Abstract
African swine fever virus (ASFV) is the causative agent of African swine fever (ASF). The virus causes an acute highly hemorrhagic disease in domestic pigs, with high mortality. Although the overall genome mutation rate of ASFV, a large DNA virus, is relatively low, ASFV exhibits genetic and antigenic diversity. ASFV can be classified into 24 genotypes on the basis of the B646L gene. Cross-protected ASFV strains can be divided into eight serogroups on the basis of antibody-mediated hemadsorption inhibition. Here, we review research progress on ASFV genotyping and serogrouping, and explain how this information assists in the rapid identification of virus origin during ASF outbreaks and will aid in the development of ASF vaccines.
Keywords: African swine fever virus, Genotype, Serogroup
Introduction
Since African swine fever (ASF) was first reported in 1921, the causative agent, African swine fever virus (ASFV) has escaped from the African continent on several occasions and spread dramatically; ASF now poses a threat to swine production worldwide [1–3]. In 2007, ASF was introduced into Georgia. Subsequently, the virus spread to the Trans-Caucasian region and reached the Russian Federation. From Russia, the virus invaded the European Union in 2014. In August 2018, the disease reached the world’s largest pig producer, China, and it is now spreading in Southeast Asian countries. The most recently affected countries are Papua New Guinea, India [4], Germany [5], the Dominican Republic [6], and Haiti [7, 8]. There currently is no commercial vaccine available for control of ASF. Factors such as genetic and antigenic diversity as well as poor cross-protective immunity hinder the development of a safe and efficacious vaccine [9].
ASFV has a linear double-stranded DNA. The genome is about 170–190 kb and divided into the left variable region (LVR, 38–48 kb), central conserved region (C region, about 125 kb), and the right variable region (RVR, 13–22 kb) [10]. Different strains may have significant differences in important positions, such as the multigene family in the LVR, the central variable region (CVR) in the C region, and the EP402R (expressing CD2v protein) genes. These variable regions are important for evolutionary analysis of ASFV. Comparative analysis of the C-terminal end of the B646L gene, which encodes the major protein p72, enables the classification of isolates into one of 24 genotypes. Isolates are also divided into eight serogroups (SG) based on phylogenetic grouping of the protein CD2v encoded by the EP402R gene. These methods allow relatively fast and easy typing of ASFV strains, and remain the first approach for identification of the origin of ASFV in case of introduction into new territories. Additionally, the analysis of other sequences, such as the CVR and the E183L gene, can help improve molecular epidemiological studies of ASFV [11, 12]. In this review, we discuss recent research progress on ASFV genotypes and SG. We hope that this report will provide a reference for further analyses of the etiology of, and diagnostic technology for, ASFV, and for vaccine development.
History of ASFV typing
From the discovery of ASFV, researchers began to consider whether pigs that recovered from ASF could gain immunity. Before ASFV cell culture was established, immunological research on ASF barely yielded any findings. The number of domestic pigs that survived natural infection was very low. However, since the 1950s, the number of attenuated strains of ASFV obtained via tissue culture has been increasing, which makes it possible to conduct immunological research on ASFV [13, 14]. Researchers have gradually found diverse ASFV antigens by using various tests such as the hemadsorption reaction [15, 16], agar diffusion precipitation test [17], complement fixation test [18], and isoelectric precipitation technology [19].
In 1963, Malmquist performed hemadsorption inhibition (HAI) experiments and cross-protection experiments and detected different antigenic ASFV types [15]. Subsequent HAI analysis of multiple strains of ASFV showed that the antigenic differences can be used to classify or type ASFV [16, 20], and ASFV strains with hemagglutinating properties were divided into three subtypes: A, B, and C [21]. However, this typing method was discontinued, probably because the high fatality rate of ASF makes it impossible to continuously obtain seroconverted domestic pigs [22].
Restriction fragment length polymorphism (RFLP) analysis was used to divide nine ASFV field isolates into four main groups [22]. In 1989, based on digestion of the ASFV genome with SalI, 23 isolates were divided into five groups, and all field isolates from Europe and America were found to have a common origin [23]. ASFV isolated from soft ticks was typed using RFLP, and the genomes of isolates from the same region were found to be more similar to each other than isolates from different regions [24]. However, RFLP is not effective in rapidly tracing the source of a single outbreak because the method is time-consuming and delivers relatively crude results; this stalled the development of ASFV typing [25].
Genotypes of ASFV
Genotyping of ASFV during outbreaks is important to reveal the origin of the virus and quickly differentiate between closely related strains [26]. Genomic research into ASFV has improved with the rapid development of gene cloning, PCR, and sequencing technology. After the successful construction of a library containing 98% of the genomic information for ASFV in 1984 [27], the number of articles on the construction of ASFV genomic libraries has gradually increased, laying the foundations for analysis. Several specific genetic targets have been used to assess ASFV genotype.
p72/VP73 (B646L) genotyping
The main capsid protein-encoding gene of ASFV (p72, VP73, or B646L) was one of the first targets for genotyping [28]. In 1990, Lopez-Otin et al. sequenced and analyzed the full length of the p72-encoding gene, laying a foundation for its study as a diagnostic gene [29]. In 1992, Steiger et al. used PCR to establish ASFV rapid diagnosis technology for the first time [30]. In 1996, Yu et al. compared the nucleic acid and protein sequences of gene from four strains isolated from different regions and found that the sequence of the gene was highly consistent among the strains (97.8% to 100% homology). This was consistent with previously published conclusions that the B646L gene sequence is relatively stable [31, 32], further establishing the importance of this gene for antigenic typing [33].
In 2001, Gonzague et al. used a partial fragment of B646L (278 bp, positions 893–1170) to demonstrate that the Malagasy ASFV isolated in 1999 was most similar to the Mozambican strain isolated in 1994 [34]. In 2003, on the basis of amino acid sequence of the C-terminal end of p72 gene (415 bp, GenBank accession no. AF301537), Bastos et al. identified 10 ASFV genotypes and established the standard ASFV genotype marker. They found up to 9.4% genetic variation in the p72 sequence indifferent ASFV strains, which is suitable for molecular epidemiological analysis, and provided a rapid and accurate method for determining the genotype of field and outbreak strains of ASFV from Southern and East African countries [25].
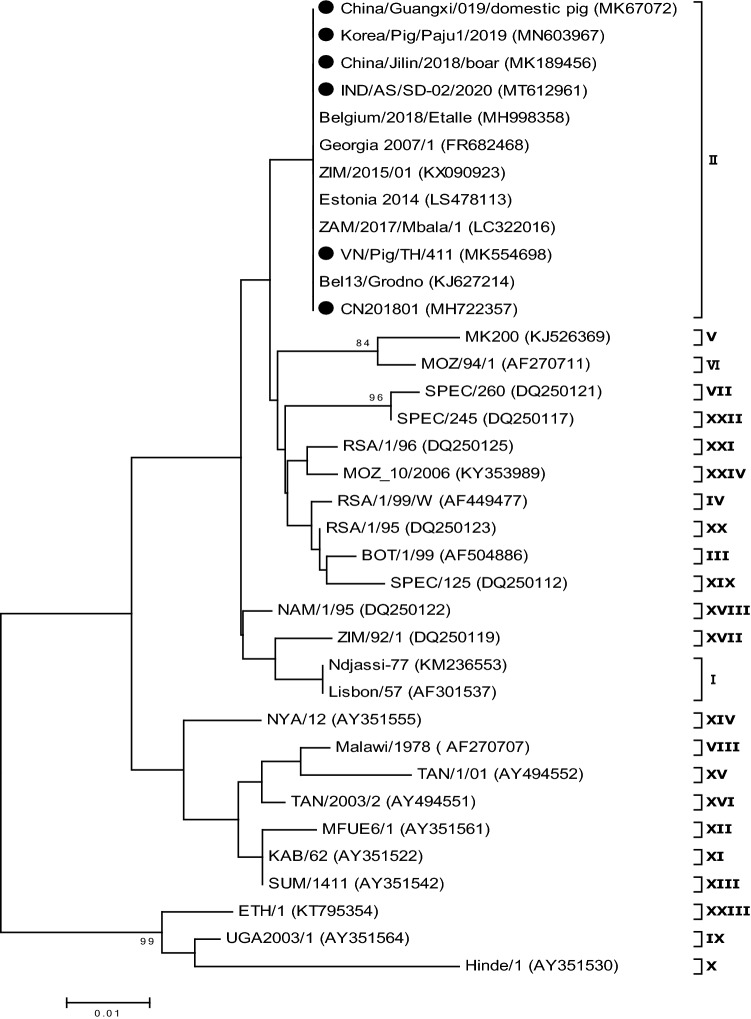
In 2005, Lubisi et al. further classified ASFV into 16 genotypes and identified strains of genotype I for the first time from sylvatic hosts in East Africa [35]. In 2007, Boshoff et al. performed a comparative analysis of 43 ASFV isolates from South Africa between 1973 and 1999 and found six novel genotypes, and they classified ASFV into 22 genotypes [36]. Since then, the evolutionary analysis of ASFV has been mainly based on the conserved B646L gene that encodes the viral protein p72, and the study of ASFV gene classification by using the B646L gene gradually became the most important typing criterion. Achenbach et al. (2017) analyzed B646L gene evolution in ASFV isolated from Ethiopia from 2011 to 2014 and identified the 23rd genotype, which is derived from the same evolutionary branch as the IX and X genotypes prevalent in East African countries and the Democratic Republic of Congo. Quembo et al. (2018) isolated 19 strains of ASFV from soft tick samples collected from Gorongosa National Forest Park, Mozambique. They found that five of the viruses belonged to a new evolutionary branch, designated the 24th genotype. Sequences of the B646L gene from the novel viruses and other known genotypes of ASFV are shown in Table 1. They were downloaded from GenBank and used to construct a phylogenetic tree, in which ASFV is divided into the 24 genotypes (Fig. 1). The results (Table 1) show that genotypic diversity is greatest in Africa; countries elsewhere with ASF outbreaks exhibit low or no genotype diversity. Generally, the current ASF genotype refers to p72 genotyping.
Table 1.
ASFV isolates used for the construction of phylogenetic tree based on partial B646L (p72) gene nucleotide sequences
| Isolate | Host species | Year of isolation | Country | GenBank accession no | p72 genotype | References |
|---|---|---|---|---|---|---|
| Ndjassi-77 | NK | 1979 | Zaire (DRC) | KM236553 | I | [37] |
| Lisbon/57 | Wild boar | 1957 | Portugal | AF301537 | I | [25] |
| China/Guangxi/2019/domestic pig | Domestic pig | 2019 | China | MK670727 | II | [38] |
| CN201801 | Domestic pig | 2018 | China | MH722357 | II | [39] |
| China/Jilin/2018/boar | Wild boar | 2018 | China | MK189456 | II | [40] |
| Korea/Pig/Paju1/2019 | Domestic pig | 2019 | South Korea | MN603967 | II | [41] |
| VN/Pig/TH/411 | Domestic pig | 2019 | Vietnam | MK554698 | II | [42] |
| IND/AS/SD-02/2020 | Domestic pig | 2020 | India | MT612961 | II | [4] |
| Belgium/2018/Etalle | Wild boar | 2018 | Belgium | MH998358 | II | [43] |
| Estonia 2014 | Wild boar | 2014 | Estonia | LS478113 | II | [44] |
| Bel13/Grodno | Domestic pig | 2013 | Belarus | KJ627214 | II | [45] |
| Georgia 2007/1 | Domestic pig | 2007 | Georgia | FR682468 | II | [8] |
| ZAM/2017/Mbala/1 | Domestic pig | 2017 | Zambia | LC322016 | II | [46] |
| ZIM/2015/01 | Domestic pig | 2015 | Zimbabwe | KX090923 | II | [47] |
| BOT/1/99 | Domestic pig | 1999 | Botswana | AF504886 | III | [48] |
| RSA/1/99/W | Warthog | 1999 | South Africa | AF449477 | IV | [25] |
| MK200 | Domestic pig | 1978 | Mozambique | KJ526369 | V | [37] |
| MOZ/94/1 | Domestic pig | 1994 | Mozambique | AF270711 | VI | [48] |
| SPEC/260 | Domestic pig | 1993 | South Africa | DQ250121 | VII | [36] |
| Malawi/1978 | Domestic pig | 1978 | Malawi | AF270707 | VIII | [25, 48] |
| UGA2003/1 | Warthog | 2003 | Uganda | AY351564 | IX | [35] |
| Hinde/1 | Wild boar | 1954 | Kenya | MK829709 | X | [35] |
| KAB/62 | Ticks | 1983 | Zambia | AY351522 | XI | [35] |
| MFUE 6/1 | Tick | 1982 | Zambia | AY351561 | XII | [35] |
| SUM/1411 | Ticks | 1983 | Zambia | AY351542 | XIII | [35] |
| NYA/12 | Ticks | 1986 | Zambia | AY351555 | XIV | [35] |
| TAN/1/01 | Wild boar | 2001 | Tanzania | AY494552 | XV | [35] |
| TAN/2003/2 | Wild boar | 2003 | Tanzania | AY494551 | XVI | [35] |
| ZIM/92/1 | Domestic pig | 1992 | Zimbabwe | DQ250119 | XVII | [36] |
| NAM/1/95 | Domestic pig | 1995 | Namibia | DQ250122 | XVIII | [36, 49] |
| SPEC/125 | Domestic pig | 1987 | South Africa | DQ250118 | XIX | [36] |
| RSA/1/95 | Domestic pig | 1995 | South Africa | DQ250123 | XX | [36] |
| RSA/1/96 | Domestic pig | 1996 | South Africa | DQ250125 | XXI | [36] |
| SPEC/245 | Domestic pig | 1992 | South Africa | DQ250117 | XXII | [36] |
| ETH/1 | Domestic pig | 2011 | Ethiopia | KT795354 | XXIII | [50] |
| MOZ_10/2006 | Tick | 2006 | Mozambique | KY353989 | XXIV | [51] |
NK not known
Fig. 1.
Evolutionary relationships of African swine fever virus (ASFV) strains on the basis of p72 (B646L) gene sequences. The phylogenetic tree was inferred using the maximum-likelihood method and Kimura 2-parameter model based on the partial B646L gene nucleotide sequences. Phylogeny was inferred using 1000 bootstrap replications, and the node values show percentage bootstrap support (only values > 80% are shown). The scale bar shows the number of substitutions per site. The sequences of Asia ASFV isolates are indicated using a closed circle (●)
CVR (9RL/B602L) genotyping
Although the genotyping method based on the B646L gene is accepted by most researchers, it does not always provide adequate typing resolution or the ability to discriminate between viruses with different biological phenotypes. Increased genotypic resolution has been achieved by the assessment of the CVR of the B602L gene [11, 12, 35, 48, 52].
Similar to other nucleocytoplasmic large DNA viruses, ASFV contains a CVR of approximately 400 bp within the relatively conserved, evolutionarily stable, central 125-kb region [53]. Irusta et al. (1996) defined the CVR of the ASFV genome and found that it encodes repeated amino acid tetramers within a late viral gene (open reading frame 9RL) in genotype I viruses. They showed that analysis of the number and composition of the repeated amino acid tetramers within the CVR may be useful for identifying and grouping ASFV isolates [54]. Researchers have found that CVR amplicons vary between 228 and 686 bp [36, 54, 55]. The differences in CVR sequence information and fragment size between different strains [12, 48, 55] make the CVR unsuitable for genotyping; however, CVR genotyping is considered to complement the standard p72 genotyping for ASFV intragenotypes [45, 55]. Owolodun et al. (2010) isolated five CVR variants from clinical samples of wild and domestic pigs at difference sampling sites in Nigeria during 2003 to 2006. Because clustered within a single phelogenetic lineage, all five CVR variants, the Ben97/1 variant (isolated from Benin, Nigeria's neighbor, 1997) and the Nigerian 2001 variant were classified into a common ancestry. Therefore, the researchers concluded that it is possible that these mutants were introduced from outside Nigeria, or mutated domestically after their introduction in 1998 [56]. The MAL/19/Karonga/1–4 virus responsible for the 2019 ASF outbreak in Malawi clustered into p72 genotype II and showed 100% nucleotide identity with ASFV previously described in Tanzania, Zambia, Georgia, China, Vietnam, Estonia, and Moldova, and the amino acid repeats encoded within the CVR of the B602L gene were 100% similar to those of the ZAM/13/Mbala ASFV isolates from Tanzania in 2017 and Zambia in 2013. It is speculated that the virus was introduced from Tanzania through transborder trade of pigs and pork products to Malawi [57]. By analyzing the amino acids encoded by the CVR within the B602L gene, Vilem et al. identified three different CVR variants of ASFV that have circulated in certain regions of Estonia since 2014, and they speculated that the epidemic of ASF in neighboring counties was responsible for the GII-CVR1/SNP1 virus identified in the county of Lääne. Although CVR analysis has been widely used to distinguish closely related ASF isolates, the low genetic diversity of the CVR means that further study of more informative genetic regions is required to confirm intragenotypic relationships [58].
p54 (E183L) genotyping
The p54-encoding gene E183L increased the intragenotypic resolution of standard p72 genotyping [11, 12, 35, 48, 52]. As early as 1995, Sun et al. confirmed that this gene can mutate between different cell passages [59]; however, few phylogenetic analyses of the gene have been conducted. Analysis of ASFV introduced into Georgia in 2007 showed that its E183L gene sequence was identical to that of five Madagascar isolates and two Mozambique isolates (Moz 1/02,Moz 2/02); however, it showed mutations in multiple locations when compared with two Mozambique strains (Moz 1/03 and Moz 1/05) isolated later. This result is important for studying the origin of the Georgian strain [60]. Gallardo et al. (2009) simultaneously used the E183L, B646L, and B602L genes to analyze the phylogeny of ASFV isolates detected in Kenya from 2006 to 2007. The results showed that the phylogenetic trees based on p54 and p72 were basically the same; however, on the largest branch of p72 genotype I, the p54 genotype could be further divided into four discrete subclusters, named Ia, Ib, Ic, and Id. Type Ia was mainly from European and American strains. Type Ib included strains from West African countries and Portuguese isolate Lisbon/57. The Portuguese isolate Lisbon/60 and South African isolate MZUKI/1979 were distributed in independent genotypes designed Ic and Id, respectively [11]. In another study using E183L nucleotide sequences, the 2015 Zambian ASFV was found to belong to genotype Id, and this virus may have originated in Zambia (or possibly the Southern African region) [61]. Further research focusing on Zambian isolates by using p54 genotyping showed that Zambian isolates divided into types Ie, If, Ig, IIa, IIb, VIIIa, VIIIb, XI, XIII, and XIV, suggesting that the endemic strains in Zambia have multiple evolutionary branches [62, 63]. To sum up, p54 typing is only used as an auxiliary index for ASFV phylogeny analysis, as the difference within E183L is not significant compared with p72 typing and the discrimination within E183L is not significant compared with CVR typing.
Other genotyping methods
To evaluate the evolutionary differences between ASFV strains, especially the evolutionary trend of strains in the same region, different genotyping methods, other than genotyping based on p72, CVR, and p54, have been established by additional assessment of the p30-encoding gene (CP204L) [60, 63, 64], tandem repeat sequences (TRS) within the intergenic region (IGR) between I73R and I329L [45, 64, 65], the CD2v-encoding gene (EP402R) [64], the thymidine kinase (TK) gene [66], the J268L [12], Bt/Sj [12], KP86R [12], and O174L genes [67], and the C315R/C147L region [68]. Sequence analysis showed that different isolates have partial differences in the length and sequence of the J268L, Bt/Sj, and KP86R genes, which can be used to distinguish between evolutionarily similar isolates [12]. Sanna et al. (2017) performed a genetic analysis of ASFV prevalent for many years in Sardinia, Italy, on the basis of the CP204L gene, EP402R gene and IGR. They found that the CP204L genes and IGR were stable, but the Sardinian strains from 1978 to 2014 could be classified into two subgroups on the basis of sequence comparison of the EP402R gene (CD2v): one group included historical isolates from 1978 to 1990, and the second was composed of the viruses collected from 1990 to 2014. Sanna et al. (2017) believed that the virus mutations were induced by the selective pressure associated with increasing controls at the farm level and improved diagnostic techniques because these measures had hampered the spread of the virus [64]. Gallardo et al. (2014) found that the sequences of ASFV isolates from Lithuania and Poland were 100% homologous with other isolates from Eastern Europe on the basis of classical genotyping (based on the variable region of the B646L gene and parts of the E183L gene); however, when they focused on analysis of TRS in the IGR between the I73R and I329L genes, the viruses from Poland and Lithuania were observed to have a TRS insertion identical to that present in ASFV isolates from Belarus and Ukraine, which is different from that present in other viruses from Eastern Europe and Russia. Thus, Gallardo et al. (2014) concluded that ASFV strains in Lithuania and Poland most probably originated in Belarus. This TRS insertion was present in ASFV isolates from Russia and Ukraine in 2012, but absent in those from Tverskaya Oblast (western Russia) in 2011 and 2012. Thus, viruses introduced into the European Union were hypothesized to originate in Russia, emerging in 2012 or even earlier, and to have been transmitted through Belarus and Ukraine [65]. These findings confirm the suitability of TRS for higher resolution ASFV genotyping. In 2018, ASF outbreaks in China were caused by the IGR II variant of ASFV, which has a TRS insertion in the IGR. In 2019, an IGR I variant (without extra TRS insertions in the IGR; only one case in wild boar) and IGR III variant (two TRS insertions in the IGR) were, respectively, isolated from Jilin and Guangxi, China (Fig. 1). These genetic data are essential for tracing the source of ASFVs and extending our knowledge of the viral evolution and epidemiology in China [38, 40]. In 2017, Onzere et al. performed an evolutionary analysis of prevalent ASFV strains on the basis of TK genes and found significant differences between the Kenyan isolates from 2011 to 2013 and a South African reference strain [66].
Previous studies suggest that other ASFV genotypes may exist and other genetic markers may be used for genotyping [45, 50]. The latest advances in whole-genome sequencing can be used to perform comprehensive genotyping and obtain data that are essential for elucidating the biology and genetic characteristics of ASFV.
Serogroups of ASFV
ASFV was considered one of the few viruses without neutralizing antibodies [69]. Malmquist (1963) first identified several ASFV antigenic SG by performing the HAI test to assess serological cross-reactivity with different ASFV isolates in vitro [70]. HAI was widely used as a tool to define the specificity of serotypes at the Pokrov Institute (Federal Research Center for Virology and Microbiology), Russia. Eight ASFV SG have been defined and thoroughly characterized using HAI. We may detect more SG if additional parameters such as hemadsorption density (the number of red blood cells per infected cell) and red blood cell contact maps are used to refine the classification of ASFV isolates [71].
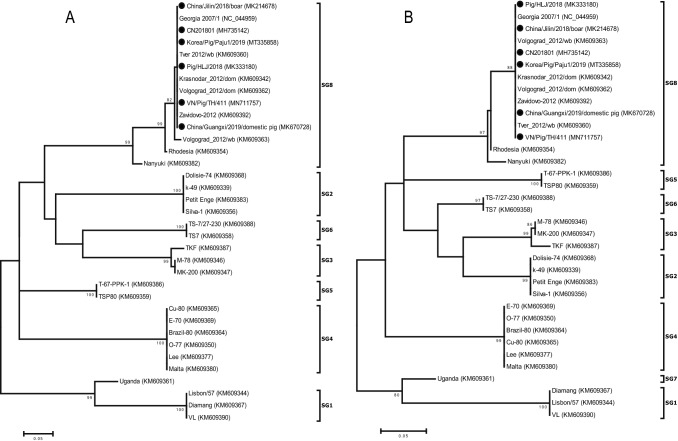
HAI assay is the gold standard for ASFV serotyping, but HAI-based serogrouping has limitations: the test requires sera from convalescent pigs, and the viral antibodies used for HAI detection are produced late and at low titers. Therefore, it is necessary to evaluate the mechanism of HAI serogrouping and find the antigenic determinants. Many researchers have confirmed that CD2v (EP402R) and C-type lectin (EP153R) are closely associated with the hemadsorption of ASFV, although these genes are the most diverse between different strains of ASFV [72–78]. Malogolovkin et al. (2015) performed phylogenetic analysis of > 80% ASFV strains and found that CD2v/C-type lectin genotype grouping is associated with HAI-based serological grouping and ASFV CD2v/C-type lectin gene interchange could induce a corresponding interchange of HAI in vitro and a cross-protection phenotype in vivo [79]. The researchers selected protein signature domains of CD2v and C-type lectin (amino acids 109–196 and 67–166, respectively) of ASFV isolate Lisbon/57 (GenBank accession no. KM609344) for phylogenetic analyses. The phylogenetic groupings based on CD2v and C-type lectin were correlated with the available eight reference SG [79]. Another method of evolutionary analysis based on short (90-nucleotide) fragments (positions 73,662–73,751, GenBank accession no. MK333180) of the EP402R sequence was first described by Thanh et al. (2021), and this method was very successful when compared with HAI assay [80]. Kim et al. clustered the first ASFV isolated from South Korea into SG8 based on a long fragment (816 nucleotides) of the EP402R gene [64, 81]. We retrieved EP402R gene sequences of ASFV isolates with known SG and novel Asia isolates from GenBank (Table 2). Phylogenetic analyses of the EP402R genes was performed using the maximum-likelihood method with 1000 bootstrap replicates in MEGA5 software. The data indicated consistency between the 88 amino acid (CD2v amino acids 109–196, corresponding to ASFV isolate Lisbon/57, GenBank accession no. KM609344) (Fig. 2A) and 90 nucleotides (Fig. 2B) of EP402R-based phylogenetic trees. Thus, CD2v/EP402Rgenotyping was demonstrated as a simple method to group ASFV by serotype.
Table 2.
ASFV isolates used for the construction of phylogenetic tree based on partial EP402R gene (CD2V) gene nucleotide sequences
| Isolate | Host species | Year of isolation | Country | GenBank accession no | Serogroup | References |
|---|---|---|---|---|---|---|
| Lisbon/57 | domestic pig | 1957 | Portugal | KM609344 | SG1 | [37] |
| Diamang | Suidae | 1982 | Angola | KM609367 | SG1 | [37] |
| VL | Suidae | 1978 | Portugal | KM609390 | SG1 | [37] |
| Petit Enge | Suidae | 1985 | Democratic Republic of the Congo | KM609383 | SG2 | [37] |
| Silva-1 | Suidae | 1982 | Angola | KM609356 | SG2 | [37] |
| Dolisie-74 | Suidae | 1974 | Democratic Republic of the Congo | KM609368 | SG2 | [37] |
| K-49 | Suidae | 1949 | Zaire | KM609339 | SG2 | [37] |
| M-78 | Suidae | 1978 | Mozambique | KM609346 | SG3 | [37] |
| MK-200 | Domestic pig | 1978 | Mozambique | KM609347 | SG3 | [37] |
| TKF | Suidae | 1981 | Tanzania | KM609387 | SG3 | [37] |
| E-70 | Domestic pig | 1970 | Spain | KM609369 | SG4 | [37] |
| Lee | Bush pig | 1955 | Kenya | KM609377 | SG4 | [37] |
| Brazil-80 | Domestic pig | 1979 | Brazil | KM609364 | SG4 | [37] |
| Cu-80 | Domestic pig | 1980 | Cuba | KM609365 | SG4 | [37] |
| O-77 | Domestic pig | 1977 | USSR | KM609350 | SG4 | [37] |
| Malta | Domestic pig | 1978 | Malta | KM609380 | SG4 | [37] |
| T-67-PPK-1 | Suidae | NK | Tanzania | KM609386 | SG5 | [37] |
| TSP80 | Suidae | 1980 | Tanzania | KM609359 | SG5 | [37] |
| TS-7/27–230 | Suidae | 1986 | Tanzania | KM609388 | SG6 | [37] |
| TS7 | Suidae | 1984 | Tanzania | KM609358 | SG6 | [37] |
| Uganda | Suidae | 1984 | Uganda | KM609361 | SG7 | [37] |
| Nanyuki | Domestic pig | 1960 | Kenya | KM609382 | SG8 | [37] |
| Rhodesia | Suidae | 1986 | Zimbabwe | KM609354 | SG8 | [37] |
| Volgograd_2012/wb | Wild boar | 2012 | Russia | KM609363 | SG8 | [37] |
| China/Guangxi/2019/ domestic pig | Domestic pig | 2019 | China | MK670728 | SG8 | [38] |
| CN201801 | Domestic pig | 2018 | China | MH735142 | SG8 | [39] |
| China/Jilin/2018/boar | Wild boar | 2018 | China | MK214678 | SG8 | [40] |
| Pig/HLJ/2018 | Domestic pig | 2018 | China | MK333180 | SG8 | [82] |
| Georigia 2007/1 | Domestic pig | 2007 | Georgia | NC_044959 | SG8 | [8] |
| Krasnodar_2012/dom | Domestic pig | 2012 | Russia | KM609342 | SG8 | [37] |
| Korea/Pig/Paju1/2019 | Domestic pig | 2019 | South Korea | MT335858 | SG8 | [83] |
| VN/Pig/TH/411 | Domestic pig | 2019 | Vietnam | MN711757 | SG8 | [42] |
| Tver_2012/wb | Wild boar | 2012 | Russia | KM609360 | SG8 | [37] |
| Volgograd_2012/dom | Domestic pig | 2012 | Russia | KM609362 | SG8 | [37] |
| Zavidovo-2012 | Wild boar | 2012 | Russia | KM609392 | SG8 | [37] |
NK not known
Fig. 2.
Phylogenetic comparison of strains on the basis of CD2v protein/gene sequences. A Eighty-eight amino acids of the CD2v protein were analyzed using MEGA5, with four rate classes, and 100 bootstrap replicates, implemented in the maximum-likelihood method. Bootstrap values > 80% are indicated at appropriate nodes. B Ninety-nucleotide sequences of the CD2v protein-encoding gene EP402R were analyzed using MEGA5 software with 1000 bootstrap replicates, implemented in the neighbor-joining method. Bootstrap values > 80% are indicated at appropriate nodes. The serogroup status of ASFV is indicated. Recent Asian isolates are indicated using a closed circle (●)
Although CD2v and C-type lectins are important for mediating cross-protective responses in vivo, they cannot provide complete serotype-specific homologous protection, suggesting that other serotype-specific protective antigens have yet to be identified. Indeed, experimental subunit vaccines and vector vaccines that use or express ASFV proteins p30, p54, and CD2v have been shown to induce protection or partial protection against strong viral attacks. However, their role in causing serotype-specific reactions is unclear [84–86]. Malogolovkin believes that, when compared with p72 genotyping, HAI serology provides a method of ASFV typing that allows better differentiation of biologically relevant phenotypes [79].
Concluding remarks
ASFV is the only member of the family Asfarviridae. The process of establishing and developing genotyping methods for ASFV is associated with varying amounts of epidemiological evidence whereby a gene is used to track the virus in a particular region. Twenty-four ASFV genotypes have been identified on the basis of the p72-encoding (B646L) gene. All 24 genotypes have been reported in Africa, mainly distributed in East Africa and South Africa. Genotypes I and II are mainly distributed in Central Africa, and genotype I is mainly distributed in some West African countries, but there has been no report of an epidemic in North Africa [87]. In the first outbreaks outside Africa, the ASFV strains once prevalent in the 1960s in Western European countries, and in the 1970s in Caribbean countries and the USSR (now Ukraine) were caused by genotype I strains. At the end of the last century, all countries outside Africa, except Italy (specifically Sardinia, where genotype I strains remained endemic), had eradicated ASFV. The second escape from Africa occurred in 2007, and the virus has since spread through large areas of Russia, Eastern Europe, China, Southeast Asia, and recently to the Dominican Republic [6] and Haiti. This has been characterized as being due to the genotype II Georgia 2007 strain. However, because the B646L (p72-encoding) gene is relatively conserved, genotypic changes do not generally occur when a strain is endemic in a certain region. Therefore, p72 genotyping is the preferred method used for the identification of ASFV origin in case of introduction into new territories, as p72 genotype clustering can be used to help trace the source of the virus at the molecular level and aid with understanding the potential transmission routes and possible modes of transmission [71]. Other genotypes, such as those based on the CVR and IGR, can help to predict molecular epidemiological changes and evolution of ASFV because the differences are more significant. For example, the IGR sequence of the first ASFV strain in Chinese wild boar (China/Jilin/2018/boar, GenBank accession no. MK189457) has 10 base deletions compared with the prevalent strain in Chinese pigs (GenBank accession no. MH735144), suggesting that this strain may have a different origin [40]. However, it is undeniable that the rapid emergence of ASFV variant strains with different IGR sequences may have happened in a short space of time and with a sympatric distribution [88].
The serotyping method based on CD2v/C-type lectin signature sequencing allows better differentiation of biologically relevant phenotypes when compared with p72 genotyping [79]. It will be more helpful in study of the diversity and antigenic variability of ASFV strains, and provide key information for vaccine design. It is believed that with continuous in-depth study of ASFV genomes and typing technology, especially whole-genome sequencing and cross-protection experiment data, we will develop a clearer understanding of the evolution of ASFV.
Author contributions
HQ and SG had idea for the article, performed the literature search and data analysis, and drafted the manuscript; XW and ZW critically revised the work.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2017YFD0501801).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving animals were in accordance with the ethical standards of China Animal Health and Epidemiology Center (No. 2019-12). All applicable international, national, and institutional guidelines for the care and use of animals were followed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hailong Qu and Shengqiang Ge have contributed equally to this article.
Contributor Information
Xiaodong Wu, Email: wuxiaodong@cahec.cn.
Zhiliang Wang, Email: wangzhiliang@cahec.cn.
References
- 1.Galindo I, Alonso C. African swine fever virus: a review. Viruses. 2017 doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cwynar P, Stojkov J, Wlazlak K. African swine fever status in Europe. Viruses. 2019 doi: 10.3390/v11040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudreault NN, Madden DW, Wilson WC, Trujillo JD, Richt JA. African swine fever virus: an emerging DNA arbovirus. Front Vet Sci. 2020;7:215. doi: 10.3389/fvets.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajukumar K, Senthilkumar D, Venkatesh G, Singh F, Patil VP, Kombiah S, Tosh C, Dubey CK, Sen A, Barman NN, Chakravarty A, Dutta B, Pegu SR, Bharali A, Singh VP. Genetic characterization of African swine fever virus from domestic pigs in India. Transbound Emerg Dis. 2021;68:2687–2692. doi: 10.1111/tbed.13986. [DOI] [PubMed] [Google Scholar]
- 5.Sauter-Louis C, Forth JH, Probst C, Staubach C, Hlinak A, Rudovsky A, Holland D, Schlieben P, Goldner M, Schatz J, Bock S, Fischer M, Schulz K, Homeier-Bachmann T, Plagemann R, Klaass U, Marquart R, Mettenleiter TC, Beer M, Conraths FJ, Blome S. Joining the club: first detection of African swine fever in wild boar in Germany. Transbound Emerg Dis. 2021;68:1744–1752. doi: 10.1111/tbed.13890. [DOI] [PubMed] [Google Scholar]
- 6.Gonzales W, Moreno C, Duran U, Henao N, Bencosme M, Lora P, Reyes R, Nunez R, De Gracia A, Perez AM. African swine fever in the Dominican Republic. Transbound Emerg Dis. 2021 doi: 10.1111/tbed.14341. [DOI] [PubMed] [Google Scholar]
- 7.Blome S, Franzke K, Beer M. African swine fever—a review of current knowledge. Virus Res. 2020;287:198099. doi: 10.1016/j.virusres.2020.198099. [DOI] [PubMed] [Google Scholar]
- 8.Chapman DA, Darby AC, Da Silva M, Upton C, Radford AD, Dixon LK. Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus. Emerg Infect Dis. 2011;17:599–605. doi: 10.3201/eid1704.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King K, Chapman D, Argilaguet JM, Fishbourne E, Hutet E, Cariolet R, Hutchings G, Oura CA, Netherton CL, Moffat K, Taylor G, Le Potier MF, Dixon LK, Takamatsu HH. Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine. 2011;29:4593–4600. doi: 10.1016/j.vaccine.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cackett G, Matelska D, Sýkora M, Portugal R, Malecki M, Bähler J, Dixon L, Werner F. The African swine fever virus transcriptome. J Virol. 2020 doi: 10.1128/jvi.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallardo C, Mwaengo DM, Macharia JM, Arias M, Taracha EA, Soler A, Okoth E, Martín E, Kasiti J, Bishop RP. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes. 2009;38:85–95. doi: 10.1007/s11262-008-0293-2. [DOI] [PubMed] [Google Scholar]
- 12.Nix RJ, Gallardo C, Hutchings G, Blanco E, Dixon LK. Molecular epidemiology of African swine fever virus studied by analysis of four variable genome regions. Adv Virol. 2006;151:2475–2494. doi: 10.1007/s00705-006-0794-z. [DOI] [PubMed] [Google Scholar]
- 13.Hess WR, Cox BF, Heuschele WP, Stone SS. Propagation and modification of African swine fever virus in cell cultures. Am J Vet Res. 1965;26:141–146. [PubMed] [Google Scholar]
- 14.Malmquist WA. Propagation, modification, and hemadsorption of African swine fever virus in cell cultures. Am J Vet Res. 1962;23:241–247. [PubMed] [Google Scholar]
- 15.Malmquist WA. Serologic and immunologic studies with African swine fever virus. Am J Vet Res. 1963;24:450–459. [PubMed] [Google Scholar]
- 16.Coggins L. A modified hemadsorption-inhibition test for African swine fever virus. Bulletin des epizooties en Afrique. 1968;16:61–64. [PubMed] [Google Scholar]
- 17.Mansi W. The study of some viruses by the plate gel diffusion precipitin test. J Comp Pathol. 1957;67:297–303. doi: 10.1016/s0368-1742(57)80029-0. [DOI] [PubMed] [Google Scholar]
- 18.Cowan KM. Immunological studies on African swine fever virus. I. Elimination of the procomplementary activity of swine serum with formalin. J Invest Med. 1961;86:465–470. [PubMed] [Google Scholar]
- 19.Stone SS, Hess WR. Separation of virus and soluble noninfectious antigens in African swine fever virus by isoelectric precipitation. Virology. 1965;26:622–629. doi: 10.1016/0042-6822(65)90325-9. [DOI] [PubMed] [Google Scholar]
- 20.Vigário JD, Terrinha AM, Bastos AL, Moura-Nunes JF, Marques D, Silva JF. Serological behaviour of isolated African swine fever virus. Brief report. Archiv fur die gesamte Virusforschung. 1970;31:387–389. doi: 10.1007/bf01253773. [DOI] [PubMed] [Google Scholar]
- 21.Vigário JD, Terrinha AM, Moura Nunes JF. Antigenic relationships among strains of African swine fecre virus. Archiv fur die gesamte Virusforschung. 1974;45:272–277. doi: 10.1007/bf01249690. [DOI] [PubMed] [Google Scholar]
- 22.Wesley RD, Tuthill AE. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev Vet Med. 1984;2:53–62. doi: 10.1016/0167-5877(84)90048-5. [DOI] [Google Scholar]
- 23.Blasco R, Agüero M, Almendral JM, Viñuela E. Variable and constant regions in African swine fever virus DNA. Virology. 1989;168:330–338. doi: 10.1016/0042-6822(89)90273-0. [DOI] [PubMed] [Google Scholar]
- 24.Dixon LK, Wilkinson PJ. Genetic diversity of African swine fever virus isolates from soft ticks (Ornithodoros moubata) inhabiting warthog burrows in Zambia. J Gen Virol. 1988;69(Pt 12):2981–2993. doi: 10.1099/0022-1317-69-12-2981. [DOI] [PubMed] [Google Scholar]
- 25.Bastos AD, Penrith ML, Crucière C, Edrich JL, Hutchings G, Roger F, Couacy-Hymann E. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Adv Virol. 2003;148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- 26.Xiong D, Zhang X, Xiong J, Yu J, Wei H. Rapid genome-wide sequence typing of African swine fever virus based on alleles. Virus Res. 2021;297:198357. doi: 10.1016/j.virusres.2021.198357. [DOI] [PubMed] [Google Scholar]
- 27.Ley V, Almendral JM, Carbonero P, Beloso A, Viñuela E, Talavera A. Molecular cloning of African swine fever virus DNA. Virology. 1984;133:249–257. doi: 10.1016/0042-6822(84)90392-1. [DOI] [PubMed] [Google Scholar]
- 28.Tabarés E, Marcotegui MA, Fernández M, Sánchez-Botija C. Proteins specified by African swine fever virus. I. Analysis of viral structural proteins and antigenic properties. Adv Virol. 1980;66:107–117. doi: 10.1007/bf01314979. [DOI] [PubMed] [Google Scholar]
- 29.López-Otín C, Freije JM, Parra F, Méndez E, Viñuela E. Mapping and sequence of the gene coding for protein p72, the major capsid protein of African swine fever virus. Virology. 1990;175:477–484. doi: 10.1016/0042-6822(90)90432-q. [DOI] [PubMed] [Google Scholar]
- 30.Steiger Y, Ackermann M, Mettraux C, Kihm U. Rapid and biologically safe diagnosis of African swine fever virus infection by using polymerase chain reaction. J Clin Microbiol. 1992;30:1–8. doi: 10.1128/jcm.30.1.1-8.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Barreno B, Sanz A, Nogal ML, Viñuela E, Enjuanes L. Monoclonal antibodies of African swine fever virus: antigenic differences among field virus isolates and viruses passaged in cell culture. J Virol. 1986;58:385–392. doi: 10.1128/jvi.58.2.385-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanz A, García-Barreno B, Nogal ML, Viñuela E, Enjuanes L. Monoclonal antibodies specific for African swine fever virus proteins. J Virol. 1985;54:199–206. doi: 10.1128/jvi.54.1.199-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Morrissy CJ, Westbury HA. Strong sequence conservation of African swine fever virus p72 protein provides the molecular basis for its antigenic stability. Adv Virol. 1996;141:1795–1802. doi: 10.1007/bf01718302. [DOI] [PubMed] [Google Scholar]
- 34.Gonzague M, Roger F, Bastos A, Burger C, Randriamparany T, Smondack S, Cruciere C. Isolation of a non-haemadsorbing, non-cytopathic strain of African swine fever virus in Madagascar. Epidemiol Infect. 2001;126:453–459. doi: 10.1017/s0950268801005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubisi BA, Bastos AD, Dwarka RM, Vosloo W. Molecular epidemiology of African swine fever in East Africa. Adv Virol. 2005;150:2439–2452. doi: 10.1007/s00705-005-0602-1. [DOI] [PubMed] [Google Scholar]
- 36.Boshoff CI, Bastos AD, Gerber LJ, Vosloo W. Genetic characterisation of African swine fever viruses from outbreaks in southern Africa (1973–1999) Vet Microbiol. 2007;121:45–55. doi: 10.1016/j.vetmic.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Malogolovkin A, Burmakina G, Titov I, Sereda A, Gogin A, Baryshnikova E, Kolbasov D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis. 2015;21:312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge S, Liu Y, Li L, Wang Q, Li J, Ren W, Liu C, Bao J, Wu X, Wang Z. An extra insertion of tandem repeat sequence in African swine fever virus, China, 2019. Virus Genes. 2019;55:843–847. doi: 10.1007/s11262-019-01704-9. [DOI] [PubMed] [Google Scholar]
- 39.Ge S, Li J, Fan X, Liu F, Li L, Wang Q, Ren W, Bao J, Liu C, Wang H, Liu Y, Zhang Y, Xu T, Wu X, Wang Z. Molecular characterization of African swine fever virus, China, 2018. Emerg Infect Dis. 2018;24:2131–2133. doi: 10.3201/eid2411.181274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Ren Z, Wang Q, Ge S, Liu Y, Liu C, Liu F, Hu Y, Li J, Bao J, Ren W, Zhang Y, Xu T, Sun C, Li L, Wang S, Fan X, Wu Z, Huang B, Guo H, Wu X, Wang Z. Infection of African swine fever in wild boar, China, 2018. Transbound Emerg Dis. 2019;66:1395–1398. doi: 10.1111/tbed.13114. [DOI] [PubMed] [Google Scholar]
- 41.Kim HJ, Cho KH, Lee SK, Kim DY, Nah JJ, Kim HJ, Kim HJ, Hwang JY, Sohn HJ, Choi JG, Kang HE, Kim YJ. Outbreak of African swine fever in South Korea, 2019. Transbound Emerg Dis. 2020;67:473–475. doi: 10.1111/tbed.13483. [DOI] [PubMed] [Google Scholar]
- 42.Tran HTT, Truong AD, Dang AK, Ly DV, Nguyen CT, Chu NT, Nguyen HT, Dang HV. Genetic characterization of African swine fever viruses circulating in North Central region of Vietnam. Transbound Emerg Dis. 2021;68:1697–1699. doi: 10.1111/tbed.13835. [DOI] [PubMed] [Google Scholar]
- 43.Garigliany M, Desmecht D, Tignon M, Cassart D, Lesenfant C, Paternostre J, Volpe R, Cay AB, van den Berg T, Linden A. Phylogeographic analysis of african swine fever virus, Western Europe, 2018. Emerg Infect Dis. 2019;25:184–186. doi: 10.3201/eid2501.181535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nurmoja I, Petrov A, Breidenstein C, Zani L, Forth JH, Beer M, Kristian M, Viltrop A, Blome S. Biological characterization of African swine fever virus genotype II strains from north-eastern Estonia in European wild boar. Transbound Emerg Dis. 2017;64:2034–2041. doi: 10.1111/tbed.12614. [DOI] [PubMed] [Google Scholar]
- 45.Gallardo C, Fernández-Pinero J, Pelayo V, Gazaev I, Markowska-Daniel I, Pridotkas G, Nieto R, Fernández-Pacheco P, Bokhan S, Nevolko O, Drozhzhe Z, Pérez C, Soler A, Kolvasov D, Arias M. Genetic variation among African swine fever genotype II viruses, eastern and central Europe. Emerg Infect Dis. 2014;20:1544–1547. doi: 10.3201/eid2009.140554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simulundu E, Sinkala Y, Chambaro HM, Chinyemba A, Banda F, Mooya LE, Ndebe J, Chitanga S, Makungu C, Munthali G, Fandamu P, Takada A, Mweene AS. Genetic characterisation of African swine fever virus from 2017 outbreaks in Zambia: Identification of p72 genotype II variants in domestic pigs. Onderstepoort J Vet Res. 2018;85:e1–e5. doi: 10.4102/ojvr.v85i1.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Heerden J, Malan K, Gadaga BM, Spargo RM. Reemergence of African swine fever in Zimbabwe, 2015. Emerg Infect Dis. 2017;23:860–861. doi: 10.3201/eid2305.161195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bastos AD, Penrith ML, Macome F, Pinto F, Thomson GR. Co-circulation of two genetically distinct viruses in an outbreak of African swine fever in Mozambique: no evidence for individual co-infection. Vet Microbiol. 2004;103:169–182. doi: 10.1016/j.vetmic.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Hakizimana JN, Nyabongo L, Ntirandekura JB, Yona C, Ntakirutimana D, Kamana O, Nauwynck H, Misinzo G. Genetic analysis of African swine fever virus from the 2018 outbreak in South-Eastern Burundi. Front Vet Sci. 2020;7:578474. doi: 10.3389/fvets.2020.578474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achenbach JE, Gallardo C, Nieto-Pelegrín E, Rivera-Arroyo B, Degefa-Negi T, Arias M, Jenberie S, Mulisa DD, Gizaw D, Gelaye E, Chibssa TR, Belaye A, Loitsch A, Forsa M, Yami M, Diallo A, Soler A, Lamien CE, Sánchez-Vizcaíno JM. Identification of a new genotype of african swine fever virus in domestic pigs from Ethiopia. Transbound Emerg Dis. 2017;64:1393–1404. doi: 10.1111/tbed.12511. [DOI] [PubMed] [Google Scholar]
- 51.Quembo CJ, Jori F, Vosloo W, Heath L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound Emerg Dis. 2018;65:420–431. doi: 10.1111/tbed.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lubisi BA, Bastos AD, Dwarka RM, Vosloo W. Intra-genotypic resolution of African swine fever viruses from an East African domestic pig cycle: a combined p72-CVR approach. Virus Genes. 2007;35:729–735. doi: 10.1007/s11262-007-0148-2. [DOI] [PubMed] [Google Scholar]
- 53.Sumption KJ, Hutchings GH, Wilkinson PJ, Dixon LK. Variable regions on the genome of Malawi isolates of African swine fever virus. J Gen Virol. 1990;71(Pt 10):2331–2340. doi: 10.1099/0022-1317-71-10-2331. [DOI] [PubMed] [Google Scholar]
- 54.Irusta PM, Borca MV, Kutish GF, Lu Z, Caler E, Carrillo C, Rock DL. Amino acid tandem repeats within a late viral gene define the central variable region of African swine fever virus. Virology. 1996;220:20–27. doi: 10.1006/viro.1996.0281. [DOI] [PubMed] [Google Scholar]
- 55.Phologane SB, Bastos AD, Penrith ML. Intra- and inter-genotypic size variation in the central variable region of the 9RL open reading frame of diverse African swine fever viruses. Virus Genes. 2005;31:357–360. doi: 10.1007/s11262-005-3254-z. [DOI] [PubMed] [Google Scholar]
- 56.Owolodun OA, Bastos AD, Antiabong JF, Ogedengbe ME, Ekong PS, Yakubu B. Molecular characterisation of African swine fever viruses from Nigeria (2003–2006) recovers multiple virus variants and reaffirms CVR epidemiological utility. Virus Genes. 2010;41:361–368. doi: 10.1007/s11262-009-0444-0. [DOI] [PubMed] [Google Scholar]
- 57.Hakizimana JN, Kamwendo G, Chulu JLC, Kamana O, Nauwynck HJ, Misinzo G. Genetic profile of African swine fever virus responsible for the 2019 outbreak in northern Malawi. BMC Vet Res. 2020;16:316. doi: 10.1186/s12917-020-02536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilem A, Nurmoja I, Niine T, Riit T, Nieto R, Viltrop A, Gallardo C. Molecular characterization of African swine fever virus isolates in Estonia in 2014–2019. Pathogens. 2020 doi: 10.3390/pathogens9070582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun H, Jacobs SC, Smith GL, Dixon LK, Parkhouse RM. African swine fever virus gene j13L encodes a 25–27 kDa virion protein with variable numbers of amino acid repeats. J Gen Virol. 1995;76(Pt 5):1117–1127. doi: 10.1099/0022-1317-76-5-1117. [DOI] [PubMed] [Google Scholar]
- 60.Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, Dwarka R, Onashvili T, Albina E, Dixon LK. African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis. 2008;14:1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thoromo J, Simulundu E, Chambaro HM, Mataa L, Lubaba CH, Pandey GS, Takada A, Misinzo G, Mweene AS. Diagnosis and genotyping of African swine fever viruses from 2015 outbreaks in Zambia. Onderstepoort J Vet Res. 2016;83:a1095. doi: 10.4102/ojvr.v83i1.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simulundu E, Chambaro HM, Sinkala Y, Kajihara M, Ogawa H, Mori A, Ndebe J, Dautu G, Mataa L, Lubaba CH, Simuntala C, Fandamu P, Simuunza M, Pandey GS, Samui KL, Misinzo G, Takada A, Mweene AS. Co-circulation of multiple genotypes of African swine fever viruses among domestic pigs in Zambia (2013–2015) Transbound Emerg Dis. 2018;65:114–122. doi: 10.1111/tbed.12635. [DOI] [PubMed] [Google Scholar]
- 63.Simulundu E, Lubaba CH, van Heerden J, Kajihara M, Mataa L, Chambaro HM, Sinkala Y, Munjita SM, Munang'andu HM, Nalubamba KS, Samui K, Pandey GS, Takada A, Mweene AS. The epidemiology of African swine fever in "nonendemic" regions of Zambia (1989–2015): implications for disease prevention and control. Viruses. 2017 doi: 10.3390/v9090236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanna G, Dei GS, Bacciu D, Angioi PP, Giammarioli M, De Mia GM, Oggiano A. Improved strategy for molecular characterization of African swine fever viruses from Sardinia, based on analysis of p30, CD2V and I73R/I329L variable regions. Transbound Emerg Dis. 2017;64:1280–1286. doi: 10.1111/tbed.12504. [DOI] [PubMed] [Google Scholar]
- 65.Goller KV, Malogolovkin AS, Katorkin S, Kolbasov D, Titov I, Höper D, Beer M, Keil GM, Portugal R, Blome S. Tandem repeat insertion in African swine fever virus, Russia, 2012. Emerg Infect Dis. 2015;21:731–732. doi: 10.3201/eid2104.141792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Onzere CK, Bastos AD, Okoth EA, Lichoti JK, Bochere EN, Owido MG, Ndambuki G, Bronsvoort M, Bishop RP. Multi-locus sequence typing of African swine fever viruses from endemic regions of Kenya and Eastern Uganda (2011–2013) reveals rapid B602L central variable region evolution. Virus Genes. 2018;54:111–123. doi: 10.1007/s11262-017-1521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazur-Panasiuk N, Woźniakowski G. The unique genetic variation within the O174L gene of Polish strains of African swine fever virus facilitates tracking virus origin. Adv Virol. 2019;164:1667–1672. doi: 10.1007/s00705-019-04224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farlow J, Donduashvili M, Kokhreidze M, Kotorashvili A, Vepkhvadze NG, Kotaria N, Gulbani A. Intra-epidemic genome variation in highly pathogenic African swine fever virus (ASFV) from the country of Georgia. Virol J. 2018;15:190. doi: 10.1186/s12985-018-1099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dimmock NJ. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 70.Malmquist WA, Hay D. Hemadsorption and cytopathic effect produced by African Swine Fever virus in swine bone marrow and buffy coat cultures. Am J Vet Res. 1960;21:104–108. [PubMed] [Google Scholar]
- 71.Malogolovkin A, Kolbasov D. Genetic and antigenic diversity of African swine fever virus. Virus Res. 2019;271:197673. doi: 10.1016/j.virusres.2019.197673. [DOI] [PubMed] [Google Scholar]
- 72.Borca MV, Kutish GF, Afonso CL, Irusta P, Carrillo C, Brun A, Sussman M, Rock DL. An African swine fever virus gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption. Virology. 1994;199:463–468. doi: 10.1006/viro.1994.1146. [DOI] [PubMed] [Google Scholar]
- 73.Borca MV, Carrillo C, Zsak L, Laegreid WW, Kutish GF, Neilan JG, Burrage TG, Rock DL. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J Virol. 1998;72:2881–2889. doi: 10.1128/JVI.72.4.2881-2889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodríguez JM, Yáñez RJ, Almazán F, Viñuela E, Rodriguez JF. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J Virol. 1993;67:5312–5320. doi: 10.1128/jvi.67.9.5312-5320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruiz-Gonzalvo F, Rodríguez F, Escribano JM. Functional and immunological properties of the baculovirus-expressed hemagglutinin of African swine fever virus. Virology. 1996;218:285–289. doi: 10.1006/viro.1996.0193. [DOI] [PubMed] [Google Scholar]
- 76.Neilan JG, Borca MV, Lu Z, Kutish GF, Kleiboeker SB, Carrillo C, Zsak L, Rock DL. An African swine fever virus ORF with similarity to C-type lectins is non-essential for growth in swine macrophages in vitro and for virus virulence in domestic swine. J Gen Virol. 1999;80(Pt 10):2693–2697. doi: 10.1099/0022-1317-80-10-2693. [DOI] [PubMed] [Google Scholar]
- 77.Galindo I, Almazán F, Bustos MJ, Viñuela E, Carrascosa AL. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology. 2000;266:340–351. doi: 10.1006/viro.1999.0080. [DOI] [PubMed] [Google Scholar]
- 78.Chapman DAG, Tcherepanov V, Upton C, Dixon LK. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J Gen Virol. 2008;89:397–408. doi: 10.1099/vir.0.83343-0. [DOI] [PubMed] [Google Scholar]
- 79.Malogolovkin A, Burmakina G, Tulman ER, Delhon G, Diel DG, Salnikov N, Kutish GF, Kolbasov D, Rock DL. African swine fever virus CD2v and C-type lectin gene loci mediate serological specificity. J Gen Virol. 2015;96:866–873. doi: 10.1099/jgv.0.000024. [DOI] [PubMed] [Google Scholar]
- 80.Thanh THT, Duc TA, Viet LD, Van HT, Thi NC, Thi CN, Thi NH, Vu DH. Rapid identification for serotyping of African swine fever virus based on the short fragment of the EP402R gene encoding for CD2-like protein. Acta Vet. 2021;71:98–106. doi: 10.2478/acve-2021-0007. [DOI] [Google Scholar]
- 81.Kim SH, Kim J, Son K, Choi Y, Jeong HS, Kim YK, Park JE, Hong YJ, Lee SI, Wang SJ, Lee HS, Kim WM, Jheong WH. Wild boar harbouring African swine fever virus in the demilitarized zone in South Korea, 2019. Emerg Microbes Infect. 2020;9:628–630. doi: 10.1080/22221751.2020.1738904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen X, He X, Zhang X, Zhang X, Liu L, Guan Y, Zhang Y, Bu Z. Genome sequences derived from pig and dried blood pig feed samples provide important insights into the transmission of African swine fever virus in China in 2018. Emerg Microbes Infect. 2019;8:303–306. doi: 10.1080/22221751.2019.1565915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HJ, Cho KH, Ryu JH, Jang MK, Chae HG, Choi JD, Nah JJ, Kim YJ, Kang HE. Isolation and genetic characterization of African swine fever virus from domestic pig farms in South Korea, 2019. Viruses. 2020 doi: 10.3390/v12111237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Argilaguet JM, Pérez-Martín E, López S, Goethe M, Escribano JM, Giesow K, Keil GM, Rodríguez F. BacMam immunization partially protects pigs against sublethal challenge with African swine fever virus. Antiviral Res. 2013;98:61–65. doi: 10.1016/j.antiviral.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Barderas MG, Rodríguez F, Gómez-Puertas P, Avilés M, Beitia F, Alonso C, Escribano JM. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Adv Virol. 2001;146:1681–1691. doi: 10.1007/s007050170056. [DOI] [PubMed] [Google Scholar]
- 86.Gómez-Puertas P, Rodríguez F, Oviedo JM, Brun A, Alonso C, Escribano JM. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology. 1998;243:461–471. doi: 10.1006/viro.1998.9068. [DOI] [PubMed] [Google Scholar]
- 87.Mulumba-Mfumu LK, Saegerman C, Dixon LK, Madimba KC, Kazadi E, Mukalakata NT, Oura CAL, Chenais E, Masembe C, Stahl K, Thiry E, Penrith ML. African swine fever: update on Eastern, Central and Southern Africa. Transbound Emerg Dis. 2019 doi: 10.1111/tbed.13187. [DOI] [PubMed] [Google Scholar]
- 88.Kim SH, Lee SI, Jeong HG, Yoo J, Jeong H, Choi Y, Son K, Jheong WH. Rapid emergence of African swine fever virus variants with different numbers of a tandem repeat sequence in South Korea. Transbound Emerg Dis. 2021;68:1726–1730. doi: 10.1111/tbed.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]