Abstract
According to the WHO 2009 classification, dengue with warning signs is at the risk of developing severe form of dengue disease. One of the most important warning signs is plasma leakage, which can be a serious complication associated with higher morbidity and mortality. We report that the frequency of CD4+CD8+ double-positive (DP) T cells is significantly increased in patients at risk of developing plasma leakage. Transcriptomic analysis demonstrated that CD4+CD8+ DP cells were distinct from CD4+ Single Positive (SP) T cells but co-clustered with CD8+ SP cells, indicating a largely similar transcriptional profile. Twenty significant differentially expressed (DE) genes were identified between CD4+CD8+ DP and CD8+ SP cells. These genes encode OX40 and CCR4 proteins as well as other molecules associated with cell signaling on the cell surface (NT5E, MXRA8, and PTPRK). While comparing the profile of gene expression in CD4+CD8+ DP cells from patients with and without warning signs of plasma leakage, similar expression profile was observed, implying a role of CD4+CD8+ DP cells in plasma leakage through a quantitative increase rather than functional alteration. This study provided novel insight into the host immune response during the acute febrile phase of DENV infection and the role of CD4+CD8+ DP T cells in the pathogenesis of plasma leakage.
Keywords: CD4+, CD8+, double positive, T cells, infectious diseases, dengue, plasma leakage, transcriptomic analysis
1. Introduction
Dengue fever (DF) is one of the major re-emerging infectious diseases and the most prevalent arthropod-borne viral disease in humans [1]. The Center for Diseases Control and Prevention (CDC) estimates that around 400 million people get infected annually, and approximately 3 billion of the world’s population live in dengue-endemic areas [2]. The 1997 WHO classification [3] distinguished four main categories of dengue disease: non-classical DF, classical DF, dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS). The diagnosis of DHF required the presence of four criteria: fever, thrombocytopenia (<100,000 platelets/mm3), as well as both hemorrhagic and plasma leakage manifestations. Several studies reported that this classification did not fully correlate with disease severity [3,4,5] and had limited sensitivity in detecting severe dengue cases that require advanced medical care [6,7]. As a consequence, a new WHO classification scheme was released [1] in 2009 that divides dengue into two categories: non-severe and severe dengue, with non-severe dengue further subcategorized as dengue without warning signs and dengue with warning signs. Plasma leakage is one of the most important warning signs, and it can be defined as abnormal extravascular accumulation of body fluids diagnosed clinically or radiologically and/or hemoconcentration (rise in hematocrit (HCT) ≥ 20% of the patient’s baseline level or a drop in HCT ≥ 20% of the baseline level following rehydration) [1].
The most dangerous complication of dengue infection is the dengue shock syndrome, a consequence of severe intravascular volume depletion from plasma leakage due to increased vascular permeability and decreased intravascular osmolarity. The severity of plasma leakage varies among patients, and failure to promptly identify and treat this warning sign of dengue is related to high mortality [8]. Thus, it is important to identify risk factors for the shock syndrome and to predict progression of dengue to more severe disease, such as plasma leakage or shock during the acute febrile phase.
Studies reported that female, infant, elderly patients, and those with comorbidities are prone to have more severe infection/shock syndrome [9,10]. Virus serotype, secondary infections, and laboratory tests, such as platelet count, serum albumin, aspartate aminotransferase (AST), and alanine aminotransferase (ALT) levels, may be also related and should be monitored during the febrile phase of illness [10]. However, these risk factors are not sensitive enough to be used in a clinical setting to predict severe disease, and more factors are yet to be determined. Thus, while several studies reported symptomatologic associations, the molecular mechanisms involved in plasma leakage progression to shock are still not well defined.
Here, we examined whether we could define changes in T-cell subset composition as a potential marker of plasma leakage progression to shock and also as a way to probe the associated underlying molecular mechanisms. We report that a population of CD4+CD8+ double-positive (DP) T cells detectable in the peripheral blood is associated with risk of plasma leakage in dengue disease. We further investigated the specific gene expression profiles associated with this subpopulation compared to other T cell subsets and as a function of disease severity.
2. Materials and Methods
2.1. Study Cohort and PBMC Isolation
The aim of this study was to investigate the association of the gene expression profiles and phenotypic attributes of CD4+CD8+ double-positive (DP) cells with plasma leakage in dengue diseases by RNA-sequencing and flow cytometry. The blood samples from patients with acute dengue virus (DENV) infection were collected upon diagnosis or admission with a median of 4 days after onset of fever (interquartile range (IQR): 4.0–6.0 days), in the North Colombo Teaching Hospital, Ragama in Gampaha District, Sri Lanka, and the National Institute of Infectious Diseases, Gothatuwa, Angoda, Sri Lanka, between 2010 and 2016. This study was approved by the appropriate local Ethics committees (see below for details). Each participant provided informed consent and was assigned a study identification number with clinical information recorded. The diagnosis of dengue infection was confirmed if at least one of the following criteria was met in acute phase serum: (1) positive reverse transcription polymerase chain reaction (RT-PCR) of DENV RNA, (2) positive serology for dengue IgM, or (3) positive dengue-specific non-structural antigen-1 (NS1). All patients were screened to ensure no history of anemia, HIV/HBV/HCV infection, or significant systemic disease. One group comprises patients that were diagnosed as dengue without warning sign of plasma leakage (D-L) cases, while the other group was classified as dengue with warning sign of plasma leakage (D+L) cases upon discharge. Plasma leakage in this study was defined as abnormal extravascular accumulation of body fluids and diagnosed according to 2009 WHO classification of dengue [1]. In the current study, no signs of plasma leakage were observed at initial diagnosis, and all the plasma leakage cases were confirmed by ultrasound several days after diagnosis/admission. The details of the patients recruited in this study were summarized in (Table 1). PBMCs were isolated at the Genetech Research Institute from whole blood as previously described [11] and cryopreserved for further analysis.
Table 1.
Demographic and clinical information of Sri Lanka dengue patients.
| Cohorts | D-L | D+L | p value | Total |
|---|---|---|---|---|
| No. of subjects | 33 | 55 | - | 88 |
| Age (years) | 26.0 (19.0–43.0) | 29.0 (23.7–35.5) | 0.465 | 29.0 (22.0–37.0) |
| Gender (% female) | 30.3% | 42.6% | 0.267 | 37.9% |
| Days of acute blood sample collection from fever onset | 5.0 (4.0–6.0) | 4.0 (4.0–6.0) | 0.246 | 4.0 (4.0–6.0) |
| Dengue tests (% positive) | ||||
| IgM | 57.6% | 66.7% | 0.371 | 63.2% |
| IgG | 72.7% | 76.4% | 0.800 | 75.0% |
| PCR | 30.3% | 49.0% | 0.118 | 41.7% |
| Secondary dengue infection (%) | 72.7% | 76.4% | 0.800 | 75.0% |
| Platelet count (×1000/mm3) | 74.0 (36.0–120.0) | 39.0 (13.0–90.0) | 0.007 ** | 59.0 (20.7–102.8) |
| Hematocrit (%) | 40.9 (37.2–44.5) | 42.3 (39.5–46.1) | 0.139 | 42.1 (37.4–44.8) |
| AST (U/L) | 113.0 (43.5–216.7) | 230.0 (96.0–309.7) | 0.017 * | 176.0 (66.5–300.1) |
| ALT (U/L) | 90.5 (32.2–168.6) | 150.0 (62.0–280.4) | 0.028 * | 111.9 (50.0–226.5) |
| %CD4+CD8+ DP | 0.77 (0.31–1.10) | 1.45 (1.13–2.07) | <0.0001 **** | 1.31 (0.61–1.67) |
D-L, dengue without plasma leakage; D+L, dengue with plasma leakage; PCR, polymerase chain reaction; AST, aspartate aminotransaminase; ALT, alanine aminotransferase; DP, double positive, -, not calculated; continuous data are expressed as median and interquartile range, p-values were compared between D-L and D+L groups, * p < 0.05, ** p < 0.01, **** p < 0.0001.
There was no difference in demographic features between the two groups. The median age was 26.0 (IQR (interquartile range): 19.0–43.0) years in the D-L group and 29.0 (IQR: 23.7–35.5) years in the D+L group. Females accounted for 30.3 % in D-L group and 42.6% in the D+L group. All blood samples were taken and sent for blood tests (shown in Table 1) during the acute febrile phase of DENV infection with a median of 5.0 (IQR: 4.0–6.0) days from fever onset in the D-L group and a median of 4.0 (IQR: 4.0–6.0) days from fever onset in the D+L group.
There were no significant differences in the results of dengue serology or PCR tests between the two groups: 57.6% of patients in the D-L group and 66.7% in the D+L group were IgM positive, 72.7% in the D-L group and 76.4% in the D+L group were IgG positive, and 30.3% in the D-L group and 46.9% in the D-L group were PCR positive. A total of 72.7% of patients in the D-L group and 76.4% in the D+L group had at least one previous episode of dengue infection.
Laboratory results showed a higher severity of disease complications in the D+L group in the acute febrile phase, including lower platelet counts (D+L: 39.0 (IQR: 13.0–90.0) ×1000/mm3 vs. D-L: 74.0 (IQR: 36.0–120.0) ×1000/mm3, p = 0.007) and worse transaminitis (AST: 230.0 (IQR: 96.0–309.7) U/L in D+L group vs. 113.0 (IQR: 43.5–216.7) U/L in D-L group, p = 0.017; ALT: 150.0 (IQR: 62.0–280.4) U/L in D+L group vs. 90.5 (IQR: 32.2–168.6) U/L in D-L group, p = 0.028). No difference in hematocrit, a marker of hemoconcentration, was seen in the acute febrile phase of DENV infection (42.3% (IQR: 39.5–46.1%) in the D+L group vs. 40.9% (37.2–44.5%) in the D-L group, p = 0.139).
2.2. Dengue Serology and PCR Tests
DENV serology tests were performed with anti-DENV IgG and IgM ELISA as previously described [12]. Reverse transcription-PCR was performed by using the DV1 and DV3 primer set [13] and the ALD 1 and ALD 2 primer set [14] in one reaction with details specified previously [15].
2.3. Flow Cytometry Analysis and Cell Sorting for RNA Sequencing
PBMCs were stained with anti-human CD3, CD4, CD8, CD14, CD19, CD56, and live/dead viability antibodies (see Table S1 for antibody details). Subsequently, live CD4+ single positive (SP) (CD14−CD19−CD56−CD3+CD4+CD8−), CD8+ SP (CD14−CD19−CD56−CD3+CD4−CD8+), and CD4+CD8+ DP (CD14−CD19−CD56−CD3+CD4+CD8+) cells were sorted into TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA) using a BD FACSAria cell sorter (BD Biosciences, San Jose, CA, USA). Flow cytometry data were analyzed by FlowJo X Software (version 10, Tree Star, Ashland, OR, USA). The gating strategy utilized were shown in Figure 1A,B, and results from representative donors from both D-L and D+L groups are shown in Figure 1B. To note that in this study, B cells, monocytes, and NK cells were removed by flowcytometry using CD19, CD14, and CD56 antibodies, and only CD3+ T cells were analyzed. However, the roles of these subsets of immune cells may be important as well and warrant further investigation because several relevant factors (e.g., NF-kB) have been associated with plasma leakage [16].
Figure 1.
Expansion of CD4+CD8+ DP population with clinical plasma leakage in dengue disease. (A) Gating strategy to identify and sort CD4+CD8+ DP T cells from human PBMCs isolated from patients who were diagnosed at the acute phase of DENV infection (4–5 days since fever onset). (B) Representative flowcytometry plots demonstrate increased percentage of CD4+CD8+ DP T cells in dengue with plasma leakage (D+L) cohort compared to those in the dengue without plasma leakage (D-L) cohort. (C) Bar graph shows the frequencies of CD4+CD8+ DP T cells in D+L cohort (red, n = 55) and D-L cohort (grey, n = 33). Error bars show median with interquartile range. Statistical analysis was performed by two-tailed Mann–Whitney test, **** p < 0.0001.
2.4. RNA Sequencing
Total RNA was purified and quantified, as previously described [17]. Purified total RNA (5 ng) was amplified following the Smart-seq2 protocol [18]. cDNA was purified using AMPure XP beads (1:1 ratio; Beckman Coulter, Carlsbad, CA, USA). From this step, 1 ng cDNA was used to prepare a standard Nextera XT sequencing library (Nextera XT DNA library preparation kit and index kit (set B and C, respectively), Illumina, San Diego, CA, USA). Samples were sequenced in 3 batches using a NovaSeq 6000 system (Illumina, San Diego, CA, USA) to obtain 50-bp paired-end reads. Both whole transcriptome amplification and sequencing library preparations were performed in a 96-well format to reduce assay-to-assay variability. Quality control steps were included to determine total RNA quality and quantity, the optimal number of PCR pre-amplification cycles, and fragment size selection using the Agilent 2100 Bioanalyzer system (Agilent, Santa Clara, CA, USA). Samples that failed quality control were eliminated from further downstream steps. Barcoded Illumina sequencing libraries (Nextera, Illumina, San Diego, CA, USA) were generated utilizing the automated platform (Biomek FXp, Beckman Coulter, Carlsbad, CA, USA). Libraries were sequenced on the NovaSeq 6000 Illumina platform to obtain 50-bp paired-end reads using the NovaSeq 6000 S4 Reagent kit v1.5 (Illumina, San Diego, CA, USA), generating a median of 25.4 million mapped reads per sample.
2.5. Transcriptomic Analysis
Interventionary studies involving animals or humans and other studies that require ethical approval must list the authority that provided approval and the corresponding ethical approval code.
The single-end reads that passed Illumina filters were filtered for reads aligning to tRNA, rRNA, adapter sequences, and spike-in controls. The reads were then aligned to UCSC hg19 reference genome using TopHat (v 1.4.1) [19]. DUST scores were calculated with PRINSEQ Lite (v 0.20.3) [20], and low-complexity reads (DUST > 4) were removed from the BAM files. The alignment results were parsed via the SAMtools [21] to generate SAM files. Read counts to each genomic feature were obtained with the HTseq-count program (v 0.6.0) [22] using the “union” option.
After removing absent features (zero counts in all samples), the raw counts from 3 batches were combined with batch effect removed by BEER v0.1.8 for R [23] and imported to R/Bioconductor package Seurat 4.0 [24] for normalization. Clustering analyses were performed using UMAP for dimensionality reduction. Differentially expressed (DE) genes among samples were identified, with p-values calculated by binomial test and adjusted for multiple comparison by the Benjamini–Hochberg algorithm [25] to control the false discovery rate. Significant DE genes were defined as both adjusted p-value of < 0.05 and the absolute value of log2 fold-change > 2, and results were presented as volcano plots and heatmaps. Information of DE genes and protein products was extracted from UniProt Consortium [26]. Enrichment analyses were performed on significant DE genes by the R package gprofiler2 [27].
2.6. Statistical Analysis
Flow cytometry and clinical data were analyzed by GraphPad Prism Version 8 (La Jolla, CA, USA). The statistical details of the experiments are provided in the respective figure legends. Normality of distribution was assessed by Shapiro–Wilk test. Comparison of the non-parametric continuous data between D+L and D-L cohorts was performed by two-tailed Mann–Whitney test, and categorical data were compared with Fisher’s exact test. Multivariate logistic regression was performed in R (version 4.0.2) (R Development Core Team, Vienna, Austria). Non-parametric data in this study are represented as median with interquartile range. p-Values < 0.05 (after adjustment if indicated) were considered statistically significant.
3. Results
3.1. Expansion of CD4+CD8+ DP T Cells Is Associated with Risk of Plasma Leakage in Dengue
A total of 88 dengue patients, including 55 with plasma leakage (D+L) and 33 without plasma leakage (D-L) were enrolled in the study and recruited from the North Colombo Teaching Hospital, Ragama in Gampaha District, Sri Lanka and the National Institute of Infectious Diseases, Gothatuwa, Angoda, Sri Lanka between 2010 and 2016. Demographic and clinical information of patient groups included in the analysis are summarized in Table 1 and described in details in methods. It is important to emphasize that the samples studied corresponded to blood draws obtained before any plasma leakage was detectable clinically or radiologically.
Initial phenotyping of peripheral blood mononuclear cells (PBMC) from the two groups (D+L and D-L) was determined by flow cytometry. Specifically, the proportions of CD4+CD8+ double-positive (DP), CD4+ single-positive (SP), and CD8+ SP T cells in acute DENV infection was assessed, using the gating strategy illustrated in Figure 1A. Representative donors from both D-L and D+L group are illustrated in Figure 1B. As shown in Figure 1C, we observed a significant higher proportion (p < 0.0001) of CD4+CD8+ DP T cells in the D+L group (median 1.45%, interquartile range (IQR): 1.13–2.07%) compared to the D-L group (median 0.77%, IQR: 0.31–1.10%), with no differences detected for CD4+ or CD8+ SP T cells (Figure S1). The detailed values of median and IQR for CD4+CD8+ DP T cells from two cohorts are summarized in Table 1.
To further evaluate the association between the proportion of CD4+CD8+ DP cells and plasma leakage, we conducted a multivariate logistic regression analysis accounting for other confounding factors, including demographic features (e.g., age, gender), previous DENV infection, as well as hematological and biochemical parameters (e.g., platelet count, hematocrit, AST, and ALT). As shown in Table 2, the percentage of CD4+CD8+ DP cells was the only significant factor (p = 0.002), positively correlated with plasma leakage (log (OR) = 1.86). These results suggested that the CD4+CD8+ DP T cell population is a novel and relevant marker of development of plasma leakage.
Table 2.
Multivariate analysis of risk factors associated with plasma leakage in dengue disease by a logistic regression model.
| Risk Factors | Estimate (β) | Standard Error | Z Score | padj Value |
|---|---|---|---|---|
| %CD4+CD8+ DP | 1.8632 | 0.5946 | 3.133 | 0.002 ** |
| Age (years) | 0.0151 | 0.0222 | 0.682 | 0.495 |
| Gender (M/F) | −0.7616 | 0.8402 | −0.907 | 0.365 |
| Secondary dengue (Y/N) | −0.1705 | 0.8201 | −0.208 | 0.835 |
| Platelet count (×1000/mm3) |
−0.0105 | 0.0082 | −1.280 | 0.200 |
| Hematocrit (%) | 0.0510 | 0.0766 | 0.666 | 0.505 |
| AST (U/L) | 0.0011 | 0.0031 | 0.369 | 0.712 |
| ALT (U/L) | 0.0001 | 0.0029 | 0.042 | 0.966 |
DP, double positive; M/F, male/female; Y/N, yes/no; AST, aspartate aminotransaminase; ALT, alanine aminotransferase, padj value, adjusted p value; Akaike information criterion (AIC): 81.106, ** p < 0.01.
3.2. The Gene Expression Profile of CD4+CD8+ DP T Cells Reveals a Higher Similarity to CD8+ T Cells as Opposed to CD4+ T Cells
To investigate the nature of CD4+CD8+ DP cells in acute DENV infection, we further characterized their transcriptomic profiles and compared with CD4+ or CD8+ single-positive (SP) cells. Cell subsets were sorted and RNA-seq performed, followed by clustering analyses using UMAP for dimensionality reduction. As shown in Figure 2A, at the transcriptomic level, CD4+CD8+ DP T cells were well resolved from CD4+ SP T cells but co-clustered with CD8+ SP T cells. This suggests that at the transcriptional level the CD4+CD8+ DP T cell population is most closely related to CD8+ SP T cells.
Figure 2.
Transcriptomic analysis of CD4+CD8+ DP T cells compared with CD4+ and CD8+ SP T cells in acute DENV infection. (A) UMAP plot of bulk RNA-seq datasets from acute DENV infection samples colored by cell group (after QC: n = 64 for CD4+CD8+ DP T cell group, n = 88 for CD4+ SP T cell group, and n = 87 for CD8+ SP T cell group), where each dot represented one RNA-seq data from one individual. (B) Volcano plot shows log2 fold change versus −log10 p-value for the comparison between CD4+CD8+ DP T cells and CD8+ SP T cells. The subset of genes with log2 fold change greater than 2 or less than −2 and adjusted p-value less than 0.05 are considered significant and indicated by dotted lines. (C) Heatmap shows the expression values after variance stabilizing transformation of the significant DE genes found between CD4+CD8+ DP T cells and CD8+ SP T cells. (D) Functional pathway enrichment analysis using g:Profiler for differentially expressed genes between CD4+CD8+ DP and CD8+ SP T cells; padj value means adjusted p value.
This result was confirmed by the analysis of which specific genes were differentially expressed (DE) between the various T-cell populations. Overall, there were 20 DE genes (absolute log2 fold change (FC) > 2, and adjusted p-value (false discovery rate, FDR) < 0.05) identified between CD4+CD8+ DP and CD8+ SP T cells (Figure 2B,C, Table 3) compared to 109 DE genes identified between CD4+CD8+ DP and CD4+ SP T cells (Figure S2 and Table S2). As expected, we observed increased expression of the CD8 (α and β) and CD4 genes by CD4+CD8+ DP T cells compared to CD4+ and CD8+ SP T cells, respectively.
Table 3.
List of DE genes found between CD4+CD8+ DP and CD8+ SP cells.
| Gene Symbol | Encoded Protein | p-Value | Log2FC | FDR | Gene Product Function * |
|---|---|---|---|---|---|
| Higher expression in CD4+CD8+ DP cells | |||||
| CD4 | T-cell surface glycoprotein CD4 | 4.11E-46 | 6.42 | 2.40E-41 | Cell differentiation antigen CD4, MHC class II receptor |
| TNFRSF4 | Tumor necrosis factor receptor superfamily member 4 (OX40) | 6.24E-11 | 3.19 | 3.64E-06 | Costimulatory molecule implicated in long-term T-cell immunity |
| CCR4 | C-C chemokine receptor type 4 (CCR4) | 5.80E-08 | 3.16 | 3.38E-03 | High-affinity binding for basophil chemoattractant, G protein coupled receptor superfamily, specific receptor for thymus and activation-regulated chemokine |
| NCF1 | Neutrophil cytosol factor 1 | 7.09E-10 | 2.08 | 4.14E-05 | Activate NADPH oxidase for superoxide production and oxygen dependent mechanism of phagocytosis |
| Reduced expression in CD4+CD8+ DP cells | |||||
| PTPRK | Receptor-type tyrosine-protein phosphatase kappa | 9.03E-08 | −2.06 | 5.27E-03 | Transmembrane receptor protein tyrosine phosphatase activity, regulation of processes involving cell contact and adhesion |
| CA6 | Carbonic anhydrase 6 | 1.58E-10 | −2.22 | 9.20E-06 | Carbonate dehydratase activity, protein binding |
| S100B | S100 Calcium-Binding Protein B | 6.52E-09 | −2.33 | 3.81E-04 | Calcium ion binding, protein homodimerization activity |
| ZNF433 | Zinc finger protein 433 | 3.47E-07 | −2.42 | 2.03E-02 | Transcriptional regulation, RNA polymerase II regulatory region sequence-specific DNA binding |
| DSEL | Dermatan-sulfate epimerase-like protein | 7.12E-08 | −2.45 | 4.15E-03 | Membrane proteins, sulfotransferase activity, isomerase activity |
| MMP28 | Matrix metalloproteinase-28 | 1.62E-07 | −2.50 | 9.48E-03 | Tissues homeostasis and repair, metalloendopeptidase activity |
| SPINK2 | Serine protease inhibitor Kazal-type 2 | 5.22E-07 | −2.67 | 3.04E-02 | Serine-type endopeptidase inhibitor activity |
| NT5E | 5′-nucleotidase | 9.12E-15 | −2.73 | 5.32E-10 | Hydrolyzes extracellular nucleotides into membrane permeable nucleosides |
| MXRA8 | Matrix remodeling-associated protein 8 | 1.56E-08 | −2.87 | 9.13E-04 | Transmembrane protein which can modulate activity of various signaling pathways |
| FBLN2 | Fibulin-2 | 2.67E-07 | −2.95 | 1.56E-02 | Extracellular matrix protein, expressed in elastic tissues including intima of blood vessels |
| CNN3 | Calponin-3 | 1.82E-11 | −2.98 | 1.06E-06 | Cytoskeletal protein involved in cell-cell adhesion, downregulated during dedifferentiation of vascular smooth muscle cell |
| PRCD | Photoreceptor disk component | 6.21E-09 | −3.15 | 3.62E-04 | Opsin binding |
| NOG | Noggin | 7.99E-13 | −3.26 | 4.66E-08 | Cytokine binding, protein homodimerization activity |
| GIPC3 | PDZ domain-containing protein | 5.15E-08 | −3.30 | 3.00E-03 | Protein binding |
| REG4 | Regenerating islet-derived protein 4 | 3.15E-15 | −3.48 | 1.84E-10 | Carbohydrate binding, signaling receptor activity |
| SDK2 | Protein sidekick-2 | 4.31E-08 | −3.62 | 2.52E-03 | Adhesion molecule |
Log2FC, log2 fold change; FDR, false-discovery rate; DE genes cut off: FDR < 0.05 and Log2FC > 2, * Gene protein products with functional annotation information was extracted from UniProt Consortium [26].
3.3. Molecular Pathways Associated with CD4+CD8+ DP T Cells
The analysis above suggested that CD4+CD8+ DP T cells are more closely related to CD8+ SP T cells in terms of their transcription profile. We next set out to examine which genes might be differentially expressed between the two T-cell compartments and might therefore define the specific gene expression pattern associated with CD4+CD8+ DP T cells. Compared to CD8+ SP T cells, we mainly observed in CD4+CD8+ DP T cells a higher expression of genes involved in T-cell activation (TNFRSF4, Tumor necrosis factor receptor superfamily member 4 or OX40), immune cell recruitment (CCR4, C-C chemokine receptor type 4), and oxygen dependent phagocytosis (NCF1, Neutrophil cytosol factor 1).
Significantly down-regulated DE genes in CD4+CD8+ DP T cells (Figure 2B,C) included PTPRK (Receptor-type tyrosine-protein phosphatase kappa), CA6 (Carbonic anhydrase 6), S100B (S100 Calcium-Binding Protein B), ZNF433 (Zinc finger protein 433), DSEL (Dermatan-sulfate epimerase-like protein), MMP28 (Matrix metalloproteinase-28), SPINK2 (Serine protease inhibitor Kazal-type 2), NT5E (5′-nucleotidase), MXRA8 (Matrix remodeling-associated protein 8), FBLN2 (Fibulin-2), CNN3 (Calponin-3), PRCD (Photoreceptor disk component), NOG (Noggin), GIPC3 (PDZ domain-containing protein), REG4 (Regenerating islet-derived protein 4), and SDK2 (Protein sidekick-2), representing intricate processes of cell differentiation and signaling in CD4+CD8+ cells (Table 3).
The functional pathway enrichment analysis using g:Profiler revealed that those DE genes were enriched in two pathways involved in cell surface receptor signaling pathway (CD4, TNFRSF4, CCR4, NT5E, MXRA8, PTPRK) and selective expression of chemokine receptors during T-cell polarization (CD4, CCR4) (Figure 2D). These results suggest that the transcriptomic profile of CD4+CD8+ DP T cells is more closely related CD8+ SP T cells with some differences detected.
3.4. Paucity of Transcriptomic Changes in CD4+CD8+ DP T Cells between Patients with and without Plasma Leakage in Dengue
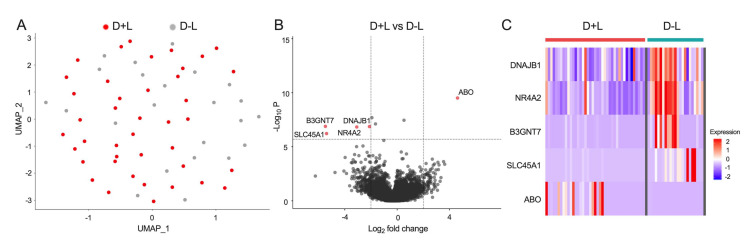
To further investigate the role of CD4+CD8+ DP T cells in the pathogenesis of plasma leakage in dengue, we compared transcriptomic profiles between the D+L and D-L groups during the acute febrile phase. CD4+CD8+ DP T cells isolated from both D+L and D-L groups displayed very similar gene expression profiles and clustered closely to one another based on UMAP analysis (Figure 3A).
Figure 3.
Comparison of transcriptomic signatures of CD4+CD8+ DP T cells from dengue with plasma leakage (D+L) cohort and dengue without plasma leakage (D-L) cohort. (A) UMAP plot of bulk RNA-seq datasets from acute DENV infection samples colored by disease groups (after QC: n = 41 for D+L group, n = 23 for D-L group), where each dot represented one RNA-seq data from one individual. (B) Volcano plot shows log2 fold change versus −log10 p-value for the comparison between CD4+CD8+ DP T cells from D+L and D-L groups. The subset of genes with log2 fold change greater than 2 or less than −2 and adjusted p-value less than 0.05 are considered significant and indicated by dotted lines. (C) Heatmap shows the expression values after variance stabilizing transformation of the significant DE genes found between CD4+CD8+ DP T cells from D+L and D-L groups.
Only five significant DE genes were identified (Figure 3B,C), which included in D+L group the down-regulated genes DNAJB1 (DnaJ homolog subfamily B member 1), NR4A2 (Nuclear receptor subfamily 4 group A member 2), B3GNT7 (UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase), and SLC45A1 (Proton-associated sugar transporter A) involved in gene expression and post-translational protein modification as well as the up-regulated gene ABO (Histo-blood group ABO system transferase) gene (Table 4).
Table 4.
List of DE genes found between CD4+CD8+ DP cells of D+L and D-L cohorts.
| Gene Symbol | Encoded Protein | p-Value | Log2FC | FDR | Gene Product Function * |
|---|---|---|---|---|---|
| Reduced expression in CD4+CD8+ DP cells of D+L cohort | |||||
| DNAJB1 | DnaJ homolog subfamily B member 1 | 1.36E-07 | −2.12 | 7.96E-03 | ATPase activator activity, transcription corepressor activity, Hsp70 protein binding, involved in protein folding |
| NR4A2 | Nuclear receptor subfamily 4 group A member 2 | 1.49E-07 | −3.08 | 8.72E-03 | RNA polymerase II regulatory region sequence-specific DNA binding |
| B3GNT7 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase | 1.31E-07 | −5.48 | 7.66E-03 | Acetylglucosaminyltransferase activity, protein binding |
| SLC45A1 | Proton-associated sugar transporter A | 6.06E-07 | −5.39 | 3.53E-02 | Sucrose:proton symporter activity |
| Higher expression in CD4+CD8+ DP cells of D+L cohort | |||||
| ABO | Histo-blood group ABO system transferase | 3.11E-10 | 4.62 | 1.82E-05 | Glycosyltransferase activity |
Log2FC, log2 fold change; FDR, false-discovery rate; DE genes cut off: FDR < 0.05 and Log2FC > 2, * Gene protein products with functional annotation information was extracted from UniProt Consortium [26].
4. Discussion
This study provided several novel observations on the host immune response during the acute phase of DENV infection. First, the report that the frequency of a novel CD4+CD8+ DP T cell subset was increased in D+L versus D-L, and it is thus associated with risk of plasma leakage in dengue disease. Second, on the basis of their transcriptomic profile, CD4+CD8+ DP T cells were most closely related to CD8+ SP T cells, but still, we were able to define 20 DE genes enriched in two functional pathways representative of selective expression of chemokine receptors and signaling on the cell surface during T-cell polarization. Third, while the transcriptomic profile of the CD4+CD8+ DP T cells was rather similar in D+L and D-L patients, five DE genes were identified.
CD4+CD8+ DP T cells have been described in several pathological conditions in comparison to normal individuals [28]; for example, expansion of DP cells has been observed in autoimmune diseases (e.g., Myasthenia gravis, Rheumatoid arthritis, multiple sclerosis, Kawasaki diseases, and systemic sclerosis), infectious diseases (e.g., HIV and EBV), and cancers (e.g., melanoma, lymphoma) [28]. However, their phenotype and role in DENV infection have not been reported. Peripheral CD4+CD8+ DP T cells are a phenotypically and functionally heterogeneous population depending on their origin and pathologic context, and their roles in the pathogenesis of autoimmune diseases, viral infections, and cancers are under ongoing debate [29]. While CD4+CD8+ DP T cells appear to function at peripheral sites as potent immune suppressors [30,31] or cells with high cytotoxic potential [32,33], a more thorough examination of their contribution to the adaptive immunity in human diseases is warranted [29].
Here, we observed that an increased frequency of CD4+CD8+ DP T cells during acute DENV infection was associated with development of plasma leakage in dengue disease. Furthermore, CD4+CD8+ DP T cells appear to be the most significant factor associated with development of plasma leakage even after adjusting for other reported risk factors, such as age, gender, platelet count, hematocrit, AST, ALT levels, and secondary dengue infection [10]. These results indicate that CD4+CD8+ DP T cells are a novel factor that warrants further investigation, especially taking into consideration of the potential implication in clinical settings and pathophysiology studies of plasma leakage (both onset and severity) in DENV infection by collecting bloods at various time points in future. In particular, the current study did not address the viral load in the blood nor the antigen specificity of the CD4+CD8+ DP T cells, and future studies will have to address their correlation with the immune response and what fraction of these CD4+CD8+ DP T cells is actually DENV specific.
In addition, the origin and function of peripheral CD4+CD8+ DP T cells in various pathological conditions are not fully understood to date. Parrot et al. reported that intra-melanoma DP T cells were transcriptome-wise closer to CD8+ SP T cells [34], while Mucida et al. found mature CD4+ T helper cells could undergo transcriptomic re-programming and generate cytotoxic CD4+CD8+ DP T cells after persistent ovalbumin antigen stimulation [35]. However, those studies only investigated limited genes by microarray, and there was no systemic RNA-sequencing (RNA-seq) data comparing transcriptomic profiles of CD4+CD8+ DP with CD4+ or CD8+ SP T cells.
Our study reports the first set of RNA-seq data of CD4+CD8+ DP T cells, and the finding that their transcriptomic profile is most closely related to CD8+ SP T cells in acute DENV infection. The principal transcripts distinguishing CD4+CD8+ DP T cells from CD8+ SP T cells encoded OX40 and CCR4, which were found up-regulated in CD4+CD8+ DP compared to CD8+ SP T cells. Interestingly, OX40 has been established as a crucial signaling molecule during persistent viral infection [36] and implicated in their pathology through maintenance of higher numbers of long-lived CD8+ effector T cells [37]. In addition, a differential regulation of the expression of the OX40 signaling pathway has been observed in asymptomatic dengue cases compared with clinical cases [38] and associated with dengue disease severity [39]. Moreover, clinical studies in endemic areas have described the associations between the levels of CC chemokines and dengue disease outcome [40,41]. In particular, the CCR4 receptor has been shown to contribute to the pathogenesis of severe conditions [42,43], including dengue [44]. Collectively, our results indicate that CD4+CD8+ DP T cells are effector T cells that are transcriptome-wise closer to CD8+ SP T cells during acute DENV infection.
While the gene expression profile of CD4+CD8+ DP T in D+L and D-L was relatively similar, DE genes were identified. Interestingly, the ABO gene was the only up-regulated gene, associated with plasma leakage during acute DENV infection. This gene encodes A and B antigens of human blood group and is autosomal dominant [45]. Studies have shown that ABO blood group plays a role in viral infection and host susceptibility, such as norovirus, rotavirus, HIV, influenza viruses, as well as SARS coronavirus (SARS-CoV) [45]. Kalayanarooj et al. found that blood group AB was associated with increased risk of severe dengue disease with plasma leakage in secondary infections [46], consistent with our findings. These differentially expressed genes, together with those discriminating CD4+CD8+ DP T cells from CD8+ SP T cells, could have clinical utility, perhaps as PCR-based markers.
Finally, it can be noted that the results presented herein do not support the previous postulate of original antigenic sin and altered responses [47,48] as key drivers of DENV immunopathology. Rather, they can be interpreted in a growing body of studies [49,50,51,52] that indicate that the T-cell responses and T-cell subsets are modulated in DENV disease and that differential disease severity is mostly associated with differences in magnitude of responses rather than phenotypic differences of different T-cell compartments modulated in the course of infection and disease. That is, severe disease is characterized by an infected cell mass that is larger than that of mild disease, and an enhanced antigen mass stimulates a greater magnitude of T-cell responses.
5. Conclusions
Plasma leakage in dengue diseases is a warning complication which requires early detection and clinical intervention. In this study, we observed increased frequency of a novel CD4+CD8+ double-positive (DP) T-cell subset in the acute febrile phase of the DENV infection is associated with risk of developing plasma leakage later in dengue disease. Further transcriptomic analysis suggested this T-cell subset is closest to CD8+ single-positive (SP) T cells. Nonetheless, the transcriptomic profile of the CD4+CD8+ DP T cells was largely similar in patients with and without plasma leakage, implying a role of CD4+CD8+ DP cells in plasma leakage through a quantitative increase rather than functional alteration. These data provide new insight into the host immune response in DENV infection and the role of CD4+CD8+ DP T cells in the pathogenesis of plasma leakage.
Acknowledgments
We wish to acknowledge all subjects for their participation and for donating their blood and time for this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v14010090/s1, Figure S1: Distribution of CD4+ and CD8+ single positive (SP) in D-L and D+L cohorts; Figure S2: Transcriptomic analysis of CD4+CD8+ DP T cells compared with CD4+ SP T cells in acute DENV infection; Table S1: List of antibodies used in the flowcytometry study; Table S2: List of DE genes found between CD4+CD8+ DP and CD4+ SP cells.
Author Contributions
Designing research studies, A.S., D.W. and E.D.Y.; investigation, E.D.Y., H.W., R.d.S.A., Y.T. and A.G.; data analysis, E.D.Y., H.W. and Y.T.; resources, R.T., S.U.A., G.P., A.W., S.P. and A.D.D.S.; manuscript writing, E.D.Y., H.W., R.d.S.A. and A.S.; supervision, A.S. and D.W.; project administration, A.F.; funding acquisition, A.S. and D.W. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number U19AI118626, HHSN272201400045C, and 75N93019C00065. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
This study was approved by the Human Subjects Protection Programs of the La Jolla Institute for Immunology (Protocols VD-085 and VD-101), the University of Colombo (IRB Protocols EC-15-002 and EC-15-095), the Nicaragua Ministry of Health (Protocol CIRE-01/10/06-13.Ver14), and the University of California Berkeley (Protocol 2010-06-1649).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database under the accession code GSE178240. The other datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors have declared that no conflict of interest exists.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization . Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. World Health Organization; Geneva, Switzerland: 2009. [PubMed] [Google Scholar]
- 2.Centre for Disease Control and Prevention About Dengue: What You Need to Know. [(accessed on 15 April 2021)]; Available online: https://www.cdc.gov/dengue/about/index.html.
- 3.Deen J.L., Harris E., Wills B., Balmaseda A., Hammond S.N., Rocha C., Dung N.M., Hung N.T., Hien T.T., Farrar J.J. The WHO dengue classification and case definitions: Time for a reassessment. Lancet. 2006;368:170–173. doi: 10.1016/S0140-6736(06)69006-5. [DOI] [PubMed] [Google Scholar]
- 4.Phuong C.X., Nhan N.T., Kneen R., Thuy P.T., van Thien C., Nga N.T., Thuy T.T., Solomon T., Stepniewska K., Wills B., et al. Clinical diagnosis and assessment of severity of confirmed dengue infections in Vietnamese children: Is the world health organization classification system helpful? Am. J. Trop. Med. Hyg. 2004;70:172–179. doi: 10.4269/ajtmh.2004.70.172. [DOI] [PubMed] [Google Scholar]
- 5.Grifoni A., Voic H., Mateus J., Fung K.M.Y., Wang A., Seumois G., De Silva A.D., Tennekon R., Premawansa S., Premawansa G., et al. Transcriptomics of acute DENV-specific CD8+ T cells does not support qualitative differences as drivers of disease severity. medRxiv. 2021 doi: 10.1101/2021.09.01.21262833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander N., Balmaseda A., Coelho I.C., Dimaano E., Hien T.T., Hung N.T., Janisch T., Kroeger A., Lum L.C., Martinez E., et al. Multicentre prospective study on dengue classification in four South-east Asian and three Latin American countries. Trop. Med. Int. Health. 2011;16:936–948. doi: 10.1111/j.1365-3156.2011.02793.x. [DOI] [PubMed] [Google Scholar]
- 7.Rigau-Perez J.G. Severe dengue: The need for new case definitions. Lancet Infect. Dis. 2006;6:297–302. doi: 10.1016/S1473-3099(06)70465-0. [DOI] [PubMed] [Google Scholar]
- 8.Rajapakse S. Dengue shock. J. Emerg. Trauma Shock. 2011;4:120–127. doi: 10.4103/0974-2700.76835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee I.K., Hsieh C.J., Lee C.T., Liu J.W. Diabetic patients suffering dengue are at risk for development of dengue shock syndrome/severe dengue: Emphasizing the impacts of co-existing comorbidity(ies) and glycemic control on dengue severity. J. Microbiol. Immunol. Infect. 2020;53:69–78. doi: 10.1016/j.jmii.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Sangkaew S., Ming D., Boonyasiri A., Honeyford K., Kalayanarooj S., Yacoub S., Dorigatti I., Holmes A. Risk predictors of progression to severe disease during the febrile phase of dengue: A systematic review and meta-analysis. Lancet Infect. Dis. 2021;21:1014–1026. doi: 10.1016/S1473-3099(20)30601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M., et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez R.J., Vazquez S. Serological diagnosis of dengue by an ELISA inhibition method (EIM) Mem. Inst. Oswaldo Cruz. 1990;85:347–351. doi: 10.1590/S0074-02761990000300012. [DOI] [PubMed] [Google Scholar]
- 13.Seah C.L., Chow V.T., Tan H.C., Can Y.C. Rapid, single-step RT-PCR typing of dengue viruses using five NS3 gene primers. J. Virol. Methods. 1995;51:193–200. doi: 10.1016/0166-0934(94)00104-O. [DOI] [PubMed] [Google Scholar]
- 14.Sudiro T.M., Ishiko H., Green S., Vaughn D.W., Nisalak A., Kalayanarooj S., Rothman A.L., Raengsakulrach B., Janus J., Kurane I., et al. Rapid diagnosis of dengue viremia by reverse transcriptase-polymerase chain reaction using 3′-noncoding region universal primers. Am. J. Trop. Med. Hyg. 1997;56:424–429. doi: 10.4269/ajtmh.1997.56.424. [DOI] [PubMed] [Google Scholar]
- 15.Kanakaratne N., Wahala W.M., Messer W.B., Tissera H.A., Shahani A., Abeysinghe N., de-Silva A.M., Gunasekera M. Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg. Infect. Dis. 2009;15:192–199. doi: 10.3201/eid1502.080926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iuvone T., D’Acquisto F., Van Osselaer N., Di Rosa M., Carnuccio R., Herman A.G. Evidence that inducible nitric oxide synthase is involved in LPS-induced plasma leakage in rat skin through the activation of nuclear factor-kappaB. Br. J. Pharmacol. 1998;123:1325–1330. doi: 10.1038/sj.bjp.0701730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seumois G., Vijayanand P., Eisley C.J., Omran N., Kalinke L., North M., Ganesan A.P., Simpson L.J., Hunkapiller N., Moltzahn F., et al. An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol. 2012;1:70–89. [PMC free article] [PubMed] [Google Scholar]
- 18.Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 19.Trapnell C., Pachter L., Salzberg S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., Wu Y., Tian W. A novel approach to remove the batch effect of single-cell data. Cell Discov. 2019;5:46. doi: 10.1038/s41421-019-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.UniProt C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parel Y., Chizzolini C. CD4+ CD8+ double positive (DP) T cells in health and disease. Autoimmun. Rev. 2004;3:215–220. doi: 10.1016/j.autrev.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Overgaard N.H., Jung J.W., Steptoe R.J., Wells J.W. CD4+/CD8+ double-positive T cells: More than just a developmental stage? J. Leukoc. Biol. 2015;97:31–38. doi: 10.1189/jlb.1RU0814-382. [DOI] [PubMed] [Google Scholar]
- 30.Das G., Augustine M.M., Das J., Bottomly K., Ray P., Ray A. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 2003;100:5324–5329. doi: 10.1073/pnas.0831037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szczepanik M., Bryniarski K., Tutaj M., Ptak M., Skrzeczynska J., Askenase P.W., Ptak W. Epicutaneous immunization induces alphabeta T-cell receptor CD4 CD8 double-positive non-specific suppressor T cells that inhibit contact sensitivity via transforming growth factor-beta. Immunology. 2005;115:42–54. doi: 10.1111/j.1365-2567.2005.02127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitchen S.G., Jones N.R., LaForge S., Whitmire J.K., Vu B.A., Galic Z., Brooks D.G., Brown S.J., Kitchen C.M., Zack J.A. CD4 on CD8(+) T cells directly enhances effector function and is a target for HIV infection. Proc. Natl. Acad. Sci. USA. 2004;101:8727–8732. doi: 10.1073/pnas.0401500101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitchen S.G., Whitmire J.K., Jones N.R., Galic Z., Kitchen C.M., Ahmed R., Zack J.A. The CD4 molecule on CD8+ T lymphocytes directly enhances the immune response to viral and cellular antigens. Proc. Natl. Acad. Sci. USA. 2005;102:3794–3799. doi: 10.1073/pnas.0406603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parrot T., Oger R., Allard M., Desfrancois J., de la Raingeard Bletiere D., Coutolleau A., Preisser L., Khammari A., Dreno B., Delneste Y., et al. Transcriptomic features of tumour-infiltrating CD4(low)CD8(high) double positive alphabeta T cells in melanoma. Sci. Rep. 2020;10:5900. doi: 10.1038/s41598-020-62664-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mucida D., Husain M.M., Muroi S., van Wijk F., Shinnakasu R., Naoe Y., Reis B.S., Huang Y., Lambolez F., Docherty M., et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boettler T., Moeckel F., Cheng Y., Heeg M., Salek-Ardakani S., Crotty S., Croft M., von Herrath M.G. OX40 facilitates control of a persistent virus infection. PLoS Pathog. 2012;8:e1002913. doi: 10.1371/journal.ppat.1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphreys I.R., Walzl G., Edwards L., Rae A., Hill S., Hussell T. A critical role for OX40 in T cell-mediated immunopathology during lung viral infection. J. Exp. Med. 2003;198:1237–1242. doi: 10.1084/jem.20030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon-Loriere E., Duong V., Tawfik A., Ung S., Ly S., Casademont I., Prot M., Courtejoie N., Bleakley K., Buchy P., et al. Increased adaptive immune responses and proper feedback regulation protect against clinical dengue. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal5088. [DOI] [PubMed] [Google Scholar]
- 39.Nikolayeva I., Bost P., Casademont I., Duong V., Koeth F., Prot M., Czerwinska U., Ly S., Bleakley K., Cantaert T., et al. A Blood RNA Signature Detecting Severe Disease in Young Dengue Patients at Hospital Arrival. J. Infect. Dis. 2018;217:1690–1698. doi: 10.1093/infdis/jiy086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bozza F.A., Cruz O.G., Zagne S.M., Azeredo E.L., Nogueira R.M., Assis E.F., Bozza P.T., Kubelka C.F. Multiplex cytokine profile from dengue patients: MIP-1beta and IFN-gamma as predictive factors for severity. BMC Infect. Dis. 2008;8:86. doi: 10.1186/1471-2334-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spain-Santana T.A., Marglin S., Ennis F.A., Rothman A.L. MIP-1 alpha and MIP-1 beta induction by dengue virus. J. Med. Virol. 2001;65:324–330. doi: 10.1002/jmv.2037. [DOI] [PubMed] [Google Scholar]
- 42.Vijayanand P., Durkin K., Hartmann G., Morjaria J., Seumois G., Staples K.J., Hall D., Bessant C., Bartholomew M., Howarth P.H., et al. Chemokine receptor 4 plays a key role in T cell recruitment into the airways of asthmatic patients. J. Immunol. 2010;184:4568–4574. doi: 10.4049/jimmunol.0901342. [DOI] [PubMed] [Google Scholar]
- 43.Yuan Q., Bromley S.K., Means T.K., Jones K.J., Hayashi F., Bhan A.K., Luster A.D. CCR4-dependent regulatory T cell function in inflammatory bowel disease. J. Exp. Med. 2007;204:1327–1334. doi: 10.1084/jem.20062076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guabiraba R., Marques R.E., Besnard A.G., Fagundes C.T., Souza D.G., Ryffel B., Teixeira M.M. Role of the chemokine receptors CCR1, CCR2 and CCR4 in the pathogenesis of experimental dengue infection in mice. PLoS ONE. 2010;5:e15680. doi: 10.1371/journal.pone.0015680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooling L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015;28:801–870. doi: 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalayanarooj S., Gibbons R.V., Vaughn D., Green S., Nisalak A., Jarman R.G., Mammen M.P., Jr., Perng G.C. Blood group AB is associated with increased risk for severe dengue disease in secondary infections. J. Infect. Dis. 2007;195:1014–1017. doi: 10.1086/512244. [DOI] [PubMed] [Google Scholar]
- 47.Mongkolsapaya J., Dejnirattisai W., Xu X.N., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S., Duangchinda T., Dong T., Rowland-Jones S., et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 48.Rothman A.L. Immunity to dengue virus: A tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011;11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 49.Tian Y., Babor M., Lane J., Schulten V., Patil V.S., Seumois G., Rosales S.L., Fu Z., Picarda G., Burel J., et al. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat. Commun. 2017;8:1473. doi: 10.1038/s41467-017-01728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Y., Babor M., Lane J., Seumois G., Liang S., Goonawardhana N.D.S., De Silva A.D., Phillips E.J., Mallal S.A., da Silva Antunes R., et al. Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles. J. Clin. Investig. 2019;129:1727–1741. doi: 10.1172/JCI123726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Y., Sette A., Weiskopf D. Cytotoxic CD4 T Cells: Differentiation, Function, and Application to Dengue Virus Infection. Front. Immunol. 2016;7:531. doi: 10.3389/fimmu.2016.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian Y., Seumois G., De-Oliveira-Pinto L.M., Mateus J., Herrera-de la Mata S., Kim C., Hinz D., Goonawardhana N.D.S., de Silva A.D., Premawansa S., et al. Molecular Signatures of Dengue Virus-Specific IL-10/IFN-gamma Co-producing CD4 T Cells and Their Association with Dengue Disease. Cell Rep. 2019;29:4482–4495.e4. doi: 10.1016/j.celrep.2019.11.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database under the accession code GSE178240. The other datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.