Abstract
Background and Objectives: Chromosomal microarray offers superior sensitivity for identification of submicroscopic copy number variants (CNVs) and is recommended for the initial genetic testing of patients with autism spectrum disorder (ASD). This study aims to determine the diagnostic yield of array comparative genomic hybridization (array-CGH) in ASD patients from a cohort of Chinese patients in Taiwan. Materials and Methods: Enrolled in this study were 80 ASD children (49 males and 31 females; 2–16 years old) followed up at Taipei MacKay Memorial Hospital between January 2010 and December 2020. The genomic DNA extracted from blood samples was analyzed by array-CGH via the Affymetrix GeneChip Genome-Wide Human single nucleotide polymorphism (SNP) and NimbleGen International Standards for Cytogenomic Arrays (ISCA) Plus Cytogenetic Arrays. The CNVs were classified into five groups: pathogenic (pathologic variant), likely pathogenic (potential pathologic variant), likely benign (potential normal genomic variant), benign (normal genomic variant), and uncertain clinical significance (variance of uncertain significance), according to the American College of Medical Genetics (ACMG) guidelines. Results: We identified 47 CNVs, 31 of which in 27 patients were clinically significant. The overall diagnostic yield was 33.8%. The most frequently clinically significant CNV was 15q11.2 deletion, which was present in 4 (5.0%) patients. Conclusions: In this study, a satisfactory diagnostic yield of array-CGH was demonstrated in a Taiwanese ASD patient cohort, supporting the clinical usefulness of array-CGH as the first-line testing of ASD in Taiwan.
Keywords: autism spectrum disorder, Taiwan, array-CGH
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder wherein patients have difficulty in communication and social interactions, stereotypical behaviors, and restricted interests. ASD has a prevalence of 1 in 161 children and is more frequent in males [1]. Its pathogenesis is multifactorial, but genetic alteration is the most important factor, with a heterogeneous change seen across the whole genome [2].
Array comparative genomic hybridization (array-CGH) and single nucleotide polymorphism (SNP) genotyping array, as chromosomal microarray analysis (CMA), are initially performed as cytogenetic diagnostic tests for ASD [3,4]. Before the development of CMA, karyotyping was the standard method to detect genetic anomalies in ASD patients. However, this could only detect large and microscopically visible chromosomal changes (>5–7 Mb), with a low diagnostic rate (3–5%) [3,5]. Fluorescence in situ hybridization (FISH) is another tool for detecting submicroscopic deletions and duplications. It could increase the diagnostic yield by 2% to 3% [3,6,7]. Nevertheless, karyotyping and FISH are not enough for evaluating the genetic etiology of ASD.
CMA can overcome the technical limitations of karyotyping and FISH as well as provide a higher resolution of the genome. The International Collaboration for Clinical Genomics, also known as the International Standard for Cytogenomic Array (ISCA) Consortium, recommends CMA as the first cytogenetic diagnostic test in non-syndromic ASD patients [3,8]. The American College of Medical Genetics (ACMG) also established the guidelines of CMA using [3,9,10]. Two studies have described the diseases related to abnormal findings in CMA. Ellison et al. reviewed 46,298 patients via CMA and found 151 disorders related to chromosomal/genetic abnormalities [3,11], with 35% of the patients having abnormal CMA findings. Riggs et al. surveyed the ISCA Consortium database and found 28,526 patients with 146 phenotypes [3,12]. Among the copy number variants (CNVs), 46% were found to be either pathogenic or likely pathogenic (1908/4125).
Many studies have described the causative role of CNVs in ASD [3,13], congenital heart diseases [3,14], epilepsy [3,15], and congenital kidney malformation [3,16]. However, the same CNVs might cause multiple diseases, and the development of disease can be attributed to many different factors. This is known as the two-hit hypothesis [3,17,18]. Due to the “two-hit hypothesis”, the clinical diagnosis, genetic counseling, and management become challenging.
In this study, we used array-CGH to evaluate ASD patients in Taiwan. The diagnostic rate was detected by array-CGH. We also analyzed the CNV characteristic and feature of these patients.
2. Materials and methods
2.1. Patients
This study assessed 80 idiopathic ASD children (49 males and 31 females) with ages ranging from 2 to 16 years. These patients were not related to each other. All of them were followed up at Taipei MacKay Memorial Hospital between January 2010 and December 2020. An autism diagnostic interview-revised (ADI-R) [19] was used to confirm the diagnosis of autism. These patients were diagnosed with idiopathic ASD, which is of unknown origin, and we excluded other potential etiologies such as neurocutaneous syndromes, other specific syndromes, and congenital or acquired infections among other common causes of autism before they had array-CGH. The intellectual level information was confirmed by the Wechsler Preschool and Primary Scale of Intelligence Fourth Edition (WPPSI-IV) and Wechsler intelligence scale for children–Fifth Edition (WISC-V) [20,21].
2.2. Array-CGH and Data Interpretation
We extracted genomic DNA from the peripheral blood according to standard protocols (Figure 1). All samples were sent to two different laboratories. The first laboratory, the National Center for Genome Medicine in Taiwan, used the Affymetrix GeneChip Genome-Wide Human SNP array 6.0 (Affymetrix, Santa Clara, CA, USA), while the second laboratory, Gene Biodesign, used the NimbleGen ISCA Plus Cytogenetic Array (Roche NimbleGen, Madison, WI, USA). The Affymetrix GeneChip Genome-Wide Human SNP array 6.0 had 50,000, 950,000, and 2,700,000 probes with resolutions ranging from 100 to 200 kb across the entire genome to detect CNVs. The Affymetrix Genotyping ConsoleTM version 3.0.1. was used to analyze the array data of 28 patients in this study. The NimbleGen ISCA Plus Cytogenetic Array contained 630,000 and 1,400,000 probes with a resolution of about 15–30 kb throughout the whole genome. The related data were represented using Nexus 6.1 (BioDiscovery, Hawthorne, CA, USA) for 12 patients in this study [3]. We handled all samples according to the manufacturers’ instructions. SPSS version 25.0 (SPSS, Inc., Chicago, IL, USA) was used to perform the statistical analysis. Statistical significance was set at p < 0.05.
Figure 1.
Basic steps involved in all DNA extraction methods.
According to ACMG guidelines [9,10], CNVs fall under one of the following five categories: pathogenic (pathologic variant), likely pathogenic (potential pathologic variant), likely benign (potential normal genomic variant), benign (normal genomic variant), and uncertain clinical significance (variance of uncertain significance (VOUS)). Pathogenic CNVs are those which cause recognized microdeletion and microduplication syndromes. These CNVs contain morbid Online Mendelian Inheritance in Man (OMIM) genes and large deletions or duplications (usually >3 Mb in size) involving many OMIM genes. They are also inherited from an affected parent and greater than 1 copy number amplification. However, it does not occur in MECP2 duplication where in some instances the parent is not affected [22]. Benign CNVs include those that are well-documented in the normal population or the public databases, not previously reported but inherited from a healthy parent, without any morbid OMIM genes, and duplications with no known dosage-sensitive genes. VOUS CNVs are those that cannot be classified as pathogenic or benign due to insufficient evidence. Recent literature does not recommend using “VOUS” to represent the “likely pathogenic” or “likely benign” categories [9]. Combining the “likely” categories and VOUS may be confusing for clinicians and patients receiving clinical reports. The cut-off value is <1.2 for loss (deletion) and >2.8 for gain (duplication). We compared the findings of our study with previous reports and evaluated the morbidity of the genes by using the following publicly available databases: Database of Genomic Variants (DGV), Database of Chromosomal Imbalance and Phenotype in Humans Using Ensemble Resources (DECIPHER), OMIM, PubMed, ClinVar, and the UCSC Genome Browser. All genomic coordinates are based on the February 2009 assembly of the Genome Reference Consortium build 37(GRCh37)/UCSC hg19.
3. Results
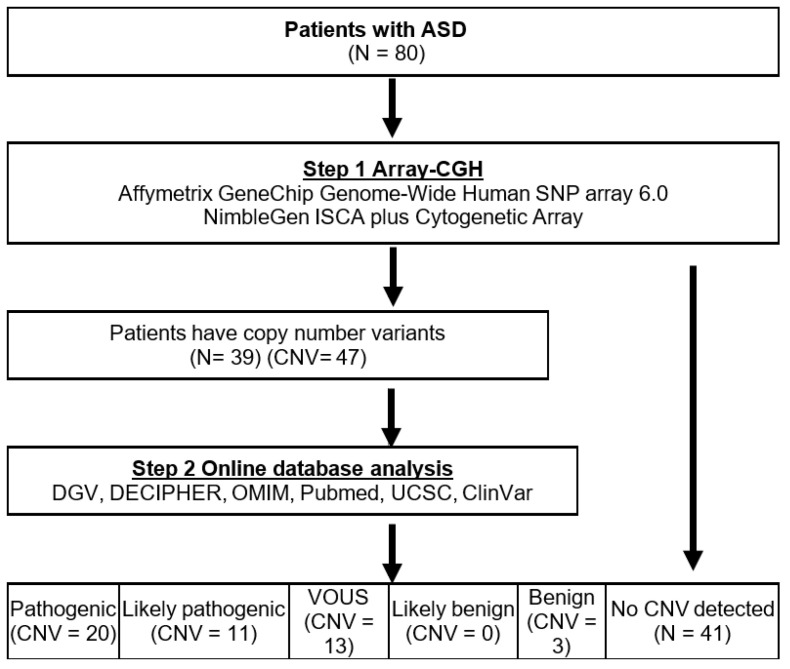
Figure 2 illustrates the diagnostic work-up of patients with ASD. A total of 47 CNVs were found in 39 ASD patients. Thirty-one patients had only one CNV and eight patients had two CNVs. Among the 47 CNVs, 32 were deletions and 15 were duplications. These CNVs were classified into the following five groups according to the clinical interpretation: 42.6% (20/47) were classified as pathogenic, 23.4% (11/47) as likely pathogenic, 27.6% (13/47) as VOUS, 0% (0/48) as likely benign, and 6.4% (3/47) as benign.
Figure 2.
Diagnostic work-up of patients with autism spectrum disorder (ASD) (N = 80). Abbreviations: ASD, autism spectrum disorder; CNV, copy number variant; VOUS, variance of uncertain significance.
The summary of patient characteristics and CNV findings is shown in Table 1. There were 47 CNVs and 80 ASD patients. The detection rate of CNVs was 58.8%. In our study, there were 24 males and 15 females with CNVs. In male and female patients, the CNV detection rates were 62.5% and 53.1%, respectively. There were 31 clinically significant CNVs in 27 patients with a diagnostic yield of 33.8%. VOUS were detected in 13 patients (16.3%). We reviewed the detected CNVs according to the published CNV map of the human genome [23].
Table 1.
Summary of patient characteristics and CNV findings; CNV, copy number variant.
| Number of Patients | 80 |
| Male | 48 |
| Female | 32 |
| Age range (median) (years) | 2–16 (6) |
| Total number of CNV | 47 |
| (Detection rate %) | (58.8) |
| Detected in male | 30 |
| (Detection rate %) | (62.5) |
| Detected in female | 17 |
| (Detection rate %) | (53.1) |
| Clinically significant CNV | 31 |
| (Diagnostic yield %) | (38.8) |
| Detected in male | 18 |
| (Diagnostic yield %) | (37.5) |
| Detected in female | 13 |
| (Diagnostic yield %) | (40.6) |
Among the 47 CNVs, 31 (65.9%) were clinically significant; 13 were duplications and 18 were deletions. The largest and smallest sizes of these significant CNVs were 17.59 Mb and 0.008 Mb, respectively. There were 22 (70.9%) CNVs smaller than 5 Mb that could not be routinely detected by karyotyping. Among the 22 CNVs, 16 (51.6%) were between 1 and 5 Mb, while 6 (19.3%) were <1 Mb.
Table 2 illustrates all clinically significant CNVs (31 CNVs) in our study. Deletions in chromosome band 15q11.2 were detected in 4 patients and these deletions were found mostly in our patients. The chromosome band 15q11.2 overlapped the Prader–Willi/Angelman region and involved the UBE3A, SNRPN, and CHRNA7 genes. Table 3 describes all 13 VOUS; 3 duplications and 10 deletions. Their sizes ranged from 0.012–148.290 Mb.
Table 2.
Clinically significant CNVs.
| Patient Number |
Gender | Array CGH Result (hg18) | Chromosome Region (Genes Associated with ASD Phenotype) |
Aberration Type |
Size (Mb) |
Clinical Significance | IQ | Additional Clinical Features |
|---|---|---|---|---|---|---|---|---|
| 1 | Male | arr15q11.2(22,842,145 − 25,235,046) × 3 | 15q11.2 (UBE3A, SNRPN, CHRNA7) |
Duplication | 2.393 | Susceptibility to ASD | N/A | Developmental delay |
| arr15q11.2q13.1(25,236,676 − 28,559,402) × 4 | 15q11.2q13.1 (UBE3A, SNRPN, CHRNA7) |
Duplication | 3.323 | |||||
| 3 | Male | arr17p11.2(16,782,546 − 20,219,464) × 1 | 17p11.2 (RAI1) |
Deletion | 3.437 | Smith-Magenis syndrome | N/A | Developmental delay and facial dysmorphism |
| 4 | Male | arr7q11.23(72,776,313 − 74,133,332) × 1 | 7q11.23 (AUTS2) |
Deletion | 1.367 | Williams syndrome | N/A | Developmental delay |
| 5 | Female | arr15q11.2q13.2(22,765,628 − 30,653,876) × 4 | 15q11.2q13.2 (UBE3A, SNRPN, CHRNA7) |
Duplication | 7.888 | Susceptibility to ASD | N/A | Developmental delay |
| arr15q13.2q13.3(30,653,877 − 32,509,926) × 3 | 15q13.2q13.3 (CHRNA7) |
Duplication | 1.856 | |||||
| 7 | Female | arr22q11.21(18,706,001 − 21,505,417) × 3 | 22q11.21 (CRKL, FGF8, TBX1) |
Duplication | 2.799 | Susceptibility to ASD | N/A | Developmental delay and facial dysmorphism |
| 8 | Male | arr4p15.1p12(28,451,191 − 47,062,229) × 4 | 4p15.1p12 (UGDH) |
Duplication | 18.611 | Susceptibility to ASD | N/A | Developmental delay |
| 9 | Female | arr22q11.23q12.1(25,695,469 − 25,903,543) × 0 | 22q11.23q12.1 (CRKL, FGF8, TBX1) |
Deletion | 0.208 | Susceptibility to ASD | N/A | Developmental delay |
| 11 | Male | arr4p16.3(72,447 − 3,848,881) × 1 | 4p16.3 (WHS) |
Deletion | 3.776 | Wolf-Hirschhorn syndrome |
33 | Developmental delay and facial dysmorphism |
| 12 | Female | arr15q11.2q13.3(22,770,421 − 32,915,593) × 1 | 15q11.2q13.3 (UBE3A, SNRPN, CHRNA7) |
Deletion | 10.145 | Angelman syndrome | N/A | Developmental delay and facial dysmorphism |
| 13 | Female | arr4p16.3(68,345 − 4,044,985) × 1.0 | 4p16.3 (WHS) |
Deletion | 3.977 | Wolf-Hirschhorn syndrome |
55 | Developmental delay and facial dysmorphism |
| 14 | Female | arr22q13.33(50,967,018 − 51,197,725) × 1 | 22q13.3 (SHANK3) |
Deletion | 0.231 | Susceptibility to ASD | N/A | Developmental delay and facial dysmorphism |
| arr4p16.3p14 (68,345 − 40,111,547) × 3 | 4p16.3p14 (WHS) |
Duplication | 40.000 | |||||
| 15 | Female | arr18p11.32p11.21(136,227 − 15,181,207) × 4 | 18p11.32 18p11.21 (SMCHD1) |
Duplication | 15.045 | Susceptibility to ASD | 57 | Developmental delay and facial dysmorphism |
| 16 | Male | arr11q13.4q14.3(71,567,724 − 89,547,851) × 4 | 11q13.4q14.3 (SHANK2) |
Duplication | 17.980 | Susceptibility to ASD | 34 | Developmental delay and facial dysmorphism |
| 18 | Female | arr2q22.1q22.3(141,332,947 − 145,948,739) × 1 | 2q22.1q22.3 (TBR1) |
Deletion | 4.161 | Susceptibility to ASD | 55 | Developmental delay and facial dysmorphism |
| 21 | Male | arr1p31.3p31.1(61,947,700 − 73,030,143) × 1 | 1p31.3p31.1 (NEGR1) |
Deletion | 11.080 | Susceptibility to ASD | N/A | Developmental delay and facial dysmorphism |
| 22 | Female | arr3q22.3q23(138,681,193 − 139,438,715) × 3 | 3q22.3q23 (ZBTB20) |
Duplication | 0.758 | Susceptibility to ASD | N/A | Developmental delay |
| 23 | Male | arr10p15.3(162,270 − 468,133) × 3 | 10p15.3 (DIP2C) |
Duplication | 0.306 | Susceptibility to ASD | 78 | Developmental delay |
| 25 | Male | arr14q21.2q22.1(45,863,061 − 50,360,747) × 0 | 14q21.2q22.1 (NIN) |
Deletion | 4.500 | Deletion of the NIN gene | N/A | Developmental delay |
| 27 | Female | arr2q23.3q24.1(150,619,633 − 157,576,339) × 1.3 | 2q23.3q24.1 (MBD5) |
Deletion | 6.957 | Susceptibility to ASD | N/A | Developmental delay |
| 28 | Male | arr18q21.33q23(60,414,497 − 78,003,508) × 1 | 18q21.33q23 (NETO1, FBXO15) |
Deletion | 17.590 | Susceptibility to ASD | N/A | Developmental delay |
| 29 | Male | arr22q11.21(18,657,470 − 21,843,336) × 1 | 22q11.21 (CRKL, FGF8, TBX1) |
Deletion | 3.190 | CATCH22 | N/A | Developmental delay |
| 30 | Male | arrXp22.31(6,450,627 − 8,141,242) × 0 | Xp22.31 (NLGN4) |
Deletion | 1.690 | Susceptibility to ASD | 80 | Developmental delay |
| arrXp22.31(8,429,167 − 8,435,863) × 0.5 | Xp22.31 (NLGN4) |
Deletion | 1.310 | |||||
| 31 | Female | arr15q11.2(20,760,484 − 23,601,857) × 1.1 | 15q11.2 (UBE3A, SNRPN, CHRNA7) |
Deletion | 2.840 | Susceptibility to ASD | 41 | Developmental delay |
| 32 | Male | arr15q11.2(22,748,697 − 23,188,522) × 1 | 15q11.2 (UBE3A, SNRPN, CHRNA7) |
Deletion | 0.440 | Susceptibility to ASD | 35 | Developmental delay |
| 33 | Male | arr15q11.2q13.1(23,614,732 − 28,536,497) × 1 | 15q11.2q13.1 (UBE3A, SNRPN, CHRNA7) |
Deletion | 4.920 | Angelman syndrome | 17 | Developmental delay |
| 34 | Male | arr9q34.3 (140,687,823 − 140,695,906) × 1 | 9q34.3 (TSC1, EHMT1) |
Deletion | 0.008 | Kleefstra syndrome | 59 | Developmental delay and facial dysmorphism |
| 36 | Male | arrXq28(152,956,854 − 155,270,560) × 2 | Xq28 (MECP2) |
Duplication | 2.310 | Susceptibility to ASD | N/A | Developmental delay |
N/A, not available; IQ, intelligence quotient; ASD, autism spectrum disorder.
Table 3.
List of variants of uncertain significance.
| Patient Number |
Gender | Array CGH Result (hg18) | Chromosome Region (Genes Associated with ASD Phenotype) |
Aberration Type |
Size (Mb) |
IQ | Additional Clinical Features |
|---|---|---|---|---|---|---|---|
| 2 | Male | arr22q11.22(22,336,268 − 22,556,733) × 1 | 22q11.22 (CRKL, FGF8, TBX1) |
Deletion | 0.220 | 72 | Developmental delay |
| 6 | Male | arr17p13.3(1693 − 2,393,788) × 1 | 17p13.3 (MDLS) |
Deletion | 2.392 | 85 | Developmental delay |
| 10 | Female | arr9p24.39p23(204,193 − 10,972,824) × 1 | 9p24.39p23 (KANK1) |
Deletion | 10.768 | N/A | Developmental delay |
| 17 | Male | arr16q22.1q22.2(69,098,865 − 72,591,930) × 1 | 16q22.1q22.2 (SCA4) |
Deletion | 3.493 | 69 | Developmental delay |
| 19 | Male | arr12p13.33p13.32(173,786 − 4,424,837) × 1 | 12p13.33p13.32 (EMG1) |
Deletion | 4.250 | 69 | Developmental delay |
| 23 | Male | arr20p12.3(8,085,389 − 8,589,571) × 1 | 20p12.3 (PLCB1) |
Deletion | 0.504 | 78 | Developmental delay |
| 24 | Male | arrXq13.1(69,228,881 − 69,240,595) × 0 | Xq13.1 (NLGN3) |
Deletion | 0.012 | N/A | Developmental delay |
| 26 | Female | arrXp21.2(29,336,996 − 29,372,188) × 1 | Xp21.2 (CDKL5) |
Deletion | 0.035 | N/A | Developmental delay |
| 36 | Male | arrXp22.33(1 − 2,196,782) × 0 | Xp22.33 (NLGN4) |
Deletion | 2.200 | N/A | Developmental delay |
| 37 | Female | arr8q21.2q21.13(51,301,121 − 54,915,042) × 1 | 8q21.2q21.13 (TCF4) |
Deletion | 3.610 | 19 | Developmental delay |
| 38 | Male | arrXq13.1q13.3(70,749,306 − 74,335,167) × 2 | Xq13.1q13.3 (NLGN3) |
Duplication | 3.590 | 72 | Developmental delay |
| 39 | Male | arr17q25.3 (77,856,839 − 78,293,128) × 2.95 | 17q25.3 (NF1) |
Duplication | 0.436 | N/A | Developmental delay |
| arrXp22.31q28(6,980,000 − 155,270,000) × 1.1 | Xp22.31q28 (NLGN4) |
Duplication | 148.290 |
N/A, not available; IQ, intelligence quotient; ASD, autism spectrum disorder.
4. Discussion
There were 27 ASD patients (33.8%) in our study with clinically significant CNVs detected by array-CGH. The rate of diagnosis is relatively high compared with other studies [24,25]. Our study showed that array-CGH could be the first-tier testing for idiopathic ASD patients due to a satisfactory diagnostic yield. Furthermore, array-CGH allows us to describe the breakpoints (BPs) of the CNV. It can also strengthen the genotype–phenotype correlation and identify candidate genes [4]. For example, a patient who had a 15q11.2 deletion at the Prader–Willi/Angelman region eventually developed autism and language delays due to a reported microdeletion at 15q11.2 between BP1 to BP2 [26,27]. 15q11.2 deletion was also the most common clinically significant CNV identified in our cohort.
In our study, the largest clinically significant CNV was a deletion within the chromosome band 18q21.33q23, and it was consistent with a previous report [28]. The 18q21.33q23 deletion had a size of 17.59 Mb and included 44 OMIM genes from PHLPP1 to PARD6G. According to a previous study, 18q deletion was associated to different phenotypes due to its remarkable genomic heterogeneity [29]. Therefore, we could not confirm diagnosis of 18q deletion by clinical characteristics; genomic analysis is necessary. Our patient had cognition delay, expressive language delay, gross and fine motor delay, hearing loss, delayed myelination of the brain, umbilical hernia, and ear canal stenosis, symptoms compatible with distal 18q deletion [30]. In previous studies, about 54% of the patients with 18q deletion had congenital cardiac anomalies [30,31,32]; however, our patient had a normal echocardiogram. The constitutional hemizygosity of 18q increases the risk of autism as well; 43% of 18q-deletion patients had autism [33]. Furthermore, if the TCF4, NETO1, and FBXO15 genes were in the region of hemizygosity, the risk of autism increased significantly [32]. Our patient had deletion of the NETO1 and FBXO15 genes. However, there was no shared region of deletion in the ASD patients with 18q deletion. Therefore, further studies are needed to confirm the genetic determinants of autism in 18q-deletion patients.
One patient in our cohort had a 14q21.2q22.1 deletion involving the NIN gene. Microcephalic primordial dwarfism disorder has been associated with compound heterozygous mutations of NIN gene [34]. However, our patient had only developmental delays without dysmorphic features. Ninein, a centrosomal protein involved in microtubule anchoring, is encoded by the NIN gene. Ninein plays an important role in microtubule stability due its influence in axonal development and bifurcation [35,36]. Disruptions of neocortex development and axon guidance are crucial factors for the development of ASD [37,38,39,40]. Thus, the NIN gene was associated with ASD possibly because of the function of ninein in axonal development and bifurcation.
The differences in certain aspects between Taiwan and European cohorts were noted by the CNV data from other studies [41,42,43,44]. According to previous reports, the most common detected CNVs in ASD occur in 16p11.2; however, this is seen in less than 1% of ASD patients [42,43,44,45]. In our study, the most frequently detected CNVs were 15q11.2 deletions, seen in 5.0% of ASD patients. To evaluate the differences between Taiwan ASD patients and other ASD cohorts, further studies are needed to assess larger Taiwan ASD cohorts compared with controls. On the other hand, VOUS comprised 13 out of 47 (27.7%) CNVs in our study. It is crucial to interpret VOUS in the context of parental data, but this information was not available during data collection. Due to their possible association with ASD, further investigations for VOUS are needed.
In our study, there were 41 patients without CNVs. However, aside from CNVs, other factors like damaging missense mutations, epigenetic alterations, environmental (in utero and early childhood), developmental factors and as-yet unknown different ways influence autism phenotype [46,47]. Based on research to date, a single condition or event could not play a major role in causing ASD. Even though syndromic or secondary autism caused by such as fragile X syndrome and tuberous sclerosis, none of these etiologies are specific to autism because these etiologies include variable proportion of patients with or without ASD [48]. New technologies in genomics and epigenomics research could uncover the epidemiology of ASD [49]. CMA has a higher resolution than conventional karyotyping. However, CMA may miss polyploidy, balanced translocations, inversion, low-level mosaicism, and marker chromosomes. Meanwhile, we could not exclude all genetic diseases by a benign CMA result. Thus, CMA should not replace the karyotyping.
There are more than 800 genes associated with autism according to case-control studies on population and animal models. In addition to three relevant CNVs and their association with ASD above, we also found other clinically significant CNVs in Table 2 and Table 3. These genes are associated with chromatin remodeling and transcriptional regulation, cell proliferation, and mostly synaptic architecture and functionality. According to the largest exome sequencing study of ASD to date [50], these genes were indicated. Other amygdala-expressed genes associated to the social pathophysiology of ASD were indicated by Herrero et al.’s survey [51].
There were some limitations in this study. Compared to the total number of ASD patients in other studies, the number of patients in our cohort was relatively small. Another limitation was that we did not have the parental samples, which could have helped determine the inheritance for VOUS. In addition, our cohort lacked cases of control CNV data from normal individuals. In other previous studies [28,52,53,54,55,56], there were also no control CNV data from normal individuals. However, according to Kousoulidou et al. study, 6 out of 50 mothers and 8 out of 50 fathers from a total of 100 parents (14%) who had ASD children appeared to carry 16 different rare variants associated with ASD [57]. From an analytical aspect, we also did not check the CNV findings using a second method. However, we reviewed all raw CNV data manually, and this matched the recommended quality parameters.
In our study, two kinds of different CMA testing were used. The methodological factors could influence the results due to different reference samples. We should use the same reference sample within one study [58]. Furthermore, according to Dana Hollenbeck et al. in 2017 [59], there are diagnostic clinical relevance of small (<500 kb) nonrecurrent CNVs during CMA clinical testing. It is necessary for careful clinical interpretation of these CNVs. These small, nonrecurrent CNVs can also facilitate the discovery of new genes involved in the pathogenesis of neurodevelopmental disorders and/or congenital anomalies. Our patient with CNV <500 kb, particularly <50 kb, did not have multiplex ligation-dependent probe amplification (MLPA) or FISH for confirmation. It should be modified in the future.
5. Conclusions
In the ASD patient cohort in Taiwan, there was a satisfying diagnostic rate by using array-CGH. Array-CGH could detect CNVs in high resolution. Comparing to the karyotyping, array-CGH could make enormous details to describe the genomic alterations in ASD patients. Therefore, array-CGH is useful for initial testing of ASD patients in Taiwan.
Author Contributions
C.-L.L. drafted the manuscript. S.-P.L., C.-P.C. and H.-Y.L. participated in the patients’ follow-up and helped in drafting the manuscript. C.-K.C., R.-Y.T. and P.-S.W. performed biochemical analyses and revised the manuscript. Y.-T.L., Y.-H.C., H.-C.C., Y.-J.C. and C.-L.C. were responsible for patient screening and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by research grants from the Ministry of Science and Technology, Executive Yuan, Taiwan (MOST-110-2314-B-195-010-MY3, MOST-110-2314-B-195-014, MOST-110-2314-B-195-029, MOST-109-2314-B-195-024, MOST-108-2314-B-195-012, and MOST-108-2314-B-195-014) and from MacKay Memorial Hospital (MMH-E-111-13, MMH-E-110-16, MMH-E-109-16, MMH-E-108-16, MMH-MM-10801, and MMH-107-82).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Mackay Memorial Hospital’s Institutional Review Board (protocol code: MMH-E-110-16, date of approval: 16 May 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data have been presented in this manuscript.
Conflicts of Interest
The authors declare no potential conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elsabbagh M., Divan G., Koh Y.J., Kim Y.S., Kauchali S., Marcín C., Montiel-Nava C., Patel V., Paula C.S., Wang C., et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang J., Gilman S.R., Chiang A.H., Sanders S.J., Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat. Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C.L., Lee C.H., Chuang C.K., Chiu H.C., Chen Y.J., Chou C.L., Wu P.S., Chen C.P., Lin H.Y., Lin S.P. Array-CGH Increased the Diagnostic Rate of Developmental Delay or Intellectual Disability in Taiwan. Pediatrics Neonatol. 2019;60:453–460. doi: 10.1016/j.pedneo.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shevell M., Ashwal S., Donley D., Flint J., Gingold M., Hirtz D., Majnemer A., Noetzel M., Sheth R.D. Practice parameter: Evaluation of the child with global developmental delay: Report of the Quality Standards Subcommittee of the American Academy of Neurology and The Practice Committee of the Child Neurology Society. Neurology. 2003;60:367–380. doi: 10.1212/01.WNL.0000031431.81555.16. [DOI] [PubMed] [Google Scholar]
- 6.Ravnan J.B., Tepperberg J.H., Papenhausen P., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L. Subtelomere FISH analysis of 11,688 cases: An evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J. Med. Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rauch A., Hoyer J., Guth S., Zweier C., Kraus C., Becker C., Zenker M., Hüffmeier U., Thiel C., Rüschendorf F., et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am. J. Med. Genet. A. 2006;140:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- 8.Moreno-De-Luca D., Sanders S.J., Willsey A.J., Mulle J.G., Lowe J.K., Geschwind D.H., State M.W., Martin C.L., Ledbetter D.H. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol. Psychiatry. 2013;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer G.B., Mendelsohn N.J. Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet. Med. 2013;15:399–407. doi: 10.1038/gim.2013.32. Correction in Genet. Med. 2013, 15, 669. [DOI] [PubMed] [Google Scholar]
- 11.Armengol L., Nevado J., Serra-Juhé C., Plaja A., Mediano C., García-Santiago F.A., García-Aragonés M., Villa O., Mansilla E., Preciado C., et al. Clinical utility of chromosomal microarray analysis in invasive prenatal diagnosis. Hum. Genet. 2012;131:513–523. doi: 10.1007/s00439-011-1095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riggs E.R., Wain K.E., Riethmaier D., Smith-Packard B., Faucett W.A., Hoppman N., Thorland E.C., Patel V.C., Miller D.T. Chromosomal microarray impacts clinical management. Clin. Genet. 2014;85:147–153. doi: 10.1111/cge.12107. [DOI] [PubMed] [Google Scholar]
- 13.Wiśniowiecka-Kowalnik B., Kastory-Bronowska M., Bartnik M., Derwińska K., Dymczak-Domini W., Szumbarska D., Ziemka E., Szczałuba K., Sykulski M., Gambin T., et al. Application of custom-designed oligonucleotide array CGH in 145 patients with autistic spectrum disorders. Eur. J. Hum. Genet. 2013;21:620–625. doi: 10.1038/ejhg.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syrmou A., Tzetis M., Fryssira H., Kosma K., Oikonomakis V., Giannikou K., Makrythanasis P., Kitsiou-Tzeli S., Kanavakis E. Array comparative genomic hybridization as a clinical diagnostic tool in syndromic and nonsyndromic congenital heart disease. Pediatric Res. 2013;73:772–776. doi: 10.1038/pr.2013.41. [DOI] [PubMed] [Google Scholar]
- 15.Bartnik M., Szczepanik E., Derwińska K., Wiśniowiecka-Kowalnik B., Gambin T., Sykulski M., Ziemkiewicz K., Kędzior M., Gos M., Hoffman-Zacharska D., et al. Application of array comparative genomic hybridization in 102 patients with epilepsy and additional neurodevelopmental disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159:60–71. doi: 10.1002/ajmg.b.32081. [DOI] [PubMed] [Google Scholar]
- 16.Sanna-Cherchi S., Kiryluk K., Burgess K.E., Bodria M., Sampson M.G., Hadley D., Nees S.N., Verbitsky M., Perry B.J., Sterken R., et al. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet. 2012;91:987–997. doi: 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C., et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girirajan S., Rosenfeld J.A., Coe B.P., Parikh S., Friedman N., Goldstein A., Filipink R.A., McConnell J.S., Angle B., Meschino W.S., et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutter M., Le Couteur A., Lord C. Autism Diagnostic Interview-Revised (ADI–R) Manual. Western Psychological Services; Los Angeles, CA, USA: 2003. [Google Scholar]
- 20.Wechsler D. Wechsler Preschool and Primary Scale of Intelligence—Fourth Edition (WPPSI-IV): Technical and Interpretive Manual. NCS Pearson, Inc.; Bloomington, IN, USA: 2012. [Google Scholar]
- 21.Wechsler D. Wechsler Intelligence Scale for Children—Fifth Edition (WISC-V): Technical and Interpretive Manual. NCS Pearson Inc.; Bloomington, MN, USA: 2014. [Google Scholar]
- 22.Sun Y., Yang Y., Luo Y., Chen M., Wang L., Huang Y., Yang Y., Dong M. Lack of MECP2 gene transcription on the duplicated alleles of two related asymptomatic females with Xq28 duplications and opposite X-chromosome inactivation skewing. Hum. Mutat. 2021;42:1429–1442. doi: 10.1002/humu.24262. [DOI] [PubMed] [Google Scholar]
- 23.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y., Dies K.A., Holm I.A., Bridgemohan C., Sobeih M.M., Caronna E.B., Miller K.J., Frazier J.A., Silverstein I., Picker J., et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125:e727–e735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnside R.D., Pasion R., Mikhail F.M., Carroll A.J., Robin N.H., Youngs E.L., Gadi I.K., Keitges E., Jaswaney V.L., Papenhausen P.R., et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: A susceptibility region for neurological dysfunction including developmental and language delay. Hum. Genet. 2011;130:517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doornbos M., Sikkema-Raddatz B., Ruijvenkamp C.A., Dijkhuizen T., Bijlsma E.K., Gijsbers A.C., Hilhorst-Hofstee Y., Hordijk R., Verbruggen K.T., Kerstjens-Frederikse W.S., et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur. J. Med. Genet. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Siu W.K., Lam C.W., Mak C.M., Lau E.T., Tang M.H., Tang W.F., Poon-Mak R.S., Lee C.C., Hung S.F., Leung P.W., et al. Diagnostic yield of array CGH in patients with autism spectrum disorder in Hong Kong. Clin. Transl. Med. 2016;5:18. doi: 10.1186/s40169-016-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heard P.L., Carter E.M., Crandall A.C., Sebold C., Hale D.E., Cody J.D. High resolution genomic analysis of 18q- using oligo-microarray comparative genomic hybridization (aCGH) Am. J. Med. Genet A. 2009;149:1431–1437. doi: 10.1002/ajmg.a.32900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cody J.D., Hasi M., Soileau B., Heard P., Carter E., Sebold C., O’Donnell L., Perry B., Stratton R.F., Hale D.E. Establishing a reference group for distal 18q-: Clinical description and molecular basis. Hum. Genet. 2014;133:199–209. doi: 10.1007/s00439-013-1364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Versacci P., Digilio M.C., Sauer U., Dallapiccola B., Marino B. Absent pulmonary valve with intact ventricular septum and patent ductus arteriosus: A specific cardiac phenotype associated with deletion 18q syndrome. Am. J. Med. Genet. A. 2005;138:185–186. doi: 10.1002/ajmg.a.30916. [DOI] [PubMed] [Google Scholar]
- 32.Van Trier D.C., Feenstra I., Bot P., de Leeuw N., Draaisma J.M. Cardiac anomalies in individuals with the 18q deletion syndrome; report of a child with Ebstein anomaly and review of the literature. Eur. J. Med. Genet. 2013;56:426–431. doi: 10.1016/j.ejmg.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 33.O’Donnell L., Soileau B., Heard P., Carter E., Sebold C., Gelfond J., Hale D.E., Cody J.D. Genetic determinants of autism in individuals with deletions of 18q. Hum. Genet. 2010;128:155–164. doi: 10.1007/s00439-010-0839-y. [DOI] [PubMed] [Google Scholar]
- 34.Dauber A., Lafranchi S.H., Maliga Z., Lui J.C., Moon J.E., McDeed C., Henke K., Zonana J., Kingman G.A., Pers T.H., et al. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J. Clin. Endocrinol. Metab. 2012;97:E2140–E2151. doi: 10.1210/jc.2012-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Tsai J.W., Imai J.H., Lian W.N., Vallee R.B., Shi S.H. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivatsa S., Parthasarathy S., Molnár Z., Tarabykin V. Sip1 downstream Effector ninein controls neocortical axonal growth, ipsilateral branching, and microtubule growth and stability. Neuron. 2015;85:998–1012. doi: 10.1016/j.neuron.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Blockus H., Chédotal A. The multifaceted roles of Slits and Robos in cortical circuits: From proliferation to axon guidance and neurological diseases. Curr. Opin. Neurobiol. 2014;27:82–88. doi: 10.1016/j.conb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Chu J., Anderson S.A. Development of cortical interneurons. Neuropsychopharmacology. 2015;40:16–23. doi: 10.1038/npp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marín O. Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 40.Robichaux M.A., Cowan C.W. Signaling mechanisms of axon guidance and early synaptogenesis. Curr. Top. Behav. Neurosci. 2014;16:19–48. doi: 10.1007/7854_2013_255. [DOI] [PubMed] [Google Scholar]
- 41.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tammimies K., Marshall C.R., Walker S., Kaur G., Thiruvahindrapuram B., Lionel A.C., Yuen R.K., Uddin M., Roberts W., Weksberg R., et al. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children with Autism Spectrum Disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 43.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z., et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girirajan S., Dennis M.Y., Baker C., Malig M., Coe B.P., Campbell C.D., Mark K., Vu T.H., Alkan C., Cheng Z., et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newschaer C.J., Croen L.A., Daniels J., Giarelli E., Grether J.K., Levy S.E., Mandell D.S., Miller L.A., Pinto-Martin J., Reaven J., et al. The Epidemiology of Autism Spectrum Disorders. Annu. Rev. Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- 47.Elsabbagh M., Mercure E., Hudry K., Chandler S., Pasco G., Charman T., Pickles A., Baron-Cohen S., Bolton P., Johnson M.H. Infant Neural Sensitivity to Dynamic Eye Gaze Is Associated with Later Emerging Autism. Curr. Biol. 2012;22:338–342. doi: 10.1016/j.cub.2011.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benvenuto A., Moavero R., Alessandrelli R., Manzi B., Curatolo P. Syndromic autism: Causes and pathogenetic pathways. World J. Pediatr. 2009;5:169–176. doi: 10.1007/s12519-009-0033-2. [DOI] [PubMed] [Google Scholar]
- 49.Masini E., Loi E., Vega-Benedetti A.F., Carta M., Doneddu G., Fadda R., Zavattari P. An Overview of the Main Genetic, Epigenetic and Environmental Factors Involved in Autism Spectrum Disorder Focusing on Synaptic Activity. Int. J. Mol. Sci. 2020;21:8290. doi: 10.3390/ijms21218290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.-Y., Peng M., Collins R., Grove J., Klei L., et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrero M.J., Velmeshev D., Hernandez-Pineda D., Sethi S., Sorrells S., Banerjee P., Sullivan C., Gupta A.R., Kriegstein A.R., Corbin J.G. Identification of amygdala-expressed genes associated with autism spectrum disorder. Mol. Autism. 2020;11:39. doi: 10.1186/s13229-020-00346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cappuccio G., Vitiello F., Casertano A., Fontana P., Genesio R., Bruzzese D., Ginocchio V.M., Mormile A., Nitsch L., Andria G., et al. New insights in the interpretation of array-CGH: Autism spectrum disorder and positive family history for intellectual disability predict the detection of pathogenic variants. Ital. J. Pediatrics. 2016;42:39. doi: 10.1186/s13052-016-0246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palka Bayard de Volo C., Alfonsi M., Morizio E., Guaciali-Franchi P., Mohn A., Chiarelli F. A 343 Italian cohort of patients analysed with array-comparative genomic hybridization: Unsolved problems and genetic counselling difficulties. J. Intellect. Disabil. Res. 2021;65:863–869. doi: 10.1111/jir.12867. [DOI] [PubMed] [Google Scholar]
- 54.Monteiro S., Pinto J., Mira Coelho A., Leão M., Dória S. Identification of Copy Number Variation by Array-CGH in Portuguese Children and Adolescents Diagnosed with Autism Spectrum Disorders. Neuropediatrics. 2019;50:367–377. doi: 10.1055/s-0039-1694797. [DOI] [PubMed] [Google Scholar]
- 55.Napoli E., Russo S., Casula L., Alesi V., Amendola F.A., Angioni A., Novelli A., Valeri G., Menghini D., Vicari S. Array-CGH Analysis in a Cohort of Phenotypically Well-Characterized Individuals with “Essential” Autism Spectrum Disorders. J. Autism Dev. Disord. 2018;48:442–449. doi: 10.1007/s10803-017-3329-4. [DOI] [PubMed] [Google Scholar]
- 56.Gümüşlü K.E., Savli H., Sünnetçi D., Çine N., Kara B., Eren Keskin S., Akkoyunlu R.U. A CGH array study in nonsyndromic (primary) autism patients: Deletions on 16p13.11, 16p11.2, 1q21.1, 2q21.1q21.2, and 8p23.1. Turk. J. Med. Sci. 2015;45:313–319. doi: 10.3906/sag-1310-81. [DOI] [PubMed] [Google Scholar]
- 57.Kousoulidou L., Moutafi M., Nicolaides P., Hadjiloizou S., Christofi C., Paradesiotou A., Anastasiadou V., Sismani C., Patsalis P.C. Screening of 50 cypriot patients with autism spectrum disorders or autistic features using 400K custom array-CGH. Biomed Res. Int. 2013;2013:843027. doi: 10.1155/2013/843027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu L., Hou Y., Bickhart D., Song J., Liu G. Comparative analysis of CNV calling algorithms: Literature survey and a case study using bovine high-density SNP data. Microarrays. 2013;2:171–185. doi: 10.3390/microarrays2030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollenbeck D., Williams C.L., Drazba K., Descartes M., Korf B.R., Rutledge S.L., Lose E.J., Robin N.H., Carroll A.J., Mikhail F.M. Clinical relevance of small copy-number variants in chromosomal microarray clinical testing. Genet. Med. 2017;19:377–385. doi: 10.1038/gim.2016.132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data have been presented in this manuscript.