Abstract
All vaccines that are prepared in chicken embryo fibroblasts (CEFs) contain a low level of particle-associated reverse transcriptase (RT) activity, which is produced from the avian cell substrate. The RNAs present in the particles have sequence homology to viral DNAs belonging to the ancient endogenous avian virus (EAV) family or to the avian sarcoma-leukosis virus (ALV)-related subgroup E endogenous virus loci. Although no replication-competent retrovirus has been associated with the RT activity produced from CEFs, there have been some theoretical safety concerns regarding potential consequences of integration of EAV and ALV sequences in human DNA, which may result from nonproductive infection with replication-defective particles or infection with EAV and ALV pseudotypes bearing measles virus envelopes. To address these possibilities, we have analyzed EAV and ALV particles in a measles virus vaccine equivalent (MVVE) preparation, obtained from a U.S. manufacturer, for integration and for replication in human peripheral blood mononuclear cells (PBMCs). The results show the absence of EAV and ALV integrants in DNA prepared from MVVE-inoculated human cells by direct DNA PCR and Alu PCR assays and no propagation of retrovirus in 18-day cultures of MVVE-inoculated human PBMCs by a highly sensitive PCR-based RT assay. These results provide further confidence regarding the safety of chicken RT activity in live viral vaccines and support the continued use of chick-cell-derived vaccines in humans.
Chicken embryo fibroblast (CEF) cultures are used for the production of live, attenuated measles virus (MV) and mumps virus vaccines licensed in the United States (19, 38, 42). These vaccines are generally administered during childhood (5) as a trivalent vaccine in combination with the attenuated rubella virus vaccine, which is grown in human diploid cells. These vaccines are currently produced by a single manufacturer in the United States and are effective in disease prevention (6). The cells used in vaccine production are required to be obtained from embryonated chicken eggs that are known or demonstrated to be free of adventitious agents pathogenic for chickens (38, 42). Recently, it was found that all chick-cell-derived vaccines, including MV and mumps virus, produced in the United States as well as in Europe contained particle-associated reverse transcriptase (RT) activity (2, 20, 30, 41). Furthermore, it was shown that the particles contained RNAs (37, 40) derived from the ancient endogenous avian virus (EAV) family designated EAV-0 (4, 28) and from subgroup E endogenous virus loci related to avian sarcoma-leukosis viruses (ALV) (9, 15). An early report showed that concentrated supernatants from CEFs contained RT activity that was not infectious for avian cells (1). The production of RT activity from CEF cultures was recently confirmed using highly sensitive PCR-based RT assays, and no replication-competent retrovirus was detected in infectivity studies using human peripheral blood mononuclear cells (PBMCs) and a variety of human and other cell lines (16, 30). These results supported the recommendation of the World Health Organization for continued use of chick-cell-derived vaccines, which was initially based upon an extensive safety record and the lack of evidence for any real health concerns regarding human use. However, theoretical concerns remained, such as those related to potential consequences of integration as a result of nonproductive infection with defective EAV and ALV particles and/or infection with EAV and ALV pseudotypes containing MV envelopes that might potentially be present in the vaccine. We have used sensitive detection assays to evaluate the infection, integration, and replication in human cells of particle-associated EAV and ALV sequences present in U.S.-manufactured MV vaccine.
MATERIALS AND METHODS
Vaccines, cells, and viruses.
Bulk lots of U.S.-licensed live attenuated MV vaccine released in 1992 (designated here as lots 12 and 29) and an MV vaccine equivalent (MVVE) preparation (a preclarified virus pool) prepared in 1996 for validation of production equipment and assessed by the manufacturer (105.1 50% tissue culture infective doses [TCID50] per 0.1 ml in Vero cells) were used in this study. It should be noted that MVVE underwent minimal freezing-thawing prior to use in this study.
Human PBMCs were from the same cryopreserved cell stock as that used in a previous study to demonstrate the absence of replication-competent retrovirus in primary CEF culture supernatants (16). Prior to infection, the cells (106 per ml) were stimulated with phytohemagglutinin (PHA; 2.5 μg per ml; Murex Diagnostics, Dartford, United Kingdom) for 72 h in RPMI complete medium (RPMI 1640 [Quality Biologicals, Gaithersburg, Md.] containing 10% fetal bovine serum [HyClone, Logan, Utah], 2 mM l-glutamine, 10 mM HEPES [pH 7.0], 100 μg of streptomycin per ml, and 100 U of penicillin per ml); then, the medium was replaced with RPMI complete medium supplemented with 10% interleukin-2 (Hemagen Diagnostics, Columbia, Md.). Amphotropic 4070A murine leukemia virus (AMLV) (14, 27) was kindly provided by Janet Hartley (National Institutes of Health, Bethesda, Md.), and a large-scale preparation of the virus was made using Mus dunni cells and stored in aliquots at −80°C. The virus titer was determined with sarcoma-positive, leukemia-negative (S+ L−) PG4 cells (1.23 × 106 focus-forming units [FFU] per ml; Biological Research Faculty and Facility, Inc., Ijamsville, Md.). Avian Rous-associated virus (RAV)-D (strain RAV-50; 5 × 106 infective doses [ID] per ml for CEF as determined by complement fixation assay) and RAV-E (strain RAV-0; 107 50% infective doses [ID50] per ml) were obtained from The American Type Culture Collection (Chantilly, Va.). RAV-50 was selected to represent subgroup D avian retroviruses, which are able to infect some mammalian cells, and RAV-0 is an endogenous avian retrovirus of subgroup E which is present in the chicken genome (26, 34).
Inoculation of cells for infectivity and integration analyses.
PHA-stimulated PBMCs were incubated for 24 h with MVVE, primary CEF culture supernatants, or retrovirus in the presence of Polybrene (4 μg per ml; to enhance retrovirus infection [8, 36]). Uninoculated cells were included as controls. For analysis of a replicating retrovirus, PBMCs (2 × 106 cells) in a 12-well plate were incubated in a total volume of 2 ml of medium with MVVE (0.2 ml), MVVE spiked with AMLV (50 μl; 6.15 × 104 FFU) without DNase treatment, and MVVE spiked with AMLV (50 μl; 6.15 × 104 FFU) with DNase (DNase I; RNase free and determined pure by fast protein liquid chromatography; Pharmacia Biotech, Uppsala, Sweden) treatment (13 U of enzyme per 100 μl) at 37°C for 30 min in 40 mM Tris-HCl (pH 7.5)–6 mM MgCl2. After 24 h, the cells were transferred to six-well plates. Subsequently, one-half of the medium was replaced every 3 or 4 days and the cells were transferred to 25-cm2 flasks and then to 75-cm2 flasks to maintain about 106 cells per ml. At each medium change, the supernatant was centrifuged at 3,000 rpm and 4°C for 10 min (GS-6KR centrifuge with a GH-3.8 horizontal rotor; Beckman, Columbia, Md.) to pellet the cells; the cells were added back to the culture, and the supernatant was filtered and stored at −80°C until culture termination. The supernatant samples were analyzed by a PCR-based RT assay and cellular DNA was analyzed by PCR as described below.
For integration analysis using MMVE, PBMCs (4 × 106) in a six-well plate were incubated with 0.4 ml of MVVE without DNase treatment or immediately after DNase treatment (16.4 U of enzyme per 100 μl under the conditions described above) in a total volume of 4 ml of medium. After 24 h, three-fourths of the cells were used to prepare DNA for direct PCR and Alu PCR analyses, and one-fourth of the cells were used to prepare MV RNA for RT-PCR analysis. DNA or RNA was prepared as described below. Additional DNA for Alu PCR analysis was prepared from PBMCs (4 × 106) in a six-well plate incubated with MVVE (0.4 ml) or AMLV (0.25 ml; 3.1 × 105 FFU) in a total volume of 4 ml of medium. At 72 h postincubation, the cells were pelleted, washed three times with 10 ml of cold phosphate-buffered saline (PBS), and resuspended in 500 μl of PBS. The cells were treated with 100 U of DNase in 10 mM MgCl2 for 1 h at 37°C and again washed with PBS. Cellular DNA was prepared as described below, except that after cell lysis, the sample was incubated at 37°C for 1 h and proteinase K (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) was inactivated by heating at 95°C for 10 min. As a positive control for direct DNA PCR analysis, PBMCs (2 × 106) were incubated in a total volume of 4 ml of medium with MVVE (0.2 ml) spiked with AMLV (100 μl; 1.23 × 105 FFU) with and without DNase treatment. PBMCs were centrifuged at 3,000 rpm (Beckman centrifuge; see above) for 10 min at 4°C and washed three times each with 10 ml of cold PBS (without Mg2+ and Ca2+). After the last wash, the cells were resuspended in 1 ml of PBS, transferred to a 1.5-ml polypropylene centrifuge tube, pelleted again at 3,000 rpm (see above), and stored at −80°C.
To evaluate infection of PBMCs with known avian retroviruses, DNA was prepared for PCR from cells (4 × 106) in a six-well plate infected with RAV-0 (103 ID50) and RAV-50 (1 × 103 ID50 and 5 × 105 ID50) in a total volume of 4 ml of medium. AMLV (6.15 × 105 FFU) was included as a positive control virus. At 72 h postinfection, cells were centrifuged at 1,200 rpm (Beckman centrifuge; see above) for 10 min at 4°C and washed three times each with 5 ml of cold PBS, and DNA was prepared as described below.
For integration analysis of retrovirus particles produced from CEFs, PBMCs (2 × 106) in a 12-well plate were incubated with primary CEF culture supernatant (0.5 ml; day-4 sample from a previous study [16]) without DNase treatment and with DNase treatment (11 U of enzyme per 100 μl) in a total volume of 2 ml of medium. As a positive control, PBMCs were incubated with CEFs spiked with AMLV (100 μl) with and without DNase treatment. At 24 h postincubation, the cells were centrifuged and washed three times each with 5 ml of PBS as described above and DNA was prepared for direct PCR analysis as described below. Additionally, frozen cell pellets (−80°C) from passage 1 and passage 5 in a previous experiment (16) in which human osteosarcoma (HOS) cells had been incubated with CEF culture supernatant or with simian foamy virus (SFV) as a positive control were analyzed for integration of EAV and SFV sequences, respectively, by direct PCR and additionally by Alu PCR for EAV sequences.
STF-PERT assay.
The TM-PERT assay (21) was modified (designated the single-tube fluorescent-product-enhanced RT [STF-PERT] assay) so that the RT reaction and the PCR were done in the same tube with AmpliWax (PE Applied Biosystems, Foster City, Calif.) by the hot-start technique. The RT standard was prepared by diluting avian myeloblastosis virus (AMV) RT (Promega, Madison, Wis.; 10 U per μl) in NZ buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 0.25 mM EDTA [pH 8.0], 0.025% Triton X-100, 50% glycerol, 0.2 mM dithiothreitol [DTT]) to prepare the initial dilution containing 30 U in 30 μl. From this dilution, serial 10-fold dilutions were made so that the highest dilution tested (10−13) contained 10 pU or 0.83 RT molecule in 10 μl and the 10−11 dilution, which contained 103 pU in 10 μl, corresponded to 83 RT molecules, or about one particle of human immunodeficiency virus type 1 (which contains about 80 RT molecules) (18) or AMV (which contains about 70 RT molecules) (25). Aliquots of the dilutions were stored at −80°C, and dilutions ranging from 10−4 to 10−13 were assayed to obtain a standard curve.
Samples were diluted 1:10 in NZ buffer; negative controls were set up in triplicate and consisted of no template and NZ buffer. MicroAmp optical tubes (PE Applied Biosystems) were set up with 25 μl of the PCR mixture (see below) and one pellet of AmpliWax PCR Gem 50 (PE Applied Biosytems). After 5 min of incubation at 60°C to melt the wax, the tubes were cooled to 37°C before 15 μl of the RT reaction mixture was added to the top of the solid wax. Ten microliters of a sample was added directly to the RT mixture, resulting in final reaction concentrations of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 0.35% Triton X-100, 200 μM each deoxynucleoside triphosphate (dNTP; PE Applied Biosystems), 2 mM freshly diluted DTT, 1.3 μM primer A (5′-GCCTTAGCAGTGCCCTGTCT-3′), 8 U of RNasin (Promega), and 300 ng of MS2 RNA template (Boehringer). The PCR mixture consisted of 10 mM Tris-HCl (pH 8.8), 200 μM each dNTP, 25 pmol of primer A, 25 pmol of primer B (5′-AACATGCTCGAGGGCCTTA-3′), 7.5 pmol of probe (5′-FAM-CCCGTGGGATGCTCCTACATGTCA-TAMRA-3′) (FAM and TAMRA amidites from PE Applied Biosystems and oligonucleotide primers A and B from the Facility for Biotechnology Resources, Center for Biologics Evaluation and Research, have been previously described [21]), 500 ng of RNase (DNase free; Boehringer), and 2.5 U of AmpliTaq Gold (PE Roche Molecular Systems, Branchburg, N.J.). The tubes were placed in a PRISM 7700 sequence detection system, and incubations were controlled by sequence detector system software (PE Applied Biosystems) with the following thermal cycler conditions: 37°C for 89 min, 95°C for 10 min, and 50 cycles at 95°C for 15 s and 56°C for 30 s. Data were analyzed using the same software.
The sensitivity of RT detection in the STF-PERT was 103 to 104 pU, which is equivalent to about 1 to 10 virions.
Cellular DNA preparation and PCR analysis.
Total cellular DNA was isolated from MVVE (1 ml) by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and then with chloroform-isoamyl alcohol (24:1). DNA was precipitated from the aqueous phase with ethanol, and the washed pellet (two washes in 70% ethanol) was resuspended in 100 μl of 10 mM Tris-HCl (pH 7.5). Five microliters was used for PCR amplification of EAV, ALV, and chicken repeat 1 element (CRE) sequences.
Cellular DNA was prepared from cryopreserved stock of uninoculated CEFs (prepared from a primary cell culture), from stored (−80°C) pellets of passage 1 and passage 5 HOS cells which had been incubated in a previous study with CEF culture supernatant or with SFV as a positive control (16), and from human PBMCs incubated in this study with MVVE, CEF supernatant, AMLV, or MVVE spiked with AMLV with and without DNase treatment (described above). Cells were washed three times with PBS (except for frozen HOS cell pellets, which had been washed prior to freezing); resuspended in buffer containing 10 mM Tris-HCl (pH 8.3), 100 mM KCl, and 2.5 mM MgCl2 (designated solution A); and lysed in an equal volume of buffer containing 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 1% Tween 20, and 1% Nonidet P-40 and to which 120 μg of proteinase K per ml had been added just prior to use. The mixture was then incubated at 55°C for 1.5 h. Proteinase K was inactivated at 95°C for 15 min, and DNA was used for PCR analysis.
The primers used for direct DNA PCR of EAV, ALV, and CRE are indicated in Table 1, and those used for direct DNA PCR of AMLV are indicated in Table 2. The PCR mixture contained 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 200 μM each dNTP, 20 pmol of each primer (ALV 21F and ALV 21R [32]; EAV F1 and EAV R1 [10]; CRE F1 and CRE R1 [13]; AMLV F1 and AMLV R1 [24]; and β-actin 5 and β-actin 3 [Clontech Laboratories, Palo Alto, Calif.]), and 2.5 U of Taq DNA polymerase (Boehringer) in a total volume of 100 μl. The cycling conditions were 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min (except in the experiment analyzing infection of PBMCs with RAV-0 and RAV-50, where 30 cycles were done). Ten microliters of the amplified material was removed and reamplified with 20 pmol of each internal primer (ALV F2 and ALV R2, EAV F2 and EAV R2, and CRE F2 and CRE R2) for 35 cycles (except in the experiment analyzing infection of PBMCs with RAV-0 and RAV-50, where 30 cycles were done) under the same reaction conditions as those described above. Primers for the detection of the human β-actin gene amplified an 838-bp DNA fragment. Ten microliters of the PCR-amplified product was analyzed on a 1.4% agarose gel, and the DNA was visualized by ethidium bromide staining of the gel.
TABLE 1.
Oligonucleotides used for DNA PCR and RT-PCR
| PCR primer | Sequence (5′ → 3′) | Nucleotide position | Fragment size (bp) |
|---|---|---|---|
| EAV F1 | GCTGTTCTTCATGTGCGAAG | 1624–1643a | 352 |
| EAV R1 | CAGTGGACGCGGTATCAACAGTAA | 1975–1951a | |
| EAV F2 | GTTCCGGGATTCTTTACAACTG | 1654–1675a | 276 |
| EAV R2 | CAGACGAGGCTCCAGCGTGAAGTC | 1929–1906a | |
| ALV 21F | TGTGTGGATTGACCAGTGG | 2789–2807b | 301 |
| ALV 21R | CGCAAGAGGAATAGAAAAGAAG | 3068–3089b | |
| ALV P1 | CCTGTCTTTGTGATCCGG | 2904–2921b | 159 |
| RSV R1 | CTTGAGGTCTAAGACCATCAG | 3062–3042c | |
| CRE F1 | GACTGGATGAGTACAAGTCTTC | 2997–3018d | 393 |
| CRE R1 | CAAGTCCAGGTAAATGACATC | 3389–3409d | |
| CRE F2 | GTCAGGCGAGGTCCCAGATGAC | 3109–3130d | 268 |
| CRE R2 | CTCATCCACCAACGCCGTCAC | 3376–3356d | |
| MNP1 | GGTTCGGATGGTTCGAGAACA | 428–448e | 477 |
| MNP2 | GGTTCATCAAGGACTCAAGTG | 884–904e | |
| MNP3 | TGAAGTGCAAGACCCTGAGGG | 465–485e | 400 |
| MNP4 | TTCATGCAGTCCAAGAGCAGG | 844–864e |
TABLE 2.
Primers used for Alu PCR
| PCR primer or probe | Sequences (5′ → 3′) | Nucleotide position (reference) |
|---|---|---|
| Primers | ||
| A3 | AGUGCCAAGUGUUUGCUGACGACUGCACUCCAGCCUGGGCGAC | (23) |
| A5 | CAGUGCCAAGUGUUUGCUGACGCCAAAGUGCUGGGAUUACAG | (23) |
| Tag3 | CAAGTGTTTGCTGACGACTGCA | (7) |
| Tag5 | CAAGTGTTTGCTGACGCCAAAG | (23) |
| EAV R10 | CCCUUCGUGUACGCCCACTGGAUGG | (40a) |
| EAV R11 | CGTGGCACGTACCTAACACATGGGGAG | (40b) |
| EAV R13 | GAGCCTGACACGTTACATCCCGAGAC | 61–86 (3c) |
| EAV F14 | AUAGGCGUGAUCGGGGUCUCGGGAUG | (40d) |
| EAV F15 | CGTGTCAGGCTCCTCCCCATGTGTTAGG | 75–101 (3c) |
| EAV F16 | GTGTCACCTCGGTATTTGGCCAAGCC | 205–230 (3c) |
| AMLV F1 | CAGGUUUAGCAAGCUAGCUUAAGUAACG | 58–85 (24e) |
| AMLV F2 | CAGAACTGAGAATAGGGAAGTTCGGATCAAG | 115–145 (24e) |
| AMLV F3 | CAGGATATCTGTGGTAAGCAGTTTCG | 181–206 (24e) |
| AMLV R1 | CGAGCUGAUUGGUUAGUUCAAAUAAG | 347–322 (24e) |
| AMLV R2 | CACAGGGTCACTTCAGGTCTTTG | 320–298 (24e) |
| AMLV R3 | CATCTGATGGGTCCCTAGAAACTGCTG | 283–257 (24e) |
| Probes | ||
| EAV F13 | ATAGGCGTGATCGGGGTCTCGGGATG | (40f) |
| EAV F18 | GCAGGCTCCCCTAAGCAACGAACATCACGGTTGCCTGCGAAAGGCAACAA | 231–280 (3c) |
RNA isolation and RT-PCR analysis.
Virion-associated RNA was isolated from 2.5 ml of MVVE. The sample was ultracentrifuged at 40,000 rpm for 2 h (Beckman TLA 45 rotor) and the pellet was resuspended in 500 μl of RNAzol B (Tel-Test, Inc., Friendswood, Tex.) and treated according to the manufacturer's protocol. RNA was treated with 10 U of DNase I (Pharmacia) in a 60-μl total volume at 37°C for 30 min. The DNase was inactivated by heating at 65°C for 10 min. cDNA was prepared by incubating RNA using a Superscript preamplification system (Life Technologies, Gaithersburg, Md.) with a random hexamer primer (250 ng) at 70°C for 10 min and chilling the sample on ice. Final reaction conditions were as follows: 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2.5 mM MgCl2, 500 μM each dNTP, and 10 mM DTT in a total volume of 20 μl. Five separate cDNA synthesis reactions were set up; three contained 200 U of Superscript IIRT, and two were set up without RT as controls. For the latter, one was spiked with 6 pg of CEF total cellular DNA to evaluate the inhibitory effect of MVVE on the amplification of EAV sequences; the other was a control to evaluate potential DNA contamination of the preparation. Samples were incubated at 25°C for 10 min and then at 42°C for 50 min. The reaction was terminated by heating at 70°C for 15 min. cDNA was precipitated with ethanol and used for PCR amplification of EAV, ALV, CRE, and MV sequences as described above. Primers for PCR amplification of MV sequences were MNP1-MNP2 and MNP3-MNP4 (12). Two of the cDNA reactions with RT were used for analysis of EAV and ALV sequences, whereas the third one was divided equally for analysis of MV and CRE sequences.
Total cellular RNA was prepared for MV analysis from human PBMCs inoculated with MVVE (with and without DNase treatment; described above). Cells were washed two times with PBS and resuspended in solution A (described above). RNA was prepared from the cell suspension using RNAzol B according to the manufacturer's protocol. After the RNA was washed with 80% ethanol, the pellet was dissolved in 20 μl of diethyl pyrocarbonate-treated water (Quality Biologicals, Inc.). Ten microliters was used for cDNA synthesis each with and without RT in a total volume of 20 μl as described above. Two microliters of the cDNA was used for PCR amplification of MV sequences with the primers described in Table 1. The PCR conditions were as described above.
Alu PCR analysis.
Integration of EAV and AMLV was analyzed by a modified Alu PCR protocol using cellular DNA prepared from human PBMCs incubated with MMVE or AMLV (described above). The primers used for the detection of Alu repeat sequences (7, 23) are indicated in Table 2. The EAV 5′-LTR and EAV 3′-LTR primers (3, 40) were used to amplify the EAV-cellular DNA junction fragments, and the AMLV 5′-LTR and AMLV 3′-LTR primers (24) were used to amplify the AMLV-cellular DNA junction fragments. PCR amplification was done using an Expand high-fidelity (HF) PCR system (Boehringer). For EAV analysis of MVVE-inoculated PBMC DNA, the hot-start technique was used; 5 μl of DNA (about 250 ng) with an oil overlay was heated at 85°C for 5 min in a mixture with 200 μM each dNTP, 10 pmol of Alu primer (A3 or A5), and 100 pmol of EAV primer (EAV R10 or EAV F14) in a total volume of 50 μl. Then, 50 μl of a mixture containing 1× Expand HF buffer (Boehringer) with 1.5 mM MgCl2 and 2.6 U of Expand enzyme mixture was added, and the sample was further heated at 85°C for 5 min. PCR was then started with the first 10 cycles at 94°C for 45 s, 57°C for 45 s, and 70°C for 6 min. DNA for AMLV analysis (about 200 ng) was amplified without the hot-start technique for the first 10 cycles at 94°C for 30 s and 60°C for 6 min in a total volume of 50 μl containing 1× Expand HF buffer (without MgCl2), 1.5 or 3.0 mM MgCl2, 200 μM each dNTP, 10 pmol of Alu primer (A3 or A5), 10 pmol of AMLV primer (AMLV R1 or AMLV F1), and 2.6 U of Expand enzyme mixture.
After completion of the 10 cycles, 1 U of uracil DNA glycosylase (Life Technologies) was added to the reaction and incubated at 37°C for 30 min. The reaction was stopped by heating at 94°C for 10 min. Then, 10 pmol each of Alu primer (Tag3 or Tag5) and EAV internal primer (EAV R11 or EAV F15) or AMLV internal primer (AMLV R2 or AMLV F2) was added, and amplifications were done by the touchdown PCR technique as described previously (23). Heminested PCR was performed with 2 μl of the amplified material, 10 pmol of Tag3 or Tag5 primer, and 10 pmol of EAV internal primer (EAV R13 or EAV F16) or with 1 μl of the amplified material and 10 pmol of AMLV internal primer (AMLV R3 or AMLV F3). Amplification was done using 2.5 U of Taq DNA polymerase (Boehringer) in a buffer containing 1.5 mM MgCl2 at 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min for 35 cycles for EAV primers and at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min for 30 cycles for AMLV primers. PCR products were separated on a 1% agarose gel, visualized by staining with ethidium bromide, and subsequently transferred to a membrane for hybridization to analyze for EAV-cellular DNA junction fragments using 32P-end-labeled oligomers F13 and F18.
Alu PCR analysis of EAV sequences was also done with DNA obtained from passage 1 and passage 5 HOS cells incubated with CEF supernatant (16) using the hot-start technique. The positive control was DNA from uninoculated passage 5 HOS cells which had been spiked with 100 copies of a cloned DNA standard (see below). A3 and EAV F14 primers were used in the first 10 cycles, and Tag3 and EAV F15 primers were used during the touchdown PCR step. The products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining of the gel. Two independent PCR analyses were performed on the samples.
DNA hybridization.
PCR-amplified products were separated by agarose gel electrophoresis, the DNA was transferred to a Nytran membrane (Schleicher & Schuell, Keene, N.H.) using the manufacturer's protocol, and the membrane was baked for 2 h at 80°C. Prehybridization was done for 3 h at 42°C in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–1% sodium dodecyl sulfate (SDS)–10× Denhardt's solution (1× Denhardt's solution is 0.02% each Ficoll, polyvinylpyrrolidone, and bovine serum albumin)–50 μg of denatured, sheared herring sperm DNA (Research Genetics Inc., Huntsville, Ala.) per ml. Hybridization was done overnight at 50°C in 6× SSPE–1% SDS using a 32P-end-labeled oligonucleotide probe (2.5 × 106 cpm per ml). After hybridization, the membrane was washed with 6× SSPE–1% SDS four times at room temperature for 10 min each time. The final wash was done with 1× SSPE–1% SDS for 3 min at 50°C. The membrane was autoradiographed using Kodak XAR film.
Construction of a plasmid DNA standard for Alu PCR.
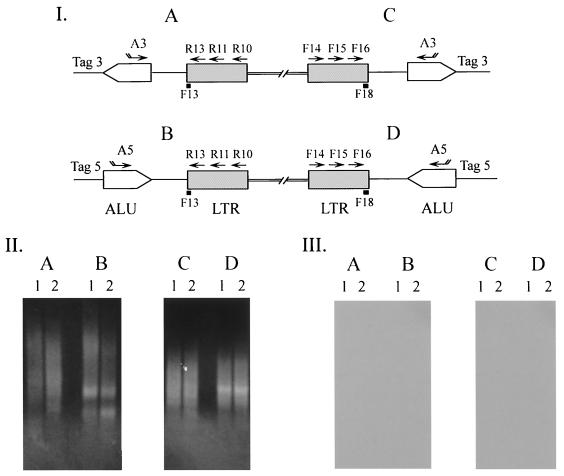
AMLV-infected PBMC DNA was amplified by Alu PCR using primers AMLV F1-A3, AMLV F2-Tag3, and AMLV F3-Tag3 as described above (see Fig. 4A, panel E). A 1.2-kb amplified fragment (see Fig. 4B, panel E, lane 2) was purified from the agarose gel using a Qiaquick column (Qiagen, Santa Clarita, Calif.). The fragment was subcloned in pCRII vector DNA (Invitrogen, Carlsbad, Calif.), and nucleotide sequences were determined. The AMLV long terminal repeat (LTR) was identified near Alu in the cellular DNA. See Fig. 4A, panel E, for the orientation of the AMLV LTR with respect to the Alu repeat. The AMLV LTR-Alu DNA was used to introduce the EAV LTR to create an AMLV-EAV two-LTR–Alu standard DNA. The EAV LTR fragment was PCR amplified from CEF total cellular DNA using primers EAV F17 and EAV R15, which contain AscI and Bsu36I restriction sites, respectively (EAV F17, 5′-AGCTCTAGAGGCGCGCCTGTTGTAATAGGCGTG-3′; EAV R15, 5′-TACCGGTACCTGAGGCTTGTTGCCTTTCGCAGC-3′). The amplified fragment was gel purified, and the EAV LTR (5′ → 3′) was cloned into AscI and Bsu36I sites present in the cellular sequences located between the AMLV LTR and Alu in the AMLV LTR-Alu DNA. See Fig. 5, panel I, C, for the orientation of the EAV LTR with respect to the Alu repeat. The two-LTR–Alu DNA was used to determine the sensitivity of Alu PCR amplification with EAV primers.
FIG. 4.
Detection of AMLV integrants by Alu PCR. (A) PCR amplification of virus-host junction fragments from AMLV-infected PBMCs was carried out using the primers described in Table 2 and indicated in the schematic diagram. (B) Panels E, F, G, and H show ethidium bromide-stained DNA fragments after electrophoresis on a 1.4% agarose gel of 10 μl of samples from the PCR reamplification of AMLV-infected PBMCs (lanes 2) or uninfected PBMCs (lanes 1). The primers used in panels E to H are indicated in the amplification strategies designated E to H, respectively, in the schematic diagram in panel A. Initially, 10 cycles of amplification were done using the Expand HF PCR system with 1.5 mM MgCl2 for AMLV F1-A3 (E) and AMLV F1-A5 (G) and 3.0 mM MgCl2 for AMLV R1-A3 (F) and AMLV R1-A5 (H). This step was followed by touchdown PCR for 40 cycles with AMLV F2-Tag3 (E), AMLV F2-Tag5 (G), AMLV R2-Tag3 (F), and AMLV R2-Tag5 (H). One microliter of the reaction was subjected to heminested PCR for 30 cycles using 2.5 U of Taq polymerase in PCR buffer containing 1.5 mM MgCl2 and AMLV F3-Tag3 (E), AMLV F3-Tag5 (G), AMLV R3-Tag3 (F) and AMLV R3-Tag5 (H). The arrowhead indicates the fragment that was isolated for nucleotide sequencing.
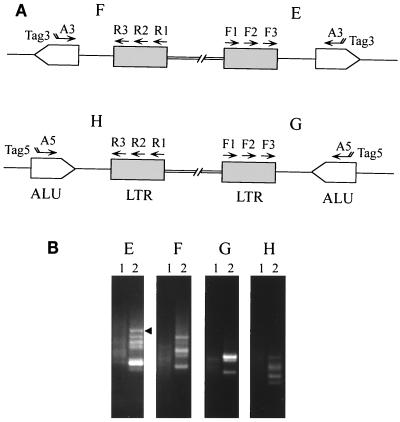
FIG. 5.
Alu PCR analysis of EAV integration in human DNA. MVVE-incubated human PBMC DNA (lanes 2) and uninoculated human PBMC DNA (lanes 1) were analyzed by the Alu PCR strategy shown in panel I. The primers shown for amplification of the virus-cell junctions and designated A to D in panel I were used in the PCR analysis in panel II, A to D, respectively. The first 10 cycles of amplification were carried out using the Expand HF PCR system with 1.5 mM MgCl2 and EAV R10-A3 (A), EAV R10-A5 (B), EAV F14-A3 (C), and EAV F14-A5 (D). This step was followed by 40 cycles of touchdown PCR with EAV R11-Tag3 (A), EAV R11-Tag5 (B), EAV F15-Tag3 (C), and EAV F15-Tag5 (D). Two microliters of the reaction was reamplified by heminested PCR for 35 cycles with Taq DNA polymerase-containing PCR buffer with 1.5 mM MgCl2. Ten microliters of this reaction was electrophoresed on a 1.0% agarose gel. After ethidium bromide staining of the gel to visualize the DNA fragments, DNA was transferred to a Nytran filter and hybridized with 32P-end-labeled F13 and F18 oligomers (black boxes in panel I). The results shown in panel III were obtained with EAV F13 (A and B) and EAV F18 (C and D).
Plasmid DNAs were grown in Escherichia coli strain DH5α (Max Efficiency; Life Technologies). Large-scale DNA preparations were made using a Qiagen purification kit.
Nucleotide sequence analysis.
PCR-amplified fragments were isolated from agarose gels using Geneclean (Bio 101, Natick, Mass.). Nucleotide sequence reactions were set up with an ABI Prism dye terminator cycle sequencing ready reaction kit and AmpliTaq DNA polymerase FS (Perkin-Elmer Cetus, Norwalk, Conn.). The sequences were determined with an ABI Prism 377 DNA sequencing system (PE Applied Biosystems).
RESULTS
Quantitative analysis of RT activity in MV vaccines.
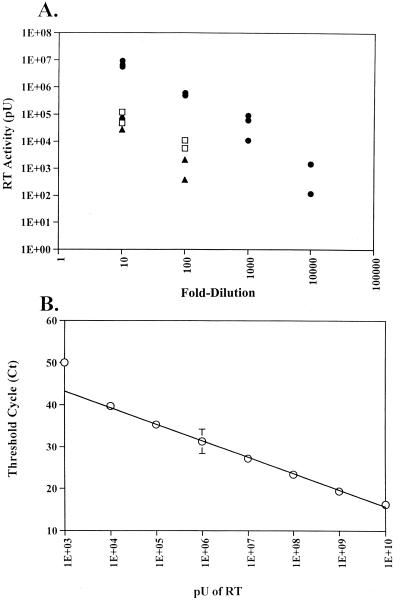
RT activity was detected in MV vaccine bulk lots 12 and 29 and in MVVE. The results are shown in Fig. 1. The RT activity was quantitated by analyzing serial dilutions of each sample in the STF-PERT assay. The number of virus particles was calculated on the basis of the RT activity at the last dilution that gave ≥104 pU of RT activity. On the basis of the AMV RT molecular weight (170,000), it was calculated that 70 RT molecules or 1 AMV virion (25) was equivalent to 1,000 pU of RT. Thus, the number of particles per microliter in undiluted MVVE was calculated on the basis of the activity of RT detected in the 103-fold dilution to be 5.5 × 103. The RT activity in lots 12 and 29 was 2 log units lower. Since the RT activity in the licensed bulk materials (lots 12 and 29) was derived from the same cell substrate source as that used in the production of MVVE by the manufacturer, further infectivity and integration studies related to particle-associated RT activity were carried out using MVVE, which was available in large quantities.
FIG. 1.
Quantitation of RT activity by the STF-PERT assay. (A) RT activity of MV vaccine lot 12 (□) and lot 29 (▴) and MVVE (●) determined in triplicate by STF-PERT analysis of serial dilutions of each sample. (B) Standard linear plot of AMV RT activity, derived by STF-PERT analysis of serial dilutions, tested in triplicate, containing the indicated amount of RT except for dilution with 1E+10 pU, which was tested in duplicate. Means and standard deviations of RT activity versus the threshold cycle (Ct) are shown.
Detection of EAV and ALV sequences in MVVE.
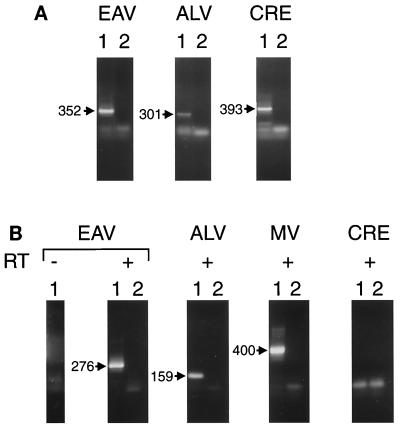
PCR amplification of DNA prepared from MVVE (Fig. 2A) revealed fragments that were similar in size to those of known EAV and ALV sequences. Fragments of similar sizes were PCR amplified from CEF DNA and confirmed as EAV or ALV related by nucleotide sequencing (data not shown). These results indicated the presence of CEF DNA in MVVE. The CRE primers amplified fragments of the expected sizes from MVVE, confirming the presence of CEF DNA in MVVE. The EAV and ALV PCR-amplified fragments from CEF DNA were cloned to create copy number standards to evaluate the sensitivity of the primers. The EAV and the ALV primer sets had similar limits of detection (about 10 DNA copies). Furthermore, analysis of CEF DNA representing different cell numbers indicated at least a 100-fold difference between the copy numbers of EAV and ALV pol-containing sequences (data not shown).
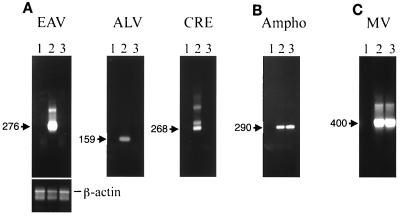
FIG. 2.
Detection of EAV and ALV sequences in MVVE. (A) Lanes 1, DNA PCR analysis of MVVE done using primers EAV F1-EAV F2, ALV 21F-ALV 21R, and CRE F1-CRE F2 for the detection of EAV, ALV, and CRE sequences, respectively. Lanes 2, PCR control without DNA. PCR products of these first amplification reactions were electrophoresed on a 1.4% agarose gel and visualized by ethidium bromide staining. The fragments expected are indicated (in base pairs). (B) RT-PCR analysis of MVVE. RNA prepared from MVVE was analyzed for DNA contamination using EAV primers (EAV F1-EAV R1 and EAV F2-EAV R2) in the absence of added RT in a cDNA synthesis reaction (leftmost lane 1). RNA prepared from MVVE was analyzed for EAV using primers EAV F1-EAV R1 and EAV F2-EAV R2, for ALV using primers ALV 21F1-ALV 21R1 and ALV P1-RSV R1, for MV using primers MNP1-MNP2 and MNP3-MNP4, and for CRE using primers CRE F1-CRE R1 and CRE F2-CRE R2 (lanes 1, + RT). The PCR control (without RNA) is shown in lanes 2. Products of the reamplification PCR were electrophoresed on a 1.4% agarose gel and visualized by ethidium bromide staining. The fragments expected are indicated (in base pairs).
Since there are numerous copies of different EAV sequences in the chicken genome, the EAV primers (F1, F2, R1, and R2) were evaluated for their detection of EAV sequences by Southern blot analysis of restriction enzyme-digested CEF DNA using 32P-end-labeled EAV primers as probes. The results indicated that the EAV primers could detect almost all the bands that hybridized to a cloned 1.4-kb EAV pol fragment, demonstrating broad detection of EAV sequences by the primers (data not shown).
PCR of RNA in virions prepared from MVVE (0.5 ml per assay) amplified EAV and ALV fragments of the expected sizes based upon the known sequences. Nucleotide sequence analysis of the gel-purified fragments confirmed their origin from the known EAV and ALV families (GenBank accession no. X59844 for EAV and J02342, J02021, and J02343 for ALV; data not shown). MV primers, used as the positive control, amplified fragments of the expected sizes from MV. No amplification was seen with CRE primers or with EAV primers in the absence of RT, indicating the absence of contaminating DNA in the RNA preparation. No inhibition of EAV amplification in an RNA preparation from 0.5 ml of MVVE was seen upon spiking with 6 pg of CEF DNA (data not shown) (three cell equivalents, based upon the avian genome size of 2 × 109 bp).
Analysis of integration of EAV and ALV sequences in MVVE.
To investigate whether the virion-associated avian retrovirus sequences present in MVVE could infect human cells, PHA-stimulated PBMCs were inoculated with MVVE with and without DNase treatment, and DNA and RNA were prepared for PCR analysis. The results shown in Fig. 3A indicate that both EAV and ALV sequences were amplified from DNA of PBMCs, inoculated with MVVE without DNase treatment (lane 2); however, no signal was detected from DNA prepared from PBMCs (250 ng per reaction) inoculated with DNase-treated MVVE (lane 3), even upon multiple PCR amplifications with EAV and ALV primers (none of five and none of six samples, respectively; data not shown). Similarly, CRE sequences were detected in an inoculum not treated with DNase but not in DNase-treated material. These results indicated that retrovirus sequences were amplified from the chick-cell-substrate-derived DNA present in the inoculum as a result of the manufacturing and were not integrated in human PBMCs as a result of exogenous virus infection.
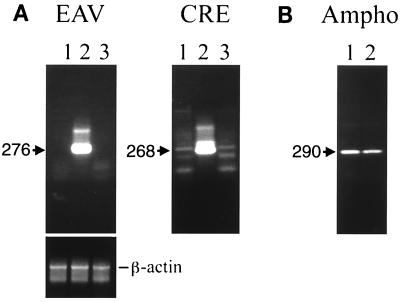
FIG. 3.
Analysis of integration of avian retrovirus sequences in MVVE-incubated human PBMCs. (A) PCR analysis done to detect the presence of EAV, ALV, and CRE sequences in DNA prepared from human PBMCs incubated with MVVE (with and without DNase treatment) at 24 h postexposure. PCR primers are indicated in the legend to Fig. 2B. (B) DNA PCR of PBMCs incubated with MVVE in the presence of AMLV as a positive control for retrovirus infection using AMLV F1-AMLV R1. (C) RT-PCR analysis of MVVE-incubated PBMCs done to demonstrate infection of PBMCs by MV using primers indicated in the legend to Fig. 2B. Lanes 1, uninoculated PBMC DNA control in panels A and B and PCR control in panel C; lanes 2, MVVE material inoculated without DNase treatment; lanes 3, MVVE material inoculated with DNase treatment. Ten microliters of amplified products (panel B and β-actin panel) or reamplified products (panels A and C) was analyzed on a 1.4% agarose gel by ethidium bromide staining of DNA. The fragments expected are indicated (in base pairs). The 838-bp fragment amplified with human β-actin primers is shown.
To demonstrate successful inoculation of PBMCs with MVVE, RT-PCR analysis for MV was done with RNA prepared from an aliquot of the same inoculated PBMCs as those used for DNA PCR analysis: the results shown in Fig. 3C indicate similar detection of MV sequences with and without DNA treatment. An additional positive control was used for retrovirus infection and integration: PCR analysis with AMLV primers of DNA prepared from PBMCs inoculated with MVVE spiked with AMLV with and without DNase treatment. The results shown in Fig. 3B indicate that AMLV and MV sequences were detected in DNase-treated MVVE without any noticeable effect of DNase treatment on virus infection. Interestingly, EAV and CRE sequences were detected in DNA obtained at day 18 from PBMCs inoculated with MVVE (see below) without DNase treatment (in three of five and two of five samples, respectively), indicating the persistence of DNA (due to the inoculum) in the cell culture. However, none were detected in the DNase-treated inoculum (none of five samples) (data not shown).
To further evaluate possible EAV and ALV infection in human PBMCs, Alu PCR was used to detect retrovirus integrants in PBMCs inoculated with MVVE; AMLV was used as a positive control. The primers used to detect LTR-cellular junctions for both 5′ and 3′ viral LTRs with either orientation of Alu are shown in Fig. 4A. The detection of AMLV integrants in human PBMC DNA is shown in Fig. 4B. Faint background signals were detected in normal human DNA; these were distinct from the intense signals specifically seen in virus-infected cell DNA. Multiple bands were detected, indicating multiple integrations. One fragment was sequenced, and the results confirmed the integration of the AMLV LTR near Alu (Fig. 4B, panel E). Similar analyses were done with EAV LTR primers to analyze for the presence of viral integrants in PBMCs inoculated with MVVE (Fig. 5, panel I). The results indicated the absence of any specific amplicon in the inoculated cell DNA (Fig. 5, panel II). Furthermore, no virus-specific bands were detected upon hybridization with 32P-labeled EAV F13 and EAV F18 oligomer probes (Fig. 5, panel III), even upon longer exposure (5 days) (data not shown). Recombinant EAV-Alu DNA was constructed to evaluate the sensitivity of detection by Alu PCR. The primers A3-EAV F14 and Tag3-EAV F15 used in Alu PCR (Fig. 5I, diagram C) could detect 100 DNA copies under the same conditions as those used for the test sample and visualization of the products on an ethidium bromide-stained agarose gel.
Analysis of integration of EAV particles in CEF supernatant.
EAV and ALV sequences were detected in supernatant from CEF cultures (data not shown) (30, 34, 40). In a previous study, to investigate the presence of replication-competent retrovirus, CEF supernatant and SFV, a positive control virus, were used to inoculate various human cells (including HOS cells); filtered culture medium was collected at various times for PERT analysis (16). The results of that study showed the absence of a replicating retrovirus associated with the EAV and ALV sequences in CEF supernatant, whereas the replication of SFV was seen at all times tested. In that study, cell pellets had been prepared from inoculated cultures at passage 1 and passage 5 (culture termination) and stored. In this study, cellular DNA was prepared from the stored pellets of the previous study (16) and analyzed for EAV and ALV sequences by direct PCR and additionally for EAV sequences by Alu PCR. The results indicated that EAV and ALV were detected at passage 1 (due to contaminating cellular DNA) but not at passage 5 upon reamplification, indicating the lack of virus replication. SFV was detected with SFV set A primers (consisting of outer primers 1-2 and inner primers 3-4) (17) at passage 1 and passage 5, indicating virus replication. To further investigate the presence of integrants, Alu PCR analysis of DNA prepared at passage 1 and passage 5 from HOS cells inoculated with CEF supernatants confirmed the absence of EAV-specific integrants, compared to the results seen for uninoculated cellular DNA. The primers used are described in Materials and Methods. Similar results were obtained in another independent PCR assay. No inhibition of amplification was seen with 100 copies of standard plasmid DNA spiked into passage 5 CEF supernatant-inoculated HOS DNA.
To investigate the potential infectivity of the virion-associated retrovirus sequences produced by CEFs in the absence of contaminating cellular DNA, PBMCs were inoculated with CEF supernatant (day 4 from the previous study [6]) with and without DNase treatment. The results indicated the presence of EAV and CRE sequences in the untreated material; however, these were not detected upon DNase treatment (Fig. 6A), indicating the absence of retrovirus integrants in the inoculated PBMC DNA. The detection of AMLV was not affected by DNase treatment (Fig. 6B).
FIG. 6.
Analysis of integration of EAV sequences present in CEF supernatants. (A) DNA prepared from CEF supernatant-incubated PBMCs was analyzed by PCR using EAV F1-EAV R1 and EAV F2-EAV R2 primers and CRE F1-CRE R1 and CRE F2-CRE R2 primers. Lanes 1, uninoculated PBMC DNA; lanes 2, CEF supernatant without DNase treatment; lanes 3, CEF supernatant with DNase treatment. (B) DNA from cells incubated with CEF supernatant spiked with AMLV was analyzed by PCR using AMLV F1-AMLV R1. Lanes 1 and 2 contain DNA prepared from cells incubated without DNase treatment and with DNase treatment, respectively. The fragments expected are indicated (in base pairs). Human β-actin primers amplified an 838-bp fragment.
Analysis of avian retrovirus infection of human cells.
To investigate the infection of human cells with known avian retroviruses, PCR analysis was done with DNA obtained from PBMCs infected with ALV-related virus strains RAV-0 and RAV-50. RAV-0 was selected because it represents an endogenous infectious locus of chick cells, and RAV-50 represents an avian retrovirus of subgroup D, which includes viruses that can infect some human cells (29, 34). PCR analysis using primers ALV 21F and ALV 21R, with reamplification using primers ALV P1 and RSV R1, indicated the absence of ALV sequences in PBMC DNA infected with RAV-0 and RAV-50, whereas AMLV sequences were detected in the DNA of AMLV-infected cells by AMLV env primers (data not shown). The absence of ALV sequences in PBMC DNA corroborated previous results demonstrating the absence of RAV-50 replication in PBMCs (16). It should be noted that cellular DNA, which was derived from the primary chick cell culture used in virus preparation, was detected in the RAV-50 stock by DNA PCR but was reduced to below-detection levels by washing of infected cells with PBS prior to PCR analysis. The RAV-0 stock was not analyzed for contaminating cellular DNA.
Infectivity studies.
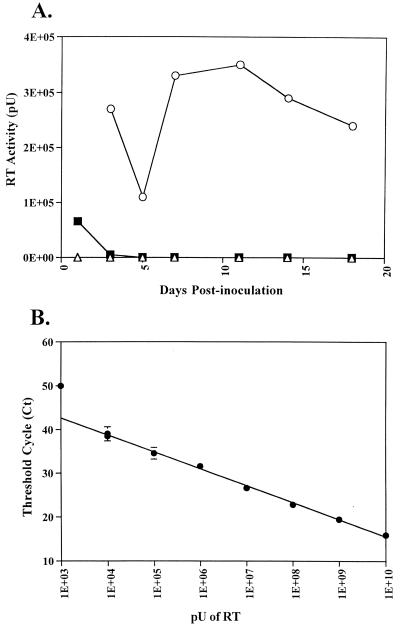
Human PBMCs were incubated with MVVE, and supernatants were collected at various times for 18 days for STF-PERT analysis to detect replication-competent retrovirus. Uninoculated cells were used as a negative control. The results shown in Fig. 7 indicate initial detection of input RT activity due to the inoculum; this activity was reduced to the level in uninoculated cells and was maintained as such during the culture period. Similar results were obtained in both STF-PERT and TM-PERT assays (data not shown). To demonstrate successful infection of human PBMCs, the filtered supernatants were analzyed for MV by titration on Vero cells (limit of detection of the assay, 100.7 TCID50 per 0.1 ml). The results indicated early and late peaks of MV replication (day 3 and day 18, respectively) in the PBMCs (data not shown). Furthermore, to demonstrate retrovirus infection in the presence of MVVE, PBMCs were incubated with MVVE spiked with AMLV in a parallel experiment. Analysis of supernatants by STF-PERT detected AMLV replication during the culture period.
FIG. 7.
Infectivity analysis of MVVE in human PBMCs. (A) PHA-stimulated human PBMCs were inoculated with MVVE (■) or MVVE spiked with AMLV (○) or left uninoculated (▵), and supernatants collected at various times were analyzed by the STF-PERT assay. (B) Linear standard curve of AMV RT standards. The threshold cycle (Ct) for each sample was plotted against the log activity of RT.
DISCUSSION
All chick-cell-derived vaccines contain a low level of RT activity, which is produced by endogenous retrovirus sequences that are present in the avian genome. The RT activity has been found to be associated with particles containing RNAs related to endogenous retrovirus sequences of the EAV family or to ALV-related endogenous virus loci. To address the primary public health safety concern of whether RT activity is associated with a replicating agent, studies were undertaken that demonstrated that the RT activity produced from CEFs, the vaccine cell substrate, was not associated with a replicating agent, based upon testing of several different human cell lines and cells of other species. However, some concerns still remained regarding the consequences of a nonproductive infection by defective, nonreplicating, RT-containing particles in the vaccine or by pseudotype virions (33, 39) that might be present in the vaccine, which could result in integration of EAV or ALV sequences in the host genome.
To address these potential public health safety concerns, we have analyzed a U.S.-manufactured MVVE preparation for potential risk associated with the RT activity present in chick-cell-derived vaccines. Analysis of MVVE confirmed the presence of particle-associated EAV and ALV sequences by RT-PCR and cellular DNA-associated EAV and ALV sequences by DNA PCR. A stronger signal was seen with EAV primers than with ALV primers, consistent with the higher copy number for EAV sequences in CEF DNA (11; this study). Additionally, the results showed (i) about 5.5 × 103 virions per microliter in MVVE by the STF-PERT assay; (ii) the absence of a replicating retrovirus or propagation of retrovirus sequences by MV in infectivity studies using human PBMCs and HOS cells inoculated with MVVE; and (iii) the absence of integration of EAV and ALV sequences in human cells, based upon direct and Alu PCR analyses of DNA prepared from MVVE- and CEF supernatant-inoculated cells.
To evaluate the results of this study as a measure of vaccine safety in humans, the amount of MVVE used in the experiments should be converted to human dose equivalents (HDE). A direct extrapolation is difficult, since the minimum titer for licensed MV vaccine at the expiration date is 103 TCID50, but the titer at the time of filling of the vials is not specified; in general, this titer is estimated to be about 104.2 TCID50. Based upon this information, it can be calculated that 14 μl of MVVE equals one HDE. Thus, 29 HDE were assayed in the infectivity study in this study for replication-competent virus. It should be noted that under the experimental conditions, the multiplicity of infection of MV was 0.1, and that of retrovirions (based upon the RT ± activity) was 0.55. The amount of the inoculum used in the infections was limited due to the presence of MV, which could result in early target cell lysis at a high multiplicity of infection, making it impossible to monitor the cultures over an extended period. It is more difficult to calculate with confidence the HDE for MVVE tested by direct PCR of inoculated cells, due to numerous assumptions that need to be made in the calculations, leading to erroneous results. At best it can be calculated that at least one HDE (about 105 particles of EAV) was tested by direct PCR for integration of retrovirus sequences. The data from the analysis of MVVE and CEF supernatants indicated the absence of a replicating agent; the absence of integrated EAV and ALV sequences in human cells provides added confidence in the safety of MV vaccine and supports the continued use of chick-cell-derived vaccines.
Retrovirus-induced tumorigenesis can involve the generation of a novel pathogenic virus by recombination between replication-competent and -defective sequences and/or activation of a cellular oncogene by an LTR due to upstream or downstream insertion of retrovirus sequences (31, 35). To address the possible integration of EAV and ALV sequences in human cells by RT-containing particles in MVVE, two PCR strategies were used: direct PCR of DNase-treated inoculum using primers from the highly conserved pol region and Alu PCR using LTR primers in conjuction with Alu primers to specifically amplify viral-cellular DNA junctions of integrants. Although the Alu PCR strategy was limited due to the detection of integrants only in or near Alu repeats (22) and was less sensitive than the direct PCR and although the direct PCR could detect new integrants only upon DNase treatment of the inoculum, the combination of both strategies provided confident results indicating the absence of avian retrovirus sequences in MVVE-inoculated human cells.
In this study, we used human PBMCs and HOS (TE-85 clone F-5) cells to evaluate the infectivity and replication of EAV and ALV particles in MVVE and the propagation of retrovirus sequences via possible EAV or ALV pseudotypes containing MV envelopes. The HOS cell line was included in the analysis since HOS cells were previously shown to be susceptible to Rous sarcoma virus (RSV) (29). These cells, however, were highly susceptible to MV lysis and were not suitable for use in extended culturing in the presence of MVVE. Integration analysis indicated that EAV and ALV sequences were not detected in HOS cells incubated with DNase-treated MVVE, indicating the absence of infection by particle-associated retrovirus sequences in MVVE. Similar results were seen with PBMCs. Furthermore, analysis of PBMC DNA from long-term cultures indicated the absence of EAV and ALV sequences in the day 18 sample with DNase-treated inoculum, confirming the absence of a propagating agent in MVVE.
The RT activity detected in MVVE was about 100 times higher than that detected in the licensed bulk lots. This result may be related to the stability of RT, which may have been affected by the longer storage time of the licensed lots (licensed in 1992) compared to that of MVVE (prepared in 1996), or to differences in handling procedures related to storage temperature and number of freeze-thaw cycles prior to analysis. The absence of an infectious retrovirus in the MVVE material used in this study and in the CEF supernatant used in the previous study (which contained levels of RT activity similar to those of MVVE, based upon comparison with the positive control AMV RT dilution series) (16) provides confidence regarding the safety of the earlier MV vaccines used in humans. The results of this study further demonstrate the absence of a known public health safety concern related to the presence of RT activity in chick-cell-derived vaccines and support World Health Organization recommendations for the continued use of chick-cell-derived vaccines in humans.
ACKNOWLEDGMENTS
We thank J. Hartley for providing AMLV; Merck & Co., Inc., for MVVE material; T. Bryan and M. Klutch for nucleotide sequencing; J. Beeler for MV titration; and W. Heneine for ALV primer sequences.
The work was supported in part by a grant from the National Vaccine Program Office to A. S. Khan.
REFERENCES
- 1.Bauer G, Friis R R, Mattersberger H, Hofschneider P H. Controlled release of particle-associated RNA-dependent DNA polymerase by primary chick embryo cell cultures. Exp Cell Res. 1978;117:383–392. doi: 10.1016/0014-4827(78)90151-9. [DOI] [PubMed] [Google Scholar]
- 2.Böni J, Stalder J, Reigel F, Schüpbach J. Detection of reverse transcriptase activity in live attenuated virus vaccines. Clin Diagn Virol. 1996;5:43–53. doi: 10.1016/0928-0197(95)00159-x. [DOI] [PubMed] [Google Scholar]
- 3.Boyce-Jacino M T, Resnick R, Faras A J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989;173:157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- 4.Boyce-Jacino M T, O'Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Immunization of adolescents: recommendations of the Advisory Committee on Immunization Practices, the American Academy of Pediatrics, the American Academy of Family Physicians, and the American Medical Association. Morb Mortal Wkly Rep. 1996;47:1–16. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Advances in global measles control and elimination: summary of the 1997 international meeting. Morb Mortal Wkly Rep. 1998;47:1–23. [PubMed] [Google Scholar]
- 7.Chumakov I M, Le Gall I, Billault A, Ougen P, Soularue P, Guillou S, Rigault P, Bui H, De Tand M F, Barillot E, et al. Isolation of chromosome 21-specific yeast artificial chromosomes from a total human genome library. Nat Genet. 1992;1:222–225. doi: 10.1038/ng0692-222. [DOI] [PubMed] [Google Scholar]
- 8.Coelen R J, Jose D G, May J T. The effect of hexadimethrine bromide (Polybrene) on the infection of the primate retroviruses SSV 1/SSAV 1 and BaEV. Arch Virol. 1983;75:307–311. doi: 10.1007/BF01314897. [DOI] [PubMed] [Google Scholar]
- 9.Coffin J M, Tsichlis P N, Conklin K F, Senior A, Robinson H L. Genomes of endogenous and exogenous avian retroviruses. Virology. 1983;126:51–72. doi: 10.1016/0042-6822(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 10.Dunwiddie C, Faras A J. Presence of retrovirus reverse transcriptase-related gene sequences in avian cells lacking endogenous avian leukosis viruses. Proc Natl Acad Sci USA. 1985;82:5097–5101. doi: 10.1073/pnas.82.15.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunwiddie C T, Resnick R, Boyce-Jacino M, Alegre J N, Faras A J. Molecular cloning and characterization of gag-, pol-, and env-related gene sequences in avian cells lacking endogenous avian leukosis viruses. J Virol. 1986;59:669–675. doi: 10.1128/jvi.59.3.669-675.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godec M S, Asher D M, Murray R S, Shin M L, Leighton L W, Gibbs C J, Jr, Gajdusek D C. Absence of measles, mumps and rubella viral genomic sequences from multiple sclerosis brain tissue by polymerase chain reaction. Ann Neurol. 1992;32:401–404. doi: 10.1002/ana.410320317. [DOI] [PubMed] [Google Scholar]
- 13.Haas N B, Grabowski J M, Sivitz A B, Burch J B. Chicken repeat 1 (CR1) elements, which define an ancient family of vertebrate non-LTR retrotransposons, contain two closely spaced open reading frames. Gene. 1997;197:305–309. doi: 10.1016/s0378-1119(97)00276-x. [DOI] [PubMed] [Google Scholar]
- 14.Hartley J W, Rowe W P. Naturally occurring murine leukemia viruses in wild mice: characterization of a new “amphotropic” class. J Virol. 1976;19:19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes S H, Toyoshima K, Bishop J M, Varmus H E. Organization of the endogenous proviruses of chicken: implications for origin and expression. Virology. 1981;108:189–207. doi: 10.1016/0042-6822(81)90538-9. [DOI] [PubMed] [Google Scholar]
- 16.Khan A S, Maudru T, Thompson A, Muller J, Sears J F, Peden K W C. The reverse transcriptase activity in cell-free medium of chicken embryo fibroblast cultures is not associated with a replication-competent retrovirus. J Clin Virol. 1998;11:7–18. doi: 10.1016/s0928-0197(98)00042-7. [DOI] [PubMed] [Google Scholar]
- 17.Khan A S, Sears J F, Muller J, Galvin T A, Shahabuddin M. Sensitive assays for isolation and detection of simian foamy retroviruses. J Clin Microbiol. 1999;37:2678–2686. doi: 10.1128/jcm.37.8.2678-2686.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimpton J, Emmerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz L E, Katz S L. Measles vaccine. In: Plotkin S A, Mortimer E A Jr, editors. Vaccines. Philadelphia, Pa: The W. B. Saunders Co.; 1994. pp. 229–276. [Google Scholar]
- 20.Maudru T, Peden K W C Cooperating Units. Analysis of a coded panel of licensed vaccines by polymerase chain reaction-based reverse transcriptase assays: a collaborative study. J Clin Virol. 1998;11:19–28. doi: 10.1016/s0928-0197(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 21.Maudru T, Peden K W C. Adaptation of the fluorogenic 5′-nuclease chemistry to a PCR-based reverse transcriptase assay. BioTechniques. 1998;25:972–975. doi: 10.2144/98256bm08. [DOI] [PubMed] [Google Scholar]
- 22.Mighell A J, Markham A F, Robinson P A. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 23.Minami M, Poussin K, Bréchot C, Paterlini P. A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics. 1995;29:403–408. doi: 10.1006/geno.1995.9004. [DOI] [PubMed] [Google Scholar]
- 24.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panet A, Baltimore D, Hanafusa T. Quantitation of avian RNA tumor virus reverse transcriptase by radioimmunoassay. J Virol. 1975;16:146–152. doi: 10.1128/jvi.16.1.146-152.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne L N. Biology of avian retroviruses. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1992. pp. 299–376. [Google Scholar]
- 27.Rasheed S, Gardner M B, Chan E. Amphotropic host range of naturally occurring wild mouse leukemia viruses. J Virol. 1976;19:13–18. doi: 10.1128/jvi.19.1.13-18.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Resnick R M, Boyce-Jacino M T, Fu Q, Faras A J. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J Virol. 1990;64:4640–4653. doi: 10.1128/jvi.64.10.4640-4653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhim J S, Trimmer R, Huebner R J, Papas T S, Jay G. Differential susceptibility of human cells to transformation by murine and avian sarcoma viruses (41441) Proc Soc Exp Biol Med. 1982;170:350–358. doi: 10.3181/00379727-170-41441. [DOI] [PubMed] [Google Scholar]
- 30.Robertson J S, Nicolson C, Riley A M, Bentley M, Dunn G, Corcoran T, Schild G C, Minor P. Assessing the significance of reverse transcriptase activity in chick cell-derived vaccines. Biologicals. 1997;25:403–414. doi: 10.1006/biol.1997.0111. [DOI] [PubMed] [Google Scholar]
- 31.Ruis B L, Benson S J, Conklin K F. Genome structure and expression of the ev/J family of avian endogenous viruses. J Virol. 1999;73:5345–5355. doi: 10.1128/jvi.73.7.5345-5355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 33.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 34.Teich N. Taxonomy of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor virus: molecular biology of tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 25–207. [Google Scholar]
- 35.Teich N, Wyke J, Mak T, Bernstein A, Hardy W. Pathogenesis of retrovirus-induced disease. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor virus: molecular biology of tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 785–998. [Google Scholar]
- 36.Toyoshima K, Vogt P K. Enhancement and inhibition of avian sarcoma viruses by polycations and polyanions. Virology. 1969;38:414–426. doi: 10.1016/0042-6822(69)90154-8. [DOI] [PubMed] [Google Scholar]
- 37.Tsang S X, Switzer W M, Shanmugam V, Johnson J A, Goldsmith C, Wright A, Fadly A, Thea D, Jaffe H, Folks T M, Heneine W. Evidence of avian leukosis virus subgroup E and endogenous avian virus in measles and mumps vaccines derived from chicken cells: investigation of transmission to vaccine recipients. J Virol. 1999;73:5843–5851. doi: 10.1128/jvi.73.7.5843-5851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.U.S. Code of Federal Regulations. Manufacture of live, attenuated measles virus vaccine. Title 21, part 630.32. U.S. Washington, D.C.: Government Printing Office; 1994. [Google Scholar]
- 39.Weiss R A, Boettiger D, Murphy H M. Pseudotypes of avian sarcoma viruses with the envelope properties of vesicular stomatitis virus. Virology. 1977;76:808–825. doi: 10.1016/0042-6822(77)90261-6. [DOI] [PubMed] [Google Scholar]
- 40.Weissmahr R N, Schüpbach J, Böni J. Reverse transcriptase activity in chicken supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J Virol. 1997;71:3005–3012. doi: 10.1128/jvi.71.4.3005-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization. Reverse trancriptase activity in chicken-cell derived vaccine. Wkly Epidemiol Rec. 1998;73:209–216. [PubMed] [Google Scholar]
- 42.World Health Organization Expert Committee on Biological Standardization. Requirements for measles, mumps and rubella vaccines and combined vaccines (live). Requirements for biological substances, no. 47. WHO Tech Rep Ser. 1994;840:100–207. [Google Scholar]