Abstract
Background:
The impact of abusive alcohol consumption on human health is remarkable. According to the World Health Organization (WHO), approximately 3.3 million people die annually because of harmful alcohol consumption (the figure represents around 5.9% of global deaths). Alcohol Use Disorder (AUD) is a chronic disease where individuals exhibit compulsive alcohol drinking and present negative emotional states when they do not drink. In the most severe manifestations of AUD, the individuals lose control over intake despite a decided will to stop drinking. Given the multiple faces and the specific forms of this disease, the term AUD often appears in the plural (AUDs). Since only a few approved pharmacological treatments are available to treat AUD and they do not apply to all individuals or AUD forms, the search for compounds that may help to eliminate the burden of the disease and complement other therapeutical approaches is necessary.
Methods:
This work reviews recent research focused on the involvement of epigenetic mechanisms in the pathophysiology of AUD. Excessive drinking leads to chronic and compulsive consumption that eventually damages the organism. The central nervous system is a key target and is the focus of this study. The search for the genetic and epigenetic mechanisms behind the intricated dysregulation induced by ethanol will aid researchers in establishing new therapy approaches.
Conclusion:
Recent findings in the field of epigenetics are essential and offer new windows for observation and research. The study of small molecules that inhibit key epienzymes involved in nucleosome architecture dynamics is necessary in order to prove their action and specificity in the laboratory and to test their effectivity and safety in clinical trials with selected patients bearing defined alterations caused by ethanol.
Keywords: Epigenetics, AUD (Alcohol Use Disorders), DNA methylation, histone modifications, noncoding RNAs, DNMT inhibitors, HDAC inhibitors, epidrugs.
1. INTRODUCTION
1.1. The Brain Aud Model. Genetic and Epigenetic Influences
Different terms referring to problematic or pathologic alcohol consumption, such as alcoholism, alcohol abuse, alcohol addiction or alcohol dependence, have been used to reach a defined diagnosis that could help apply appropriate and specific treatments. However, given the difficulty of establishing the precise diagnosis (multiple clinical manifestations and states after excessive and chronic alcohol drinking) and, due to the numerous individual factors that influence pathologies related to excessive alcohol drinking, it has been difficult to arrive at that objective. The Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5) [1], designates the term Alcohol Use Disorder (AUD) for a chronic disease where individuals exhibit compulsive alcohol intake and present negative emotional states when they do not drink. To establish this diagnosis of AUD, the person must meet at least two criteria out of eleven listed by the DSM-5. Therefore, hundreds of different combinations and, consequently, forms of AUD are possible [2]. In the most severe manifestations of AUD, the individuals lose control over consumption despite a decided will to stop drinking. However, some controversial issues have arisen as to whether the definition of AUD can help to delimit and characterize clinical entities related to abusive alcohol ingestion [3].
The impact of abusive alcohol consumption on human health is notable. According to a recent World Health Organization (WHO) report, approximately 3,3 million people (around 5,9% of global deaths) die because of harmful alcohol consumption. Besides, about 139 million DALYs losses (disability-adjusted life years), representing roughly 5% of the worldwide burden of disease and injury, were caused by alcohol ingestion [4].
Only a few pharmacological treatments are available to treat AUD (for example, disulfiram, opioid antagonists, acamprosate, baclofen, or topiramate). In many cases, patients do not respond to standard therapies or are reluctant to any treatment for different reasons. Due to the scarce availability of efficient pharmacological therapies, the search for compounds that may help to eliminate the burden of the disease and to complement other therapy approaches is necessary. It relies on a profound knowledge of the many abnormal biochemical adaptations that contribute to the development of the disorder [5, 6]. Despite limited successful results, efforts continue to find suitable drugs acting on causes and effects observed in AUD, including relapse, withdrawal, stress, or anxiety. Nowadays, clinical studies in phases I and II explore the effectiveness of drugs in the treatment of AUD. Examples of compounds investigated are modulators of monoamine neurotransmission, glutamate and GABA modulators, neuropeptide antagonists, or modulators of neuroimmune responses [7]. Some drugs administered for the treatment of mental disorders have proved their efficiency in the treatment of AUD. For example, aripiprazole, an antipsychotic drug, and agomelatine, a melatonergic antidepressant, showed beneficial effects by preventing alcohol relapse and craving [8, 9].
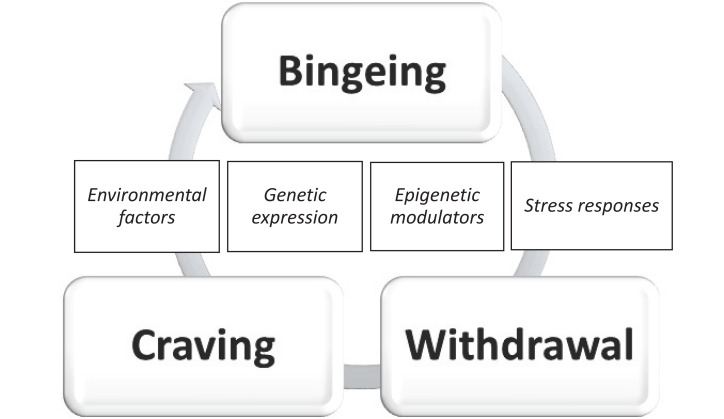
AUD can be considered a progressive and chronic disease of the brain that commences with a binge drinking pattern that stimulates brain reward areas. Prolonged and intense abuse leads to the downregulation of reward systems, deranged stress reactions, withdrawal accompanied by negative emotions, and craving (Fig. 1). Pari passu, the complex brain neurocircuitry, pathologically adapts to the stimuli and becomes altered and less efficient [10-13]. AUD often emerges in a context of comorbid mood disorders where the manifestation of anxiety, relapse, craving, or alexithymia come from an entangled pathological condition that threatens the health state of the patients and indubitably conditions treatment strategies [14, 15].
Fig. (1).
The AUD brain cycle. The figure represents the progressive and chronic character of AUD, where external factors, stress responses, gene variants, and epigenetic modulation play a crucial and interconnected role [11-16]. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
A relationship between environment and genetics is evident, though not thoroughly analyzed and understood. Alcohol intake, an environmental factor, is a condition sine qua non for the appearance of AUD. It is essential to consider that other environmental circumstances of the individual can modulate alcohol exposure (the environmental elements). Certain external stimuli, such as social control, in different forms, ameliorates genetic influence, whereas easy access to the substance or less social control tends to make more feasible the participation of the genetic impact on the development of AUD [16]. Genetic influence on AUD appearance can be assessed by analyzing global genetic weight (For example, studies on twins and relatives in a family, or ethnic groups) and investigating the participation of specific genes with molecular techniques. Both approaches add valuable information [16]. Nonetheless, the advantage of using molecular techniques to analyze some genes has the disadvantage of missing the perspective of reaching accurate information on probably hundreds of genes participating and affecting the development or maintenance of AUD coordinately. Given the polygenic character of AUD, only complementary and multidisciplinary approaches may help to unravel the mechanisms and processes implicated [17].
Genome-wide association (GWAS) studies applied to alcohol intake and AUD point that AUD is a polygenic, moderately heritable condition where dangerous drinking is a fundamental factor but not enough to be considered cause [17]. Also, since only partial correlation between alcohol dependence (according to DSM-IV diagnosis) and alcohol consumption is evident, a clear difference in the genetic influence when comparing damaging and non-damaging effects of alcohol ingestion was deducted [18].
The Association of human gene expression with the development of the alcohol-induced disease is evident. A poorly active variant of the enzyme aldehyde dehydrogenase type 2 (ALDH2), may exert a protective role against consumption since it leads to a slower metabolism of acetaldehyde (a product of oxidation of ethyl alcohol by alcohol dehydrogenase, ADH). Consequently, acetaldehyde accumulates and produces unpleasant symptoms after alcohol ingestion [19]. Other molecular studies and meta-analyses on polymorphisms affecting specific genes, in humans, rodents and insect models, associated with AUD and alcohol drinking patterns, include elements of neurotransmitter pathways. These enzymes participate in the metabolism of ethyl alcohol, both oxidative and non-oxidative. A few examples are kinases, phosphatases, phospholipases acting in signaling routes, transcription factors, transport proteins, proteins controlling circadian rhythms, proteins that participate in stress responses, insulin action, or immune responses. [18, 20-24].
Besides the role of genetic factors and their influence on damaging alcohol drinking habits and AUD phenotypes, epigenetic mechanisms play an essential task. In this work, we review recent findings on principal epigenetic mechanisms involved in AUD and alcohol drinking behavior. Besides, we will describe candidate drugs that may modulate epigenetic events associated with AUD.
2. MOLECULAR BASIS OF EPIGENETICS AND EPITRANSCRIPTOMICS
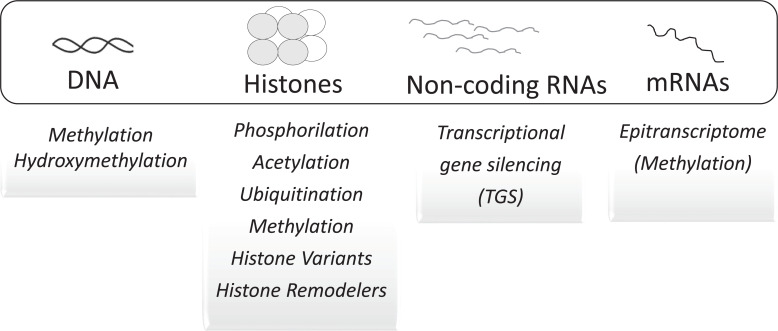
The terms epigenetics and epigenotype were coined by C.H. Waddington [25] before J.D. Watson, F.H.C. Crick, M.H.F. Wilkins, and R.E. Franklin described the molecular structure of DNA [26-28]. The terminology proposed was established after the relationship between genotype and phenotype. Its etymological origin is the word epigenesis, previously concocted by embryologists, as opposed to preformation. The term means a gradual differentiation process leading to the formation of functional organs and organ systems from seeds, fertilized eggs, or spores [29]. The concept of epigenetics has evolved [30] and today includes the molecular events that modulate the expression of genes and phenotypes of cells without altering the nucleotide sequence of DNA templates [31]. It applies to the control of gene expression through mitotic and meiotic heritable modifications (Fig. 2) that reshape the structure and organization of chromatin, without altering its nucleotide sequence [32-36]. It also applies to transcriptional and post-transcriptional control of gene expression by noncoding RNAs (ncRNAs) [37].
Fig. (2).
This figure depicts the principal epigenetic biochemical mechanisms that modulate gene expression and cellular phenotypes. These molecular mechanisms do not act as separated events, but in many cases, are related and highly coordinated [51-53]. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
Regulation of transcriptional outcomes in a cell relies on a sophisticated and coordinated net of molecular interactions that need a suited and dynamic organization of the chromatin structure. Assembling and remodeling of chromatin architecture, temporary interactions, and accessibility by different proteins, such as transcription factors (TF), histone and DNA modifiers, depends on the state and location of nucleosomes and on reversible epigenetic modifications that shape their structure and functionality. Understanding the dynamics of chromatin function requires methodological approaches to decipher nucleosome disposition and epigenetic traits that modify their conformation and performance. Some examples are single-cell Hi-C, high-throughput microscopy, Fluorescence resonance energy transfer, Cryo-electron microscopy, chemical-induced proximity assays, nucleosome occupancy, methylome sequencing, chemical and enzymatic DNA treatment, clustered regulatory interspaced short palindromic repeats-CRISPR-cas editing, hybrid site-targeting proteins and transcription activator-like effector protein (TALE)-targeting [38-47].
Observations in humans and animal experiments point to the involvement of the environment in gene expression in the so-called Developmental Origins of Health and Disease [48]. The model focuses on the prenatal period, from conception and early childhood. During this critical time of development, growth and organogenesis take place and affect future health states in life [49]. The mechanisms underlying the impact of environmental factors on the developmental program of an organism are not fully understood. Still, a modification of epigenetic modulation is considered a key element in the pathologies that originate after early exposure to adverse external conditions and toxics [50, 51].
Next, we describe the underlying molecular and functional framework of principal epigenetic mechanisms.
2.1. DNA Methylation and Demethylation
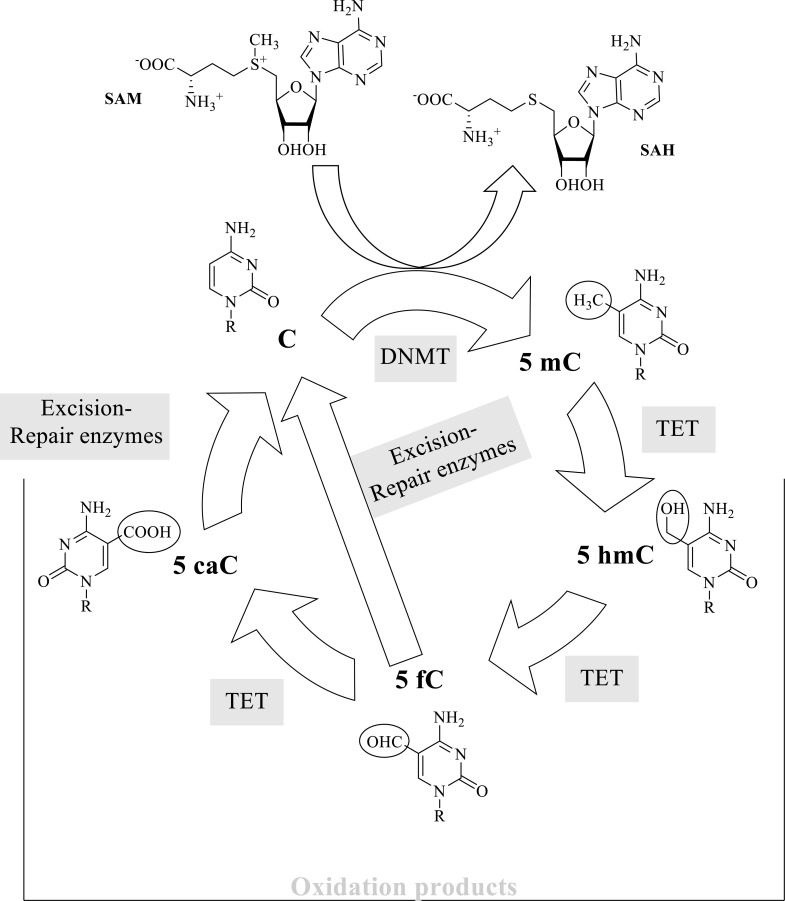
DNA methylation consists of the addition of a methyl group to a purine or pyrimidine base in DNA. In eukaryotes, it mostly occurs at the fifth position of cytosines in DNA to form 5-methylcytosine (5mC) in CpG (5´→3´direction) dinucleotides within specific DNA regions situated in a promoter, within a gene, and in repeated sequences [45, 54]. Also, cytosine methylation at other positions, not CpG islands, has physiological relevance [55, 56]. 5mC was detected as a novel pyrimidine present in DNA from thymus and named by R. Hotchkiss as “epicytosine.” He also proposed that the new peak detected in his chromatogram corresponded to methylated cytosine [57]. Cytosine modifications may alter local conformations of the double helix structure and affect the interaction of proteins that regulate DNA metabolism (see for review [58]). Methylation of cytosines is linked to essential functions, comprising inactivation of the X chromosome, genomic imprinting, silencing of retrotransposons, or structural integrity of chromosomes, among others [59-65]. The effect of DNA methylation varies depending on the context where it occurs. Methylation of enhancers and promoter regions usually silences transcription, whereas methylation of some gene sequences may induce augmented gene expression [66]. This reaction, catalyzed by DNMTs (DNA methyltransferases, EC 2.1.1.37), should be contemplated in a metabolic cycle that includes methylation, demethylation by oxidation, and excision-repair reactions. The intermediary metabolites of these reactions have a putative functional impact [62] (Fig. 3).
Fig. (3).
The biochemical process of DNA methylation and other enzymatic modifications at position 5 of a cytosine base. C, cytosine; 5mC, 5-methylcytosine and its oxidation products: 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine and 5caC, 5-carboxycytosine; DNMT, DNA methylases; TET, The Ten-Eleven Translocation family of dioxygenases; SAM, S-Adenosylmethionine (methyl donor); SAH, S-Adenosylhomocysteine [47, 59, 61-63, 65, 67].
Cytosine methylation influences chromosome stability and mostly associates with the deactivation of transcription in physiological and pathological states [45, 61, 67].
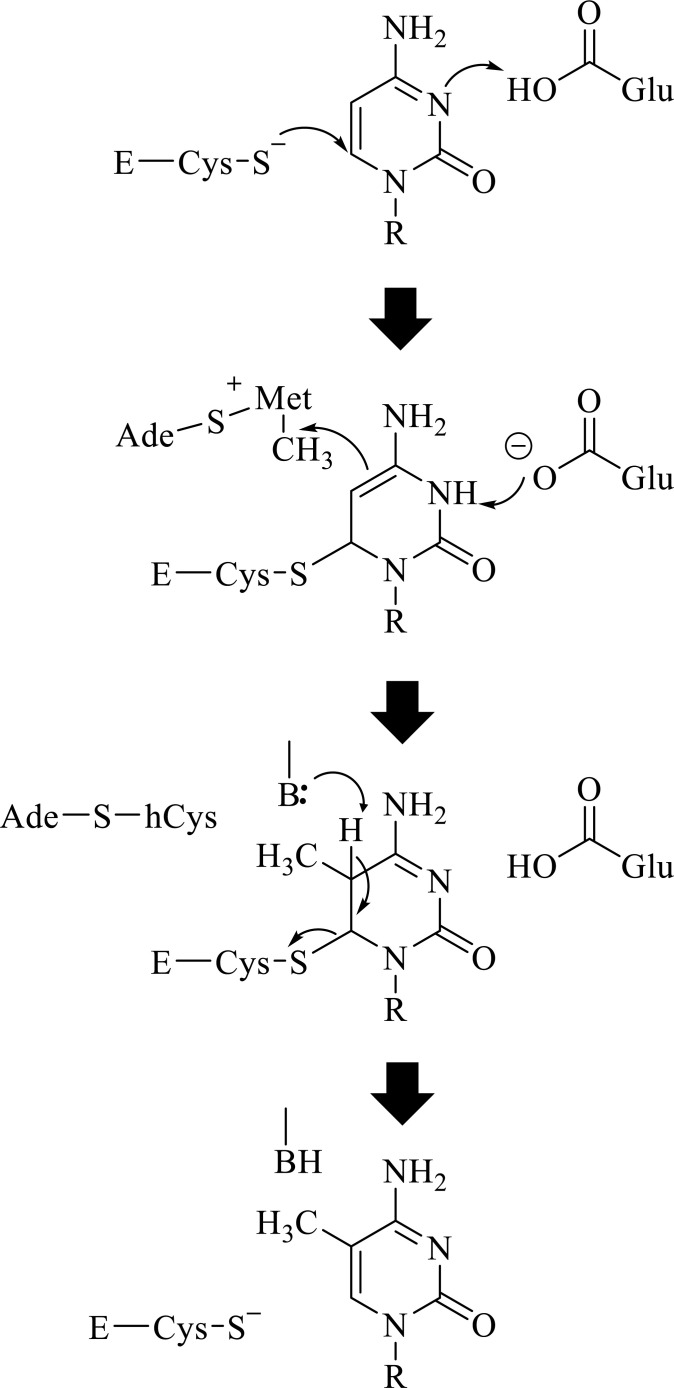
Mammals have different types of methylases: DNMT1, DNMT2, DNMT3 (A, B, C, and L) [58, 65, 67]. Functionally, they form two families: a group composed of DNMT3A, DNMT3B, and DNMT3C (which catalyzes the de novo methylation of DNA) and DNMT1 (responsible for DNA methylation maintenance). The second group displays no DNA methylation catalytic activity and includes DNMT2 and DNMT3L [58, 65, 67]. Cytosine-5 DNMTs exhibit a similar catalytic mechanism. In brief, it commences with the cysteine thiol nucleophilic attack on cytosine carbon at position 6, aided by a glutamic acid residue in the vicinity. Next, a methyl group is transferred from the donor S-adenosylmethionine to atom 5 of the cytosine rings to give 5-methylcytosine. The transfer of the methyl is also assisted by a basic group that extracts a proton from carbon 5 (Fig. 4) [67].
Fig. (4).
General catalytic mechanism of DNMTs. The figure illustrates the transfer of a methyl group from the donor, S- adenosylmethionine, to carbon 5 of the cytosine ring. The nucleophilic attack of carbon in position 6 of the pyrimidine ring by a cysteine thiol group of the enzyme facilitates the transfer. A glutamic residue and a basic residue further favor the global catalytic mechanism. Glu, glutamate residue in motif VI of the enzyme; E-Cys-S, cysteine residue from the enzyme Motif IV. Ade-S-Met, S-adenosylmethionine; Ade-S-hCys, S-adenosylhomocysteine. B represents a basic residue from the enzyme. Curved arrows indicate the movement of electrons [58, 65, 67].
DNMTs reach specific locations within promoter regions, gene bodies, or long-terminal repeat (LTR) of retrotransposons, depending on the accessibility that the chromatin environment (histone modifications and protein factors) permits [68-72]. Conservation of methylation patterns after each cell division represents the molecular basis of the epigenetic memory associated with the DNA methylation profiles [67]. Maintenance of methylation requires cycles of replication and the recognition of the hemimethylated DNA strand by UHRF1, a multi-functional protein that connects DNA methylation and histone post-translational modifications. DNMT1 methylates the newly synthesized DNA [53]. A group of MBD (Methyl Binding Domain) proteins that serve as “readers” of methylated DNA (methylases would represent the “writers”) recognize methylated cytosines within a DNA sequence. This family of proteins binds sites of methylation and also interacts with enzymes that modify histones and with noncoding RNAs [58, 73], thereby having an essential role in regulatory mechanisms of transcription and maintenance of chromatin performance and integrity [58].
DNA methylation and DNA demethylation are closely related. DNA Methylation profiles can be “lost” or “diluted” through cycles of DNA replication or specific catalyzed reactions. As the reverse reaction of methylation has no functional significance, the pathway for demethylation mostly comprises oxidation reactions of the methyl group [59] (Fig. 3). A group of TET (Ten Eleven Translocation) methyl-cytosine dioxygenase enzymes (EC 1.14.11.n2) iteratively oxidize 5mC to 5-hydroxymethylcytosine, 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Excision repair enzymes reverse the nucleobase to the unmethylated state (5hmC) [59, 62, 63]. The name TET comes after a fusion protein that appeared as a consequence of a translocation between chromosomes 10 and 11 found in a patient with acute myeloid leukemia [74]. The protein oxidizes 5mC [75, 76]. TET enzymes belong to a family of alpha-ketoglutarate and Fe(II)-dependent proteins that utilize molecular oxygen [76]. These enzymes can be considered as modifiers of DNA methylation patterns and are responsible for active demethylation that renders 5mC oxidation products [62]. Reading methylated cytosines and other modified cytosines is relevant to establish their localization and their putative role as epigenetic markers. One recognition method is the bisulfite conversion that selectively detects C. Heating DNA in the presence of sodium bisulfite deaminates cytosine to uracil but does not modify 5mC [77]. On the other hand, the use of tailored proteins, including transcription-activator-like effectors (TALEs), engineered to associate to specific nucleobases, may also aid to selectively detect relevant modifications taking place at the fifth position of the pyrimidine ring [47, 78, 79].
DNA methylation is a crucial and abundant molecular mark with multiple functional consequences depending on the localization of CpG islands, the methylation density, or the type of cell bearing the trait. Therefore, it is a complex signal which may trigger multiple outcomes [73]. Also, oxidation products may serve to alter DNA conformation and, thus, its function. For example, 5fM influences transcription processes by regulating chromatin remodeling through changes in the geometry of the helix that facilitate negative supercoiling [80].
Writers, readers, and modifiers of methylated DNA coordinate their activity and mutually influence each other on specific frames of DNA metabolism [58, 73]. The harmonized and cooperative performance, together with other epigenetic attributes, is greatly influenced by external factors that may derange the balance and lead to disease [50, 51].
2.2. Histone Modifications, Histone Variants, and Remodeling of Nucleosomes
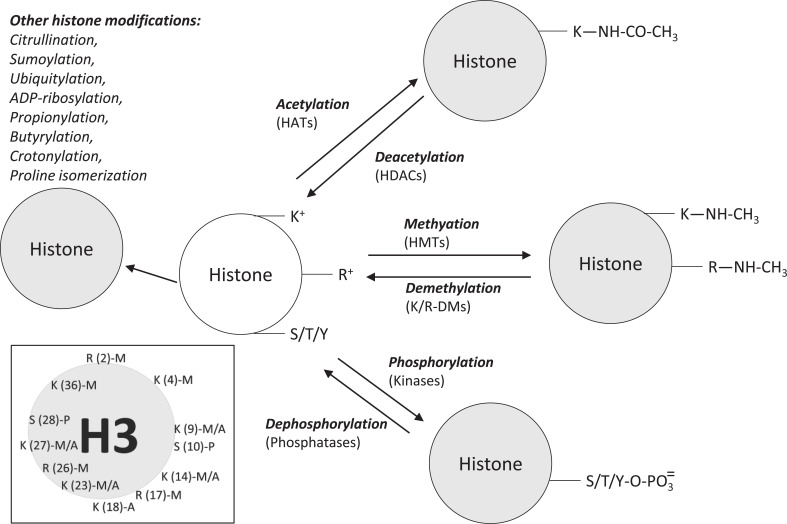
Eukaryotic chromatin is a dynamic structure with two principal components, DNA and histones. Histones group in an octamer composed of four different types of proteins in dimers: H2A, H2B, H3, and H4. H1 is a linker histone. DNA is wrapped around the octamer to form the nucleosome [81]. The structure, where DNA and histones associate is very dynamic and supports the changes and activities driven by numerous proteins that define and characterize the functional state of DNA expression on a space-time framework [82]. Different enzymatic activities are responsible for the active and inactive chromatin states [34]. Several profiles of reversible histone modifications and packaging (conditioned by the action of writers, for example, a histone acetylase; readers, such as transcription factors with domains that specifically bind (read) histone modifications and erasers, for example, a histone demethylase) would produce specific biological effects and support the basis for the existence of a so-called histone code hypothesis [83]. Next, we describe some of the main modifications of histones with epigenetic significance (Fig. 5) [84-90].
Fig. (5).
Major post-translational modifications of histones. Abbreviations: HATs, histone acetyltransferases; HDCAs, histone deacetylases; HMTs, histone methylases; K7R-DMs, histone demethylases. S, serine; K, lysine; R, arginine; Y, tyrosine. The inset shows the main modifications observed on histone 3. S(10)-P is a phosphorylated serine situated at position tenth; K(9)-M/A is lysine at position ninth that can be methylated or acetylated (see text for details) [84-90]. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
2.2.1. Histone Acetylation
With this covalent and reversible modification, an acetyl group from an acetyl-CoA molecule binds to the ε-amino group of lysine residues positioned at different sites of the protein. The histone acetyltransferases family (HATs, EC 2.3.1.48) catalyzes this reaction. HAT bi-substrate enzymes group a large family of proteins exhibiting different cellular localization, histone, and non-histone substrate specificity and catalytic mechanism of action [91]. Acetylation of basic residues decreases the positive charges and hence weakens the interaction histones-DNA and facilitates transcription. The reverse reaction is the hydrolysis of the acetyl group. This reaction increases positively charged residues. The histone deacetylases family (HDACs, EC 3.5.1.98) catalyzes this reaction by a metal-dependent hydrolysis mechanism [92]. Both enzymatic activities were landmark discoveries [93-95] that broadened the research field searching for catalytic activities affecting the functional states of euchromatin and heterochromatin [34].
The principal readers of acetylated lysins in histones are a group of proteins called the Bromodomain and Extra-Terminal (BET) family. After binding the acetylated sites, transcription factors and RNA pol II go to the assigned sites to initiate the transcription of particular genes [96-98].
2.2.2. Histone Methylation
Histone methylases (EC 2.1.1.-) catalyze the methylation of basic residues (mostly, K and R) on histones H3 and H4, by using S-adenosyl-methionine as the methyl donor. Lysine may appear mono, di, or tri-methylated at the N6 atom; arginine may be mono or di-methylated at the guanidinium group of the lateral chain [99]. Demethylases (erasers) catalyze the elimination of methyl marks. Based on sequence comparison and mechanism of action, two groups of histone demethylases have been described: FAD-dependent amine oxidases (for example, [Histone-H3]-N(6), N(6)-dimethyl-L-lysine (4) FAD-dependent demethylase, EC 1.14.99.66) and Fe (II) and alpha-ketoglutarate-dependent demethylases (EC 1.14.11.-) [100, 101].
Methylation triggers different outcomes, depending on the site it occurs. For example, methylation of lysine at position 4 of histone H3 is associated with gene activation, whereas methylation of lysine at position 9 correlated with gene inactivation [102]. These findings indicate that it is necessary to precisely locate the methylation sites on histones to signal where the activity should take place [103].
Critical methyl lysine readers include proteins with Tudor domains. These proteins interact with di- and tri-methylated sites and with some methylated arginines. They also bind chromodomains that recognize trimethylated lysine, MBT (Malignant Brain Tumor) domains that bind to mono- and demethylated sites, PWWP domains, PHDs domains (Plant homeodomain) and WDR domains [98, 104-107]. Methylation readers dock at methylated sites through specific binding domains and elicit responses such as the recruiting of other proteins that participate in the chromatin remodeling or DNA transcription [104].
2.2.3. Histone Phosphorylation
In histones, as in other proteins, phosphorylation takes place mainly at serine, threonine, and tyrosine residues. Phosphorylation is a reversible covalent modification that introduces mass and charge to the histone structure and influences, in collaboration with other histone modifications, chromatin structure, and function. For example, TRPM6-cleaved kinase phosphorylates histones. This modification is associated with a decrease in the methylation of arginine residues close to the phosphorylated sites [108]. Phosphorylation of serine in position ten on histone 3 foments acetylation of lysine at location 14 on the same histone [109]. Also, in the context of histone modifications, the action of casein kinase II is tightly connected to a deacetylation complex [110].
Many protein kinases (EC 2.7.-.-) are involved in the phosphorylation of histones. A few examples are Casein kinase II, Protein kinases ATM and ATR, PKCβ, PKCδ, JAK2, AMPK, CDK2, or PAK2. Protein phosphatases (EC 3.1.-.-) hydrolyze the phosphate ester bond. Some examples are the HTP-C phosphatase complex, EYA1/3 phosphatase, or protein phosphatase 1 [85].
Readers of phosphorylated histones recognize the phosphorylated sites and affect chromatin activity. Some readers are the 14-3-3ζ protein that binds to phosphoserine in histone 3. Other proteins that recognize phosphorylation positions are Survivin through a BIR (Baculovirus Inhibitor of apoptosis protein Repeat) domain, 53BP1, and Crb2 with a BRCT (BRCA1 C Terminus) domain, or the transcription regulator Fe65 with a PTB (Phosphotyrosine Binding) domain [98, 111, 112].
2.2.4. Other Histone Modifications
Other histone marks that add topographical and functional capacities to chromatin dynamics are also introduced after translation. Some examples are citrullination, sumoylation, ubiquitylation, ADP-ribosylation, propionylation, butyrylation, crotonylation, or proline isomerization [84, 87, 88, 113].
2.2.5. Crosstalk and Collaboration
Changes in the structure and function of chromatin do not only depend on histone post-translational modifications. Other elements also participate. Sequence variants of histone proteins [114], as well as nucleosome remodelers [115], add their action. Histone modifications are coordinated in a sophisticated and context-dependent manner and respond to the needs of the cell [86]. Writers, erasers, and readers cooperate and crosstalk to start a given outcome (gene activation or inactivation, DNA repair, or apoptosis.) [34]. The enzymes that modify the histones often group in complexes. The Polycomb (PcG) and the Trithorax (TrxG) are examples of protein groups formed by co-worker proteins that collaborate to keep “off” and “on” states of gene activity, depending on localization, cell type or temporal settings [52, 116, 117].
2.3. Noncoding RNAs (ncRNAs)
A large family of non-coding RNAs (ncRNAs) exhibits multiple functions and endeavors related to both transcription and post-transcription modulatory events. Some are short chains of less than 30 nucleobases (short interfering RNAs, siRNAs, PIWI-RNAs, piRNAs and microRNAs, miRNAs), others are medium-long chains or snoRNAs (~40-200 nucleotides), enhancer RNAs (eRNAs), polymers of up to 2000 nucleotides, and long-chain (more than 200 nucleotides) RNAs (lncRNAs), in some cases made of thousands of nucleotides [37, 118, 119]. Four main types of ncRNAs are involved in epigenetic mechanisms: siRNAs, piRNAs, miRNAs (that build up small RNA-Argonaute complexes), and lncRNAs [37].
2.3.1. Short ncRNAs (siRNAs, piRNAs, and miRNAs)
siRNAs derive from dsRNA and consist of double strands built by around 20-30 base pairs that participate in post-transcriptional control after being hydrolyzed by the endonuclease Dicer (an RNaseIII) [120]. Once processed, siRNA takes part and aids in building a complex of proteins (Argonaute-2, Dicer and TRBP, Transactivating Response RNA-Binding Protein) showing different enzymatic activities, named RISC (RNA-induced silencing complex), that targets and silences specific mRNAs [121]. Intensive research and clinical trials in various phases focus on the use of RNA interference (target delivery, safety, efficiency, dosage...) as an efficient tool to treat specific pathologies [122].
In addition to their contribution to gene silencing in the cytoplasm, siRNAs also act in the nucleus to modulate transcriptional gene silencing (TGS) in some situations. The molecular mechanisms of siRNA-dependent TGS are not entirely defined but relate to methylation of histone 3 (on Lys at positions 3 and 27) and methylation of selected DNA promoter sequences [123-125]. A long list of genes involved in pathologies such as cancer, immunological disturbances, HIV-1 infection, cardiac disease, are transcriptionally modulated by siRNAs (see more details in [124]). The transcriptional action of siRNA requires the contribution of various proteins, including Argonaute 1, DNA methyltransferase (DNMT1), and histone deacetylase (HDCA1) [126].
PIWI-interacting RNAs (piRNAs) are small nuclear RNAs that associate with PIWI (P-element induced wimpy testis) proteins, a subclass of the Argonaute family, to assemble mature transcriptional and post-transcriptional ribonucleoprotein silencing complexes [127]. The piRNAs are highly present in the germline of many organisms and are responsible for keeping the integrity of its genome and hence fertility by controlling transposon activity [127-129].
The precursors of both siRNAs and miRNAs are double-stranded RNAs. On the other hand, piRNAs, a diverse collection of thousands of different sequences, starting with a uracil base at the 5´end, result from the biogenesis process in which piRNA precursor sequences arise from single-stranded RNAs that, after trimming, bind to PIWI proteins [129, 130].
We are far to fully understand the precise role of piRNAs in transcription silencing and their transgenerational influence. Still, some mechanisms indicate their participation in transcription repression and the extension of their functionality to generations [131, 132]. piRNAs repress transcription by decreasing RNApol II activity and by increasing heterochromatin protein 1 (HP1) and trimethylated histone H3 at Lys9 [133].
The third group of small ncRNAs is microRNAs (miRNAs). Hundreds of miRNAs in various species are involved in post-transcriptional control by marking 3´UTR (untranslated) mRNA sequences [134] through base complementarity. miRNAs are first synthesized as primary miRNA transcripts and afterward modified by the nuclear ribonuclease Drosha and by cytoplasmic endonuclease Dicer. Later, these sequences bind to Argonaute proteins to build the RISC complex [134, 135].
MicroRNAs can target the activity of DNA methylases by regulating their expression, as it is the case for the control of miR29b on the expression of DNMT3 and even TET enzymes [136]. Also, they may control the synthesis of histone-modifying proteins, as is the case in the regulation of lysine-specific demethylase by miR-137 in the rat brain amygdala [137].
The implication of ncRNAs in the modulation of the genetic and epigenetic mechanisms is under extensive investigation. The expression of small ncRNAs is under genetic and epigenetic surveillance [134, 135]. Still, on the other hand, these molecules also have an active and direct role in both genetic and epigenetic events by controlling the performance of many epienzymes responsible for DNA methylation and reversible modifications on histones and RNAs [135].
2.3.2. Long ncRNAs (lncRNAs)
A large family of lncRNAs displays its activities in many organisms, including humans. According to a study based on the analysis of RNA sequences from human RNA-seq libraries, approximately 58000 transcripts corresponded to lncRNAs [119]. LncRNAs genes have different genome localizations: for example, intronic sequences of protein-coding genes, intergenic sequences, promoter sequences, antisense transcripts (complementary to a protein-coding sequence situated on the opposite strand) [37, 119, 138-140]. LncRNAs participate in many different cell activities, from the regulation of chromatin activity to transcription, maintenance of functional mRNAs, control of translation, and guiding of post-translation events [140].
LncRNAs take part in nuclear events, for example, by recruiting or obstructing the access of epienzymes that modify the chromatin (methylation ubiquitination) and, as a result, influencing the interaction between histones and specific DNA loci [141]. Additionally, lncRNAs may regulate transcription by directly interacting with mRNAs or by associating with RNA pol II transcription complexes. In the cytoplasm, lncRNAs are involved in the stability of mRNAs, regulation of translation, and post-translational events [140].
The expression “junk” DNA, generally referred to as noncoding DNA, is unfortunate today. Many not translated sequences have a central regulatory role in gene expression and are not “junk” DNA. Thousands of noncoding transcripts do not act separately but build nets of pathways of functional interaction coordinated to serve specific control and maintenance of cellular activity.
2.4. Epitranscriptomics
This term refers to covalent reversible modifications of different classes of RNAs, including methylation of adenine ring at positions 6 (m6A) and 1 (m1A), methylation of cytosine at position 5 (m5C), pseudourylation, deamination of nucleobase, and their impact on regulatory mechanisms. These modifications affect different types of RNA (rRNA, tRNA, mRNA, small nucleolar RNA, circ-RNAs, lncRNAs) and regulate gene expression by affecting mechanisms of splicing, stability, turnover, and transport of transcripts) [142]. The modifications are written, eliminated, and read by other molecules and add new pathways of influence on cell performance and fate [142-144].
The most widespread and conserved modification is methylation of N6 adenosine (m6A) that occurs on specific sequence contexts of RNAs [145]. In humans, an enzymatic heterodimer complex (the writer) that post-transcriptionally methylates adenosines is called m6A methyltransferase (MTC) and requires S-adenosyl-methionine (SAM) as the methyl donor [145, 146]. Two proteins generate the complex, the catalytic subunit Mettl3 (EC 2.1.1.348) and Metl14, the allosteric and structural adaptor protein, crucial for identifying the substrates [147]. The writing complex requires additional proteins for successful activity [148].
Oxidation reactions by RNA demethylases (EC 1.14.11.53,54), the erasers, eliminate methylation marks. Two examples are FTO (Fat mass and Obesity-associated protein) and ALKBH5 (which is part of the Fe [II]-alpha-ketoglutarate-dependent dioxygenases family) [148, 149]. FTO eliminates the methyl group by forming intermediary oxidation products, such as hydroxymethyl and formyl derivatives, whereas ALKBH5 acts on hidroxymethyladenosine to generate non-methylated adenosine [148].
Direct readers of m6A sites recognize different types of RNA and bear a YTH (YT521-B Homology) domain. Some are present in the nucleus (YTHDC1), others in the cytosol (YTHDF1, YTHDF2, YTHDF3), or in some cases, in both compartments (YTHDC2) [148]. Since RNA methylation favors the unfolding and formation of single-stranded structures, many other proteins, called indirect readers, can bind RNA and modulate its stability and turnover [148].
3. AUD AND EPIGENETICS
Changes and alterations of epigenetic mechanisms are frequently associated with human diseases (aging-coupled pathologies, inflammation, neurodegeneration, metabolic disturbances, cancer, or autoimmune disorders, among others) [32, 48, 52, 87, 150, 151]. However, the precise mechanisms at play and their collaboration with other factors, yet far from fully understanding, represent a challenge for both researchers and clinicians.
We will review the main modifications of epigenetic factors associated with alcohol consumption and alcohol-induced pathologies. Also, we will describe novel and tailored pharmacological therapies aimed at preventing or treating AUD, focusing on the allostatic alterations observed in the central nervous system.
Alcohol drinking influences epigenetic mechanisms responsible for adaptation changes of several brain circuits, mostly linked to stress management and reward, including the prefrontal cortex, the hypothalamus-hypophysis-adrenal axis (HPA), the mesolimbic dopamine pathways and the endogenous opioid pathways [10-13, 152, 153]. Methods that allow the analysis of genome-wide DNA methylation and histone modification profiles on single cells or heterogeneous samples provide valuable information to ascertain the position and functional role of modification marks on specific genes or regulatory sequences [154, 155]. Table 1 summarizes studies on the observed effect of ethanol on epigenetic mechanisms in different experimental models.
Table 1.
Representative studies that report epigenetic changes associated with alcohol exposure and alcohol brain damage (see main text).
| Subject | Sample | Main Observation | References |
|---|---|---|---|
| Various subjects | - | - | - |
| - | Different samples and exposures to ethanol | Review studies on epigenetic changes associated with AUD | [152, 157, 185, 188-195] |
| Humans | - | - | - |
| - | Post-mortem PFC (prefrontal cortex) | Methylation in 86,588 CpG islands (in males with AUDs) appeared augmented. | [196] |
| - | Post-mortem cerebellum | Downregulation of TET1 demethylase and augmented methylation of the delta subunit of GABAA (γ-Aminobutyric acid) receptor promoter. | [197] |
| - | Post-mortem brain amygdala and superior frontal cortex | Sequences of endogenous retroviruses and genes appeared hypomethylated in alcoholics. Also, histone 3 (H3K4) trimethylation increased. | [198] |
| - | Peripheral blood | AUD patients showed reduced expression of DNA methylases DNMT-3A and DNMT-3B, associated with hypermethylation of DNA. No change in DNMT1 expression was observed. | [162] |
| - | Lymphoblasts | CpG islands are hypermethylated on different locations (genes DOCK10, CENPK, HRAS, CDKR1, among others) in subjects with AUD. | [199] |
| - | Peripheral blood | The authors observed hypermethylation of promoter of the Dopamine transporter, Vasopressin, ANP (Atrial natriuretic Peptide), and GABA (γ-Aminobutyric acid) genes in AUD individuals. | [158, 200, 201] |
| - | Post-mortem temporal lobe | Increase in H3K9ac associated with prenatal alcohol exposure | [169] |
| - | Post-mortem brain regions | AUD induces down-regulation (e.g., miR-130a) and up-regulation (e.g., miR-377, miR-379, miR-604) of multiple miRNAs. | [177, 202, 203] |
| - | Post-mortem amygdala | The authors reported the up-regulation of lncRNA BDNF-AS in AUD individuals. | [181] |
| - | Peripheral blood | miRNA serum levels appeared to increase in young AUD subjects. These variations in miRNA levels are associated with brain alterations. | [174] |
| Rodents | - | - | - |
| - | Brain structures and other tissues | Ethanol intake and ethanol withdrawal-induced alterations of DNA methylation and methylation and acetylation profiles of histones associated with several genes: e.g., Arc (activity-regulated cytoskeleton-associated protein), spinophilin, postsynaptic density 95 and TrkB (tropomyosin receptor kinase B), proteoglycans and glial-cell derived neurotrophic factor (GDNF). | [168, 170-172, 204-207] |
| - | Brain amygdala | Ethanol exposure during adolescence epigenetically reduced CREB (cAMP-response element-binding) protein and increased DNMT (DNA methyltransferase) activity. NPY (neuropeptide Y) and BDNF (brain-derived neurotrophic factor) genes appear hypermethylated. | [164-167] |
| - | Hippocampus and PFC | Acetate from the oxidative metabolism of ethanol generates acetyl-CoA by the action of acetyl-CoA synthetase 2 (ACSS2) bound to chromatin and is used as a substrate to acetylate histones. Therefore, the metabolism of alcohol contributes directly to epigenetic changes in the brain. | [208] |
| Subject | Sample | Main Observation | References |
| - | Different brain regions | Down-regulation (e.g., miRNA-125a-p) and up-regulation (e.g., mir-130, miR-124 families) of multiple miRNAs after alcohol exposure. | [176, 178, 180] |
| - | Hippocampus and cortex | The authors noted reduced DNA methylation due to decreased levels of DNMT1 and DNMT3A in a mouse model of prenatal exposure to alcohol. | [161] |
| - | Brain amygdala | Binge post-natal ethanol exposure induced in adulthood increased miR-137 and down-regulated lysine demethylases Lsd1 and Lsd1+8a genes. | [137] |
| - | Pituitary | Alcohol prenatal exposure elevates mRNAs of DNA methylases (DNMT1 and DNMT3b) and histone deacetylases (DDAC2, HDAC4, and G9a). | [163] |
| Monkeys | - | - | - |
| - | Temporal lobe | A decrease in acetylation of histones H3 and H4 on different aminoacidic positions in a model of prenatal alcohol exposure | [169] |
Acute alcohol intoxication leads in general to relaxed and active chromatin due to the downregulation of DNA and histone methylation. In contrast, chronic exposure triggers adaptation mechanisms that lead to chromatin compaction. After withdrawal, chromatin tends to the condensed state due to the upregulation of DNA and histones [156].
DNA methylation condenses the structure of chromatin and usually halts transcription [51], although the location of the modification may affect the final functional outcome [66]. As far as the DNA methylation mechanisms concerns, alcohol affects this epigenetic mark in different ways, depending on the target affected, experimental model studied, route and pattern of administration (binge or continuous), time of exposure (acute or chronic), or dosage. DNA methylation is a biomarker of alcohol consumption [157]. Epigenome-wide association studies of methylation of CpG islands in different human cohorts have revealed specific methylation marks related to heavy drinking [158]. Alcohol interferes with the metabolism of SAM and SAH and thus affects DNA methylation potential. Chronic administration of ethanol to rats induced a decrease of SAH and was associated with an increased DNA methylation capacity in the cerebellum, whereas in liver tissues, DNA methylation was downregulated [159, 160]. Reduction of DNA methylases DNMT and DNMT3A in a rat model of fetal alcohol spectrum disorder (FASD), impaired DNA methylation in hippocampus and cortex tissues [161]. Other studies in human alcoholics have reported the downregulation of DNMT-3A and DNMT-3B [162]. Alcohol administration during the prenatal period activated, in adult rats pituitary, the transcription of DNMT 1 and DNMT3B genes [163]. Some studies show that exposure to ethanol during adolescence, in rodent models, upregulated DNA methyltransferase activity and induced hypermethylation of genes coding for neuropeptide Y (NPY) and BDNF (brain-derived neurotrophic factor) [164-167].
The functional state of chromatin regulates DNA transcription, and the catalytic action of histone acetylases (HACs) and histone deacetylases (HDACs) modify such a functional state [152]. Examination of HDACs gene expression in different rat tissues and organs, including the brain, showed that after intermittent alcohol administration, the expression of HDACs increased in the liver but decreased in the heart and brain amygdala. Also, binge drinking in humans and rats increased HDAC expression in monocytes from peripheral blood [168]. Prenatal alcohol contact produced both an increase in H3K9ac (acetylation of Lys9 in Histone 3) in the dentate gyrus and decreased acetylation of H3K9, H3K27, H4K12 and H4K16 in other brain regions [169]. The same authors determine that prenatal exposure to alcohol triggers a general scenery of epigenetic modifications that, depending on the brain region and development stage, includes diminished DNA and histone methylation and increased histone acetylation [169]. In the prefrontal cortex of withdrawal seizure resistant (WSR) mice, the analysis of histone methylation showed that, after alcohol intoxication, trimethylation of histone three on Lys 4 (H3K4me3) appears globally reduced, whereas H3K27me3 augmented. Both changes inhibited the expression, among others, of proteoglycan genes and genes affecting calcium signaling pathways [170]. In rats, knockdown of amygdala histone demethylase Kdm6b or Arc (activity-regulated cytoskeleton-associated protein) enhancer RNA, produced anxiety without exposition to alcohol.
Interestingly, the same molecular signatures emerge after binge ethanol exposition during adolescence [171]. After fifteen days of ethanol exposure, rats subjected to ethanol withdrawal suffered depression-like behavior connected to an increased expression of deacetylase HDCA2 and a reduction of H3K9ac (acetylated lysine 9 of histone 3) in the hippocampus [172]. In rats, chronic treatment (for fourteen days) with ethanol activated the expression of histone deacetylases HDCA2 and HDCA3. Also, a downregulation of the acetylation of H3 associated with the GABRA 1 (gamma-aminobutyric acid receptor alpha subunit) promoter region and diminished expression of the GABA A receptor alpha subunit materialize [173].
Determination of miRNA serum levels has shown a clear relationship between the concentration of some miRNAs in serum (increased) and brain structure (an increase of volumetric measurements in several brain areas) and function (neuropsychological performance altered) in AUD patients [174]. The study points to the possible use of determination of some miRNAs in peripheral blood as biomarkers of alcohol intoxication and damage. In a mouse model, Let-7, a family of miRNA, modulates the activation of several genes involved in neuroplasticity, neuroinflammation, or chromatin structural rearrangements, after exposure to ethanol [175]. MicroRNA-137 increased its level in adult rats exposed to alcohol during adolescence. Also, the expression of target genes of miR-137, such as Lsd1 (Lysine demethylase 1), decreased. Additional analysis revealed that both antagonism of miR-137 and infusion of Lsd1 siRNA into the central nucleus of the amygdala prevented the behavioral effects induced by miR-137 [137]. By using differential expression analysis, Nunez et al. [176] showed that activated expression of many miRNAs in mice and the human brains (for example, families let-7, miR-101, miR-221, miR-1952) positively correlated with activated expression of mRNAs (transcription of genes involved in synaptic function, inflammatory responses and intracellular transport of vesicles) after alcohol drinking. Analyzing the expression of noncoding miRNAs and comparing it with the coding transcriptome on specific brain locations is relevant to unveil the interaction networks of different elements of control in the context of AUD [177].
MicroRNAs may exert a neuroprotective role against cell death induced by the increased oxidative milieu generated after ethanol exposure in neurons. Both in vivo and in vitro experiments [178] reported that alcohol down-regulated the expression of miRNA-125a-5p in the rat prefrontal cortex. This reduction unleashed and augmented the expression of the ascorbic acid transporter SVCT2. Therefore, ascorbic acid entered the cell through the transporter and exerted its antioxidant and protective role against ROS (reactive oxygen species). Thus, the miRNA had a neuroprotective function. However, SVCT2 is not an exclusive target of miRNA-125a-5p, since other proteins, such as PSD-95 (postsynaptic density membrane-bound guanylate kinase) and p-38 MAPK (Map-kinase), are also regulated by this miRNA [178, 179]. Another example of miRNA down-regulation by ethanol in the prefrontal cortex concerns miR-130. This reduction closely associates with the regulation of ion channel function by interfering with the expression of proteins such as ITPR2 (inositol 1,4,5-trisphosphate receptor type 2) and ATP1A2 (alpha two isoform subunit of the N+-K+ -ATPase) [177].
Micro RNAs are critical regulators of the functional state of the transcriptome and the proteome by cooperation and support mechanisms established between their different families. A study that compared the role of miRNAs on the expression of target proteins, affected by chronic alcohol intermittent treatments, provided ex-vivo evidence that points to a global landscape of miRNA down and up-regulation that motivate different stages of AUD [180].
Long noncoding RNAs are also essential regulators of gene expression. For example, in the context of human alcohol intoxication, the up-regulation of the lncRNA antisense sequence (BDNF-AS lncRNA) ameliorates BDNF (brain-derived neurotrophic factor) expression within the amygdala. The cause of this effect is the recruitment of a histone lysine methyltransferase that trimethylates H3, situated in BDNF gene regulatory sequences, on lysine 27. In the same study, the authors established that BDNF-AS seems to be regulated by diminished adenine N-6 methylation [181]. The above is a clear example of the mutual influence of different epigenetic and epitranscriptomic mechanisms at play.
In a human genome-wide association study for alcohol drinking, the authors analyzed a lncRNA gene (locus LOC100507053), overlapping with the ADH (alcohol dehydrogenase) cluster, which presented five SNPs (single nucleotide polymorphisms) in a specific population. The findings indicate that ADH1B and LOC100507053 may regulate drinking behavior in a coordinated manner [182]. The lncRNA MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1) is a transcript retained in the nucleus that governs the synaptic formation by influencing the expression of specific genes, such as Neuroligin1 (NLGN1), a cell surface protein, and synaptic cell adhesion molecule 1 (SynCAM1) [183]. The expression of MALAT-1 is augmented in some brain regions (cerebellum, brain stem, and hippocampus) of human alcoholics and therefore makes this RNA candidate actor for synaptic adaptation after alcohol drinking [184].
Table 1 summarizes representative reports related to the alteration of epigenetic mechanisms after ethanol exposure in different experimental models. Changes in epigenetic regulators induce and maintain unbalanced the functioning of many neurotransmitter systems of the brain and feed mechanisms that adapt the brain to the presence of alcohol. Different studies show contradictory results concerning the upregulation and downregulation of epigenetic processes caused by ethanol exposure. That may be due to differences in dosage, patterns of administration, subjects, organ regions, or duration of administration. Also, epigenetic mechanisms may act differently on several targets. They can regulate activating or inhibitory genes, making intricate outcomes dependent on numerous factors that crosstalk and need both individual analysis and comprehensive integration [32, 144, 185-187].
Some players are emerging as critical regulators of the altered epigenetic landscape observed in AUD. The next chapter describes some of the drugs that control epigenetic mechanisms triggered by writers, readers, and erasers. Their modulatory role may be relevant to treat or ameliorate the damage caused by AUDs.
4. PHARMACOLOGICAL MODULATION OF EPIGENETICS: TOWARDS NEW TREATMENTS FOR AUD
This section focuses on the structure, function, and experimental testing of a few candidate drugs that may modify altered epigenetic events in AUD. The use of drugs that inhibit or regulate the epigenetic machinery has advanced in the field of cancer therapy [32, 96, 104, 209], and some epidrugs tested in this field will also be presented. As far as the AUD concerns, we need to investigate the possible application of regulatory epidrugs further.
4.1. Inhibitors of DNA Methylation
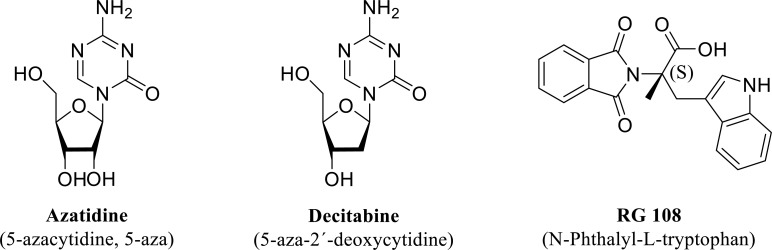
DNA methylation and demethylation are metabolic marks that regulate gene expression. Drugs that induce DNA hypomethylation are essential within the realm of cancer therapeutics. However, other pathologies may benefit from the use of drugs that control the methylation state of DNA. A family of azanucleoside DNMT inhibitors, 5´-azacytidine and decitabine (5-aza-2´-deoxycytidine) (Fig. 6), once uptaken by cells, are metabolically activated to form their respective nucleotides before incorporation into DNA by substituting cytosine nucleotides during DNA replication. DNA methyltransferase 1 recognizes the pair azacytosine-guanine and starts the nucleophilic attack of the pyrimidine ring. However, since position five corresponds to a nitrogen atom instead of a carbon atom, the reaction is not resolved, the enzyme and the base stay covalently bound, and the methylation reaction is halted [210]. These compounds are potent inhibitors of DNA methylation; however, they suffer rapid degradation by the catalytic action of cytidine deaminase [211].
Fig. (6).
Structure of the so-called first-generation nucleoside (Azacytidine and Decitabine) and non-nucleoside (RG 108) DNMT inhibitors experimentally tested in animals exposed to ethanol (see text for details).
On the other hand, the third compound showed in Fig. (6), RG 108 (N-Phthalyl-L-tryptophan) is a non-nucleoside DNMT inhibitor that binds directly to the catalytic site of DNMT1 and blocks its action [212]. This compound is a non-specific inhibitor of DNMT, serves as a substrate of cytochrome CYP2C19, and stimulates cytochromes CYP1A2 and CYP1B1 [213]. Modification of different moieties of this compound has provided a series of non-nucleoside analogs with improved inhibitory effect on DNMT1 [212].
In two experimental models of alcohol intake carried out in mice, binge-like drinking in the dark (DID) and chronic intermittent every other day (EOD), Ponomarev et al. showed that decitabine reduced the intake of alcohol and this observation associated with changes found in the brain reward pathway [214]. Also, the administration of 5-azacytidine to mice intermittently exposed to alcohol for eight weeks hampered excessive alcohol use by the rodents [215]. In a rodent model of intermittent ethanol administration during adolescence, treatment with 5-azacytidine in the adult age reversed alcohol-induced DNA hypermethylation of Npy (neuropeptide Y) and Bdnf (brain-derived neurotrophic factor) genes. It rescinded alcohol-provoked anxiety-like behavior and alcohol drinking [166]. Furthermore, the inhibitor RG108 reduced alcohol intake in alcohol-dependent rats during the abstinence period [216].
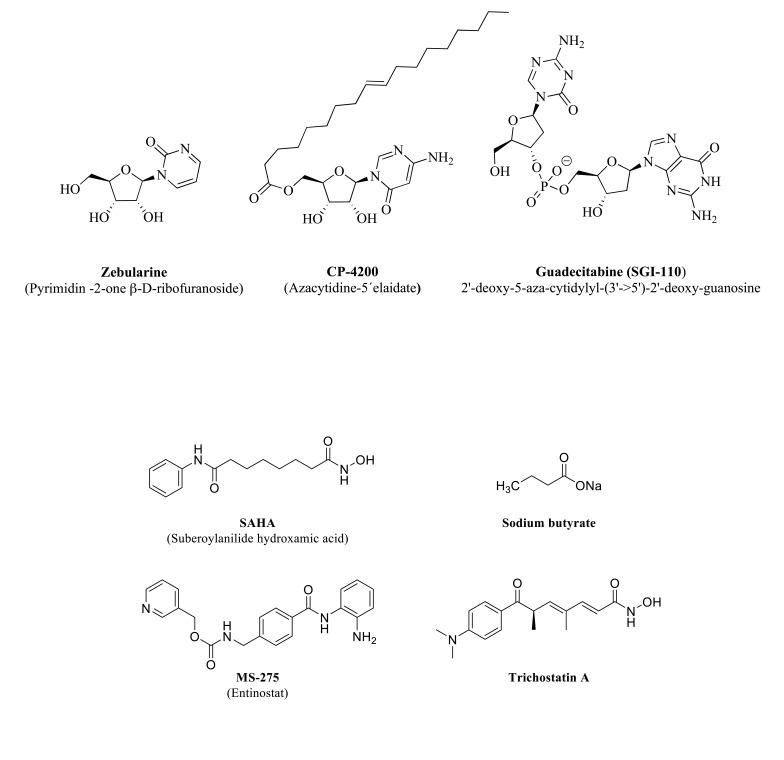
DNA methylation maybe not the only epigenetic mechanism affected by alcohol. However, given its possible involvement in the control of gene expression, it may be worth the analysis of other inhibitors. There are available new drugs tested in clinical trials in the context of therapies directed against some malignancies, with characteristics of interest, including specificity for a type of DNMT, no or little off-target activity, enough half-life, metabolic stability or optimal bioavailability [209]. Fig. (7) pictures some examples of DNMT inhibitors that can attend to the need for more directed and specific treatments to attenuate or halt ethanol intake in AUD patients. Zebularine (4 deoxyuridine), for example, inhibits both DNMTs and cytidine deaminase enzymes and gains more stability than other nucleoside compounds due to the inactivation of the hydrolytic enzyme cytidine deaminase. CP-4200 is an elaidic acid ester that behaves as an azacytidine prodrug with higher bioavailability and uptake by cells [217]. Guadecitabine (compound SGI-110) is a dinucleotide also resistant to metabolic degradation by cytidine deaminase [218].
Fig. (7).
Structure of representative DNMT inhibitors with improved characteristics concerning bioavailability, metabolic stability, compared to first-generation DNMT inhibitors, currently under experimental testing in different pathologies (see text for details).
4.2. HDAC Inhibitors
The involvement of chromatin modifications in adaptation mechanisms associated with ethanol brain damage has gained
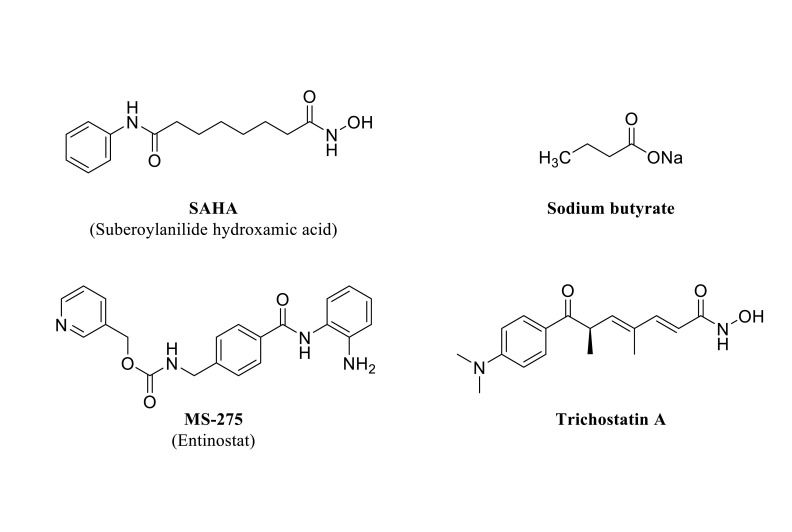
relevance with the experimental assessment of inhibitors in different experimental models. Fig. (8) shows some examples of HDCA inhibitors discussed in the text. One of the first compounds tested as an HDCA inhibitor, also known as Vorinostat, was SAHA (Suberoylanilide hydroxamic acid). This compound inhibits several HDCA isoforms acting as a chelator of the zinc atom present in the catalytic center of the Zn2+-dependent enzyme classes [219].
Fig. (8).
Structure of representative HDCA inhibitors experimentally tested in animals exposed to ethanol (see text for details).
Ethanol withdrawal induces alteration of the epigenetic landscape that may lie behind some of the neuroadaptive changes observed in this state. In an experimental model, rats were fed with ethanol for 16 days. Ethanol withdrawal increased HDCA2 protein, decreased histone H3-K9 acetylation, and caused GABA hyposensitivity in the VTA (ventral tegmental area). These observations were absent when alcohol-fed rats were previously treated with SAHA [206]. In a mouse model of chronic exposure, ethanol withdrawal also induced a reduction of H3-K9 acetylated histones and GABA (A-α1) receptor subunits in VTA that were prevented with both SAHA and Trichostatin A pretreatments [220]. Rats withdrawn form ethanol after 15 days of ethanol liquid diet administration showed depression-like behavior, increased HDCA2 mRNA expression, and reduced H3-K9 levels within the hippocampus. Treatment with SAHA during the withdrawal period significantly recovered histone acetylation to control levels and attenuated depression-like behavior [172].
The brain amygdala is a relevant center for the control of anxiety-like and alcohol drinking conducts. Trichostatin A is a compound structurally related to SAHA and presents a similar mechanism of action by reversibly inhibiting HDCA activity [221]. In a study carried out with alcohol-preferring (P-rats) and non-preferring rats (NP-rats), Trichostatin A increased histone acetylation (H3-K9 and H4-K8) and diminished anxiety-like and alcohol intake behaviors by reducing nuclear HDCA activity in the amygdala of P-rats (compared to NP-rats), associated to an increased NPY expression [222]. In mice, SAHA reduced intermittent-like alcohol drinking, without affecting saccharin or sucrose intake, and normalized increased histone H4 acetylation observed after excessive alcohol exposure [215].
Carboxylic acids also have a zinc-binding group able to inhibit HDCAs (some examples are butyric acid, valproic acid, or phenyl butyric acid) [209]. Administration of sodium butyrate to ethanol-dependent rats in a model of alcohol self-administration significantly diminished alcohol intake. Also, the short-chain fatty acid blocked ethanol drinking induced by alcohol deprivation [223]. Also, sodium butyrate effectively inhibited ethanol-induced behavioral sensitization (increased locomotor activity) associated with the regulation of specific genes in the striatum of mice [224].
Entinostat or MS-275 is another HDCA inhibitor that has a benzamide zinc-chelating motif [225]. Injected intraperitoneally, this agent reduced operant alcohol self-administration in alcohol-dependent rats [223]. Furthermore, i.c.v (intracerebroventricular) administration enhanced histone H4 acetylation within the nucleus accumbens, and the dorsolateral striatum, almost abolished ethanol self-administration, diminished motivation to drink, and reduced relapse during abstinence [226].
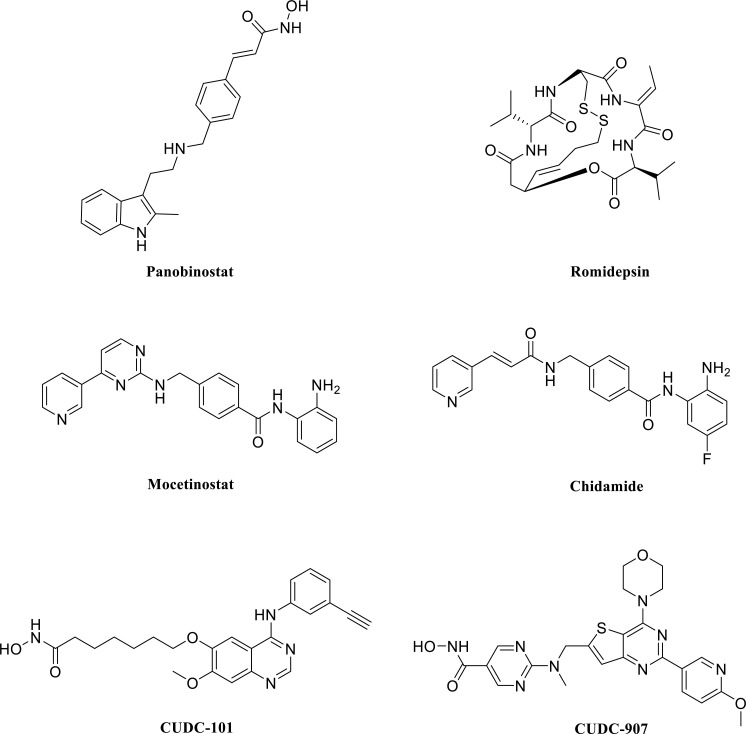
Many other HDCA inhibitors, not reviewed here, show improved characteristics related to tolerability, bioavailability, selectivity, or blood-brain barrier permeability, and are applied for the treatment of several pathologies. Some are under experimental study in different phases to tackle malignant, neurological, or immunological disorders. Therefore, the treatment of AUD can benefit from the use of these compounds that specifically block histone demethylation processes disrupted by ethanol [6, 209]. Fig. (9) shows some representative examples such as Panobinostat, a hydroxamic acid derivative already approved for the treatment of multiple myeloma [227]. Romidepsin (FK-228) is a depsipeptide with a disulfide bridge in its structure that suffers a reduction in vivo to give a thiol zinc-binding group. This agent inhibits preferentially class I HDAC [209, 228]. Mocetinostat and Chidamide are benzamide analogs that also exhibit specificity against type I HDAC [6, 209, 228].
Fig. (9).
Structure of representative HDCA inhibitors that exhibit some advantageous properties associated with bioavailability tolerability or selectivity. They are already approved for clinical use or subjected to clinical trials in different stages (see text for details).
4.3. miR Antagonists
The use of miRNA antisense oligomers to control miRNAs activity has evolved recently to treat some pathologies, especially malignancies [229]. However, this procedure confronts many challenges associated with cell entry, metabolic clearance, or delivery that require new strategies that overcome the difficulties to target specific processes with security and efficacy [230]. Oligonucleotides may be administered as free compounds or conjugated to different carriers. The control of the release and delivery of the oligonucleotide to the appropriate target is a crucial element that determines and secures its arrival and action [231].
Given the involvement of noncoding RNAs in the pathological mechanisms of adaptation associated with AUD, it seems plausible to explore candidate pharmacological tools to regulate these targets. Some experimental studies show the relevance and involvement of miRNAs in the control of the expression of genes altered by ethanol exposure, and the use of their antagonists reveals the importance of individual miRNAs in AUD pathologies. For example, adult rats intermittently administered with alcohol during the adolescence period showed increased miRNA-137 levels and direct injection of antagomiR-137 into the central nucleus of the amygdala abrogated alcohol drinking and anxiety-like behaviors and normalized Lsd and Bdnf IV expression [137]. Another miRNA antagonist, antagomiR-411, injected into the prefrontal cortex of female CB57BL/6J mice, reduced alcohol consumption when administered to mice chronically treated with ethanol. It also induced an augmented expression of glutamate AMPA 2 receptors [232]. Acute ethanol treatment in rats produced anxiolysis accompanied by a reduction of miR-494 and infusion of antagomiR-494 into the rat amygdala mimicked the anxiolytic effect of acute ethanol treatment. The antisense treatment also produced an elevated expression of CBP (Creb Binding Protein) and p300, and increased histone H3-Lys9 acetylation [233]. A study carried out in male rats subjected to two cycles of ethanol administration and withdrawal showed a deleterious effect on the brain (prefrontal and hippocampus structures) mitochondrial respiration that recovered after let-7f antagomiR treatment [234].
4.4. Other Approaches
To be accurate and achieve tailored treatments, focusing on epigenetic mechanisms that control the expression of specific genes affected in AUD appears as an acceptable procedure. Therefore, it is necessary to characterize crucial genes and how different epigenetic modifications regulate them. The field is vast and challenging, and the efforts should not only focus on DNA methylases or histone methylases (writers) but also to other targets, including the bromodomains (readers), histone deacetylases and histone demethylases (erasers) [209]. A combination of treatments and use of several inhibitors (a multi-epi-target approach) with dual or multiple effects acting simultaneously and reversibly on various epi-targets may serve to design more directed and effective therapies. An example is the two dual histone deacetylase/kinase inhibitors (compounds CUDC-101 and CUDC-907, (Fig. 9) [228]. Furthermore, joint targeting of epigenetic mechanisms and transcription factors (for example, cyclic AMP-responsive element-binding protein, CREB, nuclear factor kappa-light-chain enhancer of activated B cells, KF-kB, among others) should also be considered when exploring AUD treatments [235].
A new approach to control epigenetic mechanisms consists of taking advantage of the CRISP/Cas system to produce an inactive Cas nuclease (dead Cas, dCas) that may be used to direct transcriptional regulators (writers) to a specific gene location. This technique would give the possibility of editing the epigenetic landscape [209, 236].
Another therapy approach to pharmacologically tackle AUD would be to identify specific and predominant epi-targets within individuals and design individually customized measures. These traits would be easy to locate by determining reliable and reproducible biomarkers.
Nevertheless, it is relevant to note that combined therapies (psychotherapies, group therapies), as well as prevention interventions, can coexist with pharmacological treatments when focusing on AUD treatments (Table 2).
Table 2.
Studies that report the effect of representative epidrugs in different experimental models of alcohol administration (see main text).
| Drug Used | Experimental Model | Effect | References |
|---|---|---|---|
| DNMT inhibitors | - | - | - |
| Decitabine | Mice exposed to binge and chronic ethanol. | It reduced ethanol intake. | [211] |
| 5-azacytidine | Binge exposure of mice. | It decreased ethanol intake. | [212] |
| 5-azacytidine | Mice intermittently exposed to alcohol during adolescence. | Hypermethylation of Npy and Bdnf genes appeared reduced. Anxiety-like and alcohol drinking behavior during adulthood were reduced. |
[163] |
| RG-108 | Alcohol abstinent rats. | It decreased alcohol intake. | [213] |
| HDCA inhibitors | - | - | - |
| SAHA | Rats were chronically exposed to ethanol. | Abrogated ethanol withdrawal effects | [203] |
| SAHA | Rats were administered with a liquid diet containing ethanol. | Attenuated depression-like behavior was observed during the withdrawal period. | [169] |
| SAHA | Mice | It diminished binge-like alcohol drinking. | [212] |
| SAHA and Trichostatin A | Mice chronically exposed to ethanol | Both compounds prevented ethanol withdrawal effects. | [217] |
| Trichostatin A | Alcohol-preferring (P-rats) and non-preferring (NP-rats) |
Diminished anxiety-like behavior and alcohol intake in P-rats, compared with NP-rats | [219] |
| Sodium butyrate | Ethanol-dependent rats through alcohol self-administration. | It reduced alcohol drinking. | [220] |
| Sodium butyrate | Mice under ethanol-induced behavioral sensitization. | It inhibited ethanol-induced behavioral sensitization. | [221] |
| Entinostat | Alcohol-dependent rats. | Reduced operant alcohol administration, diminished motivation to drink, and reduced relapse during abstinence. | [220, 223] |
| miRNA antagonists | - | - | - |
| Antagomir-137 | Rats administered with ethanol during the adolescence period | The compound inhibited alcohol consumption and anxiety-like behaviors. | [133] |
| AntagomiR-411 | Female CB57BL/6J mice chronically administered with alcohol. | It diminished alcohol drinking. | [228] |
| AntagomiR-494 | Rats | It mimicked the anxiolytic effect of acute ethanol treatment. | [229] |
| Let-7f-antagomiR | Rats subjected to two cycles of ethanol administration and withdrawal | Recovered deranged mitochondrial respiration induced by ethanol treatment. | [230] |
CONCLUSION AND FUTURE RESEARCH
As many studies underlie, there is not a silver bullet to treat AUD, since this is a multifaceted pathological condition, with complex etiopathology that manifests in specific and singular forms in each individual. Nevertheless, the efforts to unveil the molecular mechanisms brought together to induce damage are continuous and will produce significant results. Genetic and epigenetic traits deeply condition AUD outcomes. We dispose of broad information about many processes that support brain damage and the bizarre manifestations of anxiety, stress, and negative emotional states related to chronic alcohol consumption. We also have accumulated knowledge associated with the mechanisms triggered in the brain after binge alcohol exposure and consolidation of use. However, we need a more integrated picture that establishes the relationship of all molecular interactions in a specific condition and individual. In this line, new findings in the field of epigenetics are, on the one hand, complicating the picture, but on the other hand, offer new windows for observation and research. The study of small molecules that inhibit key epienzymes involved in nucleosome architecture dynamics is necessary to prove their action and specificity experimentally. Moreover, to be tested appropriately in clinical trials with selected patients with defined alterations caused by deleterious ethanol consumption.
ACKNOWLEDGEMENTS
The University of Salamanca (Programa XIII, Grupo GIR Bases Moleculares del Desarrollo) supported this work. The author gratefully acknowledges Mr. Alvaro Gacho Temprano for the drawing of the chemical structures.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
REFERENCES
- 1.American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- 2.Edenberg H.J., McClintick J.N. Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: A critical review. Alcohol. Clin. Exp. Res. 2018;42(12):2281–2297. doi: 10.1111/acer.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho A.F., Heilig M., Perez A., Probst C., Rehm J. Alcohol use disorders. Lancet. 2019;394(10200):781–792. doi: 10.1016/S0140-6736(19)31775-1. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . Global Status on Alcohol and Health 2018. Geneva: WHO; 2018. [Google Scholar]
- 5.Heilig M., Augier E., Pfarr S., et al. Developing neuroscience-based treatments for alcohol addiction: A matter of choice? Transl. Psychiatry. 2019;9(1):255. doi: 10.1038/s41398-019-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourguet E., Ozdarska K., Moroy G., Jeanblanc J., Naassila M. Class I HDAC inhibitors: Potential new epigenetic therapeutics for Alcohol Use Disorder (AUD). J. Med. Chem. 2018;61(5):1745–1766. doi: 10.1021/acs.jmedchem.7b00115. [DOI] [PubMed] [Google Scholar]
- 7.Walker L.C., Lawrence A.J. Investigational drug therapies in phase I and phase II clinical trials for alcohol use disorders. Expert Opin. Investig. Drugs. 2018:1–14. doi: 10.1080/13543784.2018.1502269. [DOI] [PubMed] [Google Scholar]
- 8.Martinotti G., Orsolini L., Fornaro M., et al. Aripiprazole for relapse prevention and craving in alcohol use disorder: Current evidence and future perspectives. Expert Opin. Investig. Drugs. 2016;25(6):719–728. doi: 10.1080/13543784.2016.1175431. [DOI] [PubMed] [Google Scholar]
- 9.De Berardis D., Fornaro M., Serroni N., et al. Agomelatine beyond borders: Current evidences of its efficacy in disorders other than major depression. Int. J. Mol. Sci. 2015;16(1):1111–1130. doi: 10.3390/ijms16011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez F.D., Coveñas R. Targeting opioid and neurokinin-1 receptors to treat alcoholism. Curr. Med. Chem. 2011;18(28):4321–4334. doi: 10.2174/092986711797200444. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez F.D., Coveñas R. Targeting NPY, CRF/UCNs and NPS neuropeptide systems to treat Alcohol Use Disorder (AUD). Curr. Med. Chem. 2017;24(23):2528–2558. doi: 10.2174/0929867324666170316120836. [DOI] [PubMed] [Google Scholar]
- 12.Koob G.F., Volkow N.D. Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry. 2016;3(8):760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkow N.D., Koob G.F., McLellan A.T. Neurobiologic advances from the brain disease model of addiction. N. Engl. J. Med. 2016;374(4):363–371. doi: 10.1056/NEJMra1511480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Berardis D., Fornaro M., Valchera A., et al. Alexithymia, resilience, somatic sensations and their relationships with suicide ideation in drug naïve patients with first-episode major depression: An exploratory study in the “real world” everyday clinical practice. Early Interv. Psychiatry. 2020;14(3):336–342. doi: 10.1111/eip.12863. [DOI] [PubMed] [Google Scholar]
- 15.Fornaro M., De Berardis D., Iasevoli F., et al. Treatment adherence towards prescribed medications in bipolar-II acute depressed patients: Relationship with cyclothymic temperament and “therapeutic sensation seeking” in response towards subjective intolerance to pain. J. Affect. Disord. 2013;151(2):596–604. doi: 10.1016/j.jad.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Dick D.M., Kendler K.S. The impact of gene-environment interaction on alcohol use disorders. Alcohol Res. 2012;34(3):318–324. doi: 10.35946/arcr.v34.3.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kranzler H.R., Zhou H., Kember R.L., et al. Author Correction: Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 2019;10(1):1–3. doi: 10.1038/s41467-019-10254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters R.K., Polimanti R., Johnson E.C., et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 2018;21(12):1656–1669. doi: 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y.C., Fan J.H., Edenberg H.J., et al. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol. Clin. Exp. Res. 1997;21(7):1272–1277. doi: 10.1111/j.1530-0277.1997.tb04448.x. [DOI] [PubMed] [Google Scholar]
- 20.Farris S.P., Pietrzykowski A.Z., Miles M.F., et al. Applying the new genomics to alcohol dependence. Alcohol. 2015;49(8):825–836. doi: 10.1016/j.alcohol.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferraguti G., Pascale E., Lucarelli M. Alcohol addiction: A molecular biology perspective. Curr. Med. Chem. 2015;22(6):670–684. doi: 10.2174/0929867321666141229103158. [DOI] [PubMed] [Google Scholar]
- 22.Mayfield J., Arends M.A., Harris R.A., Blednov Y.A. Genes and Alcohol Consumption: Studies with Mutant Mice. Int. Rev. Neurobiol. 2016;126:293–355. doi: 10.1016/bs.irn.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morozova T.V., Mackay T.F., Anholt R.R. Genetics and genomics of alcohol sensitivity. Mol. Genet. Genomics. 2014;289(3):253–269. doi: 10.1007/s00438-013-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramoz N., Gorwood P. Presse Med. 2018;47(6):547–553. doi: 10.1016/j.lpm.2017.07.007. [Genetic factors in alcohol dependence]. [DOI] [PubMed] [Google Scholar]
- 25.Waddington C. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/150563a0. [DOI] [Google Scholar]
- 26.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 27.Franklin R.E., Gosling R.G. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953;172(4369):156–157. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins M.H. Physical studies of the molecular structure of deoxyribose nucleic acid and nucleoprotein. Cold Spring Harb. Symp. Quant. Biol. 1956;21:75–90. doi: 10.1101/SQB.1956.021.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Merriam-Webster I. Merriam-Webster’s medical dictionary Merriam-Webster’s medical desk dictionary. 2007. [Google Scholar]
- 30.Nicoglou A., Merlin F. Epigenetics: A way to bridge the gap between biological fields. Stud Hist Philos Biol Biomed Sci. 2017;66:73–82. doi: 10.1016/j.shpsc.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Werner R.J., Kelly A.D., Issa J.J. Epigenetics and precision oncology. Cancer J. 2017;23(5):262–269. doi: 10.1097/PPO.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berdasco M., Esteller M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019;20(2):109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 33.Stefanska B., MacEwan D.J. Epigenetics and pharmacology. Br. J. Pharmacol. 2015;172(11):2701–2704. doi: 10.1111/bph.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 35.Kundaje A., Meuleman W., Ernst J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017;18(5):275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 37.Wei J.W., Huang K., Yang C., Kang C.S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2017;37(1):3–9. doi: 10.3892/or.2016.5236. [Review]. [DOI] [PubMed] [Google Scholar]
- 38.Cuvier O., Fierz B. Dynamic chromatin technologies: From individual molecules to epigenomic regulation in cells. Nat. Rev. Genet. 2017;18(8):457–472. doi: 10.1038/nrg.2017.28. [DOI] [PubMed] [Google Scholar]
- 39.Stricker S.H., Köferle A., Beck S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017;18(1):51–66. doi: 10.1038/nrg.2016.138. [DOI] [PubMed] [Google Scholar]
- 40.Klemm S.L., Shipony Z., Greenleaf W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20(4):207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 41.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018;19(12):789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T., Dent S.Y. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat. Rev. Genet. 2014;15(2):93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plongthongkum N., Diep D.H., Zhang K. Advances in the profiling of DNA modifications: Cytosine methylation and beyond. Nat. Rev. Genet. 2014;15(10):647–661. doi: 10.1038/nrg3772. [DOI] [PubMed] [Google Scholar]
- 44.Hilton I.B., D’Ippolito A.M., Vockley C.M., et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbano A., Smith J., Weeks R.J., Chatterjee A. Gene-specific targeting of DNA methylation in the Mammalian genome. Cancers (Basel) 2019;11(10):e1515. doi: 10.3390/cancers11101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei Y., Huang Y.H., Goodell M.A. DNA methylation and de-methylation using hybrid site-targeting proteins. Genome Biol. 2018;19(1):187. doi: 10.1186/s13059-018-1566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubik G., Summerer D. TALEored epigenetics: A DNA-binding scaffold for programmable epigenome editing and analysis. ChemBioChem. 2016;17(11):975–980. doi: 10.1002/cbic.201600072. [DOI] [PubMed] [Google Scholar]
- 48.Mochizuki K., Hariya N., Honma K., Goda T. Relationship between epigenetic regulation, dietary habits, and the developmental origins of health and disease theory. Congenit. Anom. (Kyoto) 2017;57(6):184–190. doi: 10.1111/cga.12213. [DOI] [PubMed] [Google Scholar]
- 49.Mandy M., Nyirenda M. Developmental origins of health and disease: The relevance to developing nations. Int. Health. 2018;10(2):66–70. doi: 10.1093/inthealth/ihy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drake A.J., McPherson R.C., Godfrey K.M., et al. An unbalanced maternal diet in pregnancy associates with offspring epigenetic changes in genes controlling glucocorticoid action and foetal growth. Clin. Endocrinol. (Oxf.) 2012;77(6):808–815. doi: 10.1111/j.1365-2265.2012.04453.x. [DOI] [PubMed] [Google Scholar]
- 51.Jaenisch R., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 52.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 53.Xie S., Qian C. The growing complexity of UHRF1-mediated maintenance DNA methylation. Genes (Basel) 2018;9(12):e600. doi: 10.3390/genes9120600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jansz N. DNA methylation dynamics at transposable elements in mammals. Essays Biochem. 2019;63(6):677–689. doi: 10.1042/EBC20190039. [DOI] [PubMed] [Google Scholar]
- 55.Arand J., Spieler D., Karius T., et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8(6):e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patil V., Ward R.L., Hesson L.B. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9(6):823–828. doi: 10.4161/epi.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hotchkiss R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948;175(1):315–332. [PubMed] [Google Scholar]
- 58.Rausch C, Hastert FD, Cardoso MC. 2019. [DOI] [PubMed]
- 59.Bochtler M., Kolano A., Xu G.L. DNA demethylation pathways: Additional players and regulators. BioEssays. 2017;39(1):1–13. doi: 10.1002/bies.201600178. [DOI] [PubMed] [Google Scholar]
- 60.Dean W. Pathways of DNA Demethylation. Adv. Exp. Med. Biol. 2016;945:247–274. doi: 10.1007/978-3-319-43624-1_11. [DOI] [PubMed] [Google Scholar]
- 61.Cui D., Xu X. DNA methyltransferases, DNA methylation, and age-associated cognitive function. Int. J. Mol. Sci. 2018;19(5):e1315. doi: 10.3390/ijms19051315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X., Zhang Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017;18(9):517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 63.Wu S.C., Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saksouk N., Barth T.K., Ziegler-Birling C., et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell. 2014;56(4):580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 65.Chen Z., Zhang Y. Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. 2020;89(1):135–158. doi: 10.1146/annurev-biochem-103019-102815. [DOI] [PubMed] [Google Scholar]
- 66.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 67.Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19(2):81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 68.Baubec T., Colombo D.F., Wirbelauer C., et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520(7546):243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 69.Gu T., Lin X., Cullen S.M., et al. DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 2018;19(1):88. doi: 10.1186/s13059-018-1464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ooi S.K., Qiu C., Bernstein E., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma N., Pan H., Doré L.C., et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 2018;50(1):83–95. doi: 10.1038/s41588-017-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf G., Yang P., Füchtbauer A.C., et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29(5):538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ginder G.D., Williams D.C., Jr Readers of DNA methylation, the MBD family as potential therapeutic targets. Pharmacol. Ther. 2018;184:98–111. doi: 10.1016/j.pharmthera.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorsbach R.B., Moore J., Mathew S., Raimondi S.C., Mukatira S.T., Downing J.R. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11) (q22;q23). Leukemia. 2003;17(3):637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 75.Ito S., Shen L., Dai Q., et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tahiliani M., Koh K.P., Shen Y., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frommer M., McDonald L.E., Millar D.S., et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bogdanove A.J., Voytas D.F. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 79.Kubik G., Batke S., Summerer D. Programmable sensors of 5-hydroxymethylcytosine. J. Am. Chem. Soc. 2015;137(1):2–5. doi: 10.1021/ja506022t. [DOI] [PubMed] [Google Scholar]
- 80.Raiber E.A., Murat P., Chirgadze D.Y., Beraldi D., Luisi B.F., Balasubramanian S. 5-Formylcytosine alters the structure of the DNA double helix. Nat. Struct. Mol. Biol. 2015;22(1):44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 82.Luger K., Hansen J.C. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15(2):188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 83.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 84.Arnaudo A.M., Garcia B.A. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin. 2013;6(1):24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rossetto D., Avvakumov N., Côté J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7(10):1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee J.S., Smith E., Shilatifard A. The language of histone crosstalk. Cell. 2010;142(5):682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shanmugam M.K., Arfuso F., Arumugam S., et al. Role of novel histone modifications in cancer. Oncotarget. 2017;9(13):11414–11426. doi: 10.18632/oncotarget.23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamburri S., Lavarone E., Fernández-Pérez D., et al. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell. 2020;77(4):840–856.e5. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan M., Luo H., Lee S., et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young N.L., Dimaggio P.A., Garcia B.A. The significance, development and progress of high-throughput combinatorial histone code analysis. Cell. Mol. Life Sci. 2010;67(23):3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wapenaar H, Dekker FJ. 2016. [DOI] [PMC free article] [PubMed]
- 92.Lombardi P.M., Cole K.E., Dowling D.P., Christianson D.W. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 2011;21(6):735–743. doi: 10.1016/j.sbi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brownell J.E., Zhou J., Ranalli T., et al. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 94.Kuo M.H., Brownell J.E., Sobel R.E., et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383(6597):269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 95.Taunton J., Hassig C.A., Schreiber S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272(5260):408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 96.Benton C.B., Fiskus W., Bhalla K.N. Targeting histone acetylation: Readers and writers in Leukemia and cancer. Cancer J. 2017;23(5):286–291. doi: 10.1097/PPO.0000000000000284. [DOI] [PubMed] [Google Scholar]
- 97.Taniguchi Y. The Bromodomain and Extra-Terminal Domain (BET) family: Functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 2016;17(11):e1849. doi: 10.3390/ijms17111849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Musselman C.A., Lalonde M.E., Côté J., Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19(12):1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greer E.L., Beese-Sims S.E., Brookes E., et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. 2014;7(1):113–126. doi: 10.1016/j.celrep.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang M.K., Mehrazarin S., Park N.H., Wang C.Y. Epigenetic gene regulation by histone demethylases: Emerging role in oncogenesis and inflammation. Oral Dis. 2017;23(6):709–720. doi: 10.1111/odi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaniskan H.U., Martini M.L., Jin J. Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 2018;118(3):989–1068. doi: 10.1021/acs.chemrev.6b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grewal S.I., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 103.Du J., Johnson L.M., Jacobsen S.E., Patel D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16(9):519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arrowsmith C.H., Schapira M. Targeting non-bromodomain chromatin readers. Nat. Struct. Mol. Biol. 2019;26(10):863–869. doi: 10.1038/s41594-019-0290-2. [DOI] [PubMed] [Google Scholar]
- 105.James L.I., Barsyte-Lovejoy D., Zhong N., et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 2013;9(3):184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Herold J.M., Wigle T.J., Norris J.L., et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 2011;54(7):2504–2511. doi: 10.1021/jm200045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adams-Cioaba M.A., Min J. Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 2009;87(1):93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 108.Krapivinsky G., Krapivinsky L., Renthal N.E., Santa-Cruz A., Manasian Y., Clapham D.E. Histone phosphorylation by TRPM6's cleaved kinase attenuates adjacent arginine methylation to regulate gene expression. Proc. Natl. Acad. Sci. USA. 2017;114(34):E7092–E7100. doi: 10.1073/pnas.1708427114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lo W.S., Trievel R.C., Rojas J.R., et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5(6):917–926. doi: 10.1016/S1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 110.Utley R.T., Lacoste N., Jobin-Robitaille O., Allard S., Côté J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol. Cell. Biol. 2005;25(18):8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cook P.J., Ju B.G., Telese F., Wang X., Glass C.K., Rosenfeld M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 113.Gelato K.A., Fischle W. Role of histone modifications in defining chromatin structure and function. Biol. Chem. 2008;389(4):353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 114.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 115.Tsukiyama T., Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83(6):1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 116.Schwartz Y.B., Pirrotta V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013;14(12):853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 117.Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171(1):34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 118.Jiang S., Cheng S.J., Ren L.C., et al. An expanded landscape of human long noncoding RNA. Nucleic Acids Res. 2019;47(15):7842–7856. doi: 10.1093/nar/gkz621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iyer M.K., Niknafs Y.S., Malik R., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tijsterman M., Plasterk R.H. Dicers at RISC; The mechanism of RNAi. Cell. 2004;117(1):1–3. doi: 10.1016/S0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 121.Hock J., Meister G. The Argonaute protein family. Genome Biol. 2008;9(2):210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nikam R.R., Gore K.R. Journey of siRNA: Clinical developments and targeted delivery. Nucleic Acid Ther. 2018;28(4):209–224. doi: 10.1089/nat.2017.0715. [DOI] [PubMed] [Google Scholar]
- 123.Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305(5688):1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 124.Weinberg M.S., Morris K.V. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016;44(14):6505–6517. doi: 10.1093/nar/gkw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lev I., Gingold H., Rechavi O. H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. eLife. 2019;8:e40448. doi: 10.7554/eLife.40448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hawkins P.G., Santoso S., Adams C., Anest V., Morris K.V. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37(9):2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iwasaki Y.W., Siomi M.C., Siomi H. PIWI-Interacting RNA. Annu. Rev. Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 128.Brennecke J., Aravin A.A., Stark A., et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 129.Le Thomas A., Tóth K.F., Aravin A.A. To be or not to be a piRNA: Genomic origin and processing of piRNAs. Genome Biol. 2014;15(1):204. doi: 10.1186/gb4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stein C.B., Genzor P., Mitra S., et al. Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat. Commun. 2019;10(1):828. doi: 10.1038/s41467-019-08803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rechavi O. Guest list or black list: Heritable small RNAs as immunogenic memories. Trends Cell Biol. 2014;24(4):212–220. doi: 10.1016/j.tcb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rechavi O., Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 2017;27(14):R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 133.Le Thomas A., Rogers A.K., Webster A., et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27(4):390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yao Q., Chen Y., Zhou X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019;51:11–17. doi: 10.1016/j.cbpa.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 135.Moutinho C., Esteller M. MicroRNAs and epigenetics. Adv. Cancer Res. 2017;135:189–220. doi: 10.1016/bs.acr.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 136.Zhang Z., Cao Y., Zhai Y., et al. MicroRNA-29b regulates DNA methylation by targeting Dnmt3a/3b and Tet1/2/3 in porcine early embryo development. Dev. Growth Differ. 2018;60(4):197–204. doi: 10.1111/dgd.12537. [DOI] [PubMed] [Google Scholar]
- 137.Kyzar EJ, Bohnsack JP, Zhang H, Pandey SC. 2019.
- 138.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yao R.W., Wang Y., Chen L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019;21(5):542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 141.Hanly DJ, Esteller M, Berdasco M. 1748. [DOI] [PMC free article] [PubMed]
- 142.Livneh I., Moshitch-Moshkovitz S., Amariglio N., Rechavi G., Dominissini D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020;21(1):36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 143.Schwartz S. Cracking the epitranscriptome. RNA. 2016;22(2):169–174. doi: 10.1261/rna.054502.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jung Y., Goldman D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018;17(3):e12444. doi: 10.1111/gbb.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu J., Yue Y., Han D., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang H., Weng H., Chen J. The biogenesis and precise control of RNA m6A methylation. Trends Genet. 2020;36(1):44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63(2):306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 149.Zheng G., Dahl J.A., Niu Y., et al. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013;10(6):915–918. doi: 10.4161/rna.24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Règue-Guyon M., Lanfumey L., Mongeau R. Neuroepigenetics of neurotrophin signaling: Neurobiology of anxiety and affective disorders. Prog. Mol. Biol. Transl. Sci. 2018;158:159–193. doi: 10.1016/bs.pmbts.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 151.Wadhwa P.D., Buss C., Entringer S., Swanson J.M. Developmental origins of health and disease: Brief history of the approach and current focus on epigenetic mechanisms. Semin. Reprod. Med. 2009;27(5):358–368. doi: 10.1055/s-0029-1237424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Palmisano M., Pandey S.C. Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol. 2017;60:7–18. doi: 10.1016/j.alcohol.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Spanagel R., Noori H.R., Heilig M. Stress and alcohol interactions: Animal studies and clinical significance. Trends Neurosci. 2014;37(4):219–227. doi: 10.1016/j.tins.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 154.Boers R., Boers J., de Hoon B., et al. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018;28(1):88–99. doi: 10.1101/gr.222885.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yong W.S., Hsu F.M., Chen P.Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016;9:26. doi: 10.1186/s13072-016-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ciafrè S., Carito V., Ferraguti G., et al. How alcohol drinking affects our genes: An epigenetic point of view. Biochem. Cell Biol. 2019;97(4):345–356. doi: 10.1139/bcb-2018-0248. [DOI] [PubMed] [Google Scholar]
- 157.Mahna D., Puri S., Sharma S. DNA methylation signatures: Biomarkers of drug and alcohol abuse. Mutat. Res. 2018;777:19–28. doi: 10.1016/j.mrrev.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 158.Liu C., Marioni R.E., Hedman A.K., et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry. 2018;23(2):422–433. doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Auta J., Zhang H., Pandey S.C., Guidotti A. Chronic alcohol exposure differentially alters one-carbon metabolism in rat liver and brain. Alcohol. Clin. Exp. Res. 2017;41(6):1105–1111. doi: 10.1111/acer.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kamat P.K., Mallonee C.J., George A.K., Tyagi S.C., Tyagi N. Homocysteine, alcoholism, and its potential epigenetic mechanism. Alcohol. Clin. Exp. Res. 2016;40(12):2474–2481. doi: 10.1111/acer.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nagre N.N., Subbanna S., Shivakumar M., Psychoyos D., Basavarajappa B.S. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. J. Neurochem. 2015;132(4):429–442. doi: 10.1111/jnc.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bönsch D., Lenz B., Fiszer R., Frieling H., Kornhuber J., Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. (Vienna) 2006;113(9):1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- 163.Gangisetty O., Wynne O., Jabbar S., Nasello C., Sarkar D.K. Fetal alcohol exposure reduces dopamine receptor D2 and increases pituitary weight and prolactin production via epigenetic mechanisms. PLoS One. 2015;10(10):e0140699. doi: 10.1371/journal.pone.0140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Berkel T.D.M., Zhang H., Teppen T., Sakharkar A.J., Pandey S.C. Essential role of histone methyltransferase G9a in rapid tolerance to the anxiolytic effects of ethanol. Int. J. Neuropsychopharmacol. 2019;22(4):292–302. doi: 10.1093/ijnp/pyy102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Crews F.T., Robinson D.L., Chandler L.J., et al. Mechanisms of persistent neurobiological changes following adolescent alcohol axposure: NADIA consortium findings. Alcohol. Clin. Exp. Res. 2019;43(9):1806–1822. doi: 10.1111/acer.14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sakharkar A.J., Kyzar E.J., Gavin D.P., et al. Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology. 2019;157107679 doi: 10.1016/j.neuropharm.2019.107679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang H., Kyzar E.J., Bohnsack J.P., et al. Adolescent alcohol exposure epigenetically regulates CREB signaling in the adult amygdala. Sci. Rep. 2018;8(1):10376. doi: 10.1038/s41598-018-28415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.López-Moreno J.A., Marcos M., Calleja-Conde J., et al. Histone deacetylase gene expression following Binge Alcohol consumption in rats and humans. Alcohol. Clin. Exp. Res. 2015;39(10):1939–1950. doi: 10.1111/acer.12850. [DOI] [PubMed] [Google Scholar]
- 169.Jarmasz J.S., Stirton H., Basalah D., et al. Global DNA methylation and histone posttranslational modifications in human and nonhuman primate brain in association with prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 2019;43(6):1145–1162. doi: 10.1111/acer.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gavin D.P., Hashimoto J.G., Lazar N.H., Carbone L., Crabbe J.C., Guizzetti M. Stable histone methylation changes at Proteoglycan network genes following ethanol exposure. Front. Genet. 2018;9:346. doi: 10.3389/fgene.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kyzar E.J., Zhang H., Pandey S.C. Adolescent alcohol exposure epigenetically suppresses Amygdala arc enhancer RNA expression to confer adult anxiety susceptibility. Biol. Psychiatry. 2019;85(11):904–914. doi: 10.1016/j.biopsych.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen W.Y., Zhang H., Gatta E., Glover E.J., Pandey S.C., Lasek A.W. The histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) alleviates depression-like behavior and normalizes epigenetic changes in the hippocampus during ethanol withdrawal. Alcohol. 2019;78:79–87. doi: 10.1016/j.alcohol.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bohnsack J.P., Hughes B.A., O’Buckley T.K., Edokpolor K., Besheer J., Morrow A.L. Histone deacetylases mediate GABAA receptor expression, physiology, and behavioral maladaptations in rat models of alcohol dependence. Neuropsychopharmacology. 2018;43(7):1518–1529. doi: 10.1038/s41386-018-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ignacio C, Hicks SD, Burke P, et al. Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in the brain. 2015. [DOI] [PMC free article] [PubMed]
- 175.Smith M.L., Lopez M.F., Archer K.J., Wolen A.R., Becker H.C., Miles M.F. Time-course analysis of brain regional expression network responses to chronic intermittent ethanol and withdrawal: Implications for mechanisms underlying excessive ethanol consumption. PLoS One. 2016;11(1):e0146257. doi: 10.1371/journal.pone.0146257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nunez Y.O., Truitt J.M., Gorini G., et al. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during the early stages of alcohol dependence. BMC Genomics. 2013;14:725. doi: 10.1186/1471-2164-14-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang F., Gelernter J., Zhang H. Differential expression of miR-130a in postmortem prefrontal cortex of subjects with Alcohol Use Disorders. J. Addict. Res. Ther. 2013;4(155):18179. doi: 10.4172/2155,6105.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tian H., Ye X., Hou X., Yang X., Yang J., Wu C. SVCT2, a potential therapeutic target, protects against oxidative stress during ethanol-induced neurotoxicity via JNK/p38 MAPKs, NF-κB and miRNA125a-5p. Free Radic. Biol. Med. 2016;96:362–373. doi: 10.1016/j.freeradbiomed.2016.03.039. [DOI] [PubMed] [Google Scholar]
- 179.Muddashetty R.S., Nalavadi V.C., Gross C., et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42(5):673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gorini G., Nunez Y.O., Mayfield R.D. Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS One. 2013;8(12):e82565. doi: 10.1371/journal.pone.0082565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bohnsack J.P., Teppen T., Kyzar E.J., et al. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early-onset alcohol use disorders. Transl. Psychiatry. 2019;9(1):34. doi: 10.1038/s41398-019-0367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xu K., Kranzler H.R., Sherva R., et al. Genomewide association study for maximum number of alcoholic drinks in European Americans and African Americans. Alcohol. Clin. Exp. Res. 2015;39(7):1137–1147. doi: 10.1111/acer.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bernard D., Prasanth K.V., Tripathi V., et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kryger R., Fan L., Wilce P.A., Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46(7):629–634. doi: 10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 185.Berkel T.D., Pandey S.C. Emerging role of epigenetic mechanisms in alcohol addiction. Alcohol. Clin. Exp. Res. 2017;41(4):666–680. doi: 10.1111/acer.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lister R., Mukamel E.A., Nery J.R., et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pár A., Pár G. Orv. Hetil. 2019;160(14):524–532. doi: 10.1556/650.2019.31352. [Alcoholic liver disease: The roles of genetic-epigenetic factors and the effect of abstinence]. [DOI] [PubMed] [Google Scholar]
- 188.Tulisiak C.T., Harris R.A., Ponomarev I. DNA modifications in models of alcohol use disorders. Alcohol. 2017;60:19–30. doi: 10.1016/j.alcohol.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Banik A., Kandilya D., Ramya S., Stünkel W., Chong Y.S., Dheen S.T. Maternal factors that induce epigenetic changes contribute to neurological disorders in offspring. Genes (Basel) 2017;8(6):e150. doi: 10.3390/genes8060150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ungerer M., Knezovich J., Ramsay M. In utero alcohol exposure, epigenetic changes, and their consequences. Alcohol Res. 2013;35(1):37–46. doi: 10.35946/arcr.v35.1.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35(1):69–76. doi: 10.35946/arcr.v35.1.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ajonijebu D.C., Abboussi O., Russell V.A., Mabandla M.V., Daniels W.M.U. Epigenetics: A link between addiction and social environment. Cell. Mol. Life Sci. 2017;74(15):2735–2747. doi: 10.1007/s00018-017-2493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Miranda R.C. MicroRNAs and ethanol toxicity. Int. Rev. Neurobiol. 2014;115:245–284. doi: 10.1016/B978-0-12-801311-3.00007-X. [DOI] [PubMed] [Google Scholar]
- 194.Legastelois R., Jeanblanc J., Vilpoux C., Bourguet E., Naassila M. Biol. Aujourdhui. 2017;211(1):83–91. doi: 10.1051/jbio/2017014. [Epigenetic mechanisms and alcohol use disorders: A potential therapeutic target]. [DOI] [PubMed] [Google Scholar]
- 195.Mayfield R.D. Emerging roles for ncRNAs in alcohol use disorders. Alcohol. 2017;60:31–39. doi: 10.1016/j.alcohol.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang F., Xu H., Zhao H., Gelernter J., Zhang H. DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci. Rep. 2016;6:19430. doi: 10.1038/srep19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gatta E., Auta J., Gavin D.P., et al. Emerging role of one-carbon metabolism and DNA methylation enrichment on δ-containing GABAA receptor expression in the Cerebellum of subjects with Alcohol Use Disorders (AUD). Int. J. Neuropsychopharmacol. 2017;20(12):1013–1026. doi: 10.1093/ijnp/pyx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ponomarev I., Wang S., Zhang L., Harris R.A., Mayfield R.D. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Philibert R.A., Plume J.M., Gibbons F.X., Brody G.H., Beach S.R. The impact of recent alcohol use on genome wide DNA methylation signatures. Front. Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hillemacher T., Frieling H., Luber K., et al. Epigenetic regulation and gene expression of vasopressin and atrial natriuretic peptide in alcohol withdrawal. Psychoneuroendocrinology. 2009;34(4):555–560. doi: 10.1016/j.psyneuen.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 201.Hillemacher T., Frieling H., Hartl T., Wilhelm J., Kornhuber J., Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J. Psychiatr. Res. 2009;43(4):388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 202.Manzardo A.M., Gunewardena S., Butler M.G. Over-expression of the miRNA cluster at chromosome 14q32 in the alcoholic brain correlates with suppression of predicted target mRNA required for oligodendrocyte proliferation. Gene. 2013;526(2):356–363. doi: 10.1016/j.gene.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mamdani M., Williamson V., McMichael G.O., et al. Integrating mRNA and miRNA weighted gene co-expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PLoS One. 2015;10(9):e0137671. doi: 10.1371/journal.pone.0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dulman R.S., Auta J., Teppen T., Pandey S.C. Acute ethanol produces Ataxia and induces Fmr1 expression via histone modifications in the rat Cerebellum. Alcohol. Clin. Exp. Res. 2019;43(6):1191–1198. doi: 10.1111/acer.14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dong E., Guidotti A., Zhang H., Pandey S.C. Prenatal stress leads to chromatin and synaptic remodeling and excessive alcohol intake comorbid with anxiety-like behaviors in adult offspring. Neuropharmacology. 2018;140:76–85. doi: 10.1016/j.neuropharm.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.You C., Vandegrift B.J., Zhang H., Lasek A.W., Pandey S.C., Brodie M.S. Histone deacetylase inhibitor suberanilohydroxamic acid treatment reverses hyposensitivity to γ-aminobutyric acid in the ventral tegmental area during ethanol withdrawal. Alcohol. Clin. Exp. Res. 2018;42(11):2160–2171. doi: 10.1111/acer.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Maier H.B., Neyazi M., Neyazi A., et al. Alcohol consumption alters Gdnf promoter methylation and expression in rats. J. Psychiatr. Res. 2020;121:1–9. doi: 10.1016/j.jpsychires.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 208.Mews P., Egervari G., Nativio R., et al. Alcohol metabolism contributes to brain histone acetylation. Nature. 2019;574(7780):717–721. doi: 10.1038/s41586-019-1700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ganesan A., Arimondo P.B., Rots M.G., et al. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenetics. 2019;11(1):174. doi: 10.1186/s13148-019-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stresemann C., Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123(1):8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 211.Ebrahem Q., Mahfouz R.Z., Ng K.P., Saunthararajah Y. High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget. 2012;3(10):1137–1145. doi: 10.18632/oncotarget.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Asgatay S., Champion C., Marloie G., et al. Synthesis and evaluation of analogues of N-phthaloyl-l-tryptophan (RG108) as inhibitors of DNA methyltransferase 1. J. Med. Chem. 2014;57(2):421–434. doi: 10.1021/jm401419p. [DOI] [PubMed] [Google Scholar]
- 213.Schneeberger Y., Stenzig J., Hübner F., Schaefer A., Reichenspurner H., Eschenhagen T. Pharmacokinetics of the experimental non-nucleosidic DNA methyl transferase inhibitor N-Phthalyl-L-tryptophan (RG 108) in rats. Basic Clin. Pharmacol. Toxicol. 2016;118(5):327–332. doi: 10.1111/bcpt.12514. [DOI] [PubMed] [Google Scholar]
- 214.Ponomarev I., Stelly C.E., Morikawa H., Blednov Y.A., Mayfield R.D., Harris R.A. Mechanistic insights into epigenetic modulation of ethanol consumption. Alcohol. 2017;60:95–101. doi: 10.1016/j.alcohol.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Warnault V., Darcq E., Levine A., Barak S., Ron D. Chromatin remodeling--a novel strategy to control excessive alcohol drinking. Transl. Psychiatry. 2013;3(2):e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Barbier E., Tapocik J.D., Juergens N., et al. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 2015;35(15):6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Singh V., Sharma P., Capalash N. DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr. Cancer Drug Targets. 2013;13(4):379–399. doi: 10.2174/15680096113139990077. [DOI] [PubMed] [Google Scholar]
- 218.Kantarjian H.M., Roboz G.J., Kropf P.L., et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: Phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017;18(10):1317–1326. doi: 10.1016/S1470-2045(17)30576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Richon V.M., Emiliani S., Verdin E., et al. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA. 1998;95(6):3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Arora D.S., Nimitvilai S., Teppen T.L., et al. Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: Reversal by histone deacetylase inhibitors. Neuropsychopharmacology. 2013;38(9):1674–1684. doi: 10.1038/npp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yoshida M., Furumai R., Nishiyama M., Komatsu Y., Nishino N., Horinouchi S. Histone deacetylase as a new target for cancer chemotherapy. Cancer Chemother. Pharmacol. 2001;48(Suppl. 1):S20–S26. doi: 10.1007/s002800100300. [DOI] [PubMed] [Google Scholar]
- 222.Sakharkar A.J., Zhang H., Tang L., et al. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: A role in anxiety-like and alcohol-drinking behaviours. Int. J. Neuropsychopharmacol. 2014;17(8):1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Simon-O’Brien E., Alaux-Cantin S., Warnault V., Buttolo R., Naassila M., Vilpoux C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict. Biol. 2015;20(4):676–689. doi: 10.1111/adb.12161. [DOI] [PubMed] [Google Scholar]
- 224.Legastelois R., Botia B., Naassila M. Blockade of ethanol-induced behavioral sensitization by sodium butyrate: Descriptive analysis of gene regulations in the striatum. Alcohol. Clin. Exp. Res. 2013;37(7):1143–1153. doi: 10.1111/acer.12088. [DOI] [PubMed] [Google Scholar]
- 225.Suzuki T., Ando T., Tsuchiya K., et al. Synthesis and histone deacetylase inhibitory activity of new benzamide derivatives. J. Med. Chem. 1999;42(15):3001–3003. doi: 10.1021/jm980565u. [DOI] [PubMed] [Google Scholar]
- 226.Jeanblanc J., Lemoine S., Jeanblanc V., Alaux-Cantin S., Naassila M. The class I-specific HDAC inhibitor MS-275 decreases motivation to consume alcohol and relapse in heavy drinking rats. Int. J. Neuropsychopharmacol. 2015;18(9):pyv029. doi: 10.1093/ijnp/pyv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cavenagh J.D., Popat R. Optimal management of histone deacetylase inhibitor-related adverse events in patients with multiple Myeloma: A focus on Panobinostat. Clin. Lymphoma Myeloma Leuk. 2018;18(8):501–507. doi: 10.1016/j.clml.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 228.Tomaselli D., Lucidi A., Rotili D., Mai A. Epigenetic polypharmacology: A new frontier for epi-drug discovery. Med. Res. Rev. 2020;40(1):190–244. doi: 10.1002/med.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stenvang J., Petri A., Lindow M., Obad S., Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Cheng C.J., Bahal R., Babar I.A., et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Juliano R.L. Intracellular trafficking and endosomal release of Oligonucleotides: What we know and what we don’t. Nucleic Acid Ther. 2018;28(3):166–177. doi: 10.1089/nat.2018.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Most D., Salem N.A., Tiwari G.R., Blednov Y.A., Mayfield R.D., Harris R.A. Silencing synaptic MicroRNA-411 reduces voluntary alcohol consumption in mice. Addict. Biol. 2019;24(4):604–616. doi: 10.1111/adb.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Teppen T.L., Krishnan H.R., Zhang H., Sakharkar A.J., Pandey S.C. The potential role of Amygdaloid microRNA-494 in alcohol-induced anxiolysis. Biol. Psychiatry. 2016;80(9):711–719. doi: 10.1016/j.biopsych.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jung M.E., Metzger D.B. Aberrant histone acetylation promotes mitochondrial respiratory suppression in the brain of alcoholic rats. J. Pharmacol. Exp. Ther. 2015;352(2):258–266. doi: 10.1124/jpet.114.219311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Savarese A.M., Lasek A.W. Transcriptional regulators as targets for alcohol pharmacotherapies. Handb. Exp. Pharmacol. 2018;248:505–533. doi: 10.1007/164_2018_101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Geel TM, Ruiters MHJ, Cool RH, et al. 1748.