Abstract
This study aimed to investigate the human proteome profile of samples collected from whole (W) Schirmer strips (ScS) and their two parts—the bulb (B) and the rest of the strip (R)—with a comprehensive proteomic approach using a trapped ion mobility mass spectrometer, the timsTOF Pro. Eight ScS were collected from two healthy subjects at four different visits to be separated into three batches, i.e., 4W, 4B, and 4R. In total, 1582 proteins were identified in the W, B, and R batches. Among all identified proteins, binding proteins (43.4%) and those with catalytic activity (42.2%) constituted more than 80% of the molecular functions. The most represented biological processes were cellular processes (31.2%), metabolic processes (20.8%), and biological regulation (13.1%). Enzymes were the most represented protein class (41%), consisting mainly of hydrolases (47.5%), oxidoreductases (22.1%), and transferases (16.7%). The bulb (B), which is in contact with the conjunctiva, might collect both tear and cell proteins and therefore promote the identification of more proteins. Processing B and R separately before mass spectrometry (MS) analysis, combined with the high data acquisition speed and the addition of ion-mobility-based separation in the timsTOF Pro, can bring a new dimension to biomarker investigations of a limited sample such as tear fluid.
Keywords: proteome, tears, Schirmer strip, timsTOF Pro, signaling pathways
1. Introduction
The tear film, composed of the secretions of lacrimal glands, Meibomian glands, and goblet cells, is essential for many vital functions of the ocular surface (OS), such as protection, lubrication, and nutrition [1,2]. A healthy tear film represents a barrier between the eye and the environment [3]. By coating the OS, the tear film provides an optically smooth surface, which is necessary for the refraction of light onto the retina [4,5]. Tear fluid (TF) is an important medium for the evaluation of OS disease (OSD), and it is suitable for prognostic and diagnostic purposes [6]. Despite its small volume, TF offers several advantages for biochemical analysis, biomarker discovery, and development of new drugs [5,6,7,8,9]. TF can be collected with various instruments such as microcapillary tubes, absorbent cellulose acetate filters, polyester wicks, and Schirmer strips (ScS) [10,11], the latter of which is used by clinicians to perform the Schirmer’s tear test (STT). The STT is a standard ophthalmologic test that measures tear production based on the wetted length of the strip. It is one of the most commonly used clinical tests for the evaluation of dry eye disease (DED) [12,13]. Likewise, it is relatively convenient, rapid and reliable, and therefore, despite a slight and brief tingling when placed into the conjunctival cul-de-sac, it is used more frequently than the glass capillary tube method, which requires a delicate procedure on the open eye [14]. Apart from being more convenient to collect tear samples, especially in patients with DED, the ScS offers the opportunity to collect, through the bulb (the upper part of the ScS), superficial conjunctival cells [11]. In other words, secreted proteins can be collected using the capillary method, while a combination of cellular and soluble proteins can be collected using ScS [15]. These collection methods are of major interest in identifying biomarker candidates in OSD such as DED, using powerful “omics” technologies. Indeed, tear proteomics is increasingly employed for such investigations [16,17]. In the last two decades, new insights have been achieved in tear proteomics studies using various systems such as Q-TOF or Orbitrap [18,19,20]. The first major proteomics dataset for tears was created by de Souza et al. in 2006, with the identification of 491 proteins, and in the last decade, several new proteomics approaches have been used, creating large datasets of the tear proteome by using various forms of advanced mass spectrometry (MS) [16,18,20,21,22]. These studies, revealing the datasets for the healthy tear proteome are summarized in Table 1.
Table 1.
Overview of proteomics studies that have created the largest datasets for the healthy human tear proteome using LC-MS/MS from 2006 to 2020. NIPs, number of identified proteins; HSs, healthy subjects; MS, mass spectrometry; FDR, false discovery rate.
| Group, Year | Goal | Tear Sampling Method | Sample Preparation | MS Technology | Protein Identification Criteria |
NIPs in HSs |
|---|---|---|---|---|---|---|
| de Souza et al., Genome Biol., 2006 [22] | Characterization of the protein content of the human tear fluid from a HSs | Microcapillary method | With pre-fractionation of proteins with one-dimensional SDS-PAGE (13 fractions) or without (in-solution digestion of the whole samples) | Hybrid linear ion trap–Fourier-transform (LTQ-FT) and linear ion trap-Orbitrap (LTQ-Orbitrap) |
Two peptides with Mascot scores of >35, Two peptides with Mascot scores of >27 (p ≤ 0.01), or one peptide with an Mascot score of >54 (p ≤ 0.0001), when MS3 was performed |
491 |
| Zhou et al., J. Proteomics, 2012 [18] | Analysis of the human tear proteome from HSs | Schirmer strips | Offline SCX fractionation of peptides (6 fractions) | TripleTOF 5600 system | FDR <1% for peptides | 1543 |
| Aass et al., Anal. Biochem., 2015 [19] | Optimizing extraction method from Schirmer strips to study the tear proteome | Schirmer strips | Offline SCX fractionation of peptides (16 fractions) | LTQ-Orbitrap XL hybrid | Peptide and protein with FDRs of <1% (high) and 5% (relaxed) | 1526 |
| Kandhavelu et al., J. Proteomics, 2016 [20] | Comparison of tear proteins in control and fungal keratitis patients | Capillary method | N-linked glycoprotein enrichment or one-dimensional SDS-PAGE pre-fractionation of proteins (26 fractions) | LTQ-Orbitrap Velos Pro | One peptide with an FDR of <5% | 1873 |
| Dor et al., Exp. Eye Res., 2019 [21] | Characterization of healthy human tear protein composition | Schirmer strips | Off-gel electrophoresis of peptides (12 fractions) | LTQ-Orbitrap Velos Pro TripleTOF 5600+ in SWATH-MS mode |
Two peptides with FDRs of <1% | 1351 |
| Nättinen et al., Trans. Vis. Sci. Tech., 2020 [23] | Investigation of protein profile differences between capillary and Schirmer strip tear fluid samples | Schirmer strips | No pre-fractionation (in-solution digestion of whole samples) | TripleTOF 5600 + in SWATH-MS mode | FDR <1% | 908 |
| Capillary method | 404 | |||||
| Hua et al., BMC Ophthalmol., 2020 [24] | Quantification of potential candidate biomarkers for HSV-1 epithelial keratitis | Microcapillary method | No pre-fractionation (in-solution digestion of whole samples) | LTQ-Orbitrap XL | FDR <1% | 949 |
| Ponzini et al., Int. J. Mol. Sci., 2021 [25] | Demonstration of feasibility of single-tear quantitative proteomics | Capillary method | No pre-fractionation (in-solution digestion of whole samples) | Orbitrap fusion | One peptide with an FDR of <1.0% | 932 |
| Zysset-Burri et al., Inv. Ophthalmol. Vis. Sci., 2021 [26] | Exploring the interplay between the ocular surface microbiome and the tear proteome | Schirmer strips | With pre-fractionation of (5 factions) proteins with one-dimensional SDS-PAGE | QExactive HF | FDR <1% | 2172 |
Interest in MS continues to grow, and new technologies are emerging for TF analysis [27]. These more efficient techniques have promoted the reliable identification of large numbers of proteins from TF samples [28,29,30]. Thus, MS has been implemented in several studies exploring the modulation or dysregulation of proteins in OSD [15,18,31,32,33,34]. Nano-scale liquid chromatography (nano-LC), coupled to tandem MS (MS/MS), provides the improved chromatographic separation of peptides, higher sensitivity, and extended dynamic ranges to identify a large number of proteins [35,36]. Therefore, nano-LC-MS/MS has become the first choice for proteomic analysis of small samples such as TF [37]. The timsTOF Pro mass spectrometer (Bruker Daltonics, Bremen, Germany), which combines the sensitivity of trapped ion-mobility spectrometry (TIMS) and a fast quadrupole time-of-flight (Q-TOF), has been providing a large dataset for exploring the proteome of several biological fluids [38,39]. After peptide separation with nano-LC, peptides are subsequently ionized and trapped in a dual TIMS [40]. This dual TIMS technology traps ions in the first section and separates them according to their mobility in the second section [41], functioning as a third dimension of separation [39]. Thanks to the dual TIMS technique, the parallel accumulation-serial fragmentation (PASEF) acquisition method was developed, providing a duty cycle of nearly 100% (nearly no ion loss) and a high sensitivity [39]. PASEF corresponds to a mass selective release of peptide ions from TIMS devices for MS/MS at a high acquisition speed, increasing MS/MS scan rates up to 100 Hz [42,43]. In conclusion, the timsTOF Pro provides extremely high speeds and sensitivity to reach new depths in proteomics analysis using a small amount of samples.
The current study sought to perform a comprehensive tear proteomics investigation from samples collected with ScS using nanoElute ultra-high-pressure liquid chromatography (UHPLC) coupled to the timsTOF Pro. The protein composition of the various sections of the ScS can be considered to characterize the human ScS-extracted proteome (SEP) and describe in detail the signaling pathways involved.
2. Results
2.1. Number of Identified Proteins (NIPs) and Their Distributions in the Sections of the ScS
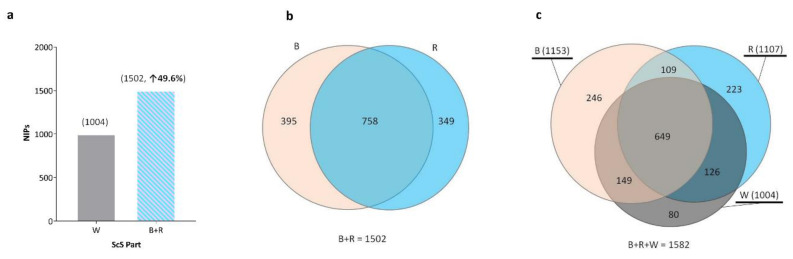
In the whole ScS (W), the NIPs was 1004. When the two sections of the ScS were analyzed separately, 1153 proteins were identified in the upper part (or bulb (B)) and 1107 were in the rest of the ScS (R). Therefore, processing sections B and R of the ScS as two different sample batches increased the NIPs from 1004 to 1502, for a 49.6% increase in identified proteins compared to in the entire strip (W) (Figure 1a). Of these 1502 proteins identified in sections B and R, a higher number of proteins, 758, were common, compared to 395 and 349 in sections B and the R, respectively (Figure 1b). Moreover, among the identified proteins in the 3 batches (W, B, and R), 649 proteins were common, whereas a smaller number of unique proteins, 80, 246, and 223 in W, B, and R, respectively, were identified (Figure 1c). The 20 most abundant proteins identified with the highest mean MS/MS spectral count in W, B, and R are shown in Tables S2–S4.
Figure 1.
Number of identified proteins (NIPs) and their distributions in the various sections of the Schirmer strips (ScS). (a) Effect of processing two different sections of the ScS (B + R) on the NIPs. (b) Comparison of the NIPs between the two sections of the ScS and section R. (c) Venn diagram displaying the comparison of the NIPs in the whole strip (W), bulb (B), and the rest of the strip (R).
In total, 1582 proteins were identified by summing the 3 batches with no pre-fractionation during sample preparation. The raw data was represented in Table S1, and the MS proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030334. Moreover, we compared these results with those from four proteomics studies that used advanced MS technologies in healthy human tear proteome [19,20,21,25] in order to enrich the SEP dataset. Among the 1183 proteins identified in our study, 266 were detected in study (A), 972 were detected in study (B), 487 were detected in study (C), and 543 were detected in study (D) (Figure S1). More interestingly, we identified 399 new proteins that were not identified in these studies.
2.2. Distribution within ScS Sections and Spectral Counts for the Previously Described Tear Proteins
The tear proteins identified in this study and previously reported in the literature for their role in OSD are presented with their spectral counts (Table 2). This classification also highlights the specific and common proteins identified in ScS batches. Some of these common proteins, such as PIP, HSPG2, and C3, were among the 20 most abundant proteins identified with the highest mean of spectral count in W, B, and R (Tables S2–S4). Interestingly, when considering clinically relevant OSD biomarkers in tears, such as lysozyme C, lactotransferrin, and lipocalin-1, our results showed that their mean spectral counts were rather similar in the three batches. However, the spectral count for another relevant OSD biomarker, serum albumin, was different among batches, with 101.7, 204.3, 67 in W, B, and R, respectively. This difference of the spectral counts between batches was also found for ALDH1A3 and α-enolase, with a higher abundance in B, for mammaglobin-B with a higher abundance in W, and HSPG2 with a higher abundance in R. Additionally, some proteins were either unique only to two batches or even just one. For instance, MUC5AC and IL-18 were detected with a higher spectral count in B compared to W but were undetected in the R section of the ScS. Finally, matrix metalloproteinase (MMP)-9 was detected only in B, with a low quantity, whereas Serpin B3 and B4 were specifically identified in R.
Table 2.
List of identified proteins previously described in ocular surface disease. The mean spectral count and standard deviation (SD) of each protein in the whole strip (W), the bulb (B), and the rest (R) of the strip are presented. The proteins are classified as those common in all batches (1), two batches (2), or one batch (3) and are ranked according to their mean of spectra in W. *, proteins reported in dry eye disease studies. Accession number refers to UniProt identification.
| Accession Number | Protein Name | W | B | R |
|---|---|---|---|---|
| Mean ± SD | ||||
| (1) Common in all batches | ||||
| P12273 | Prolactin-induced protein [1] | 142.3 ± 14 | 108 ± 15.4 | 121.7 ± 2.5 |
| P98160 | Heparan sulfate proteoglycan 2 [1,29] | 113.3 ± 5.5 | 56.3 ± 4.9 | 123.7 ± 1.5 |
| P61626 | Lysozyme C * [6,29,44] | 111.3 ± 6.7 | 104 ± 4.6 | 108.7 ± 5.5 |
| P01876 | Ig alpha-1 chain C region [6,29] | 107 ± 2.6 | 61.7 ± 3.2 | 74 ± 4.4 |
| P02768 | Serum albumin * [6,45] | 101.7 ± 2.1 | 204.3 ± 8.1 | 67 ± 4 |
| P02788 | Lactotransferrin * [29,44] | 100.7 ± 5.6 | 130.3 ± 7.1 | 101.7 ± 3.5 |
| P01024 | Complement C3 [22,44] | 72.7 ± 3.2 | 69.7 ± 1.5 | 62.7 ± 2.1 |
| Q9UGM3 | Salivary agglutinin [1,44] | 50.7 ± 4 | 48 ± 2 | 54.3 ± 0.6 |
| P31025 | Lipocalin-1 * [6,29,44] | 45.3 ± 2.1 | 50 ± 3.6 | 36.7 ± 2.3 |
| P00352 | Retinal dehydrogenase 1 [23,44] | 35.3 ± 2.1 | 46 ± 4 | 24.7 ± 2.1 |
| O75556 | Mammaglobin-B [1,44] | 35.3 ± 0.6 | 18 ± 1 | 6.3 ± 1.2 |
| P06733 | Alpha-enolase [1,44] | 31.3 ± 1.2 | 52 ± 4.6 | 27.3 ± 0.6 |
| P02787 | Serotransferrin [46] | 24.7 ± 3.1 | 59 ± 4 | 26.7 ± 1.5 |
| P07900 | Heat shock protein HSP 90-alpha [23,47] | 22.3 ± 1.5 | 8 ± 2.6 | 24.3 ± 1.5 |
| P04083 | Annexin A1 [23,44] | 22.3 ± 0.6 | 24.7 ± 0.6 | 18 ± 1 |
| P06702 | Protein S100-A9 [1] | 19 ± 1 | 28.7 ± 2.1 | 22 ± 1 |
| P07858 | Cathepsin B [1] | 18.3 ± 1.5 | 19.7 ± 1.5 | 22 ± 1.7 |
| P47895 | Aldehyde dehydrogenase family 1 member A3 [23,48] | 18 ± 2.6 | 23.7 ± 1.5 | 6.3 ± 2.1 |
| P00738 | Haptoglobin [1] | 18 ± 2 | 19 ± 1.7 | 14 ± 0 |
| P30740 | Serpin B1 [22,23] | 15.7 ± 2.1 | 24.3 ± 1.2 | 8.7 ± 3.2 |
| P05109 | Protein S100-A8 [1] | 15.7 ± 0.6 | 22.3 ± 3.2 | 22 ± 2.6 |
| P25311 | Zinc-alpha-2-glycoprotein [1] | 12.7 ± 1.5 | 14.3 ± 1.5 | 16.3 ± 1.5 |
| P07355 | Annexin A2 [23,44] | 12.7 ± 1.2 | 36.7 ± 3.5 | 25 ± 1 |
| P18510 | Interleukin 1 receptor antagonist protein [49] | 9.7 ± 1.5 | 12.3 ± 1.5 | 7 ± 1 |
| Q8WUM4 | Programmed cell death 6-interacting protein [50,51] | 9 ± 0 | 2.7 ± 0.6 | 5 ± 1.7 |
| P23528 | Cofilin-1 [1] | 8.7 ± 3.8 | 15.3 ± 2.1 | 8.7 ± 2.1 |
| P07476 | Involucrin [52] | 6 ± 1 | 3 ± 1.7 | 20.3 ± 2.3 |
| P01023 | Alpha-2-macroglobulin [22,52] | 5.3 ± 1.5 | 15.7 ± 2.3 | 7.7 ± 1.2 |
| Q6UXB2 | C-X-C motif chemokine ligand 17 [53] | 1 ± 1 | 0.7 ± 1.2 | 1 ± 1 |
| (2) Common to two batches | ||||
| P98088 | Mucin-5AC [44,54,55] | 10.3 ± 0.6 | 54.3 ± 3.8 | 0 |
| Q14116 | Interleukin 18 [56] | 0.7 ± 0.6 | 2 ± 1 | 0 |
| P02778 | C-X-C motif chemokine ligand 10 [57] | 2 ± 0 | 0 | 0.3 ± 0.6 |
| (3) Unique to one batch | ||||
| Q9UHD0 | Interleukin 19 [58] | 0.7 ± 1.2 | 0 | 0 |
| P14780 | Matrix metallopeptidase 9 * [49,59,60] | 0 | 0.3 ± 0.6 | 0 |
| P29508 | Serpin B3 [61] | 0 | 0 | 23.7 ± 2.5 |
| P48594 | Serpin B4 [61] | 0 | 0 | 12 ± 1 |
| Q9UHA7 | Interleukin 36 alpha [62] | 0 | 0 | 0.3 ± 0.6 |
2.3. Functional Annotation Analysis in the Various Sections of the ScS
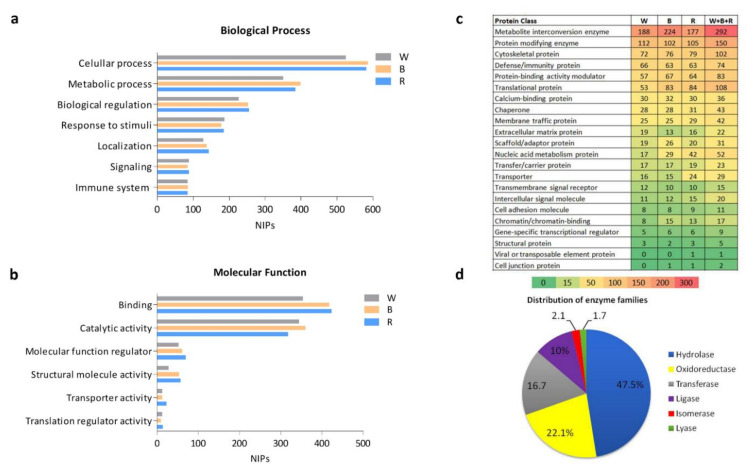
Despite the several unique proteins identified in each batch (W, B, and R), no major difference was observed in biological processes or molecular functions, as shown in Figure 2a,b. Over 800 identified proteins were related to cellular processes, and 565 identified proteins were associated to binding as a molecular function. With regard to total identified proteins, cellular processes, metabolic processes, biological regulation, and responses to stimuli formed the major subgroups, with percentages of 31.3%, 20.9%, 13%, and 8.9%, respectively, of the biological processes (Table S5). Concerning molecular function, binding and catalytic activity represented the major subgroups, with percentages of 44% and 38.3%, respectively (Table S6).
Figure 2.
Functional analysis and structural classification of identified proteins. (a) Subgroups of the biological process. (b) Subgroups of molecular function. The NIPs illustrated in the x-axis show the numbers of proteins involved in (a,b). (c) The list and number of protein classes in each ScS section and all batches. (d) Distribution and classification of all identified enzymes.
In addition to biological process and molecular function classification, the protein classes in the healthy SEP are shown in Figure 2c. The metabolite interconversion enzyme class was the most represented group, with 224 in B and less in the two other parts (177 in R and 188 in W). This was followed by protein-modifying enzymes, cytoskeletal proteins, and immunity proteins. In other protein classes, the numbers of proteins in the three batches were not particularly different (Figure 2c). Interestingly, when all identified proteins from the three batches were considered, enzymes formed almost the majority (41%) of the SEP, with 480 identified proteins. Furthermore, the numbers of these enzymes were distributed differently among various protein classes, for example metabolite interconversion enzyme class (292) could include protein modifying enzymes (150), translational proteins (14), transporters (9), nucleic acid metabolism (9), and protein-binding activity modulators (6). The distribution of the sub-families of all identified enzymes are shown in Figure 2d. The largest enzyme families consisted of hydrolases (47.5%) followed by oxidoreductases (22.1%) and transferases (16.7%). Isomerase and lyase were less represented in the healthy SEP. Inversely, when the unique proteins of each batch were considered, numbering 80 in W, 246 in B, and 223 in R (Figure 1c), the differences among subclasses of molecular function, biological processes, and protein classes were more distinctive in the three batches (Figure S2). Indeed, in biological process classification, the proportion of the response to stimulus proteins, signaling, and immune system processes was higher in W (Figure S2a). As for molecular function classification, the proteins with catalytic activity formed the highest percentage (55%) in W (Figure S2b). Nevertheless, the percentage of binding proteins in W (31.7%) was lower than in B or R. Additionally, the classification of unique proteins in each batch (Figure S2c) highlighted a major representation of metabolite interconversion enzymes in B, with 61, 33 and 14 proteins in B, R, and W, respectively. Moreover, nucleic acid metabolism proteins were preferentially found in R, with 20, 8, and 1 proteins in R, B, and W, respectively. Finally, unique proteins in the protein-binding activity modulator and transporter protein classes were not identified in W.
2.4. Signaling Pathways in the ScS-Extracted Proteome
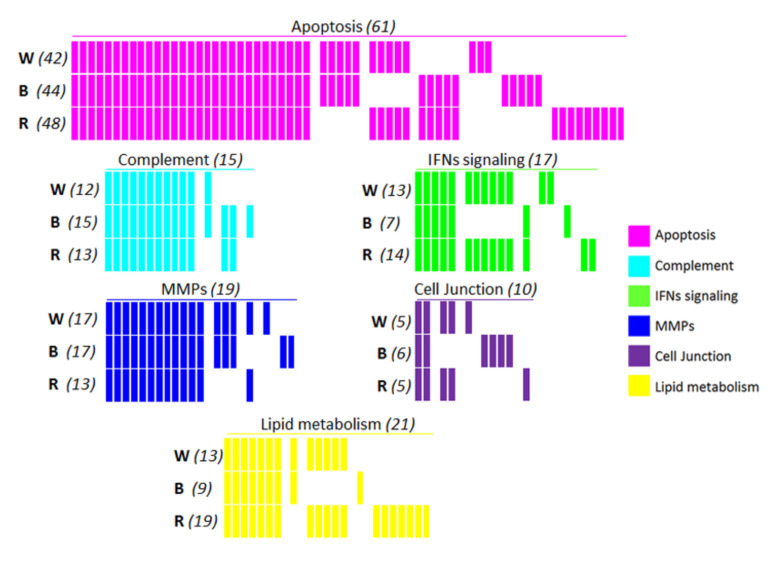
Six major signaling pathways, consisting of identified proteins in W, B, and R, are illustrated in Figure 3, as a barcode highlighting the overall distribution of molecular effectors for each batch. The corresponding proteins are listed in Table S7. Apoptosis was the most represented signaling pathway among these. Nearly half of the proteins involved in apoptosis (29) were common amongst the three batches. In apoptosis, 17 unique proteins, mostly related to proteasome subunits (PSM), were detected. These specific cellular effectors numbered 3 for W (PSMD5, BID, and TNFSF10), 5 for B (PSMB7, PSMD9, PSMF1, DFFA, and FNTA), and 9 for R (PSMC2, -4, and -6, PSMD1, -6, -12, PSME3, LMNB1, and STK24). In the complement cascade, the majority of the proteins were common among the three batches, representing both the alternative and classical pathways. Complement factor C1QB, a molecular effector of the classical pathway in response to antigen-antibody response, was only detected in B. Proteins involved in interferon (IFN) signaling pathways, such as Eukaryotic Translation Initiation Factors EIF4-A1, -A2, -A3, and -G1, were identified in both R and W, while EIF4E was specifically identified in R. Only one among the interferon-stimulating genes (ISGs), MX1, was specifically identified in the W batch. Among the 23 proteins involved in MMP pathways, 12 were common among the three batches, and only three unique proteins, including MMP-9, were in B. In the cell junction signaling pathway, only two proteins, nectin cell adhesion molecule 4 (PVRL4) and cadherin (CDH1), were common to the three batches, whereas more proteins were related to B, with four specifically identified, i.e., ACTN1, FLNA, PLEC, and VASP. Finally, the majority of the lipid metabolism-related proteins were identified in R, with 20 molecular effectors mostly represented by the phospholipase A2 (PLA2) family, including PLA2G2A, -G4B, -G4E, and -G4D, related to phospholipid metabolism. Only one enzyme, 3-ketodihydrosphingosine reductase (KDSR), related to de novo sphingolipid biosynthesis, was specifically identified in B. Likewise, a fatty acid metabolism effector, hydroxyacyl-CoA dehydrogenase (HADHA), was specifically detected in R.
Figure 3.
Barcodes of the major signaling pathways and NIPs in each ScS batch. The six signaling pathways were distributed as apoptosis, complement, interferons (IFNs) signaling, matrix metalloproteinases (MMPs), cell junction, and lipid metabolism in the whole strip (W), the bulb (B), and the rest (R). Each bar represents one protein. The total NIPs (W + B + R) are indicated between the bracket above each pathway barcode, and the corresponding NIPs for each batch are indicated between brackets on the left.
3. Discussion
The importance of the biological function of TF in the protection of the OS justifies the interest in deepening our knowledge of its composition and the role of its molecular effectors in reacting to changes in the OS [63]. In this study, nano-LC-MS/MS analysis was performed using nanoElute UHPLC coupled to a timsTOF Pro mass spectrometer, which enables tunable high performance for differentiating many isobaric and isomeric molecules present within biological systems [64]. The addition of ion mobility separation to the chromatographic-mass separation increases sensitivity and reduces spectral complexity in this MS [40]. The timsTOF Pro enables the analysis of samples with a minimal sample size less than 200 ng with a sequencing speed exceeding 100 Hz [39]. Tear samples can be collected using various methods [65]. However, the ScS approach remains the most common method due to its advantages over the other methods [14]. In order to use this powerful MS technology, we sought to optimize sample preparation. Thus, we investigated the various sections of the ScS, considering the whole strip (W) and its two parts, the bulb (B), and the rest of the ScS (R). The upper section (B), in contact with the conjunctiva, has the feature of cell adhesion lacking in the remaining part (R). The healthy SEP was investigated using different parts of the ScS with the timsTOF Pro to enrich the protein dataset while revealing the differences among protein profiles in W, B, and R. Indeed, processing B and R separately increased the total NIPs, even though the protein quantity of each batch (W, B, and R) was initially normalized before the MS/MS analysis. The composition of the protein content in W should be more varied and complex than in B and R, as it includes proteins from the totality of the strip. However, new and specific proteins for each piece of the strip were identified, when the strip was cut into two pieces. Thus, factors such as the ScS area and the mechanical action during the protein elution step seem to participate in the protein composition as well as the extraction yield. This suggests that the complexity of the protein composition and the pre-analytical phases of sample preparation greatly influence the nano-LC-MS/MS data analysis. As compared to other tear proteome studies [19,20,21], the large number of proteins identified in this study has increased our knowledge and contributed to enriching the tear proteome dataset to better decipher all the functions of tears. This might also have facilitated the detection of proteins due to their lower quantities in B and R. Eluting these two regions separately and pooling them before one-run MS/MS analysis could improve protein identification. Interestingly, as previously reported, enzymes represented the largest group of proteins in the SEP. The composition of enzymes in the healthy SEP revealed that hydrolases, oxidoreductases, and transferases formed the main enzyme families [21]. In our study, the number of identified enzyme families enriched the proteome dataset, where the percentage of identified hydrolases increased from 41% to 47.5% and the percentage of oxidoreductases increased from 15% to 22.1% compared to the results of this study. Nevertheless, the percentage of transferases decreased from 27% to 16.7% [21]. Likewise, another tear enzyme family, with antioxidant enzymes such as superoxide dismutases (SOD-1, -2, and -3), glutathione peroxidases (GPX-1, -3, and -4), catalase (CAT), and glutathione reductase (GSR), is responsible for the elimination of excessive amounts of reactive oxygen species [66]. In addition to the enrichment in tear enzyme identification, several calcium-binding proteins as Annexin A and S100A family members have been identified. These enzymes have a broad range of intracellular and extracellular regulatory functions encompassing the regulation of cellular apoptosis, energy metabolism, protein phosphorylation, and inflammation, among others [67]. Interestingly, the regulation of these proteins of interest is known to be associated with OSD [1]. The specificity of this study was derived from combining the timsTOF analysis with the elution of three different areas of the ScS, revealing major common proteins as well as unique proteins identified with a single batch. Additionally, although common proteins were identified between two or three batches, they differed by their mean of spectral counts. For instance, mucin-5AC, the major mucin secreted by the conjunctival goblet cells [55], was found in both the entire ScS (W) and the proximal section (B) of the ScS, which is in contact with the conjunctival mucosa, but was not found in the distal portion (R) of the ScS, which does not contain cells and is wetted only by capillary action. The additional recovery observed from the proximal portion (B) might be explained by the relationship between the molecular weight and the membrane pore size of the ScS, where a diameter of 20–25 µm allows large proteins such as mucin-5AC (641 kDa) to be preferentially retained [45]. Thus, migration to the remaining section of the ScS (R) might be hampered. This specific identification in the whole ScS and the proximal section (B), with different spectral means, was also observed for Mucin-5A and Mucin-5B. Therefore, the STT seems to be the most convenient collection method for tear film mucins, supported by the fact that the quantity of Mucin-5AC is higher in ScS than in basal tears or eye-flush tears [54].
The different relative abundance depending on sample preparation enables improvements in protein identification, either for comprehensive proteomic studies or for routine assays such as Western blotting and ELISA. For example, myeloperoxidase (MPO), involved in inflammation, host defense, and neutrophil function [68], was specifically found in B, but not in R. Thus, the sample preparation of B alone will probably be beneficial for the enrichment of the analytical tear sample. In the report of the Tear Film Subcommittee of the TFOS DEWS II, 102 extracellular and 20 intracellular proteins were referenced [1]. Of these 20 intracellular proteins, 12 of them were found in this study. The spectral means of these 12 proteins were higher in B than in R and W. This result logically suggests that B concentrates intracellular proteins. Likewise, among the 102 extracellular proteins, albumin, lactotransferrin, complement C3, lipocalin-1 enolase-1, and annexin A2 displayed a higher spectral count in B compared to in R. It could be hypothesized that these tear proteins accumulate in B and, a lesser extent, migrate to R via capillary action.
Interestingly, the mean spectral count of the most unique proteins in B and R was higher than that of the unique proteins in W. Indeed, the mean spectral count of 80 unique proteins in W was less than 2, while several unique proteins in B and R had mean spectral counts reaching 6.7 and 19.3, respectively. This finding could suggest that the unique proteins in B and R could be more informative compared to the unique proteins in W. Despite the widespread use of MS in proteomic studies, the identification of low-abundance proteins such as cytokines remains a challenge, and multiplex technology is still used for their quantification [69,70]. This is also supported by the fact that the concentrations of cytokines in tears are very low [57]. However, the resolutive specificity of the timsTOF Pro enables the identification of these molecules. Indeed, seven cytokines, consisting of interleukins and chemokines, were identified in the various batches. The specificities of identification were also reported according to the ScS sections, such as for IL-19 in W and IL-36γ, IL-36Ra, and IL-36α, which were identified only in R. CXCL17 and IL1-RN were identified in all three batches. IL-18 was shared by both W and B, and CXCL10 was shared by both W and R.
IL-1RN, IL-18, CXCL17, and CXC10 were previously detected in a few tear proteomic studies, while IL-36α, IL-36γ, and IL-36Ra were identified only in our study, as opposed to the four other studies [19,20,21,24]. IL-36,a new member of the IL-1 cytokine family, contains IL-36α, IL-36β, IL-36γ, and IL-36Ra and plays a crucial role in inflammation and immunity [71]. The modulation of IL-36α has been reported in Sjögren’s syndrome [62]. IL-36γ induces Mucin 5AC expression [72]. The role of IL-36 members in OSD is of great interest for further investigation. These results highlight the interest to study separately the various remaining sections of the ScS for exploring cytokine modulations in OSD.
The enrichment of the SEP dataset also allows the description of the multiple signaling pathways in healthy human tears. Six major signaling pathways have been reported for the OS, and proteins involved in these pathways have been determined. These pathways involve the following: (1) apoptosis; (2) complement; (3) IFN signaling; (4) MMPs; (5) cell junction; and (6) lipid metabolism. In apoptosis pathways, more proteins were identified than in other pathways. Apoptosis, as a response to physiological stimuli to maintain homeostatic mechanisms during cellular development, aging, and injuries, is highly significant in OS changes [73,74]. Proteasome 20S and 26S subunits (PSMA/B and C/D, respectively) and proteasome subunit activator 3 (PSME3) constitute almost half of the proteins from this pathway. Additionally, the well-described apoptotic enzymes such as CASP-3/7 [74] were specifically identified in W and B. The complement system is a crucial mediator of the innate immune response, and it prevents the spread of infection to other cells and tissues through opsonization, attracting immune cells or direct lysis, removing damaged cells/tissues and preventing the development of chronic inflammation [75]. Furthermore, the protein composition linked to the complement system differs according to the collection method and the size of the palpebral fissure. Thus, it has been reported that the expression levels of these proteins are higher in tear samples collected via the capillary method in comparison to by the STT [23]. Another study demonstrated the identification of C1Q, C3, CFB, C4, C5, and C9 in closed-eye tears and only the identification of C3, CFB, and C4 in open-eye and reflex tears [76]. The methodology used in this study enriched the dataset of complement cascades by adding C1R, C1QC, C6, C4BPA, C7, C8A, CFD, CFH, and CFI complement factors. The interferon-signaling pathway is also an important cellular response in OSD, which supports further investigations to understand the regulation of these molecular effectors [60,77,78,79]. Proteins identified in interferon signaling pathways, such as eukaryotic translation-initiation factors (EIF4A-1, -2, and -3, EIF4G1, and EIF4E), important regulators of mRNA translation [80], were identified in R and W, but not in B. Additionally, the human leukocyte antigen (HLA) class I proteins (HLA-A/B/C) could be identified in each region of the strip, except for HLA-E, which is unique to R. Regarding the extracellular matrix (ECM) and cell junction, two pathways were identified according to the identified proteins in each section of the ScS. The enzymes involved in the proteolysis of the ECM, such as MMPs, play a great role in wound healing and inflammation, and their increased activity has been reported in OSD [81]. As previously reported, MMP-9 in human corneal epithelial cells correlates positively with increasing osmolarity, which is mediated by the activation of c-Jun N-terminal kinases (JNKs) and stress-activated protein kinase (SAPK) pathways [82]. Furthermore, the MMP-9 test is a valuable diagnostic tool in DED [83]. The majority of identified protein components of the MMP pathway were shared by all batches. Interestingly, MMP-9 was identified only in B, but with a very low spectral mean, which could be explained by the healthy status of the donors. This finding should guide the choice of ScS preparation for further MMP-9 investigation. Additionally, collagen alpha chain proteins such as COL6A1, COL14A1, and COL9A3 were mostly found in all batches, except for COL9A3, which was not identified in B. A comparison with previous comprehensive proteomic studies [19,20,21,24] identified new collagen alpha chain components in tears—COL6A1 and COL9A3. These molecular effectors involved in the integrity of cellular membranes in the allergic response, infectious conditions, and other OSDs [84] could provide new insight for exploring potential targets of interest.
Finally, lipids are essential components in cellular functions, signaling networks, and energy metabolism [85]. The members of this molecular family are key mediators in intercellular and intracellular processes in both inflammatory processes and cell death phenomena [86,87]. Interestingly, these bioactive lipids and their modulation were also described in studies of an in vitro DED model [88,89]. This metabolic involvement is supported by the identification of several enzymes. The secretory calcium-dependent phospholipase A2 family, including PLA2-G4B, -G4D, and -G4E, was specifically identified in R. These enzymes are involved in the inflammatory response and host antimicrobial defense and play a role in the glycerophospholipid catabolic process/phospholipid metabolic process [90]. Likewise, several other lipid metabolism pathways were identified with tear enzymes, such as glycosphingolipid metabolism and fatty acid metabolism (HADHA). De novo sphingolipid biosynthesis pathways were also identified, based on KDSR, a specific enzyme identified only in R. Among the identified molecules involved in lipid metabolism, 12 of them were newly identified in our study as opposed to the other three comprehensive tear proteomic studies that we compared [19,20,21].
Lastly, analyzing tear and cellular proteins in the various sections of the ScS increased the NIPs as well as our knowledge of the SEP. Eluting these two portions before MS analysis provides additional protein characterization information compared to eluting the entire strip. High NIPs such as enzymes and other molecular effectors might help to enrich the investigation of signaling pathway regulation and cellular processes in OS homeostasis. This proteome profile could be also slightly enriched with the use of a larger cohort.
4. Materials and Methods
4.1. Sample Collection and Processing
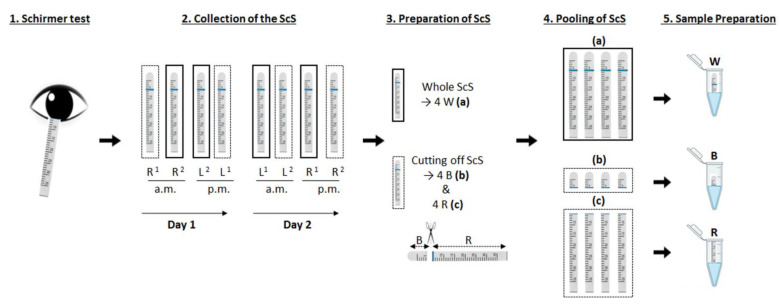
The study was performed according to the tenets of the Declaration of Helsinki and GCP, and written consent was obtained from all subjects after explaining the protocol and the scope of the study. The study was approved by the Ethics Committee CPP–Ile-de-France (number: 2018-A02800-55). Two healthy subjects (HSs) who participated in this study were volunteers. Two healthy volunteers were males with age 32 and 33, respectively. All ScS collected for this study reached the full length of wetness within 5 min of the sampling time. They did not wear contact lenses, used no systemic or ocular medication, had no past history of systemic or ophthalmic diseases known to alter tear production and had not undergone any ophthalmic surgeries. Both HSs in this study had to pass an ocular examination including tear break-up time (TBUT), corneal fluorescein staining (CFS), the evaluation of conjunctival hyperemia, and a STT to exclude the presence of any clinical pathology.
The bulb (B) of the ScS (Schirmer-plus®, Gecis; Neung Sur Beuvron, France) was gently inserted into the lower conjunctival cul-de-sac without local anesthesia, after which the subjects closed their eyes. The ScS were collected after 5 minutes, unless the entire strip was fully saturated with tears before. Sample collection from the 2 HSs and processing is illustrated in Figure 4. For each subject, ScS samples were collected from both eyes in the morning (a.m.) and afternoon (p.m.) for two consecutive days. Collected strips were placed in a sterile plastic microtube and immediately stored at −80 °C until the analytical procedure. To batch the samples, 8 wetted strips were separated into two batches. In the first batch (a), the 4 whole strips were pooled (W). In the second batch, the whole ScS was cut into two parts to provide the upper part containing the bulb (B) and the remaining part (R). This homogeneous sectioning provided two batches of samples, (b) and (c), with 4 bulbs and 4 rests, respectively. As for the W samples, the ScS were also pooled. Each pooled batch (a–c) was eluted in 1 mL of 100 mM ammonium bicarbonate (AmBic; Sigma-Aldrich, St. Louis, MO, USA) containing protease inhibitor cocktail (P2714; Sigma-Aldrich, St. Louis, MO, USA), placed on an IKA® VXR basic Vibrax® (IKA®-Werke GmbH & Co. KG, Staufen, Germany) orbital shaker at 1500 motions/minute at 4 °C for 4 h. The samples were centrifuged at 14,000 rpm at 4 °C for 5 min, and the supernatants were collected. Protein quantification assay was carried out using a BCA Protein Assay Kit (Thermo Scientific, Pierce, IL, USA) and using Spark® Multimode Microplate Reader (Tekan, Männedorf, Switzerland).The samples were stored at −80 °C until MS analysis.
Figure 4.
Illustrations of tear sample collection with ScS and sample processing. Eight ScS were collected from two HSs without anesthesia in 4 different sampling periods over 2 days. Two equivalent groups of the ScS were obtained by pooling them. In the first group, whole strips (W) were used; in the second group, the strips were cut to separate the bulbs (B) and the rest of the strips (R). The 4 pooled W strips (a), 4 B strips (b), and 4 R strips (c) were extracted in ammonium bicarbonate to prepare an analytical sample for protein quantification and MS analysis. R, right eye; L, left eye; a.m., in the morning; p.m., in the afternoon; 1, healthy subject-1; 2, healthy subject-2; ScS, Schirmer strips; W, the whole strip; B, the bulb; R, the rest of the strip.
4.2. Sample Preparation for Nano-LC-MS/MS Analysis
Sample preparation was performed on 10 µg of proteins for each pooled sample, adjusted to the same volume and same protein concentration. Then, dithiothreitol (#3483-12-3; Sigma-Aldrich, St. Louis, MO, USA) was added to the samples up to 5 mM as a final concentration to reduce the proteins in a 37 °C water bath for 30 min. Subsequently, alkylation was performed by adding iodoacetamide (#144-48-9; Sigma-Aldrich, St. Louis, MO, USA) up to 15 mM for 30 min in the dark and at room temperature. The samples were then subjected to two-step in-solution digestion, first by adding 200 ng of Lys-C protease (#90307; Thermo Fisher Scientific) (protease/protein mass ratio equal to 1:50) and incubating the samples for 120 min at 37 °C and then by adding 200 ng of trypsin (#90058; Thermo Fisher Scientific; protease/protein mass ratio equal to 1:50) for overnight incubation at 37 °C. Following the digestion, the automated StageTips desalting of the peptides was performed using DigestProMSi (Intavis, Cologne, Germany). The eluted peptides were dried using a vacuum centrifuge and resuspended in 0.1% formic acid (#F0507; Sigma-Aldrich, St. Louis, MO, USA) and 2% acetonitrile (#271004; Sigma-Aldrich, St. Louis, MO, USA). For each sample, 500 ng of peptides were injected in triplicate into a nanoElute UHPLC spectrometer (Bruker, Champs-sur-Marne, France) coupled to a timsTOF Pro mass spectrometer (Bruker, Champs-sur-Marne, France). The peptides were directly loaded and separated on an Aurora2 RP-C18 analytical column (25 cm, 75 μm i.d., 120 Å, 1.6 µm; IonOpticks, Fitzroy, VIC Australia) at a flow rate of 400 nL/min at 40 °C. Mobile phase A consisted of 2% acetonitrile and 0.1% formic acid, and mobile phase B consisted of 99.9% acetonitrile containing 0.1% formic acid. A 90 min elution gradient was performed by increasing mobile phase B from 0% B to 3% over 1 min, from 3% B to 15% over 56 min, then from 15% B to 23% over 21 min, and from 23% B to 32% over 13 min. MS acquisition was run in data-dependent acquisition (DDA) mode with PASEF. The accumulation time was set to 100 ms in the TIMS tunnel. The capillary voltage was set to 1.6 kV, the mass ranged from 100 to 1700 m/z in MS and MS/MS, and the mobility ranged from 0.6 to 1.6 cm2/(Vs). Dynamic exclusion was activated for ions within a mass of 0.015 m/z and a mobility of 0.015 cm2/(Vs) and released after 0.4 min. Exclusion criteria was defined as 4 times higher ion intensity of a precursor. Low-abundance precursors below the target value of 20,000 arbitrary units (a.u.) and intensity of 2500 a.u. were selected several times for PASEF-MS/MS, until the target value was reached. Parent ion selection was achieved by using a two-dimensional m/z and 1/k0 selection area filter, allowing the exclusion of singly charged ions. The total cycle time was 1.1 s with 10 PASEF cycles.
4.3. Nano-LC-MS/MS Data Analysis
Raw MS/MS data were analyzed with Bruker Compass Data Analysis (version 5.1) and further processed with MaxQuant software (version 1.6.8.0) for protein identification [91]. The search parameters were set as follows: mass tolerances of 25 ppm for MS1 and 40 ppm for MS2, carbamidomethyl as fixed modification, acetyl (protein N-terminal), and oxidation (on M residue) as variable modifications; up to 2 missed cleavages were allowed. Protein identifications were performed against the Uniprot Homo sapiens database. The Uniprot homo sapiens database version used was UP000005640_9606 in 2019 (https://www.uniprot.org/, accessed on 2 August 2019) with 20,672 entries on 2 August 2019, while the p-values of peptides and proteins were adjusted to obtain the corresponding FDRs of <1%, with a minimum of 1 peptide per protein. The semi-quantitative estimations of the protein content of the samples were performed, taking into account the numbers of matched spectra per protein. The spectral counting method was used for the comparison. This method is defined as the total number of spectra identified for a protein, and it has been accepted as a practical, label-free, semi-quantitative measure of protein abundance in proteomics studies [92].
To identify new tear proteins using a timsTOF Pro in this study, a non-supervised comparison analysis was conducted with four recent studies using advanced MS technologies, i.e., Kandhavelu, et al. [20], (B) Dor, et al. [21], Aass, et al. [20], and Ponzini, et al. [25] (Figure S1). These three studies analyzing tears from HSs were selected for their high numbers of identified proteins.
4.4. Functional Annotation and Signaling Pathway Network Analysis
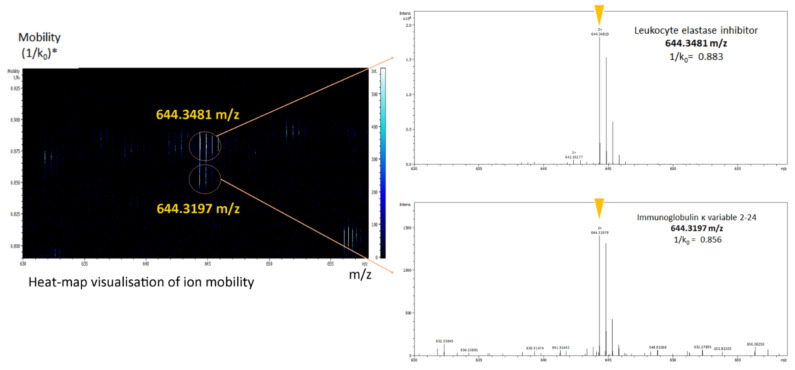
All identified proteins from the three groups (W, B, and R) were examined by comparing gene ontology (GO) terms. The proportions of the associated proteins in each group were obtained from the PANTHER Classification System (http://www.pantherdb.org/, accessed on 3 May 2021) [93]. Among 1582 identified proteins, Panther software could classify 1562 proteins according to their functional classification. The Reactome database was used for signaling pathway analysis (https://reactome.org/, accessed on 16 March 2021) [94]. As an example of the powerful abilities of this new-generation MS device, the timsTOF Pro, to separate different molecules with the same mass-to-charge ratio (m/z), Figure 5 presents how the peptide characterization was assessed by sequencing two of the targeted peptides possessing almost the same mass—leukocyte elastase inhibitor and immunoglobulin kappa variable 2-24 (644.3438 and 644.3144 m/z for HNSSGSILFLGR and FSGSGAGTDFTLK, respectively). These proteins were co-eluted at the same retention time and with a very similar m/z, but the peptides were differentiated by the timsTOF Pro, thanks to its third dimension ion-mobility feature.
Figure 5.
Heat-map visualization of ion mobility. The peptide ions of two different proteins with almost the same mass-to-charge ratio (m/z) at a single time point were differentiated by the timsTOF Pro analysis. The circled lines indicate the m/z and mobility positions of two precursor ions selected for fragmentation by parallel accumulation-serial fragmentation (PASEF). *; (1/k0), the drift time.
5. Conclusions
By merging three samples corresponding to different sections of the ScS, 1582 proteins were identified in this study. Enzymes constituted the largest protein group among the identified proteins. Investigating two portions of the ScS separately enabled the identification of a number of additional proteins. Eluting sections B and R separately before analysis with this robust, highly sensitive MS device, the timsTOF Pro, might offer a holistic view of the main proteins present on the OS, from both cellular and soluble components, akin to combining a conjunctival imprint and a tear sample. The timsTOF Pro might bring a new dimension to the profiling of the tear proteome in OSD due to its unique sensitivity, enabling in-depth proteomic analysis from a small tear sample. The dataset created will contribute to the tear proteome as well as to the description and comparison of multiple signaling pathways associated with the OS.
This study may be the basis for new extensive studies on ScS specimens collected from both HSs and patients with various OSD. Collecting tear specimens with the ScS and eluting proteins from B and R separately before MS analysis might be the best method for future clinical studies of the OS proteome.
Acknowledgments
The authors thank Kevin Clark, for editing advice.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/metabo12010002/s1. Table S1: Raw data of all identified proteins provided by the ScS extract of the 3 batches (W + B + R); Table S2: List of identified proteins with the highest mean of the spectral count in the whole strip (W); Table S3: List of identified proteins with the highest mean of the spectral count in the ScS bulb (B); Table S4: List of identified proteins with the highest mean of the spectral count in the rest (R) of the ScS; Table S5: Biological processes associated with identified proteins in the whole strip (W), bulb (B), rest of the strip (R), and all identified proteins (W + B + R); Table S6: Molecular functions of identified proteins in the whole strip (W), bulb (B), rest of the strip (R), and all identified proteins; Table S7: List of proteins identified in the 6 chosen signaling pathways. Figure S1: Venn diagram comparing other four comprehensive proteomics studies; Figure S2: Functional analysis and structural classification of unique proteins in each batch. (a) Subgroups of biological processes. (b) Subgroups of molecular function. (c) The list and number of protein classes in each ScS section.
Author Contributions
Conceptualization: K.K., F.B.-B., C.B. and M.A.A.; methodology, K.K., M.A.A., F.B.-B., C.B., C.P., S.C., I.K. and R.M.; validation, K.K., M.A.A., F.B.-B., C.B., R.M., C.P., S.C., and I.K.; formal analysis, K.K., M.A.A., F.B.-B., C.B., R.M., I.K., S.C. and C.P., writing—original draft preparation, K.K., M.A.A., F.B.-B., C.B., R.M., C.P., S.C. and R.M.; writing—review and editing, K.K., M.A.A., F.B.-B., C.B., R.M., C.P., S.C., R.M. and I.K.; visualization, K.K., M.A.A., F.B.-B., C.B., R.M., S.C., C.P., R.M. and I.K.; supervision, K.K., M.A.A., F.B.-B. and C.B.. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Sorbonne Université and the Institut National de la Santé et de la Recherche Médicale, ANR, LabEx LIFESENSES (ANR-10-LABX-65), and IHU FOReSIGHT (ANR-18-IAHU-01). M.A.A. was funded by a H2020-MSCA-ETN program (IT-DED3; grant agreement: 765608).
Institutional Review Board Statement
The study was approved by the Ethics Committee CPP–Ile-de-France (number: 2018-A02800-55).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study according to the Ethics Committee CPP–Ile-de-France (number: 2018-A02800-55).
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030334 (accessed on 12 December 2021).
Conflicts of Interest
C.B. is a consultant for Aerie, Alcon, Allergan, Horus Pharma, Santen, Théa; C.B. and F.B.-B. have received research grants from Horus Pharma, Santen, and Théa.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Willcox M.D.P.P., Argüeso P., Georgiev G.A., Holopainen J.M., Laurie G.W., Millar T.J., Papas E.B., Rolland J.P., Schmidt T.A., Stahl U., et al. TFOS DEWS II Tear Film Report. Ocul. Surf. 2017;15:366–403. doi: 10.1016/j.jtos.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopacz D., Niezgoda Ł., Fudalej E., Nowak A., Maciejewicz P. Ocular Surface Diseases—Some Current Date on Tear Film Problem and Keratoconic Diagnosis. Vol. 32. IntechOpen; Warsaw, Poland: 2021. Tear Film—Physiology and Disturbances in Various Diseases and Disorders; pp. 137–144. [DOI] [Google Scholar]
- 3.Craig J.P., Nelson J.D., Azar D.T., Belmonte C., Bron A.J., Chauhan S.K., de Paiva C.S., Gomes J.A.P., Hammitt K.M., Jones L., et al. TFOS DEWS II Report Executive Summary. Ocul. Surf. 2017;15:802–812. doi: 10.1016/j.jtos.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Cwiklik L. Tear Film Lipid Layer: A Molecular Level View. Biochim. Biophys. Acta Biomembr. 2016;1858:2421–2430. doi: 10.1016/j.bbamem.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Versura P., Campos E.C. Disease Update on Human Tear Proteome. Eur. Ophthalmic Rev. 2013;07:36. doi: 10.17925/EOR.2013.07.01.36. [DOI] [Google Scholar]
- 6.Azkargorta M., Soria J., Acera A., Iloro I., Elortza F. Human Tear Proteomics and Peptidomics in Ophthalmology: Toward the Translation of Proteomic Biomarkers into Clinical Practice. J. Proteom. 2017;150:359–367. doi: 10.1016/j.jprot.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Pinto-Fraga J., Enríquez-de-Salamanca A., Calonge M., González-García M.J., López-Miguel A., López-de la Rosa A., García-Vázquez C., Calder V., Stern M.E., Fernández I. Severity, Therapeutic, and Activity Tear Biomarkers in Dry Eye Disease: An Analysis from a Phase III Clinical Trial. Ocul. Surf. 2018;16:368–376. doi: 10.1016/j.jtos.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Enríquez-De-Salamanca A., Bonini S., Calonge M. Molecular and Cellular Biomarkers in Dry Eye Disease and Ocular Allergy. Curr. Opin. Allergy Clin. Immunol. 2012;12:523–533. doi: 10.1097/ACI.0b013e328357b488. [DOI] [PubMed] [Google Scholar]
- 9.Kuo M.-T., Fang P.-C., Chao T.-L., Chen A., Lai Y.-H., Huang Y.-T., Tseng C.-Y. Tear Proteomics Approach to Monitoring Sjögren Syndrome or Dry Eye Disease. Int. J. Mol. Sci. 2019;20:1932. doi: 10.3390/ijms20081932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieczyński J., Szulc U., Harazna J., Szulc A., Kiewisz J. Tear Fluid Collection Methods: Review of Current Techniques. Eur. J. Ophthalmol. 2021;31:2245–2251. doi: 10.1177/1120672121998922. [DOI] [PubMed] [Google Scholar]
- 11.Saraygord-Afshari N., Naderi-Manesh H., Naderi M. Increasing Proteome Coverage for Gel-Based Human Tear Proteome Maps: Towards a More Comprehensive Profiling. Biomed. Chromatogr. 2015;29:1056–1067. doi: 10.1002/bmc.3392. [DOI] [PubMed] [Google Scholar]
- 12.Brott N.R., Ronquillo Y. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2021. Schirmer test. [PubMed] [Google Scholar]
- 13.Kim Y.H., Graham A.D., Li W., Radke C.J., Lin M.C. Human Lacrimal Production Rate and Wetted Length of Modified Schirmer’s Tear Test Strips. Transl. Vis. Sci. Technol. 2019;8:40. doi: 10.1167/tvst.8.3.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posa A., Bräuer L., Schicht M., Garreis F., Beileke S., Paulsen F. Schirmer Strip vs. Capillary Tube Method: Non-Invasive Methods of Obtaining Proteins from Tear Fluid. Ann. Anat. Anat. Anz. 2013;195:137–142. doi: 10.1016/j.aanat.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Green-Church K.B., Nichols K.K., Kleinholz N.M., Zhang L., Nichols J.J. Investigation of the Human Tear Film Proteome Using Multiple Proteomic Approaches. Mol. Vis. 2008;14:456–470. [PMC free article] [PubMed] [Google Scholar]
- 16.You J., Willcox M.D., Madigan M.C., Wasinger V., Schiller B., Walsh B.J., Graham P.H., Kearsley J.H., Li Y. Advances in Clinical Chemistry. Volume 62. Elsevier Inc.; Amsterdam, The Netherlands: 2013. Tear Fluid Protein Biomarkers; pp. 151–196. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z., Du C.X., Pan X.D. The Use of In-Strip Digestion for Fast Proteomic Analysis on Tear Fluid from Dry Eye Patients. PLoS ONE. 2018;13:e0200702. doi: 10.1371/journal.pone.0200702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L., Zhao S.Z., Koh S.K., Chen L., Vaz C., Tanavde V., Li X.R., Beuerman R.W. In-Depth Analysis of the Human Tear Proteome. J. Proteom. 2012;75:3877–3885. doi: 10.1016/j.jprot.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 19.Aass C., Norheim I., Eriksen E.F., Thorsby P.M., Pepaj M. Single Unit Filter-Aided Method for Fast Proteomic Analysis of Tear Fluid. Anal. Biochem. 2015;480:1–5. doi: 10.1016/j.ab.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Kandhavelu J., Demonte N.L., Namperumalsamy V.P., Prajna L., Thangavel C., Jayapal J.M., Kuppamuthu D. Aspergillus Flavus Induced Alterations in Tear Protein Profile Reveal Pathogen-Induced Host Response to Fungal Infection. J. Proteom. 2017;152:13–21. doi: 10.1016/j.jprot.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Dor M., Eperon S., Lalive P.H., Guex-Crosier Y., Hamedani M., Salvisberg C., Turck N. Investigation of the Global Protein Content from Healthy Human Tears. Exp. Eye Res. 2019;179:64–74. doi: 10.1016/j.exer.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 22.de Souza G.A., Godoy L.M.F., Mann M. Identification of 491 Proteins in the Tear Fluid Proteome Reveals a Large Number of Proteases and Protease Inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nättinen J., Aapola U., Jylhä A., Vaajanen A., Uusitalo H. Comparison of Capillary and Schirmer Strip Tear Fluid Sampling Methods Using SWATH-MS Proteomics Approach. Transl. Vis. Sci. Technol. 2020;9:16. doi: 10.1167/tvst.9.3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H., Yang X., Wang Y., Zheng X., Zhang Y., Shao Y. Comparative Analysis of the Tear Protein Profile in Herpes Simplex Virus Type 1 Epithelial Keratitis. BMC Ophthalmol. 2020;20:355. doi: 10.1186/s12886-020-01626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponzini E., Ami D., Duse A., Santambrogio C., De Palma A., Di Silvestre D., Mauri P., Pezzoli F., Natalello A., Tavazzi S., et al. Single-tear proteomics: A feasible approach to precision medicine. Int. J. Mol. Sci. 2021;22:10750. doi: 10.3390/ijms221910750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zysset-Burri D.C., Schlegel I., Lincke J.B., Jaggi D., Keller I., Heller M., Lagache S.B., Wolf S., Zinkernagel M.S. Understanding the interactions between the ocular surface microbiome and the tear proteome. Investig. Ophthalmol. Vis. Sci. 2021;62:8. doi: 10.1167/iovs.62.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y.N., Di D., Yuan Z.C., Wu L., Hu B. Schirmer Paper Noninvasive Microsampling for Direct Mass Spectrometry Analysis of Human Tears. Anal. Chem. 2020;92:6207–6212. doi: 10.1021/acs.analchem.9b05078. [DOI] [PubMed] [Google Scholar]
- 28.Nättinen J., Jylhä A., Aapola U., Mäkinen P., Beuerman R., Pietilä J., Vaajanen A., Uusitalo H. Age-Associated Changes in Human Tear Proteome. Clin. Proteom. 2019;16:11. doi: 10.1186/s12014-019-9233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L., Beuerman R.W., Choi M.C., Shao Z.Z., Xiao R.L., Yang H., Tong L., Liu S., Stern M.E., Tan D. Identification of Tear Fluid Biomarkers in Dry Eye Syndrome Using ITRAQ Quantitative Proteomics. J. Proteome Res. 2009;8:4889–4905. doi: 10.1021/pr900686s. [DOI] [PubMed] [Google Scholar]
- 30.Nättinen J., Jylhä A., Aapola U., Parkkari M., Mikhailova A., Beuerman R.W., Uusitalo H. Patient Stratification in Clinical Glaucoma Trials Using the Individual Tear Proteome. Sci. Rep. 2018;8:12038. doi: 10.1038/s41598-018-30369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grus F.H., Podust V.N., Bruns K., Lackner K., Fu S., Dalmasso E.A., Wirthlin A., Pfeiffer N. SELDI-TOF-MS ProteinChip Array Profiling of Tears from Patients with Dry Eye. Investig. Ophthalmol. Vis. Sci. 2005;46:863–876. doi: 10.1167/iovs.04-0448. [DOI] [PubMed] [Google Scholar]
- 32.Li N., Wang N., Zheng J., Liu X.M., Lever O.W., Erickson P.M., Li L. Characterization of Human Tear Proteome Using Multiple Proteomic Analysis Techniques. J. Proteome Res. 2005;4:2052–2061. doi: 10.1021/pr0501970. [DOI] [PubMed] [Google Scholar]
- 33.Nättinen J., Jylhä A., Aapola U., Enríquez-de-Salamanca A., Pinto-Fraga J., López-Miguel A., González-García M.J., Stern M.E., Calonge M., Zhou L., et al. Topical Fluorometholone Treatment and Desiccating Stress Change Inflammatory Protein Expression in Tears. Ocul. Surf. 2018;16:84–92. doi: 10.1016/j.jtos.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Tong L., Zhou L., Beuerman R.W., Zhao S.Z., Li X.R. Association of Tear Proteins with Meibomian Gland Disease and Dry Eye Symptoms. Br. J. Ophthalmol. 2011;95:848–852. doi: 10.1136/bjo.2010.185256. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh E.J., Bereman M.S., Durand S., Valaskovic G.A., MacCoss M.J. Effects of Column and Gradient Lengths on Peak Capacity and Peptide Identification in Nanoflow LC-MS/MS of Complex Proteomic Samples. J. Am. Soc. Mass Spectrom. 2013;24:148–153. doi: 10.1007/s13361-012-0508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J.Y.W., Sze Y.H., Bian J.F., Lam T.C. Critical Role of Mass Spectrometry Proteomics in Tear Biomarker Discovery for Multifactorial Ocular Diseases (Review) Int. J. Mol. Med. 2021;47:83. doi: 10.3892/ijmm.2021.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaspari M., Cuda G. Nano LC–MS/MS: A robust setup for proteomic analysis. In: Toms S.A., Weil R.J., editors. Methods in Molecular Biology. Vol. 790. Humana Press; Totowa, NJ, USA: 2011. pp. 115–126. [DOI] [PubMed] [Google Scholar]
- 38.Macron C., Lavigne R., Núñez A.G., Affolter M., Pineau C., Dayon L. Exploration of Human Cerebrospinal Fluid: A Large Proteome Dataset Revealed by Trapped Ion Mobility Time-of-Flight Mass Spectrometry. Data Br. 2020;31:105704. doi: 10.1016/j.dib.2020.105704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier F., Brunner A.D., Koch S., Koch H., Lubeck M., Krause M., Goedecke N., Decker J., Kosinski T., Park M.A., et al. Online Parallel Accumulation–Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteom. 2018;17:2534–2545. doi: 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier F., Brunner A.D., Frank M., Ha A., Bludau I., Voytik E., Kaspar-Schoenefeld S., Lubeck M., Raether O., Bache N., et al. DiaPASEF: Parallel Accumulation–Serial Fragmentation Combined with Data-Independent Acquisition. Nat. Methods. 2020;17:1229–1236. doi: 10.1038/s41592-020-00998-0. [DOI] [PubMed] [Google Scholar]
- 41.Hamada S., Pionneau C., Parizot C., Silvie O., Chardonnet S., Marinach C. In-depth Proteomic Analysis of Plasmodium Berghei Sporozoites Using Trapped Ion Mobility Spectrometry with Parallel Accumulation-serial Fragmentation. Proteomics. 2021;21:2000305. doi: 10.1002/pmic.202000305. [DOI] [PubMed] [Google Scholar]
- 42.Yates J.R. Recent Technical Advances in Proteomics. F1000Research. 2019;8:1–8. doi: 10.12688/f1000research.16987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasilopoulou C.G., Sulek K., Brunner A.-D., Meitei N.S., Schweiger-Hufnagel U., Meyer S.W., Barsch A., Mann M., Meier F. Trapped Ion Mobility Spectrometry and PASEF Enable In-Depth Lipidomics from Minimal Sample Amounts. Nat. Commun. 2020;11:331. doi: 10.1038/s41467-019-14044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong L., Zhou X.Y., Jylha A., Aapola U., Liu D.N., Koh S.K., Tian D., Quah J., Uusitalo H., Beuerman R.W., et al. Quantitation of 47 Human Tear Proteins Using High Resolution Multiple Reaction Monitoring (HR-MRM) Based-Mass Spectrometry. J. Proteom. 2015;115:36–48. doi: 10.1016/j.jprot.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Denisin A.K., Karns K., Herr A.E. Post-Collection Processing of Schirmer Strip-Collected Human Tear Fluid Impacts Protein Content. Analyst. 2012;137:5088–5096. doi: 10.1039/c2an35821b. [DOI] [PubMed] [Google Scholar]
- 46.Zhou L., Wei R., Zhao P., Koh S.K., Beuerman R.W., Ding C. Proteomic Analysis Revealed the Altered Tear Protein Profile in a Rabbit Model of Sjögren’s Syndrome-Associated Dry Eye. Proteomics. 2013;13:2469–2481. doi: 10.1002/pmic.201200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung J.H., Ji Y.W., Hwang H.S., Oh J.W., Kim H.C., Lee H.K., Kim K.P. Proteomic Analysis of Human Lacrimal and Tear Fluid in Dry Eye Disease. Sci. Rep. 2017;7:13363. doi: 10.1038/s41598-017-13817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erkelens M.N., Mebius R.E. Retinoic Acid and Immune Homeostasis: A Balancing Act. Trends Immunol. 2017;38:168–180. doi: 10.1016/j.it.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Liang H., Kessal K., Rabut G., Daull P., Garrigue J.S., Parsadaniantz S.M., Docquier M., Baudouin C., Brignole-Baudouin F. Correlation of Clinical Symptoms and Signs with Conjunctival Gene Expression in Primary Sjögren Syndrome Dry Eye Patients. Ocul. Surf. 2019;17:516–525. doi: 10.1016/j.jtos.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Gottenberg J.E., Cagnard N., Lucchesi C., Letourneur F., Mistou S., Lazure T., Jacques S., Ba N., Ittah M., Lepajolec C., et al. Activation of IFN Pathways and Plasmacytoid Dendritic Cell Recruitment in Target Organs of Primary Sjögren’s Syndrome. Proc. Natl. Acad. Sci. USA. 2006;103:2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murgoci A.N., Cardon T., Aboulouard S., Duhamel M., Fournier I., Cizkova D., Salzet M. Reference and Ghost Proteins Identification in Rat C6 Glioma Extracellular Vesicles. iScience. 2020;23:101045. doi: 10.1016/j.isci.2020.101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirai N., Kawasaki S., Tanioka H., Connon C.J., Yamasaki K., Yokoi N., Komuro A., Kinoshita S. Pathological Keratinisation in the Conjunctival Epithelium of Sjögren’s Syndrome. Exp. Eye Res. 2006;82:371–378. doi: 10.1016/j.exer.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Hernández-Ruiz M., Zlotnik A., Llorente L., Hernandez-Molina G. Markedly High Salivary and Lacrimal CXCL17 Levels in Primary Sjögren’s Syndrome. Jt. Bone Spine. 2018;85:379–380. doi: 10.1016/j.jbspin.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 54.Ablamowicz A.F., Nichols J.J. Concentrations of MUC16 and MUC5AC Using Three Tear Collection Methods. Mol. Vis. 2017;23:529–537. [PMC free article] [PubMed] [Google Scholar]
- 55.Gipson I.K., Argüeso P. Role of Mucins in the Function of the Corneal and Conjunctival Epithelia. Int. Rev. Cytol. 2003;231:1–49. doi: 10.1016/S0074-7696(03)31001-0. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., Deng F., Zheng J., Yin J., Huang R., Liu W., Lin Q., Gao Y., Gao X., Yu X., et al. High Circulating Level of Interleukin-18 in Patients with Primary Sjögren’s Syndrome Is Associated with Disease Activity. Mod. Rheumatol. 2016;26:156–158. doi: 10.3109/14397595.2015.1021409. [DOI] [PubMed] [Google Scholar]
- 57.Yoon K.C., Park C.S., You I.C., Choi H.J., Lee K.H., Im S.K., Park H.Y., Pflugfelder S.C. Expression of CXCL9, -10, -11, and CXCR3 in the Tear Film and Ocular Surface of Patients with Dry Eye Syndrome. Investig. Ophthalmol. Vis. Sci. 2010;51:643–650. doi: 10.1167/iovs.09-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuzawa-Carballeda J., Sánchez-Guerrero J., Betanzos J.L., Enriquez A.B., Avila-Casado C., Llorente L., Hernández-Molina G. Differential Cytokine Expression and Regulatory Cells in Patients with Primary and Secondary Sjögren’s Syndrome. Scand. J. Immunol. 2014;80:432–440. doi: 10.1111/sji.12224. [DOI] [PubMed] [Google Scholar]
- 59.Daull P., Barabino S., Feraille L., Kessal K., Docquier M., Parsadaniantz S.M., Baudouin C., Garrigue J.S. Modulation of Inflammation-Related Genes in the Cornea of a Mouse Model of Dry Eye upon Treatment with Cyclosporine Eye Drops. Curr. Eye Res. 2019;44:476–485. doi: 10.1080/02713683.2018.1563197. [DOI] [PubMed] [Google Scholar]
- 60.Tsubota K., Pflugfelder S.C., Liu Z., Baudouin C., Kim H.M., Messmer E.M., Kruse F., Liang L., Carreno-Galeano J.T., Rolando M., et al. Defining Dry Eye from a Clinical Perspective. Int. J. Mol. Sci. 2020;21:9271. doi: 10.3390/ijms21239271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomes S., Marques P.I., Matthiesen R., Seixas S. Adaptive Evolution and Divergence of SERPINB3: A Young Duplicate in Great Apes. PLoS ONE. 2014;9:e104935. doi: 10.1371/journal.pone.0104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciccia F., Accardo-Palumbo A., Alessandro R., Alessandri C., Priori R., Guggino G., Raimondo S., Carubbi F., Valesini G., Giacomelli R., et al. Interleukin-36α Axis Is Modulated in Patients with Primary Sjögren’s Syndrome. Clin. Exp. Immunol. 2015;181:230–238. doi: 10.1111/cei.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pflugfelder S.C., Stern M.E. Biological Functions of Tear Film. Exp. Eye Res. 2020;197:108115. doi: 10.1016/j.exer.2020.108115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spraggins J., Djambazova K., Rivera E., Migas L., Neumann E., Fuetterer A., Suetering J., Goedecke N., Ly A., Van De Plas R., et al. High Performance Molecular Imaging with MALDI Trapped Ion Mobility Time-of-Flight (TimsTOF) Mass Spectrometry. Anal. Chem. 2019;91:14552–14560. doi: 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rentka A., Koroskenyi K., Harsfalvi J., Szekanecz Z., Szucs G., Szodoray P., Kemeny-Beke A. Evaluation of Commonly Used Tear Sampling Methods and Their Relevance in Subsequent Biochemical Analysis. Ann. Clin. Biochem. 2017;54:521–529. doi: 10.1177/0004563217695843. [DOI] [PubMed] [Google Scholar]
- 66.Cecerska-Heryć E., Surowska O., Heryć R., Serwin N., Napiontek-Balińska S., Dołęgowska B. Are Antioxidant Enzymes Essential Markers in the Diagnosis and Monitoring of Cancer Patients—A Review. Clin. Biochem. 2021;93:1–8. doi: 10.1016/j.clinbiochem.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Marenholz I., Heizmann C.W., Fritz G. S100 Proteins in Mouse and Man: From Evolution to Function and Pathology (Including an Update of the Nomenclature) Biochem. Biophys. Res. Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 68.Aratani Y. Myeloperoxidase: Its Role for Host Defense, Inflammation, and Neutrophil Function. Arch. Biochem. Biophys. 2018;640:47–52. doi: 10.1016/j.abb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Stenken J.A., Poschenrieder A.J. Bioanalytical Chemistry of Cytokines—A Review. Anal. Chim. Acta. 2015;853:95–115. doi: 10.1016/j.aca.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou L., Beuerman R.W. Tear Analysis in Ocular Surface Diseases. Prog. Retin. Eye Res. 2012;31:527–550. doi: 10.1016/j.preteyeres.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 71.Chen W., Yu X., Yuan X.-R., Chen B., Cai N., Zeng S., Sun Y., Li H. The Role of IL-36 in the Pathophysiological Processes of Autoimmune Diseases. Front. Pharmacol. 2021;12:2643. doi: 10.3389/fphar.2021.727956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bae C.H., Choi Y.S., Na H.G., Song S.Y., Kim Y.D. Interleukin (IL) 36 Gamma Induces Mucin 5AC, Oligomeric Mucus/Gel-Forming Expression via IL-36 Receptor–Extracellular Signal Regulated Kinase 1 and 2, and P38–Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells in Human Airway Epithelial Cells. Am. J. Rhinol. Allergy. 2018;32:87–93. doi: 10.1177/1945892418762844. [DOI] [PubMed] [Google Scholar]
- 73.Yeh S., Song X.J., Farley W., Li D.-Q., Stern M.E., Pflugfelder S.C. Apoptosis of Ocular Surface Cells in Experimentally Induced Dry Eye. Investig. Ophthalmol. Vis. Sci. 2003;44:124–129. doi: 10.1167/iovs.02-0581. [DOI] [PubMed] [Google Scholar]
- 74.McComb S., Chan P.K., Guinot A., Hartmannsdottir H., Jenni S., Dobay M.P., Bourquin J.P., Bornhauser B.C. Efficient Apoptosis Requires Feedback Amplification of Upstream Apoptotic Signals by Effector Caspase-3 or -7. Sci. Adv. 2019;5:eaau9433. doi: 10.1126/sciadv.aau9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cedzyński M., Thielens N.M., Mollnes T.E., Vorup-Jensen T. Editorial: The Role of Complement in Health and Disease. Front. Immunol. 2019;10:1869. doi: 10.3389/fimmu.2019.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willcox M.D., Morris C.A., Thakur A., Sack R.A., Wickson J., Boey W. Complement and Complement Regulatory Proteins in Human Tears. Investig. Ophthalmol. Vis. Sci. 1997;38:1–8. [PubMed] [Google Scholar]
- 77.Kessal K., Liang H., Rabut G., Daull P., Garrigue J.S., Docquier M., Parsadaniantz S.M., Baudouin C., Brignole-Baudouin F. Conjunctival Inflammatory Gene Expression Profiling in Dry Eye Disease: Correlations with HLA-DRA and HLA-DRB1. Front. Immunol. 2018;9:2271. doi: 10.3389/fimmu.2018.02271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stern M.E., Schaumburg C.S., Pflugfelder S.C. Dry Eye as a Mucosal Autoimmune Disease. Int. Rev. Immunol. 2013;32:19–41. doi: 10.3109/08830185.2012.748052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Chauhan S.K., Saban D.R., Sadrai Z., Okanobo A., Dana R. Interferon-γ-Secreting NK Cells Promote Induction of Dry Eye Disease. J. Leukoc. Biol. 2011;89:965–972. doi: 10.1189/jlb.1110611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Platanias L.C. Mechanisms of Type-I- and Type-II-Interferon-Mediated Signaling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 81.Chotikavanich S., de Paiva C.S., Li D.Q., Chen J.J., Bian F., Farley W.J., Pflugfelder S.C. Production and Activity of Matrix Metalloproteinase-9 on the Ocular Surface Increase in Dysfunctional Tear Syndrome. Investig. Ophthalmol. Vis. Sci. 2009;50:3203–3209. doi: 10.1167/iovs.08-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li D.Q., Chen Z., Song X.J., Luo L., Pflugfelder S.C. Stimulation of Matrix Metalloproteinases by Hyperosmolarity via a JNK Pathway in Human Corneal Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2004;45:4302–4311. doi: 10.1167/iovs.04-0299. [DOI] [PubMed] [Google Scholar]
- 83.Messmer E.M., von Lindenfels V., Garbe A., Kampik A. Matrix Metalloproteinase 9 Testing in Dry Eye Disease Using a Commercially Available Point-of-Care Immunoassay. Ophthalmology. 2016;123:2300–2308. doi: 10.1016/j.ophtha.2016.07.028. [DOI] [PubMed] [Google Scholar]
- 84.Mantelli F., Mauris J., Argüeso P. The Ocular Surface Epithelial Barrier and Other Mechanisms of Mucosal Protection. Curr. Opin. Allergy Clin. Immunol. 2013;13:563–568. doi: 10.1097/ACI.0b013e3283645899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hannun Y.A., Obeid L.M. Sphingolipids and Their Metabolism in Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Magtanong L., Ko P.J., Dixon S.J. Emerging Roles for Lipids in Non-Apoptotic Cell Death. Cell Death Differ. 2016;23:1099–1109. doi: 10.1038/cdd.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pettus B.J., Chalfant C.E., Hannun Y.A. Ceramide in Apoptosis: An Overview and Current Perspectives. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2002;1585:114–125. doi: 10.1016/S1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- 88.Magny R., Kessal K., Regazzetti A., Ben Yedder A., Baudouin C., Mélik Parsadaniantz S., Brignole-Baudouin F., Laprévote O., Auzeil N. Lipidomic Analysis of Epithelial Corneal Cells Following Hyperosmolarity and Benzalkonium Chloride Exposure: New Insights in Dry Eye Disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158728. doi: 10.1016/j.bbalip.2020.158728. [DOI] [PubMed] [Google Scholar]
- 89.Magny R., Regazzetti A., Kessal K., Genta-Jouve G., Baudouin C., Mélik-Parsadaniantz S., Brignole-Baudouin F., Laprévote O., Auzeil N. Lipid Annotation by Combination of UHPLC-HRMS (Ms), Molecular Networking, and Retention Time Prediction: Application to a Lipidomic Study of in Vitro Models of Dry Eye Disease. Metabolites. 2020;10:225. doi: 10.3390/metabo10060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Birts C.N., Barton C.H., Wilton D.C. Catalytic and Non-Catalytic Functions of Human IIA Phospholipase A2. Trends Biochem. Sci. 2010;35:28–35. doi: 10.1016/j.tibs.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 91.Cox J., Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 92.Lundgren H.D., Hwang S.-I., Wu L., Han K.D. Role of Spectral Counting in Quantitative Proteomics. (Report) Pub: Expert. Expert Rev. Proteom. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 93.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-Scale Gene Function Analysis with the Panther Classification System. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Joshi-Tope G., Gillespie M., Vastrik I., D’Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: A Knowledgebase of Biological Pathways. Nucleic Acids Res. 2005;33:428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030334 (accessed on 12 December 2021).