Abstract
The aim of this study was to compare the recently developed typing approach for methicillin-resistant Staphylococcus aureus (MRSA) based on the DNA sequencing of the protein A gene polymorphic region (spaA typing) with a combination of three well-established molecular typing techniques: ClaI-mecA vicinity polymorphisms, ClaI-Tn554 insertion patterns, and SmaI pulsed-field gel electrophoresis (PFGE) profiles. In order to evaluate the applicability of this typing technique in different types of studies, two groups of MRSA clinical isolates were analyzed: a collection of 185 MRSA isolates circulating in Hungary recovered from 17 hospitals in seven cities during a 3-year period (1994 through 1996), and a selection of 53 MRSA strains isolated in a single hospital in Hungary between 1997 and 1998. The 238 MRSA clinical strains from Hungary were first classified in clonal types (defined as ClaI-mecA::ClaI-Tn554::SmaI-PFGE patterns), and 65 of the 238 strains, representing major MRSA clones and some sporadic clones, were further analyzed by spaA typing. Our results showed that the lineages most recently introduced in the hospital setting showed little variability in spaA types, whereas the MRSA clones circulating for a longer period of time and spread among several hospitals showed a higher degree of variability. The implementation of the spaA typing method was straightforward, and the results obtained were reproducible, unambiguous, and easily interpreted. This method seems to be adequate for outbreak investigations but should be complemented with other techniques in long-term surveillance or in studies comparing distant clonal lineages.
Three molecular typing techniques (5) have been largely used for the characterization of clones of methicillin-resistant Staphylococcus aureus (MRSA) and enabled the detection of widely spread MRSA lineages, such as the Iberian, Brazilian, New York/Tokyo, and pediatric MRSA clones (1, 7, 11, 17, 20, 21, 24). The combined methods consist of (5, 10) Southern blot analysis of chromosomal ClaI digests with a mecA DNA probe (ClaI-mecA polymorphisms) and with a Tn554 transposon probe (ClaI-Tn554 insertion patterns) and restriction fragment length polymorphism analysis of chromosomal DNA generated after cleavage with SmaI and pulsed-field gel electrophoresis (PFGE) (SmaI-PFGE). ClaI-mecA polymorphisms are a consequence of the variability in the vicinity of the mecA gene, the central element of methicillin resistance, and ClaI-Tn554 patterns reflect the location and copy number of the transposon Tn554, present in most MRSA clinical isolates (10). PFGE provides fine fingerprinting of the chromosomal background with high discriminatory power and has been suggested as the gold standard for the molecular typing of MRSA (25, 26).
DNA sequencing-based typing techniques are being developed with obvious advantages in speed, unambiguous data interpretation, simplicity of large-scale database creation, and standardization among laboratories (8). Recently, DNA sequencing of the spaA gene (protein A determinant) polymorphic region for typing of MRSA strains was evaluated (22). The polymorphic X region is involved in attachment to the cell wall and consists of a variable number of 24-bp repeats, short sequence repeats, which seem to arise from deletion and duplication of the repetitive units and also by point mutation (9, 27). The existence of well-conserved regions flanking the X region coding sequence in spaA allows the use of primers for PCR amplification and direct sequence typing. Shopsin and colleagues (22) have shown that, despite its high degree of polymorphism, the X region of protein A has a variation rate low enough to provide suitable discrimination for outbreak investigations or strain collections restricted to one location and recovered within a short period of time.
In this study we evaluated the discriminatory power of spaA typing to differentiate MRSA clones and assessed the correlation between this sequencing typing method and the combined molecular typing methods ClaI-mecA, ClaI-Tn554, and SmaI-PFGE patterns referred to above. Using these three combined methods, we have studied two groups of MRSA strains with different characteristics recovered from Hungarian hospitals: a representative collection of MRSA strains circulating in Hungarian hospitals during a 3-year period (1994 to 1996) and a collection of MRSA isolates recovered from a single hospital during a 2-year period (1997 to 1998). Fifty-six strains representing the most important clones spread in Hungarian hospitals between 1994 and 1998 and also some sporadic clones were further analyzed by spaA typing to analyze the correlation between clonal types defined as ClaI-mecA polymorphisms::ClaI-Tn554 patterns::SmaI-PFGE profiles and spaA types.
(Part of this study was presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, American Society for Microbiology, abstr. E170, 1998.)
MATERIALS AND METHODS
Clinical isolates.
The 238 MRSA clinical isolates from Hungary included in this study comprise (i) a collection of 185 isolates recovered between 1994 and 1996 at 17 different hospitals located in seven different cities, which were chosen to be representative of MRSA epidemic clones circulating in Hungarian hospitals, and (ii) 53 isolates recovered at a single hospital from January to October 1997 and from January to December 1998, chosen to illustrate a short-term type of study. Antibiograms were performed by the clinical laboratories using the Kirby-Bauer technique, according to the published recommendations and definitions (14). The panel of antibiotics was different from hospital to hospital, but the great majority of the isolates were multiresistant to the antimicrobial agents tested, such as penicillin, oxacillin, erythromycin, tetracycline, ciprofloxacin, and gentamicin. All isolates were susceptible to vancomycin and teicoplanin.
Molecular typing.
ClaI-mecA polymorphism, ClaI-Tn554 insertion pattern, and SmaI-PFGE profile analyses were performed and interpreted as previously described (3, 5, 10, 26).
spaA typing.
spaA typing was performed essentially as previously described (22). Chromosomal DNA for PCR was prepared (2) and diluted to approximately 0.5 ng/μl. Primers for amplification and sequencing of the X region of the spaA gene were designed based on the published sequence (accession no. J01786) and purchased from Gibco-BRL (Life Technologies, Grand Island, N.Y.): SpaF1, GAC GAT CCT TCG GTG ACG, nucleotides 1096 to 1113, and SpaR1, CAG CAG TAG TGC CGT TTG C, nucleotides 1534 to 1516. PCR amplification was performed in a GeneAmp PCR System 9600 thermocycler (Perkin-Elmer Cetus [PE], Branchburg, N.J.), with 2.5 ng of DNA, 5 μl of 10× PCR buffer II (PE), 4 μl of deoxynucleoside triposphate mix at 10 mM (PE), 2.5 U of AmpliTaq Gold DNA polymerase (PE), 1.5 mM MgCl2 (PE), and 10 mmol of each primer in a final reaction volume of 50 μl in 0.2-ml PCR tubes (PE). Thermal cycling parameters were as follows: predenaturation for 10 min at 95°C; 30 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 45 s, postextension for 10 min at 72°C; and soaking at 4°C. PCR products (2 μl) were visualized by conventional minigel electrophoresis and purified with the Wizard PCR-prep DNA purification system (Promega, Madison, Wis.). DNA cycle sequencing reactions were performed with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE) in a final reaction volume of 10 μl: 1 μl of amplified and purified DNA (20 to 30 ng); 2 μl of SpaF1 or SpaR1 at 2 pmol/μl; 4 μl of Ready Reaction mix; and 3 μl of H2O MilliQ. Amplification parameters were as follows: 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, and soaking at 4°C. DNA was precipitated at room temperature for 15 min with 50 μl of 95% ethanol–10 μl of H2O MilliQ–2 μl of 3 M sodium acetate (pH = 4.6), centrifuged in a microcentrifuge for 20 min at 13,000 rpm, washed with 250 μl of 70% ethanol, centrifuged for 10 min at 13,000 rpm, and dried for 1 min at 90°C. DNA sequences were determined by electrophoresis in an ABI Prism 377 DNA sequencer (PE) according to the manufacturer's instructions at the DNA sequencing facility located at Instituto Gulbenkian de Ciência, Oeiras, Portugal. The assembly of both sequences was performed with SeqMan software (DNAStar software package; Lasergene, Madison, Wis.). Consensus sequences were sought for the previously defined 24-bp repeat polymorphisms (22), using specific software (GeneSearch, designed by Ludwig Krippahl). The output (spaA type) consists of a sequence of letters that correspond to the succession of the different 24-bp repeats within the polymorphic region of the spaA gene.
RESULTS
Clonal analysis of Hungarian MRSA.
Tables 1 and 2 summarize the clonal types found in the 1994 to 1996 and 1997 to 1998 collections, respectively. The clonal types were compared to those of a previous study in which the application of these techniques to a collection of 48 MRSA clinical isolates recovered from six provincial hospitals located hundreds of kilometers apart in Hungary between 1993 and 1994 demonstrated the existence of a unique epidemic MRSA clone, the Hungarian MRSA clone (clone III::B::A), which was present in 81% of the isolates (6). In the present study, this lineage, characterized by PFGE pattern A, was still present in both collections, but its prevalence had decreased to 70% in 1994 to 1996 and its variability, expressed by the panoply of PFGE subtypes (37 subtypes were found for PFGE pattern A), ClaI-mecA types, and ClaI-Tn554 insertion patterns, had increased, as shown by the presence of clonal type III::B::A in only 33% of the isolates in 1994 to 1996. The observed variability in the ClaI-mecA polymorphisms (patterns III, IX, XI, and III′) was characterized by small shifts in the hybridization fragment size of the mecA downstream vicinity which were detected in strains isolated after 1993 and that were recently explained (16) as being caused by different copy numbers of the direct repeat unit (dru) within the hypervariable region downstream of mecA (19). Pattern IX has 11 dru copies, pattern XI has 10 copies, pattern III has 9 copies, and pattern III′ has 8 copies (16). Therefore, it is reasonable to consider that clonal types III::B::A, IX::B::A, XI::B::A, and III′::B::A are equivalent, and altogether they accounted for 42% of the clonal types in 1994 to 1996. ClaI-Tn554 insertion patterns are a consequence of the location and copy number of a transposon, by definition a mobile element, and within the PFGE pattern A cluster as many as 16 different insertion patterns were found, all characterized by the presence of multiple copies of transposon Tn554. However, since PFGE is the technique with the highest discriminatory power, strains sharing PFGE pattern A were classified as closely related and accounted for 70% of the clonal types in 1994 to 1996 and 40% in the 1997 to 1998 collection. This high degree of variability within an MRSA clone has never been reported before and seems to be a particular characteristic of the Hungarian clone. It is not observed, for example, in the highly epidemic Iberian and Brazilian clones (H. de Lencastre, unpublished observations).
TABLE 1.
Molecular typing of 185 MRSA clinical strains isolated between 1994 and 1996 at 17 hospitals located in seven cities in Hungary
| Clone type | City (county) or county | Yr of isolation | No. of isolates | Clonal typea | Total no. of isolates (% of total) |
|---|---|---|---|---|---|
| Epidemic A | Szekesfehervar (Fejer) | 1994, 1996 | 12 | III::B::A | 61 (33.0) |
| Dunaujvaros (Fejer) | 1994 | 3 | |||
| Debrecen (Hajdu) | 1994 | 2 | |||
| Miskolc (Borsod-A. Z.) | 1996 | 15 | |||
| Somogy County | 1994, 1996 | 22 | |||
| Budapest | 1995, 1996 | 7 | |||
| Dunaujvaros (Fejer) | 1996 | 9 | III′::B::A | 12 (6.5) | |
| Debrecen (Hajdu) | 1994 | 2 | |||
| Budapest | 1995 | 1 | |||
| Somogy County | 1994, 1996 | 2 | IX::B::A | 2 | |
| Debrecen (Hajdu) | 1994 | 1 | XI::B::A | 2 | |
| Miskolc (Borsod-A. Z.) | 1995 | 1 | |||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::M::A | 26 (14.1) | |
| Somogy County | 1996 | 6 | |||
| Budapest | 1995 | 19 | |||
| Dunaujvaros (Fejer) | 1996 | 3 | III′::M::A | 4 (2.1) | |
| Budapest | 1995 | 1 | |||
| Miskolc (Borsod-A. Z.) | 1995 | 3 | III::W::A | 4 (2.1) | |
| Somogy County | 1996 | 1 | |||
| Budapest | 1995 | 1 | III::DD::A | ||
| Budapest | 1995 | 1 | III::F::A | ||
| Budapest | 1996 | 2 | III::A::A | ||
| Budapest | 1996 | 1 | III::B2::A | ||
| Budapest | 1996 | 1 | III::k::A | ||
| Budapest | 1996 | 1 | XI::k::A | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::W1::A | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::i::A | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::m::A | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::n::A | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::t::A | ||
| Miskolc (Borsod-A. Z.) | 1996 | 1 | III::λλ::A | ||
| Somogy County | 1994 | 1 | IX::E::A | ||
| Somogy County | 1994 | 1 | III::i::A | ||
| Somogy County | 1996 | 1 | XI::B1::A | ||
| Somogy County | 1996 | 1 | III::M1::A | ||
| Szekesfehervar (Fejer) | 1996 | 1 | III::M::A | ||
| Szekesfehervar (Fejer) | 1996 | 1 | III::i::A | ||
| Total | 130 (70.2) | ||||
| Epidemic D | Budapest | 1994, 1996 | 18 | II::q::D | 18 (9.7) |
| Somogy County | 1994 | 1 | II::D::D | 9 (4.9) | |
| Budapest | 1995, 1996 | 8 | |||
| Budapest | 1996 | 1 | II::A1::D | 1 | |
| Total | 28 (15.1) | ||||
| Sporadic | Budapest | 1995 | 1 | III::B::Q | |
| Budapest | 1995 | 1 | II::A1::R | ||
| Budapest | 1996 | 4 | II::A::E | ||
| Budapest | 1996 | 1 | II::D::E | ||
| Budapest | 1995 | 1 | XI::B2::O | ||
| Budapest | 1996 | 2 | VI::F::P | ||
| Miskolc (Borsod-A. Z.) | 1995 | 1 | III::W::H | ||
| Miskolc (Borsod-A. Z.) | 1996 | 1 | III::B::B | ||
| Miskolc (Borsod-A. Z.) | 1996 | 1 | III::W::B | ||
| Miskolc (Borsod-A. Z.) | 1996 | 1 | III′::M1::F | ||
| Miskolc (Borsod-A. Z.) | 1996 | 1 | III′::αα::G | ||
| Szekesfehervar (Fejer) | 1994 | 1 | III::B::M | ||
| Szekesfehervar (Fejer) | 1994 | 1 | III::j::M | ||
| Szekesfehervar (Fejer) | 1994 | 2 | III::B::N | ||
| Szekesfehervar (Fejer) | 1994 | 1 | III::j::N | ||
| Szekesfehervar (Fejer) | 1996 | 1 | III::B1::B | ||
| Dunaujvaros (Fejer) | 1994 | 1 | III::n::B | ||
| Dunaujvaros (Fejer) | 1994 | 1 | IX::B::K | ||
| Somogy County | 1996 | 1 | III::B::I | ||
| Somogy County | 1996 | 1 | III::x::J | ||
| Debrecen (Hajdu) | 1994 | 1 | III::p::L | ||
| Szombathely (Vas) | 1994 | 1 | III::M::N | ||
| Total | 27 (14.6) |
Clonal types were defined on the basis of ClaI-mecA polymorphisms::ClaI-Tn554 patterns::SmaI-PFGE restriction profiles. A total of 185 strains were examined. ClaI-mecA polymorphisms with a prime and ClaI-Tn554 insertion patterns with number codes (e.g., B1) contain small variations from previously described patterns. ClaI-Tn554 insertion patterns with lowercase letters are new patterns not described in previous studies.
TABLE 2.
Molecular typing of 53 MRSA clinical strains isolated from a single hospital in Hungary between 1997 and 1998
| PFGE pattern type | Yr of isolation | No. of isolates | Clonal typea | Total no. of isolates (% of total) |
|---|---|---|---|---|
| A | 1997 | 7 | III::M1::A | 9 (17.0) |
| 1998 | 2 | |||
| 1997 | 1 | III::B2::A | 5 (9.4) | |
| 1998 | 4 | |||
| 1997 | 2 | III::B1::A | 6 | |
| 1998 | 2 | XI::B::A | ||
| 1998 | 1 | III′::B3::A | ||
| 1998 | 1 | III′::q::A | ||
| Total | 20 (37.8) | |||
| D | 1997 | 4 | II::q::D | 5 |
| 1997 | 1 | II::q1::D | ||
| Total | 5 (9.4) | |||
| S | 1997 | 12 | II::E1::S | 15 (28.3) |
| 1998 | 3 | |||
| Varied | 1997 | 2 | III′::B2::B | 13 (24.5) |
| 1997 | 1 | III::B2::B | ||
| 1997 | 1 | VIII::aa::M | ||
| 1997 | 1 | III::M4::T | ||
| 1997 | 1 | III::M1::Y | ||
| 1998 | 1 | III′::n::B | ||
| 1998 | 2 | I::NH::W | ||
| 1998 | 1 | III::B2::N | ||
| 1998 | 1 | VII::ρ1::V | ||
| 1998 | 1 | V::W1::X | ||
| 1998 | 1 | III::ββ1::Z | ||
| Total | 13 (24.5) |
See Table 1, footnote a.
In the 1994 to 1996 collection, a new family of clones were found in Hungarian hospitals, PFGE pattern D-related clones II::q::D, II::A1::D, and II::D::D (15% of the isolates), which were also detected in 9% of the isolates recovered from the single hospital studied in 1997 to 1998. In this same hospital in 1997 to 1998, a previously undetected lineage (II::E1::S) was present in 28% of the isolates. Both lineages showed significantly less variability than PFGE pattern A-related clones, which may be explained by its more recent introduction in Hungarian hospitals and, in the case of clone II::E1::S, by the fact that it was detected in isolates from a single hospital. These lineages do not appear (by the typing methods used) to be related to other MRSA clones spread in other countries and previously identified in this laboratory. Figure 1 shows some subtypes of the most important PFGE patterns found in this study, PFGE patterns A, D, and S.
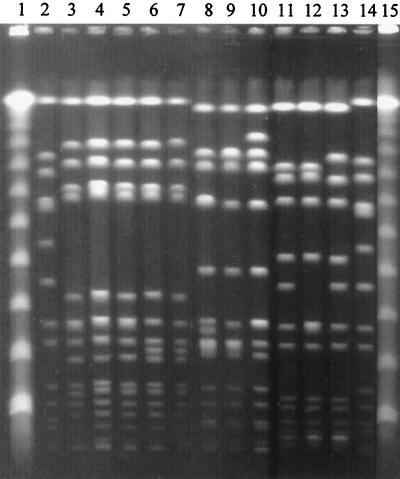
FIG. 1.
Representative gel of some subtypes of the most frequent SmaI-PFGE profiles and one sporadic clone. Strain codes indicate the period of isolation: HUSA, 1993 to 1994 (4); HU, 1994 to 1996; and HUR, 1997 to 1998. Samples are as follows: lanes 1 and 15, molecular size markers (lambda DNA ladder; New England Biolabs); lanes 2 and 14, reference strain NCTC8325; lanes 3 to 5, pattern A1 (HUSA67, HU1, and HUR36); lane 6, pattern A36 (HU221); lane 7, pattern A37 (HUR1); lane 8, pattern D1 (HU181); lane 9, pattern D2 (HU164); lane 10, pattern D3 (HU150); lane 11, pattern S1 (HUR9); lane 12, pattern S4 (HUR95); lane 13, pattern S5 (HUR94).
spaA typing.
Fifty-six strains representing the most important clonal types detected in Hungarian hospitals in 1994 to 1996 and 1997 to 1998 were studied by spaA typing. For each lineage (defined according to PFGE patterns A, D, and S), several strains with different ClaI-mecA and ClaI-Tn554 patterns were selected from different hospitals, cities, and periods of isolation. Nine strains belonging to sporadic clones were also included. Nine different spaA types were found among the 65 five strains studied (Table 3).
TABLE 3.
spaA typing of selected MRSA strains isolated in Hungary in 1994 to 1996 and 1997 to 1998
| spaA typea | Clonal typeb (no. of strains) |
|---|---|
| WGKAKBAOKAOMQ | III::B1::A (2), IX::B::A (2), III::M::A (1) |
| WGKAKAOKAOMQ | III::B::A (5), III′::B::A (2), III::B2::A (1), III′::B2::A (2), XI::B::A (1), III::M::A (2), III::M1::A (6), III::W::A (2), III::B′::B (1), III::N::B (1), III::W::B (1) |
| WGKAOKAOMQ | III::B::A (1), III′::B3::A (1), III::M::A (1), III′::q::A (1), V::W1::A (1), III::B::B (1), III::M4::T (1), III::M′::H (1) |
| XKAOKAOMQ | XI::B::A (2), III′::n::A (1) |
| WGKAQAQQ | II::q::D (7), II::q1::D (1), II::D::D (3), II::A′::D (1) |
| TIMBMDMGMK | II::E1::S (9) |
| TIMBME | II::E1::S (1) |
| WGKAOMQ | III::ββ1::Z (1) |
| YHFGFMBQBLO | I::NH::W (2) |
spaA types were assigned according to Shopsin and colleagues (22). Each letter corresponds to a different polymorphism of the 24-bp repeat; the sequence of letters correponds to the sequence of the 24-bp repeats within the spaA gene polymorphic X region.
Clonal types were defined on the basis of ClaI-mecA polymorphisms::ClaI-Tn554 patterns::SmaI-PFGE restriction profiles.
Four related spaA types sharing the KAOMQ motif (spaA types WGKAKBAOKAOMQ, WGKAKAOKAOMQ, WGKAOKAOMQ, and XKAOKAOMQ) were found among the isolates belonging to PFGE pattern A-related clones. However two of these spaA types (WGKAKAOKAOMQ and WGKAOKAOMQ) were also found among strains belonging to the sporadic clones characterized by PFGE patterns B, T, and H.
The spaA type WGKAQAQQ was specific for the 12 isolates studied belonging to PFGE pattern D-related clones. The spaA type TIMBMDMGMK was specific for 9 of 10 isolates studied belonging to clonal type II::E1::S, and the other isolate was characterized by the related spaA type TIMBME. The remaining two spaA types, YHFGFMBQBLO and WGKAOMQ, were specific for strains belonging to the sporadic clones I::NH::W and III::ββ1::Z, respectively.
DISCUSSION
We have applied the recently developed MRSA typing technique based on DNA sequencing of the protein A gene polymorphic region, spaA typing (22), to characterize two distinct MRSA collections representing two different kinds of studies: a representative collection of MRSA clinical isolates circulating in Hungarian hospitals during a 3-year period and a collection of clinical strains recovered from a single hospital during a 2-year period, also in Hungary. spaA types were compared to the results obtained using other molecular typing techniques (ClaI-mecA, ClaI-Tn554, and SmaI-PFGE patterns) in order to evaluate the discriminatory power of spaA typing and its use to characterize MRSA clones circulating in a particular country or hospital. In addition, we wanted to assess the ease of implementation and execution of this method in our laboratory, since we have been interested on the molecular typing of clinical strains of MRSA, and therefore, spaA typing was potentially useful in our studies.
Implementation of spaA typing technique.
The implementation of the spaA typing technique, essentially according to the published procedure (22), was straightforward and easy to establish in our laboratory, requiring only some expertise in PCR and DNA sequencing techniques, which are easily achieved with modern reagent kits, thermocyclers, and sequencers. To evaluate the reproducibility of the spaA types, four strains were typed twice, and in all cases the spaA types obtained were exactly the same. The stability of spaA types was also evaluated for one strain, which was daily diluted in fresh medium over a 3-week period (approximately 15 × 109 generations), and no changes in spaA type were detected. In order to make a first evaluation of spaA typing, a small collection of 20 MRSA isolates representative of well-characterized and internationally spread clones, such as the Iberian (7, 21), Brazilian (24), and New York/Tokyo (1, 11) clones, was tested, and it was found that spaA typing was able to discriminate among the different MRSA lineages: Iberian clone spaA type YHFGFMBQBLO; Brazilian clone spaA type XKAOMQ; and New York/Tokyo clone spaA type TJMBMDMGMK (Oliveira et al., unpublished data).
Comparison between spaA typing and other molecular typing techniques.
In this study, spaA typing was excellent in discriminating the clonal lineages more recently introduced in Hungarian hospitals (PFGE pattern D clones and clone II::E1::S), which were characterized by the specific spaA types WGKAQAQQ and TIMBMDMGMK/TIMBME, respectively.
However, the isolates belonging to the clonal lineage characterized by PFGE pattern A, circulating in Hungarian hospitals at least since 1993, were characterized by a cluster of four spaA types with the KAOMQ motif, and two of these spaA types were also found among strains belonging to the sporadic clones characterized by PFGE patterns B, T, and H. These findings suggest that these sporadic clones (III::B::B, III::B′::B, III::W::B, III::M4::T, and III::M′::H) may have evolved from clone III::B::A, so that a less discriminative technique like spaA typing might not be able to differentiate them. This hypothesis is supported by the fact that these sporadic clones show the same ClaI-mecA and ClaI-Tn554 types as PFGE pattern A-related clones. Moreover, another sporadic clone analyzed by spaA typing (clone I::NH::W), with nothing in common with clone III::B::A, displayed the unrelated and specific spaA type YHFGFMBQBLO, whereas the sporadic clone III::ββ1::Z (sharing ClaI-mecA type III) was characterized by spaA type WGKAOMO, with the KAOMQ motif. The variability of spaA types among the PFGE pattern A-related clones parallels the variability also detected by the other typing techniques, suggesting that the mutation rate of the spaA gene polymorphic region is comparable to the variability rate of ClaI-mecA polymorphisms, ClaI-Tn554 insertion patterns, and PFGE subtypes.
The application of spaA typing also provided interesting clonal relationships among MRSA. The sporadic clone I::NH::W found in Hungary in 1998 (Table 2) showed exactly the same spaA type as the Iberian clone-related strains isolated since the mid-1980s and also strain DEN2125 isolated in Denmark in 1964 (results not shown), confirming the previous finding (4) that the Iberian clone, first described in Spain (7) and since then shown to be widely spread throughout Europe (12, 13, 21) and the United States (17, 18), may have in strain DEN2125 a evolutionary precursor. The I::NH::W clone may also be an Iberian clone derivative that has just been introduced in Hungary and will eventually disseminate among Hungarian hospitals. Analysis of Fig. 2, in which the PFGE profiles of these strains are displayed, showed that these strains do not have the same PFGE pattern, although these patterns seem to be related (10 band differences). The stability in the spaA type of Iberian clone-related strains over a span of at least three decades contrasts with the variability found in the spaA type related to the Hungarian clone strains and was somehow unexpected, since the molecular basis for spaA type is precisely the polymorphic X region within the spaA gene. These findings suggest that there are highly stable alleles of the X region, presumably very efficient in determining pathogenesis and/or adherence mechanisms.
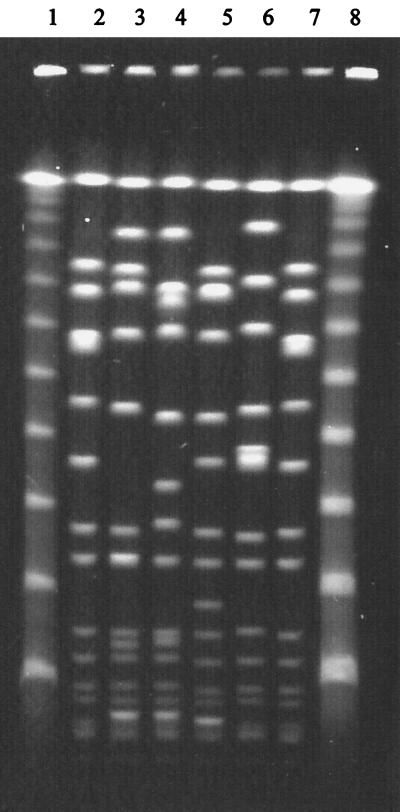
FIG. 2.
SmaI-PFGE profiles of strains with the Iberian spaA type. Strains are as follows: lanes 1 and 8, molecular size markers (lambda DNA ladder; New England Biolabs); lanes 2 and 7, reference strain NCTC8325; lane 3, PER34 (Iberian clone representative strain [7]); lane 4, HUC191 (Iberian clone closely related strain [15]); lane 5, DEN2125 (archaic clone [1]); and lane 6, HUR97 (sporadic clone isolated in Hungary).
As previously suggested by Shopsin and colleagues (22) and also confirmed by Tang and colleagues (23), DNA sequencing of the protein A gene polymorphic region as a typing technique seems to be a powerful technique for MRSA typing. This is especially true for MRSA isolates restricted to one location and to a short period of isolation (that is, for outbreak investigations), as was shown in this study by the excellent discrimination of the clonal lineages more recently introduced in Hungarian hospitals (clones related to PFGE patterns D and S). spaA typing also had a satisfactory capacity for discrimination in more diverse collections, as illustrated in this study with the PFGE pattern A clonal lineage. However, in these cases, other typing techniques and a more careful interpretation of the spaA types may be needed. In our study, we found spaA types to be stable and reproducible, and the spaA typing technique was easy to implement and provided unambiguous results.
ACKNOWLEDGMENTS
We thank the late Anna Marton for the gift of some MRSA clinical isolates and L. Krippahl (Faculdade de Ciências e Technologia, Universidade Nova de Lisboa, Monte da Caparica, Portugal) for writing the GeneSearch computer program.
Partial support for this study was provided by projects PRAXIS XXI/2/2.2/SAU/1295/95 and PRAXIS XXI/P/SAU/14052/98 from Fundação para a Ciência e Tecnologia, Lisbon, Portugal, and Project 31 CEM/NET from IBET, Oeiras, Portugal, awarded to H. de Lencastre. The 1997 to 1998 collection was recovered under Project RESIST, with a grant from Rhône-Poulenc Rorer S.A. to A. Tomasz and H. de Lencastre. D. C. Oliveira and M. Aires-de-Sousa were supported by grants BD/4162/96 and BD/13731/97, respectively, from Fundação para a Ciência e Tecnologia, Lisbon, Portugal, and C. R. Alves was supported by grant 001/99/BIC/P from ITQB, Oeiras, Portugal. P. Major was supported by Fundação Calouste Gulbenkian for his CEM/NET Fellowship Project at ITQB/UNL, Oeiras, Portugal.
REFERENCES
- 1.Aires-de-Sousa M, de Lencastre H, Santos-Sanches I, Kikuchi K, Totsuka K, Tomasz A. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus (MRSA) isolates widely spread in New York City and in a hospital in Tokyo, Japan. Microb Drug Resist. 2000;6:253–258. doi: 10.1089/mdr.2000.6.253. [DOI] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa M, Santos-Sanches I, van Belkum A, van Leeuwen W, Verbrugh H, de Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb Drug Resist. 1996;2:331–341. doi: 10.1089/mdr.1996.2.331. [DOI] [PubMed] [Google Scholar]
- 3.Chung M, de Lencastre H, Matthews P, Tomasz A the Multilab Project Collaborators: I. Adamsson, M. Aires-de-Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. Santos Sanches, J.-H. Song, P. T. Tassios, and P. Villari. Molecular typing of methicillin resistant Staphylococcus aureus by pulsed field gel electrophoresis: Comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 4.de Lencastre H, Chung M, Westh H. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb Drug Resist. 2000;6:1–10. doi: 10.1089/mdr.2000.6.1. [DOI] [PubMed] [Google Scholar]
- 5.de Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 6.de Lencastre H, Severina E P, Milch H, Thege M K, Tomasz A. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin Microbiol Infect. 1997;3:289–296. doi: 10.1111/j.1469-0691.1997.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez M A, de Lencastre H, Linares J, Tomasz A. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright M C, Day N P, Davies C E, Peacock S J, Spratt B G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guss B, Uhlen M, Nilsson B, Lindberg M, Sjoquist J, Sjodahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 10.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 11.Kreiswirth B N, Lutwick S M, Chapnick E K, Gradon J D, Lutwick L I, Sepkowitz D V, Eisner W, Levi M H. Tracing the spread of methicillin-resistant Staphylococcus aureus by Southern blot hybridization using gene-specific probes of mec and Tn554. Microb Drug Resist. 1995;1:307–313. doi: 10.1089/mdr.1995.1.307. [DOI] [PubMed] [Google Scholar]
- 12.Mato R, Sanches I S, Venditti M, Platt D J, Brown A, de Lencastre H. Spread of the multiresistant Iberian clone of methicillin-resistant Staphylococuus aureus (MRSA) to Italy and Scotland. Microb Drug Resist. 1998;4:107–112. doi: 10.1089/mdr.1998.4.107. [DOI] [PubMed] [Google Scholar]
- 13.Melter O, Santos-Sanches I, Schindler J, Aires-de-Sousa M, Mato R, Kovarova V, Zemlickova H, de Lencastre H. Methicillin-resistant Staphylococcus aureus clonal types in the Czech Republic. J Clin Microbiol. 1999;37:2798–2803. doi: 10.1128/jcm.37.9.2798-2803.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2–A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 15.Oliveira D, Santos-Sanches I, Tamayo M, Ribeiro G, Mato R, Costa D, de Lencastre H. Virtually all MRSA infections in the largest Portuguese hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin Microbiol Infect. 1998;4:373–384. doi: 10.1111/j.1469-0691.1998.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira D C, Wu S W, de Lencastre H. Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphs of the antibiotic resistance gene. Antimicrob Agents Chemother. 2000;44:1906–1910. doi: 10.1128/aac.44.7.1906-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts R B, de Lencastre A, Eisner W, Severina E P, Shopsin B, Kreiswirth B N, Tomasz A. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. MRSA Collaborative Study Group. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 18.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 19.Ryffel C, Bucher R, Kayser F H, Berger-Bachi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173:7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sá-Leão R, Santos-Sanches I, Dias D, Peres I, Barros R M, de Lencastre H. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J Clin Microbiol. 1999;37:1913–1920. doi: 10.1128/jcm.37.6.1913-1920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanches I S, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol. 1995;33:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shopsin B, Gomez M, Montgomery S O, Smith D H, Waddington M, Dodge D E, Bost D A, Riehman M, Naidich S, Kreiswirth B N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y W, Waddington M G, Smith D H, Manahan J M, Kohner P C, Highsmith L M, Li H, Cockerill F R, 3rd, Thompson R L, Montgomery S O, Persing D H. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2000;38:1347–1351. doi: 10.1128/jcm.38.4.1347-1351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira L A, Resende C A, Ormonde L R, Rosenbaum R, Figueiredo A M, de Lencastre H, Tomasz A. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, et al. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A, a gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]