mL was adjusted to 0.5–2.5 × 103 for yeasts (C. albicans and C. neoformans) and 0.4–5 × 104 CFU/mL for filamentous fungi (T. rubrum e A. fumigatus), by comparison with a standard curve previously stablished in our laboratory.
The peptides PS1, PS2, PS, PS3, PS4, PS5 and PS6 at 1 mM in acetic acid at 0.01% were diluted in RPMI 1640 culture media at concentrations ranging from 1 to 250 μM. Amphotericin B (AMB) at concentrations below 15 μg/mL (16 μM) and acetic acid from 0.0002 to 0.0025% was used as control.
The fungi suspensions were added to a 96 well plate (100 µL/well) and the samples (100 µL) in different concentrations were added to each well. The plate was incubated at 28 °C for 24–72 h, and 24 h before the end of the assay 25 µL of the rezazurin dye at 0.02% were added to each well. The minimal inhibitory concentration (MIC90) was defined as the lowest concentration that prevents the rezazurin’s change in color from blue to pink due to the inhibition of at least 90% of the microorganism’s growth. The assays were performed in three independent experiments.
Author Contributions
Conceptualization: K.F.M.P. and N.O.; Investigation: P.G.C., I.L.R., A.O.d.S.; Formal analysis: K.F.M.P., N.O.; Funding acquisition: N.O.; Writing—original draft: N.O.; Writing—review & editing: A.O.d.S., K.F.M.P., N.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by São Paulo Research Foundation (www.fapesp.br)—FAPESP: 2015/00003-5 (N.O.), 2017/11735-2 (fellowship of I.L.R.); 2008/06524-3 (A.O.S).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contributions
We tested many snake β-defensins against bacteria from oral flora, Micrococcus luteus, and Escherichia coli. This work is the first testing of the antibacterial activity of snake β-defensins besides crotamine. A bioisosterism approach was used to design the derived peptides from β-defensins from Lachesis muta, Bothrops jararaca, and Crotalus durissus snakes. Our results have shown that (i) the defensin’s C-terminal portion seems to be crucial against bacteria; (ii) the presence of tryptophan residue into the derived peptide’s sequence plays an important role for the antibacterial activity; and (iii) the Cys to Ser substitution has abolished the derived peptides’ antifungal activity.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mahlapuu M., Håkansson J., Ringstad L., Björn C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlig T., Kyprianou T., Martinelli F.G., Oppici C.A., Heiligers D., Hills D., Calvo X.R., Verhaert P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014;4:58–69. doi: 10.1016/j.euprot.2014.05.003. [DOI] [Google Scholar]
- 3.Gil-Gil T., Laborda P., Sanz-García F., Hernando-Amado S., Blanco P., Martínez J.L. Antimicrobial resistance: A multifaceted problem with multipronged solutions. Microbiologyopen. 2019;8:e945. doi: 10.1002/mbo3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrès E. Cationic antimicrobial peptides in clinical development, with special focus on thanatin and heliomicin. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:881–888. doi: 10.1007/s10096-011-1430-8. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Hong J., Liu X., Yang H., Liu R., Wu J., Wang A., Lin D., Lai R. Snake Cathelicidin from Bungarus fasciatus Is a Potent Peptide Antibiotics. PLoS ONE. 2008;3:e3217. doi: 10.1371/journal.pone.0003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blower R.J., Barksdale S.M., van Hoek M.L. Snake Cathelicidin NA-CATH and Smaller Helical Antimicrobial Peptides Are Effective against Burkholderia thailandensis. PLoS Negl. Trop. Dis. 2015;9:e0003862. doi: 10.1371/journal.pntd.0003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao F., Lan X.-Q., Du Y., Chen P.-Y., Zhao J., Zhao F., Lee W.-H., Zhang Y. King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates. Zool. Res. 2018;39:87–96. doi: 10.24272/j.issn.2095-8137.2018.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barros E., Gonçalves R.M., Cardoso M.H., Santos N.C., Franco O.L., Cândido E.S. Snake Venom Cathelicidins as Natural Antimicrobial Peptides. Front. Pharmacol. 2019;10:1415. doi: 10.3389/fphar.2019.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlile S.R., Shiels J., Kerrigan L., Delaney R., Megaw J., Gilmore B.F., Weldon S., Dalton J.P., Taggart C.C. Sea snake cathelicidin (Hc-cath) exerts a protective effect in mouse models of lung inflammation and infection. Sci. Rep. 2019;9:6071. doi: 10.1038/s41598-019-42537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Peinado C., Dias A.S., Mendonça D.A., Castanho M.A.R.B., Veiga A.S., Andreu D. Structural determinants conferring unusual long life in human serum to rattlesnake-derived antimicrobial peptide Ctn [15–34] J. Pep. Sci. 2019;25:e3195. doi: 10.1002/psc.3195. [DOI] [PubMed] [Google Scholar]
- 11.Dean S.N., Bishop B.M., van Hoek M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011;11:114. doi: 10.1186/1471-2180-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajbakhsh M., Akhavan M.M., Fallah F., Karimi A. A Recombinant Snake Cathelicidin Derivative Peptide: Antibiofilm Properties and Expression in Escherichia coli. Biomolecules. 2018;8:118. doi: 10.3390/biom8040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samy R.P., Gopalakrishnakone P., Stiles B.G., Girish K.S., Swamy S.N., Hemshekhar M., Tan K.S., Rowan E.G., Sethi G., Chow V.T.K. Snake Venom Phospholipases A2: A Novel Tool against Bacterial Diseases. Curr. Med. Chem. 2012;19:6150–6162. doi: 10.2174/0929867311209066150. [DOI] [PubMed] [Google Scholar]
- 14.Izidoro L.F.M., Sobrinho J.C., Mendes M.M., Costa T.R., Grabner A.N., Rodrigues V.M., da Silva S.L., Zanchi F.B., Zuliani J.P., Fernandes C.F.C., et al. Snake Venom L-Amino Acid Oxidases: Trends in Pharmacology and Biochemistry. Biomed. Res. Int. 2014;2014:196754. doi: 10.1155/2014/196754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samy R.P., Gopalakrishnakone P., Chow V.T.K., Ho B. Viper Metalloproteinase (Agkistrodon halys Pallas) with Antimicrobial Activity against Multi-Drug Resistant Human Pathogens. J. Cell. Physiol. 2008;216:54–68. doi: 10.1002/jcp.21373. [DOI] [PubMed] [Google Scholar]
- 16.Yount N.Y., Kupferwasser D., Spisni A., Dutz S.M., Ramjan Z.H., Sharma S., Waring A.J., Yeaman M.R. Selective reciprocity in antimicrobial activity versus cytotoxicity of hBD-2 and crotamine. Proc. Natl. Acad. Sci. USA. 2009;106:14972–14977. doi: 10.1073/pnas.0904465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oguiura N., Boni-Mitake M., Affonso R., Zhang G. In vitro antibacterial and hemolytic activities of crotamine, a small basic myotoxin from rattlesnake Crotalus durissus. J. Antibiot. 2011;64:327–331. doi: 10.1038/ja.2011.10. [DOI] [PubMed] [Google Scholar]
- 18.Yamane E.S., Bizerra F.C., Oliveira E.B., Moreira J.T., Rajabi M., Nunes G.L.C., de Souza A.O., da Silva I.D.C.G., Yamane T., Karpel R.L., et al. Unraveling the antifungal activity of a South American rattlesnake toxin Crotamine. Biochimie. 2013;95:231–240. doi: 10.1016/j.biochi.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Lomonte B., Ângulo Y., Moreno E. Synthetic Peptides Derived from the C-Terminal Region of Lys49 Phospholipase A2 Homologues from Viperidae Snake Venoms: Biomimetic Activities and Potential Applications. Curr. Pharm. Des. 2010;16:3224–3230. doi: 10.2174/138161210793292456. [DOI] [PubMed] [Google Scholar]
- 20.Chen W., Yang B., Zhou H., Sun L., Dou J., Qian H., Huang W., Mei Y., Han J. Structure–activity relationships of a snake cathelicidin-related peptide, BF-15. Peptides. 2011;32:2497–2503. doi: 10.1016/j.peptides.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Almeida J.R., Mendes B., Lancellotti M., Marangoni S., Vale N., Passos O., Ramos M.J., Fernandes P.A., Gomes P., Da Silva S.L. A novel synthetic peptide inspired on Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom active against multidrug resistant clinical isolates. Eur. J. Med. Chem. 2018;149:248–256. doi: 10.1016/j.ejmech.2018.02.055. [DOI] [PubMed] [Google Scholar]
- 22.Falcao C.B., Radis-Baptista G. Crotamine and crotalicidin, membrane active peptides from Crotalus durissus terrificus rattlesnake venom, and their structurally-minimized fragments for applications in medicine and biotechnology. Peptides. 2020;126:170234. doi: 10.1016/j.peptides.2019.170234. [DOI] [PubMed] [Google Scholar]
- 23.Oguiura N., Boni-Mitake M., Rádis-Baptista G. New view on crotamine, a small basic polypeptide myotoxin from South American rattlesnake venom. Toxicon. 2005;46:363–370. doi: 10.1016/j.toxicon.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Coronado M.A., Gabdulkhakov A., Georgieva D., Sankaran B., Murakami M.T., Arni R.K., Betzel C. Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus. Acta Cryst. 2013;D69:1958–1964. doi: 10.1107/S0907444913018003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rádis-Baptista G., Kubo T., Oguiura N., Silva A.R.B.P., Hayashi M.A.F., Oliveira E.B., Yamane T. Identification of crotasin, a crotamine-related gene of Crotalus durissus terrificus. Toxicon. 2004;43:751–759. doi: 10.1016/j.toxicon.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Corrêa P.G., Oguiura N. Phylogenetic analysis of β-defensin-like genes of Bothrops, Crotalus and Lachesis snakes. Toxicon. 2013;69:65–74. doi: 10.1016/j.toxicon.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira Y.S., Corrêa P.G., Oguiura N. Beta-defensin genes of the Colubridae snakes Phalotris mertensi, Thamnodynastes hypoconia, and T. strigatus. Toxicon. 2018;146:124–128. doi: 10.1016/j.toxicon.2018.02.048. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y., Cai Y., Bommineni Y.R., Fernando S.C., Prakash O., Gilliland S.E., Zhang G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006;281:2858–2867. doi: 10.1074/jbc.M507180200. [DOI] [PubMed] [Google Scholar]
- 29.Patani G.A., LaVoie E.J. Bioisosterism: A Rational Approach in Drug Design. Chem. Rev. 1996;96:3147–3176. doi: 10.1021/cr950066q. [DOI] [PubMed] [Google Scholar]
- 30.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palomino J.C., Martin A., Camacho M., Guerra H., Swings J., Portaels F. Resazurin microtiter assay plate: Simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehghani R., Sharif M.R., Moniri R., Sharif A., Kashani H.H. The identification of bacterial flora in oral cavity of snakes. Comp. Clin. Pathol. 2016;25:279–283. doi: 10.1007/s00580-015-2178-9. [DOI] [Google Scholar]
- 33.Dehghani1 R., Sharif A., Assadi M.A., Kashani H.H., Sharif M.R. Fungal flora in the mouth of venomous and non-venomous snakes. Comp. Clin. Pathol. 2016;25:1207–1211. doi: 10.1007/s00580-016-2330-1. [DOI] [Google Scholar]
- 34.Jorge M.T., Ribeiro L.A. Infections in the bite site after envenoming by snakes of the Bothrops genus. J. Venom. Anim. Toxins. 1997;3:264–272. doi: 10.1590/S0104-79301997000200002. [DOI] [Google Scholar]
- 35.Garcia-Lima E., Laure C.J. A study of bacterial contamination of rattlesnake venom. Rev. Soc. Bras. Med. Trop. 1987;20:19–21. doi: 10.1590/S0037-86821987000100004. [DOI] [PubMed] [Google Scholar]
- 36.Bercovici D., Chudzinski A.M., Dias W.O., Esteves M.I., Hiraichi E., Oishi N.Y., Picarelli Z.P., Rocha M.C., Ueda C.M.P.M., Yamanouye N., et al. A systematic fraction of Crotalus durissus terrificus venom. Mem. Inst. Butantan. 1987;49:69–78. [Google Scholar]
- 37.Sharma H., Nagaraj R. Human β-Defensin 4 with Non-Native Disulfide Bridges Exhibit Antimicrobial Activity. PLoS ONE. 2015;10:e0119525. doi: 10.1371/journal.pone.0119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., Zhou L., Li J., Suresh A., Verma C., Foo Y.H., Yap E.P.H., Tan D.T.H., Beuerman R.W. Linear Analogues of Human b-Defensin 3: Concepts for Design of Antimicrobial Peptides with Reduced Cytotoxicity to Mammalian Cells. ChemBioChem. 2008;9:964–973. doi: 10.1002/cbic.200700560. [DOI] [PubMed] [Google Scholar]
- 39.Huang X.-X., Gao C.-Y., Zhao Q.-J., Li C.-L. Antimicrobial Characterization of Site-Directed Mutagenesis of Porcine Beta Defensin 2. PLoS ONE. 2015;10:e0118170. doi: 10.1371/journal.pone.0118170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samy R.P., Kandasamy M., Gopalakrishnakone P., Stiles B.G., Rowan E.G., Becker D., Shanmugam M.K., Sethi G., Chow V.T.K. Wound Healing Activity and Mechanisms of Action of an Antibacterial Protein from the Venom of the Eastern Diamondback Rattlesnake (Crotalus adamanteus) PLoS ONE. 2014;9:e80199. doi: 10.1371/journal.pone.0080199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samy R.P., Stiles B.G., Chinnathambi A., Zayed M.E., Alharbi S.A., Franco O.L., Rowan E.G., Kumar A.P., Lim L.H.K., Sethi G. Viperatoxin-II: A novel viper venom protein as an effective bactericidal Agent. FEBS Open Bio. 2015;5:928–941. doi: 10.1016/j.fob.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudarshan S., Dhananjaya B.L. Antibacterial Potential of a Basic Phospholipase A2 (VRV_PL_V) of Daboia russellii pulchella (Russell’s Viper) Venom. Biochemistry. 2014;79:1237–1244. doi: 10.1134/S000629791411011X. [DOI] [PubMed] [Google Scholar]
- 43.Sudarshan S., Dhananjaya B.L. Antibacterial Potential of a Basic Phospholipase A2 (VRV_PL_VIIIa) of Daboia russellii pulchella (Russell’s Viper) Venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015;21:17. doi: 10.1186/s40409-015-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudarshan S., Dhananjaya B.L. The Antimicrobial Activity of an Acidic Phospholipase A2 (NN-XIa-PLA2) from the Venom of Naja naja naja (Indian Cobra) Appl. Biochem. Biotechnol. 2015;176:2027–2038. doi: 10.1007/s12010-015-1698-8. [DOI] [PubMed] [Google Scholar]
- 45.Corrêa E.A., Kayano A.M., Diniz-Sousa R., Setúbal S.S., Zanchi F.B., Zuliani J.P., Matos N.B., Almeida J.R., Resende L.M., Marangoni S., et al. Isolation, structural and functional characterization of a new Lys49 phospholipase A2 homologue from Bothrops neuwiedi urutu with bactericidal potential. Toxicon. 2016;115:13–21. doi: 10.1016/j.toxicon.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 46.Guo C., Liu S., Yao Y., Zhang Q., Sun M.-Z. Past decade study of snake venom L-amino acid oxidase. Toxicon. 2012;60:302–311. doi: 10.1016/j.toxicon.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Muñoz L.J.V., Estrada-Gomez S., Núñez V., Sanz L., Calvete J.J. Characterization and cDNA sequence of Bothriechis schlegelii l-aminoacid oxidase with antibacterial activity. Int. J. Biol. Macromol. 2014;69:200–207. doi: 10.1016/j.ijbiomac.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 48.Costa T.R., Menaldo D.L., da Silva C.P., Sorrechia R., de Albuquerque S., Pietro R.C.L.R., Ghisla S., Antunes L.M.G., Sampaio S.V. Evaluating the microbicidal, antiparasitic and antitumor effects of CR-LAAO from Calloselasma rhodostoma venom. Int. J. Biol. Macromol. 2015;80:489–497. doi: 10.1016/j.ijbiomac.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Falcao C.B., de La Torre B.G., Perez-Peinado C., Barron A.E., Andreu D., Radis-Baptista G. Vipericidins: A novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids. 2014;46:2561–2571. doi: 10.1007/s00726-014-1801-4. [DOI] [PubMed] [Google Scholar]
- 50.Wei L., Gao J., Zhang S., Wu S., Xie Z., Ling G., Kuang Y.-Q., Yang Y., Yu H., Wang Y. Identification and Characterization of the First Cathelicidin from Sea Snakes with Potent Antimicrobial and Antiinflammatory Activity and Special Mechanism. J. Biol. Chem. 2015;290:16633–16652. doi: 10.1074/jbc.M115.642645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M.L., Tan N.H., Fung S.Y., Sekaran S.D. Antibacterial action of a heat-stable form of L-amino acid oxidase isolated from king cobra (Ophiophagus hannah) venom. Comp. Biochem. Physiol. C. 2011;153:237–242. doi: 10.1016/j.cbpc.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Okubo B.M., Silva O.N., Migliolo L., Gomes D.G., Porto W.F., Batista C.L., Ramos C.S., Holanda H.H.S., Dias S.C., Franco O.L., et al. Evaluation of an Antimicrobial L-Amino Acid Oxidase and Peptide Derivatives from Bothropoides mattogrosensis Pitviper Venom. PLoS ONE. 2012;7:e33639. doi: 10.1371/journal.pone.0033639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phua C.S., Vejayan J., Ambu S., Ponnudurai G., Gorajana A. Purification and antibacterial activities of an L-amino acid oxidase from king cobra (Ophiophagus hannah) venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2012;18:198–207. doi: 10.1590/S1678-91992012000200010. [DOI] [Google Scholar]
- 54.Skarnes R.C. L-amino acid oxidase, a bacterial system. Nature. 1970;225:1072. doi: 10.1038/2251072a0. [DOI] [PubMed] [Google Scholar]
- 55.Accary C., Hraoui-Bloquet S., Hamze M., Mallem Y., El Omar F., Sabatier J.-M., Desfontis J.-C., Fajloun Z. Protein Content Analysis and Antimicrobial Activity of the Crude Venom of Montivipera bornmuelleri; a Viper from Lebanon. Infect. Disord. Drug Targets. 2014;14:49–55. doi: 10.2174/1871526514666140522114754. [DOI] [PubMed] [Google Scholar]
- 56.Lai Y., Gallo R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alibardi L. Granulocytes of Reptilian Sauropsids Contain Beta-Defensin-like Peptides: A Comparative Ultrastructural Survey. J. Morphol. 2013;274:877–886. doi: 10.1002/jmor.20143. [DOI] [PubMed] [Google Scholar]
- 58.Alibardi L., Celeghin A., Dalla Valle L. Wounding in lizards results in the release of beta-defensins at the wound site and formation of an antimicrobial barrier. Dev. Comp. Immunol. 2012;36:557–565. doi: 10.1016/j.dci.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 59.Dorin J.R., Barratt C.L.R. Importance of β-defensins in sperm function. Mol. Hum. Reprod. 2014;20:821–826. doi: 10.1093/molehr/gau050. [DOI] [PubMed] [Google Scholar]
- 60.Craik D.J., Fairlie D.P., Liras S., Price D. The Future of Peptide-based Drugs. Chem. Biol. Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 61.Hoover D.M., Wu Z., Tucker K., Lu W., Lubkowski J. Antimicrobial Characterization of Human β-Defensin 3 Derivatives. Antimicrob. Agents Chemother. 2003;47:2804–2809. doi: 10.1128/AAC.47.9.2804-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudheendra U.S., Dhople V., Datta A., Kar R.K., Shelburne C.E., Bhunia A., Ramamoorthy A. Membrane disruptive antimicrobial activities of human β-defensin-3 analogs. Eur. J. Med. Chem. 2015;91:91–99. doi: 10.1016/j.ejmech.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scudiero O., Nigro E., Cantisani M., Colavita I., Leone M., Mercurio F.A., Galdiero M., Pessi A., Daniele A., Salvatore F., et al. Design and activity of a cyclic mini-β-defensin analog: A novel antimicrobial tool. Int. J. Nanomed. 2015;10:6523–6539. doi: 10.2147/IJN.S89610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saravanan R., Li X., Lim K., Mohanram H., Peng L., Mishra B., Basu A., Lee J.-M., Bhattacharjya S., Leong S.S.J. Design of Short Membrane Selective Antimicrobial Peptides Containing Tryptophan and Arginine Residues for Improved Activity, Salt-Resistance, and Biocompatibility. Biotechnol. Bioeng. 2014;111:37–49. doi: 10.1002/bit.25003. [DOI] [PubMed] [Google Scholar]
- 65.Wessolowski A., Bienert M., Dathe M. Antimicrobial activity of arginine- and tryptophan-rich hexapeptides: The effects of aromatic clusters, d-amino acid substitution and cyclization. J. Pept. Res. 2004;64:159–169. doi: 10.1111/j.1399-3011.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- 66.Dal Mas C., Pinheiro D.A., Campeiro J.D., Mattei B., Oliveira V., Oliveira E.B., Miranda A., Perez K.R., Hayashi M.A.F. Biophysical and biological properties of small linear peptides derived from crotamine, a cationic antimicrobial/antitumoral toxin with cell penetrating and cargo delivery abilities. Biochim. Biophys. Acta Biomembr. 2017;1859:2340–2349. doi: 10.1016/j.bbamem.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Ponnappan N., Budagavi D.P., Chugh A. CyLoP-1: Membrane-active peptide with cell-penetrating and antimicrobial properties. Biochim. Biophys. Acta. 2017;1859:167–176. doi: 10.1016/j.bbamem.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Mandal M., Jagannadham M.V., Nagaraj R. Antibacterial activities and conformations of bovine β-defensin BNBD-12 and analogs:structural and disulfide bridge requirements for activity. Peptides. 2002;23:413–418. doi: 10.1016/S0196-9781(01)00628-3. [DOI] [PubMed] [Google Scholar]
- 69.Hilpert K., Elliott M.R., Volkmer-Engert R., Henklein P., Donini O., Zhou Q., Winkler D.F.H., Hancock R.E.W. Sequence requirements and an optimization strategy for short antimicrobial peptides. Chem. Biol. 2006;13:1101–1107. doi: 10.1016/j.chembiol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 70.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020;19:311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 71.Stone K.L., Williams K.R. Enzymatic digestion of proteins in solutions and in SDS polyacrylamide gel. In: Walker J.M., editor. The Protein Protocol Handbook. Human Press Inc.; Totowa, NJ, USA: 1996. pp. 415–421. [Google Scholar]
- 72.Boni-Mitake M., Costa H., Spencer P.J., Vassillieff V.S., Rogero J.R. Effects of 60Co gamma radiation on crotamine. Braz. J. Med. Biol. Res. 2001;34:1531–1538. doi: 10.1590/S0100-879X2001001200004. [DOI] [PubMed] [Google Scholar]
- 73.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 74.Breneman C.M., Wiberg K.B. Determining atom-centered monopoles from molecular electrostatic potentials.The need for high sampling density in formamide conformational analysis. J. Comput. Chem. 1990;11:361–373. doi: 10.1002/jcc.540110311. [DOI] [Google Scholar]
- 75.Clinical and Laboratory standards Institute (CLSI) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4th ed. Clinical and Laboratory standards Institute; Wayne, PA, USA: 2017. CLSI Standard M27. [Google Scholar]