Abstract
For many years group A streptococci of T type 28 (T28) have been common in southern Sweden; however, since 1995 resistance to both macrolide-lincosamide-streptogramin B (MLS) antibiotics and tetracycline was observed among T28 isolates, which prompted the present studies on clonal relatedness of antibiotic-resistant T28 strains. By extended T typing, 95 of 100 examined tetracycline-resistant strains showed the combination T9-T13-T28; of these, 94 belonged to M type 77 (M77) and one belonged to M73. Three strains were T28-M28 and two were T28-M nontypeable. The serological M77 was confirmed by PCR capture enzyme-linked immunosorbent assay, emm amplicon restriction profiling, and emm sequence typing. Fifty strains were examined for superantigen genes: speA was detected in three blood isolates only, whereas all isolates harbored speB, and only two of the strains were negative for speC. Eighty-nine of the 100 strains were also macrolide resistant, of which 59 were inducibly MLS resistant (IR) and 21 were constitutively MLS resistant (CR), 6 were noninducibly resistant (NI), and 3 had novel subphenotypes recently reported by our group. Resistance genes were determined by PCR and hybridization methods. Eighty-four of the 100 strains harbored tetM. ermB was detected in all CR and IR strains, and mefA was found in all NI strains; both ermB and mefA were identified in two strains with novel subphenotypes. Pulsed-field gel electrophoresis showed that these antibiotic-resistant M77 strains belonged to at least five different clones.
Group A streptococci (GAS) are a major cause of acute pharyngotonsillitis and impetigo, common infections which may be followed by the nonsuppurative complications acute rheumatic fever and acute glomerulonephritis. GAS may also give rise to severe invasive manifestations such as erysipelas, endometritis, sepsis, necrotizing fasciitis, and a toxic shock-like syndrome with multiorgan failure. On the basis of this background, and due to a high contagiousness, there is a consensus that most streptococcal infections should be treated by antibiotics (2).
GAS are still highly susceptible to beta-lactam drugs (17), but alternative options are limited. Resistance to macrolides, advocated for GAS infections primarily in cases of beta-lactam allergy or intolerance, has been reported from many countries following overuse of these drugs (20); the resistance may also involve lincosamides, which are of high value both in failures of penicillin therapy of pharyngotonsillitis and for severe infections (16). Resistance to tetracycline, which is not recommended in the treatment of GAS infections, has also been reported from many areas (10, 15).
Oligoclonal spread of macrolide-resistant GAS has often been documented. In Japan, epidemic occurrence of M type 12 (M12) strains, resistant to macrolide-lincosamide-streptogramin B (MLS) antibiotics as well as tetracycline, was reported in the 1970s (14). In both Finland and Sweden, erythromycin-resistant GAS of T types 4 and 12 (T4 and T12) showed a high prevalence during the late 1980s (9). Though triggered by a high macrolide consumption, these outbreaks might subside without major changes of antibiotic prescription, perhaps due to an increasing herd immunity to the actual type.
Streptococcal resistance to macrolides is often due to target methylation and may be constitutive, comprising all MLS antibiotics, or inducible (25). Two methylase genes, ermA and ermB, have been detected in GAS (13). During the last decade, noninducible, selective resistance to macrolides due to an efflux mechanism has been increasingly noted (21); in GAS, such resistance is mediated by gene mefA, encoding a transporter protein (24). Furthermore, novel subphenotypes of MLS resistance, sometimes due to the concomitant presence of erm and mef genes, have been recently described (8, 9)
Recently, T9-T13-T28 GAS strains resistant to both MLS drugs and tetracycline have appeared in our area and, for a few years, accounted for a majority of macrolide-resistant isolates. In the present study, M type, clonal relatedness, and macrolide resistance phenotypes of these multidrug-resistant strains have been analyzed.
MATERIALS AND METHODS
Bacterial isolates.
The strains used in this study were tetracycline-resistant clinical isolates of T28 from our diagnostic laboratory, collected from 1995 to 1999. Eighty-nine of the strains were also erythromycin resistant. Three strains were blood isolates, whereas the remaining strains were throat isolates. Strains were maintained frozen at −80°C in calf serum.
Study areas.
Skåne, the southern-most province of Sweden, has a population of approximately 1 millon inhabitants, whereas the Lund, Landskrona, and Orup part of Skåne has 191,000 inhabitants.
Media and drugs.
Erythromycin powder was purchased from Sigma, and disks containing antibiotics were from Biodisk AB (Solna, Sweden). Mutanolysin, lysozyme chloride, and hyaluronidase were from Sigma.
Determination of antibiotic susceptibility.
The in vitro susceptibility to antibiotics was tested by disk diffusion on PDM agar (Progressive Diagnostics Manufacturers) in accordance with instruction provided by the Swedish Reference Group for Antibiotics (home page: www.srga.org). MICs for resistant strains were determined by the E-test (Biodisk AB) in accordance with the recommendations of the manufacturer. The phenotype of MLS resistance was tested by disk diffusion (21).
Epidemiology of antibiotic resistance.
Surveillance data on antibiotic resistance from the Clinical Diagnostic Laboratory, Lund University Hospital, since 1991 were available.
Isolation of total DNA.
Streptococcal DNA was extracted by using the method described by Seppälä et al. (22).
Detection of antibiotic resistance genes by PCR.
PCR was performed with a volume of 50 μl containing 4 μl of (5 mM) deoxynucleoside triphosphate mixture, 5 μl of 10× reaction buffer, 0.2 μl (1 U) of Taq polymerase, 2.5 μl (20 pmol) of each primer, 2 μl of streptococcal DNA, and sufficient double-distilled water for the 50-μl total volume. In the thermal reactor, a total of 35 cycles, comprising denaturation at 94°C for 20 s, annealing at 42°C for 1 min, and synthesis at 72°C for 1 min, were carried out. For detection of ermA, ermB, mefA, and tetM, previously described primers were used (3, 18, 22). The PCR products were separated by electrophoresis in a 1% agarose gel, stained with ethidium bromide, and photographed with Polaroid film under UV light. Table 1 shows primers and probes.
TABLE 1.
Primers used
| Primera | Sequence (5′–3′) | Expected size of product (bp) |
|---|---|---|
| ermB1 | AAA(C/T)TGATTTTT(A/T)GTAAA | 530 |
| ermB2 | AGGTAAAGGGCATTT | |
| ermA1 | AGGTTATAATGAAACAGA | 208 |
| ermA2 | GCATGACATAAACCTTCA | |
| mefA1 | CTATGACAGCCTCAATGCG | 1,400 |
| mefA2 | ACCGATTCTATCAGCAAAG | |
| tetM(f)1 | ATAGACACGCCAGGACATAT | 1,116 |
| tetM(f)2 | GTTTATCACGGAAGTGCAA | 686 |
| tetM(r)3 | GGAGCCCAGAAAGGATTCGG | |
| speA1 | CTTAAGAACCAAGAGATGGC | 200 |
| speA2 | ATAGGCTTTGGATACCATCG | |
| speB1 | TTCTAGGATACTCTACCAGC | 300 |
| speB2 | ATTTGAGCAGTTGCAGTAGC | |
| speC1 | CATCTATGGAGGAATTACGC | 246 |
| speC2 | TGTGCCAATTTCGATTCTGC | |
| MF1 | ATAAGGAGCATAAAAATGGCT | |
| MR1 | AGCTTAGTTTTCTTCTTTGCG | 500–1,100b |
| MF2 | TATT(C/G)GCTTAGAAAATTAA | |
| MR2 | GCAAGTTCTTCAGCTTGTTT |
f, forward primer; r, reverse primer.
Depending on the emm gene.
Detection of resistance genes by DNA hybridization.
DNA probes representing the different resistance genes were produced from the respective PCR products of known strains, generated as described above. After the band was cut from the gel and the DNA was transferred to a nylon membrane, nonradioactive DNA labeling with digoxigenin (Boehringer Mannheim) was performed. Hybridization and detection were according to the manufacturer's instruction.
Serological typing.
T typing was performed by slide agglutination according to previously documented methods (12). Since, at the Lund laboratory, only a subset of T antisera were available, more-extended typing was performed at the Public Health Laboratory Service (PHLS), London, United Kingdom. Serological M typing and opacity factor (OF) typing (OF neutralization test) were according to standard procedures at the PHLS/World Health Organization (WHO) Streptococcal Reference Unit, London, United Kingdom (11).
PCR-linked immunosorbent assay (PCR capture enzyme-linked immunosorbent assay [ELISA]) for emm typing.
The method followed our previous description (10).
Fast preparation of streptococcal genomic DNA.
A few colonies from a fresh overnight culture on blood agar were inoculated into 1 ml of phosphate-buffered saline (PBS) buffer (pH 7.4). The tube was incubated for 30 min at 60°C and centrifuged for 1 min at 18,000 × g. The pellet was washed two times with 1 ml of PBS, resuspended in 100 μl of PBS containing 10 U of mutanolysin, and incubated for 30 min at 37°C. The suspension was boiled for 5 min and centrifuged for 10 min at 18,000 × g. The supernatant, containing genomic DNA, was used for amplification of emm and spe genes.
PFGE.
Selected strains were typed by pulsed-field gel electrophoresis (PFGE) in accordance with standard methods at the PHLS/WHO Streptococcal Reference Unit (10).
emm amplicon restriction profiling (ERP) analysis.
To compare isolates within the same serological M type, the emm amplicons were subjected to restriction enzyme cleavage analysis by previously documented methods (1).
emm gene sequence typing.
The primers used are shown in Table 1. PCR was done as described above. The product was purified by using the Qia Quick PCR purification kit (Qiagen) as described by the manufacturer. emm sequencing was performed directly from the purified product using a maximum of 6 μl of product per reaction. Four microliters of M forward primer, 8 μl of “premix” for the ABI 310 sequencer, and 20 μl of aqua ad injectionem were mixed for cycling, and the following parameters were then used: preheating at 96°C for 2 min and then 25 cycles with denaturation at 96°C for 15 s, annealing at 50°C for 15 s, and elongation at 60°C for 4 min. The product was removed from the cycler and precipitated with 2 μl of 3 M sodium acetate (pH 5.2) in 50 μl of 95% ethanol and kept on ice for 10 min. After centrifugation at full speed (18,000 × g) for 30 min the supernatant was removed and the pellet was washed with 250 μl of 70% ethanol. The supernatant was again removed by centrifugation at full speed for 15 min, and the pellet was dried in a speed vacuum for 2 to 3 min. The pellet was suspended in a genetic analysis tube with 20 μl of template suspension reagent and heated for 2 min at 90°C and then chilled on ice for 2 min. Ten plates were sequenced in an ABI Prism 310 DNA sequencer. The sequence obtained was compared with the nucleotide sequences encoding the N-terminal hypervariable portion of streptococcal M proteins for a similarity search against the National Institutes of Health DNA database. The degree of similarity required for emm gene designation was >95% identity for a portion of at least 200 bp within the 5′ region of the respective gene using the latest information via the Internet (www.cdc.gov/ncidod/biotech/strep/strains/emmtypes.html).
Superantigen gene detection.
The presence of the genes for erythrogenic toxins A, B, and C (speA, speB and speC, respectively) was investigated using primers speA1 and -A2, speB1 and -B2, and speC1 and -C2, respectively, as shown in Table 1. PCR was performed in a volume of 50 μl, containing 5 μl of deoxynucleoside triphosphate mixture (5 mM), 10 μl of 10 × reaction buffer 3 μl of DNA template, 0.2 μl (1 U) of Taq polymerase, 2.5 μl (20 pmol) of each primer, 5 μl of MgCl2 (50 mM), and sufficient double-distilled water for the 50-μl total volume. In the thermal reactor, a total of 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and synthesis at 72°C for 1 min were carried out.
Antibiotic consumption.
Information on sales of antibiotics from The National Corporation of Pharmacies was kindly provided by Helena Lundahl, Lund, Sweden. Data on national, regional, and municipal levels are available and were given as defined daily doses per 1,000 inhabitants (DDD/TID).
Nucleotide sequence accession numbers.
Ten of the emm sequences obtained in this study were submitted to GenBank and were assigned accession no. AF 261033 to 261039, AF 240471 and 240472, and AF 210432.
RESULTS
Phenotype determination.
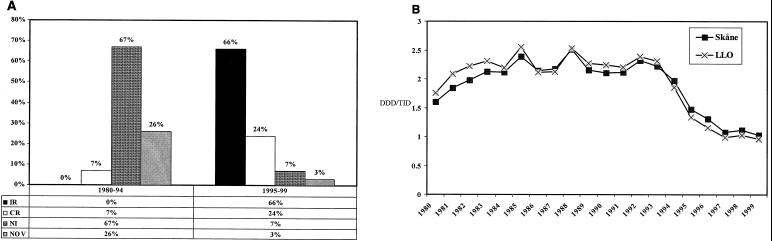
Out of 100 tetracycline-resistant strains tested 89 were also resistant to erythromycin; of the latter, 59 (66%) were inducibly resistant (IR) and 21 (24%) were constitutively resistant (CR) to MLS and 6 (7%) had the noninducibly resistant (NI) phenotype. Three strains were highly resistant to erythromycin but susceptible or weakly resistant to clindamycin; their phenotype agreed with previously described, novel subphenotypes (9). As shown in Fig. 1A, the distribution of MLS phenotypes differed markedly from that recorded among 200 randomly selected strains from 1980 to 1994, earlier reported (9).
FIG. 1.
MLS resistance phenotypes and antibiotic consumption in southern Sweden from 1980 to 1999. (A) Distribution of MLS antibiotic resistance phenotypes among GAS isolates from 1980 to 1994 (n = 200) and 1995 to 1999 (n = 89). NOV, novel MLS resistance subphenotype. (B) Macrolide consumption in the province Skåne and the area Lund-Landskrona-Orup (LLO).
Demonstration of streptococcal erythromycin resistance determinants.
By PCR, the resistance determinant tetM was identified in 54 of 70 tetracycline-resistant strains tested (Table 2). As expected, a single band of 1.4 kbp was detected in each of 6 NI. strains using the primers for mefA, whereas all 59 IR and 21 CR strains examined gave a band of 530 bp when tested for ermB. Two of the strains with novel MLS phenotypes were positive for both ermB and mefA, whereas one strain was negative. All strains were negative for ermA. Eighty-four of the 100 strains hybridized with the tetM probe. All 21 CR and 59 IR isolates hybridized with the ermB probe, whereas the NI strains gave no hybridization signal. In contrast, each of the NI strains were positive with the probe created from mefA (Table 2).
TABLE 2.
Resistance determinants of 100 GAS isolates from 1995 to 1999
| Resistance phenotype | Total no.b | No. of strains positive by PCR for:
|
No. of strains that hybridized with:
|
||||
|---|---|---|---|---|---|---|---|
| ermB | mefA | tetMc | ermB | mefA | tetM | ||
| IR | 59 | 59 | 0 | 24/30 | 59 | 0 | 52 |
| CR | 21 | 21 | 0 | 17/20 | 21 | 0 | 19 |
| NI | 6 | 0 | 6 | 0/6 | 0 | 6 | 0 |
| NOVa | 3 | 2 | 2 | 2/3 | 2 | 2 | 2 |
| Tet | 11 | 0 | 0 | 11/11 | 0 | 0 | 11 |
| Total | 100 | 82 | 8 | 54/70 | 82 | 8 | 84 |
Two strains were positive for ermB as well as mefA, whereas one strain was negative for both. NOV, novel MLS resistance subphenotype.
Total number of strains with the indicated phenotype.
Number of strains positive for tetM/number of tetracycline-resistant strains.
Serological and molecular typing.
All the 100 antibiotic-resistant T28 strains were further typed. Ninety-four of the strains were of type T9-T13-T28 and belonged to M77, one was T9-T13-T28 and M73. Of remaining strains, three were of type T28-M28 whereas two were T28-M nontypeable. Concordant results for the emm type were obtained by PCR capture ELISA (data not shown).
ERP analysis.
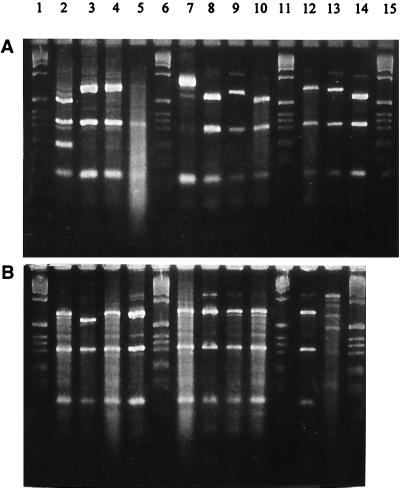
Among 50 M77 strains tested, ERP showed three different patterns. As demonstrated in Fig. 2 for 17 of the strains, three major bands were invariably present, with the molecular weight of one of these different for different patterns.
FIG. 2.
emm gene polymorphism tests of 17 M77, one M28, and one M73 GAS strains. Lanes 1, 6, 11, and 15 (A) and 1, 6, 11, and 14 (B), molecular weight marker (1-kbp DNA ladder; GIBCO); lane 2 (A), M28; lane 13 (B), M73. The remaining lanes show M77 strains.
emm gene sequence typing.
The 50 M77 strains mentioned above were subjected to emm gene sequencing. All the strains showed >97% homology to the Centers for Disease Control and Prevention emm77 sequence, and homology within the different patterns of ERP was >95%. Due to the high similarity between the 50 emm sequences obtained, only 10 of these were submitted to GenBank.
PFGE.
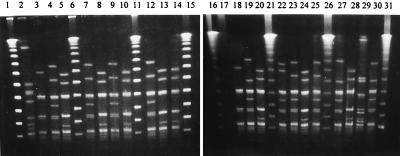
The same 50 M77 isolates, representing different antibiotic resistance phenotypes, were subjected to PFGE typing. A limited clonal heterogeneity was suggested by the identification of five different pulsotypes (arbitrarily designated 1 to 5), as shown in Fig. 3 for 23 of the strains. All 11 strains with resistance only to tetracycline belonged to a common pulsotype. The IR and CR strains were distributed between two pulsotypes, whereas the NI strains had the same pulsotype. One strain with a novel MLS subphenotype, exhibiting both ermB and mefA, showed a unique pulsotype. Distinct pulsotypes were also recorded for the M28 and M73 strains (Table 3).
FIG. 3.
PFGE patterns of 23 GAS strains of different antibiotic resistance phenotypes. Lanes 1, 6, 11, 15, 16, 21, 26, and 31, DNA ladder (lambda PFGE ladder; Bio-Rad); lanes 2 and 17, PF1; lane 29, PF2; lanes 23, and 25, PF3; lanes 4, 7, 9, 12, 19, 27, and 30, PF4; lanes 5, 8, 10, 13, 14, 20, 24, and 28, PF5; lanes 3, and 18, PF6; lane 22, PF7. See Table 3 for relations between pulsotype, phenotype, and M type.
TABLE 3.
Summary of PFGE analysis of 53 selected antibiotic-resistant S. pyogenes isolates from southern Sweden
| Antibiotic resistance phenotypea | M type | Pulsotype | No. of isolates (n = 53) |
|---|---|---|---|
| IR, Tet | 28 | PF1 | 2 |
| IR, Tet | 73 | PF2 | 1 |
| Tet | 77 | PF3 | 11 |
| IR, Tet | 77 | PF4, PF5 | 22 |
| CR, Tet | 77 | PF4, PF5 | 11 |
| NI, Tet | 77 | PF6 | 5 |
| NOV, Tet | 77 | PF7 | 1 |
NOV, novel MLS resistance subphenotype; Tet, tetracycline resistance.
Superantigen gene detection.
The 50 M77 strains were also tested for the presence of superantigen genes. All exhibited speB, and only two were negative for speC. However, speA was detected in three strains only, each of them a blood isolate.
Antibiotic consumption.
As shown in Fig. 1B, the local consumption of macrolides increased gradually during the 1980s, from 1.60 to 2.55 DDD/TID. A gradual decline was noted from 1994 until 1999, the consumption reaching a level of 0.96 DDD/TID.
Antibiotic resistance data.
The annual prevalence in our area of antibiotic resistance among GAS increased in the early 1990s to 12%, whereas from 1994 on it did not exceed 2.3%. Clindamycin resistance was below 0.2% until 1998, when a level of 0.6% was noted. In contrast, the prevalence of tetracycline resistance rose significantly during the decade, from 2 to 15%.
DISCUSSION
In this study, the common occurrence of T9-T13-T28 among recent tetracycline- and macrolide-resistant Swedish (southern region) Streptococcus pyogenes strains is reported. The results of serological M typing demonstrated that almost all these strains were M77. Furthermore, by PCR capture ELISA and emm gene sequence typing, their type identities were confirmed in all cases. Though ERP analysis revealed three patterns, the difference between these resided in the molecular weight of one band only and was probably due to a variation in the number of repeat sequences well known for the streptococcal M protein (7). Conventionally, the M77 is associated with T13 and/or T28, whereas T9 and the T antigen complex T9-T13-T28 to our knowledge have not been previously found (see www.cdc.gov/ncidod/biotech/strep/strains/emmtypes.html). Since M typing has not been previously performed at our laboratory, we do not know whether M77 has occurred earlier in Sweden.
Three of the present strains were blood isolates, demonstrating that M77 is an invasive serotype. Interestingly, speA, the gene of a major superantigen often ascribed a role in severe GAS infections (6), was detected in these three strains but not in a number of noninvasive isolates. Since T28 has been a common type accounting for invasive disease in Sweden (23), it will be of interest to examine the M types of additional T28 invasive isolates.
By PFGE five different pulsotypes among the M77 antibiotic-resistant strains were revealed, showing that several clones accounted for their spread. All the strains selected for the present study, as mentioned, were tetracycline resistant, and the majority harbored the resistance determinant tetM. Notably, tetM was detected in most of the IR and CR strains but not in any of the six noninducibly macrolide-resistant strains. Possibly some other tet determinant was carried by the latter strains, apparently forming a single clone. As expected, methylase gene ermB was demonstrated in all CR and IR strains whereas in the NI strains mefA was identified. The presence of macrolide resistance in almost all the tetracycline-resistant strains studied may suggest that the corresponding genes occurred on a common, mobile element, as has been earlier reported for streptococcal species (4).
We have recently reported on three novel subphenotypes of MLS resistance among GAS of other M types (9); of the present M77 strains three exhibited such novel subphenotypes, implying high-level erythromycin resistance and low resistance of susceptibility to clindamycin, and two of the strains carried both ermB and mefA. The combination of these determinants has also been detected in Italian GAS isolates (8).
Since in Sweden tetracycline is used in the treatment of chlamydial and mycoplasmal infections rather than streptococcal infections, the level of tetracycline resistance among GAS clinical isolates, 13 to 16%, appeared comparatively high. However, in certain countries much higher rates were recently reported (5, 10). From 1994 on the pattern of resistance phenotypes in erythromycin-resistant GAS strains in southern Sweden changed from a predominance of the NI phenotype to the classical IR and CR MLS phenotypes. Only 6% of the present strains had the NI phenotype. In parallel with this change a marked decrease of both macrolide consumption and level of erythromycin resistance among GAS (from 12 to 1.8%) was noted. Recently, a similar trend was reported from Finland (19). A relation between occurrence of the different macrolide resistance phenotypes and total consumption of macrolide antibiotics therefore appears conceivable.
In conclusion, among current Swedish, multiantibiotic-resistant, T9-T13-T28 GAS isolates, M77 was established for almost all strains by serological M typing, PCR capture ELISA, ERP, and emm gene sequencing. Furthermore, five pulsotypes were distinguished, and clonal heterogeneity was also emphasized by the occurrence of four different MLS resistance phenotypes among the strains. Nevertheless, in our area the total level of erythromycin resistance among GAS is currently low, presumably an effect of declining usage of macrolides in the treatment of respiratory tract infections.
ACKNOWLEDGMENTS
The study was supported by Pharmacia & Upjohn, Abbott Scandinavia AB, the Royal Physiographical Society, the Alfred Österlund Foundation, the Medical Faculty of Lund University, and the Erik Philip Sörensen Foundation.
We are grateful to Janet Philipsson and Britt-Marie Thulin for collecting clinical strains.
REFERENCES
- 1.Beall B, Facklam R R, Elliott J A, Franklin A R, Hoenes T, Jackson D, LaClaire L, Thompson T, Viswanathan R. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J Med Microbiol. 1998;47:893–898. doi: 10.1099/00222615-47-10-893. [DOI] [PubMed] [Google Scholar]
- 2.Bisno A L, Gerber M A, Gwaltney J M J, Kaplan E L, Schwartz R H. Diagnosis and management of group A streptococcal pharyngitis: a practice guideline. Infectious Diseases Society of America. Clin Infect Dis. 1997;25:574–583. doi: 10.1086/513768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clancy J, Dib H F, Petitpas J W, Yuan W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother. 1997;41:2719–2723. doi: 10.1128/aac.41.12.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont D, Delbos F, de Cespedes G, Horaud T. Old and new (Tn3708) mobile chromosomal elements in streptococci and enterococci. Dev Biol Stand. 1995;85:55–61. [PubMed] [Google Scholar]
- 5.el Bocer M, Fendri C, Ben Hassen A, Kamoun A, Boudabbous A, Ben Redjeb S. Study of antibiotic sensitivity of Streptococcus pyogenes isolated in the hospital milieu. Med Trop. 1993;53:13–17. [PubMed] [Google Scholar]
- 6.Eriksson B K, Andersson J, Holm S E, Norgren M. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J Infect Dis. 1999;180:410–418. doi: 10.1086/314872. [DOI] [PubMed] [Google Scholar]
- 7.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovanetti E, Montanari M P, Mingoia M, Varaldo P E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–1940. doi: 10.1128/aac.43.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasir A, Schalén C. Survey of macrolide resistance phenotypes in Swedish clinical isolates of Streptococcus pyogenes. J Antimicrob Chemother. 1998;41:135–137. doi: 10.1093/jac/41.1.135. [DOI] [PubMed] [Google Scholar]
- 10.Jasir A, Tanna A, Noorani A, Mirsalehian A, Efstratiou A, Schalén C. High rate of tetracycline resistance in Streptococcus pyogenes in Iran: an epidemiological study. J Clin Microbiol. 2000;38:2103–2107. doi: 10.1128/jcm.38.6.2103-2107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson D R. Laboratory diagnosis of group A streptococcal infactions. Geneva, Switzerland: World Health Organization; 1996. [Google Scholar]
- 12.Johnson D R, Kaplan E L. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J Med Microbiol. 1993;38:311–315. doi: 10.1099/00222615-38-5-311. [DOI] [PubMed] [Google Scholar]
- 13.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama S, Yoshioka H, Fujita K, Takimoto M, Satake Y. Sensitivity of group A streptococci to antibiotics. Prevalence of resistance to erythromycin in Japan. Am J Dis Child. 1979;133:1143–1145. doi: 10.1001/archpedi.1979.02130110051007. [DOI] [PubMed] [Google Scholar]
- 15.Orden B, Martinez R, Lopez-de-los M A, Franco A. Antibiotic resistance to erythromycin, clindamycin and tetracycline of 573 strains of Streptococcus pyogenes (1992-1994) Enferm Infecc Microbiol Clin. 1996;14:86–89. [PubMed] [Google Scholar]
- 16.Orrling A, Stjernquist D A, Schalén C, Kamme C. Clindamycin in persisting streptococcal pharyngotonsillitis after penicillin treatment. Scand J Infect Dis. 1994;26:535–541. doi: 10.3109/00365549409011811. [DOI] [PubMed] [Google Scholar]
- 17.Orrling A, Stjernquist D A, Schalén C, Kamme C. Treatment failure in streptococcal pharyngotonsillitis. An attempt to identify penicillin tolerant Streptococcus pyogenes. Scand J Infect Dis. 1996;28:143–147. doi: 10.3109/00365549609049065. [DOI] [PubMed] [Google Scholar]
- 18.Roberts M C, Pang Y, Riley D E, Hillier S L, Berger R C, Krieger J N. Detection of Tet M and Tet O tetracycline resistance genes by polymerase chain reaction. Mol Cell Probes. 1993;7:387–393. doi: 10.1006/mcpr.1993.1057. [DOI] [PubMed] [Google Scholar]
- 19.Seppälä H, Klaukka T, Vuopio V J, Muotiala A, Helenius H, Lager K, Huovinen P. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. Finnish Study Group for Antimicrobial Resistance. N Engl J Med. 1997;337:441–446. doi: 10.1056/NEJM199708143370701. [DOI] [PubMed] [Google Scholar]
- 20.Seppälä H, Nissinen A, Järvinen H, Huovinen S, Henriksson T, Herva E, Holm S E, Jahkola M, Katila M L, Klaukka T, et al. Resistance to erythromycin in group A streptococci. N Engl J Med. 1992;326:292–297. doi: 10.1056/NEJM199201303260503. [DOI] [PubMed] [Google Scholar]
- 21.Seppälä H, Nissinen A, Yu Q, Huovinen P. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J Antimicrob Chemother. 1993;32:885–891. doi: 10.1093/jac/32.6.885. [DOI] [PubMed] [Google Scholar]
- 22.Seppälä H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strömberg A, Romanus V, Burman L G. Outbreak of group A streptococcal bacteremia in Sweden: an epidemiologic and clinical study. J Infect Dis. 1991;164:595–598. doi: 10.1093/infdis/164.3.595. [DOI] [PubMed] [Google Scholar]
- 24.Sutcliffe J, Tait K A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisblum B. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression—a review. J Antimicrob Chemother. 1985;16(Suppl A):63–90. doi: 10.1093/jac/16.suppl_a.63. [DOI] [PubMed] [Google Scholar]