Abstract
To investigate whether there are methicillin-resistant Staphylococcus aureus (MRSA) strains with reduced susceptibility to vancomycin in Thailand, a total of 155 MRSA strains isolated from patients hospitalized between 1988 and 1999 in university hospitals in Thailand were tested for glycopeptide susceptibility. All the strains were classified as susceptible to vancomycin and teicoplanin when judged by NCCLS criteria for glycopeptide susceptibility using the agar dilution MIC determination. Vancomycin MICs at which 50 and 90% of the isolates tested were inhibited (MIC50 and MIC90, respectively) were 0.5 and 1 μg/ml, respectively, with a range of 0.25 to 2 μg/ml. For teicoplanin, MIC50 and MIC90 were 2 μg/ml, with a range of 0.5 to 4 μg/ml. However, one-point population analysis identified three MRSA strains, MR135, MR187, and MR209, which contained subpopulations of cells that could grow in 4 μg of vancomycin per ml. The proportions of the subpopulations were 2 × 10−4, 1.5 × 10−6, and 4 × 10−7, respectively. The subsequent performance of a complete population analysis and testing for the emergence of mutants with reduced susceptibility to vancomycin (MIC ≥ 8 μg/ml) confirmed that these strains were heterogeneously resistant to vancomycin. Two of these strains caused infection that was refractory to vancomycin therapy. Pulsed-field gel electrophoresis showed that the two strains had identical SmaI macrorestriction patterns and that they were one of the common types of MRSA isolated in the hospital. This is the first report of heterogeneous resistance to vancomycin in Thailand and an early warning for the possible emergence of vancomycin resistance in S. aureus in Southeast Asia.
Vancomycin is a useful antibiotic against gram-positive pathogens. However, with an increased use of the antibiotic, resistance has been noticed in various species of bacteria such as enterococci (18, 19), Staphylococcus haemolyticus (27), and Staphylococcus epidermidis (9). In 1996, the first Staphylococcus aureus strain with reduced susceptibility to vancomycin, designated VRSA for vancomycin-resistant S. aureus (13) or GISA for glycopeptide-intermediate S. aureus (26), was isolated from a Japanese patient who contracted vancomycin-refractory surgical incision site infection (4). Subsequently, a total of five similar strains were reported from the United States (5, 6), France (23), and Korea (16). The vancomycin MIC for these strains is 8 μg/ml. Vancomycin therapy was unsuccessful with all of these infected patients. The emergence and spread of such resistant strains are expected to raise the morbidity and mortality rates of nosocomial infection significantly. Fortunately, so far only a few isolates have been reported in the world. However, the putative precursor strains for vancomycin resistance, designated hetero-VRSA (13) or hetero-VISA for heterogeneously vancomycin-intermediate S. aureus (7), are reported to be disseminated not only in Japan but also in various other countries in the world (2, 10, 11, 13–15, 17, 24, 29; Z. Gulay, T. Atay, M. Kucukguven, and N. Yulug, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-136, 1998). Heterogeneously resistant (heteroresistant) strains spontaneously generate mutants with reduced susceptibilities to vancomycin at a frequency of 1 in 1 million or more (13). In this study, the status of the glycopeptide susceptibility of Thai methicillin-resistant S. aureus (MRSA) strains was analyzed as the initiation step of an annual surveillance program for the emergence of glycopeptide resistance in Thai S. aureus clinical isolates.
CASE REPORT
Three hetero-VRSA strains (MR135, MR187, and MR209) were isolated from the following clinical patients. The first two patients were hospitalized in the same urban hospital in Bangkok but in different wards at different periods of time. The third patient was admitted to the medical ward of a regional hospital of Thailand. These two hospitals were separated from each other by about 600 km.
Case 1 (strain MR135).
The case 1 patient was a 68-year-old woman who was a resident of Bangkok. She had diabetes and developed a long-term surgical-site MRSA infection at her knee after her second total knee replacement operation in March 1998. The patient was continuously treated with vancomycin from 14 May to 27 July (75 days) and with teicoplanin for 7 days from 29 July to 4 August 1998. However, MRSA was repeatedly isolated from synovial fluid in her knee seven times during the time period from 6 May to 31 July. MR135 was isolated from the synovial fluid taken on the third day of teicoplanin therapy (31 July 1998). The patient became febrile and developed a purulent discharge from the surgical site at her knee. The patient expired on 7 August 1998.
Case 2 (strain MR187).
The case 2 patient was a 16-year-old woman who resided in Bangkok. From 13 May to 9 September in 1996, the patient had chronic retroperitoneal infection with MRSA. Multiple MRSA strains were isolated from various body sites before vancomycin treatment was given: they were isolated from her sputum on 2 June with Pseudomonas aeruginosa, from a blood specimen on 5 June, and from an exploration biopsy sample from her abdomen on 14 June. The patient subsequently received long-term treatment with vancomycin (for 49 days, from 21 June to 9 August). However, MRSA was repeatedly isolated from the drainage of her intra-abdominal abscess during the course of vancomycin therapy. MR187 was isolated on 4 July (14 days after the initiation of vancomycin therapy). Vancomycin treatment was discontinued, and laparoscope-assisted surgical excisions of the intra-abdominal abscess with removal of necrotic tissue and drainage of pus were performed several times in August. The drainage fluid from her abdomen became sterile on 6 September, and the recovered patient was discharged from the hospital.
Case 3 (strain MR209).
MR209 was isolated on 17 June 1999 from the sputum of a 61-year-old man in Khon Kaen, in the northern part of Thailand, who developed nosocomial pneumonia with Acinetobacter sp., Klebsiella pneumoniae, and MRSA. The patient had diabetes melitus, liver cirrhosis, hepatic encephalopathy, and cerebral infarction as underlying diseases. He was treated with cefotaxime for 5 days from 17 to 21 June but was not treated with any glycopeptide antibiotic. His condition deteriorated, and he died at home shortly after he was discharged from the hospital.
MATERIALS AND METHODS
Bacterial strains and antibiotics.
S. aureus strains for which the oxacillin MIC was 4 μg/ml or above were defined as MRSA according to the National Committee for Clinical Laboratory Standards (NCCLS) (20). A total of 155 MRSA strains isolated from the patients hospitalized between 1988 and 1999, including the three strains MR135, MR187, and MR209, were analyzed: 148 strains, including MR135 and MR187, were from the urban university hospital, and 7 strains, including MR209, were from the regional hospital. They were isolated from the respiratory tract (85 strains), pus or exudative fluids (62 strains), blood (6 strains), and urine (2 strains).
Nine of the vancomycin-susceptible MRSA strains were subjected to pulsed-field gel electrophoresis (PFGE) genotyping together with the three vancomycin-heteroresistant strains (MR135, MR187, and MR209). The nine strains were isolates of the urban hospital. Five of them were isolates from five patients who stayed in the same surgical ward of the urban hospital in the same time period (from 1998 to 1999) with the case 1 patient from whom strain MR135 was isolated. The rest (four strains) were from four patients who stayed in the medical intensive care unit of the regional hospital in the same time period (from 1996 to 1997) with the case 2 patient from whom strain MR187 was isolated. FDA209P (ATCC 6538P), an S. aureus type strain, was purchased from the Japanese National Institute of Health and Disease Prevention. Mu3 (ATCO 700698) is an MRSA strain isolated in Japan in 1996 with heterogeneous resistance to vancomycin (13). Mu50 (ATCC 700699), isolated in Japan in 1996, is the first MRSA strain with reduced susceptibility to vancomycin (12, 26).
Vancomycin standard powder was purchased from Sigma (Steinheim, Germany). Teicoplanin was provided by Hoechst-Marion-Roussell Co. (Tokyo, Japan).
Screening of MRSA with reduced susceptibility to glycopeptides.
The screening was performed by one-point population analysis (13), as follows. Overnight culture in brain heart infusion (BHI) broth was adjusted to an optical density at 578 nm of 0.3 (about 108 CFU/ml) and then diluted 10-fold (about 107 CFU/ml). One hundred microliters of the cell suspension was spread onto a BHI agar plate containing 4 μg of vancomycin per ml. The plate was incubated at 37°C for 48 h, and cell growth was inspected at 24 and 48 h. The strain was judged to be susceptible to vancomycin if the cell growth was not apparent within 48 h. The strain was judged to be a possible VRSA if confluent cell growth was seen within 24 h and a possible heteroresistant strain if a countable number of colonies was apparent within 48 h. Confirmation of vancomycin resistance of the strain was based on the vancomycin MIC being equal to or greater than 8 μg/ml, and that of heteroresistance was based on a lower MIC (≤ 4 μg/ml), a population curve with a heterogeneous pattern, and a positive result in a resistant mutant emergence test (13).
Analysis of resistant subpopulations of bacteria (population analysis).
Population analysis was performed by spreading 100 μl of the starting cell suspension (made by adjusting an overnight culture in BHI broth to an optical density at 578 nm of 0.3 [about 108 CFU/ml] with fresh BHI broth) and 10-fold dilutions of this suspension onto BHI agar plates containing various concentrations of vancomycin (13, 21). After incubation at 37°C for 48 h, the number of colonies grown on each plate was counted. The number of resistant cells contained in 100 μl of the starting cell suspension was calculated and plotted on a semilogarithmic graph. Population analysis was performed with both vancomycin and teicoplanin.
Genotyping.
Genotypes of hetero-VRSA and related MRSA strains were analyzed by PFGE (28). PFGE was performed for 22 h using a contour-clamped homogeneous electric field apparatus DRII (Bio-Rad) with a pulse time of 5 to 40 s. SmaI macrorestriction patterns of the three heteroresistant strains (MR135, MR187, and MR209) and nine related vancomycin-susceptible MRSA strains were compared. PFGE patterns were compared by visual examination and interpreted as follows. Isolates with no difference in banding patterns of chromosomal fragments were defined as indistinguishable, those with three or fewer fragment differences were defined as closely related, and those with four or greater fragment differences were defined as different (25).
RESULTS
Glycopeptide susceptibility of Thai strains.
Based on the NCCLS criteria and routine clinical laboratory testing, all the isolates were judged to be susceptible to vancomycin (MIC ≤ 4 μg/ml) and teicoplanin (MIC ≤ 8 μg/ml). Vancomycin MICs at which 50 and 90% of the isolates were inhibited (MIC50 and MIC90, respectively) were 0.5 and 1 μg/ml, respectively, with a range of 0.25 to 2 μg/ml. The teicoplanin MIC50 and MIC90 were both 2 μg/ml, with a range of 0.5 to 4 μg/ml.
Analysis of glycopeptide-resistant subpopulations of the three MRSA strains.
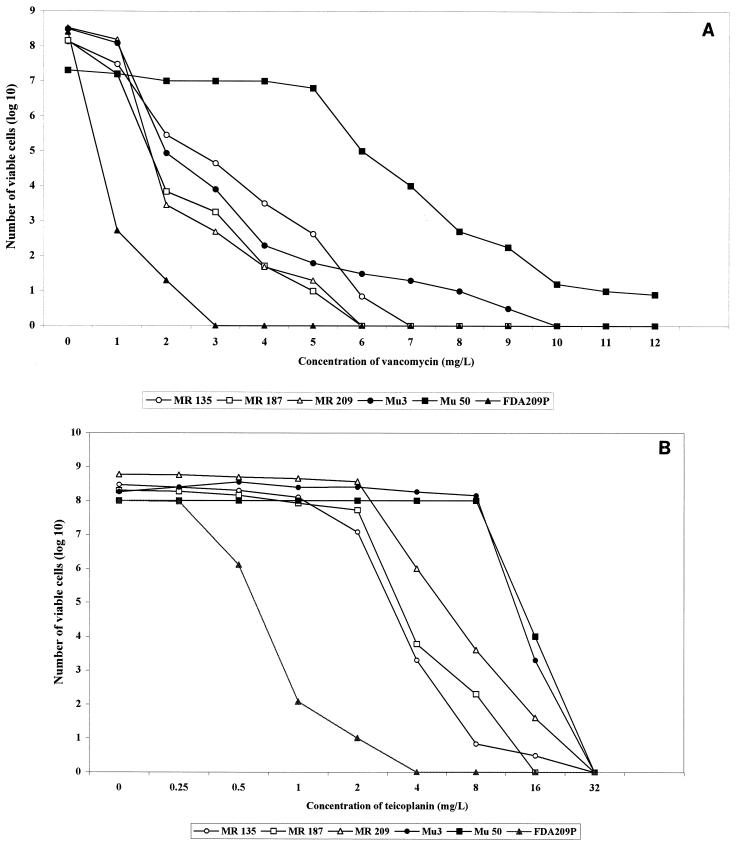
With an inoculum size of 107 CFU, one strain, MR135, exhibited confluent growth of cells on BHI agar containing 4 μg of vancomycin per ml within 24 h. Two other strains, MR187 and MR209, showed positive growth on the screening agar plate within 48 h, with formation of 15 and 4 colonies, respectively. All three strains had glycopeptide MICs in the susceptible range: a vancomycin MIC of 1 μg/ml for all the strains and teicoplanin MICs of 2 μg/ml for MR135, 1 μg/ml for MR187, and 4 μg/ml for MR209, respectively. Figure 1 illustrates the population analysis curves of MR135, MR187, and MR209 in comparison with those of Mu3, Mu50, MR126 (one of the glycopeptide-susceptible Thai strains), and S. aureus type strain FDA209A: Figure 1A shows population curves for resistance to vancomycin. The sizes of the subpopulations of the three Thai strains that were resistant to 2 to 5 μg of vancomycin per ml were smaller than those of Mu50 and were comparable to those of Mu3. Strain MR135 had greater resistant subpopulations than Mu3 to 2 to 5 μg of vancomycin per ml (Fig. 1A). MR187 and MR209 were more susceptible to vancomycin than Mu3, but both isolates also contained resistant subpopulations of cells that could grow in the presence of 4 and 5 μg of vancomycin per ml. All three Thai strains had smaller sizes of subpopulations resistant to 6 to 9 μg of vancomycin ml than those of Mu3.
FIG. 1.
Analysis of resistant subpopulations of Thai MRSA strains to vancomycin (A) and teicoplanin (B). Three Thai strains, MR135, MR187, and MR209, contained subpopulations resistant to 4 to 6 μg of vancomycin per ml (A) and 8 μg of teicoplanin per ml (B), in contrast to Thai strain MR126 and S. aureus type strain FDA209P. When compared to Japanese strain Mu3, Thai strains did not contain subpopulations resistant to 7 to 9 μg of vancomycin per ml but MR135 had larger sizes of subpopulations resistant to 2 to 5 μg of vancomycin per ml. Mu50 is a Japanese MRSA strain for which the vancomycin MIC is 8 μg/ml.
A resistant-mutant emergence test was performed with strains MR135, MR187, and MR209 by the following procedure. One of the colonies of each strain grown on the BHI agar plate containing 4 μg of vancomycin per ml was picked and subjected to colony purification to get rid of contaminated vancomycin-susceptible but nondead cells, as follows. The picked colony was streaked onto a fresh BHI agar plate without vancomycin, and one of the formed colonies was picked again to establish it as the one-step resistant mutant of the strain. The mutant strain thus established was cultivated overnight without antibiotic and subjected to MIC determination. Mutants for which the vancomycin MIC was 8 μg/ml were obtained from all three strains by this one-step selection procedure.
Figure 1B illustrates the teicoplanin-resistant subpopulation profiles of MR135, MR187, and MR209 in comparison with those of Mu3, Mu50, FDA209A, and MR126. The three strains had similar resistant-subpopulation patterns, but they contained relatively smaller sizes of resistant subpopulations than those of Mu3 and Mu50. Almost 100% of the cell populations of Mu3 and Mu50 grew in the presence of 8 μg of teicoplanin per ml, whereas only 10−7 to 10−4 fractions of the populations grew in the Thai strains.
Genotypes of the three heteroresistant strains.
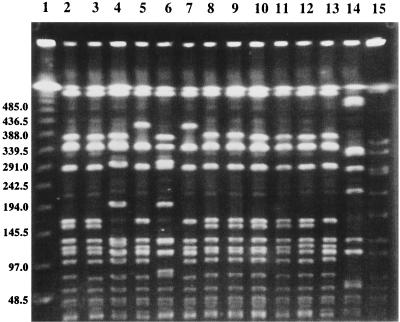
PFGE banding patterns of MR135, MR187, and MR209 are shown in Fig. 2 in comparison with those of vancomycin-susceptible S. aureus strains and Mu3. The Thai heteroresistant strains (lanes 2 to 4) and the related vancomycin-susceptible MRSA strains isolated from the same ward and during overlapping time periods (lanes 5 to 13) had very similar PFGE banding patterns. The Thai heteroresistant strains, however, had at least seven band differences from Mu3, a Japanese isolate (lane 14). Strains MR135 (lane 2) and MR187 (lane 3) were isolated from different units of the urban hospital in different years (MR135 from a surgery unit in 1998 and MR187 from an intensive care unit in 1996), but they had identical SmaI macrorestriction patterns. The vancomycin-susceptible MRSA strains (lanes 5 to 13) had either identical (lanes 8 to 12) or very similar PFGE banding patterns compared with those of MR135 (lane 2) and MR187 (lane 3). The other heteroresistant strain, MR209 (lane 4), isolated from the regional hospital, had a different PFGE banding pattern than those of the other two strains, but it was noticed that it had a pattern similar to that of one of the vancomycin-susceptible MRSA strains in the urban hospital (lane 6).
FIG. 2.
PFGE banding patterns of SmaI-digested chromosomal DNAs of Thai MRSA strains with and without reduced susceptibilities to glycopeptides. Lanes: 1, lambda concatemer standard marker (molecular sizes in kilobases are indicated on the left); 2 to 4, strains MR135, MR187, and MR209, respectively; 5 to 9, vancomycin-susceptible MRSA strains isolated from four patients hospitalized in the same surgical ward during the same period (from 1998 to 1999) as the patient from whom strain MR135 was isolated; 10 to 13, vancomycin-susceptible MRSA strains isolated from five patients admitted to the same medical intensive care unit during the same period (from 1996 to 1997) as the patient from whom strain MR187 was isolated; 14, Mu3; 15, SmaI-digested total DNA from NCTC 8325, used as a standard.
DISCUSSION
This is the first report of infection due to MRSA strains with reduced susceptibility to vancomycin in Thailand. Although the three isolates were heterogeneously resistant to vancomycin, this report is an early warning that S. aureus strains with full resistance to vancomycin might emerge in the future, emphasizing the importance of a laboratory capability of identifying heterogeneous vancomycin resistance. The disk diffusion method is inadequate to detect S. aureus strains with reduced susceptibilities to vancomycin (26). Although well-standardized microdilution MIC determination can detect S. aureus clinical isolates with reduced susceptibilities to vancomycin, it cannot detect heteroresistance. Based on the MICs, all three isolates reported in this study are judged to be susceptible to glycopeptide antibiotics (20). Strains MR135 and MR187, however, were isolated from patients whose MRSA infection did not respond favorably to long-term vancomycin treatment. Other researchers also reported that vancomycin treatment failure was associated with infection caused by MRSA heteroresistant to vancomycin (1, 13, 15, 17, 23, 29). Efforts to detect heteroresistant strains, therefore, may be warranted not only to monitor the progression of glycopeptide resistance acquisition by local MRSA strains, but also to predict the clinical effectiveness of glycopeptide treatment of the patients infected with MRSA.
MRSA strains with reduced susceptibilities to vancomycin in Japan and in the United States were isolated after prolonged exposure to the antibiotic (4, 12, 24), as were two Thai strains. MR135 was isolated from a patient treated with vancomycin for 75 days and with teicoplanin for 3 days, and MR187 was isolated after 14 days of vancomycin therapy. Strain MR209 was an exception. The strain was isolated from a patient who had not been treated with any glycopeptide antibiotic. In this case, the strain might have been transmitted to the patient from another patient who had undergone glycopeptide treatment of MRSA infection. This possibility may explain why the strain had the lowest vancomycin resistance of the three strains, since the vancomycin resistance phenotype is unstable in the absence of antibiotic pressure (3). MR209 was marginal with regard to heteroresistance if heteroresistance is defined as having resistant subpopulations whose number is greater than one in 106 (13).
The genotypes of three heteroresistant strains were quite distinct from that of the Japanese strain Mu3 (Fig. 2), but they were indistinguishable from the genotypes other Thai MRSA strains retaining vancomycin susceptibility. Therefore, as recently demonstrated in in vitro experiments (22), it is likely that vancomycin resistance is acquired by MRSA strains with diverse genetic backgrounds besides that of the Japanese MRSA clonotype II-A (13). In this regard, however, it was noted that Thai strains had a different pattern of resistance from that of Mu3. In contrast to Mu3, the Thai strains did not contain larger resistant subpopulations capable of growth in 6 to 9 μg of vancomycin per ml (Fig. 1A). They were also less resistant to teicoplanin than Mu3 or Mu50 (Fig. 1B). These phenotypic differences in the resistance patterns indicate that the mechanism of glycopeptide resistance in Thai strains is different from that in Japanese MRSA strains (8). Research is under way to clarify the genetic mechanism behind Thai strains expressing reduced susceptibilities to glycopeptide antibiotics.
ACKNOWLEDGMENTS
This work was a part of the Nosocomial Infections and Drug-Resistant Microorganisms project, which was supported by the Core University System Exchange Programme under the Japan Society for the Promotion of Science, coordinated by the University of Tokyo Graduate School of Medicine and Mahidol University. The work was also supported by an unrestricted grant from Wyeth Lederle Japan Ltd.
We thank Rachada Sathitmathakul and Siriporn Sripalakit, Centre for Nosocomial Infection Control, Faculty of Medicine, Siriraj Hospital, Mahidol University.
REFERENCES
- 1.Ariza J, Pujol M, Cabo J, Pena C, Fernandez N, Linares J, Ayats J, Godiol F. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet. 1999;353:1587–1588. doi: 10.1016/s0140-6736(99)01017-x. [DOI] [PubMed] [Google Scholar]
- 2.Bierbaum G, Fuchs K, Lenz W, Szekat C, Sahl H G. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur J Clin Microbiol Infect Dis. 1999;18:691–696. doi: 10.1007/s100960050380. [DOI] [PubMed] [Google Scholar]
- 3.Boyle-Vavra S, Berke S K, Lee J C, Daum R S. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother. 2000;44:272–277. doi: 10.1128/aac.44.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Reduced susceptibility of Staphylococcus aureus to vancomycin—Japan 1996. Morbl Mortal Wkly Rep. 1997;46:624–626. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbl Mortal Wkly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbl Mortal Wkly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 7.Chesneau O, Morvan A, El Solh N. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J Antimicrob Chemother. 2000;45:887–890. doi: 10.1093/jac/45.6.887. [DOI] [PubMed] [Google Scholar]
- 8.Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Contribution of thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob Agents Chemother. 2000;44:2276–2285. doi: 10.1128/aac.44.9.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett D O, Jochimsen E, Murfitt K, Hill B, McAllister S, Nelson P, Spera R V, Sall R K, Tenover F C, Johnston J, Zimmer B, Jarvis W R. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect Control Hosp Epidemiol. 1999;20:167–170. doi: 10.1086/501605. [DOI] [PubMed] [Google Scholar]
- 10.Geisel R, Schmitz F J, Thomas L, Berns G, Zetsche O, Ulrich B, Fluit A C, Labischinsky H, Witte W. Emergence of heterogeneous intermediate vancomycin resistance in Staphylococcus aureus isolates in the Düsseldorf area. J Antimicrob Chemother. 1999;43:846–848. doi: 10.1093/jac/43.6.846. [DOI] [PubMed] [Google Scholar]
- 11.Haraga I, Nomura S, Nagayama A. The effects of vancomycin and beta-lactam antibiotics on vancomycin-resistant Staphylococcus aureus. N Engl J Med. 1999;341:1624–1625. doi: 10.1056/NEJM199911183412117. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 14.Howe R A, Bowker K E, Walsh T R, Feest T G, MacGowan A P. Vancomycin-resistant Staphylococcus aureus. Lancet. 1998;351:602. doi: 10.1016/S0140-6736(05)78597-4. [DOI] [PubMed] [Google Scholar]
- 15.Kantzanou M, Tassios P T, Tseleni-Kotsovil A, Legakis N J, Vatopoulos A C. Reduced susceptibility to vancomycin of nosocomial isolates of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1999;43:729–731. doi: 10.1093/jac/43.5.729. [DOI] [PubMed] [Google Scholar]
- 16.Kim M-N, Pai C H, Woo J H, Ryu J S, Hiramatsu K. Vancomycin-intermediate Staphylococcus aureus in Korea. J Clin Microbiol. 2000;38:3879–3881. doi: 10.1128/jcm.38.10.3879-3881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchese A, Balistreri G, Tonoli E, Dabbia E A, Schito G C. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J Clin Microbiol. 2000;38:866–869. doi: 10.1128/jcm.38.2.866-869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayhall C G. Prevention and control of vancomycin resistance in Gram-positive coccal microorganisms: fire prevention and fire fighting. Infect Control Hosp Epidemiol. 1996;17:353–355. doi: 10.1086/647315. [DOI] [PubMed] [Google Scholar]
- 19.Morris J G, Jr, Shay D K, Hebden J N, et al. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Establishment of endemicity in a university medical center. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7–A5. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 21.Peterson L R, Shanholtzer C J. Tests for bactericidal effects of antimicrobial agents: technical performance and clinical relevance. Clin Microbiol Rev. 1997;5:697–701. doi: 10.1128/cmr.5.4.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeltz R F, Singh V K, Schmidt J L, Batten M A, Baranyk C S, Nadakavukaren M J, Jayaswal R K, Wilkinson B J. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob Agents Chemother. 2000;44:294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploy M C, Grélaud C, Martin C, de Lumley L, Dennis F. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet. 1998;351:1212. doi: 10.1016/s0140-6736(05)79166-2. [DOI] [PubMed] [Google Scholar]
- 24.Siradzki K, Roberts R B, Harber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veach L A, Pfaller M A, Barrett M, Koontz F P, Wenzel R P. Vancomycin resistance in Staphylococcus haemolyticus causing colonization and bloodstream infection. J Clin Microbiol. 1990;28:2064–2068. doi: 10.1128/jcm.28.9.2064-2068.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada A, Katayama Y, Hiramatsu K, Yokota T. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1991;176:1319–1325. doi: 10.1016/0006-291x(91)90430-f. [DOI] [PubMed] [Google Scholar]
- 29.Wong S S Y, Wong P, Ho P L, Woo P C Y, Yuen K Y. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin Infect Dis. 1999;29:760–767. doi: 10.1086/520429. [DOI] [PubMed] [Google Scholar]