Abstract
Dermatomycoses are very common infections caused mainly by dermatophytes. Scytalidiosis is a differential mycological diagnosis, especially in tropical and subtropical areas. Since a culture-based diagnosis takes 2 to 3 weeks, we set up a PCR-restriction fragment length polymorphism (RFLP) method for rapid discrimination of these fungi in clinical samples. The hypervariable V4 domain of the small ribosomal subunit 18S gene was chosen as the target for PCR. The corresponding sequences from 19 fungal species (9 dermatophytes, 2 Scytalidium species, 6 other filamentous fungi, and 2 yeasts) were obtained from databases or were determined in the laboratory. Sequences were aligned to design primers for dermatophyte-specific PCR and to identify digestion sites for RFLP analysis. The reliability of PCR-RFLP for the diagnosis of dermatomycosis was assessed on fungal cultures and on specimens from patients with suspected dermatomycosis. Two sets of primers preferentially amplified fungal DNA from dermatophytes (DH1L and DH1R) or from Scytalidium spp. (DH2L and DH1R) relative to DNA from bacteria, yeasts, some other filamentous fungi, and humans. Digestion of PCR products with EaeI or BamHI discriminated between dermatophytes and Scytalidium species, as shown with cultures of 31 different fungal species. When clinical samples were tested by PCR-RFLP, blindly to mycological findings, the results of the two methods agreed for 74 of 75 samples. Dermatophytes and Scytalidium spp. can thus be readily discriminated by PCR-RFLP within 24 h. This method can be applied to clinical samples and is suited to rapid etiologic diagnosis and treatment selection for patients with dermatomycosis.
Dermatophytes, which belong to the genera Trichophyton, Microsporum, and Epidermophyton, are extremely widespread fungi that infect human skin, hair, and nails. They are responsible for most superficial fungal infections, causing 94.7% of cases of tinea pedis and 81.9% of cases of onychomycosis in the United States (13). Scytalidium hyalinum and Scytalidium dimidiatum are molds responsible for skin lesions and onychomycoses, which mimic those due to Trichophyton rubrum. These infections are frequent in tropical and subtropical areas. For example, S. dimidiatum accounts for 39% of dermatomycoses in Thai soldiers, whereas dermatophytes account for only 5% (6). In Gabon, S. dimidiatum was responsible for 34.2% of such cases, either alone or jointly with a dermatophyte or Candida albicans (14).
Laboratory diagnosis of dermatomycosis is based on the demonstration of hyphae by direct microscopic examination of clinical samples, followed by species identification by culture. Microscopic examination is rapid, but it can be difficult to differentiate hyphae from dermatophytes or molds. Culture requires at least 2 to 3 weeks to obtain typical macroscopic and microscopic features for specific dermatophyte identification. In rare cases, identification is hindered by the absence of specific macroscopic and microscopic characteristics; subculture on specific media is then required, further delaying the diagnosis by several weeks. Thus, a simple, rapid, and specific method able to confirm the presence or absence of dermatophytes or Scytalidium would be useful in choosing the appropriate treatment (Scytalidium spp. are resistant to most antifungal drugs). PCR is a candidate method, but its ability to discriminate between dermatophytes and other fungi that may be present on human skin remains to be demonstrated. In previous studies, Kappe et al. (12) and Bock et al. (2, 3) used a set of primers (TR1-TR2) that could amplify DNA from seven dermatophyte species but not DNA from several other fungi (mainly yeasts), plants, animals, or humans. These authors performed hybridization with probes specific for Candida albicans, Aspergillus fumigatus, or dermatophytes to confirm the specificity of the reaction. This PCR technique had limitations, since only seven dermatophyte species were studied. In addition, bioinformatic analysis of corresponding DNA sequences from data banks of various fungi indicated a high possibility of obtaining the 581-bp amplicon from the 18S gene when using the TR1-TR2 primers set with many nondermatophyte fungal species. We confirmed that these primers could amplify DNA from other common dermatophytes (Trichophyton soudanense, T. tonsurans, T. violaceum, and Microsporum canis) but also from nondermatophyte fungi (Scopulariopsis brevicaulis, S. hyalinum, S. dimidiatum, Aspergillus niger, A. fumigatus, and Trichosporon cutaneum) (data not shown).
This prompted us to develop a new molecular-biology-based approach combining two techniques. First, we used PCR amplification of the variable V4 region of 18S ribosomal DNA (rDNA), a method that can readily and preferentially amplify DNA of dermatophytes and Scytalidium spp. relative to other filamentous fungi and yeasts. Then, PCR amplicons were submitted to restriction fragment length polymorphism (RFLP) analysis to confirm the diagnosis and to differentiate dermatophytes from Scytalidium spp. The performance of this ribotyping method was assessed both experimentally and on samples from patients with dermatomycoses.
MATERIALS AND METHODS
Strain origins and fungal isolates.
This study focused on fungi of medical importance in dermatology, i.e., dermatophytes, S. dimidiatum, and S. hyalinum, and several yeasts or other filamentous fungi. Nineteen fungal species were selected to set up the PCR and RFLP techniques (reference numbers at the Pasteur Institut Collection, Paris, France, are given in parentheses): T. rubrum (IP2537.00), T. mentagrophytes var. mentagrophytes (IP2538.00), T. mentagrophytes var. interdigitale (IP2539.00), T. violaceum (IP2472.98), T. tonsurans (IP2549.00), T. soudanense (IP2516.99), Microsporum canis (IP2540.00), M. audouinii var. langeronii (IP2517.99), Epidermophyton floccosum (IP2542.00), S. dimidiatum (IP1278.81), S. hyalinum (IP1517.83), Geotrichum candidum (IP2547.00), Fusarium oxysporum (IP625.72), Scopulariopsis brevicaulis (IP2518.99), A. fumigatus (IP2544.00), A. niger (IP2550.00), Acremonium rosegruseum (IP2242.94), C. albicans (IP2546.00), and Trichosporon cutaneum (IP2548.00). Each strain had been subcultured at 27°C on Sabouraud-chloramphenicol agar (bioMérieux, Marcy l'Etoile, France). To assess the PCR and RFLP techniques, we selected the following 12 additional species of dermatophytes, filamentous fungi, and yeasts: Trichophyton equinum (IP2252.94), T. schoenleinii (IP1255.81), T. ochraceum (IP2208.94), T. erinacei (IP2251.94), M. audouinii (IP1960.90), M. gypseum (IP2143.93), M. ferrugineum (IP1821.88), M. praecox (IP2551.00), M. nanum (IP1820.88), Candida parapsilosis (IP2545.00), Aspergillus flavus (IP2543.00), and A. nidulans (IP17.60), and 22 other isolates collected from the mycology laboratory of The Hôpital Saint-Louis: 5 T. rubrum, 1 T. mentagrophytes var. interdigitale, 2 T. soudanense, 1 E. floccosum, 2 S. dimidiatum, 1 S. hyalinum, 5 C. albicans, 1 Trichosporon cutaneum, 1 A. fumigatus, 1 Acremonium sp., 1 G. candidum, and 1 Fusarium sp. strains. Human and bacterial DNAs (from Pseudomonas aeruginosa) were used as controls.
Human samples.
Seventy-five samples collected from patients with suspected dermatomycosis (9 hair, 37 skin, and 29 nail samples) were obtained from the Hôpital Saint-Louis mycology laboratory. Part of each sample was processed using classical mycological techniques, i.e., direct microscopic examination in black chlorazol solution, followed by culture for specific identification, and the remainder was tested by PCR-RFLP as described below, blindly vis-à-vis the results of mycological examinations.
Experimental design.
The design of specific primers (Fig. 1) comprised the following sequential steps: (i) analysis of target sequences (18S rDNA gene) in the literature and several databases; (ii) sequencing of the unknown V4 domains of five fungi; (iii) alignment of these sequences and selection of the most discriminatory primers for dermatophytes, Scytalidium and yeasts, and other contaminants; and (iv) identification and testing of fungus-specific polymorphic endonuclease sites within the V4 domain of pathogenic species.
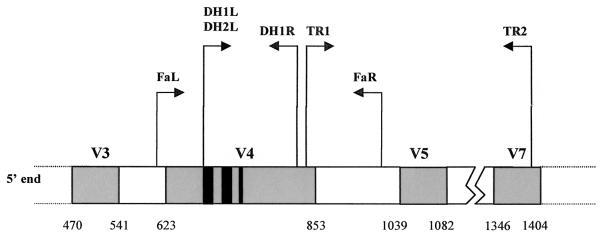
FIG. 1.
Schematic representation of the small ribosomal subunit 18S gene. The hatched boxes correspond to the polymorphic region from V3 to V7. The black boxes correspond to the highly polymorphic region of V4 (nucleotides 679 to 690, 697 to 717, and 725 to 731 in Saccharomyces cerevisiae numbering). Primer locations: i) TR1 and TR2 primers, described by Bock et al. as dermatophyte specific (2, 3); FaR and FaL, used to sequence the V4 domain of T. soudanense, M. langeronii, Scopulariopsis brevicaulis, S. dimidiatum, and S. hyalinum; and DH1L-DH1R and DH2L-DH1R, used respectively to amplify dermatophytes and Scytalidium spp.
Database consultation for 18S fungal DNA sequences.
The Genome, Japan, Urbana, Ribosomal Project, and EMBL databases were consulted. We obtained specific information on the 19 selected fungal species, including representative and reference sequences of the small ribosomal subunit (V4 domain). When the available sequences were polymorphic or different for a given fungus, the consensus sequence was studied. When identical sequences were available for a given fungus, we selected a leader sequence. Each sequence was associated with a GenBank accession number (Table 1). All of the sequences in FASTA format were aligned with the MULTALIGN program (4); homology was studied with the BLAST-N program (1), and restriction sites were identified with the CLUSTER program.
TABLE 1.
GenBank accession numbers of the 18S sequences of the studied organisms
| Organism | Accession no. |
|---|---|
| Dermatophytes | |
| Epidermophyton floccosum | Z34923 |
| Microsporum canis | Z34925 |
| Microsporum audouinii var. langeronii | AF194083 |
| Trichophyton mentagrophytes | Z34926 |
| Trichophyton rubrum | X58570 |
| Trichophyton soudanense | AF195572 |
| Trichophyton tonsurans | Z34929 |
| Trichophyton violaceum | Z34930 |
| Filamentous fungi | |
| Acremonium alternatum | U43970 |
| Acremonium chrysogenum | U43971 |
| Acremonium furcatum | U43972 |
| Acremonium kiliense | U43973 |
| Acremonium mucorum | U43966, U57663 |
| Acremonium rutilum | U43967 |
| Acremonium strictum | U43968 |
| Aspergillus flavus | D63696, X78537 |
| Aspergillus fumigatus | M60300 |
| Aspergillus nidulans | X78539 |
| Aspergillus niger | D63697, X78538 |
| Aspergillus terreus | AB008409, X78540 |
| Endomyces geotrichum | U00974 |
| Fusarium merismoides | AF141950 |
| Galactomyces geotrichum | X69842 |
| Scopulariopsis brevicaulis | AF19557 |
| Scytalidium dimidiatum | AF204293 |
| Scytalidium hyalinum | AF204294 |
| Yeasts | |
| Candida albicans | M60302 |
| Trichosporon cutaneum | X60182 |
| Human | U13369 |
DNA extraction.
DNA from cultures was purified by using the sodium dodecyl sulfate (SDS) standard protocol followed by phenol-chloroform extraction (10) in the presence of 0.5 M KCl. The extract was then treated with RNase (Sigma, St. Louis, Mo.) for 30 min at 37°C and a final concentration of 10 μg/ml. To eliminate SDS and black pigments from S. dimidiatum, which are known to be strong PCR inhibitors, we used potassium acetate buffer to precipitate the SDS-protein complexes and activated charcoal to eliminate the pigments (7, 15).
DNA was extracted from clinical samples with three different procedures: (i) standard phenol-chloroform extraction; (ii) a Chelex 100 extraction technique (Bio-Rad, Hercules, Calif.) as described by Jung et al. (9); or (iii) the High-Pure-PCR-Template preparation kit (16) (Boehringer-Mannheim, Mannheim, Germany). The absence of contaminating RNA was checked by electrophoresis on ethidium bromide-agarose gel. DNA concentrations were determined with a Hoefer TKO 100 minifluorometer (Pharmacia Biotech, Orsay, France) using Hoechst 33258 fluorescent dye.
PCR.
PCR was performed in a reaction mixture of 25 μl containing 1.5 mM MgCl2, 50 μM concentrations (each) of various deoxynucleoside triphosphates, 25 pmol of each primer, 1 U of Taq DNA polymerase (Eurogentec, Seraing, Belgium), and 25 ng of extracted DNA. Amplification conditions varied according to the primer set. One set of primers, FaL and FaR, was used to amplify and sequence the V4 domain of five fungi for which no data on this domain were available in the databases, namely, Scopulariopsis brevicaulis, S. dimidiatum, S. hyalinum, T. soudanense, and M. audouinii var. langeronii. The nucleotide structures were 5′-GTAATTCCAGCTCCAATAGCG-3′ for FaL and 5′GTATCTGATCGTCTTCGATC-3′ for FaR (5′ ends at positions 577 and 1006, respectively, according to the 18S rDNA sequence Z75578 of Saccharomyces cerevisiae [8]). For these primers the PCR conditions consisted of denaturation for 3 min at 94°C, followed by 35 amplification cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, and one final cycle at 72°C for 5 min. A second set of primers, DH1L and DH1R, was used for the specific dermatophyte identification protocol. The nucleotide structures were 5′-TGCACTGGTCCGGCTGGG-3′ for DH1L and 5′-CGGCGGTCCTAGAAACCAAC-3′ for DH1R (5′ ends at positions 631 and 813 respectively, according to the 18S rDNA sequence X58570 of T. rubrum). A third primer set, DH2L and DH1R, was used for the specific identification of Scytalidium spp. The nucleotide structure of DH2L was 5′-TGTACTGGTCCGGCCGGG-3′ (5′ end at position 631 according to the 18S rDNA sequence X58570 of T. rubrum). The amplification conditions consisted of 3 min at 94°C; 30 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 40 s; and one final cycle at 72°C for 5 min. Next, 5 μl of PCR products was electrophoresed in 2% agarose gel in the presence of ethidium bromide and visualized under UV light.
Sequencing of PCR products.
The FaR-FaL PCR products of Scopulariopsis brevicaulis, S. dimidiatum, S. hyalinum, T. soudanense, and M. audouinii var. langeronii were purified with Microcon 100 column (Amicon, Bedford, Mass.). The two strands of amplified DNA were sequenced using the PCR primers and the BigDye Terminator kit (Applied Biosystems) on an automated sequencer (Applied Biosystems 377XL).
RFLP analysis.
DH1L-DH1R and DH2L-DH1R PCR products were digested with EaeI and BamHI, respectively (Boehringer-Mannheim) for 1 h at 37°C in a total volume of 20 μl containing 5 μl of the specific PCR product and 5 U of the enzyme. The digested samples were analyzed on 3.5% Nusieve BET-agarose electrophoresis gels (FMC Bioproducts, Rockland, Maine).
Nucleotide sequence accession numbers.
The 18S sequences of S. Brevicaulis, S. dimidiatum, S. hyalinum, T. Soudanense, and M. audouinii var. langeronii were submitted to GenBank under accession numbers AF19557, AF204293, AF204294, AF195572, and AF194083, respectively.
RESULTS
DNA sequences of the V4 region.
Most sequences of the fungal 18S ribosomal subunits available in databases are described as polymorphic variable regions, particularly the V4 domain, except for S. brevicaulis, S. dimidiatum, S. hyalinum, T. soudanense, and M. audouinii var. langeronii. For these fungi, we sequenced the relevant domain by using FaR and FaL as PCR and sequencing primers. The resulting original sequences were aligned with those of other organisms. S. dimidiatum and S. hyalinum had an identical V4 domain nucleotide structure. The nucleotide structural homology was 95.9% between the dermatophytes and the tested Scytalidium species. The percentage homology between dermatophytes, Scytalidium, and other fungal sequences (yeasts and other filamentous fungi in Table 1) was 83.1% (78.2% when S. cerevisiae and human 18S sequences were included).
PCR amplification of dermatophyte and Scytalidium DNA.
Despite the higher sequence homology between dermatophytes, S. hyalinum, S. dimidiatum, yeasts, and other filamentous fungi in the V4 domain, two sets of primers were designed for preferential amplification of dermatophytes or Scytalidium spp. In the 5′ end, the DH1L and DH2L primers are located in a highly polymorphic region of the V4 domain; in the 3′ end, the DH1R primer is located in a semipolymorphic V4 region.
We first used these primers on DNA extracted from cultures of 19 different fungal species. DNA (25 ng) from all of the tested dermatophytes and Scytalidium was preferentially amplified with DH1L-DH1R and DH2L-DH1R, respectively, and PCR products had the expected size of 183 or 184 bp. Human and bacterial DNA were not amplified. However, the high sequence homology of the V4 domain among the fungi resulted in weak amplification of nondermatophyte fungi, A. fumigatus, A. niger, T. cutaneum, G. candidum, C. albicans, F. oxysporum, Acremonium sp., and S. brevicaulis. Each of these fungi yielded a faint band with both sets of primers, but they could nonetheless be clearly differentiated from the signal obtained with dermatophytes (Fig. 2 and 3). Elimination of PCR inhibitors by precipitation of SDS-protein complexes with potassium acetate and the use of activated charcoal to eliminate pigments from black fungi markedly improved the sensitivity, although the pigments were not fully eliminated.
FIG. 2.
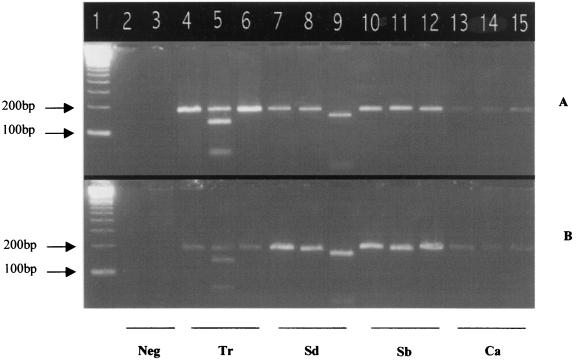
Amplification and restriction patterns of four representative fungal strains. (A) Results of PCR and RFLP obtained with the DH1L-DH1R primers. (B) Results of PCR and RFLP obtained with the DH2L-DH1R primers. Lane 1, 100-bp ladder; lane 2, human DNA (Neg); lane 3, PCR without DNA (Neg); lanes 4, 5, and 6, T. rubrum (Tr); lanes 7, 8, and 9, S. dimidiatum (Sd); lanes 10, 11, and 12, Scopulariopsis brevicaulis (Sb); lanes 13, 14, and 15, C. albicans (Ca). Lanes 4, 7, 10, and 13 show nondigested products; lanes 5, 8, 11, and 14 show partial EaeI digestion (130 and 53 to 54 nucleotides); lanes 6, 9, 12, and 15 show BamHI digestion (150 and 34 nucleotides).
FIG. 3.
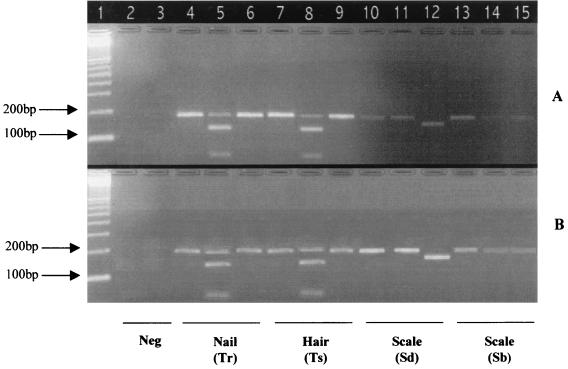
Amplification and restriction patterns of human samples. (A) Results of PCR and RFLP obtained with the DH1L-DH1R primers. (B) Results of PCR and RFLP obtained with the DH2L-DH1R primers. Lane 1, 100-bp ladder; lane 2, human DNA (Neg); lane 3, PCR without DNA (Neg); lanes 4, 5, and 6, nail sample infected with T. rubrum (Tr); lanes 7, 8, and 9, hair sample infected with T. soudanense (Ts); lanes 10, 11, and 12, scale sample infected with S. dimidiatum (Sd); lanes 13, 14, and 15, scale sample infected with S. brevicaulis (Sb). Lanes 4, 7, 10, and 13 show nondigested products; lanes 5, 8, 11, and 14 show EaeI partial digestion (130 and 53 to 54 nucleotides); lanes 6, 9, 12, and 15 show BamHI digestion (150 and 34 nucleotides).
RFLP analysis for specific discrimination.
The DH1L-DH1R primer set was designed to inhibit the EaeI restriction site located at position 73 or 74 of the V4 domain of some nondermatophyte fungi, including Scytalidium spp., but not the EaeI restriction site present in all dermatophytes at position 113 or 114 of the V4 domain (Fig. 2). EaeI digestion resulted in two fragments of different sizes (130 bp and 53 to 54 bp) with dermatophyte PCR products. The DH1L-DH1R PCR products of the nondermatophyte fungi A. fumigatus, A. niger, T. cutaneum, G. candidum, F. oxysporum, Acremonium sp., and Scopulariopsis brevicaulis, and also S. dimidiatum and S. hyalinum, were not digested by EaeI, since they do not possess a restriction site for this enzyme. DH2L-DH1R was designed to amplify a target region which contains one BamHI restriction site in Scytalidium spp. but no restriction site in dermatophytes and other fungi. With S. dimidiatum and S. hyalinum, BamHI digestion resulted in two bands of different sizes (150 and 34 bp) (Fig. 2). As expected, a single nondigested band was observed with other fungi, including dermatophytes, since they do not contain a restriction site for this enzyme (Fig. 2).
The reproducibility of PCR-RFLP was examined in 50 repeat experiments performed on T. rubrum and S. dimidiatum. Each experiment yielded similar PCR and digestion patterns.
Assessment of PCR and PCR-RFLP with other fungal isolates and clinical specimens.
The specificity of PCR and PCR-RFLP was checked on cultures of 12 additional species and 22 fungal cultures routinely obtained in the laboratory. As with the isolates used to set up the procedures, PCR resulted in a strong signal with dermatophytes, S. dimidiatum, and S. hyalinum and a weak band with some other fungi (data not shown).
Before testing clinical samples, we examined three extraction procedures on a few selected skin, hair, and nail samples. The phenol-chloroform technique was successful but time-consuming; the Chelex 100 protocol was rapid and simple but unreliable because of the persistence of PCR inhibitors, requiring repeat PCR on diluted DNA samples. The High-Pure-PCR-Template preparation kit was less time-consuming and easier to use than the two other procedures; moreover, the DNA extracts did not contain PCR inhibitors. This technique was used to examine the 75 clinical samples. Each sample was tested in duplicate. Each run included T. rubrum (IP2537.00) and S. dimidiatum (IP1278.81) DNA as positive PCR controls and human DNA as the negative PCR control. As with fungal cultures, PCR-RFLP yielded typical patterns showing the presence or absence of dermatophytes and Scytalidium (Fig. 3). The PCR-RFLP results agreed with those of classical mycological examination in 74 of 75 cases (99%) (Table 2).
TABLE 2.
Correlation between mycological examination and PCR-RFLP for dermatophyte or Scytalidium spp. detection in human samples
| Type of sample (n) | No. of samples | Mycological examination result (direct examination/culture) | PCR-RFLP finding |
|---|---|---|---|
| Nails (29) | 10 | Negative/Sterilea | Negative |
| 15 | Fungal hyphae/T. rubruma | Dermatophyte | |
| 1 | Fungal hyphae/T. soudanense | Dermatophyte | |
| 1 | Fungal hyphae/T. rubrum and S. dimidiatumb | Dermatophyte | |
| 1 | Fungal hyphae/F. oxysporumc | Negative | |
| 1 | Fungal hyphae/S. dimidiatum | Scytalidium | |
| Skin (37) | 14 | Negative/sterile | Negative |
| 13 | Fungal hyphae/T. rubrum | Dermatophyte | |
| 2 | Fungal hyphae/T. mentagrophytes var. interdigitale | Dermatophyte | |
| 1 | Fungal hyphae/T. soudanense | Dermatophyte | |
| 2 | Fungal hyphae/steriled | Dermatophyte | |
| 4 | Fungal hyphae/S. dimidiatum | Scytalidium | |
| 1 | Fungal hyphae/S. hyalinum | Scytalidium | |
| Hair (9) | 6 | Negative/sterile | Negative |
| 2 | Endothrix/T. soudanense | Dermatophyte | |
| 1 | Ectothrix/M. langeronii | Dermatophyte |
Some of these samples were contaminated in culture by nonpathogenic molds or yeasts.
Sterile, PCR-RFLP failed to detect S. dimidiatum.
F. oxysporum was pathogenic and was responsible for the onychomycosis.
These two samples were collected from the soles of patients with tinea pedis. Dermatophytosis was diagnosed because T. rubrum was detected on other sites of the feet.
In one case of concurrent infection by S. dimidiatum and T. rubrum, PCR-RFLP failed to detect the Scytalidium sp. The small size of this nail sample prevented us from repeating the extraction step and the PCR analysis. We assessed one nail sample for the absence of Scytalidium spp. and dermatophytes, but we confirmed the presence of Fusarium or Acremonium sp. since EaeI digestion of the DH2L-DH1R product yielded 172- and 11-nt digests (data not shown), a pattern corresponding to the specific digestion site of these fungi. The culture approach identified F. oxysporum as responsible for the onychomycosis.
DISCUSSION
These results show that dermatophytes and Scytalidium spp. can be readily discriminated by PCR-RFLP and that this method is suitable for the routine diagnosis with clinical samples. Molecular biology techniques such as PCR combined with hybridization have already been used to distinguish some dermatophytes from A. fumigatus (12) or C. albicans (3), but they suffered from cross-reactivity with various filamentous fungi. This was probably due to the lack of information on dermatophyte genomes: until 1994, only the 18S sequence of T. rubrum was known. The marked increase in available fungal DNA sequences in recent years has facilitated new molecular approaches to the diagnosis of dermatophyte infections.
The first aim of our study was to identify new targets that can be used for specific amplification of dermatophytes and Scytalidium spp., with no cross-reactivity with other fungi or bacteria present on human skin. We focused on the V4 domain of the 18S ribosomal gene because of its high degree of polymorphism among fungal families and species (11). Sequences of most fungi responsible for dermatomycoses and of bacterial and fungal contaminants were found in the databases or were sequenced (M. audounii var. langeronii, T. soudanense, Scopulariopsis brevicaulis, S. dimidiatum, and S. hyalinum).
A method based on PCR followed by RFLP was then developed to discriminate between dermatophytes and Scytalidium spp. The primers were designed to preferentially amplify dermatophyte DNA relative to other fungi with the DH1L-DH1R set and Scytalidium spp. with the DH2L-DH1R set. EaeI and BamHI were used to differentiate dermatophytes from Scytalidium and from other fungi, yielding the first molecular method for identifying Scytalidium spp. The reliability of our PCR-RFLP method was assessed on DNA extracted from cultures of 31 different fungal species. PCR-RFLP always gave the expected patterns and clearly distinguished between dermatophytes, Scytalidium spp., and other fungi. Results were reproducible on different isolates of the same species. Finally, the performance of PCR-RFLP was assessed on 75 clinical samples that were tested blindly vis-à-vis the results of mycological examination. Using the High-Pure-PCR-Template preparation kit, which was found to be the most reliable and most simple extraction technique, we obtained 99% agreement between the results of PCR-RFLP and mycological examination. There were no false-positive reactions with negative controls or samples contaminated by molds. Each case of single-agent infection by a dermatophyte or Scytalidium sp. was correctly identified, even in presence of other nonpathogenic yeasts or molds. We failed to detect Scytalidium in a nail sample coinfected by T. rubrum and S. dimidiatum. Cross-inhibition of PCR was unlikely, since we used two different sets of primers. This negative result may be related to a very low degree of infection by Scytalidium and/or the small size of the sample.
Since accurate and rapid diagnosis of dermatomycosis can guide the treatment choice, our PCR-RFLP technique may provide significant clinical benefits. Differentiation of dermatophytes from Scytalidium spp. is crucial, since S. dimidiatum and S. hyalinum are not responsive to most antifungal drugs (5). In classical mycological diagnosis, microscopic examination cannot differentiate between hyphae from dermatophytes and from Scytalidium spp., and culture requires at least 3 weeks for specific identification. This PCR-RFLP method offers the unique advantage of being directly applicable to clinical samples, discriminating dermatophytes from Scytalidium spp. within 24 h. Since this method is simple and reliable, it can be used in laboratories with no mycological specialization for rapid etiologic diagnosis and treatment selection.
ACKNOWLEDGMENTS
This study was supported by a grant from the Ministère de l'Enseignement de la Recherche et de la Technologie for M. Machouart-Dubach.
We thank Catherine Massart and Christelle Vaury for help in sequencing the fungal DNA and David Young for reviewing the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock M, Maiwald M, Kappe R, Nickel P, Näher H. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses. 1994;37:79–84. doi: 10.1111/j.1439-0507.1994.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 3.Bock M, Nickel P, Maiwald M, Kappe R, Näher H. Diagnosis of dermatomycoses with polymerase chain reaction. Hautarzt. 1997;48:175–180. doi: 10.1007/s001050050566. [DOI] [PubMed] [Google Scholar]
- 4.Brodskii L I, Ivanov V V, Kalaidzidis I L, Leontovich A M, Nikolaev V K, Feranchuk S I, Drachev V A. GeneBee-NET: an internet-based server for biopolymer structure analysis. Biokhimiya. 1995;60:1221–1230. [PubMed] [Google Scholar]
- 5.Elewski B E, Greer D L. Hendesonula toruloidea and Scytalidium hyalinum. Arch Dermatol. 1991;127:1041–1044. [PubMed] [Google Scholar]
- 6.Elewski B, Hay R J. International summit on cutaneous antifungal therapy. J Am Acad Dermatol. 1995;33:816–822. doi: 10.1016/0190-9622(95)91838-8. [DOI] [PubMed] [Google Scholar]
- 7.Gelfand D H. Taq DNA polymerase. In: Erlich H A, editor. PCR technology. M. New York, N.Y: Stockton Press; 1989. pp. 17–22. [Google Scholar]
- 8.James S A, Cai J, Roberts I N, Collins M D. A phylogenetic analysis of the genus Saccharomyces based on 18S rRNA gene sequences: description of Saccharomyces kunashirensis sp. nov. and Saccharomyces martiniae sp. nov. Int J Syst Bacteriol. 1997;47:453–460. doi: 10.1099/00207713-47-2-453. [DOI] [PubMed] [Google Scholar]
- 9.Jung J M, Comey C T, Baer D B, Budowle B. Extraction strategy for obtaining DNA from bloodstains for PCR amplification and typing of the HLA-DQ alpha gene. Int J Legal Med. 1991;104:145–148. doi: 10.1007/BF01369719. [DOI] [PubMed] [Google Scholar]
- 10.Kac G, Bougnoux M E, Feuilhade de Chauvin M, Sene S, Derouin F. Genetic diversity among Trichophyton mentagrophytes isolates using random amplified polymorphic DNA method. Br J Dermatol. 1999;140:839–844. doi: 10.1046/j.1365-2133.1999.02812.x. [DOI] [PubMed] [Google Scholar]
- 11.Kappe R, Fauser C, Okeke N, Maiwald M. Universal fungus-specific primers systems and group specific hybridization oligonucleotides for 18S rDNA. Mycoses. 1996;39:25–30. doi: 10.1111/j.1439-0507.1996.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 12.Kappe R, Okeke N, Fauser C, Maiwald M, Sonntag H G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 13.Kemna M E, Elewski B E. A U.S. epidemiologic survey of superficial fungal diseases. J Am Acad Dermatol. 1996;35:539–542. doi: 10.1016/s0190-9622(96)90675-1. [DOI] [PubMed] [Google Scholar]
- 14.Kombila M, Martz M, Gomez de Diaz M, de Bièvre C, Richard-Lenoble D. Hendersonula toruloidea as an agent of mycotic foot infection in Gabon. J Med Vet Mycol. 1990;28:215–223. doi: 10.1080/02681219080000281. [DOI] [PubMed] [Google Scholar]
- 15.Roeijmans H J, De Hoog G S, Tan C S, Figge M J. Molecular taxonomy and GC/MS of metabolites of Scytalidium hyalinum and Nattrassia mangiferae (Hendersonula toruloidea) J Med Vet Mycol. 1997;35:181–188. [PubMed] [Google Scholar]
- 16.Vogelstein B, Gillepsie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]