Abstract
We developed a new method capable of detecting point mutations in the 23S rRNA gene of Helicobacter pylori using a LightCycler. Our method can detect a mutation in this gene in less than 1 h and can process many samples at once, thereby contributing to the selection of patients suitable for clarithromycin-based therapy.
The resistance of Helicobacter pylori to clarithromycin is a major cause of failure in eradication therapies (8); furthermore, mutations in the 23S rRNA gene (an A-to-G transition at nucleotide position 2143 [A2143G] or 2144 [A2144G]) have been found to confer resistance to clarithromycin to H. pylori (3, 8, 10, 12). Reports indicate that the rate of eradication of H. pylori with clarithromycin treatment is only 40% for mutant strains, whereas it is 85% for wild-type strains (6).
To date, the detection of mutations in the H. pylori 23S rRNA gene is mainly performed by PCR, followed by digestion with restriction enzymes. The resulting DNA fragments are separated on agarose or acrylamide gels, and the presence of a mutation is determined by the pattern of the restriction fragment length polymorphism (RFLP) by PCR-RFLP analysis (6, 7). This detection procedure is, however, time-consuming and requires optimization of the PCR. In addition, contamination of the amplified product can also cause false-positive results. Recently, a new high-speed thermal cycler that uses glass capillaries (LightCycler; Roche Diagnostics, Tokyo, Japan) has been introduced. Using this recently introduced machine along with attached fluorescenct probes, we have established a new method that detects point mutations in the 23S rRNA gene of H. pylori. We further examined the sensitivity and specificity of this method by comparison with PCR-RFLP analysis as a “gold standard” and analysis of clinical samples (gastric tissue) for the presence of mutant H. pylori strains.
MATERIALS AND METHODS
Gastric tissues.
Gastric tissue was taken from 186 patients with epigastralgia during gastric endoscopy examination, and a rapid urease test was performed with a Helicocheck kit (Otsuka Pharmaceutical, Tokyo, Japan). DNA was isolated from each tissue with a QIAamp DNA mini kit (Qiagen, Tokyo, Japan) according to the manufacturer's protocol. The isolated DNA was then dissolved in 20 μl of 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and stored at −20°C until use.
By using specimens obtained from the antrums of another 84 patients who were Helicocheck test positive, H. pylori was isolated and cultured on Columbia agar with 5% (vol/vol) horse blood and Dent antibiotic supplement (Oxford, Basingstoke, United Kingdom) at 37°C for 5 days under microaerobic conditions by using the Campy-Pak system (BBL, Cockeysville, Md.). Identification of H. pylori was based on colony morphology, microscopy, and positive urease, catalase, and oxidase activities. The MIC of clarithromycin was determined by the agar dilution method with agar plates containing clarithromycin at concentrations ranging from 0.03 to 100 mg/liter. The MIC was defined as the minimum concentration of clarithromycin at which bacterial growth was inhibited. H. pylori strains were classified as resistant to clarithromycin when the MIC was >1 mg/liter.
Amplification and melting analysis of 23S rRNA gene of H. pylori using the LightCycler.
The primers used for PCR of the 23S rRNA gene of H. pylori (GenBank accession no. U27270) were CRFL-1 (5′-ATGAATGGCGTAACGAGAT-3′; nucleotides 2051 to 2070) and CRRL-2 (5′-ACACTCAACTTGCGATTCC-3′; nucleotides 2410 to 2391), and the result was a 360-bp product (Fig. 1). In addition, two kinds of probes (detection probe MP-W and anchor probe AP-2186) were included in the PCR mixture. The detection probe was 5′ labeled with LC-Red 640 and 3′ phosphorylated, and the anchor probe was 3′ labeled with fluorescein isothiocyanate. The latter was located downstream of the former at a distance of 1 nucleotide. The locations of these primers and probes are shown in Fig. 1.
FIG. 1.
Locations of primers and hybridization probes used for our method. Primers used are shown in italics at each 5′ end (primer CRFL-1; nucleotides 2051 to 2070) and 3′ end (primer CRRL-2; nucleotides 2410 to 2391). The amplified product is 360 bp. Both detection (MP-W) and anchor (AP-2186) probes are shown midproduct. MP-W is 3′ phosphorylated and 5′ labeled with LC-Red 640, and AP-2186 is labeled with fluorescein isothiocyanate at its 3′ end. Other kinds of detection probes are also shown in the lower part of the figure.
With a LightCycler, PCR was performed in 20-μl volumes in glass capillaries (Roche Diagnostics). Twenty microliters of PCR mixture contained 2 μl of sample DNA, 13.2 μl of H2O, 1.6 μl of MgCl2 (25 mM), 0.4 μl each of CRFL-1 and CRRL-2 (25 mM each), 0.2 ml each of MP-W (40 mM) and AP-2151 (20 mM), and 2 μl of DNA-Master Hybridization Probes (Roche Diagnostics) containing Taq DNA polymerase, reaction buffer, a deoxynucleoside triphosphate mixture, and 10 mM MgCl2 in a 10× concentrate. Cycling conditions consisted of an initial denaturation at 94°C for 2 min, followed by 45 cycles with denaturation at 94°C for 0 s, annealing at 55°C for 10 s, and extension at 72°C for 15 s, with a ramping time of 20°C/s.
After completion of the amplification process, samples of the reaction mixture were denatured at 94°C for 0 s, held at 50°C for 5 s, and then slowly heated to 80°C at a ramp rate of 0.1°C/s. During this process, declining fluorescence was continuously monitored, and melting curves were constructed automatically with software attached to the LightCycler. Melting curves were converted to melting peaks by plotting the negative derivative of the fluorescence with respect to temperature (−dF/dT) (see Fig. 2).
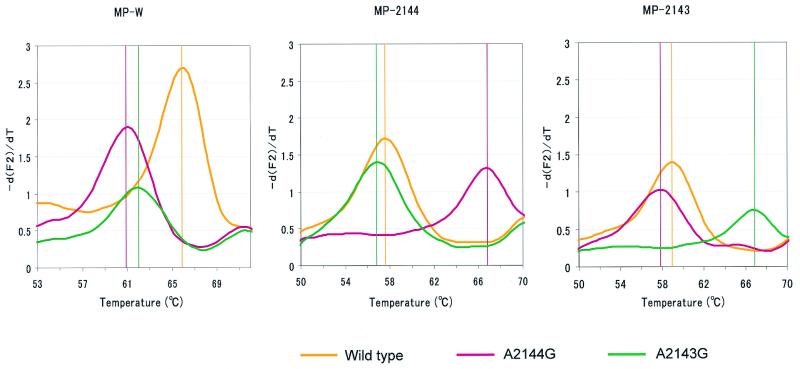
FIG. 2.
Melting analysis was performed with templates (DNAs from the wild type and mutants with the A2144G and A2143G mutations) synthesized in vitro. Melting curves were converted to melting peaks by plotting the negative derivative of the fluorescence (−dF/dT) with respect to temperature. The melting peak for each strain is shown at a different temperature. In addition to the MP-W probe, two other probes (each specific to the mutant with either the A2144G or the A2143G mutation) were used. The probes used are shown above each graph. The melting peaks for each strain (wild type, yellow; mutant with the A2144G mutation, pink; mutant with the A2143G mutation, green) are shown in the same figure. The vertical lines in each graph represent the melting temperatures of each template.
Standard DNA.
Standard DNAs were synthesized in vitro by PCR (with primers CRFL-1 and CRR1-2) with DNAs from wild-type strains and strains with the A2143G and A2144G mutations as templates. The sequences of these standard DNAs were in complete accordance with those reported for the 23S rRNA gene of H. pylori in GenBank (accession number U27270). These DNAs were used for each experiment.
Melting analysis using DNA standards.
With each standard DNA (DNAs of the wild type and the strains with the A2143G and A2144G mutations), a melting analysis was done with wild-type-specific detection probes (MP-W) as well as A2143G or A2144G mutation-specific detection probes (MP-2143 and MP-2144, respectively) (see Fig. 2).
Sensitivity in detecting mutant strains either by RFLP analysis or with the Light Cycler.
In order to clarify the sensitivity of detection of mutant H. pylori among wild-type strains mutant DNA (from strains with either the A2143G or the A2144G mutations), synthesized as described above, was mixed with the DNA of the wild-type strain at ratios of 1:1, 1:1/2, 1:1/10, 1:1/20, and 1:1/50. These mixtures were processed for melting analysis with the LightCycler.
PCR-RFLP analysis.
PCR-RFLP analysis was performed as described by Maeda et al. (6) with primers CRF-4 (5′-AGT GGA GGT GAA AATTCC-3′; nucleotides 2105 to 2122) and CRR-1 (5′-TAA GAG CCA AAG CCC TTA C-3′; nucleotides 2239 to 2221), resulting in a 135-bp product. PCR was performed in a thermal cycler (GeneAmp PCR system 9600; Applied Biosystems) with a 50-μl volume containing 2 μl of sample DNA, 5 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3], 15 mM MgCl2, 0.01% [wt/vol] gelatin), 0.5 μl each of CRF-4 and CRR-1 at 25 mM, 0.4 μL of a 25 mM deoxynucleoside triphosphate mixture, and 1 U of Taq DNA polymerase (Applied Biosystems). After heating of the mixture at 94°C for 5 min, PCR was performed for 34 cycles, with denaturation at 94°C for 30 s and extension at 57°C for 30 s, followed by incubation at 72°C for 4 min. The PCR product was digested with either the BsaI or the MboII restriction enzyme and was then electrophoresed on an acrylamide gel. The presence of the mutant was detected by examination of the patterns of the restricted fragments. While PCR products derived from the mutant were digested with either BsaI (mutant with the A2144G mutation) or MboII (mutant with the A2143G mutation), this was not possible for the wild-type strain.
RESULTS
Melting analysis of wild-type strain and two kinds of mutants with mutations in the 23S rRNA gene.
By using standard DNAs, including those of the wild type and mutants with the A2143G and A2144G mutations, a melting analysis was performed. By using M-WP, the fluorescence emitted by the mutant strains declined more rapidly than that emitted by the wild-type strain. Figure 2 shows the melting peaks of each standard DNA, with distinct peaks at different temperatures. In addition, by using MP-2144 or MP-2143 as the detection probe, the presence of each mutant strain could be clarified (Fig. 2).
Detection of mutants among wild-type H. pylori.
As shown in Fig. 3, the mutant with the A2143G mutation, as well as the mutant with the A2144G mutation (data not shown), could be detected when it contaminated a mixture with the wild type at a level of as little as 10%.
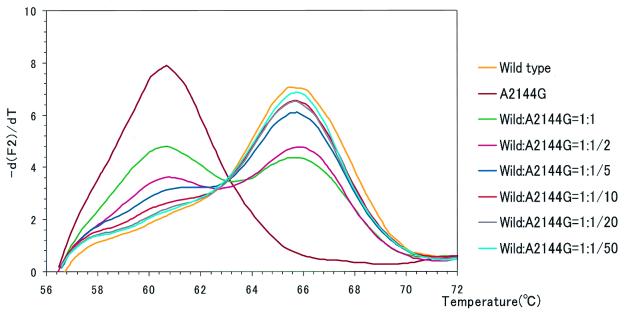
FIG. 3.
Using a mixture of standard DNAs (from the wild type and the mutant with the A2144G mutation), the sensitivity of detection of a mutant among wild-type H. pylori strains was examined. The template for the mutant with the A2144G mutation was mixed with the template for the wild type at ratios (wild type to mutant) of 1:1, 1:1/2, 1:1/5, 1:1/10, 1:1/20, and 1:1/50. Both wild type-only and mutant-only templates were included as controls. The detection probe used was MP-W. As the ratio of the mutant DNA as a contaminant of the wild-type DNA increased, another peak appeared at a temperature in accordance with the melting temperature of the mutant DNA. These peaks can be recognized until the ratio becomes as low as 1:1/10.
MIC for H. pylori and detection of mutants by melting analysis.
Table 1 shows the correlation between the genotypes of the H. pylori strains and the MIC of clarithromycin. All mutant H. pylori isolates were resistant to clarithromycin.
TABLE 1.
Correlation between H. pylori isolates and MIC of clarithromycin
| MIC (mg/liter) | No. of strains detected by melting analysis
|
Wild type + mutant with A2144G mutation | |
|---|---|---|---|
| Wild type | Mutant with A2144G mutation | ||
| >1a | 0 | 5 | 11 |
| ≤1 | 68 | 0 | 0 |
As described in Materials and Methods, H. pylori strains were classified as resistant to clarithromycin when the MIC was more than 1 mg/liter.
Detection of H. pylori mutants by PCR-RFLP analysis and melting analysis.
Of 186 patients, H. pylori was detected in 151 patients with the Helicocheck kit. The 23S rRNA gene of H. pylori was amplified from all 151 Helicocheck-positive patients but not the 35 Helicocheck-negative patients.
If the existence of a mutant was suspected, the melting analysis was repeated with mutant-specific detection probes (MP-2143 or MP-2144), for clarification of the mutant type (Fig. 2). Using a combination of three kinds of probes, we examined strains for the presence of the mutant 23S rRNA gene. One hundred nineteen patients had wild-type strains only, 19 patients had mutant strains only, and 13 patients had both wild-type and mutant strains (Table 2). These results were in accordance with the results obtained by PCR-RFLP analysis.
TABLE 2.
Results of melting analysis among 186 patients examined with the LightCycler
| Results obtained by melting analysis
|
Results obtained by RFLP analysisa
|
||||||
|---|---|---|---|---|---|---|---|
| Wild type | A2144G | A2143G | No. (%) of patientsb | BsaI | MboI | Uncut band | No. of patients |
| + | − | − | 119 (78.8) | − | − | + | 119 |
| − | + | − | 17 (11.2) | + | − | − | 17 |
| − | − | + | 1 (0.66) | − | + | − | 1 |
| + | + | − | 10 (6.6) | + | − | + | 10 |
| + | − | + | 1 (0.66) | − | + | + | 1 |
| − | + | + | 1 (0.66) | + | + | − | 1 |
| + | + | + | 2 (1.33) | + | + | + | 2 |
| Subtotal | 151 | 151 | |||||
| − | − | − | 35 | − | − | − | 35 |
| Total | 186 | 186 | |||||
BsaI, PCR products can be cut by BsaI (presence of the mutant with the A2144G mutation); MboI, PCR products can be cut by MboI (presence of the mutant with the A2143G mutation); uncut band, presence of uncut band even after treatment with both restriction enzymes (presence of the wild type).
Values in parentheses indicate the percentage of patients in whom H. pylori was detected with the LightCycler.
DISCUSSION
For the LightCyler melting analysis, we used a long anchor probe (a 36-mer) along with a second shorter probe (an 18-mer) for the purpose of recognizing adjacent sequences with the shorter probe lying over the mutation site. When these probes hybridize with the PCR product within a 1-nucleotide interval, fluorescence is emitted by fluorescence resonance energy transfer (4). After completion of the PCR, the temperature was slowly raised from 50 to 80°C. Due to the great stability of the anchor probe, a loss of fluorescence occurs as the shorter probe melts off the template. Meanwhile, the wild-type-specific detection probe dissociates from the mutant-specific detection probe at a temperature lower than that for the wild-type sequence. A single-base mismatch under the detection probe results in a shift in the melting temperature to a lower temperature (1). This shift leads to another peak at a temperature lower than that for the wild-type strain (Fig. 2). By changing the kinds of detection probes, we can determine the presence of mutants with the A2143G and A2144G mutations.
The A2143C mutation, recently reported by Stone et al. (9) and Occhialini et al. (8), was not searched for in the present study because Matsuoka et al. (7) and Maeda et al. (5) reported that the mutation was not detected in either 82 or 412 strains, respectively, isolated from gastric tissue derived from Japanese patients. However, by using a detection probe which is hybridized with the strain with the A2143C mutation, the mutation could be detected by our method. We are preparing the probe and examining whether the mutation was detected among strains from Japanese patients.
As shown in Fig. 3, LightCycler melting analysis can detect mutants when they are present among wild-type strains at a level of as little as 10%. In addition, the ratio of contaminant mutants among wild-type strains can be estimated by comparing the melting curves with those obtained by using a mixture of standard DNAs (Fig. 3).
The sensitivity of detection of the mutant between PCR-RFLP analysis and LightCycler melting analysis was compared with 186 gastric tissue specimens. The results obtained by either method were comparable for all specimens. Our method could be as useful as PCR-RFLP analysis.
By using a LightCycler, the whole process was completed within 40 min. Due to the quick adaptation of temperature in the glass capillary (high surface-to-volume ratio), PCR can occur under quicker cycling conditions. Moreover, different from the PCR-RFLP technique, we need not deal with the amplified product once the reaction mixture is set in the LightCycler. Therefore, this simple procedure results in less chance of contamination and eliminates the need to perform a digestion with restriction enzymes.
As shown at Table 1, mutant H. pylori isolates were all resistant to clarithromycin. Therefore, our method could predict the presence of a clarithromycin-resistant strain before cultural data for H. pylori are obtained.
The coexistence of wild-type and mutant strains of H. pylori has been reported in several papers (2, 7, 11, 13, 14). Also, reportedly, clarithromycin-resistant and clarithromycin-sensitive strains do coexist in the same patient (13, 14), even among patients with no history of clarithromycin exposure (7). On the other hand, it has been observed that failed therapy with clarithromycin-based regimens may cause antimicrobial resistance in H. pylori (2, 11). Although the mechanism through which the clarithromycin-resistant strain appears during treatment remains to be clarified, our method may help to further elucidate this mechanism. In addition, the possibility of the existence of a heterozygous strain, with only one of two 23S rRNA genes containing a base substitution, could not be denied.
In conclusion, our method can easily detect the presence of a clarithromycin-resistant strain and can also process many samples at once, thereby contributing to the identification of those patients suitable for clarithromycin-based therapy for the treatment of H. pylori infection.
REFERENCES
- 1.Aslanidis C, Nauck M, Schmitz G. High-speed prothrombin G-A 20210 and methylenetetrahydrofolate reductase C-T 677 mutation detection using real-time fluorescence PCR and melting curves. BioTechniques. 1999;27:234–238. doi: 10.2144/99272bm04. [DOI] [PubMed] [Google Scholar]
- 2.Enroth H, Bjorkholm B, Engstrand L. Occurrence of resistance mutation and clonal expansion in Helicobacter pylori multiple-strain infection: a potential risk in clarithromycin-based therapy. Clin Infect Dis. 1999;28:1305–1307. doi: 10.1086/514796. [DOI] [PubMed] [Google Scholar]
- 3.Hulten K, Gibreel A, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livak K J, Flood S J A, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 5.Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, Omata M. Detection of clarithromycin-resistant Helicobacter pyroli strains by a preferential homoduplex formation assay. J Clin Microbiol. 2000;38:210–214. doi: 10.1128/jcm.38.1.210-214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pyroli specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317–321. doi: 10.1136/gut.43.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuoka M, Yoshida Y, Hayakawa K, Fukuchi S, Sugano K. Similtaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut. 1999;45:503–507. doi: 10.1136/gut.45.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone G G, Shortridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka S K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 10.Talor N S, Fox J G, Akopyrants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vakil N, Hahn B, McSorley D. Clarithromycin-resistant Helicobacter pylori in patients with duodenal ulcer in the United States. Am J Gastroenterol. 1998;93:1432–1435. doi: 10.1111/j.1572-0241.1998.455_t.x. [DOI] [PubMed] [Google Scholar]
- 12.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G E, Jiang Q, Taylor D E. Genotypic characterization of clarithromycin-resistant and -susceptible Helicobacter pylori strains from the same patient demonstrates existence of two unrelated isolates. J Clin Microbiol. 1998;36:2730–2731. doi: 10.1128/jcm.36.9.2730-2731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G E, Rahman M S, Humayun M Z, Taylor D E. Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:683–685. doi: 10.1128/aac.43.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]